Abstract
Understanding the signaling differences that distinguish human HER2-amplified (HER2-positive (HER2+)) breast cancers from other breast cancer subtypes may help to identify protein drug targets for the specific treatment of HER2+ breast cancers. We performed two kinome-wide small interfering RNA (siRNA) screens on five HER2+ breast cancer cell lines, seven breast cancer cell lines in which HER2 was not amplified and two normal breast cell lines. To pinpoint the main kinases driving HER2 signaling, we performed a comprehensive siRNA screen that identified loss of the HER2/HER3 heterodimer as having the most prominent inhibitory effect on the growth of HER2+ breast cancer cells. In a second siRNA screen focused on identifying genes that could sensitize HER2+ cells to trastuzumab treatment, we found that loss of signaling members downstream of phosphatidylinositol 3 kinase (PI3K) potentiated the growth inhibitory effects of trastuzumab. Loss of HER2 and HER3, as well as proteins involved in mitogenic and environmental stress pathways inhibited the proliferation of HER2+ cells only in the absence of trastuzumab, suggesting that these pathways are inhibited by trastuzumab treatment. Loss of essential G2/M cell cycle mediators or proteins involved in vesicle organization exerted inhibitory effects on HER2+ cell growth that were unaffected by trastuzumab. Furthermore, the use of a sensitization index (SI) identified targeting the PI3K pathway to sensitize to trastuzumab treatment. Antagonism using the SI identified MYO3A, MYO3B and MPZL1 as antagonizers to trastuzumab treatment among HER2+ cell lines. Our results suggest that the dimerization partners of HER2 are important for determining the activation of downstream proliferation pathways. Understanding the complex layers of signaling triggered downstream of HER2 homodimers and heterodimers will facilitate the selection of better targets for combination therapies intended to treat HER2+ breast cancers.
Introduction
Amplification of HER2 is observed in approximately 20% of all invasive breast cancers in humans, and is associated with an increased risk of disease progression and poor overall survival.1 HER2-positive (HER2+) breast cancers show elevated expression of HER2 mRNA caused by amplification of the HER2 gene, as well as overexpression of the HER2 protein. The HER2 transmembrane receptor transduces growth signals and acts as a driver of cell proliferation.2 The observation that HER2 overexpression is the critical oncogenic driver in HER2+ breast tumors has led to the development of therapies targeting this protein, including the monoclonal antibody trastuzumab.3, 4
The HER2 protein forms both homodimers and heterodimers with other epidermal growth factor receptor family receptor tyrosine kinases, and they exhibit variations in downstream signaling. For example, the heterodimer composed of epidermal growth factor receptor plus HER2 activates the RAS/mitogen-activated protein kinase (MAPK), phosphatidylinositol 3 kinase (PI3K) and phospholipase C, gamma (PLCγ) signaling pathways.5, 6 In contrast, the HER2 homodimer lacks phosphorylated tyrosines in its cytoplasmic tail, preventing it from docking to the PI3K pathway adaptor protein p85 and activating PI3K signaling. Instead, engagement of the HER2 homodimer primarily activates the GRB2 and SHC adaptor proteins that initiate RAS/MAPK signaling.7 A heterodimer containing HER2 plus HER3 relies on HER3 for its signaling. Unlike HER2, HER3 can bind to p85 and triggers strong activation of the PI3K/AKT pathway.8
To identify novel regulators of HER2 signaling, we performed two unbiased RNA interference loss-of-function screens. High-throughput RNA interference screens using small interfering RNA (siRNA) treatment of cancer cell lines have previously been utilized to identify novel drug targets.9, 10 We used a library of siRNAs directed specifically at kinases to examine 12 breast cancer cell lines, five of which were HER2+. Our first approach (‘comprehensive screen') was designed to identify kinases whose inhibition specifically reduced growth of HER2+ cell lines but had no effect on growth of non-HER2+ cells or normal breast cells. This was performed by Comparative Marker Selection (CMS) (Gene Pattern, MIT), where permutation testing revealed the least number of genes (in our case, siRNAs) able to distinguish two predetermined groups (HER2 amplified and HER2 non-amplified). In our second approach (‘sensitizer screen'), we identified siRNAs that sensitized HER2+ cells to trastuzumab treatment; that is, we pinpointed kinases whose inhibition in combination with trastuzumab further reduced the proliferation of HER2+ cells. The results of these screens show that HER2 and HER3 are the strongest drivers of oncogenic HER2 signaling. We also demonstrate that targeting downstream members of the PI3K pathway in combination with trastuzumab leads to a greater degree of inhibition of HER2+ cell proliferation. Our data suggest several potential combination therapies that may provide more effective treatment of HER2+ breast cancers.
Results
Identification of kinases whose inhibition reduces breast cancer cell line proliferation
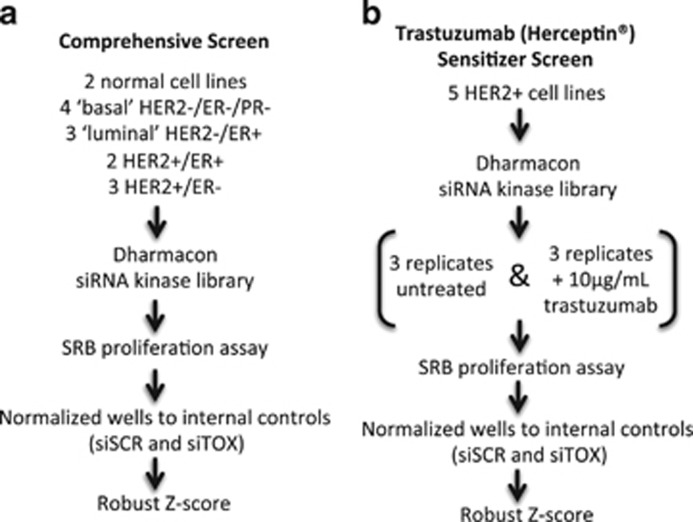
Fourteen cell lines comprising various breast cancer subtypes and normal breast cell line controls were selected for high-throughput siRNA screening with an siRNA library targeting 778 kinases and kinase-related genes (Supplementary Tables S1, S2). These cell lines included four lacking the estrogen receptor (ER), progesterone receptor (PR) and HER2 expression (Triple-negative breast cancers; ER−/PR−/HER2−); three that were (ER+; ER+/PR±/HER2−); five that were HER2+ (HER2+; ER±/PR±) and two that were derived from normal breast tissue. Schematic diagrams illustrating our comprehensive and sensitizer screens are shown in Figure 1. The comprehensive approach sought to screen all 14 cell lines to identify kinases whose inhibition had an effect on the growth of only HER2+ breast cancer cells (Figure 1a). Kinases affecting other breast cancer subtypes are to be described in separate article. The sensitizer approach sought to identify kinases whose inhibition further decreased the proliferation of trastuzumab-treated HER2+ cells (Figure 1b). The failure of an siRNA-transfected cell line to proliferate was detected as a decrease in total protein content as determined by the sulforhodamine B assay. The data were standardized using a robust Z-score statistic11 and interpreted such that a Z-score of 0 indicated that the inhibition of a particular kinase had no effect on cell proliferation, and a negative Z-score indicated that inhibition of that kinase decreased cell proliferation (Supplementary Figure S1).
Figure 1.
siRNA kinome screening strategy. (a) Comprehensive screen strategy. Two normal breast cell lines and 12 breast cancer cell lines of the indicated molecular subtypes were transfected with the siRNA kinase library and cultured for 3 days. Cells were fixed in 10% trichloroacetic acid (TCA) and stained with sulforhodamine B (SRB) to determine proliferation. SRB assay absorbance values were normalized to internal controls (siSCR and siTOX). Robust Z-scores were calculated for each siRNA. (b) Sensitizer screen strategy. Five HER2+ breast cancer cell lines were transfected with the siRNA kinase library in six replicates. Three replicates were treated with trastuzumab (10 μg/ml), whereas the other three remained untreated. Cells were harvested at 3 days post trastuzumab and processed as described in a.
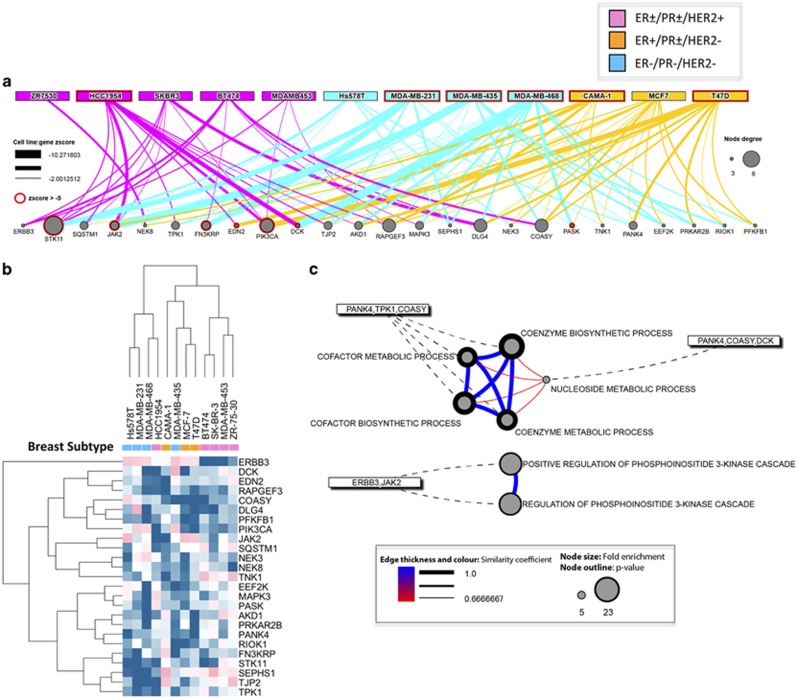
The strongest suppressive effects on proliferation of three or more breast cancer cell lines are summarized in the network interaction diagram and heatmap shown in Figures 2a and b. Hits that were common to the normal breast cell lines (MCF10A and MCF12A), which included many enzymes involved in the regulation of cell cycle entry (PLK1, CHEK1 and WEE1), were excluded from the hit list (Supplementary Table S3). The most significant hit was STK11/LKB1. LKB1 serves a link between polarity and metabolic status of the cell by the LKB1–AMPK–mTOR signaling pathway.12 Other strong candidates, included PIK3CA, DLG4, RAPGEF3 and COASY genes, involved in PI3K signaling and cell polarity.13, 14, 15 Gene enrichment analysis of the 25 hits was conducted by DAVID.16, 17 Hits identified were involved in coenzyme metabolic process (Figure 2c). The genes in the PI3K signaling pathway were also enriched but did not reach significance (Figure 2c, Supplementary Table S4).
Figure 2.
Comprehensive kinase screen. (a) Network interaction diagram of the top siRNAs that decreased proliferation across cell lines (comprehensive screen). Hits common to normal breast cell lines (MCF10A and MCF12A) not shown. Node size is proportional to number of cell lines in which a given node was a hit. Edges are color coded by breast cancer subtype (pink, ER±/PR±/HER2+ blue, ER−/PR−/HER2− orange, ER+/PR±/HER2−). Edge thickness represents intensity of proliferation decrease (Z-score). (b) Heatmap of siRNA from (a) that inhibit proliferation of breast cancer cell lines. (c) GO and pathway enrichment analysis (DAVID) of siRNA-targeted genes that decreased breast cancer cell line proliferation in the comprehensive screen. Circular nodes describe the class term examined for enrichment, with the size of the node scaled by the fold enrichment for the class term across the siRNA library screened. The thickness of the node outline represents P-value.
Identification of kinases whose inhibition preferentially blocks the proliferation of HER2+ breast cancer cell lines
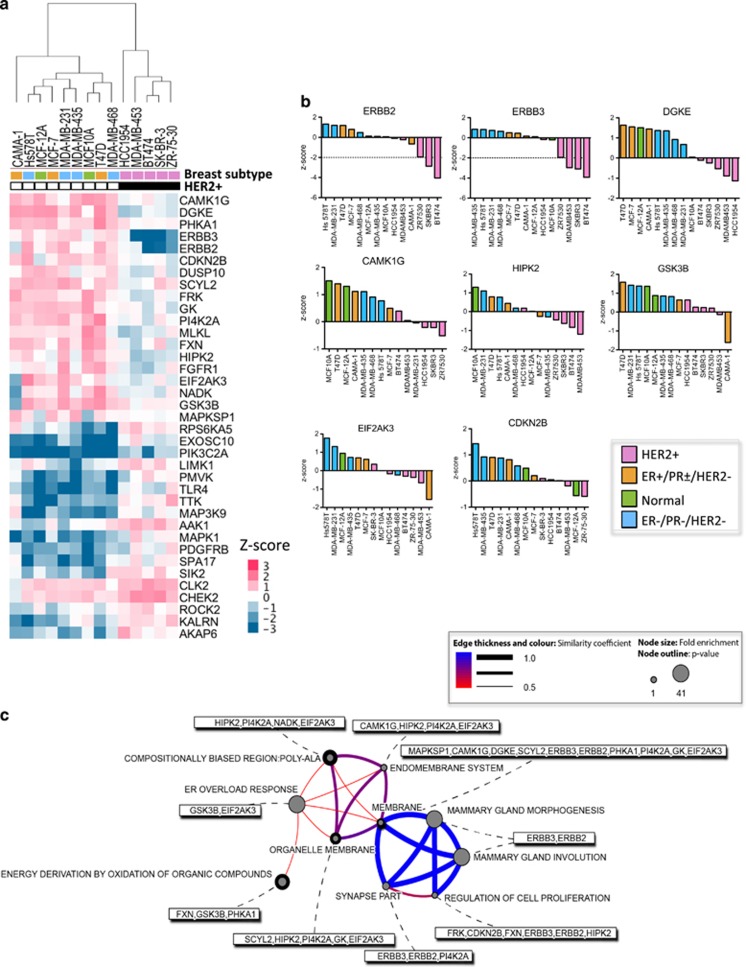
To identify kinases whose inhibition preferentially blocks the proliferation of HER2+ cells, we performed a CMS analysis of our comprehensive screen data. Permutation testing revealed the least number of genes (in our case, siRNAs) able to distinguish two predetermined groups among our 14 breast cell lines: HER2+ cell lines, and cell lines in which HER2 was not amplified. Inhibition of ERBB3/HER3 or ERBB2/HER2 produced the strongest blocks in HER2+ cell proliferation (Figure 3a), with both hits having Z-scores of <−2. Other siRNAs had more subtle inhibitory effects that were still unique to HER2+ cells. Loss of CAMK1G, DGKE and HIPK2 reduced the growth of HER2+ cells but stimulated the proliferation of non-HER2-amplified cells (Figure 3b).
Figure 3.
Identification of kinases whose inhibition preferentially blocks the proliferation of HER2+ breast cancer cell lines. (a) Heatmap of unbiased selection of siRNAs that differ in their effects on HER2+ and non-HER2-amplified breast cancer cell lines (comprehensive screen). CMS based on HER2 status was performed using Gene Pattern software. Only statistically significant effects are shown (P<0.01, permutation test, median T-test). Supervised hierarchical clustering analysis of siRNAs and cell lines was performed using Cluster 3.0 software (Michael Eisen). Data were visualized using Java TreeView. (b) Waterfall plots of the indicated kinases from (a) whose siRNA-mediated inhibition selectively blocked the proliferation of HER2+ cell lines (pink bars). (c) Gene enrichment analysis (DAVID) of siRNA-targeted genes identified in a. Analysis was performed as for Figure 2c.
Gene enrichment analysis by DAVID18 of hits from Figure 3a highlighted genes involved in mammary gland morphogenesis (ERBB2, ERBB3; P=0.045), organelle membranes (SCYL2, HIPK2, PI4K2A, GK, EIF2AK3; P=0.033), the endomembrane system—a collection of membranous structures involved in vesicle transport within the cell (PI4K2A, EIF2AK3, GSK3B, CAMK1G, HIPK2, PHKA1; P=0.040), as well as the ER overload response—the signaling response to accumulation of normal or misfolded proteins (GSK3B, EIF2AK3; P=0.045) (Figure 3c, Supplementary Table S5).
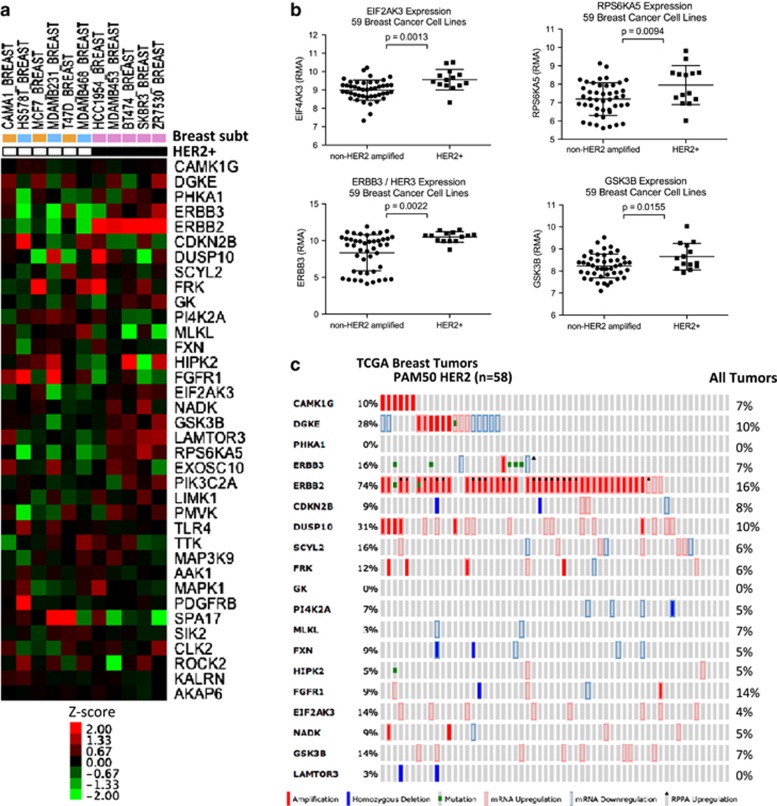
Using the Cancer Cell Library Encyclopedia (CCLE),19 assessment of the basal mRNA expression levels of candidate genes among the cell lines screened, as well as all breast cancer lines in CCLE showed increased expression of HER2, HER3, EIF2AK3, RPS6KA5 and GSK3B in HER2+ breast cancer cell lines (Figures 4a and b) (Student's t-test, P<0.05). Expression of candidate genes across 59 breast cancer cell lines and the correlation with HER2 expression was shown in Supplementary Figures S2 and S3. Furthermore, OncoPrint visualization of genomic alterations (copy number alterations, somatic mutations, mRNA expression and protein phosphorylation) across 58 breast tumors defined as part of the HER2 subtype by PAM50 expression profiling was performed.20, 21, 22 The PAM50 HER2 subtype showed consistent increase in expression of DUSP10 (28%) EIF2AK3 (14%), GSK3B (14%) and ERBB2 (71%) (Figure 4c). The group also showed amplification in CAMK1G (10%), DGKE (10%), DUSP10 (10%) and ERBB2 (67%) (Figure 4c). No change in overall survival was observed (data not shown).22
Figure 4.
Genetic characterization of pathways whose inhibition blocks proliferation preferentially in HER2+ breast cancer cell lines. (a) mRNA expression analysis of the hits from 11 of 14 breast cell lines screened. Data from MCF10A, MCF12A were not available from CCLE.19 MDA-MB-435 was excluded from analysis because of its annotation as a melanoma cell line. (b) Increased expression of EIF2AK3, RPS6KA5, GSK3B and HER3 in HER2+ cell lines (n=14) compared with non-HER2-amplified cell lines (n=35) using CCLE19 (Student's t-test, P<0.05). Expression values were expressed as log-transformed Robust Multi-array Average (RMA).52 (c) Visualization of genomic alterations (copy number alterations, somatic mutations and mRNA expression) across 58 breast tumors identified to the PAM50 HER2 subtype.22
Identification of kinases whose inhibition sensitizes HER2+ breast cancer cells to trastuzumab treatment
We next sought to identify kinases whose inhibition would further reduce the proliferation of HER2+ cell lines treated with trastuzumab. Our five HER2+ cell lines were transfected with the siRNA library and treated (or not) with trastuzumab. For each cell line, the effects on proliferation of siRNA treatment alone, or treatment with a combination of a specific siRNA with trastuzumab, were analyzed to construct a ranking of siRNAs from strongest to weakest inhibitory capacity (Supplementary Tables S6 and S7). The top 5% (38/778) of ‘siRNA alone' hits that decreased the growth of each HER2+ cell line tested were compared with ‘siRNA plus trastuzumab' hits to generate three classes of siRNA hits.
Some siRNAs were strong inhibitors of HER2+ cell proliferation only without trastuzumab (‘untreated') and lost this capacity in the presence of trastuzumab, whereas other siRNAs were equally strong inhibitors of HER2+ cell proliferation in the presence or absence of trastuzumab (‘neutral'). The most interesting siRNAs potentiated the ability of trastuzumab to inhibit HER2+ cell proliferation (‘treated'). The effects of the top 5% of hits in each of these three categories were analyzed (Figure 5, Supplementary Figures S4–S7, Supplementary Tables S8–S10). The kinases represented by these hits were investigated in an attempt to identify genes and pathways whose inhibition would best sensitize HER2+ cancer cells to trastuzumab treatment.
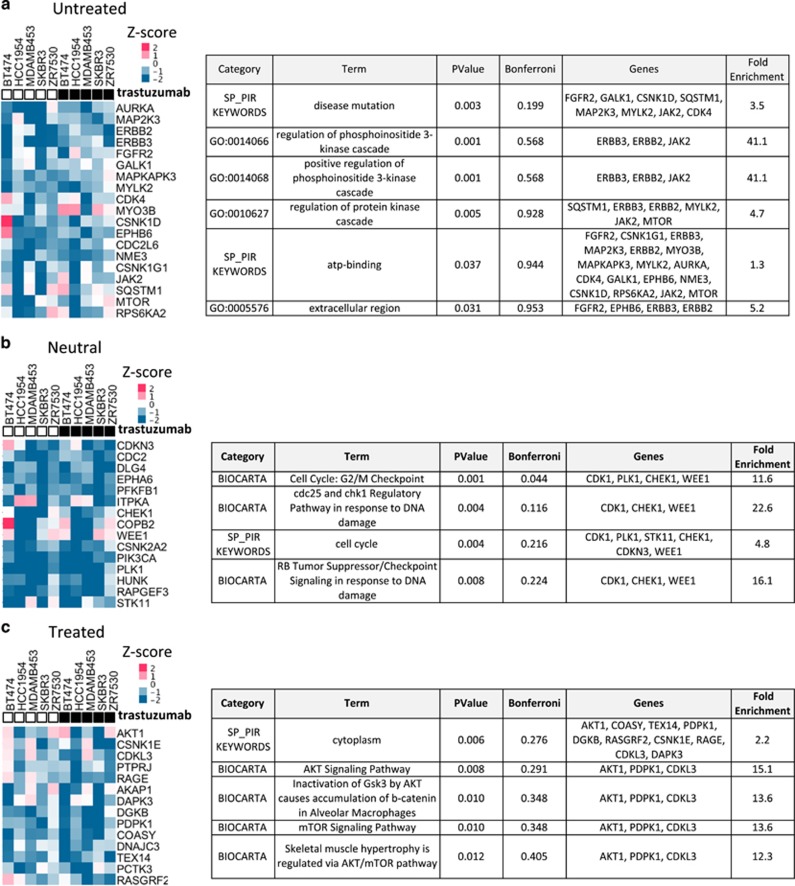
Figure 5.
Identification of kinases whose inhibition sensitizes HER2+ breast cancer cells to trastuzumab treatment. Heatmaps of the top 5% of siRNAs that decreased the proliferation of trastuzumab-treated cells of three or more HER2+ breast cancer cell lines. Kinases were classified into the indicated three categories: (a) ‘untreated', which inhibited proliferation only in the absence of trastuzumab; (b) ‘neutral', which inhibited proliferation regardless of trastuzumab treatment; and (c) ‘treated', which inhibited proliferation only in the presence of trastuzumab; GO and pathway enrichment of each group of hits was performed as in Figure 2c. All terms had P<0.05 and were sorted by increasing Bonferroni values.
Untreated category
The ‘untreated' category of siRNAs in the sensitizer screen showed effective inhibitors of HER2+ cell proliferation only in the absence of trastuzumab treatment (Figure 5a). In other words, if the transfection of a given siRNA and trastuzumab treatment were combined, the ability of the siRNA to inhibit HER2+ cell growth was reduced. Hits particularly enriched in this group by DAVID included genes associated with the extracellular region of the cell (P=0.031) and positive regulation of PI3K (P=0.001) (Figure 5a, Supplementary Table S8). These processes included namely ERBB2/HER2, ERBB3/HER3, FGFR2, EPHB6, JAK2 and MTOR genes. Other hits were mediators in mitogenic and environmental stress pathways, namely MAP2K3/MKK3, MAPKAPK3, RPS6KA2/p90RSK and ERBB2/HER2. Functional interaction partners of hits showed enrichment for ErbB and epidermal growth factor receptor signaling pathways (Supplementary Figure S5). These data show that loss of signaling by receptor tyrosine kinases and MAPKs decreases the proliferation of HER2+ cells only in the absence of trastuzumab.
Neutral category
Many of the same hits inhibiting the growth of both normal and breast cancer cells identified in the comprehensive screen also emerged in the sensitizer screen as siRNAs that efficiently blocked HER2+ cell growth regardless of trastuzumab treatment (Figures 2 and 5b, Supplementary Figure S6; Supplementary Table S3). Such hits included siRNAs targeting: G2/M cell cycle transition regulators (CHEK1, PLK1, WEE1, CDKN3/CDK3 and CDC2/CDK1); vesicle organization (COPB2 and DLG4); cell polarity and cell-cell junction proteins (COPB2, DLG4, STK11/LKB1, RAPGEF3 and CSNK2A2); HUNK, a HER2 target gene mediating cell survival; and PIK3CA. These results suggest that, as expected, the inactivation of G2/M checkpoint transition regulators and the loss of vesicle organization proteins can significantly inhibit cell proliferation irrespective of trastuzumab treatment.
Treated category
The siRNAs that potentiated the inhibitory effects of trastuzumab on HER2+ cells included those targeting several genes acting downstream of PI3K, including PDPK1, AKT1 and COASY13 (Figure 5c). Gene enrichment analysis showed enrichment for genes involved in AKT signaling (P=0.008) and MTOR signaling (P=0.01) pathways (Figure 5c, Supplementary Table S10). In addition to annotation by DAVID, other hits were PTPRJ, a phosphatase and negative regulator of AKT signaling;23 RASGRF2, a guanine exchange factor (GEF) activating both RAS and the cytoskeleton protein RAC1; the anchor-associated protein regulating PKA (AKAP1); and DGKB, a regulator of DAG signaling that also acts downstream of G-protein coupled receptors. Functional interaction partners of the hits showed enrichment for pathways signaling by nerve growth factor and the regulation of glucose uptake and transport (Supplementary Figure S7). These data suggest that targeting downstream elements of the PI3K pathway can potentiate the effects of trastuzumab treatment.
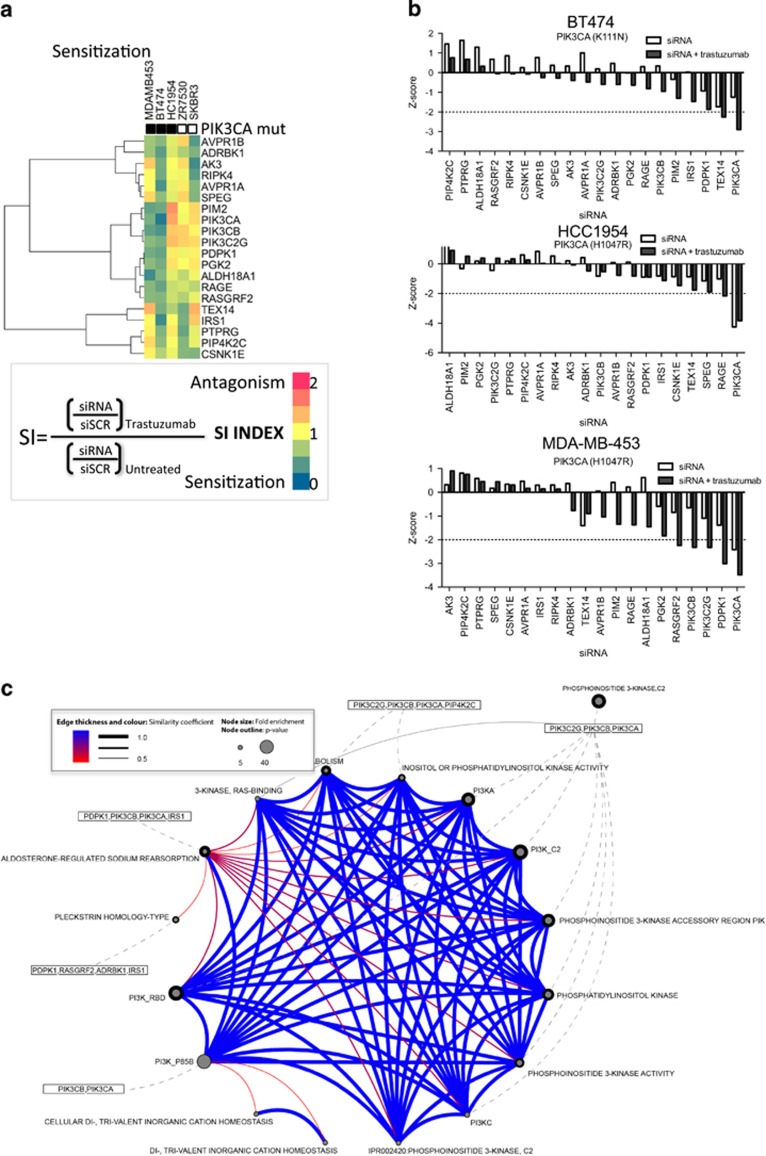
Sensitization to trastuzumab
To confirm our identification of siRNAs that sensitize HER2+ cells to trastuzumab, we used a ‘sensitization index' (SI) that could pinpoint highly efficient sensitizer siRNAs. Using an established ratio,24 we defined siRNAs that strongly sensitized HER2+ cells to trastuzumab treatment as having an SI<0.75 in two or more cell lines (Figures 6a and b, Supplementary Table S11). A group of siRNAs with SI values lower than this threshold was tabulated for HER2+ cell lines and gene enrichment was performed (Supplementary Table S12). The siRNAs best able to sensitize HER2+ cells to trastuzumab treatment were those targeting genes encoding elements of the PI3K pathway (PIK3C2G, PIK3CA, PIK3CB, PIP4K2C and PDPK1) and Ca2+ signaling pathways (AVPR1A/B, ADRBK1, IRS1 and PIK3CB) (Figure 6c, Supplementary Table S12). No hits were common ‘sensitizers' among all five cell lines. Many PI3K-related hits were found among cell lines with PIK3CA mutations (Figure 6b, Supplementary Figure S8). When SI hits were compared with the best 14 ‘treated' of trastuzumab-mediated inhibition of proliferation as measured by the top 5% ranked Z-scores in Figure 5c, five hits overlapped with the hits identified using the SI method (CSNK1E, PDPK1, RAGE, RASGRF2 and TEX14). Both approaches were thus successful in identifying siRNAs that can sensitize HER2+ cells to trastuzumab treatment, and their overlapping results reinforce our confidence in their validity.
Figure 6.
Inhibition of PI3K kinases results in sensitization to trastuzumab treatment. (a) Heatmap of siRNAs that sensitize cells to trastuzumab treatment, as defined by an SI value of <0.75. (b) Proliferation effect of ‘sensitization' hits in HER2+ breast cancer cell lines with mutations in PIK3CA. (c) GO and pathway enrichment analysis (DAVID) of sensitization hits shows enrichment for PI3K activity (P<0.05).
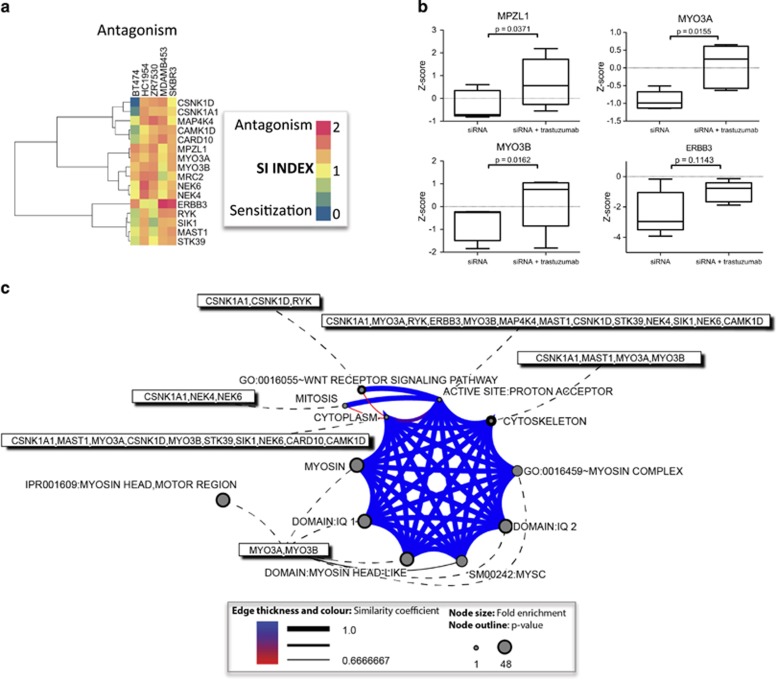
Identification of kinases whose inhibition results in resistance to or antagonism of trastuzumab
To identify kinases whose inhibition lessened the effect of trastuzumab treatment, we used the SI index.24 Using the established ratio of SI>1.25 in three or more cell lines, we identified antagonistic hits to trastuzumab (Figure 7a, Supplementary Table S13). No siRNA showed antagonism among all cell lines tested (Figure 7a). The strongest antagonism among all cell lines was seen among MPZL1, MYO3A and MYO3B (Student's t-test, P<0.05) (Figure 7b, Supplementary Figure S9). MPZL1 is a cell surface receptor. Its activity is dependent on both its cytoplasmic immunoreceptor tyrosine inhibitory motif and its interaction with the tyrosine protein phosphatase, src homology phosphatase-2.25, 26 Class III myosins (MYO3A and MYO3B) are unique members of the myosin superfamily in that they contain both a motor and kinase domain.27 Gene enrichment analysis and functional annotation of the highlighted genes involved Wnt receptor signaling (CSNK1A1, CSNK1D and RYK), mitosis (NEK4, NEK6 and CSNK1A1), myosin signaling (MYO3A and MYO3B) and cytoskeleton (CSNK1A1, MAST1, MYO3A and MYO3B) (Figure 7c, Supplementary Table S14). These data suggest that mechanisms of resistance to trastuzumab treatment vary between cell lines.
Figure 7.
Inhibition of MPZL1, MYO3A and MYO3B kinases results in antagonism to trastuzumab treatment. (a) Heatmap of siRNAs that antagonize trastuzumab treatment, as defined by an SI value of >1.25. (b) Boxplots of siRNA that are antagonistic to trastuzumab treatment (Student t-test, significance P<0.05). (c) GO and pathway enrichment analysis (DAVID) of antagonism hits shows enrichment for myosin proteins, Wnt receptor, mitosis and the cytoskeleton (P<0.05).
Discussion
HER2 and HER3 are both important for HER2+ cell proliferation
Our comprehensive screen was designed to identify kinases whose siRNA-mediated inhibition preferentially reduced the proliferation of HER2+ compared with non-HER2-amplified breast cancer cell lines. Our analysis pinpointed both HER2 and HER3 as prominent hits (Figure 3a), in that the siRNAs targeting these genes were able to independently and selectively decrease HER2+ cell proliferation. This finding suggests that both HER2 and HER3 have essential roles in driving the oncogenic proliferation of HER2+ breast cancers. Our findings are therefore consistent with previous work showing that HER3 is essential for HER2 receptor signaling.28
HER2 and HER3 are both required for trastuzumab effectiveness
Although both HER2- and HER3-targeting siRNAs were able to inhibit HER2+ cell proliferation in the absence of trastuzumab, these agents were not as effective in the presence of trastuzumab (Figure 5a). The identification of HER2 as a hit only in the absence of trastuzumab suggests that the mechanism of action of trastuzumab cannot operate when HER2 expression is downregulated. The identification of HER3 as a strong target only in the absence of trastuzumab was somewhat unexpected, since Ghosh et al.29 have previously proposed that trastuzumab binds only to HER2 homodimers. We had therefore expected that loss of HER3 would remain a strong inhibitor of proliferation even in the presence of trastuzumab. Instead, we found that siRNA-mediated loss of HER3 even antagonized the anti-proliferative effects of trastuzumab (Figure 7), suggesting that HER3 signaling is also required for trastuzumab effectiveness. These results imply that although trastuzumab may bind preferentially to HER2 homodimers, it does not act exclusively on these structures. In support of our argument, studies by Juntilla et al.30 have suggested that at least part of trastuzumab's effect is due to the disruption of the HER2/HER3/PI3K complex.
Tumor cells can be sensitized to trastuzumab by targeting elements downstream of PI3K
Our sensitizer screen was designed to identify kinases whose siRNA-mediated inhibition sensitized HER2+ cells to trastuzumab treatment. We pinpointed several downstream targets of PI3K signaling that showed stronger inhibition upon treatment with trastuzumab (‘treated') (Figure 5c). Many of these kinases act directly downstream of PI3K, including PDPK1 and AKT1. PDPK1 and AKT1 are frequently activated in HER2+ breast cancers.31, 32, 33 The acetyl-CoA synthase COASY, which binds to the regulatory unit of PI3K and influences its downstream signaling,13 was present as well in this group. Our SI ‘sensitization' approach also identified many PI3K subunits, as well as elements of the PI3K pathway potentiated the inhibitory effects of trastuzumab particularly in PIK3CA mutated cell lines (Figure 6). Thus, despite the fact that PIK3CA itself emerged from the ‘neutral' screen, we believe that PIK3CA strongly influences signaling downstream of the HER2/PI3K-mediated signaling driving HER2+ breast cancer cell proliferation.
PIK3CA mutations have been previously associated with HER2 overexpression and are correlated with poor patient outcome.34, 35 TCGA data have shown that 42% of HER2 subtype breast cancers have mutations in PIK3CA.22 Interestingly, we found that PI3K-activating mutations were strongly represented among the HER2+ cell lines screened in our study (Supplementary Table S1). Overall, our findings indicate that PI3K has a vital role in oncogenic signaling downstream of HER2, and that the targeting of PI3K or its downstream mediators must be combined with HER2/HER3 inhibition to successfully block HER2+ cell proliferation. Thus far, murine xenograft models have shown that mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies.36 Garnett et al.37 have shown that the combination inhibition of HER2-HER3 dimer, in addition to a PIK3 inhibition, blocks the PI3K signaling and growth of HER2+ breast cancers in murine xenograft models.
A caveat of any PI3K inhibition strategy is that prolonged PI3K shutdown triggers a negative feedback loop that activates AKT,38 an event that could restore cancer cell proliferation. In addition, extended PI3K inhibition in cultured HER2-overexpressing cells leads to increased dependence on extracellular signal-regulated kinase (ERK) signaling, as well as upregulation and activation of HER3.39 Taken together, these observations suggest that the combined inhibition of HER2, HER3 and downstream PI3K signaling elements is necessary to ensure abrogation of the proliferation of HER2+ cancer cells.
MAPK signaling is downstream of HER2 signaling
We identified several siRNAs that decreased cancer cell proliferation only in the absence of trastuzumab (‘untreated', Figure 5a). We speculated that many of these siRNAs inhibited kinases that were direct targets of trastuzumab (HER2 and HER3) or downstream of HER2. We propose that the siRNA blocking these molecules may have a growth inhibitory effect similar to that of trastuzumab. Among these siRNAs were several targeting members of MAPK signaling pathways, including MAP2K3/MKK3, MAPKAPK3 and RPS6KA2/p90RSK. Previous work has established that MAP2K3 is activated in response to mitogens and environmental stress stimuli and subsequently phosphorylates p38. MAPKAPK3, the second MAPK-related kinase identified in our screen, is also involved in mitogen and stress stimulation, and is phosphorylated by ERK, p38 MAPK and c-Jun N-terminal kinase (JNK).40 RPS6KA2/p90RSK is a downstream effector of ERK signaling. These data suggest that the HER2-containing complexes targeted by trastuzumab depend on downstream MAPK signaling to drive cancer cell proliferation.
The inhibition of the MAP2K3 protein has been previously observed to preferentially block the growth of HER2+ breast cancers.41 Our screen confirmed that siRNA targeting MAP2K3 strongly impaired the proliferation of three HER2+ cell lines, but our work unexpectedly revealed that the growth of triple-negative and ER+ breast cancer cell lines is also reduced when MAP2K3 is inhibited (Supplementary Table S3).
The identification of these particular siRNAs as hits in the sensitizer screen suggests that, in the presence of trastuzumab, these downstream components of the MAPK signaling pathway may already be downregulated. Thus, siRNA-mediated knockdown of these proteins would not enhance any growth inhibition caused by trastuzumab. Our data also imply that p38/JNK/ERK signaling downstream of HER2 may have a greater role in breast cancer cell proliferation than previously thought, and that trastuzumab likely targets and suppresses stress response MAPK signaling. More work is needed to better understand the p38/JNK/ERK signaling pathway in this context and whether its deregulation contributes to trastuzumab resistance.
siRNA that causes resistance to trastuzumab
The identification of kinases whose siRNA-mediated inhibition caused resistance to trastuzumab proved to be difficult to achieve. The siRNAs that were the strongest stimulators of HER2+ cell proliferation on their own were unaffected by the addition of trastuzumab (Supplementary Table S15). Antagonism by SI (SI>1.25) identified the knockdown of MPZL1, MYO3A and MYO3B resulting in increased proliferation of trastuzumab-treated HER2+ cells. Functional annotation of the antagonism results showed enrichment for myosin proteins, Wnt receptor signaling, mitosis and cytoskeleton. The MYO3A and MYO3B genes code for actin-based motor proteins with protein kinase activity.42 These myosins interact via their myosin head domain with actin filaments and link signal transduction to the actin cytoskeleton.43 Myosin proteins have yet to be associated with HER2 breast cancer signaling. Our combined results suggest that the moderate inhibitory effect on HER2+ cell proliferation caused by the loss of myosin proteins or changes in the Wnt receptor signaling via casein kinase 1 can be reverted to induce the proliferation when cells are treated with trastuzumab.
Conclusion
The results of this study have highlighted the dynamic nature of signaling downstream of HER2, signaling that can greatly influence the outcome of treatment of HER2+ breast cancers. Our data indicate that it will be important to examine how different pharmacologic HER2 inhibitors affect downstream HER2 signaling to maximize their growth inhibitory effects on HER2+ breast cancers. Our findings also demonstrate the crucial roles of signaling downstream of PI3K activation and stress-induced MAPK activation to HER2+ cell proliferation. Importantly, we have shown that HER2+ breast cancer signaling relies not only on both HER2 and HER3 but also on PI3K and MAPK pathway activation. Thus, we have identified signaling pathways that are potentially dominant in HER2+ breast cancer cell lines and thus prime candidates for inhibitory targeting. New strategies for combined inhibition of multiple signaling pathways downstream of various HER2 dimerization partners may have the potential to more effectively treat HER2+ breast cancers.
Materials and methods
Breast cell lines
Twelve breast cancer cell lines (HER2+: BT474, SKBR3, MDA-MB-453, HCC1954; ER+: CAMA-1, MCF7, T47D; ER−/PR−/HER2−: MDA-MB-231, MDA-MB-435, MDA-MB-468, HS578T) and two normal breast cell lines (MCF10A and MCF12A) were obtained from ATCC (Manassas, VA, USA) and maintained according to the supplier's instructions. Cell line seeding density was determined empirically, and cells were passaged as needed.
siRNA kinase library
The ‘comprehensive' and ‘sensitizer' screens were performed using a SMARTpool siRNA kinase library from Dharmacon (G-003500-20, Lafayette, CO, USA) that contained 778 siRNA pools comprising 518 kinases and 260 ‘kinase-related genes'. Each siRNA pool consisted of four distinct siRNA species targeting different sequences of a single kinase target transcript. The positive control for cell death in the screen was siTOX (D-001500-01; Dharmacon), a proprietary siRNA that targets genes responsible for apoptosis and cell death. The negative control was ON-TARGET siSCR (D-001810-10; Dharmacon). It should be noted that, since our purchase of this library in 2005, 49 annotated genes for which siRNAs were present in the library have lost their NCBI accession numbers. These genes are indicated with an asterisk in the data analyses.
siRNA transfection and sulforhodamine B proliferation assay
Using the Tecan Freedom EVO robotic workstation (Mannedorf, Switzerland), exponentially growing cells were plated in 96-well plates at 30% confluence in 80 μl of medium as recommended by ATCC. Cells were allowed to adhere overnight and then transfected with 20 pmol of a single siRNA library pool suspended in 20 μl OPTI-MEM with 0.1 μl Lipofectamine2000 (LF2000; Life Technologies, Carlsbad, CA, USA). Each 96-well plate contained 80 siRNA pools; 3 negative, non-targeting scrambled control (siSCR) wells; and 3 positive control (siTOX) wells. After overnight incubation, the medium in all wells was changed. For the sensitizer screen, trastuzumab (10 μg/ml) was added to the appropriate wells. At 4 days post transfection, cells were fixed with 10% trichloroacetic acid and stained with sulforhodamine B (Sigma-Aldrich, St Louis, MO, USA) to quantitate total cellular protein content, which was taken as a measure of cell proliferation.44
Normalization of screen data
To account for possible plate-to-plate differences during siRNA library transfection, sulforhodamine B absorbance results were normalized to the values of the siTOX and siSCR wells of each plate. Normalized data were log transformed and the median and median absolute deviation (mad) were used to compute the Z-score of each kinase in relation to the results for the entire 778 kinase library for a particular cell line.11
Clustering methods
Data were subjected to hierarchical clustering using a Pearson correlation measure and a complete linkage-clustering algorithm with Cluster 3.0 (Michael Eisen, Stanford University, Stanford, CA, USA). Heatmaps of Z-scores were drawn using Java TreeView.45
Candidate gene selection
CMS analysis based on cell line characteristics was performed using Gene Pattern software (MIT, Boston, MA, USA). Supervised analyses identified genes belonging to one of two groups (for example, HER2 amplified vs non-HER2 amplified). The absolute difference between the medians of the two groups was calculated. Permutation testing was used to determine if the observed difference between the sample medians was large enough to reject the null hypothesis, which was that the two groups came from the same probability distribution. A cutoff value of P<0.05 was used. No corrections were made for multiple comparisons.
Enrichment networks
Gene enrichment and functional analyses were completed using DAVID version 6.716, 17 with the Dharmacon SMARTpool 778 kinase siRNA library as the background set. Enriched terms (in our case, siRNAs) were considered significant when P<0.05. Multiple comparison values were included but not used in making cutoff decisions because of the size of the background set.
Enrichment analyses were visualized in network interaction diagrams using NAViGaTOR.46, 47 Node outline indicated P-values, size of node represented fold enrichment of class terms. Edges describe the relationships between enriched terms using the overlap coefficient.48
Protein–protein interaction networks
Protein–protein interaction networks were visualized using NAViGaTOR version 2.3.1.46, 47 Both the Interologous Interaction Database (I2D; http://ophid.utoronto.ca/i2d) version 249, 50 and Reactome Functional Interactions 2012 (functional interactions predicted from Reactome)51 were used to define protein–protein interaction networks. Interaction evidence from two or more sources was used to highlight edges (thick edge). Screen hits were used as primary nodes, and interactions joining primary nodes and their neighbors served as the basis for the analysis. Shortest paths were calculated between all primary nodes. Nodes falling on shortest paths of length 1 or 2 were highlighted, and remaining edges were made partially transparent to reduce network density. To map the remaining disconnected primary nodes, genes on the shortest path connecting them to an existing node in the network were included.
Acknowledgments
We thank Dr Mary Saunders for insightful scientific editing. VL was supported by a doctoral fellowship from the Canadian Institutes of Health Research and a PhD fellowship from the Canadian Breast Cancer Foundation—Ontario Chapter. EAS and IJ were supported in part by grants from Ontario Research Fund (GL2-01-030), Canada Foundation for Innovation (CFI #12301, #203373, #29272, #225404) and the Canada Research Chair Program (CRC #203373 and #225404).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Sjogren S, Inganas M, Lindgren A, Holmberg L, Bergh J. Prognostic and predictive value of c-erbB-2 overexpression in primary breast cancer, alone and in combination with other prognostic markers. J Clin Oncol. 1998;16:462–469. doi: 10.1200/JCO.1998.16.2.462. [DOI] [PubMed] [Google Scholar]
- Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61:1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- Venter DJ, Tuzi NL, Kumar S, Gullick WJ. Overexpression of the c-erbB-2 oncoprotein in human breast carcinomas: immunohistological assessment correlates with gene amplification. Lancet. 1987;2:69–72. doi: 10.1016/s0140-6736(87)92736-x. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Graus-Porta D, Beerli RR, Rohrer J, Gay B, Hynes NE. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol. 1998;18:5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peles E, Levy RB, Or E, Ullrich A, Yarden Y. Oncogenic forms of the neu/HER2 tyrosine kinase are permanently coupled to phospholipase C gamma. EMBO J. 1991;10:2077–2086. doi: 10.1002/j.1460-2075.1991.tb07739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltoff SP, Carraway KL, 3rd, Prigent SA, Gullick WG, Cantley LC. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol. 1994;14:3550–3558. doi: 10.1128/mcb.14.6.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorns E, Turner NC, Elliott R, Syed N, Garrone O, Gasco M, et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 2008;13:91–104. doi: 10.1016/j.ccr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screen. 2011;16:775–785. doi: 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- Mirouse V, Swick LL, Kazgan N, St, Johnston D, Brenman JE. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Breus O, Panasyuk G, Gout IT, Filonenko V, Nemazanyy I. CoA synthase is in complex with p85alphaPI3K and affects PI3K signaling pathway. Biochem Biophys Res Commun. 2009;385:581–585. doi: 10.1016/j.bbrc.2009.05.102. [DOI] [PubMed] [Google Scholar]
- Ludford-Menting MJ, Thomas SJ, Crimeen B, Harris LJ, Loveland BE, Bills M, et al. A functional interaction between CD46 and DLG4: a role for DLG4 in epithelial polarization. J Biol Chem. 2002;277:4477–4484. doi: 10.1074/jbc.M108479200. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Sand C, Jakobs KH, Michel MC, Weernink PA. Epac and the cardiovascular system. Curr Opin Pharmacol. 2007;7:193–200. doi: 10.1016/j.coph.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Clague MJ, Prior IA. Phosphatome profiling reveals PTPN2, PTPRJ and PTEN as potent negative regulators of PKB/Akt activation in Ras-mutated cancer cells. Biochem J. 2010;426:65–72. doi: 10.1042/BJ20091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, Marani M, Pardo O, Warne PH, Kelly G, Sahai E, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Zannettino AC, Roubelakis M, Welldon KJ, Jackson DE, Simmons PJ, Bendall LJ, et al. Novel mesenchymal and haematopoietic cell isoforms of the SHP-2 docking receptor, PZR: identification, molecular cloning and effects on cell migration. Biochem J. 2003;370:537–549. doi: 10.1042/BJ20020935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubelakis MG, Martin-Rendon E, Tsaknakis G, Stavropoulos A, Watt SM. The murine ortholog of the SHP-2 binding molecule, PZR accelerates cell migration on fibronectin and is expressed in early embryo formation. J Cell Biochem. 2007;102:955–969. doi: 10.1002/jcb.21334. [DOI] [PubMed] [Google Scholar]
- Komaba S, Inoue A, Maruta S, Hosoya H, Ikebe M. Determination of human myosin III as a motor protein having a protein kinase activity. J Biol Chem. 2003;278:21352–21360. doi: 10.1074/jbc.M300757200. [DOI] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Narasanna A, Wang SE, Liu S, Chakrabarty A, Balko JM, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011;71:1871–1882. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Tokunaga E, Kimura Y, Oki E, Ueda N, Futatsugi M, Mashino K, et al. Akt is frequently activated in HER2/neu-positive breast cancers and associated with poor prognosis among hormone-treated patients. Int J Cancer. 2006;118:284–289. doi: 10.1002/ijc.21358. [DOI] [PubMed] [Google Scholar]
- Park SS, Kim SW. Activated Akt signaling pathway in invasive ductal carcinoma of the breast: correlation with HER2 overexpression. Oncol Rep. 2007;18:139–143. [PubMed] [Google Scholar]
- Lin HJ, Hsieh FC, Song H, Lin J. Elevated phosphorylation and activation of PDK-1/AKT pathway in human breast cancer. Br J Cancer. 2005;93:1372–1381. doi: 10.1038/sj.bjc.6602862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, et al. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110alpha inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer Res. 2013;73:6013–6023. doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, et al. 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol Cell Biol. 1996;16:6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, et al. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero OA, Moore JE, Unrath WC, Manor U, Salles FT, Grati M, et al. Intermolecular autophosphorylation regulates myosin IIIa activity and localization in parallel actin bundles. J Biol Chem. 2010;285:35770–35782. doi: 10.1074/jbc.M110.144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler M. Are class III and class IX myosins motorized signalling molecules. Biochimica et Biophysica Acta. 2000;1496:52–59. doi: 10.1016/s0167-4889(00)00008-2. [DOI] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natil Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, et al. NAViGaTOR: network analysis, visualization and graphing Toronto. Bioinformatics. 2009;25:3327–3329. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrello C, Otasek D, Fortney K, Agapito G, Cannataro M, Shirdel E, et al. Visual data mining of biological networks: one size does not fit all. PLoS Comput Biol. 2013;9:e1002833. doi: 10.1371/journal.pcbi.1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.