Abstract
Primary sex determination “switches” evolve rapidly, but Doublesex (DSX) related transcription factors (DMRTs) act downstream of these switches to control sexual development in most animal species. Drosophila dsx encodes female- and male-specific isoforms (DSXF and DSXM), but little is known about how dsx controls sexual development, whether DSXF and DSXM bind different targets, or how DSX proteins direct different outcomes in diverse tissues. We undertook genome-wide analyses to identify DSX targets using in vivo occupancy, binding site prediction, and evolutionary conservation. We find that DSXF and DSXM bind thousands of the same targets in multiple tissues in both sexes, yet these targets have sex- and tissue-specific functions. Interestingly, DSX targets show considerable overlap with targets identified for mouse DMRT1. DSX targets include transcription factors and signaling pathway components providing for direct and indirect regulation of sex-biased expression.
Introduction
Genetically encoded sexual dimorphism allows males and females to differ in appearance, physiology, and behavior. Differences in gamete morphology and systems that ensure they meet are often obvious, but there are subtle aspects of sex differentiation impacting organs and physiology throughout the body. Controlling the sexual development of a broad range of cell types is a challenge since sex-biased gene expression advantageous in one tissue may be detrimental in another. Sex determination systems must therefore provide organism-level, sex-specific modulation of gene expression simultaneously compatible with a range of tissue-specific requirements. Sex-specific and tissue-specific gene expression must be tightly integrated, but how this occurs is not well understood.
Primary sex determination signals vary, but doublesex and mab-3 related transcription factors (DMRTs) control sex determination and differentiation in many species (Zarkower, 2013). For example, XY humans with deletions of 3 DMRT genes exhibit sex reversal (Raymond et al., 1999). In Drosophila melanogaster, doublesex (dsx) is required for sexually dimorphic morphology, physiology, and behavior. Transformer (TRA) and Transformer 2 (TRA2) regulate female-specific alternative splicing of dsx to encode DSXF protein. Without TRA, male-specific splicing of dsx pre-mRNA occurs, and this transcript encodes DSXM (Burtis and Baker, 1989; Nagoshi et al., 1988). The DSXF and DSXM isoforms have the same DNA-binding and dimerization domains but have different C-termini (Bayrer et al., 2005; Zhang et al., 2006). Intersex (IX) binds the C-terminus of DSXF and is required for DSXF function (Yang et al., 2008) suggesting that the sex-specific C-termini are effector domains interacting with co-factors to modulate gene expression. DSXF and DSXM are required for proper sexual development, and loss of dsx function results in an intersexual phenotype. DSXF and DSXM have opposing effects on gene expression (Coschigano and Wensink, 1993). Thus, expressing both isoforms in the same fly results in an intersexual phenotype similar to dsx loss of function (Nagoshi and Baker, 1990).
In addition to regulation by alternative splicing, dsx is expressed highly tissue-specifically indicating that cells are on a “need to know” basis for sex (Hempel and Oliver, 2007; Lee et al., 2002; Rideout et al., 2010; Robinett et al., 2010). dsx is expressed in subsets of neurons, gut cells, gonadal somatic cells, and in adipose and hepatic tissues. These cell types derive from all primary germ layers and have diverse roles in metabolism, gametogenesis, morphology, and behavior. While the transcriptional inputs to dsx expression are not fully understood, Drosophila HOX and other patterning genes regulate dsx in at least some tissues (Foronda et al., 2012; Tanaka et al., 2011; Wang et al., 2011; Wang and Yoder, 2012; Yoder, 2012).
Although DSX has been studied for 50 years, there are still few defined DSX targets and these cannot fully explain the sexually dimorphic morphologies and behaviors regulated by dsx. The known DSX target genes were identified on a case-by-case basis (Burtis et al., 1991; Shirangi et al., 2009; Williams et al., 2008). There have been large numbers of genome-wide expression studies on the sexes but few attempts to link this expression directly to DSX (Lebo et al., 2009). One study identified genes with sex-biased and dsx-dependent expression in genital discs, but did not address whether these were directly or indirectly regulated (Chatterjee et al., 2011). DSXF occupancy was examined genome-wide and filtered using a precise 13-mer to predict 23 direct target genes (Luo et al., 2011), but did not capture known DSX targets and is therefore unlikely to be complete. We combined an extensive DSX occupancy study on both DSXF and DSXM isoforms in multiple tissues with comparative genomic analyses (20 Drosophila species and mouse), expression profiling of a tissue during an acute switch in DSX isoform, and an unbiased dsx genetic interaction screen. We also determined the roles of predicted DSX targets in dsx-expressing cells. Our analyses reveal that DSX is bound to many of the same targets in males and females and in different tissues indicating that DSX action is regulated downstream of DSX binding. Further, we find a striking conservation of DSX targets in the Drosophila genus including orthologs of mouse DMRT1 targets (Murphy et al., 2010) suggesting that control of sexual dimorphism may be similar in diverse animal species.
Results
DSX occupancy
To determine where DSX binds in the D. melanogaster genome, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) on S2 cells expressing tagged DSXM or DSXF. We also performed DSXM or DSXF DNA adenine methyltransferase identification (DamID) on adult ovary and adult female and male fat body in transgenic flies followed by either sequencing (DamID-seq) or hybridization to microarrays (DamID-chip). We chose adult fat body and ovary since dsx plays a role in maintaining sexually dimorphic gene expression in both organs. We confirmed nuclear expression of tagged DSX and unfused Dam control by immunohistochemistry (Figure S1). Further, expression of Dam-dsxF in males using dsx-GAL4 feminized appropriate tissues (e.g. sex combs, reproductive tract, and gonads) indicating these constructs were functional (Figure S1, over expression of Dam-dsxM was lethal). We conducted DamID experiments using low, basal expression in the absence of a GAL4 driver to avoid known toxicity associated with Dam expression and artifacts due to DSX over expression. For all samples, we explored the continuous distribution of DSX occupancy using background-subtracted values to control for general chromatin accessibility. We identified peaks of occupancy using a stringent 1% FDR cutoff (Supplemental Experimental Procedures).
The first occupancy analysis step was at the level of peaks. We expected DSX occupancy near known DSX targets (Figure 1A,B). Indeed, the (Yolk Protein 1) and 2 loci (Yp1 and Yp2) showed strong DSX occupancy in the fat body and ovary, where these genes are expressed at high levels, along with weak occupancy in S2 cells. In contrast, the bric-a-brac 1 (bab1) locus showed strong DSX occupancy in all samples. While we observed occupancy at the previously identified Yp1/2 and bab1 DSX response elements, we also found a strongly occupied region upstream of bab1 that may represent an additional DSX-dependent enhancer.
Figure 1. DSX occupancy and binding sites.
(A–B) Scaled read density plots (background subtracted, arbitrary scale) from five replicated occupancy experiments (as labeled) for (A) the Yp1, Yp2, and (B) bab1. FlyBase gene models showing transcription start sites (bent arrows), coding exons (thick rectangles), non-coding regions (thin rectangles), introns (lines), and known DSX response elements (DSX-RE). (C) Heatmap of k-means clustering of background-subtracted, ranked occupancy scores (color scale on the left) for all D. melanogaster genes (optimal k value k = 5). (D) Box plots of gene-level occupancy scores averaged from 6 occupancy data sets in each occupancy cluster.
We next associated DSX binding sites with nearby genes and generated a DSX occupancy score. Yp1/2 and bab1 were typical examples of DSX occupancy patterns with a strong preference for occupancy in the gene body +1 kb upstream of transcription start (Figure S2). Therefore, we assigned DSX peaks to genes either using peaks occurring within this region or by using a 2 kb window centered on the annotated transcription start. Both methods limit artificial contributions of nearby upstream genes; however, the fixed-range method uncouples gene length from occupancy yet misses binding at many intronic enhancers. The gene body +1 kb definition captures intronic enhancers, but biases towards longer genes. We elected to use gene body +1 kb as this captured genes with intronic enhancers, such as bab1; however, both methods produced similar results (Table S1).
We determined gene occupancy strength using the strongest peak (peakmax) or the sum of all peaks (peaksum). We elected not to normalize for gene length as it introduced bias against long genes, such as bab1, with discrete, strong DSX binding. The two occupancy strength methods produced similar results (Spearman’s r > 0.9), and we chose the peaksum method to favor genes with multiple regions of strong occupancy.
There are many ways to examine the gene/sample relationships in DSX occupancy patterns. Supervised clustering (k-means, where k = 5) of genes’ ranked occupancy scores (Figure 1C, Table S1) revealed clusters of DSX occupancy patterns among genes that exhibit very low (cluster 4), tissue non-specific (clusters 3 and 5), and tissue- and/or technique-specific occupancy (clusters 1 and 2). In this analysis, the bona fide DSX target bab1 was in cluster 5 while the Yp1/2 genes were in cluster 3 due to modest occupancy in S2 cells. Genes ranking in the top 10% of occupancy were almost exclusively in cluster 5. Genes outside of clusters 3 and 5 had low occupancy values, although there were a few with strong occupancy in each cluster (Figure 1D).
Interestingly, DSXF and DSXM proteins had similar occupancy patterns (Figure 1C), suggesting the sex-specific effector domains and sex-biased chromatin environments had little impact on where DSX binds. However, there are transcriptional “hotspots” that are known to bind a host of different factors (Negre et al., 2011). To determine if the tissue non-specific occupancy and common DSXM/F patterns were due to non-specific binding at accessible chromatin or hotspots, we correlated our results with modENCODE occupancy experiments and found that DSXF and DSXM occupancy pattern similarity is not explained by either chromatin accessibility or hotspots (Figure S3). Additionally, removing peaks associated with hotspots prior to analysis did not influence the overall occupancy patterns (not shown).
We conclude that the strongest DSX binding occurs in a largely sex and tissue non-specific manner. This observation focused our attention on genes following this pattern, but there are genes with tissue-specific or isoform-specific occupancy patterns that may be extremely interesting for future work. Since DSX has diverse roles in different sexes and tissues, focusing on these genes allowed us to address a previously unexplored question of how DSX integrates with tissue-specific factors rather than regulation simply by where DSX binds.
Sequence analysis of DSX binding sites
We hypothesized that the observed occupancy pattern would be due to direct DSX binding while other contacts may be indirect due to 3D structures such as looping. One simple prediction is that DSX-occupied regions should contain a DSX binding site. DSX DNA binding specificity has been defined biochemically (Erdman et al., 1996; Murphy et al., 2007; Yi and Zarkower, 1999). We were able to identify a motif statistically similar to the DSX PWM (Tomtom E-value<0.01) by de novo motif finding under occupied ChIP- seq regions (MEME-ChiP E-value<0.01), as well as enrichment of sequences matching the DSX PWM under peaks (p<0.01; Fisher’s Exact Test).
A major problem with transcription factor studies is that binding sites are common in the genome and can be bound in both functional and non-functional contexts (Fisher et al., 2012). To enhance predictions of functional binding sites, we used comparative genomics to analyze DSX binding site conservation among 20 species of Drosophila (Adams et al., 2000; Chen et al., 2014; Drosophila 12 Genomes et al., 2007; Richards et al., 2005). While conservation is not always predictive of function (Villar et al., 2014), and some non-conserved sites may be interesting species-specific targets, evolutionarily conserved sites are likely to regulate the vast array of genes showing sex-biased expression in the genus (Chen et al., 2014; Zhang et al., 2007).
The dsx sex-specific splicing pattern and encoded DNA binding domain was highly conserved across ~34 million years of Drosophila evolution (Figure S2). Therefore, we used the same biochemically-defined DSX PWM and 100 position shuffled PWMs as controls to scan the D. melanogaster genome and 19 other species in the Drosophila genus. We extracted the D. melanogaster sites using the same gene body +1 kb definition above except that we excluded coding sequence to avoid confounding DSX site and codon conservation. We assigned each identified DSX or control site in D. melanogaster a conservation index (CI) score based on the evolutionary distance at which sites could still be identified in the homologous gene using a combination of sequence and distance from the first coding exon (Table S2). Gene-level CI scores were calculated by summing site-level CIs (Table S1). We also extracted the well defined and gene length corrected PHylogenetic Analysis with Space/Time (PhastCons) sequences (Siepel et al., 2005) and calculated the mean PhastCons score for DSX sites. Briefly, a high CI or PhastCons score indicates a conserved site or the presence of a de novo site with similar sequence at the same relative position.
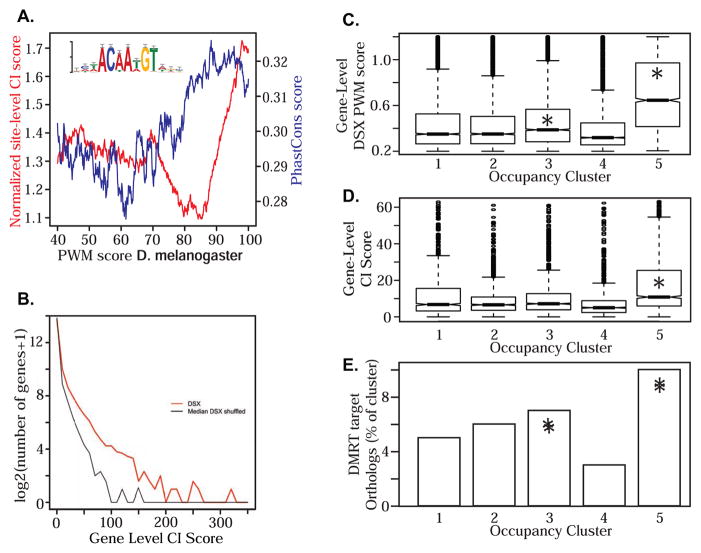
As expected, sites more closely matching the PWM were more likely to have deeper evolutionary conservation (Figure 2A). We observed a clear increase in the correlation between PWM score and normalized site-level CI score with a prominent “break” above the 90th percentile of PWM scores. The PhastCons scores also showed a break but at a lower PWM score. This indicates that strong scoring sites show high evolutionary conservation, and are therefore more likely to be functional. Interestingly, both methods showed poorest conservation in moderately strong D. melanogaster sites. The meaning of this distribution is unclear but may suggest selection against sites with modest affinity for DSX that could potentially result in deleterious sex-specific regulation. At the gene-level, DSX CI scores were significantly more conserved (KS test p < 2.2e-16) across evolutionary distance than shuffled PWM CI scores (Figure 2B). For this study, we chose to focus our attention on genes with conserved DSX sites rather than D. melanogaster-specific due to either species-specific function or chance.
Figure 2. DSX occupancy and binding-site evolution.
(A) Normalized site-level CI scores plotted against PWM percentile rank score (red line) and PhastCons scores for DSX motifs (blue line). (B) Histogram of gene-level conservation index scores for DSX (red line) and the median of 100 shuffled DSX motifs (black line). (C–E) For each occupancy cluster the distribution of gene level DSX PWM scores (C), gene-level CI scores (D), and % genes in each cluster that are orthologs of mouse DMRT1 targets (E) are shown. Significant (p < 0.01) enrichment using Kruskal-Wallis (B,C) or Fisher’s exact tests (D) is indicated (*).
Comparing in vivo occupancy with sequence analysis
We combined datasets to focus on genes most likely to be functional DSX targets. As described above, genes in clusters 3 and 5 had the highest occupancy scores (Figure 1D). Interestingly, genes in clusters 3 and 5 also had significantly higher gene-level PWM scores than other clusters (Figure 2C). Genes in cluster 5 also showed significantly higher gene-level CI scores (Figure 2D), indicating that genes strongly occupied in D. melanogaster had better conservation of DSX binding sites in the Drosophila phylogeny. To determine if occupancy is also conserved, we asked if genes occupied by DSX had orthologs occupied by the mouse DSX ortholog, DMRT1 (Murphy et al., 2010). Strikingly, these orthologs were enriched in DSX occupancy (Figure 2E). This is somewhat surprising given the tremendous differences in sexual dimorphism between species. Perhaps this reflects the fact that DSX/DMRT1 orthologs control sexual dimorphism across the animal kingdom and act primarily in gonads where sexually dimorphic development is more similar in different species. Overall, our occupancy and sequence analysis are strongly concordant. We therefore focused much of our attention on genes with strong occupancy, strong PWM scores, and strong conservation.
Finally, we examined enrichment of gene ontology terms (GO terms) among occupied genes and occupancy clusters (Table S5). We found strong enrichment for many different coherent groups of genes in ontologies supporting the idea that DSX controls a wide-range of pathways and functions.
DSX-regulated expression in fat body
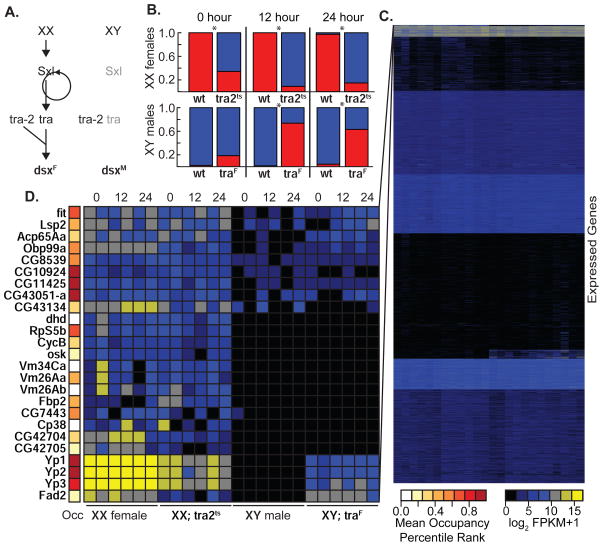
The above indicates that many DSX targets exhibit widespread occupancy independent of sex or tissue. However, to control sex-specific functions of distinct tissues, we expect that DSXF and DSXM should act on a subset of bound genes in any given tissue. To test this hypothesis, we examined expression in the adult fat body where we directly assayed DSX occupancy and where DSX-dependent expression of the Yp1/2 genes occurs. We induced an acute switch in DSX isoform (DSXF to DSXM or vice versa) using temperature-sensitive alleles of tra2 or a heat inducible tra system (UAS-traF; tub-GAL4/tub-GAL80ts) and performed expression profiling by sequencing (RNA-seq) following temperature shifts (Figure 3, Table S1). We reasoned that switching between DSX isoform states would provide a greater net change in expression than loss of DSX function since DSXM and DSXF are thought to have opposing roles in target gene regulation. We measured expression genome-wide and performed k-means clustering to illustrate the overall pattern of expression in fat body (Figure 3C). 25 genes showed the strongest sex-biased expression, but only Yp1/2/3, and Fad2 showed an increase in expression correlating with higher DSXF relative to DSXM (Figure 3D). The Yp1/2 response was expected based on known DSX regulation, thus confirming that we manipulated known DSX outputs. The Fad2 locus encodes a female-specific sterol desaturase involved in sex pheromone signaling (Chertemps et al., 2006) that is directly regulated by DSX in oenocytes (Shirangi et al., 2009). Our data indicate that DSX also regulates Fad2 in the fat body, although we observed poor DSX occupancy raising the possibility of indirect regulation. There were a few genes, such as CG10924, CG11425, CG43051-a, showing sex-biased expression and strong occupancy, whose expression did not change upon DSX isoform switch. Perhaps these genes are regulated by DSX during development or in another context but are dsx-independent in adult fat body. Despite DSX occupancy at thousands of genes in the adult fat body, astonishingly few were transcriptionally regulated by DSX in this tissue. This suggests that many genes are poised to respond to DSX but that additional cues (temporal, spatial, nutritional, and/or hormonal) are also required. We conclude that DSX regulatory specificity depends both on where DSX is bound and the ability to coordinate with other sex-, tissue-, or condition-specific factors.
Figure 3. Tissue-specific DSX function.
(A) Sex determination in female (XX) and male (XY) flies. Functional mRNAs (black) and non-functional mRNAs (grey) are indicated. (B) dsxM (blue) and dsxF (red) mRNA isoform usage in control and experimental (genotypes below) in adult fat body following temperature shifts (time post shift above). Genes are listed (left). Significant differences (p<0.001, Fisher’s Exact Test) are shown (*). (C,D) Heatmap of gene expression (sample order fixed as labeled in D) and genes (rows). (D) The top cluster from (C). Mean occupancy scores (Occ) from fat body DamID-seq and DamID-array samples (color-coded).
Dose-dependent genetic interactions with dsx
If DSX binds to many genes independent of sex or tissue, then we would expect only a subset of targets would be relevant in any given sex and tissue. As a test of this hypothesis, we conducted an unbiased genetic screen to identify genomic regions that interact with dsx (Figure 4A–E, Table S3). To compromise dsx function, we used the dsxD allele which only produces DSXM. Consequently, XX; dsxD/+ animals produce both DSXF (from dsx+) and DSXM (from dsxD) resulting in an intersexual phenotype similar to dsx- (Figure 4A–C, Figure S4). We tested 101 heterozygous deletions of the 2nd chromosome (~33% of the genome) in the XX; dsxD/+ background for modifications of external sexual morphology (i.e. genitalia, abdomen and sex combs) to determine if genetic interactions showed tissue-specificity (Table S3).
Figure 4. Tissue-specific genetic interactions with dsxD.
DSX isoform in wildtype: (A) XX females, (B) XY males, and (C) XX; dsxD/+ intersexes. (D) Feminized XX; dsxD/+ intersexes in Df(2R)BSC109/+. Scanning electron micrographs (SEMs) of genitalia (below) showing a major female feature (vaginal plate, red) and a major male feature (genital arch, blue) in false color. Scale bar = 100μm. (E) The 2nd chromosome with tested regions feminizing (red), masculinizing (blue), feminizing and masculinizing (purple), or having no effect (grey) on the intersexual phenotype in genitalia, abdominal pigmentation (Abd Pig), or sex combs (rows).
These experiments revealed extensive tissue-specific genetic interactions (Figure 4E). For example, in XX; Df(2R)BSC109/+; dsxD/+ flies, all male-like genital structures were missing and female genital structures were more pronounced including a larger, fully-closed vaginal plate replete with teeth (Figure 4D); however, there were no changes in sex comb morphology, tergite number, or abdominal pigmentation. These data suggest that a gene(s) in the Df(2R)BSC109 region is required, in conjunction with dsx, for male genital disc development but not in other tested tissues. Of 101 deletions tested, 19 deficiencies defining 17 unique genomic intervals modified the dsxD/+ external phenotype (Figure 4E). We also observed an enrichment of predicted target genes within regions exhibiting genetic interaction with dsx compared to those that did not (p < 0.01, Fisher’s exact test).
Strikingly, only a single region affected sex differentiation in more than one tissue, and this region includes intersex, which encodes a DSXF-binding protein important for all known aspects of DSXF function (Garrett-Engele et al., 2002). The remaining 16 interacting regions modified the dsxD/+ phenotype in a single tissue. Assuming that regions interacting with dsxD are randomly distributed in the genome, ~50 such “large-effect” regions exist. However, additional loci with smaller effects and loci altering internal sexual morphology, physiology or behavior are likely, suggesting that this is an underestimate. We conclude that genes interacting with dsx do so in a highly tissue-specific manner despite tissue non-specific DSX binding at many genes.
Tissue-specific effects of predicted DSX targets
Since most loci interacting with dsx do so in a highly tissue-specific manner, we wanted to determine if this was true for specific DSX target genes. We selected 80 genes and examined their roles in dsx-expressing tissues using dsx-GAL4 (Rideout et al., 2010; Robinett et al., 2010) to drive UAS-RNAi (Table S4). Genes were selected primarily due to high occupancy, PWM scores, and conservation. We also biased the set to named genes with existing alleles and selected some genes based on other criteria such as localization to a region interacting with dsxD (Table S4). This was not a random screen but still allows us to analyze tissue- and sex-specificity in likely targets.
As in the dsxD interaction screen, we observed striking tissue-specific phenotypes in 16 sexually dimorphic tissues (Table S4). For example, thickveins knockdown (tkvRNAi) or dsxRNAi both resulted in increased male-like abdominal pigmentation in females (Figure 5A), but tkvRNAi had no effect in any other tissue in either sex. In gonads, abdominal-A (abd-ARNAi) females exhibited disorganized ovaries that did not attach to the rudimentary genital tract but no testis phenotype, while bunched (bunRNAi) males had a bulbous testis with no ovary phenotype (Figure 5B). Another clear tissue-specific sex transformation occurred in neuralized (neurRNAi) females (Figure 5C), which had the male-specific large central bristle. In males, chameau (chmRNAi) resulted in pointed sex comb teeth, as observed in females, but sex combs showed male thickness, rotation, and pigmentation (Figure 5C) indicating that multiple pathways regulate the wildtype male sex comb phenotype. In addition to the ovary phenotype, abd-ARNAi females displayed recessed vaginal plates with reduced teeth number. Similarly, bunRNAi males were missing the penis apparatus and most clasper teeth (Figure 5D) in addition to the testis phenotype. Interestingly, we also observed female defects in one tissue and male defects in another. For example, longitudinals lacking (lolaRNAi) females were almost entirely lacking external genitalia, while males had wide, bulbous testes (Table S4). Thus, the RNAi results demonstrate that genes bound by DSX in multiple tissues can have a striking combination of sex- and tissue-specific functions in sex differentiation.
Figure 5. Tissue-specific functions of DSX target genes.
All images left to right. (A) Abdominal pigmentation in wildtype female, male, dsxRNAi female, and tkvRNAi female. (B) Gross anatomy of gonads from wildtype female, abd-ARNAi female, wildtype male, and bunRNAi male. Terminal filaments and hubs (anti-N-Cad in green), somatic gonadal cells (anti-traffic jam, TJ in blue), and germ cells (anti-Vasa in red) are shown. Scale bar = 50μm. (C) First leg tarsal segments from wildtype female, male, neurRNAi female, dsxRNAi male, and chmRNAi male. The male-specific central bristle is indicated (arrowhead). (D) SEMs of genitalia from wildtype female, abd-ARNAi female, wildtype male, and bunRNAi male. Scale bar = 50μm.
DOT1 complex
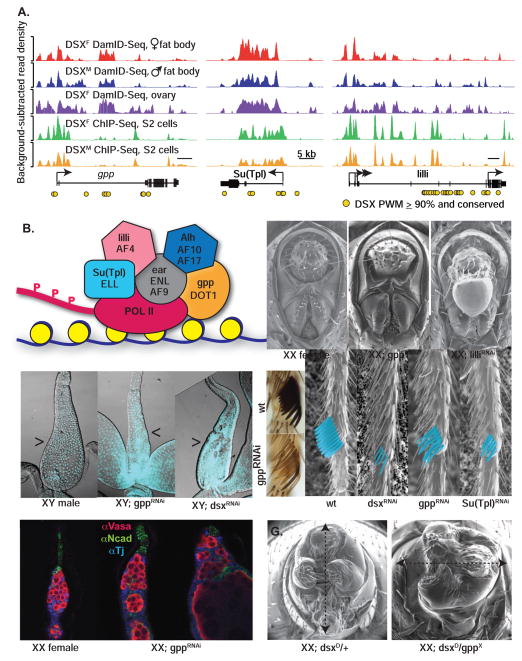
We would expect that multiple genes in a complex co-regulated by DSX would exhibit similar loss of function phenotypes. Many genes encoding the Disruptor Of Telomeric silencing-1 (DOT1) complex(es) are positive transcriptional regulators that methylate histone H3 at lysine 79 (Nguyen and Zhang, 2011) (Figure 6A). In Drosophila, DOT1 is encoded by grappa (gpp) (Shanower et al., 2005) and members of the complexes are encoded by lilliputian (lilli), ENL/AF9-related (ear), Alhambra (Alh), and Supressor of triplolethal (Su(Tpl)) (Figure 6B). We observed strong DSX occupancy at gpp, Su(Tpl), lilli, and Alh, but not ear (Figure 6A, Table S1). DSXF occupancy at Alh and lilli was previously reported (Luo et al., 2011). Furthermore, the DSX binding sites for these genes were well conserved in the Drosophila phylogeny, and the mouse orthologs of Su(Tpl) and lilli are occupied by mouse DMRT1 (Murphy et al., 2010) suggesting DOT1-containing complexes are evolutionarily conserved targets of DMRTs. Given that these proteins function together in a variety of complexes, mutations should result in similar sex-transformation phenotypes.
Figure 6. Function of DOT1 in sex differentiation.
(A) DSX occupancy (see Figure 1) for the gpp, Su(Tpl), and lilli loci (see Figure 1). Positions of high scoring (≥90th percentile PWM score) DSX binding sites conserved in at least one other Drosophila species (yellow circles). (B) Model of DOT1 and associated yeast proteins (capital letters) and Drosophila orthologs (italics) loaded onto elongating RNA polymerase. (C–G) Images left to right unless indicated: (C) SEMs of wildtype, gppRNAi, and lilliRNAi female genitalia with vaginal plate and teeth highlighted (dotted). Scale bar = 100μm. (D) Wildtype, gppRNAi, and dsxRNAi male ejaculatory ducts (arrowheads) stained with DAPI (light blue). Scale bar = 100μm. (E) Wildtype (left top) and gppRNAi XY sex combs (left bottom). SEMs (last four panels) of wildtype, dsxRNAi, gppRNAi, and Su(Tpl)RNAi XY sex combs (teeth false colored). (F) Wildtype and two examples of gppRNAi female germline niches (see Figure 5 for antibodies). Scale bar = 10μm. G) XX; dsxD/+ control and XX; dsxD/gppX genitalia, showing the incomplete axis of rotation (dotted). Scale bar = 100μm.
When we knocked down the DOT1 complex, we observed sex- and tissue-specific phenotypes (except for AlhRNAi). We found reduced vaginal teeth numbers in gppRNAi, Su(Tpl)RNAi, and lilliRNAi females, while males were missing lateral lobes, claspers, and penis apparati (Figure 6C, Table S4). Additionally, male-specific genital disc rotation (Adam et al., 2003) was incomplete in gppRNAi, Su(Tpl)RNAi, and lilliRNAi males (Table S4). Parovaria and spermathecae were missing from gppRNAi female reproductive tracts, while gppRNAi resulted in a narrow ejaculatory duct in males, similar to dsxRNAi (Figure 6D). The female and male internal reproductive structures derive from different segments of the genital disc (Estrada et al., 2003), suggesting that gpp has sex- and segment-specific roles in both internal and external genital development. In males, gppRNAi, Su(Tpl)RNAi, lilliRNAi, or earRNAi resulted in decreased sex comb pigmentation (Figure 6E, Table S4). Additionally, gppRNAi and Su(Tpl)RNAi reduced sex comb bristle number and resulted in feminized (thinner and pointed) bristles (Figure 6E). Lastly, gppRNAi altered the morphology of the ovarian niche where we observed collapsed terminal filaments and excessive numbers of early stage germ cells (Figure 6F), while the male niche was unaffected. In summary, the defects observed in these RNAi experiments indicate that members of the DOT1 complex(es) have similar sex- and tissue-specific functions in dsx-expressing cells.
The fact that knockdown of DOT1 complex members results in sex- and tissue-specific phenotypes is consistent with them being DSX target genes. Alternatively, DOT1’s general role in gene regulation could result in phenotypes unrelated to dsx function. To address this, we examined genetic interactions between dsx and alleles of DOT1 complex members. In the dsxD background, heterozygosity for gppX reduced male genitalia structures like in gppRNAi and lilliRNAi (Figure 6G, Table S4). In addition, XX; dsxD/+ gonads had either male (hub) or female (terminal filament) germline niche structures (14% hub, n=106, Figure S4). We observed increased hub frequency in XX; dsxD/+ gonads when heterozygous gpp (gppX, 62% hub, n=37) or lilli (lilliA17–2, 39% hub, n=36). Thus, both RNAi and genetic interaction experiments suggest that DOT1 is involved in sex-specific niche development regulated by DSX.
Discussion
Identifying genes directly regulated by a transcription factor is complicated because transcription factors recognize short sequences that can arise by chance. The use of multiple genome-wide techniques helps winnow potential targets. To understand how DSX contributes to sex- and tissue-specific development, we undertook a series of genome-wide experiments and analyses to determine: where DSX is bound in different cell types, which sites are evolutionarily conserved, the relationship between site strength and occupancy, which genes respond to acute changes to DSXF/DSXM isoform abundance, and how many genomic regions genetically interact with dsx. We then performed RNAi knockdown of candidate targets and found striking tissue- and sex-specific transformations of sexually dimorphic structures. This rich set of targets will be useful for understanding the sex differentiation network in the powerful Drosophila system, and enrichment for orthologs of mouse DMRT1 targets among DSX targets strongly suggests that some of this network will be conserved in mammals.
The logic of DSX regulation
The sex-specific developmental programs of the gonadal mesoderm, the leg or genital imaginal discs, the fat body, and the nervous system are all likely to be highly divergent, yet all depend on DSX. How is this achieved? DSXF and DSXM could be recruited to different loci. While we do find a few genes with sex-specific occupancy patterns, this model is not well supported. DSX could bind different genes in different tissues. While we found examples of tissue-biased DSX occupancy, most genes are occupied regardless of tissue or sex. Finally, DSX could always bind a given target gene, but regulation would depend on the combinatorial activity of other gene-specific factors. Our work provides strongest evidence for the last model. There is also support for this combinatorial model in the literature. The bab1 locus is regulated by an enhancer that bears both DSX and homeobox protein binding sites to control sex-specific expression along the anterior/posterior axis (Williams et al., 2008). We did not examine occupancy and expression changes throughout development, but our results predict that for a given target there are both positive and negative transcriptional responses to DSXF/M that vary among tissues of the sexes throughout development.
A large number of target genes might suggest that DSX acts as a “micro-manager” of sexual development, regulating the expression of many or most terminal sex-differentiation genes. However, our unbiased screen to identify genes interacting with dsx predicts a smaller number (~50) of “major effect” loci acting in the dsx pathway. How can we reconcile the disparity between the large numbers of potential DSX target genes with many fewer predicted to have “major” effects? DSX may delegate regulatory function to pattern formation pathways that lead to sex-specific development of organ systems. This would explain the large number of transcriptional regulators that show DSX-dependent, sex-biased expression (Chatterjee et al., 2011) predicted to be DSX targets. In addition, many genes regulated by DSX might provide subtle, but evolutionarily significant, “minor” polygenic effects on development or physiology. DSX regulation of these minor effect loci could help explain the effects of genetic background on sex-related phenotypes. These major and minor effect genes would both be strongly selected for in the course of evolution. Among predicted targets, we found enrichment for GO terms for transcription (adjusted p-value=1.85E-7) and signaling (adjusted p-value=1.44E-53), suggesting that DSX regulates gene expression of terminal differentiation factors by direct and indirect mechanisms (Table S5).
Types of DSX targets
The types of target genes predicted by our analyses illustrate how DSX is able to exhibit such powerful effects on developmental pathways. One group of predicted target genes are involved in short-range (e.g. WNT, EGF, and DPP), and long-range (e.g. Insulin and Ecdysone) signaling. Thus, DSX expression could have far-reaching effects on the development of surrounding cells and beyond. Indeed, DSX modulates short-range signaling pathways in both the genital disc (Ahmad and Baker, 2002; Gorfinkiel et al., 2003; Keisman et al., 2001) and gonad (DeFalco et al., 2008; Oliver et al., 1993; Wawersik et al., 2005). Also, YP expression requires hormonal communication in addition to DSX (Bownes et al., 1996), and titers of the steroid ecdysone are highly female-biased and germline-dependent in adults (Parisi et al., 2010), consistent with a physiological loop in which DSX as a direct transcriptional regulator of hormonal signaling pathways. This also provides a mechanism for cells to “consult” on sex-specific developmental paths and allows for reinforced and maintained sexual decisions. Such signaling mechanisms are common in sex determination. In C. elegans, the secreted factor HER-1 is a component of the primary sex determination cascade (Zarkower, 2006), and WNT and FGF signaling reinforces/specifies sexual differentiation in mammals (Eggers and Sinclair, 2012) and flies (Ahmad and Baker, 2002; DeFalco et al., 2008). These overlapping modules of gene interactions suggest significant commonalities between Drosophila and mammals.
Another major class of potential DSX targets encode transcriptional regulators, many of which have sex-specific expression patterns (Barmina et al., 2005; Chatterjee et al., 2011; Williams et al., 2008). By activating/repressing transcription factors, DSX could delegate regulation to activate pathways that proceed largely without further input by DSX. There is clear evidence for this mode of action. In the absence of dsx function, both male and female reproductive structures are found. This is opposed to the absence of all sexual structures expected for a “micro-manager” of their development. Further, in dsx- (N. Camara, CW and MVD, in revision) or XX; dsxD/+ gonads, either male or female stem cell niches form stochastically. We would expect these structures to be absent if dsx was required for their formation. These data are consistent with DSX being a regulator of other regulators that control female- and male-specific development.
We also found epigenetic transcriptional regulators among potential DSX target genes suggesting a function in fine-tuning and/or memory. For example, the DOT1 epigenetic machine mediates H3K79 methylation (Steger et al., 2008). While DSX control of DOT1 could act as cellular memory system and/or generally boost expression of many genes, it may also function to regulate gene expression of a few genes that contribute to sexually dimorphic phenotypes. For example, gpp- males have a partial female-specific abdominal segment 7, reduced segment 5/6 pigmentation, and genital rotation defects (in Abd-B interactions), all of which are consistent with either segment identity change (Shanower et al., 2005) or feminization.
Feedback systems and cross-regulation also affect the output and stability of genetic pathways. Strikingly, members of the sex determination hierarchy also appear to be DSX targets. dsx and fru are bound both by DSX (this study) and FRU (Neville et al., 2014). Sex lethal (Sxl), which regulates tra, also has conserved DSX binding sites. There is precedent for feedback in sex determination as TRA is a feedback regulator of Sxl (Siera and Cline, 2008). Similarly, predicted targets of DSX such as Sex combs reduced, Abdominal-B (Abd-B), and others encode transcription factors known to regulate dsx expression (Chatterjee et al., 2011; Devi and Shyamala, 2013; Tanaka et al., 2011; Wang and Yoder, 2012). Recently, micro-RNAs have been shown to modulate sex determination (Weng et al., 2013) suggesting we are far from understanding even the basic framework for sex determination and differentiation. Even providing sexual information is more complicated than we anticipated as gpp is required for development of vaginal teeth in females and sex-combs in males. This suggests that the sexual directionality of DSXF and DSXM regulation of gpp depends on tissue-specific co-factors. We suggest that sex differentiation occurs via a set of context-dependent networks -- replete with rich auto-regulation, cross-regulation, and feedback -- not a hierarchy.
In summary, the wiring diagram surrounding dsx may be quite complex as DSX directly or indirectly regulates a broad set of transcription factor genes including some that regulate dsx expression. If DSX is regulated by, and a regulator of, a broad array of transcription factors widely deployed during development, then inappropriate expression of DSX could be deleterious. Indeed, ectopic expression of dsx results in widespread changes in morphology and lethality (Jursnich and Burtis, 1993), suggesting that dsx must be tightly regulated. The dsx gene is expressed in a highly tissue-specific manner consistent with the idea that only those tissues with sex-specific developmental programs express dsx suggesting that dsx expression must be tightly regulated. Understanding the logic by which DSX acts to control dimorphic developmental outcomes in different tissues in the context of multiple highly integrated networks is a key question in sex determination.
Conclusions
In over 100 years of studying sex determination and differentiation, only a few key genes have been identified. We provide a rich set of DSX targets for future studies and broadly outline the DSX mechanism of action. We conclude that DSX binding confers the possibility of sex-specific regulation where context-specific factors determine the consequences of binding. Resulting complex and context-dependent expression patterns mean that DSXF can act as a positive regulator of a gene in one tissue, and DSXM can act a positive regulator of the same locus in another. DSX acts by a combination of delegating control to transcription factors and by directly micromanaging terminal differentiation genes in a tightly integrated dance of regulatory inputs.
Experimental Procedures
Fly Stocks
Fly stocks were obtained from the Bloomington Drosophila Stock Center (Cook et al., 2010), the Transgenic RNAi Project (Ni et al., 2011), and from the B.S. Baker lab and other generous members of the Drosophila community. See FlyBase for gene and allele descriptions (Marygold et al., 2013) and Tables S3-S4 further information.
ChIP-seq, DamID-seq, DamID-array, RNA-seq
Transgenic DamID flies were made using sex-specific dsx cDNAs (gift of Gyunghee Lee) in pUASt-att-NDamMyc integrated into attP2 on chromosome 3L using φC31 site-directed integration (Bischof et al., 2007). S2 cells were transfected with pMT5.1-DSXM-V5-HisB or pMT5.1-DSXF-V5-HisB (Garrett-Engele et al., 2002) and pCoBlast (Invitrogen, Carlsbad, CA, USA) as the selection plasmid using Effectene (Qiagen, Valencia, CA, USA). Chromatin immunoprecipitation was performed with anti-V5 tag monoclonal antibody (Inivitrogen, Carlsbad, CA, USA) on Protein G coupled Dynabeads (Invitrogen, Carlsbad, CA, USA).
ChIP-seq libraries were constructed with the Genomic DNA sample preparation kit and were sequenced on a GA1 (Illumina, San Diego, CA, USA). RNA-seq libraries were made with the TruSeq RNA Sample Preparation Kit v2 and were sequenced on the HiSeq 2000 (Illumina, San Diego, CA, USA). Reads were mapped to FlyBase 5.46 using Tophat 1.4.1(Trapnell et al., 2009) and/or Bowtie 0.12.7 (Langmead et al., 2009). HTSeq 0.5.1p2 and DESeq 1.12.0 were used to count DNA-seq reads in 500 base pair bins (Anders and Huber, 2010; Anders et al., 2014). Transcript abundance was determined using Cufflinks 2.1.1 (Trapnell et al., 2012). k-means clustering of FPKM values was performed using the kmeans package in R (Gentleman et al., 2004). Splice junction counts were obtained using Spanki 0.4.2 (Sturgill et al., 2013). The WTD method of peak calling from the ChIP-seq analysis program SPP (version 1.11) was used to call peaks with an FDR of 0.01 (Kharchenko et al., 2008). Nimblegen performed DNA labeling and array hybridization (Roche NimbleGen, Madison WI, USA). Adjacent selected bins or probes were combined into features to produce peaks using BEDTools v2.16.2 (Quinlan and Hall, 2010). Gene level occupancy scores were calculated by summing signal in 500 bp bins under all called peak regions within the gene body + 1 kb upstream of the transcription start site. All experiments were performed in replicate. DSX occupancy data (ChIP-seq, DamID-seq, DamID-array) and RNA-seq data are available under GEO (Barrett et al., 2013) series accession GSE49480.
Sequence Analysis
We found motifs de novo using MEME-ChIP (Machanick and Bailey, 2011) and compared de novo identified motifs to the DSX PWM in TOMTOM (Gupta et al., 2007). We used the PWM for DSX in JASPAR format (Mathelier et al., 2013) to search genomes (Adams et al., 2000; Chen et al., 2014; Drosophila 12 Genomes et al., 2007; Richards et al., 2005) with the Bio.Site.search_pwm method in BioPython (Cock et al., 2009). Non-melanogaster sites were aligned to D. melanogaster using liftover chain file (Chen et al., 2014) and we summed log odds by position. Site-level conservation scores were computed by summing the substitution/site distance of each species for which a conserved sequence exists (Chen et al., 2014). Orthologs of mouse DMRT1 targets (Murphy et al., 2010) were identified using Ensembl biomart (Flicek et al., 2013). k-means clustering of FPKM values was performed using the kmeans package in R (Gentleman et al., 2004). See Supplemental Experimental Procedures for further details.
Supplementary Material
Figure S1: Expression of Dam-DSX fusion protein and resultant phenotypes, related to Figure 1. S2 cells carrying the pMT5.1-DSXM-V5-His construct before (A, A′) and after (B, B′) 60 hour induction. S2 cells carrying the pMT5.1-DSXF-V5-His construct before (C, C′) and after (D, D′) 60 hour induction. Scale bar = 10μm. A, B, C, and D are the V5 channel (white), and A′, B′, C′, and D′ are the DAPI channel (blue). Third instar larval fatbody from dsx-GAL4/+ (E, E′) dsx-GAL4/UAS-Dam-myc (F, F′) dsx-GAL4/UAS-Dam-myc-dsxM (G, G′) and dsx-GAL4/UAS-Dam-myc-dsxF (H, H′). E,F,G, and H are merged images of anti-myc (green) and DAPI (blue). E′, F′, G′ and H′ is a split of only the anti-myc signal (white). Testes (I,J) and ejaculatory ducts (K,L) were dissected and stained with DAPI. Light microscopy images of sex combs from control dsx-GAL4/TM6 (M) and dsx-GAL4/UAS-Dam-myc-dsxF (N). Scale bar = 50μm. The Dam fusion proteins include the myc epitope incorporated at the C-terminus of the Dam coding sequence such that Dam fusion proteins can be detected with anti-myc antibodies.
Figure S2: Conservation of the DSX DNA binding domain and sex-specific splicing, related to Figures 1, 2, and 3.
(A) Diagram of the DSXM and DSXF proteins (above) and amino acid sequence alignment of the DSX DNA binding domain from 20 Drosophila species (below). Cysteine and histidine residues in the Zn-binding site are highlighted in tan. The evolutionary distance from D. melanogaster is indicated in substitutions/site (ss) (Chen et al., 2014). Color-coding of D. melanogaster amino acids represent mutations that do not affect DSX activity (green), partially affect activity (orange), or impair activity (red) (adapted from (Zhang et al., 2006)). (B) Bar graphs representing the percentage of dsx splicing events resulting in production of female (red) or male (blue) isoform from RNA-seq data obtained from adult females (F) or males (M) from 7 Drosophila species (Chen et al., 2014). (C) The normalized (% of maximum average occupancy) distribution of DSX occupancy values along a generic gene model using +1.5Kb upstream of transcription start (bent arrow), the gene body (rectangle), where the first 0.5Kb and last 0.5Kb are at base level, and the middle 0.5Kb is scaled from all gene models, and the -1.5Kb downstream region are shown.
Figure S3. DSXF- and DSXM- occupied regions are not correlated with other transcription factors, related to Figure 1 and 2. Hierarchically clustered heatmap of pairwise similarity metrics between all 255 available ChIP-chip and ChIP-seq experiments and DSX ChIP-seq (highlighted in red) as well as DSX DamID-seq/chip (highlighted in blue). Brighter colors indicate higher similarity (higher fraction of sites with p < 0.05); DSXF and DSXM (highlighted in red (ChIP-seq) and blue (DamID)) are more similar to each other than they are to any other assayed factor. Self-self comparisons along the diagonal are indicated in gray. Colored blocks along the left side indicate broad clusters. See methods for details. Table S6 contains the source and description of all occupancy data sets tested for correlation with DSX occupancy data.
Figure S4: XX; d s xD/+ gonad phenotypes, related to Figure 6. Representative images of gonads dissected from XX; d s xD/+ adults having either female-like terminal filaments (A) or male-like hubs (B). Terminal filaments and hubs are marked with anti-N-Cad (green), hubs are marked with anti-FasIII (blue) and anti-N-cad (green), and germ cells with anti-Vasa (red). Scale bar = 50μm.
An .xlsx workbook file containing all gene-level occupancy scores, binary scores for occupied versus unoccupied, gene-level conservation index (CI) scores, gene-level DSX position weight matrix scores, k-means cluster ID for occupancy score clustering, binary scores for mouse DMRT1 target orthology, and RNA-seq FPKM values for each RNA-seq sample. See sheet titled “README” for more information and definition of column headings.
An .xlsx workbook file containing DSX position weight matrix (PWM) scores for all 566,628 DSX motif-related sequences (sheet “PWM_scores”), conservation index scores, and PhastCons scores for all 173,775 sites with a positive PWM scores located within a D. melanogaster gene body plus 1 kb upstream excepting those in coding sequence (sheet “CI_scores”), conservation index (CI) scores for the 17,380 sites in the top 10% of all sites with any relationship to the DSX PWM that occur within a gene excluding coding sequence or 1Kb upstream of a gene (sheet “top_10_percent_PWM_CI_scores”), and the DSX position weight matrix along with all shuffled position weight matricies (“PWMs”). See sheet titled “README” for more information and definition of column headings.
A .xlsx workbook file containing results from the unbiased genetic interaction tests between dsxD and 101 deficiencies of the 2nd chromosome, 17 genomic intervals interacting with dsxD, and the tested genomic intervals that did not interact with dsxD. For each deficiency tested for genetic interaction with dsxD, a description of the genetic interaction phenotype is provided (“Phenotype Description”) along with the FlyBase deficiency name and aberration ID (“FlyBase ID”). The known or estimated cytological location (“Deleted Segment”) as well as chromosome base position (FlyBase release 5) for the left and right breakpoints are also provided.
For each gene tested by RNAi knockdown, the FlyBase gene name and ID (“FlyBase Identifier”) are provided along with a text description summarizing phenotypic findings after RNAi knockdown (“Phenotype Descriptions”). There is also a text summary of DSX occupancy, occupancy clustering, gene-level PWM score, gene-level conservation index, and other criteria used to select the gene for RNAi test (“Criteria for Selection”).
An .xlsx workbook file containing enriched gene ontology terms in the 3,717 occupied genes identified in this work. The ontology domains molecular function and biological process were examined, with output from each domain occupying a separate sheet in this file. See sheet titled “README” for more information and definition of column headings.
An .xlsx workbook file containing a source and description for all occupancy data sets tested for correlation with DSX occupancy data. See sheet titled “README” for more information and definition of columns headings.
Acknowledgments
We thank the Developmental Studies Hybridoma Bank, Drosophila RNAi Screening Center, Bruce Baker, Ken Howard, Gyunghee Lee, Ruth Lehmann, Tony Southall, and Phil Garrett-Engele for stocks and reagents and Shaad Ahmad for comments on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, NIDDK (BO) and NCBI (TP). CW and EJ were supported by NIH grant 5R21HD066244-02 awarded to MVD, and EJ was supported by NIH grant 5F31HD076558-02. MCN and HJP were supported by grants from the Wellcome Trust to SFG (WT085521MA and WT082987MF). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Author Contributions
E.C., C.W., L.H., M.N., and H.P. performed DSX molecular biology and occupancy experiments. C.W., E.J., E.C., and D.S. performed RNA-seq. C.W., E.J., and E.C. performed molecular genetics experiments. Y.-A.K., Z.-X.C, R.D., and E.C. performed conservation and meta-analysis. R.D. and D.S. performed bioinformatics analysis. H.S. sequenced samples. E.C. was consortium coordinator. C.W, B.O., and M.V.D. wrote the manuscript. T.P., S.G., M.V.D., and B.O. supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam G, Perrimon N, Noselli S. The retinoic-like juvenile hormone controls the looping of left-right asymmetric organs in Drosophila. Development (Cambridge, England) 2003;130:2397–2406. doi: 10.1242/dev.00460. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science (New York, NY) 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ahmad SM, Baker BS. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell. 2002;109:651–661. doi: 10.1016/s0092-8674(02)00744-4. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq A Python framework to work with high-throughput sequencing data. 2014. bioRxiv preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmina O, Gonzalo M, McIntyre LM, Kopp A. Sex- and segment-specific modulation of gene expression profiles in Drosophila. Developmental biology. 2005;288:528–544. doi: 10.1016/j.ydbio.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrer JR, Zhang W, Weiss MA. Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation. The Journal of biological chemistry. 2005;280:32989–32996. doi: 10.1074/jbc.M507990200. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M, Ronaldson E, Mauchline D. 20-Hydroxyecdysone, but not juvenile hormone, regulation of yolk protein gene expression can be mapped to cis-acting DNA sequences. Developmental biology. 1996;173:475–489. doi: 10.1006/dbio.1996.0041. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. The EMBO journal. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SS, Uppendahl LD, Chowdhury MA, Ip PL, Siegal ML. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development (Cambridge, England) 2011;138:1099–1109. doi: 10.1242/dev.055731. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, et al. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome research. 2014;24:1209–1223. doi: 10.1101/gr.159384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueyama M, Wicker-Thomas C. A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behaviour in Drosophila melanogaster. Insect Mol Biol. 2006;15:465–473. doi: 10.1111/j.1365-2583.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics (Oxford, England) 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KR, Parks AL, Jacobus LM, Kaufman TC, Matthews KA. New research resources at the Bloomington Drosophila Stock Center. Fly. 2010;4:88–91. doi: 10.4161/fly.4.1.11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes & development. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- DeFalco T, Camara N, Le Bras S, Van Doren M. Nonautonomous sex determination controls sexually dimorphic development of the Drosophila gonad. Developmental cell. 2008;14:275–286. doi: 10.1016/j.devcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi TR, Shyamala BV. Male- and female-specific variants of doublesex gene products have different roles to play towards regulation of Sex combs reduced expression and sex comb morphogenesis in Drosophila. Journal of biosciences. 2013;38:455–460. doi: 10.1007/s12038-013-9348-1. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes C. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Eggers S, Sinclair A. Mammalian sex determination-insights from humans and mice. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2012;20:215–238. doi: 10.1007/s10577-012-9274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Casares F, Sanchez-Herrero E. Development of the genitalia in Drosophila melanogaster. Differentiation; research in biological diversity. 2003;71:299–310. doi: 10.1046/j.1432-0436.2003.03017.x. [DOI] [PubMed] [Google Scholar]
- Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic acids research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D, Martin P, Sanchez-Herrero E. Drosophila Hox and sex-determination genes control segment elimination through EGFR and extramacrochetae activity. PLoS genetics. 2012;8:e1002874. doi: 10.1371/journal.pgen.1002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Engele CM, Siegal ML, Manoli DS, Williams BC, Li H, Baker BS. intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development (Cambridge, England) 2002;129:4661–4675. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Sanchez L, Guerrero I. Development of the Drosophila genital disc requires interactions between its segmental primordia. Development (Cambridge, England) 2003;130:295–305. doi: 10.1242/dev.00214. [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome biology. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel LU, Oliver B. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC developmental biology. 2007;7:113. doi: 10.1186/1471-213X-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jursnich VA, Burtis KC. A positive role in differentiation for the male doublesex protein of Drosophila. Developmental biology. 1993;155:235–249. doi: 10.1006/dbio.1993.1021. [DOI] [PubMed] [Google Scholar]
- Keisman EL, Christiansen AE, Baker BS. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginal disc. Developmental cell. 2001;1:215–225. doi: 10.1016/s1534-5807(01)00027-2. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature biotechnology. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo MS, Sanders LE, Sun F, Arbeitman MN. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC genomics. 2009;10:80. doi: 10.1186/1471-2164-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Hall JC, Park JH. Doublesex gene expression in the central nervous system of Drosophila melanogaster. J Neurogenet. 2002;16:229–248. doi: 10.1080/01677060216292. [DOI] [PubMed] [Google Scholar]
- Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development (Cambridge, England) 2011;138:2761–2771. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics (Oxford, England) 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. FlyBase: improvements to the bibliography. Nucleic acids research. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic acids research. 2013 doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13360–13365. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC molecular biology. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi RN, Baker BS. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes & development. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville MC, Nojima T, Ashley E, Parker DJ, Walker J, Southall T, Van de Sande B, Marques AC, Fischer B, Brand AH, et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Current biology : CB. 2014;24:229–241. doi: 10.1016/j.cub.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes & development. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B, Kim YJ, Baker BS. Sex-lethal, master and slave: a hierarchy of germ-line sex determination in Drosophila. Development (Cambridge, England) 1993;119:897–908. doi: 10.1242/dev.119.3.897. [DOI] [PubMed] [Google Scholar]
- Parisi MJ, Gupta V, Sturgill D, Warren JT, Jallon JM, Malone JH, Zhang Y, Gilbert LI, Oliver B. Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC genomics. 2010;11:346. doi: 10.1186/1471-2164-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Human molecular genetics. 1999;8:989–996. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome research. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature neuroscience. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS biology. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS biology. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siera SG, Cline TW. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics. 2008;180:1963–1981. doi: 10.1534/genetics.108.093898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Molecular and cellular biology. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D, Malone JH, Sun X, Smith HE, Rabinow L, Samson ML, Oliver B. Design of RNA splicing analysis null models for post hoc filtering of Drosophila head RNA-Seq data with the splicing analysis kit (Spanki) BMC bioinformatics. 2013;14:320. doi: 10.1186/1471-2105-14-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS biology. 2011;9:e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans - mechanisms and functional implications. Nature reviews Genetics. 2014;15:221–233. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kidd BJ, Carroll SB, Yoder JH. Sexually dimorphic regulation of the Wingless morphogen controls sex-specific segment number in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11139–11144. doi: 10.1073/pnas.1108431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoder JH. Hox-mediated regulation of doublesex sculpts sex-specific abdomen morphology in Drosophila. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:1076–1090. doi: 10.1002/dvdy.23791. [DOI] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–567. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R, Chin JS, Yew JY, Bushati N, Cohen SM. miR-124 controls male reproductive success in Drosophila. eLife. 2013;2:e00640. doi: 10.7554/eLife.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang W, Bayrer JR, Weiss MA. Doublesex and the regulation of sexual dimorphism in Drosophila melanogaster: structure, function, and mutagenesis of a female-specific domain. The Journal of biological chemistry. 2008;283:7280–7292. doi: 10.1074/jbc.M708742200. [DOI] [PubMed] [Google Scholar]
- Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development (Cambridge, England) 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- Yoder JH. Abdominal segment reduction: development and evolution of a deeply fixed trait. Fly. 2012;6:240–245. doi: 10.4161/fly.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D. WormBook : the online review of C elegans biology. 2006. Somatic sex determination; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D. DMRT genes in vertebrate gametogenesis. Current topics in developmental biology. 2013;102:327–356. doi: 10.1016/B978-0-12-416024-8.00012-X. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li B, Singh R, Narendra U, Zhu L, Weiss MA. Regulation of sexual dimorphism: mutational and chemogenetic analysis of the doublesex DM domain. Molecular and cellular biology. 2006;26:535–547. doi: 10.1128/MCB.26.2.535-547.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Expression of Dam-DSX fusion protein and resultant phenotypes, related to Figure 1. S2 cells carrying the pMT5.1-DSXM-V5-His construct before (A, A′) and after (B, B′) 60 hour induction. S2 cells carrying the pMT5.1-DSXF-V5-His construct before (C, C′) and after (D, D′) 60 hour induction. Scale bar = 10μm. A, B, C, and D are the V5 channel (white), and A′, B′, C′, and D′ are the DAPI channel (blue). Third instar larval fatbody from dsx-GAL4/+ (E, E′) dsx-GAL4/UAS-Dam-myc (F, F′) dsx-GAL4/UAS-Dam-myc-dsxM (G, G′) and dsx-GAL4/UAS-Dam-myc-dsxF (H, H′). E,F,G, and H are merged images of anti-myc (green) and DAPI (blue). E′, F′, G′ and H′ is a split of only the anti-myc signal (white). Testes (I,J) and ejaculatory ducts (K,L) were dissected and stained with DAPI. Light microscopy images of sex combs from control dsx-GAL4/TM6 (M) and dsx-GAL4/UAS-Dam-myc-dsxF (N). Scale bar = 50μm. The Dam fusion proteins include the myc epitope incorporated at the C-terminus of the Dam coding sequence such that Dam fusion proteins can be detected with anti-myc antibodies.
Figure S2: Conservation of the DSX DNA binding domain and sex-specific splicing, related to Figures 1, 2, and 3.
(A) Diagram of the DSXM and DSXF proteins (above) and amino acid sequence alignment of the DSX DNA binding domain from 20 Drosophila species (below). Cysteine and histidine residues in the Zn-binding site are highlighted in tan. The evolutionary distance from D. melanogaster is indicated in substitutions/site (ss) (Chen et al., 2014). Color-coding of D. melanogaster amino acids represent mutations that do not affect DSX activity (green), partially affect activity (orange), or impair activity (red) (adapted from (Zhang et al., 2006)). (B) Bar graphs representing the percentage of dsx splicing events resulting in production of female (red) or male (blue) isoform from RNA-seq data obtained from adult females (F) or males (M) from 7 Drosophila species (Chen et al., 2014). (C) The normalized (% of maximum average occupancy) distribution of DSX occupancy values along a generic gene model using +1.5Kb upstream of transcription start (bent arrow), the gene body (rectangle), where the first 0.5Kb and last 0.5Kb are at base level, and the middle 0.5Kb is scaled from all gene models, and the -1.5Kb downstream region are shown.
Figure S3. DSXF- and DSXM- occupied regions are not correlated with other transcription factors, related to Figure 1 and 2. Hierarchically clustered heatmap of pairwise similarity metrics between all 255 available ChIP-chip and ChIP-seq experiments and DSX ChIP-seq (highlighted in red) as well as DSX DamID-seq/chip (highlighted in blue). Brighter colors indicate higher similarity (higher fraction of sites with p < 0.05); DSXF and DSXM (highlighted in red (ChIP-seq) and blue (DamID)) are more similar to each other than they are to any other assayed factor. Self-self comparisons along the diagonal are indicated in gray. Colored blocks along the left side indicate broad clusters. See methods for details. Table S6 contains the source and description of all occupancy data sets tested for correlation with DSX occupancy data.
Figure S4: XX; d s xD/+ gonad phenotypes, related to Figure 6. Representative images of gonads dissected from XX; d s xD/+ adults having either female-like terminal filaments (A) or male-like hubs (B). Terminal filaments and hubs are marked with anti-N-Cad (green), hubs are marked with anti-FasIII (blue) and anti-N-cad (green), and germ cells with anti-Vasa (red). Scale bar = 50μm.
An .xlsx workbook file containing all gene-level occupancy scores, binary scores for occupied versus unoccupied, gene-level conservation index (CI) scores, gene-level DSX position weight matrix scores, k-means cluster ID for occupancy score clustering, binary scores for mouse DMRT1 target orthology, and RNA-seq FPKM values for each RNA-seq sample. See sheet titled “README” for more information and definition of column headings.
An .xlsx workbook file containing DSX position weight matrix (PWM) scores for all 566,628 DSX motif-related sequences (sheet “PWM_scores”), conservation index scores, and PhastCons scores for all 173,775 sites with a positive PWM scores located within a D. melanogaster gene body plus 1 kb upstream excepting those in coding sequence (sheet “CI_scores”), conservation index (CI) scores for the 17,380 sites in the top 10% of all sites with any relationship to the DSX PWM that occur within a gene excluding coding sequence or 1Kb upstream of a gene (sheet “top_10_percent_PWM_CI_scores”), and the DSX position weight matrix along with all shuffled position weight matricies (“PWMs”). See sheet titled “README” for more information and definition of column headings.
A .xlsx workbook file containing results from the unbiased genetic interaction tests between dsxD and 101 deficiencies of the 2nd chromosome, 17 genomic intervals interacting with dsxD, and the tested genomic intervals that did not interact with dsxD. For each deficiency tested for genetic interaction with dsxD, a description of the genetic interaction phenotype is provided (“Phenotype Description”) along with the FlyBase deficiency name and aberration ID (“FlyBase ID”). The known or estimated cytological location (“Deleted Segment”) as well as chromosome base position (FlyBase release 5) for the left and right breakpoints are also provided.
For each gene tested by RNAi knockdown, the FlyBase gene name and ID (“FlyBase Identifier”) are provided along with a text description summarizing phenotypic findings after RNAi knockdown (“Phenotype Descriptions”). There is also a text summary of DSX occupancy, occupancy clustering, gene-level PWM score, gene-level conservation index, and other criteria used to select the gene for RNAi test (“Criteria for Selection”).
An .xlsx workbook file containing enriched gene ontology terms in the 3,717 occupied genes identified in this work. The ontology domains molecular function and biological process were examined, with output from each domain occupying a separate sheet in this file. See sheet titled “README” for more information and definition of column headings.
An .xlsx workbook file containing a source and description for all occupancy data sets tested for correlation with DSX occupancy data. See sheet titled “README” for more information and definition of columns headings.