Abstract
Killer-cell immunoglobulin-like receptors (KIRs) on natural killer (NK) cells have been linked to a wide spectrum of health conditions such as chronic infections, autoimmune diseases, pregnancy complications, cancers, and transplant failures. A small subset of effector memory T cells also expresses KIRs. Here, we use modern analytic tools including genome-wide and multiplex molecular, phenotypic, and functional assays to characterize the KIR+ T cells in human blood. We find that KIR+ T cells primarily reside in the CD56+ T population that is distinctively DNAM-1high with a genome-wide quiescent transcriptome, short telomere, and limited TCR excision circles. During cytomegalovirus (CMV) reactivation in bone marrow transplant recipients, KIR+CD56+ T cells rapidly expanded in real-time, but not KIR+CD56− T cells or KIR+ NK cells. In CMV+ asymptomatic donors, as much as 50% of CD56+ T cells are KIR+, and most are distinguishably KIR2DL2/3+NKG2C+CD57+. Functionally, the KIR+CD56+ T-cell subset lyses cancer cells and CMVpp65-pulsed target cells in a dual KIR-dependent and TCR-dependent manner. Analysis of metabolic transcriptome confirms the immunological memory status of KIR+CD56+ T cells, in contrast to KIR−CD56+ T cells that are more active in energy metabolism and effector differentiation. KIR−CD56+ T cells have >25-fold higher level of expression of RORC than the KIR+ counterpart and are a previously unknown producer of IL-13 rather than IL-17 in multiplex cytokine arrays. Our data provide fundamental insights intoKIR + T cells biologically and clinically.
INTRODUCTION
Human natural killer (NK) cells are part of the innate immune system and recognize microbe-infected cells and tumor cells through a combination of activating and inhibitory receptors that do not require somatic gene rearrangement, such as the killer-cell immunoglobulin-like receptor (KIR) family (1). T cells, in contrast, mediate adaptive immune response to major histocompatibility complex (MHC)–bound antigens through recognition by rearranged T-cell receptors (TCRs) (2). KIR generates diversity through variable haplotype gene content, allele polymorphism, and stochastic expression (3, 4), whereas TCR recombines αβ or γδ chains during development (5). Both KIR and TCR generate specificity and are useful developmental markers. KIR+ NK cells are usually CD56dim, cytotoxic, and developmentally more mature than KIR−CD56bright cytokine-secreting NK cells (6). γδ T cells bear more innate-like attributes and appear earlier in the thymus than αβ T cells (5). TCR is never found in NK cells, but a subset of terminallydifferentiated effector memory T cells expresses KIR (7–9).
KIR+ T cells were first identified two decades ago (10), and were found in the CD8+, CD4+, TCR γδ+, and αβ+ T-cell fractions (8, 11–15). Most KIR+ T cells are αβ+CD8+, possess a memory phenotype, and are generated upon TCR recognition of HLA-E–associated viral peptides after monoclonal or oligoclonal expansion (16–18). Continuous TCR engagement sustains their KIR expression with resultant resistance to apoptosis (19–23). These cells are important in the control of infections such as cytomegalovirus (CMV) and hepatitis C virus infections (14, 15, 18, 24, 25). KIR expression and function are fundamentally different in T cells and NK cells (26). For instance, the KIR repertoire in NK cells is different from that in T cells from the same individual (27, 28). While KIR acquisition during NK cell development is stochastic, essential for licensing and tuning of responsiveness to self-MHC, KIR is acquired in T cells after TCR rearrangement and antigen encounter, and its repertoire is independent of self-MHC (9, 28, 29). The KIR promoters on NK cells have a minimal size of 120–250 bp, are regulated by all-or-none methylation, and involve transcriptional factors such as YY1, CRE/ATF, RUNX3, and Sp1 (30–34). In contrast, the KIR promoter in T cells has a minimal size of 60 bp, patchy methylation, and involvement of different sets of transcriptional factors (35, 36). While inhibitory KIRs have similar suppressive function in NK cells and T cells (37–39), activating KIRs appear unable to trigger T cells directly and serve rather in a co-stimulatory role without consistent DAP12 expression (40–42).
Similar to KIR, CD56 is another NK-related receptor that is expressed on a small subset of T cells characterized by reduced proliferative potential because of upregulation of P16 and P53 (43, 44). Most KIR+ T cells are CD56+, and the CD56 expression level correlates well with both NK and CD8+ CTL functions (45). In this study, we used modern genome-wide and high-throughput multiplex assays to characterize the KIR+ T cells in human blood. We found that KIR+ T cells primarily resided in the CD56+ T cell population that was distinctively DNAM-1high with a unique genome-wide quiescent transcriptome substantially different from that of the CD56−T cells, NK cells, and Vα24 +Vβ11+iNKT cells. In addition, the KIR+ subset of CD56+ T cells was cytotoxic to cancer cells and CMV-infected cells in a dual KIR- & TCR-dependent manner, whereas the KIR− subset was a unique metabolically active RORC+ cytokine producer of IL-13. In a bone marrow transplant cohort, KIR+CD56+ T cells expanded rapidly during CMV reactivation, and these KIR+ cells remained prevalent in asymptomatic CMV+ donors with a characteristic phenotype of KIR2DL2/3+NKG2C+CD57+. Our findings provide a fundamental understanding of the biology of these unique subsets of T lymphocytes and have significant implications in medicine and cellular therapy.
MATERIALS AND METHODS
Cell purification and flow cytometry
Healthy donor peripheral blood was obtained after informed consent approved by the St. Jude Children’s Research Hospital institutional review board. Lymphocyte subsets were sorted with FACSAriaII (BD Biosciences, San Jose, CA) using anti-CD3, anti-CD56, anti-CD14, anti-KIR2DL1, anti-KIR2DL2/3, and anti-KIR3DL1. The purity of CD3+CD56− T cells, CD3−CD56+ NK cells, CD3+CD56+ T cells, CD3+CD56+KIR− T cells, and CD3+CD56+KIR+ T cells was more than 98% in all experiments. iNKT cells were cloned from PBMCs using a modified protocol (46). Briefly, the PBMCs were cultured with 100 ng/mL α-galactosylceramide (αGC) and 50 ng/mL IL-2 for 3 weeks. The iNKT cells were flow sorted for TCRVα24 (clone 6B11). Then, the cells were restimulated with allogeneic PBMCs, fresh IL-2, and αGC every week. Before each experiment, cells were re-sorted for TCRVα24. The purities of iNKT for TCRVα24 and CD1d-tetramer were >95% and >90%, respectively.
Phenotypes of cell subsets were analyzed by flow cytometry with LSRII (BD Biosciences) and FlowJo 8.8.6 software (Tree Star, Ashland, OR). The following clones of antibodies were used: anti-KIR2DL1 (EB6.B), anti-KIR2DL2/3 (CH-L, GL183), anti-KIR3DL1 (DX9), anti-CD95 (UB2), anti-DNAM-1 (DX11), anti-CD11a (HI111), anti-NTBA (292811), anti-CD244 (2-69), anti-CD54 (84H10), anti-CD155 (TX21), anti-CD48 (J4-57), anti-CD16 (B73.1), anti-CD4 (MT310), anti-CD8 (DK25, SK1), anti-TCRαβ (WT31), anti-TCRγδ (11F2), anti-TCRVα24 (6B11), anti-CD45RO (UCHL1), anti-CD45RA (L48), anti-CD3 (SK7, UCHT1), anti-CD25 (2A3), anti-CD38 (T16), anti-CD69 (L78), anti-CD90 (5E10), anti-granzyme B (GB10), anti-NKG2a (Z199), anti-NKp30 (Z25), anti-NKp44 (Z231), anti-NKp46 (BAB281), anti-NKG2D (1D11), anti-CD253 (RIK-2), anti-TRAILR1 (DJR1), anti-TRAILR2 (71908), anti-CD27 (CLB-27/1), anti-CD122 (TM-Beta1), anti-CD94 (HP-3B1), anti-CD178 (14C2), anti-CD56 (MY31, N901), anti-CD137 (1AH2), anti-CD152 (BN13), anti-CD127 (R34.34), anti-CD73 (AD2), anti-CD44 (MEM85), anti-CD62L (DREG56), anti-CD28 (CD28.2), anti-CD14 (MphiP9), anti-NKG2C (134591), anti-CD57 (HNK-1) and anti-HLA-E (3D12).
Patient and transplant information
Thirty patients who underwent allogeneic stem cell transplantation at St. Jude were included. Transplant approaches have been described previously (47). These patients or their parents had given informed consent approved by our institutional review board before enrolling in the institutional transplant protocol. After the completion of a preparative regimen, blood samples were obtained weekly for CMV surveillance using quantitative PCR until 100 days after transplantation and then every 2–4 weeks thereafter. Enumeration of KIR+ and KIR− CD56+ T cells was performed monthly for 3 months and then every 2–4 months thereafter.
CMV experiments
Blood samples from CMV-seropositive asymptomatic healthy donors were screened for HLA-A*0201. Percentages of CMVpp65-tetramer+ cells were measured using HLA-A*0201-CMVpp65 tetramer-PE (iTag, Beckman Coulter, Brea, CA). The CMVpp65-tetramer+ CD56+ T cells were expanded in SCGM medium (CellGenix GmbH, Freiburg, Germany) in the presence of IL-2 (500 U/mL, Cellgenix), IL-15 (10 ng/mL, Cellgenix), OKT3 (10 ng/mL, Biolegend, San Diego, CA), 1 μg/mL CMVpp65 peptide (Anaspec, CA), and 5% human AB serum (Lonza, Basel, Switzerland) for 14 days. The cells were replenished with fresh medium and cytokines every 3 days. In selected experiments, CMVpp65-tetramer+ cells in the KIR− and KIR+ subsets of CD56+ T cells were sorted by flow cytometry with >95% purity.
TCR Vβ spectratyping
The CDR3-encoding region of the TCRVβ gene was amplified using 25 TCRVβ subfamily–specific primers and a carboxyfluorescein (FAM)-conjugated TCRVβ constant region–specific primer (48). The PCR products were denatured with Hi-Di formamide (Applied Biosystems, Carlsbad, CA) and electrophoresed along with Gene Scan-500 LIZ size standard (Applied Biosystems) on a 3130xl Genetic Analyzer (Applied Biosystems). The overall complexity of TCRVβ in each subset was calculated by summation of the total number of peaks in each subfamily.
Affymetrix expression analysis
RNA pre-amplification, labeling and hybridization on Human Genome U133Plus 2.0 GeneChip array were performed in the St. Jude Hartwell Center for Bioinformatics & Biotechnology microarray core facility according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA). Expression values were summarized using the MAS5 algorithm as implemented in the GCOS v1.4 software (Affymetrix). Signals were variance adjusted by log-transformation prior to statistical analysis. Analysis of variance (ANOVA) was performed using Partek Genomics Suite v6.4. The false discovery rate was controlled at a level of 0.05 unless otherwise stated. Gene ontology and pathway analysis was performed using the DAVID bioinformatics database. Gene set enrichment analysis was performed using the KEGG canonical pathways database as described (49). The data have been deposited in the Gene Expression Omnibus, NCBI and are accessible through GEO Series accession number GSE47855 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47855).
Cytotoxicity assay
Cytotoxicity assays were performed using DELFIA BATDA reagent (PerkinElmer Life and Analytical Sciences, Waltham, MA) according to the manufacturer’s instructions. BATDA-labeled target cells were cocultured with effector cells for 2 h at 37°C. The fluorescence was measured using a Wallac Victor 2 Counter Plate Reader (PerkinElmer Life and Analytical Sciences) (4). For KIR blocking experiments, cells were pretreated with KIR antibody CH-L or pan-HLA-ABC antibody W6/32, IgG2 clone that has minimal binding affinity to human FcγIIIa receptor CD16 (50, 51).
Multiplex cytokine measurement
The production of TH-1, TH-2, TReg, and TH-17 cytokines, after stimulation with 1 of the 20 stimuli for 3 days, was measured by the Luminex system using a Milliplex MAP 7-plex Panel I kit for IFN-γ, IL-4, IL-5, IL-6, IL-10, IL-13, and IL-17; Panel II kit for IL-21; and Panel III kit for TGFβ (MilliPore, Billerica, MA). Cells were plated at a density of 5×104 cells per well. Stimuli included TLR ligands 1-9 (Human TLR1-9 agonist kit, Invivogen, San Diego, CA), EBV (B95-8 Type 1 purified viral lysate), HSV-1 (MacIntyre Purified, all from Advanced Biotechnologies Inc., Columbia, MD), phytohemagglutinin, phorbol myristate acetate, concanavalin A (all from Sigma, St. Louis, MO), IFN-α, IFN-β, IL-2, IL-6, and TGF-β.
Real-time PCR
Transcripts were quantified using Applied Biosystems pre-designed Taqman Gene Expression assay on an ABI PRISM 7900HT (Applied Biosystems) according to the manufacturer’s instructions. For rorc quantification, the Hs01076112_m1 probe was used (52). Relative expression of the target transcript was calculated by the cycling threshold (CT) method as 2−ΔΔCTrelative to th e expression in NK cells.
TH-17 differentiation
CD4+ cells from various lymphocyte subsets were sorted by flow cytometry (Aria II, BD Biosciences) and cultured under TH-17 differentiation conditions as previously described (53). Cells plated at 5×104 per well were primed with immobilized CD3 and CD28 antibodies for 3 days and then incubated with IL-2 (20 U/mL, R&D Systems, Minneapolis, MN), IL-1β (10 ng/mL, R&D Systems), IL-6 (50 ng/mL, R&D Systems), IL-23 (20 ng/mL, R&D Systems), neutralizing anti-IFN-γ (10 μg/mL, BD Biosciences), and anti-IL-4 (10 μg/mL, BD Biosciences) for 5 days. The cells were allowed to proliferate for 7 more days with the addition of low-dose IL-2 (10 U/mL, R&D Systems) and IL-23 (20 ng/mL, R&D Systems).
CD107 degranulation assay
Lymphocyte subsets were activated for 3 days with either 500 U/mL rhIL-2 and 10 ng/mL IL-15 (R&D Systems), or anti-CD3 (OKT3) and CD28 (CD28.2) antibodies (Biolegends) and then used in a CD107 mobilization assay with K562 cells as the target (4). A redirected assay was performed with P815 cells (ATCC, Manassas, VA) that were pre-incubated with the indicated antibodies (5 μg/mL) and washed twice before being used as the target cells.
Statistical analysis
The exact Wilcoxon signed rank test was used to compare samples from the same donor, and the exact Wilcoxon rank sum test was used for independent samples. The relationships of various lymphocyte subsets were revealed using dendrograms by cluster analysis with the complete linkage method based on surface phenotype similarity. All analyses were performed using Rsoftware (version 2.10). No adjustments were made for multiple testing.
RESULTS
KIR+ T cells primarily reside in the CD56+ T cell population with limited TCRVβ repertoire, TREC, and telomere length
In adult blood, about half of the NK cells were KIR+; in contrast, only about 1% of CD3+ T cells were KIR+, and they primarily resided in the CD56+ rather than the CD56− fraction (Figures 1A and 1B). CD56+ T cells accounted for 4.8% of the lymphocytes in adult blood (Figure S1A), but they were rare in cord blood (0.41% on average; Figure S1B). In iNKT cells, a substantial percentage was CD56+ (Figure S1C, average 36.9% ± 21.7%) but they were all KIR−.
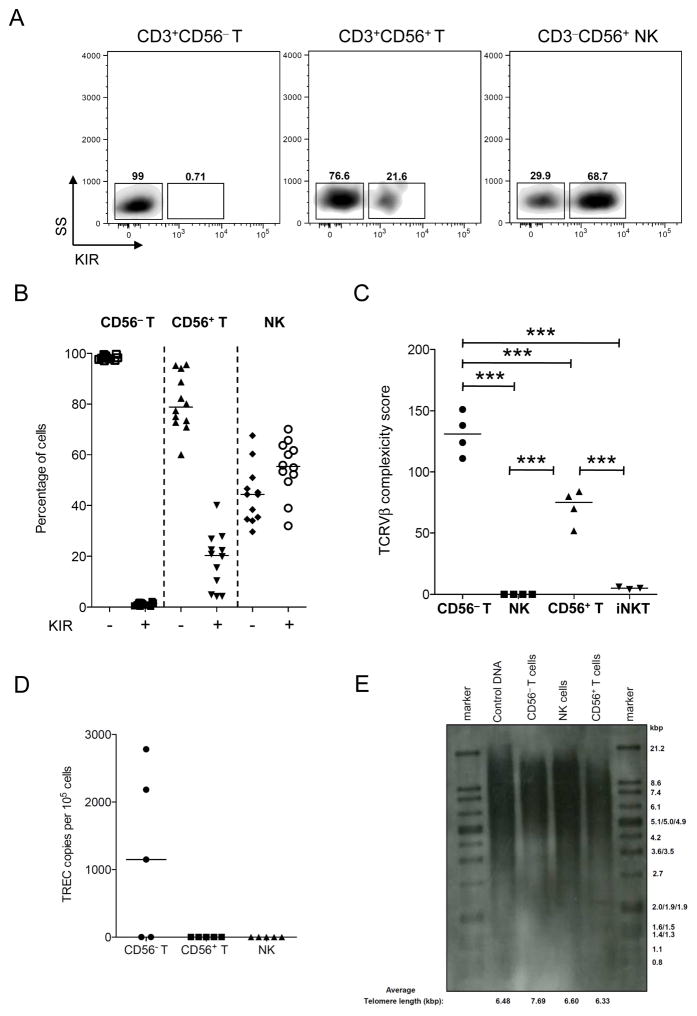
Figure 1. KIR, TCRVβ, TREC and telomere lengths.
(A) Flow cytometry for KIRs on CD56− T, CD56+ T, and NK cells. Numbers indicate the percentage of cells in each region. Data are from representative experiment of 12 individuals. (B) Comparisons of percentages of KIR− and KIR+ subsets in CD56− T, CD56+ T, and NK cells. Data are from 12 individuals. (C) TCRVβ diversity. (D) Low number of TREC in CD56+ T cells. (E) Telomere length of CD56+ T cells was shorter than those of CD56− T and NK cells. DNA from an immortal cell line (lane 2) was used as a positive control. Data are representative of three independent experiments. The bars in the dot-plot (B–D) represent the median. ***p<0.001.
Compared with CD56− T cells, CD56+ T cells had a less diverse TCRVβ repertoire and a skewed Gaussian pattern (Figure S1D). As expected, iNKT cells were solely TCRVβ11+, and NK cells did not express any TCRβ. Thus, CD56+ T cells have a lower TCRVβ complexity score than CD56− T cells but higher than that of NK and iNKT cells (Figure 1C). CD56+ T cells also had lower copy numbers of the signal joint T-cell receptor excision circle (TREC) than CD56− T cells (Figure 1D), indicating that CD56+ T cells had undergone extensive post-thymic proliferation. This finding supported the observation that the telomeres of CD56+ T cells were shorter than those of CD56− T and NK cells (Figure 1E).
CD56+ T cells have distinct surface phenotype and genome-wide transcriptional pattern
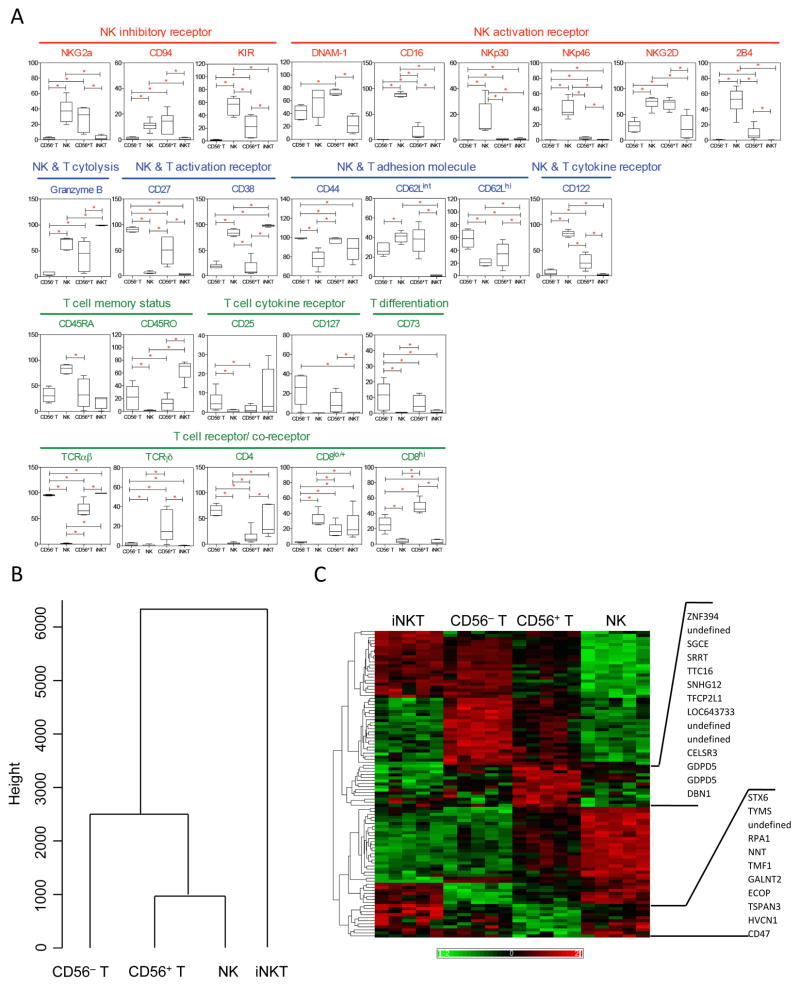
To distinguish CD56+ T cells from CD56− T, NK, and iNKT cells, we performed an immunophenotyping array using 41 NK and T biomarkers related to activation, costimulation, inhibition, cytotoxicity, adhesion, and development (Figure 2A and Table S1). Compared with CD56− T cells, CD56+ T cells expressed higher levels of NK markers including CD16, CD94/NKG2a, NKG2D, CD122, DNAM-1, and granzyme B, as well as TCRγδ and CD8. In contrast, CD56− T cells expressed more CD4, CD27, and TCRαβ than CD56+ T cells. Compared with NK cells, CD56+ T cells expressed more CD4, CD8, and activation markers CD27 and CD44 but less NK-associated molecules including CD16, NKp30, NKp46, 2B4, and CD122. Remarkably, no marker was expressed more in both CD56− T and NK cells than in CD56+ T cells. Compared with iNKT cells, CD56+ T cells expressed more CD122, CD8, CD45RA, CD27, and CD62L, as well as NK receptors CD16, NTB-A, NKG2D, DNAM-1, NKp46, 2B4, NKG2a, and CD94, but less CD4, CD38, CD25, CD45RO, and CD69. Notably, CD56+ T cells expressed the most DNAM-1, CD8, and TCRγδ among the 4 cell types. In a dendrogram generated based on data from all 41 biomarkers using cluster analysis with the complete linkage method (Figure 2B), CD56+ T and NK cells were in the same branch but were disparate from CD56− T cells and farthest from iNKT cells in surface phenotype.
Figure 2. CD56+ T cells are distinct from NK, CD56− T, and iNKT cells in surface phenotype.
(A) Box-plots summarize the percentage of cells testing positive in CD56− T, NK, CD56+ T, and iNKT populations. Markers without substantial differences are not shown in the figure: NKp44, NTB-A, FasL, TRAIL-R1, TRAIL-R2, TRAIL, Fas, CD48, PVR, 4-1BB, ICAM-1, CD69, CLTA-4, CD11a, and CD90. (B) Dendrogram shows the relationship among CD56− T, CD56+ T, NK, and iNKT cells in surface phenotype using all 41 biomarkers. (C) Hierarchical clustering of 101 transcripts with differential expression between CD56+ T cells and other cell types. Red indicates relative high expression and green indicates relative low expression across the data set. Each column represents a sample from different individuals. Names of the genes significantly upregulated or downregulated in CD56+ T cells are listed on the right. Data in (A–C) are from 5–6 individuals.
Since phenotypic analyses using antibodies and flow cytometry in this and prior studies could provide only limited data, we further characterized the CD56+ T cells at a genome-wide level by comparing the transcriptome in the 4 cell types using microarray profiling. A large number of genes (9,366) were differentially transcribed (p=0.0056, false discovery rate [FDR]<0.01). Pairwise comparisons between CD56+ T cells and each other cell type revealed 101 CD56+ T “signature genes” that were differentially expressed with p<0.0024 and FDR<0.3 (Figure 2C). Among those transcripts, 9 genes were unidentified, and 30% were enriched compared with CD56− T, NK, and iNKT cells, especially those involved in transcription repression in mitogen-activated protein kinase pathways, such as znf394, gdpd5, tfcp2l1, and snhg12. Genes that were preferentially downregulated in CD56+ T cells were those involved in metabolism, such as stx6, nnt, galnt2, and hvcn1; in DNA replication, such as tyms, rpa1 tmf1, and ecop; and in adhesion, such as CD47 and tspan3. Analysis using the KEGG canonical pathway database (49) showed that the activity of 29 pathways was significantly suppressed in CD56+ Tcells compared with the other subgroups. Among the top pathways were those associated with cell division and metabolic activity, including cell cycle, DNA and nucleotide synthesis, oxidative phosphorylation, fatty acid biosynthesis, amino acid metabolism, and allograft rejection. Notably, no pathways were significantly enriched in CD56+ T cells, indicating definitively their relative transcriptional quiescence and defining ultimately their molecular identity apart from other lymphocyte subsets.
CD56+T cells are cytotoxic in proinflammatory milieu through the KIR +subset
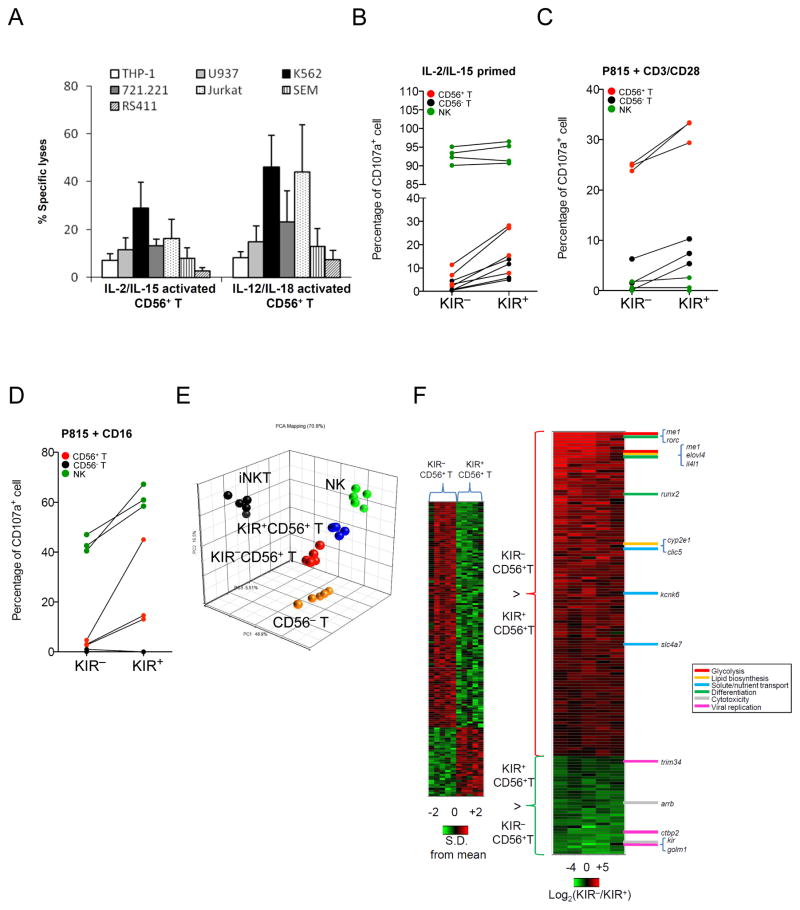
Although CD56+ T cells have been called “NK-like”, surprisingly we found that highly purified resting CD56+ T cells were tolerant to standard NK-susceptible targets, such as K562, THP-1, U937, Jurkat, and 721.221 cells (Figure S1E), compatible with our genome-wide hypoactive transcriptome data. In addition, they are tolerant to standard iNKT-susceptible targets such as CD1d-restricted αGC-loaded Jurkat cells (Figure S1F). To confirm that the non-reactivity of CD56+ T cells to αGC-loaded Jurkat cells was not a unique target cell-line effect, we generated a CD1d-transduced cell line using NK-resistant RS4;11 cells to avoid any false readout by contaminating NK cells. We found that the αGC-loaded RS4;11-CD1d mutant was indeed susceptible to killing by iNKT cells but not by CD56+T cells.
Based on the genome-wide quiescent transcriptional pattern, we hypothesized that CD56+ T cells required a pro-inflammatory milieu to exhibit their NK-like activity. Indeed, CD56+ T cells stimulated with IL-2 and IL-15 or IL-12 and IL-18 could kill all NK-susceptible targets (Figure 3A). Because the KIR+ rather than KIR− subset of NK cells is primarily cytotoxic, we further hypothesized that a similar distinction existed in CD56+ T cells. In a single-cell degranulation assay, significantly more K562-reactive cells were found in KIR+ than in KIR−CD56+ T cells (Figure 3B). Similarly, a significantly higher percentage of KIR+CD56+ T cells degranulated in the presence of FcγRIIIa+ murine target cells coated with antibodies specific to CD3/CD28 (Figure 3C), even though both subsets were CD3+ and the KIR+CD56+ T cells expressed significantly less CD28 and CD90 (Figure S2A). Using anti-CD16 antibody–coated P815 cells, we found that CD107a+ cells were confined to the KIR+CD56− T subset, whereas <5% of the KIR−CD56+ T had degranulation (Figure 3D). In CD56− T cells, the TCRγδ+ rather than the TCRαβ+ population is naturally cytotoxic (54). In CD56+ T cells, we found that the TCRγδ+ population was enriched with the KIR2DL2/3+and KIR3DL1 +cytotoxic subset (Figure S2B).
Figure 3. KIR+CD56+ T cells are more cytotoxic and phenotypically disparate from the KIR− subset.
(A) Cytotoxicity of CD56+ T cells stimulated ex vivo with IL-2 and IL-15 or IL-12 and IL-18 for 3 days. Data are mean and S.E.M. (B) CD107a degranulation assay in KIR+ versus KIR− subsets of CD56− T (black), CD56+ T (red) and NK cells (green) against K562 after IL-2/IL-15 priming, or against P815 cells with (C) CD3/CD28 redirection or (D) CD16 redirection. (E) Principal component analysis plot using ~3,900 transcripts among iNKT (black), CD56−T (orange), KIR−CD56+T (red), KIR +CD56+T (blue), and NK (green) cells filtered with FDR 0.01 and 10% present calls. (F) Heat map representation of gene expression profile that differed between KIR+ and KIR− subsets in CD56+ T cells (ANOVA p<0.001). In left panel, expression is depicted in standard deviation units relative to the mean across all samples for each transcript. In right panel, expression is presented as relative transcript level between KIR− and KIR+ subset in a log2-scale, and the genes are ordered by mean difference observed. Genes of interest annotated to specific functional ontologies are shown to the right of the heat map. Data are from 5 (A, E, and F), 4 (B), and 3 (Cand D) experiments. * p<0.05.
KIR+CD56+ T cells are phenotypically distinct and more NK-like than KIR−CD56+ T cells
In comparison to KIR−CD56+ T cells, TCR and flow analyses revealed considerably restricted TCRVβ spectrum (Figure S2C) and higher expression of NKp46, NKG2D, 2B4, CD16, CD38, CD122, CD11a and granzyme B in KIR+CD56+ T cells (Figure S2D). We then extended the comparison to genome-wide level by expression profiling of KIR+ and KIR− CD56+ T cells in the context of those in NK, CD56− T, and iNKT cells. As shown in the principal component analysis plot (Figure 3E), the KIR+ subset clustered closer to NK cells, whereas the KIR− subset was closer to CD56− T cells. Among the 146 genes differentially expressed between the KIR+ and KIR− subsets of CD56+ T cells (p<0.001, FDR<0.28, Figure 3F), genes in glycolysis, glutaminolysis and ATP synthesis were enriched in the KIR− subset. NADP+-dependent cytosolic malic enzyme (me1), which converts malic acid to pyruvate during fatty acid biosynthesis, had the highest fold difference between the two subsets with 42.37-fold higher expression in KIR− population. Other genes differentially expressed in the KIR− subset regulated lipid metabolism (e.g. elov4 and cyp2e1), solute/nutrient transport (e.g. clic5, kcnk6 and slc4a7) and differentiation (e.g. rorc, il4l1, ltk and runx2). In contrast, the KIR+ subset was characterized by the lack of transcripts for glycolysis and glutaminolysis, but increased expression of genes related to cytotoxicity regulation (e.g. arrb) and viral infection (e.g. ctbp2, golm1 and trim6-34). It is known that during T cell activation and memory development, metabolic transcription program could alter cell fate and differentiation (55); thus while activated T cells increase nutrient uptake, ATP synthesis, and energy utilization mainly through glycolysis and glutaminolysis, memory T cells by contrast maintain house-keeping energy conversion process through lipid oxidation (56, 57). Collectively, our gene expression data support the hypothesis that KIR−CD56+ T cells are metabolically active T subset poised to effector differentiation and reprograming, whereas KIR+ CD56+ T cells are metabolically quiescent memory cells equipped with cytotoxic pathways set for long-term pathogen control.
KIR+CD56+T cells expand in human cytomegalovirus infection
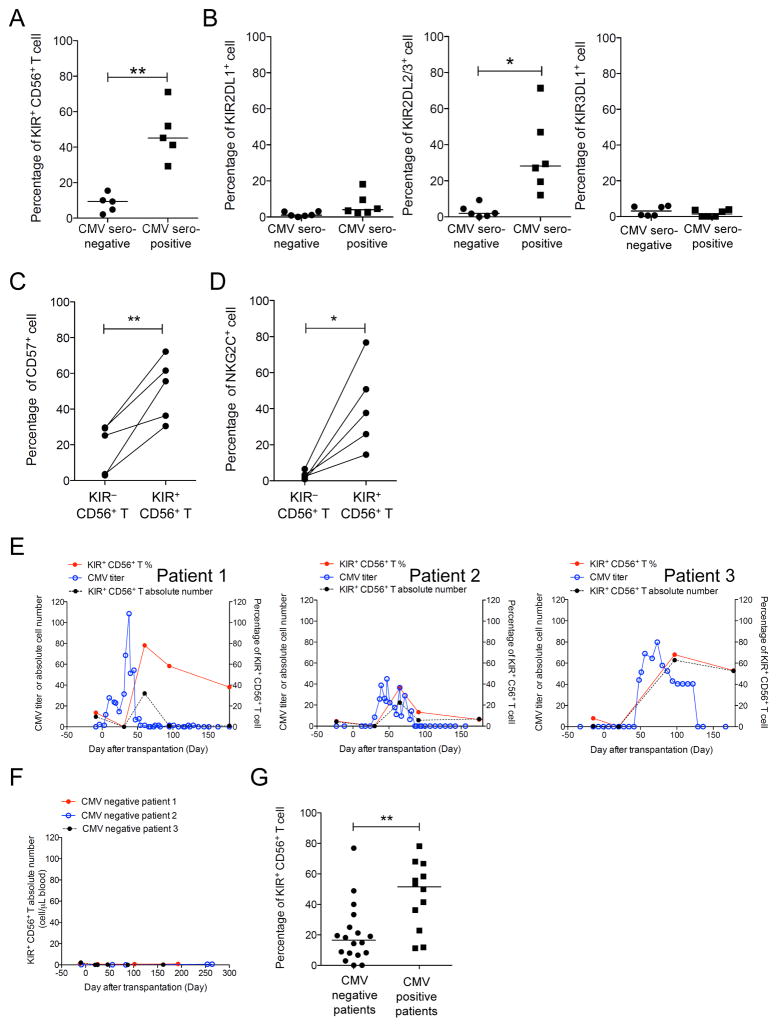
Previous studies have shown that some KIR+ T cells are CMV-specific (15, 18, 24, 25), but the kinetics of their response to CMV reactivation is unclear. Therefore, we examined a cohort of donors and recipients of hematopoietic stem cell transplantation. We found that almost half of the CD56+ T cells were KIR+ in healthy asymptomatic CMV-seropositive donors, which was significantly more than in CMV-seronegative donors (Figure 4A; p=0.0018). Notably, KIR2DL2/3 was overrepresented in CMV+ donors but not KIR2DL1 or KIR3DL1; thus, the majority of KIR positivity in CD56+ T cells was related solely to KIR2DL2/3 expression (Figure 4B). Furthermore in CMV+ donors, expression of CD57 in KIR+CD56+ T cells was higher than that in KIR−CD56+ T cells (Figure 4C), whereas nearly all NKG2C+CD56+ T cells resided in the KIR+ rather than the KIR− population (median 37.7% versus 5%; Figure 4D). Thus, KIR2DL2/3+CD57+NKG2C+ was a signature phenotype of the KIR+CD56+ T cells.
Figure 4. KIR+ subset of CD56+ T cells in human CMV infection.
(A) Comparison of percentage of KIR+ cells or (B) single KIR2DL1+, KIR2DL2/3+, or KIR3DL1+ cells in the CD56+ T cell population between CMV-seropositive and -seronegative asymptomatic donors. (C) Comparisons of percentages of CD57+ or (D) NKG2C+ cells in KIR− and KIR+ CD56+ T cells from CMV-seropositive donors. (E) CMV titer in copy per microgram of DNA (blue), absolute number of KIR+CD56+ T cells per microliter of blood (black), and percentage of the KIR+ subset in the CD56+ T population (red) from 3 representative patients were plotted against time. (F) Constant low levels of KIR+ CD56+ T cells were observed from patients without CMV infection/reactivation. Day 0 is the day of transplantation. Each line represents one patient. (G) Comparison of the median percentage of the KIR+ subset in CD56+ T cells during the first 100 days after transplantation between groups of patients with or without CMV viremia. The bars are the median. * p<0.05; ** p<0.01.
Among the 30 stem cell transplant recipients, 12 (40%) had CMV reactivation detectable in blood by real-time PCR in the first 100 days after transplantation. Notably, all had an increase in the percentage of the KIR+ subset in CD56+ T cells at the time of CMV reactivation. When the CMV titers started to rise, both the percentage and absolute number of KIR+CD56+ T cells increased (Figure 4E). Once the viremia was under control, the KIR+CD56+ T population gradually decreased but remained generally higher than before viral reactivation. In contrast, patients without CMV reactivation during transplant courses had constant KIR+CD56+ T cell numbers (Figure 4F). Thus, overall, the median percentage of KIR+CD56+ T cells during the first 100 days after transplantation was significantly higher (p=0.0021) in patients with CMV reactivation than in those without (Figure 4G).
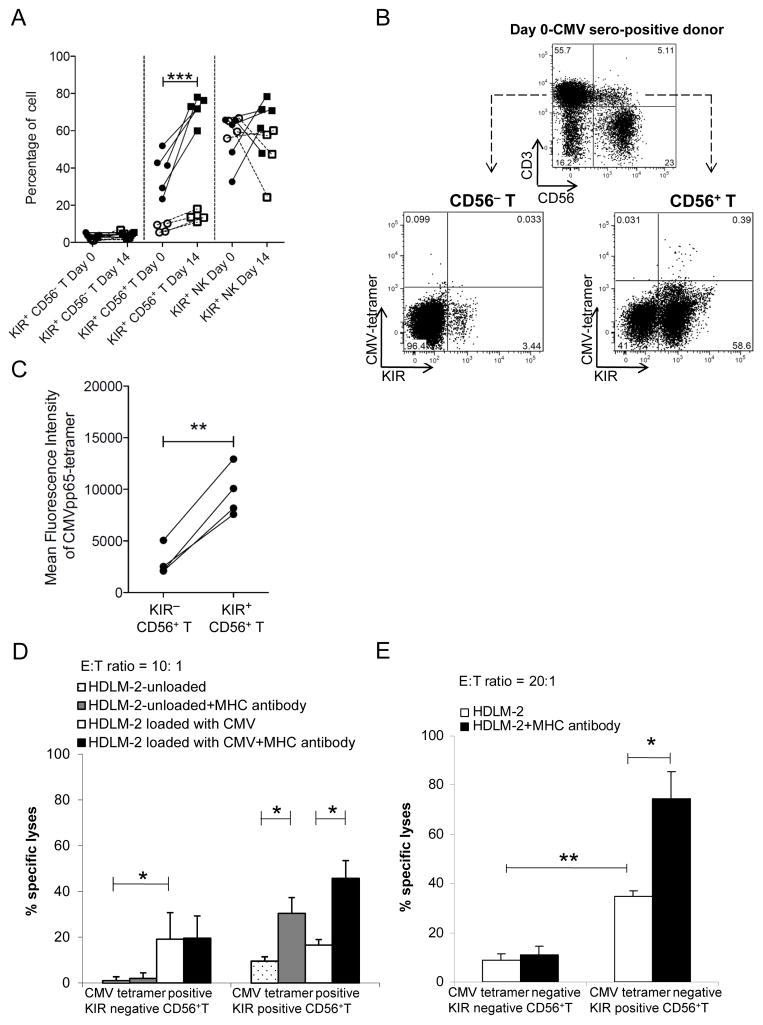
During CMV reactivation in the transplant recipients, we did not observe a similar response in the KIR+CD56− T cell or KIR+CD56+ NK cell populations. When these 3 cell populations from seropositive donors were investigated on their potential for memory response to CMV in vitro after 2 weeks of activation using 1 μg/mL of CMVpp65 peptide, the percentage of the KIR+ subset increased only in the CD56+ T population (from median 41.3% to 73%) and not in CD56− T cells (p<0.05, Figure 5A). Furthermore, expansion was not observed in KIR+CD56+ T cells obtained from CMV-seronegative donors, indicating that the expansion in seropositive donors was a memory response.
Figure 5. Memory and lytic response by KIR+CD56+ T cells.
(A) Percentage of KIR+ cells in CD56− T, CD56+ T, and NK cells from CMV-seropositive (closed symbols with solid lines, n=5) and seronegative (open symbols with dotted lines, n=4) donors before and after 14-day pulsing with CMVpp65. (B) Flow gating strategy for CMVpp65 tetramer against KIR expression on CD56+ and CD56− T cells. Numbers in quadrants indicate the percentage of positive cells. Results are representative of 4 independent experiments. (C) Paired comparison of mean fluorescence intensity of HLA-A*02:01 CMVpp65-tetramer+ in KIR+ versus KIR− subsets of CD56+ T cells. (D) CMVpp65-peptide–specific CD56+ T cells were tested against CMVpp65-peptide–loaded HDLM-2 cells pretreated with pan-MHCI antibody (black bar) or untreated (white bar). Unloaded HDLM-2 cells with (gray bar) or without (dotted white bar) pan-MHCI antibody were used as control. (E) Cytotoxicity of CMVpp65-tetramer–negative KIR+CD56+ T versus KIR−CD56+ T cells against HDLM-2 cells with or without pan-MHCI antibody. Data represent 4 (C-E) independent experiments. * p<0.05; ** p<0.01.
The median frequency of HLA-A*02:01 CMVpp65 tetramer–positive cells was 0.39% in KIR+CD56+ T cells, which was 4-fold higher than that in KIR−CD56− T cells and 1 log higher than that in KIR−CD56+ T or KIR+CD56− T cells in seropositive donors (Figure 5B). Notably, the mean fluorescence intensity was much higher in the KIR+ than in the KIR− CD56+ T cells (Figure 5C). No binding was observed in either KIR− or KIR+ CD56+ T cells from seronegative donors.
Using IL-2, IL-15 and OKT3, we successfully expanded ex vivo and subsequently flow-sorted enough CMVpp65-specific KIR+ subsets of CD56+ T cells for further functional experiments. We tested whether KIR+CD56+ T cells, which were obtained from HLA-A*02:01+ CMV-seropositive donors and proliferated ex vivo for 2 weeks in response to CMVpp65 peptide, had variable cytotoxicity to CMV-infected cells in a KIR-dependent manner. The NK cell–resistant HDLM-2 lymphoblastic cell line, which was HLA-A*02:01+ and HLA-C1+ (ligand for inhibitory KIR2DL2/3), was used as the target. For CMVpp65-tetramer–positive CD56+ T cells (Figure 5D), the KIR−CD56+ T cells had only a modest effect on the CMVpp65-peptide–loaded HDLM-2 cells, and the killing did not increase with KIR-ligand blockade. In contrast, CMVpp65-tetramer–positive KIR+CD56+ T cells did not kill peptide-loaded HDLM-2 cells well, but the cytotoxicity increased significantly with KIR-ligand blockade. This suggested that KIR expression and signaling modulated the memory response of human CD56+ T cells to specific pathogens such as CMV. As expected, the cytotoxicity of KIR−CD56+ and KIR+CD56+ cells against peptide-unloaded HDLM-2 cells was lower than that with peptide-loaded cells.
We then assessed the effects of KIR signaling on non–CMV-specific cytotoxicity. For CMVpp65-tetramer–negative CD56+ T cells, the KIR+ subset killed HDLM-2 cells significantly better than its KIR− counterpart (40% vs. 10% on average; Figure 5E). With KIR-ligand blockade, the KIR+ subset’s cytotoxicity further increased. Similar effect was observed with KIR2DL2/3 antibody blockade (Figure S2E). Taken together, these data suggested that KIR was broadly functional in regulating non–CMV-specific immunity in CD56+T cells as well.
KIR−CD56+T cells are Rorc+IL -13 secretor
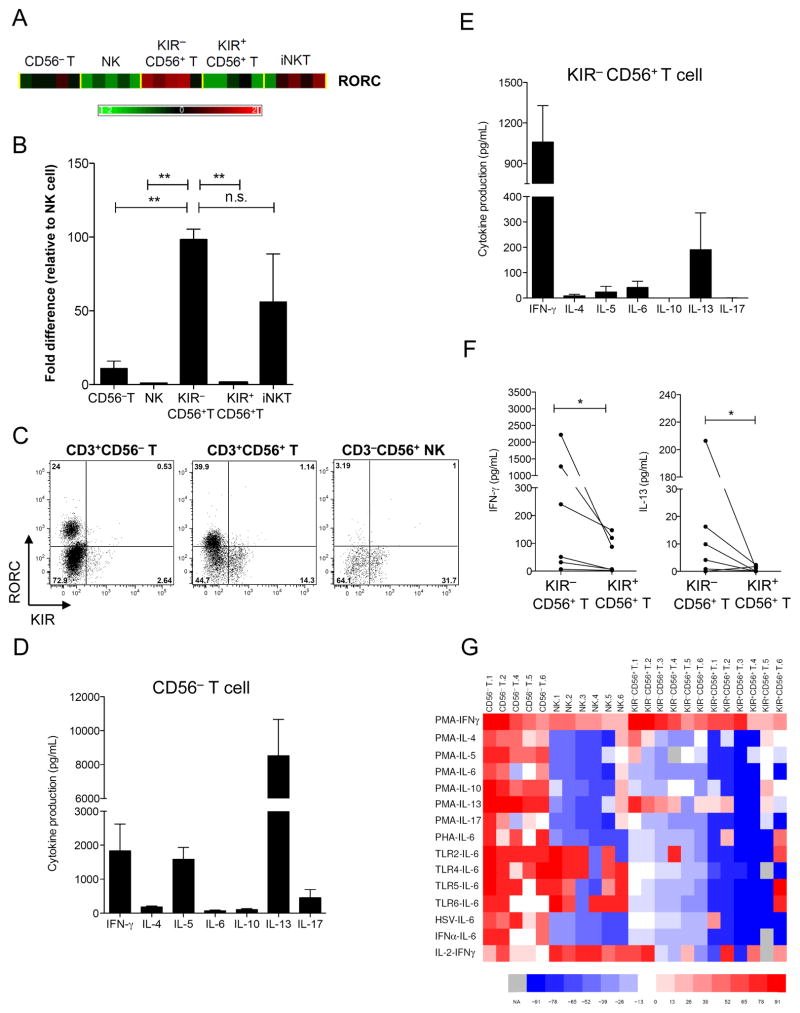
Rorc was among the top 5 of 146 genes differentially expressed between KIR− and KIR+ subsets of CD56+ T cells in our genome-wide expression analysis (Figure 3F). The KIR− subset had a 27.7-fold higher expression (Figure 6A), which was confirmed by real-time PCR (Figure 6B). Rorc mRNA was found in CD56− T and iNKT cells as well, but the level of Rorc was significantly higher in KIR−CD56+ T cells than in CD56− T, NK, and KIR+CD56+T cells.
Figure 6. KIR−CD56+ T cells are RORC+ IL-13 secretor.
(A) Heat map of Rorc expression in CD56− T, KIR−CD56+ T, KIR+CD56+ T, NK, and iNKT cells. (B) Quantitative PCR of RorcmRNA transcript in different subsets. The data presented arenormalized to that of NK cells. The results are from 5 independent experiments. (C) Intracellular staining of RORC in CD4+-enriched fraction of T, NK, and CD56+ T cells after TH-17 differentiation culture conditions. Numbers in quadrants indicate the percentage of cells positive. The results are representative of 3 independent experiments. (D) Luminex array of cytokine production from CD56− T and (E) KIR−CD56+ T cells after culturing in TH-17 differentiation conditions and restimulation with anti-CD3 and PMA for 36 h. Data represent mean and SD from 2 independent experiments. (F) Pairwise comparison of IFN-γ and IL-13 production upon PMA stimulation between KIR− and KIR+ CD56+ T subsets. (G) Luminex analysis of cytokine production after immune stimulation. The results are displayed as a heat map for stimulus-product combinations that were different across CD56− T, NK, KIR−CD56+ T and KIR+CD56+ T cells. The color coding on the heat map is according to the mean fluorescence intensity from multiplex beads coated with antibodies against indicated cytokines after log10-transformation, ranking, and fitting into a color scale of red to blue. Spectraof red and blue represent values higher and lower than the mean rank, respectively. Column on the left labels the cytokine produced with the indicated stimulus. Numbers beside the population names indicate the donor number. TLR2: HKLM; TLR4: LPS E. coli K12; TLR5: flagellin S. typhimurium; TLR6: FSL1; HSV: herpes simplex virus; PHA: phytohemagglutinin; PMA: phorbol 12-myristate 13-acetate; IFN-α: interferon-α; and IL-2: interleukin-2. NA: not available and shown as gray. Kruskal-Wallis rank sum test was used to compare the ranks of all cytokines in CD56− T, NK, KIR−CD56+ T and KIR+CD56+ T cells, and significant ranks are displayed in the heat map.
Rorc is the human homologue of murine transcription factor Rorγt, which is involved in IL-17 induction in subsets of αβ and γδ T cells (53, 58). Rorc also directs the production of IL-17 in iNKT cells (59) and IL-17 and IL-22 in lymphoid tissue-inducer cells and NK-like cells (52). In contrast, mature NK cells express little or no RORC (52). We questioned whether the expression of rorc in the KIR− CD56+ T subset was related to IL-17 production, similar to that in CD56− T lineages. We first investigated whether the RORC protein could be induced and maintained in CD56+ T cells. Under TH-17 differentiation conditions with IL-1β, IL-6, IL-2, and IL-23 (53), a prominent population of RORC+ cells was found in both the KIR−CD56+ TH and KIR−CD56− TH populations but not in NK cells (Figure 6C). Notably, the KIR+CD56+ T cell subset remained negative for RORC at both the transcript and protein level. Using CD56− T cells as control, we found that KIR−CD56+ T cells were able to secrete IFN-γ and IL-13 upon PMA stimulation (Figure 6D and E). In comparison with KIR+CD56+ T cells, KIR−CD56+ T cells produced significantly more IFN-γ and IL-13 (Figure 6F).
To further assess the global cytokine profile of KIR−CD56+ T cells in innate and adaptive immunity, we performed multiplex cytokine arrays using 200 stimulation-cytokine combinations challenging KIR−CD56+ T cells with 20 common stimuli, including toll-like receptor 1–9 agonists, viral lysates, mitogens, and cytokines that favor various TH polarizations. The production of 10 TH-1, TH-2, TReg, and TH-17 signature cytokines was measured. One-way non-parametric ANOVA showed that 16 of the 200 stimulation-cytokine combinations were statistically different across T, NK, KIR−CD56+ T and KIR+CD56+ T cells (Figure 6G). As expected, CD56− T cells contained all subpopulations capable of response to various stimuli by producing all types of TH cytokines. NK cells also, as expected, secreted IFN-γ (with PMA or IL-2) and IL-6 (with TLR agonists). However, KIR−CD56+ T cells did not produce IL-5, IL-6, IL-10, or IL-17 upon stimulation with any TLRs, viral lysates, mitogens, or cytokines tested. All these data suggest that KIR−CD56+ T cells are not broad cytokine secretors but rather a unique population of previously unknown RORC+ IL-13–producing cells.
DISCUSSION
In this study, we confirmed prior findings that KIR+CD56+ T cells are rare (<1%) in umbilical cord blood and account for <20% of T cells in CMV-seronegative adults (60, 61). However, during CMV reactivation in an immunocompromised state such as that in transplant recipients, we found that the KIR+CD56+ T population markedly expanded in real time corresponding to viral replication. Notably, after expansion, the proportion of these cells declined as the viral copy number in blood decreased but remained higher than that in the pre-reactivation phase. Because CMV persists throughout life, it is conceivable that repeated subclinical reactivations may mechanistically account for the expansion of the KIR+ fraction over time (62). This growth may occur at the expense of naïve T cells to maintain homeostasis in the total T-cell number (62). In this regard, the KIR+CD56+ T subset had a more restricted TCR repertoire than the KIR−CD56+ cells. In CMV-seropositive donors, as much as 50% of the CD56+ T cells were KIR+. These cells have a unique signature of being CD57+NKG2C+KIR2DL2/3+ but KIR2DL1/KIR3DL1−. Phenotypically similar CD57+NKG2C+ NK cells have been observed previously during CMV infection and reactivation (63, 64). Thus, both T cells and NK cells may preferentially use some shared activating receptors such as NKG2C to recognize HLA-E that is altered during CMV infection (25, 65). The biological reason for the preferential expression of KIR2DL2/3 over KIR2DL1 or KIR3DL1 in CD56+ T cell response to CMV is unknown, although similar predilection has also been observed in NK cells in chronic viral hepatitis and acute chikungunya or CMV infection (64, 66, 67). One plausible speculation in evolutionary terms might be better viral protection by C1-specific KIRs, as the C1 epitope preceded the C2 epitope by several million years and C1-receptors are universally expressed in all people (68, 69). Notably, we found that KIR+CD56− T cells did not have similar memory response to human CMV, suggesting that the T memory response to CMV reactivation in humans is primarily mediated through the unique subset of KIR+CD56+T cells.
Although both KIR+ T cells and CD56+ T cells have been called NK-like T cells, our genome-wide transcriptional assays clearly showed for the first time that they are remarkably distinct from conventional NK cells and iNKT cells. Even though the multi-parameter surface-molecule dendrogram and gene-expression array placed CD56+ T cells and KIR+CD56+ T cells closer to NK cells than to CD56− T cells and KIR−CD56+ T cells, the 2 former cell populations have a signature quiescent transcriptome. In this regard, KIR− and KIR+CD56+ T cells possess distinct metabolomics: while KIR−CD56+ cells highly express genes for glycolysis, energy conversion, nutrients uptakes, and DNA metabolism (thus providing the essential fuel for the function of effector genes) (55, 56), KIR+CD56+ cells are relatively quiescent metabolically with transcriptions limited to house-keeping genes and genes related to cytotoxicity and anti-viral pathways. In line with these findings, functional assays of KIR+CD56+ T cells revealed minimal cytokine secretion capability as shown by cytokine array analyses with multiple stimuli, as well as their negligible degranulation and cytotoxicity during steady state against standard NK-and iNKT -sensitive targets. However, upon priming with proinflammatory cytokines, cross-linking of CD3 and CD28, or triggering of CD16, KIR+CD56+ T cells could be cytotoxic to cancer cells or responsive to CMV in a dual KIR-dependent and TCR-dependent manner. Thus, KIR+CD56+ memory T cells may use both KIR and TCR to monitor tumor transformation or viral reactivation with or without MHCI downregulation. While the TCRs on the KIR+CD56+ T subset may enable antigen specificity and memory responses (9), the KIRs may survey against immune escape through downregulation of MHC and simultaneously prevent over-aggression toward normal cells by prematurely terminating the TCR immune synapse (70). Furthermore, the DNAM-1+ and CD16+ subsets of CD56+ T cells was predominately KIR+, thus allowing both natural cytotoxicity and antibody-dependent cytotoxicity to be self-modulated by KIR.
Another novel finding of this study is that KIR can uncouple the effector function of CD56+ T cells. Specifically, while the KIR+ subset is cytotoxic and responsive to CMV infection and cancer cells, the KIR− subset is a previously unknown RORC+ IL-13 producer. The high level of RORC expression uncovered initially by the genome-wide array in the KIR−CD56+ T cell population was surprising, as RORC is a transcription factor typically present in TH-17 cells and other IL-17-producing cells, including γδ T cells, iNKT cells, and innate lymphoid cells (71). Instead of producing IL-17, this novel RORC+KIR−CD56+ T subset produces IL-13, which may paradoxically inhibit TH-17 cells in production of IL-17 through STAT6 and GATA3 (72, 73). Thus, our finding discloses a potential, unexpected role of RORC in negative regulation of IL-17–associated pathways.
In summary, the 2 populations of KIR+CD56+ and KIR−CD56+ T cells are molecularly, phenotypically, functionally and metabolically distinct from the other well-characterized lymphocyte subsets. Whereas the KIR+CD56+ T cytotoxic effector cells may be valuable for immunotherapy of cancers and CMV infections in lymphopenic hosts, the KIR−CD56+ IL-13 producing cells may be useful for the treatment of IL-17–associated health conditions such as chronic inflammatory diseases and graft-versus-host disease (74, 75). Both populations can be easily isolated for future immunologic studies and clinical use.
Supplementary Material
Acknowledgments
We thank Jim Houston for cell sorting; David Galloway for scientific editing; and Dr. Douglas R. Green for critical review of the manuscript.
Footnotes
Disclosures
The authors have no conflicting financial interests.
References
- 1.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothenberg EV. Lineage determination in the immune system. Immunol Rev. 2010;238:5–11. doi: 10.1111/j.1600-065x.2010.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 4.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat Rev Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- 6.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips JH, Gumperz JE, Parham P, Lanier LL. Superantigen-dependent, cell-mediated cytotoxicity inhibited by MHC class I receptors on T lymphocytes. Science. 1995;268:403–405. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- 8.Mingari MC, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, Demarest J, Pantaleo G, Fauci AS, Moretta L. Human CD8+ T lymphocyte subsets that express HLA class I-specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proc Natl Acad Sci U S A. 1996;93:12433–12438. doi: 10.1073/pnas.93.22.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential expression of leukocyte receptor complex-encoded Ig-like receptors correlates with the transition from effector to memory CTL. J Immunol. 2001;166:3933–3941. doi: 10.4049/jimmunol.166.6.3933. [DOI] [PubMed] [Google Scholar]
- 10.Ferrini S, Cambiaggi A, Meazza R, Sforzini S, Marciano S, Mingari MC, Moretta L. T cell clones expressing the natural killer cell-related p58 receptor molecule display heterogeneity in phenotypic properties and p58 function. European journal of immunology. 1994;24:2294–2298. doi: 10.1002/eji.1830241005. [DOI] [PubMed] [Google Scholar]
- 11.Battistini L, Borsellino G, Sawicki G, Poccia F, Salvetti M, Ristori G, Brosnan CF. Phenotypic and cytokine analysis of human peripheral blood gamma delta T cells expressing NK cell receptors. J Immunol. 1997;159:3723–3730. [PubMed] [Google Scholar]
- 12.Snyder MR, Muegge LO, Offord C, O’Fallon WM, Bajzer Z, Weyand CM, Goronzy JJ. Formation of the killer Ig-like receptor repertoire on CD4+CD28null T cells. J Immunol. 2002;168:3839–3846. doi: 10.4049/jimmunol.168.8.3839. [DOI] [PubMed] [Google Scholar]
- 13.Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D, Pitard V, Merville P, Moreau JF, Delfraissy JF, Dechanet-Merville J, Bonneville M, Venet A, Vivier E. Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol. 2004;173:7223–7229. doi: 10.4049/jimmunol.173.12.7223. [DOI] [PubMed] [Google Scholar]
- 14.Bjorkstrom NK, Gonzalez VD, Malmberg KJ, Falconer K, Alaeus A, Nowak G, Jorns C, Ericzon BG, Weiland O, Sandberg JK, Ljunggren HG. Elevated numbers of Fc gamma RIIIA+ (CD16+) effector CD8 T cells with NK cell-like function in chronic hepatitis C virus infection. J Immunol. 2008;181:4219–4228. doi: 10.4049/jimmunol.181.6.4219. [DOI] [PubMed] [Google Scholar]
- 15.van Bergen J, Kooy-Winkelaar EM, van Dongen H, van Gaalen FA, Thompson A, Huizinga TW, Feltkamp MC, Toes RE, Koning F. Functional killer Ig-like receptors on human memory CD4+ T cells specific for cytomegalovirus. J Immunol. 2009;182:4175–4182. doi: 10.4049/jimmunol.0800455. [DOI] [PubMed] [Google Scholar]
- 16.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol. 2004;4:190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 17.Mingari MC, Pietra G, Moretta L. Human cytolytic T lymphocytes expressing HLA class-I-specific inhibitory receptors. Current opinion in immunology. 2005;17:312–319. doi: 10.1016/j.coi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, Moretta L, Mingari MC. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci U S A. 2003;100:10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huard B, Karlsson L. KIR expression on self-reactive CD8+ T cells is controlled by T-cell receptor engagement. Nature. 2000;403:325–328. doi: 10.1038/35002105. [DOI] [PubMed] [Google Scholar]
- 20.Ugolini S, Arpin C, Anfossi N, Walzer T, Cambiaggi A, Forster R, Lipp M, Toes RE, Melief CJ, Marvel J, Vivier E. Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat Immunol. 2001;2:430–435. doi: 10.1038/87740. [DOI] [PubMed] [Google Scholar]
- 21.Marti F, Xu CW, Selvakumar A, Brent R, Dupont B, King PD. LCK-phosphorylated human killer cell-inhibitory receptors recruit and activate phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 1998;95:11810–11815. doi: 10.1073/pnas.95.20.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chwae YJ, Chang MJ, Park SM, Yoon H, Park HJ, Kim SJ, Kim J. Molecular mechanism of the activation-induced cell death inhibition mediated by a p70 inhibitory killer cell Ig-like receptor in Jurkat T cells. J Immunol. 2002;169:3726–3735. doi: 10.4049/jimmunol.169.7.3726. [DOI] [PubMed] [Google Scholar]
- 23.Gati A, Guerra N, Gaudin C, Da Rocha S, Escudier B, Lecluse Y, Bettaieb A, Chouaib S, Caignard A. CD158 receptor controls cytotoxic T-lymphocyte susceptibility to tumor-mediated activation-induced cell death by interfering with Fas signaling. Cancer research. 2003;63:7475–7482. [PubMed] [Google Scholar]
- 24.van der Veken LT, Campelo MD, van der Hoorn MA, Hagedoorn RS, van Egmond HM, van Bergen J, Willemze R, Falkenburg JH, Heemskerk MH. Functional analysis of killer Ig-like receptor-expressing cytomegalovirus-specific CD8+ T cells. J Immunol. 2009;182:92–101. doi: 10.4049/jimmunol.182.1.92. [DOI] [PubMed] [Google Scholar]
- 25.van Stijn A, Rowshani AT, Yong SL, Baas F, Roosnek E, ten Berge IJ, van Lier RA. Human cytomegalovirus infection induces a rapid and sustained change in the expression of NK cell receptors on CD8+ T cells. J Immunol. 2008;180:4550–4560. doi: 10.4049/jimmunol.180.7.4550. [DOI] [PubMed] [Google Scholar]
- 26.van Bergen J, Koning F. The tortoise and the hare: slowly evolving T-cell responses take hastily evolving KIR. Immunology. 2010;131:301–309. doi: 10.1111/j.1365-2567.2010.03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjorkstrom NK, Beziat V, Cichocki F, Liu LL, Levine J, Larsson S, Koup RA, Anderson SK, Ljunggren HG, Malmberg KJ. CD8 T cells express randomly selected KIRs with distinct specificities compared to NK cells. Blood. 2012;120:3455–65. doi: 10.1182/blood-2012-03-416867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhrberg M, Valiante NM, Young NT, Lanier LL, Phillips JH, Parham P. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166:3923–3932. doi: 10.4049/jimmunol.166.6.3923. [DOI] [PubMed] [Google Scholar]
- 29.Vely F, Peyrat M, Couedel C, Morcet J, Halary F, Davodeau F, Romagne F, Scotet E, Saulquin X, Houssaint E, Schleinitz N, Moretta A, Vivier E, Bonneville M. Regulation of inhibitory and activating killer-cell Ig-like receptor expression occurs in T cells after termination of TCR rearrangements. J Immunol. 2001;166:2487–2494. doi: 10.4049/jimmunol.166.4.2487. [DOI] [PubMed] [Google Scholar]
- 30.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 31.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. The Journal of experimental medicine. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 33.Presnell SR, Zhang L, Ramilo CA, Chan HW, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. International immunology. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 34.Trompeter HI, Gomez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, Uhrberg M. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. 2005;174:4135–4143. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Vallejo AN, Jiang Y, Weyand CM, Goronzy JJ. Distinct transcriptional control mechanisms of killer immunoglobulin-like receptors in natural killer (NK) and in T cells. The Journal of biological chemistry. 2005;280:24277–24285. doi: 10.1074/jbc.M500727200. [DOI] [PubMed] [Google Scholar]
- 36.Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood. 2009;114:3422–3430. doi: 10.1182/blood-2009-01-200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Andrea A, Chang C, Phillips JH, Lanier LL. Regulation of T cell lymphokine production by killer cell inhibitory receptor recognition of self HLA class I alleles. The Journal of experimental medicine. 1996;184:789–794. doi: 10.1084/jem.184.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerra N, Michel F, Gati A, Gaudin C, Mishal Z, Escudier B, Acuto O, Chouaib S, Caignard A. Engagement of the inhibitory receptor CD158a interrupts TCR signaling, preventing dynamic membrane reorganization in CTL/tumor cell interaction. Blood. 2002;100:2874–2881. doi: 10.1182/blood-2002-02-0643. [DOI] [PubMed] [Google Scholar]
- 39.De Maria A, Ferraris A, Guastella M, Pilia S, Cantoni C, Polero L, Mingari MC, Bassetti D, Fauci AS, Moretta L. Expression of HLA class I-specific inhibitory natural killer cell receptors in HIV-specific cytolytic T lymphocytes: impairment of specific cytolytic functions. Proc Natl Acad Sci U S A. 1997;94:10285–10288. doi: 10.1073/pnas.94.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandelboim O, Davis DM, Reyburn HT, Vales-Gomez M, Sheu EG, Pazmany L, Strominger JL. Enhancement of class II-restricted T cell responses by costimulatory NK receptors for class I MHC proteins. Science. 1996;274:2097–2100. doi: 10.1126/science.274.5295.2097. [DOI] [PubMed] [Google Scholar]
- 41.Mandelboim O, Kent S, Davis DM, Wilson SB, Okazaki T, Jackson R, Hafler D, Strominger JL. Natural killer activating receptors trigger interferon gamma secretion from T cells and natural killer cells. Proc Natl Acad Sci U S A. 1998;95:3798–3803. doi: 10.1073/pnas.95.7.3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder MR, Nakajima T, Leibson PJ, Weyand CM, Goronzy JJ. Stimulatory killer Ig-like receptors modulate T cell activation through DAP12-dependent and DAP12-independent mechanisms. J Immunol. 2004;173:3725–3731. doi: 10.4049/jimmunol.173.6.3725. [DOI] [PubMed] [Google Scholar]
- 43.Ortaldo JR, Winkler-Pickett RT, Yagita H, Young HA. Comparative studies of CD3- and CD3+ CD56+ cells: examination of morphology, functions, T cell receptor rearrangement, and pore-forming protein expression. Cellular immunology. 1991;136:486–495. doi: 10.1016/0008-8749(91)90369-m. [DOI] [PubMed] [Google Scholar]
- 44.Lemster BH, Michel JJ, Montag DT, Paat JJ, Studenski SA, Newman AB, Vallejo AN. Induction of CD56 and TCR-independent activation of T cells with aging. J Immunol. 2008;180:1979–1990. doi: 10.4049/jimmunol.180.3.1979. [DOI] [PubMed] [Google Scholar]
- 45.Pittet MJ, Speiser DE, Valmori D, Cerottini JC, Romero P. Cutting edge: cytolytic effector function in human circulating CD8+ T cells closely correlates with CD56 surface expression. J Immunol. 2000;164:1148–1152. doi: 10.4049/jimmunol.164.3.1148. [DOI] [PubMed] [Google Scholar]
- 46.Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B, Kronenberg M, Seeger RC. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J Immunol. 2001;167:3114–3122. doi: 10.4049/jimmunol.167.6.3114. [DOI] [PubMed] [Google Scholar]
- 47.Leung W, Campana D, Yang J, Pei D, Coustan-Smith E, Gan K, Rubnitz JE, Sandlund JT, Ribeiro RC, Srinivasan A, Hartford C, Triplett BM, Dallas M, Pillai A, Handgretinger R, Laver JH, Pui CH. High success rate of hematopoietic cell transplantation regardless of donor source in children with very high-risk leukemia. Blood. 2011;118:223–230. doi: 10.1182/blood-2011-01-333070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung W, Neale G, Behm F, Iyengar R, Finkelstein D, Kastan MB, Pui CH. Deficient innate immunity, thymopoiesis, and gene expression response to radiation in survivors of childhood acute lymphoblastic leukemia. Cancer Epidemiol. 2010;34:303–308. doi: 10.1016/j.canep.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 51.Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, Romeo E, Cognet C, Martinetti M, Maccario R, Mingari MC, Vivier E, Moretta L, Locatelli F, Moretta A. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009;113:3119–3129. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 52.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ, Spits H. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 53.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira LM. Gammadelta T Cells: Innately Adaptive Immune Cells? International reviews of immunology. 2013 doi: 10.3109/08830185.2013.783831. [DOI] [PubMed] [Google Scholar]
- 55.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 58.Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. Type 17 T helper cells-origins, features and possible roles in rheumatic disease. Nat Rev Rheumatol. 2009;5:325–331. doi: 10.1038/nrrheum.2009.80. [DOI] [PubMed] [Google Scholar]
- 59.Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, Eberl G, Leite-de-Moraes MC. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mingari MC, Ponte M, Cantoni C, Vitale C, Schiavetti F, Bertone S, Bellomo R, Cappai AT, Biassoni R. HLA-class I-specific inhibitory receptors in human cytolytic T lymphocytes: molecular characterization, distribution in lymphoid tissues and co-expression by individual T cells. International immunology. 1997;9:485–491. doi: 10.1093/intimm/9.4.485. [DOI] [PubMed] [Google Scholar]
- 61.Hermann E, Berthe A, Truyens C, Alonso-Vega C, Parrado R, Torrico F, Carlier Y, Braud VM. Killer cell immunoglobulin-like receptor expression induction on neonatal CD8(+) T cells in vitro and following congenital infection with Trypanosoma cruzi. Immunology. 2010;129:418–426. doi: 10.1111/j.1365-2567.2009.03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pawelec G. Accumulations of KIR+ T cells in the elderly. Blood. 2009;114:3360–3361. doi: 10.1182/blood-2009-08-235259. [DOI] [PubMed] [Google Scholar]
- 63.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Verges S, Lanier LL, Weisdorf D, Miller JS. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012;119:2665–2674. doi: 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 66.Petitdemange C, Becquart P, Wauquier N, Beziat V, Debre P, Leroy EM, Vieillard V. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS pathogens. 2011;7:e1002268. doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, Theodorou I, Martinot M, Debre P, Bjorkstrom NK, Malmberg KJ, Marcellin P, Vieillard V. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. European journal of immunology. 2012;42:447–457. doi: 10.1002/eji.201141826. [DOI] [PubMed] [Google Scholar]
- 68.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leung W, Iyengar R, Triplett B, Turner V, Behm FG, Holladay MS, Houston J, Handgretinger R. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- 70.Henel G, Singh K, Cui D, Pryshchep S, Lee WW, Weyand CM, Goronzy JJ. Uncoupling of T-cell effector functions by inhibitory killer immunoglobulin-like receptors. Blood. 2006;107:4449–4457. doi: 10.1182/blood-2005-06-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cornelissen F, Aparicio Domingo P, Reijmers RM, Cupedo T. Activation and effector functions of human RORC+ innate lymphoid cells. Current opinion in immunology. 2011;23:361–367. doi: 10.1016/j.coi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Newcomb DC, Boswell MG, Huckabee MM, Goleniewska K, Dulek DE, Reiss S, Lukacs NW, Kolls JK, Peebles RS., Jr IL-13 regulates Th17 secretion of IL-17A in an IL-10-dependent manner. J Immunol. 2012;188:1027–1035. doi: 10.4049/jimmunol.1102216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newcomb DC, Zhou W, Moore ML, Goleniewska K, Hershey GK, Kolls JK, Peebles RS., Jr A functional IL-13 receptor is expressed on polarized murine CD4+ Th17 cells and IL-13 signaling attenuates Th17 cytokine production. J Immunol. 2009;182:5317–5321. doi: 10.4049/jimmunol.0803868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu S, Qian Y. IL-17/IL-17 receptor system in autoimmune disease: mechanisms and therapeutic potential. Clinical science. 2012;122:487–511. doi: 10.1042/CS20110496. [DOI] [PubMed] [Google Scholar]
- 75.Serody JS, Hill GR. The IL-17 differentiation pathway and its role in transplant outcome. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2012;18:S56–61. doi: 10.1016/j.bbmt.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.