Abstract
Nearly two decades since the first retrieval of Neanderthal DNA, recent advances in next-generation sequencing technologies have allowed the generation of high-coverage genomes from two archaic hominins, a Neanderthal and a Denisovan, as well as a complete mitochondrial genome from remains which probably represent early members of the Neanderthal lineage. This genomic information, coupled with diversity exome data from several Neanderthal specimens is shedding new light on evolutionary processes such as the genetic basis of Neanderthal and modern human-specific adaptations—including morphological and behavioural traits—as well as the extent and nature of the admixture events between them. An emerging picture is that Neanderthals had a long-term small population size, lived in small and isolated groups and probably practised inbreeding at times. Deleterious genetic effects associated with these demographic factors could have played a role in their extinction. The analysis of DNA from further remains making use of new large-scale hybridization-capture-based methods as well as of new approaches to discriminate contaminant DNA sequences will provide genetic information in spatial and temporal scales that could help clarify the Neanderthal's—and our very own—evolutionary history.
Keywords: Neanderthal, demography, evolutionary history, extinction
1. Introduction
The best way to understand our evolutionary history as modern humans is comparing our own genome with those of our closest relatives. The genetic bases of the traits that we do not share with them are going to be those that define our singularity as a species. Until recently, we only had the chimpanzees for such comparisons; however, our lineage and that leading to them probably separated more than 6 million years ago and thus, they constitute a very distant relative. Let us take language for instance, our unique ability to communicate abstract ideas that is often inferred to set us apart from the rest of the natural world. Chimpanzees do not speak, not only because they have a different brain and a different genetic make-up, but also because they do not have the vocal tract that enables us to produce the sounds we use for it. Therefore, it is quite clear that, for understanding adaptive processes that probably took place not at the origin of the hominin lineage, but millions of years afterwards, the chimpanzee represents a rather poor reference.
Depending on which adaptive processes are addressed, an obvious source of comparison would be to obtain genetic data from fossils that represent remains of our hominin relatives. Given that Neanderthals are our closest and best-known relatives, in addition to their prevalence up to the late Pleistocene (giving more chances for DNA preservation), this makes them ideal candidates to identify those traits that might have originated within our own evolutionary lineage.
2. Neanderthal mitochondrial DNA sequences
The first Neanderthal sequence was obtained in 1997 by a team led by Svante Pääbo. They were able to recover the mitochondrial DNA (mtDNA) hypervariable region 1 by polymerase chain reaction (PCR) from the Neanderthal holotype specimen from Feldhofer cave in Germany. By comparing it against a panel of worldwide present-day human mtDNA sequences, the data indicated that Neanderthals were a sister group to anatomically modern humans, providing no evidence of interbreeding between Neanderthals and modern humans, at least to a level sufficient to result in Neanderthal mtDNA introgression into the modern human mtDNA gene pool [1].
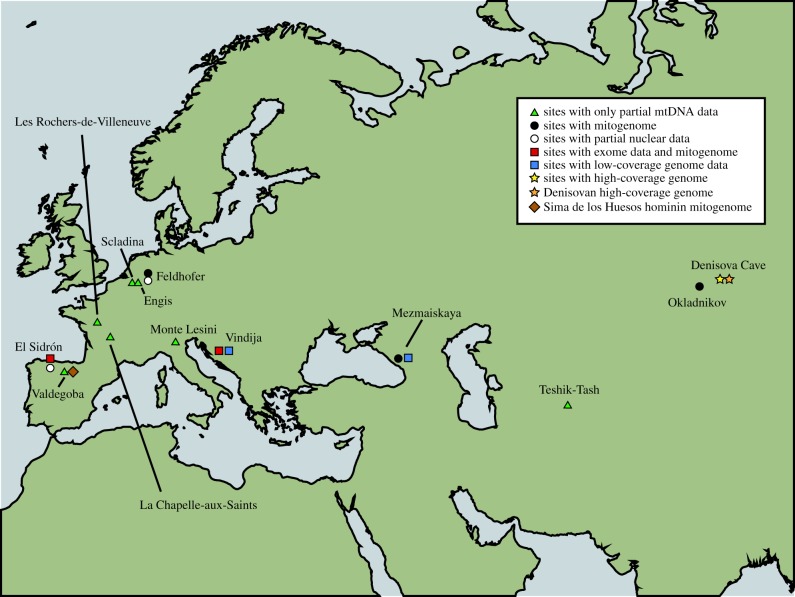
During the 15 years following this publication, other Neanderthal sequences from different sites such as Mezmaiskaya (Russia) and Vindija (Croatia) in 2000, Engis (Belgium), La Chapelle-aux-Saints (France), Les Rochers-de-Villeneuve (France) in 2004, El Sidrón (Spain) in 2005, 2006 and 2011, Monti Lessini (Italy) and Scladina (Belgium) in 2006, Teshik-Tash (Uzbekistan) and Okladnivok (Russia) in 2007 and Valdegoba (Spain) in 2012 were successfully amplified with the same technical approach [2–12] (figure 1). A common observation of all these studies was that Neanderthal mtDNA sequences were similar to each other—suggesting a general low diversity—and different to any reported modern human mtDNA. Furthermore, some studies began analysing a possible phylogeographic structure; the basal sequences in the phylogenetic trees were from the easternmost Neanderthals (located in Central Asia) or from the oldest ones (Valdegoba and Scladina) [12]. This seems to support an east–west genetic cline and also the existence of temporal bottlenecks that shaped the mtDNA diversity. Recent western European Neanderthals (roughly less than 50 000 years) constitute a tightly defined group with low mitochondrial genetic variation in comparison with both eastern and older (more than 50 000 years) European Neanderthals. Eastern and western Neanderthals seem to have diverged approximately 55 000–70 000 years ago followed by an extinction of western Neanderthals throughout most of their range and a subsequent recolonization of the region [12].
Figure 1.
Geographical map showing Neanderthal and Denisovan sites with different types of genetic data (partial mitochondrial, complete mitogenomes, exomes, partial nuclear data or complete genomes) retrieved.
However, to explore these migration patterns across time and space, we need to have a basic understanding of the Neanderthals' demography. Fortunately, there is a Neanderthal site that can provide such information because it may represent a family group. The Spanish site of El Sidrón is thought to be a synchronic accumulation of at least 12 Neanderthals including three female and three male adults, three adolescents, two juveniles and one infant. Complete and partial mtDNA sequences from all the available individuals suggest that Neanderthals there formed small kinship-structured bands that practised patrilocal mating behaviour and had relatively long inter-birth intervals (ca 3 years) when compared with modern human populations. In addition to providing intriguing anthropological insights into a Neanderthal social group—similar features have also been described in modern hunter–gatherers—such information may help in choosing demographic parameters when generating models of Neanderthal population dynamics [1].
3. Mitochondrial genomes and the advent of the new sequencing technologies
With the introduction of next-generation sequencing (NGS) technologies to the field of ancient DNA (aDNA), it was possible for the first time to retrieve complete mitochondrial genomes, first by shotgun sequencing of a sample from Vindija cave [13] and later with targeted hybridization-capture enrichment methods [14]. The whole mtDNA genome allowed a more precise estimate of the divergence time between modern human and Neanderthal mtDNA lineages, which was reported to be 660 000 years considering all sites of the mtDNA [13] or close to 400 000 years considering only third codon sites of the mtDNA [15]. Another striking observation was that the ratio of non-synonymous to synonymous evolutionary rates was significantly higher on the Neanderthal lineage, a result that would fit with Neanderthals having a smaller effective population size, and thus evolving under lower selective constraints than modern humans [13]. By 2009, the analysis of six complete Neanderthal mtDNA genomes indicated that the variation among Neanderthals was approximately one-third of that estimated for present-day humans worldwide, suggesting a female effective population size of less than 3500 individuals [14]. This finding was surprising given that the Neanderthal sequences stem from several distinct time points spanning thousands of years across a wide geographical range, and thus it appears to be a conservative estimate with respect to sampling at a contemporaneous time period. The most recent common ancestor (MRCA) of the Neanderthal samples analysed was estimated to have lived approximately 110 000 years ago, which is much less than the age estimated for modern human mtDNAs [3].
Furthermore, these new sequencing technologies allowed precise estimates of modern human contamination in the high-coverage mtDNA genomes obtained, but also the description of misincorporation patterns related to cytosine deaminations at the edge of the sequencing reads that is characteristic of aDNA sequences, and increases with time [16,17]. In subsequent studies, these patterns allowed the identification of authentic Neanderthal sequences and opened up the possibility of analysing Neanderthal samples that were previously discarded for genetic studies due to their high level of present-day human contamination [18].
4. The first nuclear DNA sequences
As Neanderthal mitochondrial diversity was being studied, attention also turned to nuclear loci. Although challenging, given the lower proportion of nuclear DNA compared with mtDNA, researchers were thrilled by the idea as it unlocked the possibility of assessing whether emblematic functional and phenotypic modern human traits were shared by Neanderthals.
Between 2007 and 2009, by amplifying small nuclear regions encompassing functional variants, researchers found that some Neanderthals were probably red-haired and pale skinned [19], they had bitter taste perception ability [20] and presented the ABO blood type O [21]. In addition, having the same functional variants as modern humans in the FOXP2—a gene that when mutated generates a speech and language impediment—suggested that Neanderthals might have been able to communicate with similar language capabilities to ours, or at least they had the genetic basis to do so [22]. Nonetheless, recent studies found differences between most modern humans and Neanderthals in a regulatory element near the FOXP2 gene that could have functional implications [23].
While recovering short pieces of nuclear DNA became possible in well-preserved and uncontaminated specimens, the sequencing of a whole Neanderthal genome remained a difficult challenge, owing to the low amount of nuclear DNA sequences relative to environmental sequences, and the limitations of the available technology. Two pioneer studies managed to recover 65 kb of nuclear DNA and 1 Mb of sequence of Neanderthal nuclear DNA by cloning and sequencing short fragments of DNA [24] or by metagenomic sequencing [25], respectively. They estimated coalescence times between modern humans and Neanderthals to be roughly between 700 000 and 500 000 years ago. However, it was subsequently demonstrated that a significant fraction of the data generated by the second study derived from modern human contaminant DNA [26]. As a result of this early pitfall, more stringent measures were taken while constructing the sequencing libraries, eliminating potential environmental and modern human contamination [27,28].
5. The Neanderthal and Denisovan draft genomes
The year 2010 saw not only the publication of the long-expected Neanderthal draft genome [28] but also that of a previously unknown hominin, called Denisovan, named after the cave in the Altai Mountains where the remains were discovered [29]. Currently only two teeth and a finger bone (the latter with extraordinary levels of DNA preservation, approx. 70% of endogenous DNA) have been attributed to the Denisovans. Both nuclear and mtDNA extracted from these remains suggest that Denisovans were as genetically diverse as two present-day humans from different continents and more diverse than Neanderthals from throughout their range, suggesting that their effective population size was relatively large [30] (see also a later discussion in [31]). By employing a user-defined hybridization-capture method, a high-coverage mtDNA genome from the Denisovan finger bone was retrieved [32], and it was estimated that it diverged from the common ancestor of modern humans and Neanderthals around 1 million years ago [33]. Moreover, as both nuclear archaic genomes were sequenced, clearer phylogenetic relationships were established for the first time. The MRCA of modern humans, Neanderthals and Denisovans was found to have lived at least 800 000 years ago, whereas the Denisovan and Neanderthal genomes were more closely related to each other—as sister species—and their divergence time was around 600 000 years ago.
In addition to the general hominin phylogeny, the analysis of five present-day humans from different continental areas suggested that non-Africans shared 1–4% more derived alleles with Neanderthals than with sub-Saharan Africans [28], whereas present-day Melanesians also seemed to share 4–6% of their DNA with the Denisovan individual. The Neanderthal signal was later also observed in African populations, which is likely the result of back-to-Africa migrations [34–36]. These results were interpreted as evidence of Neanderthals interbreeding with the ancestors of all non-Africans and subsequently a Denisovan-like population mainly with the ancestors of South East Asians [37]; however, marginal Denisovan admixture has also been reported in continental Asian populations [31,38], further entangling this later admixture scenario. This notwithstanding, the proportions of admixture are probably overestimates if some degree of structure was present among ancient humans in Africa, as already pointed out in [28,39–41]. If this were the case, incomplete lineage sorting and not introgression could explain some genetic similarities between modern non-African humans and Neanderthals, although certainly not all of them.
6. High-coverage genomes
A major technical breakthrough in 2012 involved a novel library preparation method that exploited single-stranded DNA and greatly increased the yield of sequencing from ancient samples. Briefly, instead of building the libraries exclusively from double-stranded DNA—where only sequences without ‘nicks' or single-strand breaks can be incorporated into NGS libraries—the new method first denatures DNA fragments and incorporates the single strands of DNA into NGS libraries, allowing for the recovery of significantly more DNA molecules than hitherto possible. By applying this new method, a 30X coverage genome from the same Denisovan sample [42] and a 54X coverage genome from a female Neanderthal toe bone [31] also from Denisova Cave—known as the Altai Neanderthal—were generated.
Having high-quality genome data not only offers refined insights into Neanderthal relatedness to modern humans, but also allows us to start addressing questions concerning their diversity and demographic history, something that could not be done with low coverage data. For instance, under a no gene flow scenario, the date of the split of the archaic and modern human populations, which by necessity is more recent than sequence divergence, can be estimated. Recently, mutation rates have also been a subject of debate [43]. Based on a mutation rate of 1.03 × 10−9 derived from the fossil record (which is essentially two times faster than the genealogical one), the population split between Denisovans, Neanderthals and modern humans probably occured between 383 000 and 257 000 years ago, whereas the populations that evolved into Neanderthals and Denisovans separated roughly 236 000–190 000 years ago [31].
A more precise idea of how and when the admixture with archaic humans occured is also beginning to emerge. By coupling high-coverage archaic and present-day human genomes, the amount of DNA introgressed from Neanderthals into non-sub-Saharan Africans has been refined to a range of 1.5–2.1% of Neanderthal ancestry in present-day populations [44]. It has also been observed that Neanderthal-derived DNA in all non-Africans is more closely related to a low coverage genome from the Mezmaiskaya skeleton in the Caucasus than to the Altai or to the Vindija genome [31]. The linkage disequilibrium pattern of haplotypes of suspected Neanderthal origin suggests a date of admixture between 37 000 and 82 000 years ago [45]. Altogether, these observations seemed to indicate that a currently unsampled Middle Palaeolithic Neanderthal population living in the Levant and/or western Asia encountered modern humans as they migrated out of Africa, subsequently spreading the signature of introgression as they populated the rest of the world.
Furthermore, it has recently been shown that East Asians and native Americans may have between 1.7 and 2% more Neanderthal admixture than other non-African populations, which suggests that a second introgression event took place after European and Asians populations diverged [42,46]. This latter finding was unexpected given the archaeological evidence of a long-term occupation of Neanderthals in Europe and a possible late overlap with early modern human migrations into Europe. Moreover, Late Palaeolithic and Mesolithic modern human genomes have so far failed to demonstrate a closer relatedness to Neanderthals [47,48]. High-coverage genomes of Late Pleistocene Europeans—and also from other populations—will be needed to estimate accurately if other admixture events could have occured with Neanderthals or Denisovans. Interestingly, some lines of evidence suggest that interbreeding may have been limited by genetic incompatibilities (below) and thus a short-lived increase in Neanderthal admixture would only be observed close to the interbreeding events(s) [44].
In addition to determining the phylogenetic relationships among hominins, a potentially interesting application of the high-coverage genomes is to investigate in detail the introgressed regions and see whether they harbour genetic variants that could be beneficial to modern humans. Several recent publications suggest that some archaic variants could have been advantageous or at least functionally relevant after being introgressed into modern humans [49–53]. For instance, Neanderthal haplotypes in European and East Asians are enriched for genes harbouring keratin filaments—a protein expressed in skin, hair and nails—suggesting that skin or hair adaptation to non-African environments was enhanced after the admixture event [53]. Inversely, there seem to be large ‘deserts' of Neanderthal ancestry, which implies that selection may have acted to remove genetic material derived from Neanderthals [44,54]. Furthermore, genes that are more highly expressed in testes than in any other tissue are especially reduced in Neanderthal ancestry, and there is an approximately fivefold reduction of Neanderthal ancestry on the X chromosome [44]; these observations can be interpreted as selection eliminating Neandertal-derived genes that may have reduced male fertility. Furthermore, the known differences in effective population size between East Asians and Europeans could have resulted in less efficient selection to remove Neanderthal-derived deleterious alleles and thus be the cause for the excess of Neanderthal signal observed in East Asians populations [44], although others suggested it was more probably attributable to further interbreeding in the East [54], as suggested earlier.
7. Neanderthal genomic diversity and demographic trends
The opportunity to analyse large genomic regions from different Neanderthal specimens opens the possibility of studying diversity patterns that could be related to specific demographic and evolutionary processes, and that can also shed light on their extinction process.
The recent advent of the high-coverage exomes of two Neanderthals, one from Vindija 33.15 (40X) in Croatia and the other from El Sidrón SD1253 in Spain (12X) [55] (figure 1), has allowed a start in addressing those subjects. Together with the exome regions of the Altai and the Denisovan genomes, the Neandertal exomes have been compared with the same regions from three modern individuals from Africa, Europe and Asia/Pacific. Interestingly, it was found that the average heterozygosity—the number of nucleotide differences within an individual per thousand base pairs—among the three Neanderthals was 0.128, which is approximately a third of what is seen in present-day humans. The three Neanderthals have longer runs of homozygosity than modern humans. The Altai individual has been reported to have an inbreeding coefficient of one-eighth—indicating that the parents were as closely related as half-siblings. Additionally, possible weaker consanguinity signals are also present in the Vindija and El Sidrón material. Additional samples would be of paramount importance to see whether the homozygosity tracks increase in length over time, and whether this correlates with the extinction process. Considering the two individuals securely dated (approx. 44 000 years ago for Vindija 33.15 and approx. 49 000 years ago for El Sidrón 1253), the homozygosity tracks longer than 200 kb almost double in about 5000 years [55]. In addition, the genetic differentiation among individuals is larger among Neanderthals than among present-day humans. This suggests that Neanderthals lived in small and relatively isolated populations, which probably caused them to become more differentiated from each other when compared with modern humans.
Furthermore, inferences from the high-coverage Neanderthal and Denisovan genomes [31] suggest that some time after 0.5–1.0 million years ago their ancestral populations decreased in size for hundreds of thousands of years. A low population size over a long time would reduce the efficacy of purifying selection and contribute to a larger fraction of likely deleterious alleles, particularly at low frequency. In accordance with what would be expected of a long-term low population size, the Neanderthal exomes show that the proportion of all derived SNPs that are inferred to change amino acids and to be deleterious—assessed from alleles expected to affect the protein function or that occured in conserved positions—is larger than in modern human populations. Among derived amino acid-changing alleles likely to be at low frequency in Neanderthals, not only a higher proportion is inferred to alter protein function, but also they seem to be the functional variants with the most deleterious consequences when compared with SNPs at lower frequency in the modern human populations. However, it is interesting to note that these results seem not to affect the deleterious load per individual, since the number of genes associated with non-dominant traits with heterozygous- or homozygous-derived alleles inferred to be deleterious, is not different between Neanderthal and present-day individuals [56]. Therefore, susceptibility of Neanderthals to any specific genetic disorder cannot be inferred from these data [55].
8. Modern human- and Neanderthal-specific traits
The high-coverage Neanderthal and Denisova genomes now provide a sound basis to identify genomic changes specific to modern humans and, with that, a list of substitutions accountable for ‘what makes us modern humans' has emerged [30,31,38].
Moreover, the exomes of the three Neanderthals and the Denisovan individual allow us, for the first time, to identify derived amino acid changes shared by three Neanderthals as well as the Denisovan individual that are not seen, or only occur at a very low frequency, in present-day humans. Such changes are of interest since they may underlie phenotypes specific to the archaic populations. By calculating the fraction of all amino acid changes specific to either the archaic or modern human lineages for each phenotype category of genes in the Human Phenotype Ontology database, an estimation of the enrichment of amino acid changes in phenotypes in each archaic lineage has been obtained [55]. The authors find that genes involved in skeletal morphology may have changed more on the Neanderthal and Denisova lineages than on the preceding lineage from the common ancestor shared with chimpanzees. These genetic changes could underlie some skeletal Neandertal traits such as a reduced lordosis—the curvature of the lumbar and cervical spine; unfortunately, the fact that there is so far little morphological evidence from Denisovans hinders corroborating further associations between genetic changes and morphological traits in the lineage specific to archaic humans. In the modern human lineage, there is an overrepresentation of some behavioural genes; intriguingly, some of these genes have been related to traits such as ‘hyperactivity’ or ‘aggressive behaviour’ [55].
Thus, most of our understanding of the biology of ancient humans will no longer be limited by the inaccessibility of the data but by our functional interpretation of modern human genomes [57]. Moreover, regulatory changes have also been shown to be of importance in recent human evolution [58], and thus not only coding variants should be taken into account when reconstructing the biology of archaic humans from genetic data.
Nevertheless, functional studies will be essential to better understand the function and importance not only of genetic variants already discovered and specific to the modern human lineage, but also the Neanderthal-lineage-specific changes.
A recent study has decoded the ancient methylation patterns from NGS data to infer the gene expression of a Palaeo-Eskimo individual approximately 4000 years old [59]. Moreover, further work [60] suggests that even though archaic and modern humans share more than 99% of their genetic sequence, there seem to be methylation differences between these hominin groups that are twice as likely to occur in genes implicated in disease, especially brain disease-associated regions, than in genes that are not associated with illness. Methylation differences are also found in HOXD, a gene cluster that regulates limb development, suggesting that some of these epigenetic patterns may explain why, for example, Neanderthals had short distal limb segments in comparison with many modern humans. However, in order to assess what the observed epigenetic differences mean in terms of biology, further functional experiments are necessary. Nonetheless, both of these publications suggest that it will be possible to track epigenomic information through time, and thus they have set up the foundations for yet another new discipline: palaeoepigenetics [59,60].
9. Super-archaic DNA
The mtDNA genome of a ca 400 000-year-old hominin from the Sima de los Huesos in Atapuerca (Spain; figure 1) has been sequenced recently [61]. Interestingly, the skeletal remains had previously been classified as H. heidelbergensis and dated to approximately 600 000 years ago, but both the classification and the date were the subject of dispute [62], and given that the remains exhibit a number of derived Neanderthal traits they have been postulated as the ancestors of Neanderthals. A recent analysis of 27 individuals from this palaeontological site (now dated to ca 430 000 years ago) shows that these ‘Sima de los Huesos' hominins present many Neanderthal-derived traits in their face and teeth, whereas the braincase still retained ‘primitive’ conditions [63]; it seems that late Neanderthal braincase shapes are not found in Europe before approximately 200 000 years ago. Thus, these data suggest that Neanderthal features did not evolve as a block but rather they were fixed at different rates and paces in different parts of the anatomy. Moreover, and further complicating the scenario, the only Sima mtDNA sequence obtained so far seems to be phylogenetically most similar to that of Denisovans [61], found thousands of miles away, and much younger in age. Although nuclear genome sequences of these specimens would be needed to ascertain their precise relationship to archaic and modern humans, this study provides evidence that aDNA techniques have become sensitive enough to recover and analyse DNA from Middle Pleistocene hominin remains, even from non-permafrost environments.
Furthermore, although morphological evidence suggests that Neanderthal features were already present in European fossils over 400 000 years ago, and that by 130 000 years ago their characteristic suite of traits was fully established [64], no genetic information has yet been recovered from samples older than 100 000 years. It seems obvious that many relevant evolutionary processes took place between these two dates, perhaps related to dramatic climatic events and triggered by the action of genetic drift [64]. Moreover, it is not clear yet whether Neanderthals from other geographical areas or time periods are genetically similar to the ones that have already been analysed. While there are clearly differences between early and late members of the Neanderthal lineage, opinions vary over the unity of European and Asian varieties of this hominin group [64,65]. It will be interesting to address how Neanderthals from different time points related to each other and to what extent climatic conditions or other factors contributed to shape their genetic diversity through adaptation and also demographic reductions and expansions.
Furthermore, having Neanderthal serial time data will enable us to move from a primarily descriptive basis of their demographic history and population dynamics to estimate genetic parameters, for instance, their mutation rate, precise temporal population sizes or local diversity patterns.
10. Extinction process
Almost 20 years of Neanderthal palaeogenetics and palaeogenomics have shown us that Neanderthals, an extinct human population, shared a common evolutionary history with modern humans until approximately 0.5 million years ago. Recent studies suggest that after their lineages separated, their demographic trajectories differed: while the Neanderthal population decreased in size for hundreds of thousands of years (as did that of Denisovans), the ancestors of present-day humans stabilized or increased in number [31]. This observation makes sense in the light of what we know about their genetic diversity, which was no more than a third of what has been estimated for modern humans worldwide. In addition, their coding gene patterns show evidence of reduced efficiency of purifying selection and a larger fraction of probably deleterious alleles, particularly at low frequency. Although we are beginning to grasp general patterns of Neanderthal genetic diversity, we cannot completely understand the consequences of their particular demography and population dynamics unless more specimens contemporaneous with each other are analysed. These data will be paramount in helping us understand to what extent Neanderthals were affected by their small population size, relative isolation and inbreeding practices. For example, the new data might allow us to observe whether they displayed a significant accumulation of variants associated with recessive disorders in comparison with modern humans. While this is just a hypothesis, it could be that an accumulation of genetic deleterious effects associated with decreased effective population size, exacerbated by inbreeding practices in the last Neanderthals, may have contributed to their final demise.
11. Future developments
To address some of the previously mentioned unsolved questions about the Neanderthals' evolutionary history, extensive sampling of new fossils will be needed, and even though ongoing archaeological excavations will hopefully continue to produce material for aDNA studies, it is clear that a number of Neanderthal samples of interest may be stored within museums under less than ideal conditions, or may not have been excavated and handled with enough care to prevent contamination [66] (figure 2). Two main caveats arise from this: many specimens will probably have low endogenous DNA contents, and might have been contaminated significantly with modern human DNA.
Figure 2.

An anti-contamination protocol developed at El Sidrón Neanderthal site in Spain to properly handle ancient samples for DNA analysis. The specimens are excavated with sterile laboratory gear and immediately frozen.
As samples from older periods are screened in the search for precious genetic material, even sequencing a mitochondrial genome may require significant amounts of bone tissue [61], which may conflict with conservation purposes. Furthermore, target capture techniques have proved to be most efficient in accessing samples with low endogenous DNA; however, only certain genomic regions (e.g. mtDNA or exomes) have been retrieved with high-coverage using this approach [32,55,67]. Recently, a whole genome capture method that uses home-made biotinylated RNA probes as bait (which significantly reduces the cost of probe design) has been developed [68]. While this approach sounds attractive, it seems to introduce a bias against shorter DNA molecules, which is something that will have to be addressed before it can be fruitfully applied to samples of very degraded (and therefore short) Neanderthal DNA [69].
Moreover, regardless of whether samples have been recently excavated or handled without proper anti-contamination measures, as older specimens or samples stemming from a large range of latitudes and site-specific conditions are analysed, a significant proportion of present-day human contamination can be expected. At present, contamination is efficiently estimated, but only two in silico approaches have been developed to putatively separate endogenous from contaminant material [18,70]. However, neither of them precludes the sequencing of contaminant material, which might not be suitable if a high number of poorly handled and preserved samples have to be screened.
Even though very well-preserved samples have been found, it is unlikely that we will discover very ancient samples with an elevated content of endogenous DNA. Therefore, new methodological approaches for enriching the amount of endogenous material, by retaining only informative damaged molecules, will need to be developed to make large screenings economically feasible. Nonetheless, aDNA studies will still be limited by the amount of endogenous DNA present in the sample. Until new methodological approaches are available, target capture and even shotgun sequencing will no doubt continue to be used, depending on the nature of the samples and the scientific questions being addressed. However, it remains to be seen whether single molecule sequencing technologies can, efficiently and without error, transform the field of aDNA and hominin palaeogenomics.
Given that most historic Neanderthal samples are of great value to understand key aspects of their population dynamics and biology across time, new experimental and computational methods will be crucial to access the endogenous DNA required to fully explain Neanderthal and our own evolutionary histories.
Acknowledgements
We are grateful to Pierre Luisi, Hannes Schroeder and Maria Ávila Arcos for helpful comments on the manuscript and Iñigo Olalde for technical support to produce the map figure.
Funding statement
The authors are supported by the FEDER and Spanish Government Grant BFU2012–34157.
References
- 1.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pa S. 1997. Neandertal DNA sequences and the origin of modern humans. 90, 19–30. [DOI] [PubMed] [Google Scholar]
- 2.Ovchinnikov I, Götherström A, Romanova G, Kharitonov V, Lidén K, Goodwin W. 2000. Molecular analysis of Neanderthal DNA from the northern Caucasus. 404, 490–493. [DOI] [PubMed] [Google Scholar]
- 3.Krings M, et al. 2000. A view of Neandertal genetic diversity. Nat. Genet. 26, 144–146. ( 10.1038/79855) [DOI] [PubMed] [Google Scholar]
- 4.Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S. 2004. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2, E57 ( 10.1371/journal.pbio.0020057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordes J, Cochard D, Couchoud I, Dubrasquet D, Trinkaus E, Rochers-de-villeneuve L. 2005. A late Neandertal femur from Les. Proc. Natl Acad. Sci. USA 102, 7085–7090. ( 10.1073/pnas.0502656102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalueza-Fox C, et al. 2005. Neandertal evolutionary genetics: mitochondrial DNA data from the Iberian peninsula. Mol. Biol. Evol. 22, 1077–1081. ( 10.1093/molbev/msi094) [DOI] [PubMed] [Google Scholar]
- 7.Lalueza-Fox C, et al. 2006. Mitochondrial DNA of an Iberian Neandertal suggests a population affinity with other European Neandertals. Curr. Biol. 16, R629–R630. ( 10.1016/j.cub.2006.07.044) [DOI] [PubMed] [Google Scholar]
- 8.Lalueza-Fox C, et al. 2011. Genetic evidence for patrilocal mating behavior among Neandertal groups. Proc. Natl Acad. Sci. USA 108, 250–253. ( 10.1073/pnas.1011553108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caramelli D, et al. 2006. A highly divergent mtDNA sequence in a Neandertal individual from Italy. Curr. Biol. 16, R630–R632. ( 10.1016/j.cub.2006.07.043) [DOI] [PubMed] [Google Scholar]
- 10.Orlando L, Darlu P, Toussaint M, Bonjean D, Otte M, Hänni C. 2005. Revisiting Neandertal diversity with a mtDNA sequence. Curr. Biol. 16, 400–402. ( 10.1016/j.cub.2006.05.019) [DOI] [PubMed] [Google Scholar]
- 11.Krause J, et al. 2007. Neanderthals in central Asia and Siberia. Nature 449, 902–904. ( 10.1038/nature06193) [DOI] [PubMed] [Google Scholar]
- 12.Dalén L, et al. 2012. Partial genetic turnover in Neandertals: continuity in the East and population replacement in the West. Mol. Biol. Evol. 29, 1893–1897. ( 10.1093/molbev/mss074) [DOI] [PubMed] [Google Scholar]
- 13.Green S, et al. 2008. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426. ( 10.1016/j.cell.2008.06.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs AW, et al. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321. ( 10.1126/science.1174462) [DOI] [PubMed] [Google Scholar]
- 15.Endicott P, Ho SYW, Stringer C. 2010. Using genetic evidence to evaluate four palaeoanthropological hypotheses for the timing of Neanderthal and modern human origins. J. Hum. Evol. 59, 87–95. ( 10.1016/j.jhevol.2010.04.005) [DOI] [PubMed] [Google Scholar]
- 16.Briggs AW, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14 616–14 621. ( 10.1073/pnas.0704665104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. 2012. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 7, e34131 ( 10.1371/journal.pone.0034131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, Jakobsson M. 2014. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl Acad. Sci. USA 111, 2229–2234. ( 10.1073/pnas.1318934111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalueza-Fox C, et al. 2007. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science 318, 1453–1455. ( 10.1126/science.1147417) [DOI] [PubMed] [Google Scholar]
- 20.Lalueza-Fox C, Gigli E, de la Rasilla M, Fortea J, Rosas A. 2009. Bitter taste perception in Neanderthals through the analysis of the TAS2R38 gene. Biol. Lett. 5, 809–811. ( 10.1098/rsbl.2009.0532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalueza-Fox C, Gigli E, de la Rasilla M, Fortea J, Rosas A, Bertranpetit J, Krause J. 2008. Genetic characterization of the ABO blood group in Neandertals. BMC Evol. Biol. 8, 342 ( 10.1186/1471-2148-8-342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause J, et al. 2007. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr. Biol. 17, 1908–1912. ( 10.1016/j.cub.2007.10.008) [DOI] [PubMed] [Google Scholar]
- 23.Maricic T, et al. 2013. A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Mol. Biol. Evol. 30, 844–852. ( 10.1093/molbev/mss271) [DOI] [PubMed] [Google Scholar]
- 24.Noonan JP, et al. 2006. Sequencing and analysis of Neanderthal genomic DNA. Science 314, 1113–1118. ( 10.1126/science.1131412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green RE, et al. 2006. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336. ( 10.1038/nature05336) [DOI] [PubMed] [Google Scholar]
- 26.Wall JD, Kim SK. 2007. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 3, 1862–1866. ( 10.1371/journal.pgen.0030175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green RE, Briggs AW, Krause J, Prüfer K, Burbano HA, Siebauer M, Lachmann M, Pääbo S. 2009. The Neandertal genome and ancient DNA authenticity. EMBO J. 28, 2494–2502. ( 10.1038/emboj.2009.222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328, 710–722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. ( 10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pennisi E. 2010. More genomes from Denisova cave show mixing of early human groups. Science 340, 799. [DOI] [PubMed] [Google Scholar]
- 31.Prüfer K, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49. ( 10.1038/nature12886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE 5, e14004 ( 10.1371/journal.pone.0014004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Pääbo S. 2010. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897. ( 10.1038/nature08976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sánchez-Quinto F, Botigué LR, Civit S, Arenas C, Avila-Arcos MC, Bustamante CD, Comas D, Lalueza-Fox C. 2012. North African populations carry the signature of admixture with Neandertals. PLoS ONE 7, e47765 ( 10.1371/journal.pone.0047765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell JK, Patterson N, Loh P-R, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D. 2014. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl Acad. Sci. USA 111, 2632–2637. ( 10.1073/pnas.1313787111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Lachance J, Tishkoff SA, Hey J, Xing J. 2013. Apparent variation in Neanderthal admixture among African populations is consistent with gene flow from non-African populations. Genome Biol. Evol. 5, 2075–2081. ( 10.1093/gbe/evt160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich D, et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89, 516–528. ( 10.1016/j.ajhg.2011.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoglund P, Jakobsson M. 2011. Archaic human ancestry in East Asia. Proc. Natl Acad. Sci. USA 108, 18 301–18 306. ( 10.1073/pnas.1108181108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eriksson A, Manica A. 2012. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc. Natl Acad. Sci. USA 109, 13 956–13 960. ( 10.1073/pnas.1200567109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowery RK, Uribe G, Jimenez EB, Weiss MA, Herrera KJ, Regueiro M, Herrera RJ. 2013. Neanderthal and Denisova genetic affinities with contemporary humans: introgression versus common ancestral polymorphisms. Gene 530, 83–94. ( 10.1016/j.gene.2013.06.005) [DOI] [PubMed] [Google Scholar]
- 41.Eriksson A, Manica A. 2014. The doubly-conditioned frequency spectrum does not distinguish between ancient population structure and hybridisation. Mol. Biol. Evol 31, 1618–1621. ( 10.1093/molbe/msu103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer M, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226. ( 10.1126/science.1224344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scally A, Durbin R. 2012. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 13, 745–753. ( 10.1038/nrg3295) [DOI] [PubMed] [Google Scholar]
- 44.Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357. ( 10.1038/nature12961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. 2012. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 8, e1002947 ( 10.1371/journal.pgen.1002947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wall JD, et al. 2013. Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics 194, 199–209. ( 10.1534/genetics.112.148213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raghavan M, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91. ( 10.1038/nature12736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olalde I, et al. 2014. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228. ( 10.1038/nature12960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abi-Rached L, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334, 89–94. ( 10.1126/science.1209202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendez FL, Watkins JC, Hammer MF. 2012. A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 91, 265–274. ( 10.1016/j.ajhg.2012.06.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendez FL, Watkins JC, Hammer MF. 2013. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol. Biol. Evol. 30, 798–801. ( 10.1093/molbev/mst004) [DOI] [PubMed] [Google Scholar]
- 52.Williams AL, et al. 2014. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506, 97–101. ( 10.1038/nature12828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huerta-Sánchez E, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197. ( 10.1038/nature13408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021. ( 10.1126/science.1245938) [DOI] [PubMed] [Google Scholar]
- 55.Castellano S, et al. 2014. Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl Acad. Sci. USA 111, 6666–6671. ( 10.1073/pnas.1405138111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simons YB, Turchin MC, Pritchard JK, Sella G. 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46, 220–224. ( 10.1038/ng.2896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawks J. 2013. Significance of Neandertal and Denisovan genomes in human evolution. Annu. Rev. Anthropol. 42, 433–449. ( 10.1146/annurev-anthro-092412-155548) [DOI] [Google Scholar]
- 58.Fraser HB. 2013. Gene expression drives local adaptation in humans. Genome Res. 23, 1089–1096. ( 10.1101/gr.152710.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen JS, et al. 2014. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 24, 454–466. ( 10.1101/gr.163592.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gokhman D, Lavi E, Prüfer K, Fraga MF, Riancho JA, Kelso J, Pääbo S, Meshorer E, Carmel L. 2014. Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science 344, 523–527. ( 10.1126/science.1250368) [DOI] [PubMed] [Google Scholar]
- 61.Meyer M, et al. 2014. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406. ( 10.1038/nature12788) [DOI] [PubMed] [Google Scholar]
- 62.Stringer C. 2012. The status of Homo heidelbergensis (Schoetensack 1908). Evol. Anthropol. 21, 101–107. ( 10.1002/evan.21311) [DOI] [PubMed] [Google Scholar]
- 63.Arsuaga JL, et al. 2014. Neandertal roots: cranial and chronological evidence from Sima de los Huesos. Science 344, 1358–1363. ( 10.1126/science.1253958) [DOI] [PubMed] [Google Scholar]
- 64.Weaver TD. 2009. The meaning of Neandertal skeletal morphology. Proc. Natl Acad. Sci. USA 106, 16 028–16 033. ( 10.1073/pnas.0903864106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawks J. 2012. Dynamics of genetic and morphological variability within Neandertals. J. Anthropol. Sci. 90, 81–97. ( 10.4436/jass.90019) [DOI] [PubMed] [Google Scholar]
- 66.Fortea J, de la Rasilla M, García-Tabernero A, Gigli E, Rosas A, Lalueza-Fox C. 2008. Excavation protocol of bone remains for Neandertal DNA analysis in El Sidrón Cave (Asturias, Spain). J. Hum. Evol. 55, 353–357. ( 10.1016/j.jhevol.2008.03.005) [DOI] [PubMed] [Google Scholar]
- 67.Burbano HA, et al. 2010. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725. ( 10.1126/science.1188046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carpenter ML, et al. 2013. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864. ( 10.1016/j.ajhg.2013.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avila-Arcos MS, et al. 2014. Comparative performance of two whole genome capture methodologies on ancient DNA illumina libraries. bioRxiv ( 10.1101/007419) [DOI] [Google Scholar]
- 70.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]