Abstract
Recently, the study of ancient DNA (aDNA) has been greatly enhanced by the development of second-generation DNA sequencing technologies and targeted enrichment strategies. These developments have allowed the recovery of several complete ancient genomes, a result that would have been considered virtually impossible only a decade ago. Prior to these developments, aDNA research was largely focused on the recovery of short DNA sequences and their use in the study of phylogenetic relationships, molecular rates, species identification and population structure. However, it is now possible to sequence a large number of modern and ancient complete genomes from a single species and thereby study the genomic patterns of evolutionary change over time. Such a study would herald the beginnings of ancient population genomics and its use in the study of evolution. Species that are amenable to such large-scale studies warrant increased research effort. We report here progress on a population genomic study of the Adélie penguin (Pygoscelis adeliae). This species is ideally suited to ancient population genomic research because both modern and ancient samples are abundant in the permafrost conditions of Antarctica. This species will enable us to directly address many of the fundamental questions in ecology and evolution.
Keywords: ancient DNA, population genomics, Adélie penguin, evolutionary rates
1. Introduction
The field of ancient DNA (aDNA) research has witnessed remarkable growth and achievements in its relatively short history. In its first two decades, aDNA research focused primarily on the recovery of short fragments of DNA amplified through the polymerase chain reaction (PCR). Typically, results were limited to mitochondrial loci, and often sequences were collected from only one or a small number of samples [1]. Nonetheless, informative DNA sequences were reported from a wide range of species, including ancient humans [2] and Late Pleistocene and Holocene megafauna [3,4]. Over the last decade, however, there has been a period of great expansion in aDNA research as the field has incorporated improved DNA extraction techniques, second-generation sequencing technologies, and more recently, whole-genome capture methods [5]. The most prominent publications in aDNA research during this period have exploited second-generation sequencing [6]. These publications have rapidly shifted the focus on the field to the recovery of full mitochondrial and nuclear genomes of targeted species, and have included draft nuclear genomes of human and woolly mammoth samples from the Holocene [7–9], Neanderthal and Denisovan specimens from the Late Pleistocene [10,11], and, astoundingly, a 700 000 year old horse genome [12]. Simultaneously, such efforts are exploring the limits of DNA preservation, recovery and sequencing [13]. Conversely, numerous smaller studies have used improved aDNA extraction and amplification methods to recover informative loci from relatively large numbers of preserved samples within a targeted species [14,15]. It is clear that both strategies have merit. For example, studies of ancient humans will probably always be limited to a small number of available samples [16]. Yet the value of these data in understanding the history of our own species is important, and consequently, substantial resources have been dedicated to the production of individual draft hominin genomes [9–11]. Alternatively, in taxa with larger numbers of preserved samples, more extensive sampling focusing on single or several loci can allow direct testing of hypotheses about a species' history [14]. At the same time, neither strategy can be considered ideal, and it is a difficult challenge to balance the number of individuals sampled and the number of loci sequenced in order to produce the most robust and informative datasets within economic constraints.
To date, there have been no published examples that incorporate strategies featuring the application of both improved DNA recovery methods and high-throughput sequencing technologies to large numbers of ancient samples. Such examples can provide insights into questions regarding genomic responses to environmental pressures and provide useful data for directly testing evolutionary hypotheses, for example, whether the molecular signatures observed in the data could be explained by selection or by neutral mutational process.
In this paper, we present the initial results of genomic sequencing of large numbers of ancient samples of the Adélie penguin (Pygoscelis adeliae Hombron and Jacquinot). Modern and ancient breeding colonies of Adélie penguins are abundant in the Ross Sea region of Antarctica, allowing large numbers of both modern and ancient samples to be collected. In addition, the cold and dry Antarctic environment provides ideal conditions for the preservation of bone and soft tissue, and at the same time, decreases the rate of degradation of endogenous DNA and contamination of sample tissues by fungi and bacteria. Last but not least, Adélie penguin colonies feature typically high rates of natal return [17], such that single colonies can provide stratigraphic records spanning thousands of years.
2. The benefits of a population genomics approach
The concept of population genomics has been foreshadowed in a number of reviews [18,19] and has been discussed in relation to conservation biology [20], adaptation [21], landscape-evolution effects [22] speciation [23,24] and molecular ecology [25], among others. In general, the strengths of a population genomics versus a population genetics approach are twofold. First, as a direct extension of population genetics, population genomics offers increased accuracy and an improved ability to detect outlier loci for traditional population genetic estimations, such as effective population size and relatedness of populations [19,26]. Second, population genomics allows for more effective exploration of questions of ‘how?’ and ‘why?’ rather than simply ‘what?’ and ‘when?’. For example, since the entire or large parts of the genome can be interrogated at once, it is possible to test broadly for regions of the genome under selection, and from these tests form hypotheses relating adaptation to life-history traits and historic environmental and climate trends (e.g. [27]). Alternatively, broad patterns of heterozygosity at variant sites across the genome may be used to interrogate long-term demographic trends, which in turn may be used to better understand historic genetic admixture patterns [28]. Ultimately, greater exploration and application of a species' nuclear genome should result in finer resolution of population demographic parameters, more robust interpretations of species' or population histories, and a wider range of questions that can be addressed in research efforts [19]. A population genomics approach should greatly benefit aDNA research in particular, as to date the great majority of population genetic studies using historic samples have been limited to very small regions of the (primarily) mitochondrial and nuclear genomes [14,15]. Inclusion of a broader sampling of the nuclear genome in these systems will alleviate dependence on single to several loci and should increase the statistical robustness of results.
Aside from financial considerations, perhaps the greatest challenge in the broad application of a population genomics approach to both modern and ancient taxa is the lack of reference genomes in non-model taxa. This challenge has been addressed with numerous strategies [25], and certainly will be alleviated by degrees as an increasing number and diversity of genomes is sequenced. Still, the inherent characteristics specifically of ancient nucleic acids limit the generation of genome-scale data in taxa without a reference genome. For example, consistent de novo assembly of shotgun-sequenced degraded DNA fragments is unlikely at the genome scale [29]. The highly fragmented nature of aDNA combined with typically low endogenous contents would also pose great challenges to many reduced representation library approaches, such as restriction-site associated DNA sequencing [30]. Furthermore, poor conservation of RNA molecules, generally precludes generation of transcriptome sequence (although see [31]). Alternatively, more effective starting points are either using a genomic assembly available from one or more closely related taxa (i.e. use of the human genome for assembly of the Neanderthal genome [10]) or de novo genomic sequencing and assembly in a contemporary representative of a targeted taxon. In either case, significant portions of the genome can be identified and targeted for sequencing and/or assembly through targeted enrichment or shotgun sequencing strategies in ancient samples.
3. Developing strategies to counter low endogenous DNA in ancient samples
From the earliest reports of aDNA sequencing, it has been clear that most ancient samples contain low amounts of both total and endogenous DNA [32,33]. This is due both to spontaneous damage (e.g. oxidation, hydrolysis) resulting in fragmentation and difficulty in enrichment of endogenous sequences, and also to colonization of tissues by degrading fungi and bacteria [34,35]. Prior to second-generation sequencing technologies, it was difficult to determine the endogenous DNA contents of ancient samples. Rather, efforts were made at quantifying whole-DNA extractions typically through fluorescence measurements (e.g. [36]). Second-generation sequencing platforms, which rapidly produce millions to billions of base pairs (bp) of sequence data in typically short (tens to hundreds of bp) fragments, are well suited both for aDNA sequencing in general and for quantifying endogenous DNA contents of ancient samples specifically. Application of these platforms to aDNA studies has greatly facilitated direct measurement of endogenous DNA contents from a variety of samples.
In a broad sense, expected trends have been supported, for example, specimens preserved under relatively optimal conditions (e.g. permafrost) tend to have higher endogenous DNA contents. Notably, Middle Holocene woolly mammoth and Palaeo-Eskimo samples have yielded very high (45–94%) levels of endogenous DNA [7,8,37]. On the other hand, selected maize samples aged to ca 700 years and from temperate environments have also yielded high (greater than 90%) endogenous DNA contents [38], and Denisovan samples from a cave in southern Siberia aged 30 000–50 000 years contained up to approximately 70% endogenous DNA contents [11,39]. More typically, however, levels of endogenous DNA in ancient samples range from ca less than 1 to 10%. For example, efforts with Neanderthal specimens have reported from ca 0.2 to 6% endogenous contents [10,40,41], while levels from Pleistocene cave bear samples have ranged from 1.1 to 5.8% [16]. It is also important to note that many of these values result from ‘best case scenarios’. For example, the levels of endogenous Neanderthal aDNA reported by Green et al. (ca 6%) [40] represented the most promising sample among over 70 bone and tooth samples tested.
The direct quantitation of endogenous DNA contents in ancient samples has helped spur the recent development and application of enrichment and targeting strategies associated with second-generation sequencing. Perhaps the simplest approach involved application of several restriction enzymes to Neanderthal genomic libraries to selectively cleave contaminating bacterial sequences [10], effectively resulting in a four- to sixfold increase in the proportion of endogenous DNA. Targeted enrichment strategies using multiplex PCR and both array- and solution-based hybridization have also been applied to selected nuclear loci and organelle genomes [38,42–44], the nuclear exome [41] and, most recently, full chromosomes and nuclear genomes [5,45]. Finally, an innovative single-stranded library preparation technique has recently been developed [46], which increases representation of intact strands of highly degraded DNA. To some degree, all of these advances have been in response to limitations in sample quality or quantity, or to economize data generation. For taxa that consistently yield high levels of endogenous DNA from ancient samples, it is possible that these strategies will not be necessary. Ultimately, any given project will require a balance between the amount and quality of samples available, financial constraints and the amount of data appropriate to pursue individual research questions.
4. The Adélie penguin system
Adélie penguins represent the dominant feature of the Antarctic terrestrial fauna during the austral summer. With a total population size estimated around 2.5–3 million breeding pairs [47], Adélie penguins comprise approximately 90% of the avian biomass of Antarctica [48]. Breeding colonies are irregularly distributed around the Antarctic continent, and are almost exclusively restricted to ice-free coastlines. For example, around 30% of the total breeding population is found along the coastline of the Ross Sea, while single large colonies (greater than 100 000 breeding pairs) are present in similar habitat on both the Antarctic Peninsula and Rauer Island [47]. There is direct evidence that individuals of the species exhibit strong natal return over significant periods of time [17], although this pattern may be disrupted by environmental alterations, such as the presence of mega-icebergs [49]. Trophic interactions involving the Adélie penguin are relatively simple, as adults are preyed upon almost exclusively by leopard seals [50]. In turn, the great proportion of the Adélie penguin diet consists of krill (Euphausia spp.) and silverfish (Pleuragramma antarcticum) [51]. Adélie penguin eggs and young are also preyed upon by large seabirds, most prominently by the South Polar skua (Catharacta maccormicki) [52].
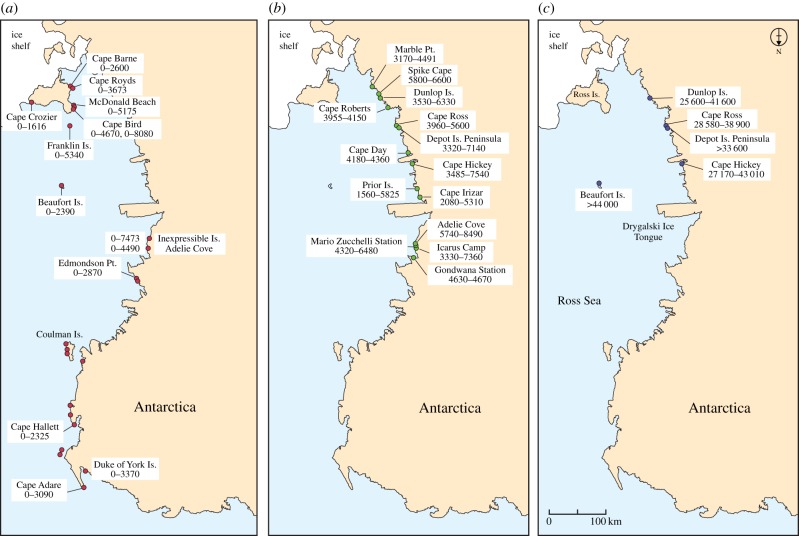
In the Ross Sea area, there are a large number of modern colonies of Adélie penguins (figure 1a). In addition, abandoned colonies in this region were occupied in the Holocene period (figure 1b). Finally, there are a smaller number of locations that were occupied in the Late Pleistocene period, which are found south of the Drygalski Ice Tongue (figure 1c). The ages and occupation periods of many of these colonies have been determined by radiocarbon methods [53–55]. In general, these records indicate that the Adélie is both a resilient species and capable of colonizing new areas. Several colonies, for example those found at Inexpressible Island, Franklin Island and Adélie Cove, have supported Adélie penguin occupation for periods up to approximately 7500 years, while most modern colonies are less than 2000 years old [54] and some abandoned colonies appear to have been used over time intervals of only several hundred years [53]. Historic demographic patterns have been largely correlated with changing glacial and sea ice conditions, which are in turn linked to climatic trends, and reflect the dynamic nature and environmental sensitivity of the Adélie penguin species documented in studies of contemporary colonies [56–58].
Figure 1.
Distribution of modern and ancient Adélie penguin colonies in the Ross Sea region of Antarctica. (a) Dots indicate presently occupied colonies and approximate periods of occupancy determined from radiocarbon dating. (b) Location and period of occupancy of breeding colonies occupied during the Holocene period. (c) Location and ages of breeding colonies of Adélie penguins from the Late Pleistocene period. (Online version in colour.)
Supported by this intricate understanding of Adélie and the environmental history of the Ross Sea area, aDNA sequences obtained from ancient Adélie penguin samples have been applied to questions of broad evolutionary significance. The mitochondrial locus hypervariable region 1 as well as complete mitochondrial genomes has been used to directly measure rates of evolution in comparison to phylogenetic-based estimates [59–61], and to determine coalescence times between two distinct and highly variable mitochondrial lineages [62]. Nuclear microsatellite loci, on the other hand, have been used to demonstrate and quantify microevolutionary change [49] over a period of several thousand years. Although studies such as these reveal important insight into evolutionary patterns, thus far results have been limited by PCR-based methodology to a relatively small amount of sequence. Application of second-generation sequencing to well-preserved Adélie samples could not only strengthen results such as these, but also broaden the scope of testable evolutionary hypotheses through generation of increased numbers and types of sequenced loci.
5. Results
(a). Adélie library characteristics and genomic coverage
We prepared 56 indexed ancient Adélie penguin libraries from samples collected from the Ross Sea area of Antarctica and C14 aged from several hundred to ca 7000 years, using methods optimized for aDNA extraction and sequencing [63,64]. Preliminary sequencing of these samples in a full flow cell of the Illumina HiSeq2000 (eight samples per lane) was performed in order to estimate genomic coverage and endogenous DNA contents and potentially recover high-copy loci, and resulted in an average of 15.5 million total sequence reads per sample (3.2–24.7 million reads range).
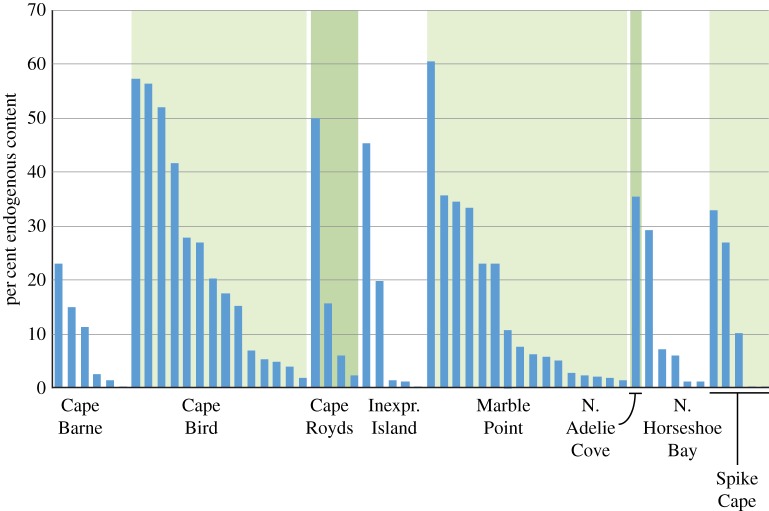
In the case of the Adélie penguin, limited, but high quality, genomic resources are available, as this species' genome has been sequenced through a larger consortium project (http://phybirds.genomics.org.cn/Phylogenomics analysis of Birds). The genome consists of 1.203 billion bp of sequenced DNA assembled in 752 scaffolds and 165 fully contiguous sequences. Approximately, 15 300 mRNA sequences have been annotated, consisting of over 140 000 coding sequences, although less than two-thirds of these have been assigned putative functions. In addition, several fully sequenced mitochondrial genomes for the Adélie penguin are available in Genbank. From mapping sequence reads in our prepared libraries to the draft nuclear genome and a representative mitochondrial genome (Genbank ID KC875855.1), endogenous contents ranged from 0.01 to 60.55% (16.83% average), and were highly variable within sampling locations (figure 2). As expected, the great majority of endogenous reads (greater than 99.6% average) mapped to the nuclear genome.
Figure 2.
Endogenous DNA contents of samples from Holocene Adélie penguin colonies in the Ross Sea region of Antarctica. Endogenous values represent percentages of sequence reads mapping to nuclear and mitochondrial reference genomes divided by the total number of reads for each sample. (Online version in colour.)
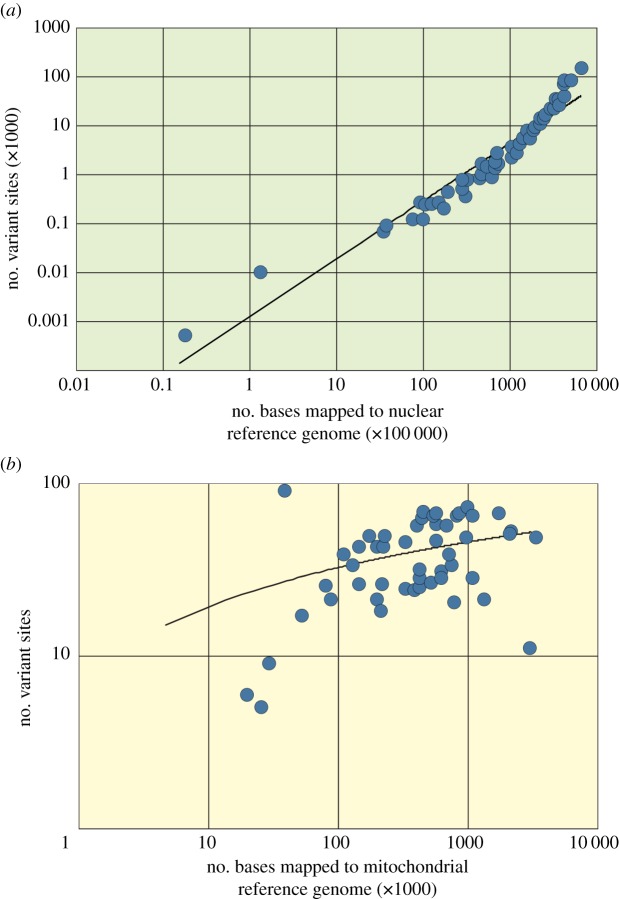
The average length of reads aligning to the reference genomes was calculated at just over 56 bp, with reads aligning to the mitochondrial genome slightly longer than reads aligning to the nuclear genome (58.23 bp compared with 56.34 bp, respectively; p = 0.0141; table 1). On average, approximately 10% of the nuclear genome was covered by one or more reads (123.7 million positions on average), while 16.1 thousand positions of the 17.5 kbp mitochondrial genome were covered. The resulting average coverage depths were 0.136 and 33.39 for the nuclear and mitochondrial reference genomes, respectively (table 1). We found that neither endogenous DNA content nor read length were strongly correlated to the age of the sample, nor was average read length correlated to the endogenous DNA content (r2 < 0.05 in all cases). Conversely, the total number of aligned bases (i.e. the total number of aligned reads multiplied by read length) was positively correlated to the number of variant sites identified when mapping reads to either the nuclear or mitochondrial reference genome (figure 3). This relationship was relatively weak for the mitochondrial genome due to saturation effect owing to its small size. Saturation was also probably responsible for higher rates of duplication seen in reads mapping to the mitochondrial versus nuclear genome (4.74% compared to 20.13% average, respectively; p < 0.001), as there was a stronger correlation between coverage depth and duplication levels for reads aligned to the mitochondrial genome than the nuclear genome (y = 0.4059x + 6.5749, r2 = 0.84906 versus y = 14.02x + 2.8374, r2 = 0.48116, respectively).
Table 1.
Summary of ancient Adélie penguin sequence reads aligned to nuclear and mitochondrial reference genomes. (Average values of 56 ancient libraries are shown, with standard deviations in parentheses. All values except for last column were calculated after removal of duplicates from mapped read pools.)
| reference genome | number of aligned reads, millions | per cent endogenous reads aligned | length of aligned reads, bp | average coverage depth | number of genome positions covered, ≥1× | number of variant sites | duplication, % of original aligned read pool |
|---|---|---|---|---|---|---|---|
| nuclear | 2.58 (2.63) | 16.77 (17.27) | 56.34 (11.21) | 0.136 (0.141) | 123 755 026 (117 895 958) | 13 883 (26 877) | 4.74 (2.85) |
| mitochondrial | 0.01 (0.01) | 0.06 (0.06) | 58.23 (11.90) | 33.39 (37.18) | 16 104 (3649) | 38.09 (20.41) | 20.13 (16.46) |
Figure 3.
Relationships between number of bases mapped in ancient Adélie penguin samples and number of variant sites identified for (a) nuclear (trendline shown for y = 2 × 10−6 x1.1781, r2 = 0.93652) and (b) mitochondrial reference genomes (trendline shown for y = 5.6658 ln(x)−32.917, r2 = 0.22572). All axes shown in logarithmic scale. (Online version in colour.)
(b). Full mitochondrial genomes add insight into rates of evolution
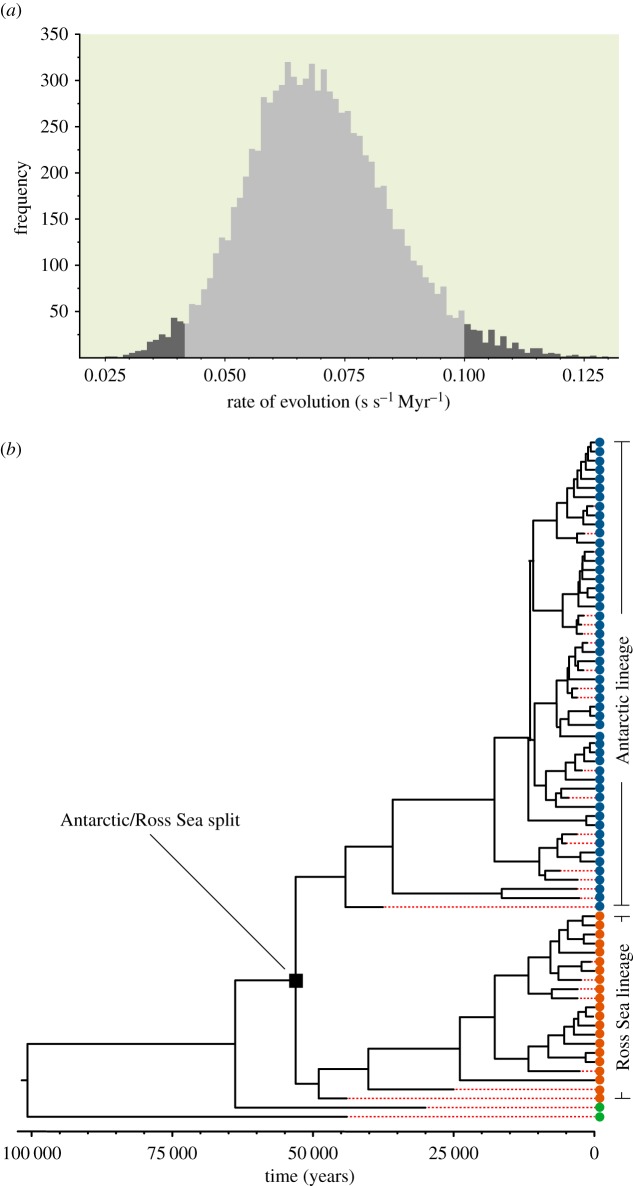
We also sequenced complete mitochondrial genomes of 22 modern Adélie penguin individuals to an average coverage depth greater than 20×. Combined with the assemblies from ancient samples discussed above and previously published mitochondrial genomes, we analysed a total of 29 selected ancient and 46 modern Adélie penguin mitogenomes to estimate the rate of mitochondrial sequence evolution. The Bayesian statistics based Markov chain Monte Carlo method employed in the software BEAST was used for this purpose [65]. The best model of sequence evolution was determined by the program Modeltest [66]. We used an uncorrelated lognormal molecular clock to estimate the rate. We first used synonymous codon positions to estimate the neutral rate of evolution. Our analyses produced a rate of 0.07 (highest posterior density, HPD 0.042–0.100) substitutions per site per million years (s s−1 Myr−1) (table 2; figure 4a) under either a constant or exponential population growth model. We also examined rates of evolution in constrained sites such as non-synonymous sites and RNAs using methods described previously [61]. The rate at non-synonymous sites was nearly an order of magnitude slower than that at neutral sites (0.007 s s−1 Myr−1; HPD 0.002–0.012). In turn, the rate for rRNA loci (0.01 s s−1 Myr−1) was close to that of non-synonymous sites, while the rate for tRNA loci (0.02 s s−1 Myr−1) was approximately twice that of the former. Our rate analysis using neutral synonymous sites revealed that the coalescence age of Adélie penguin populations used in this study is 101 000 years (HPD 55–167 kya; figure 4b). The time of divergence between the two major Adélie penguin mitochondrial haplotypes, the Antarctic and Ross Sea lineages, was estimated to be 53 kya (HPD 44–68 kya).
Table 2.
Rates of evolution estimated using ancient and modern penguin genomes. (Highest posterior density values shown in parentheses.)
| type of positions | rate of evolution (s s−1 Myr−1) |
|
|---|---|---|
| Subramanian et al. [61] | this study | |
| synonymous sites | 0.073 (0.025–0.123) | 0.070 (0.042–0.100) |
| non-synonymous sites | 0.007 (0.002–0.012 | 0.011 (0.006–0.015) |
| tRNA | 0.020 (0.007–0.034) | 0.023 (0.014–0.034) |
| rRNA | 0.010 (0.003–0.017) | 0.019 (0.011–0.027) |
| full mitochondrial genome | 0.024 (0.008–0.040) | 0.028 (0.017–0.040) |
Figure 4.
Molecular rates for mitochondrial sequences and times of divergence of mitochondrial lineages. (a) Posterior distribution of rates of evolution estimated using a relaxed log normal molecular clock and the HKY + gamma model of evolution. (b) Bayesian chronogram showing divergence/coalescence times among modern and ancient Adélie penguin populations used in this study. Dotted horizontal lines denote cessation of branch lengths corresponding to the ages of samples. (Online version in colour.)
6. Discussion
Our understanding of the state of preservation of many ancient Adélie penguin remains preserved over serial time points, together with the availability of modern samples suggests this species is ideal for a comprehensive ancient population genomic study. Most importantly, the presence of large numbers of well-preserved samples contributes to sequencing success and to the ability to perform robust tests of evolutionary hypotheses. In this study, we used modifications to DNA extraction procedures and an improved genomic library building method [63,64] to efficiently build a large number of genomic libraries from ancient Adélie penguin samples for sequencing on the Illumina platform. These samples contained relatively high levels of endogenous DNA (ca 17% on average) and are amenable to increased sequencing efforts. Even with the preliminary sequencing effort reported here (eight samples per lane of an Illumina HiSeq), these libraries produced 56 mitochondrial genomes, at an average coverage depth of over 20× and over 90% completion. Our efforts also produced on average over 120 million bp of nuclear data per sample, with consistently low rates of sequence read duplication. Because of the relative high diversity of sequence reads in these genomic libraries, it is likely that increased sequencing effort will allow the entire nuclear genome of most of these samples to be sequenced. Alternatively, it is likely that a targeted enrichment strategy could be effectively applied in Adélie penguins to efficiently standardize the portion of the nuclear genome sequenced. This may be particularly important as older samples are incorporated into our sequencing efforts, for example from preserved remains aged prior to the last glacial maximum [61].
The evolutionary rates estimated using 75 complete mitochondrial genomes were very similar to our previous estimates using a much smaller dataset of 20 mitogenomes (table 2) [61]. For instance, the rate at synonymous sites estimated in this study (0.07 s s−1 Myr−1—HPD 0.042–0.1) was nearly identical to our previous estimate (0.073 s s−1 Myr−1—HPD 0.025–0.123). However, the increased dataset used in this study resulted in a narrower HPD interval. These estimates offer further support for our claim of higher mitogenomic rates in birds and penguins, in particular [61]. Similarly, these results are consistent with our previous estimates of coalescence times within and between the two distinct mitochondrial lineages of the Adélie penguin (the Antarctic and Ross Sea lineages) based on sampling of a smaller portion of the mitochondrial genome [61,62]. The divergence time between Antarctic and Ross Sea mitochondrial lineages estimated in this study (53 kya; HPD 44–68 kya) overlaps with that estimated previously (ca 63 kya). However the ages of the Antarctic (44 kya) and Ross Sea (50 kya) lineages estimated here were much higher than those estimated in our earlier work (18 and 19 kya, respectively). As previous estimates were based only on modern samples, it is probable that the observed older ages of these lineages were due to inclusion of ancient penguin mitochondrial genomes. This suggests the extinction of a number of ancient penguin populations belonging to these two lineages.
We have reported here only the mitochondrial genomes generated in this study. We intend to increase sequencing efforts to produce complete nuclear genomes of ancient and modern Adélie genomes. The large number of genome-wide population polymorphisms identified by this work will provide new insights on the genetics and evolution of Adélie penguins over time. For example, estimates of rates and patterns of genetic variation throughout the genome will help to identify regions showing significant deviations with respect to mutation and recombination, including non-coding regions under non-neutral evolution. This will further help to quantify and localize episodes of purifying selection and adaptive evolution in Adélie penguin genomes, and identify conserved regulatory elements. Furthermore, single nucleotide polymorphism in coding genes could be used to identify protein functional pathways influenced by positive selection. This might elucidate adaptive functional responses to increased temperatures in the Antarctic. Although previous studies have examined some of the patterns mentioned above they were generally based on one or several genes [59,60]. A major drawback in such studies is that they fail to distinguish the gene-specific from genome-wide evolutionary patterns. As we will examine all or most of the genes in the Adélie genome we will be able to quantify the mean genome-level mutation or selection patterns and hence could measure how individual genes deviate from the genomic mean.
Although results from ancient Adélie penguin samples are promising, there are probably many other ancient systems that will also prove highly suitable for the application of high-throughput sequencing for large numbers of samples. For example, previous work on large Arctic and subarctic mammals, such as the Beringian steppe bison, brown and cave bears, and woolly mammoth, reported the recovery of DNA from nearly 30 to over 400 samples [67–70]. At the same time, it is also likely that taxa from more temperate environments will provide useful data. Previous suggestions to pursue aDNA sequencing in brine shrimp from hypersaline lakes or extinct avian taxa from Polynesian islands ([71], see also [1,72,73]) seem prescient in the light of second-generation sequencing advancements, as large numbers of chronologically dated samples are potentially available. Alternatively, sample-rich temperate systems might be revisited with updated technologies, such as the New Zealand brown kiwi [74] or various rodent species ([75,76], see also [77]). Finally, while the majority of aDNA research has traditionally targeted humans and other animals [78], certainly other lineages, including plants and bacteria, should not be overlooked. The successful sequencing of nuclear loci from a preserved conifer species [79] suggests that ancient woody plant samples may be directly sampled and could be excellent targets for high-throughput aDNA studies.
7. Conclusion
In concert with the development of increasingly high-throughput sequencing technologies and improved aDNA extraction techniques, the field of aDNA is poised to make great and robust empirical contributions to evolutionary biology as it moves into its fourth decade. In particular, if focus is shifted toward ideal taxa (i.e. those with large numbers of high-quality samples), we will rapidly gain a better understanding of genomic evolutionary changes from the Pleistocene and Holocene periods to the present.
Acknowledgements
The authors would like to thank the following people and groups: Indy Siva from Griffith University, Edan Scriven from the University of Queensland, and the Queensland Cyber Infrastructure Foundation for computing resources and assistance, Yvette Wharton for field assistance, Vivian Ward for graphics, and Tim Heupink and Leon Huynen for valuable comments on the manuscript.
Funding statement
This research was funded by the Australian Research Council in the form of a linkage grant LP110200229: ‘How will animals respond to climate change? A genomic approach’. Logistic support was provided by Antarctica New Zealand.
References
- 1.Wayne RK, Leonard JA, Cooper A. 1999. Full of sound and fury: the recent history of ancient DNA. Ann. Rev. Ecol. Syst. 30, 457–477. ( 10.2307/221692) [DOI] [Google Scholar]
- 2.Handt O, et al. 1994. Molecular genetic analyses of the Tyrolean Ice Man. Science 264, 1775–1778. ( 10.1126/science.8209259) [DOI] [PubMed] [Google Scholar]
- 3.Cooper A, Mourer-Chauviré C, Chambers GK, von Haeseler A, Wilson AC, Pääbo S. 1992. Independent origins of New Zealand moas and kiwis. Proc. Natl Acad. Sci. USA 89, 8741–8744. ( 10.1073/pnas.89.18.8741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagelberg E, Thomas MG, Cook CE, Sher AV, Baryshnikov GF, Lister AM. 1994. DNA from ancient mammoth bones. Nature 370, 333–334. ( 10.1038/370333b0) [DOI] [PubMed] [Google Scholar]
- 5.Carpenter ML, et al. 2013. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 1–13. ( 10.1016/j.ajhg.2013.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millar CD, Huynen L, Subramanian S, Mohandesan E, Lambert DM. 2008. New developments in ancient genomics. Trends Ecol. Evol. 23, 386–393. ( 10.1016/j.tree.2008.04.002) [DOI] [PubMed] [Google Scholar]
- 7.Miller W, et al. 2008. Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456, 387–390. ( 10.1038/nature07446) [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen M, et al. 2010. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762. ( 10.1038/nature08835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller A, et al. 2012. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 ( 10.1038/ncomms1701) [DOI] [PubMed] [Google Scholar]
- 10.Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328, 710–722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer M, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226. ( 10.1126/science.1224344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlando L, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78. ( 10.1038/nature12323) [DOI] [PubMed] [Google Scholar]
- 13.Millar CD, Lambert DM. 2013. Ancient DNA: towards a million-year-old genome. Nature 499, 34–35. ( 10.1038/nature12263) [DOI] [PubMed] [Google Scholar]
- 14.Ramakrishnan UMA, Hadly EA. 2009. Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Mol. Ecol. 18, 1310–1330. ( 10.1111/j.1365-294X.2009.04092.x) [DOI] [PubMed] [Google Scholar]
- 15.de Bruyn M, Hoelzel AR, Carvalho GR, Hofreiter M. 2011. Faunal histories from Holocene ancient DNA. Trends Ecol. Evol. 26, 405–413. ( 10.1016/j.tree.2011.03.021) [DOI] [PubMed] [Google Scholar]
- 16.Noonan JP. 2010. Neanderthal genomics and the evolution of modern humans. Genome Res. 20, 547–553. ( 10.1101/gr.076000.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainley DG, LeResche RE, Sladen WJL. 1983. Breeding biology of the Adelie penguin. Berkeley, CA: University of California Press. [Google Scholar]
- 18.Feder ME, Mitchell-Olds T. 2003. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 4, 649–655. ( 10.1038/nrg1128) [DOI] [PubMed] [Google Scholar]
- 19.Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4, 981–994. ( 10.1038/nrg1226) [DOI] [PubMed] [Google Scholar]
- 20.Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma R, Hedrick PW. 2010. Conservation genetics in transition to conservation genomics. Trends Genet. 26, 177–187. ( 10.1016/j.tig.2010.01.001) [DOI] [PubMed] [Google Scholar]
- 21.Stapley J, et al. 2010. Adaptation genomics: the next generation. Trends Ecol. Evol. 25, 705–712. ( 10.1016/j.tree.2010.09.002) [DOI] [PubMed] [Google Scholar]
- 22.Lowry DB. 2010. Landscape evolutionary genomics. Biol. Lett. 6, 502–504. ( 10.1098/rsbl.2009.0969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernatchez L, et al. 2010. On the origin of species: insights from the ecological genomics of lake whitefish. Phil. Trans. R. Soc. B 365, 1783–1800. ( 10.1098/rstb.2009.0274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feder JL, Egan SP, Nosil P. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350. ( 10.1016/j.tig.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 25.Ekblom R, Galindo J. 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107, 1–15. ( 10.1038/hdy.2010.152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black WC, IV, Baer CF, Antolin MF, DuTeau NM. 2001. Population genomics: genome-wide sampling of insect populations. Annu. Rev. Entomol. 46, 441–469. ( 10.1146/annurev.ento.46.1.441) [DOI] [PubMed] [Google Scholar]
- 27.Yi X, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. ( 10.1126/science.1190371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496. ( 10.1038/nature10231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staats M, Erkens RHJ, van de Vossenberg B, Wieringa JJ, Kraaijeveld K, Stielow B, Geml J, Richardson JE, Bakker FT. 2013. Genomic treasure troves: complete genome sequencing of herbarium and insect museum specimens. PLoS ONE 8, e69189 ( 10.1371/journal.pone.0069189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guy PL. 2013. Ancient RNA? RT-PCR of 50-year-old RNA identifies peach latent mosaic viroid. Arch. Virol. 158, 691–694. ( 10.1007/s00705-012-1527-0) [DOI] [PubMed] [Google Scholar]
- 32.Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. 1984. DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282–284. ( 10.1038/312282a0) [DOI] [PubMed] [Google Scholar]
- 33.Pääbo S. 1985. Molecular cloning of ancient Egyptian mummy DNA. Nature 314, 644–645. ( 10.1038/314644a0) [DOI] [PubMed] [Google Scholar]
- 34.Pääbo S, et al. 2004. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679. ( 10.1146/annurev.genet.37.110801.143214) [DOI] [PubMed] [Google Scholar]
- 35.Hebsgaard MB, Phillips MJ, Willerslev E. 2005. Geologically ancient DNA: fact or artefact? Trends Microbiol. 13, 212–220. ( 10.1016/j.tim.2005.03.010) [DOI] [PubMed] [Google Scholar]
- 36.Pääbo S. 1989. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl Acad. Sci. USA 86, 1939–1943. ( 10.1073/pnas.86.6.1939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poinar HN, et al. 2006. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science 311, 392–394. ( 10.1126/science.1123360) [DOI] [PubMed] [Google Scholar]
- 38.Avila-Arcos MC, et al. 2011. Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA. Nat. Sci. Rep. 1, 74 ( 10.1038/srep00074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. ( 10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green RE, et al. 2006. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336. ( 10.1038/nature05336) [DOI] [PubMed] [Google Scholar]
- 41.Burbano HA, et al. 2010. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725. ( 10.1126/science.1188046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briggs AW, et al. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321. ( 10.1126/science.1174462) [DOI] [PubMed] [Google Scholar]
- 43.Stiller M, Knapp M, Stenzel U, Hofreiter M, Meyer M. 2009. Direct multiplex sequencing. (DMPS): a novel method for targeted high-throughput sequencing of ancient and highly degraded DNA. Genome Res. 19, 1843–1848. ( 10.1101/gr.095760.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lari M, et al. 2011. The complete mitochondrial genome of an 11,450-year-old aurochsen (Bos primigenius) from Central Italy. BMC Evol. Biol. 11, 32 ( 10.1186/1471-2148-11-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S. 2013. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227. ( 10.1073/pnas.1221359110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gansauge M-T, Meyer M. 2013. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748. ( 10.1038/nprot.2013.038) [DOI] [PubMed] [Google Scholar]
- 47.Ainley DG. 2002. The Adelie penguin: bellwether of climate change. New York, NY: Columbia University Press. [Google Scholar]
- 48.Croxall JP, Prince PA. 1979. Antarctic seabird and seal monitoring studies. Polar Rec. 19, 573–595. ( 10.1017/S0032247400002680) [DOI] [Google Scholar]
- 49.Shepherd LD, Millar CD, Ballard G, Ainley DG, Wilson PR, Haynes GD, Baroni C, Lambert DM. 2005. Microevolution and mega-icebergs in the Antarctic. Proc. Natl Acad. Sci. USA 102, 16 717–16 722. ( 10.1073/pnas.0502281102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ainley DG, Ballard G, Karl BJ, Dugger KM. 2005. Leopard seal predation rates at penguin colonies of different size. Antarct. Sci. 17, 335–340. ( 10.1017/S0954102005002750) [DOI] [Google Scholar]
- 51.Polito M, Emslie SD, Walker W. 2002. A 1000-year record of Adelie penguin diets in the southern Ross Sea. Antarct. Sci. 14, 327–332. ( 10.1017/S0954102002000184) [DOI] [Google Scholar]
- 52.Tenaza R. 1971. Behavior and nesting success relative to nest location in Adelie penguins (Pygoscelis adeliae). Condor 73, 81–92. ( 10.2307/1366127) [DOI] [Google Scholar]
- 53.Baroni C, Orombelli G. 1994. Abandoned penguin rookeries as Holocene paleoclimatic indicators in Antarctica. Geology 22, 23–26. () [DOI] [Google Scholar]
- 54.Emslie SD, Coats L, Licht K. 2007. A 45,000 yr record of Adélie penguins and climate change in the Ross Sea, Antarctica. Geology 35, 61–64. ( 10.1130/g23011a.1) [DOI] [Google Scholar]
- 55.Hu Q-H, Sun L-G, Xie Z-Q, Emslie SD, Liu X-D. 2013. Increase in penguin populations during the Little Ice Age in the Ross Sea, Antarctica. Nat. Sci. Rep. 3, 2472 ( 10.1038/srep02472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor RH, Wilson PR. 1990. Recent increase and southern expansion of Adelie penguin populations in the Ross Sea, Antarctica, related to climatic warming. NZ J. Ecol. 14, 25–29. [Google Scholar]
- 57.Kato A, Ropert-Coudert Y, Naito Y. 2002. Changes in Adélie penguin breeding populations in Lützow-Holm Bay, Antarctica, in relation to sea-ice conditions. Polar Biol. 25, 934–938. ( 10.1007/s00300-002-0434-3) [DOI] [Google Scholar]
- 58.LaRue MA, Ainley DG, Swanson M, Dugger KM, Lyver POB, Barton K, Ballard G. 2013. Climate change winners: receding ice fields facilitate colony expansion and altered dynamics in an Adelie penguin metapopulation. PLoS ONE 8, e60568 ( 10.1371/journal.pone.0060568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambert DM, Ritchie PA, Millar CD, Holland B, Drummond AJ, Baroni C. 2002. Rates of evolution in ancient DNA from Adélie penguins. Science 295, 2270–2273. ( 10.1126/science.1068105) [DOI] [PubMed] [Google Scholar]
- 60.Millar CD, Dodd A, Anderson J, Gibb GC, Ritchie PA, Baroni C, Woodhams MD, Hendy MD, Lambert DM. 2008. Mutation and evolutionary rates in Adelie penguins from the Antarctic. PLoS Genet. 4, e1000209 ( 10.1371/journal.pgen.1000209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian S, Denver DR, Millar CD, Heupink T, Aschrafi A, Emslie SD, Baroni C, Lambert DM. 2009. High mitogenomic evolutionary rates and time dependency. Trends Genet. 25, 482–486. ( 10.1016/j.tig.2009.09.005) [DOI] [PubMed] [Google Scholar]
- 62.Ritchie PA, Millar CD, Gibb GC, Baroni C, Lambert DM. 2004. Ancient DNA enables timing of the Pleistocene origin and Holocene expansion of two Adélie penguin lineages in Antarctica. Mol. Biol. Evol. 21, 240–248. ( 10.1093/molbev/msh012) [DOI] [PubMed] [Google Scholar]
- 63.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 64.Dabney J, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15 758–15 763. ( 10.1073/pnas.1314445110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drummond A, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. ( 10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 67.Shapiro B, et al. 2004. Rise and fall of the Beringian steppe bison. Science 306, 1561–1565. ( 10.1126/science.1101074) [DOI] [PubMed] [Google Scholar]
- 68.Hofreiter M, Münzel S, Conard NJ, Pollack J, Slatkin M, Weiss G, Pääbo S. 2007. Sudden replacement of cave bear mitochondrial DNA in the late Pleistocene. Curr. Biol. 17, R122–R123. ( 10.1016/j.cub.2007.01.026) [DOI] [PubMed] [Google Scholar]
- 69.Valdiosera CE, et al. 2007. Staying out in the cold: glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 16, 5140–5148. ( 10.1111/j.1365-294X.2007.03590.x) [DOI] [PubMed] [Google Scholar]
- 70.Debruyne R, et al. 2008. Out of America: ancient DNA evidence for a New World origin of late Quaternary woolly mammoths. Curr. Biol. 18, 1320–1326. ( 10.1016/j.cub.2008.07.061) [DOI] [PubMed] [Google Scholar]
- 71.Clegg JS, Jackson SA. 1997. Significance of cyst fragments of Artemia sp. recovered from a 27,000 year old core taken under the Great Salt Lake, Utah, USA. Int. J. Salt Lake Res. 6, 207–216. ( 10.1007/bf02449925) [DOI] [Google Scholar]
- 72.Sorenson MD, Cooper A, Paxinos EE, Quinn TW, James HF, Olson SL, Fleischer RC. 1999. Relationships of the extinct Moa-Nalos, flightless Hawaiian waterfowl, based on ancient DNA. Proc. R. Soc. Lond. B 266, 2187–2193. ( 10.1098/rspb.1999.0907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Djamali M, et al. 2010. A 200,000-year record of the brine shrimp Artemia (Crustacea: Anostraca) remains in Lake Urmia, NW Iran. Int. J. Aquat. Sci. 1, 14–18. [Google Scholar]
- 74.Shepherd LD, Lambert DM. 2008. Ancient DNA and conservation: lessons from the endangered kiwi of New Zealand. Mol. Ecol. 17, 2174–2184. ( 10.1111/j.1365-294X.2008.03749.x) [DOI] [PubMed] [Google Scholar]
- 75.Hardy C, Callou C, Vigne JD, Casane D, Dennebouy N, Mounolou JC, Monnerot M. 1995. Rabbit mitochondrial DNA diversity from prehistoric to modern times. J. Mol. Evol. 40, 227–237. ( 10.1007/bf00163228) [DOI] [PubMed] [Google Scholar]
- 76.Hadly EA, Ramakrishnan UMA, Chan YL, van Tuinen M, O'Keefe K, Spaeth PA, Conroy CJ. 2004. Genetic response to climatic change: insights from ancient DNA and phylochronology. PLoS Biol. 2, e290 ( 10.1371/journal.pbio.0020290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R, Moritz C. 2013. Unlocking the vault: next-generation museum population genomics. Mol. Ecol. 22, 6018–6032. ( 10.1111/mec.12516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gugerli F, Parducci L, Petit RJ. 2005. Ancient plant DNA: review and prospects. New Phytol. 166, 409–418. ( 10.1111/j.1469-8137.2005.01360.x) [DOI] [PubMed] [Google Scholar]
- 79.Tani N, Tsumura Y, Sato H. 2003. Nuclear gene sequences and DNA variation of Cryptomeria japonica samples from the postglacial period. Mol. Ecol. 12, 859–868. ( 10.1046/j.1365-294X.2003.01779.x) [DOI] [PubMed] [Google Scholar]