Abstract
We compared DNA, pollen and macrofossil data obtained from Weichselian interstadial (age more than 40 kyr) and Holocene (maximum age 8400 cal yr BP) peat sediments from northern Europe and used them to reconstruct contemporary floristic compositions at two sites. The majority of the samples provided plant DNA sequences of good quality with success amplification rates depending on age. DNA and sequencing analysis provided five plant taxa from the older site and nine taxa from the younger site, corresponding to 7% and 15% of the total number of taxa identified by the three proxies together. At both sites, pollen analysis detected the largest (54) and DNA the lowest (10) number of taxa, but five of the DNA taxa were not detected by pollen and macrofossils. The finding of a larger overlap between DNA and pollen than between DNA and macrofossils proxies seems to go against our previous suggestion based on lacustrine sediments that DNA originates principally from plant tissues and less from pollen. At both sites, we also detected Quercus spp. DNA, but few pollen grains were found in the record, and these are normally interpreted as long-distance dispersal. We confirm that in palaeoecological investigations, sedimentary DNA analysis is less comprehensive than classical morphological analysis, but is a complementary and important tool to obtain a more complete picture of past flora.
Keywords: ancient DNA, plant macrofossils, pollen, barcoding, Weichselian interstadial, Holocene
1. Introduction
Over the past three decades, researchers have obtained authentic ancient DNA (aDNA) from a variety of Late Quaternary fossil samples, providing answers to important evolutionary and palaeoecological questions. Despite this, plants still receive little attention compared with animals. What are required for plant aDNA studies are (i) a well-preserved source of aDNA information of local origin, (ii) abundant and well-dated fossil material, and (iii) powerful molecular techniques to extract aDNA information efficiently. Pollen, macrofossils and sedimentary DNA preserved in peat and lake sediments have proven to be an optimal source of plant DNA information in the recent past [1–5].
Before the advent of aDNA studies, scientists relied on morphological information from the fossil record, complemented recently by observations of spatial genotypic variation of living taxa, to understand past plant community distribution. These records have indicated that plants have expanded and contracted their ranges many times during the last glacial–interglacial cycles in both hemispheres. A weakness of the fossil pollen record is, however, that the absence of pollen in a sediment sample does not rule out the possibility of small low-density populations [6,7]. In these cases, plant macrofossils, such as bark, leaves and needles, offer stronger evidence of the local presence of taxa, thus complementing pollen analysis [8–11]. However, macrofossils contain DNA [12], and if preserved under optimal conditions, as in peat and lake sediments from high latitudes and cold environments, are likely to provide a major proportion of the DNA contained within the sediment (sedimentary DNA or sedDNA [1]). Nonetheless, whether macrofossils or other plant tissues like pollen are the primary source of sedDNA is not clear and this can only be tested by performing comparative studies with sedDNA, macrofossils and pollen from the same sediment settings. Jørgensen et al. [3] conducted such a comparative survey and used the metabarcoding technique (identification of taxa from environmental samples such as sedDNA against a database/library of reference sequences [13]) on ancient permafrost samples from northern Siberia spanning the Late Pleistocene. Using generic primers specifically designed for plant-degraded DNA recovered from sediments (trnL, [2]), they showed that the three proxies (pollen, macrofossils and sedDNA) are complementary rather than overlapping, but that sedDNA shared a greater overlap with macrofossils, suggesting that it predominantly originates from these remains. Similar studies have also shown that DNA signals from pollen-producer taxa can be detected even when a few individuals are present in the environment, or when they are abundant but produce little pollen (e.g. during harsh environmental conditions) [5,14–16]. Similarly, the DNA can reveal pollen-limited taxa or taxa with soft and easily degraded tissues, such as aquatic or semi-aquatic plants, that are often under-represented in the fossil records. These studies also show little overlap between proxies, but a higher similarity between macrofossils and DNA.
Here, we used three different palaeobotanical proxies (sedDNA, plant macrofossils and pollen) to describe past floristic compositions of peat/peaty layers collected from northern Europe. The studied samples represent two different time periods, with Weichselian interstadial samples (age more than 40 kyr) collected from northern Finland (NF) and Holocene samples (11.7–0 kyr) from north-eastern European Russia (NER). In particular, we investigated how well DNA signals reflect the fossil plant assemblages preserved in non-limnic depositional environments, and whether the age of the deposits has an important effect on the preservation of DNA. The focus was on a comparison of the proxies rather than preparing detailed palaeovegetation reconstructions for these sites. We used metabarcoding analysis on sedDNA to assess the presence of plant taxa through time and the relative contribution of macrofossils and pollen to the DNA signal at each site.
2. Material and methods
(a). Study sites
Peat samples were collected from two sites. The Kaarreoja site (NF) is situated in Finnish Lapland, at the current alpine tree line zone dominated by mountain birch Betula pubescens subsp. czerepanovii (syn. tortuosa; 68°31′ N, 26°48′ E; figure 1). The Weichselian 20–30 cm thick peat layer was underlain by ca 30 cm of silt and overlain by 2 m of sandy till, followed by a shallow deposit of fluvial sand and a modern organic soil on the top. The Seida site (NER) is located in a discontinuous permafrost region in north-eastern European Russia (67°35′ N, 62°56′ E). The top 40 cm is the active layer that melts during summer. Below that the peat remains frozen throughout the year. The site is located just beyond the current latitudinal tree line, and some individual trees, namely mountain birch, remain present near the coring site. Otherwise, the peat plateau represents a treeless tundra dominated by dwarf shrubs, lichens and bryophytes.
Figure 1.
Locations of the two study sites, Kaarreoja (NF) and Seida (NER).
For the NER site, the Holocene regional climate and vegetation histories are relatively well established [17–22], whereas the Weichselian interstadial palaeoenvironmental conditions at the NF site are less well constrained [10,23–27].
(b). Peat sampling
NF sediments were collected in summer 2012 from an exposed and cleaned sediment wall section. The till layer overlying the peat layer was removed by an excavator, but the final cleaning of the sample surface was carried out manually. During collection, the sediment column was not exposed to the open air; thus any contamination by modern pollen is unlikely. To avoid exposure to air, a column of sediment was extracted using a metal box (ca 0.5 m long) open on one side only. The open side was pushed against the sediment wall, pulled back and immediately covered after extraction. The sediment surface that was briefly exposed to the air was later removed in a laboratory that specializes in optical luminescence analyses. The collected sediment sections were cut into 1-cm slices at the University of Oulu. Subsamples required for pollen analyses remained in Oulu, whereas the rest of the peat was transported to the Department of Environmental Sciences in the University of Helsinki where the sliced samples were stored in airproof plastic bags in a freezer in a laboratory free from DNA research.
NER sediments were collected in summer 2012 when the pollination season was over, so that there was no risk of contamination by modern pollen. The active peat layer (from the surface to 40 cm depth) was collected with a Russian peat corer, and the frozen, permafrost part of the sequence (below 40 cm) with a motorized drill. The collected peat was cut into 2-cm slices in the field. The slices were placed in airproof plastic bags, stored in non-transparent plastic bags and kept cold during train transport to Komi Science Centre in Syktyvkar. Here, the samples were stored in a freezer until final transportation in styrofoam boxes to the University of Helsinki, where they were stored in a freezer.
(c). Subsampling for DNA analysis
Subsampling from the two cores was performed on frozen material at the Department of Environmental Sciences at the University of Helsinki. We changed disposable tools between samples to avoid cross-contamination and collected in total seven samples of ca 30 g wet-weight (three from NF and four from NER) of different ages (table 1). The external 2-cm part from the surface of the sediment core was discarded to avoid contamination. The samples were stored in sterile plastic bags at –20°C until DNA extraction.
Table 1.
Sediment samples used for palaeoecological and barcoding analyses at Kaarreoja (NF) and Seida (NER) study sites, with summary of the results obtained after PCR amplifications of the trnL fragment (g/h). Depths are given in centimetres from the top of the sequences and ages in calendar years before present (BP). For the NF site, it was not possible to assign sample-specific ages, but the age of a ca 53 cm thick organic layer is 42 000–52 000 years.
| sample | depth | age | PCR runs | fragments | clones | assigned plant sequences | unknown plant sequences |
|---|---|---|---|---|---|---|---|
| NF1 | 278–279 | 42 000–52 000 | 16 | 3 | 36 | — | — |
| NF2 | 256–257 | 42 000–52 000 | 16 | 2 | 36 | 38 | — |
| NF3 | 228–229 | 42 000–52 000 | 15 | 7 | 133 | 61 | — |
| NF tot | 47 | 12 | 205 | 99 | — | ||
| NER1 | 2–4 | ∼6000 | 30 | 11 | 118 | 67 | — |
| NER5 | 42–44 | ∼7000 | 32 | 9 | 108 | 64 | 16 |
| NER10 | 152–154 | ∼8200 | 14 | 10 | 108 | 39 | — |
| NER12 | 164–166 | ∼8500 | 12 | 7 | 84 | 49 | — |
| NER tot | 88 | 37 | 418 | 219 | 16 | ||
| total | 135 | 49 | 623 | 318 | 16 | ||
| 334 | |||||||
(d). Chronology
The NF peat section was dated using radiocarbon (14C) and optically stimulated luminescence (OSL) dating at the Tallinn University of Technology and LUOMUS (former Dating Laboratory of Helsinki), University of Helsinki. The NER peat section was radiocarbon dated in the Poznan Radiocarbon Laboratory using bulk peat samples. The 14C dates were calibrated in the CALIB software v. 7.0.0 [28], using the IntCal13 calibration curve [29]. An age–depth model was calculated for the NER site using the method of Heegaard et al. [30] in ‘R’ (v. 2.15.0) [31].
(e). Plant macrofossil analyses
Macrofossil analyses were carried out on 20 cm3 (NF) and 5 cm3 (NER) subsamples, respectively. The samples were cleaned under running water, and the material retained on a 140 µm sieve analysed under a stereomicroscope.
(f). Pollen analyses
Three samples were analysed for pollen content at the NF site. Pollen samples were prepared using heavy liquids modified from Zabenskie et al. [32] and from Zabenskie & Gajewski (http://www.lpc.uottawa.ca/resources/pollen%20-%20heavy%20liquid.html). Lithium heteropolytungstate solution was used instead of sodium polytungstate and without HF treatment. A minimum of 500 pollen grains and spores of terrestrial vascular plants were counted, with aquatic species, bryophyte spores and Pediastrum green algae excluded from the pollen sum. Identification was based on Moore et al. [33] and Beug [34], and the reference collection held at the Department of Palynology and Climate Dynamics, University of Göttingen.
At the NER site, we followed the standard methods described by Bennett & Willis [35] for pollen concentration. A minimum of 1000 pollen grains and spores of terrestrial vascular plants were counted. Subsequently, new pollen taxa were recorded together with a reference taxon (Picea abies) to calculate percentage values for any new taxa found [36]. Aquatic species, bryophyte spores and Pediastrum green algae were excluded from the pollen sum. Pollen taxonomy follows Bennett [36] modified for Sweden using the checklist by Karlsson [37].
(g). aDNA analyses and taxonomic assignment
DNA extractions were performed in dedicated aDNA facilities at the Centre for GeoGenetics at the University of Copenhagen, following established aDNA precautions [38]. We extracted DNA from the seven samples in two batches (NF and NER samples) using 2 g of sediment. Each batch included a negative control (extraction control monitoring for contamination during extraction), which was treated identically to the sediment samples. We used a combined Sergey Bulat protocol and Cambio PowerMax Soil DNA isolation kit protocol (MoBio Laboratories, Cambridge, UK), which employs a silica clean-up method [39]. Following extraction, DNA was purified using the PowerMax Soil DNA isolation kit protocol. Polymerase chain reaction (PCR) amplifications of the trnL fragment with the g and h primers [2] were performed at Uppsala University, in a clean aDNA room, physically separated from modern DNA laboratories. The length of the trnL fragment varies from 13 to 158 base pairs (bp) in the Arctic database [42]. We followed established aDNA methodologies during amplification from aDNA extracts [38] and standard procedures for cloning and sequencing [14]. On the seven samples, we ran 135 PCRs in 10 batches that comprised between nine and 22 PCRs and two negative controls (an extraction control and a PCR control to monitor for contamination during PCR). For amplification, we used the Qiagen multiplex PCR kit, increasing amplification cycles to 40 and following procedures as in Parducci et al. [14]. After amplification, 5 µl of PCR products was screened on 2% agarose gel (110 V for 45 min), and from amplifications visible on gel, we purified 1–2 µl using ExoSAP-IT (Affymetrix) and used this for cloning with the CloneJet PCR cloning kit (Fermentas). We sequenced between 10 and 25 clones per fragment with inserts of expected sizes using the Macrogen DNA Sequencing service (The Netherlands).
Taxonomic assignment was performed as in Parducci et al. [14]. Briefly, trimmed sequences were aligned using BioEdit v. 7.1.3.0 [40] to identify errors owing to base call mis-identifications and post-mortem DNA damage [41]. We constructed a database that included a subset of the trnL sequences from the Arctic database [42] and all trnL sequences available in GenBank for known boreal taxa, lichens and other plant taxa reported from palsa and peat mires of continental Europe (electronic supplementary material, table S1). Using BLAST software, we first compared the sequences with GenBank and then with our database for a final assignment. Taxonomic assignments were strict, and only sequences with a maximum of two or three nucleotide differences from those matching with the databases were considered. We assigned sequences carrying only type 1 and type 2 transition substitutions that are typically present in fragments amplified from damaged aDNA templates [43,44]. Sequences with more than two nucleotide differences were considered as unassigned (unknown plant origin), even if they were identified to family or higher taxonomic levels.
All three proxy analyses were always performed from the same sample slice.
3. Results
(a). Sediment characteristics and chronologies
For the NF interstadial peat layer, a bulk peat sample yielded an uncalibrated 14C age of ca 30 200, and a wood sample of more than 45 000 yr BP. The combined OSL (on adjacent mineral layers; Hel-TL04274) and 14C dating (Hela-2693) procedure provided an approximate minimum (45 kyr BP) and maximum (52 ± 12 kyr) age range for the interstadial layer (table 1). The studied section consists of coarse-grain peaty material probably deposited in an environment where the water level seasonally fluctuated. The material was much less humified than the Holocene peat from the Russian site. Plant remains were well preserved and large pieces, for example, of Equisetum and birch (bark and leaves), and birch seeds with wings still attached were consistently detected.
The top part of the Holocene NER peat section has been eroded away, with 14C datings suggesting an age of ca 5900 cal yr BP for the peat surface, and 8500–5900 cal yr BP for the entire sequence (table 1). The peat was highly humified, and the amount of unidentified organic matter high throughout the section.
(b). Plant macrofossil assemblages
All three NF samples contained large amounts of identifiable vegetative plant remains and were taxonomically rich (NF3 and NF1 also contained several species of fen bryophytes). The plant assemblages indicate a moist minerotrophic environment typical, for instance, of floodplains or shorelines. Some true aquatic species were also detected, indicating the presence of a standing water body.
At the NER site, the plant stratigraphy indicates a gradual succession from an Early Holocene nutrient-rich fen environment—supporting several herb species, tree birch and spruce—towards a nutrient-poor and drier peat–plateau environment, where very few identifiable plant remains were detected. The site was forested at least until ca 6800 cal yr BP, which is in agreement with earlier vegetation and climate reconstructions [21,45]. Cooling triggered permafrost aggradation after 5000 cal yr BP [17,46], but our peat record is lacking for this period, probably owing to associated peat erosion dynamics.
(c). Pollen stratigraphy
Herbaceous taxa dominate the three pollen samples at the NF site, with Cyperaceae, Poaceae, Caryophyllaceae and Ranunculaceae as the most frequent taxa. Tree pollen is dominated by Betula, with Pinus present in minor percentages. Aquatic species and green algae Pediastrum are present in the bottommost sample. The preliminary pollen results suggest that the environment was a wetland surrounded by open birch forest and possibly tundra vegetation.
At the NER site Betula sp., Picea and Cyperaceae are the dominant pollen taxa throughout the sequence, together constituting 80–90% of all pollen. In addition, Equisetum and Filipendula reach very high values of 50–60% in single samples. These peaks likely represent highly localized, massive spore/pollen input and coincide with abundant occurrences of Equisetum vegetative remains and Filipendula ulmaria seeds in the macrofossil record.
(d). Plant DNA analysis
We successfully amplified and sequenced the trnLg/h fragment from six of seven samples (table 1), with extraction and PCR controls remaining clean in seven of 10 PCR batches. We performed 135 PCR amplifications yielding 12 PCR fragments from the older site (NF) and 37 from the younger site (NER) with no bands in the corresponding control blanks. After cloning, we obtained 40–100 colonies per fragment, and we screened and sequenced 623 colonies (ca 89 colonies per fragment). After DNA sequencing and filtering analysis, 99 plant sequences at NF and 235 at NER remained (total 334), of which 318 were assigned to known plant sequences and 16 to unassigned plant sequences (table 2).
Table 2.
Number of DNA sequences obtained from the NF and NER study sites with respective taxonomic assignment.
| NF1 | NF2 | NF3 | NER1 | NER5 | NER10 | NER12 | sum | family | genus | species |
|---|---|---|---|---|---|---|---|---|---|---|
| 67 | 56 | 29 | 5 | 157 | Dicranaceae | Dicranella | D. cerviculata | |||
| 3 | 3 | Equisetaceae | Equisetum | E. fluviatile/E. arvense/ E. sylvaticum | ||||||
| 2 | 2 | Pinaceae | Pinus | P. sylvestris | ||||||
| 1 | 1 | Picea | P. abies | |||||||
| 37 | 1 | 38 | Betulaceae | Betula | B. pubescens/B. nana | |||||
| 36 | 11 | 47 | Nymphaeaceae | Nymphaea/Nuphar | N. tetragona/ | |||||
| N. pumila | ||||||||||
| 3 | 6 | 9 | Ericaceae | Rhododendron | ||||||
| 2 | 2 | Rosaceae | Sorbus | S. aucuparia | ||||||
| 2 | 2 | Anacardiaceae | Anacardium | A. occidentale | ||||||
| 22 | 20 | 42 | Fagaceae | Quercus/Castanopsis/Castanea | ||||||
| 2 | 2 | Solanaceae | Solanum | |||||||
| 5 | 1 | 6 | Capsicum | |||||||
| 7 | 7 | Brassicaceae | Brassica | |||||||
| 16 | 16 | unassigned plant sequences (moss-like) | ||||||||
| 0 | 38 | 61 | 67 | 80 | 39 | 49 | 334 | |||
| 99 | 235 | 334 | ||||||||
(e). The three proxies
At both sites, results from the three proxies recognized 59 taxa within five different plant groups: trees and shrubs, herbs, graminoids, ferns and sporophytes, and aquatic plants. The taxa were identified at different taxonomic levels by the three proxies, from species to family (table 3 and figure 2).
Table 3.
Taxa detected from both study sites (NF + NER), from NF and NER with pollen (P, yellow), macrofossil (M, green) and DNA barcoding (DNA, blue) analysis. An asterisk indicates taxa detected by all three proxies at each site. In blue are reported taxa detected by DNA only and in red those likely originated from contamination. The latter are not included in the total count for DNA.
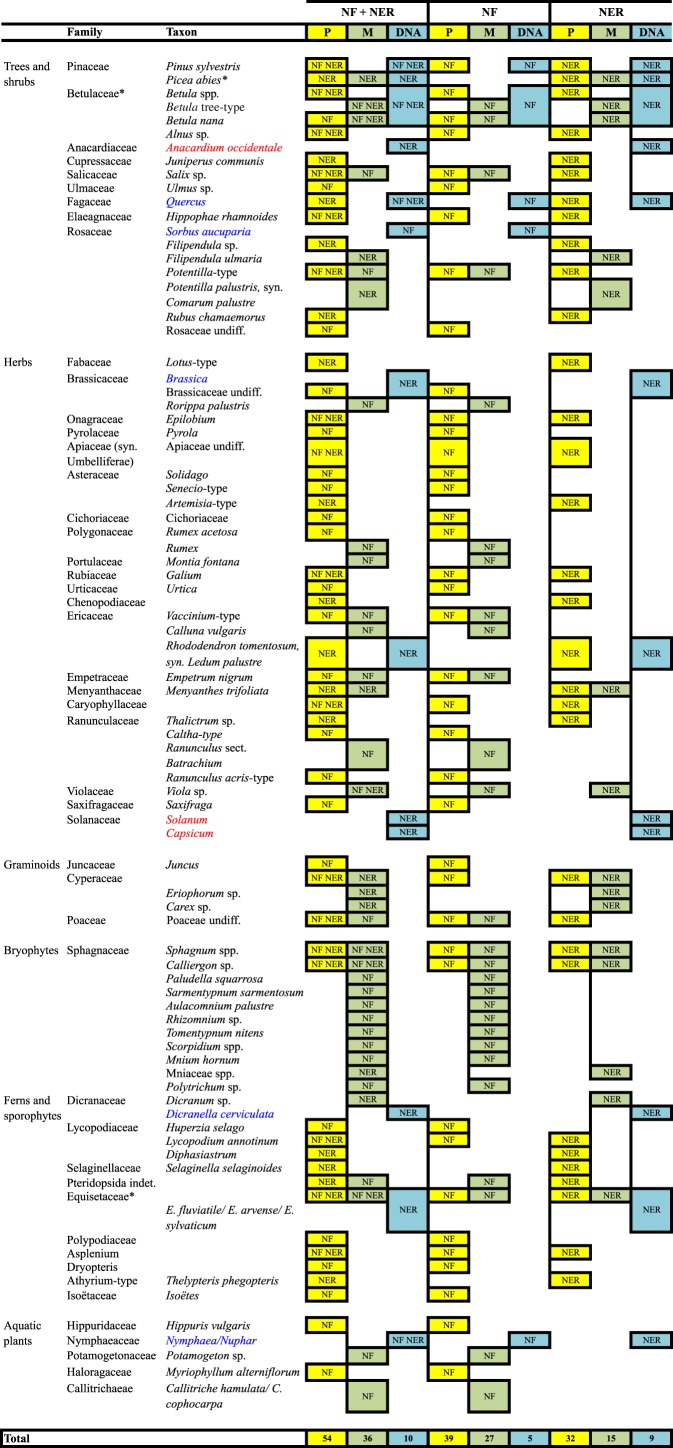 |
Figure 2.
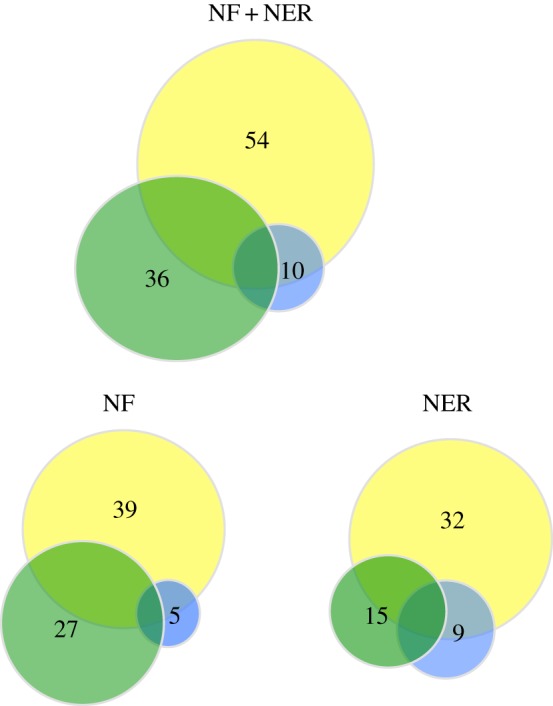
Number of plant taxa detected in peat sediments from Kaarreoja (NF) and Seida (NER) study sites using the three proxies: pollen (yellow), macrofossils (green) and sedDNA (blue). See table 3 for details. (Online version in colour.)
4. Discussion
(a). Authenticity of aDNA sequences
Monitoring for contamination is crucial when working with aDNA. Contamination can never be avoided completely, although it can be controlled and recognized, and it should be always reported. A general consensus in aDNA research has initially been that specific criteria should be used to distinguish recent from aDNA sequences [47]. However, the scientific community also agrees that researchers should explain how their data were obtained and why they are authentic rather than following criteria dictated by others [48]. In this study, in order to minimize pre- and post-excavation contamination, we avoided periods of pollen release of extant vegetation during sample processing; we stored samples in sterile packaging, and we worked in dedicated aDNA facilities during the post-excavation processes (DNA extractions and PCR set-up). Sampling and manipulation of samples for molecular work was performed in clean rooms, using disposable and sterilized tools. DNA extractions were carried out at Centre for GeoGenetics in Copenhagen, whereas amplifications were set up at Uppsala University in facilities specifically designed for aDNA research. Nevertheless, contamination could not be completely avoided and in NER samples we detected six Capsicum and two Anacardiaceae sequences that are likely attributable to contamination from food (the Anacardiaceae family includes food species like cashew (Anarcadium) and mango (Mangifera)) and/or plastic materials used for sterile laboratory consumables (table 2).
Overall, the following lines of evidence suggest our aDNA sequences were authentic. First, there was an inverse relationship between sample age and amplification success (25% PCR success rate in the Weichselian interstadial samples and 42% in the Holocene). Second, the samples behaved consistently in all PCR batches (e.g. NF3 worked consistently better than NF2, and NER1 and the younger sample NER5 better than the older NER10 and NER12). Third, we used sequences only from batches where PCR controls remained clean (seven of 10 batches), and the sequences obtained from the three positive controls showed either no insertion and/or PCR by-products, or no match with those from the sediments (one Persea americana sequence (avocado) and two Nymphaea spp. sequences). With these three exceptions, all plant taxa detected by DNA were identified in modern vegetation surveys and/or were present in our plant database (electronic supplementary material, table S1).
(b). Proxy power and limitations
We can summarize our results as follows: (i) the three proxies gave complementary results with limited overlap; (ii) pollen and macrofossil assemblages included the largest number of taxa; (iii) fewer taxa were identified from DNA, but many were not included in the fossil assemblages; (iv) macrofossils from the NF interstadial samples were better preserved than in the NER Holocene samples, suggesting quicker burial and subsequent anaerobic conditions; (v) taxa identified by DNA were both terrestrial and aquatic and assigned to low taxonomic levels (genera or species); (vi) plant macrofossil data compare well with previous studies from each region, respectively (NF: [26] and NER: [17,46]).
On the whole, our results were similar to those obtained in recent comparative multi-proxy studies from sediments in Siberia [3], Scandinavia [7] and Greenland [5], showing how the DNA proxy offers a complementary tool for identification of taxa not represented in the fossil record (in our case: Sorbus aucuparia, Quercus spp., Brassica spp., Dicranella cerviculata and Nymphaea/Nuphar spp.). Only a few taxa were detected by all three proxies (figure 2), and in our case, DNA shared more taxa with pollen than with macrofossils at both sites. The latter result differs from a previous data comparison based on lacustrine sediments [3] and may indicate that in peat sediments DNA preservation from plant remains is lower. Yet, in many cases, the DNA provided identification at a low taxonomic level (genus or species), whereas many of the taxa found by pollen could be identified at family/group/type level only. DNA also suggested local presence of important taxa such as S. aucuparia at NF (not detected by pollen or macrofossils) and Pinus sylvestris at both sites (detected only by pollen that is, however, often transported over long distances). DNA data, therefore, even if representing a small subset of the total flora present in each period, provided important information that otherwise would go undetected. As such, our findings confirm that barcoding is an important complementary tool that should be used in combination for pollen and macrofossil analyses to achieve a detailed palaeovegetational reconstruction in ancient environments.
(c). Different proxies give different results
Our DNA results from peat sediments seem to be at odds with our previous hypothesis based on lacustrine environments, which proposed that the DNA signal comes principally from plant tissues such as bark, leaves or fruits (macrofossils) rather than from pollen. Instead, our DNA record revealed many strong pollen-producer taxa, and we found a greater overlap between DNA and pollen than between DNA and macrofossils.
The number of taxa we found with the DNA approach compared with what was found with pollen and macrofossils was not high, but comparable to what has been found in recent studies. For the pollen data, our finding of 59 taxa and 33 for the macrofossils was also similar to previous studies. Pollen and macrofossil results were also consistent with previous palaeoecological studies conducted in the same regions (NF [26] and NER [17,46]), showing that at NF the vegetation was dominated by boreal trees, telmatic plants and minerotrophic fen species, and at NER by fen/swamp species with spruce and birch. The DNA record also largely confirmed such flora compositions.
A few Quercus spp. pollen grains were present in the NER pollen samples. Thermophilous species such as Quercus do not currently occur at these latitudes. In addition, previous Holocene pollen records from the same region have detected low amounts of Quercus pollen. However, as small amounts have also been detected from the Late Holocene layers (ca 1500 cal yr BP) [17], a reasonable conclusion has been that pollen represents long-distance dispersal from lower latitudes. On the other hand, the available climate reconstructions from north-eastern European Russia have shown that summer temperatures may have been sufficiently high for Quercus to grow (3°C higher than at present, e.g. [17,23,49,50]) during the earliest part of the Holocene until ca 5000 cal yr BP. In addition, Holocene pollen data recently obtained from the nearby Lake Kharinei showed frequent pollen grains from other thermophilous species, such as Ulmus and Tilia [21] during the mid-Holocene, and because these species, especially Tilia, are mostly insect-pollinated, it was assumed that the source was relatively close. Interestingly, our molecular analysis detected Quercus spp. DNA signals at both study sites (NF3 and NER12; 42–52 kyr and 8400 cal yr BP, respectively). Possible options as to how to interpret this finding are (i) Quercus spp. was growing at the sampling point; (ii) some Quercus remains have been re-deposited from an older deposit located nearby; (iii) DNA comes from pollen; (iv) DNA comes from contamination with modern pollen. We cannot fully exclude contamination, but this seems unlikely for the reasons we presented in §2b,c,g. A local presence of Quercus spp. seems also unlikely as, based on macrofossil data, both sites were wooded fen (peatland/wetland) habitats, which does not seem a natural habitat for this species. In the case of NF, where fluvial activity was apparently present, redeposition of older material or material originating from further distances cannot be ruled out completely, whereas in the NER peatland environment, this is less probable. We conclude, therefore, that at the moment our DNA evidence is not sufficient to suggest the local presence of this species at this latitude and additional DNA and fossil studies are necessary to validate any hypothesis.
(d). Proxy biases and differences
In terms of number of taxa detected, our DNA data differed at the two sites and although overall correspondence between proxies was not very high in both cases, we found a better complementarity at the younger NER site. There are several reasons why the DNA and the traditional palaeoecological approaches produce different results and these have been recently discussed by Parducci et al. [7]. One reason may be that the pollen, macrofossil and DNA originate from different plant tissues with only partial mixing and may even represent different plant communities depending on the study environment that is under investigation (lakes or peat). For example, it may be that plants are better preserved in lakes than in peats (as possibly is their DNA), and therefore we see a greater overlap between macrofossils and DNA proxies in lakes than in peats. A second reason is that barcoding is biased in favour or against certain species owing to primer binding site homology, as the trnL primers are not completely conserved between species (they are quasi-universal). A third important aspect is linked with the taxonomic resolution that also differs between proxies. In the DNA approach, the level of taxonomic resolution varies depending on the availability of trnL sequences in the databases used for assignment.
Taphonomy underlies all palaeoecology (pollen and macrofossils) and, in our opinion, is the underlying factor that influences DNA results from sediments. Unfortunately, we still know little about the decay processes that occur in sedimentary environments. We know that optimal conditions for aDNA survival are cold environments, but we still do not fully understand the correlation between age and DNA content of the sediments and whether such correlation differs in peat versus lake settings. Our results, compared with previous data from lakes, indicate a similar level of DNA preservation for peat and lake sediments and better preservation at younger sites compared with older sites. However, we know little about the physical processes occurring in these sedimentary sequences through time and how these influence the movement of DNA molecules. Vertical movement of water and oxygen are a possibility in unfrozen peat cores such as NF, but not in permafrost peat cores such as NER and our palaeoecological results suggest that the NF site may have been in a fluvial active environment at time of deposition. Nevertheless, it is difficult to reconstruct the post-depositional processes at the NF site as subsequent continental glaciation (during MIS 2) may have eroded away the top part of the original layer.
What is also uncertain is the source of the plant DNA. It is very likely that bulk peat sediments contain DNA from a large variety of plant sources (seed, roots, leaves, fruits, pollen, etc.). However, this and other recent studies seem to provide contrasting results regarding the relative contribution of pollen and macrofossils to the total DNA yield, and how this is preserved. Seeds, needles and bud scales can be blown or washed over long distances [51], especially in open and/or ice-covered landscapes, but do not travel as far as pollen. This has important implications in relation to the possible local origin of the DNA signal. Previous studies from lakes failed to detect DNA from major tree taxa that produce large amounts of pollen (e.g. Picea, Pinus, Corylus, Betula and Alnus) [3,7], thereby agreeing with previous assumptions that DNA is local in origin and does not move over long distances [1,3,14]. Our study on peat, however, showed more similarity between the molecular and the pollen record. This finding, related to the varying levels of decay processes in the different settings and to the different approaches used for taxonomic classification of the DNA sequences, does inevitably cause differences between the taxa recovered at different sites and ages, and consequently also produces differences in results between proxies.
5. Conclusion
Our study largely showed limited overlap between proxies and, therefore, permitted a more thorough investigation of the plant composition in the two study sites. The three proxies also provided different levels of taxonomic resolution and when combined, revealed more detailed information on plant composition than could be achieved by each proxy individually. Overall, this proxy complementarity is important for palaeofloristic reconstructions, particularly as DNA and macrofossil analysis permits detection of local taxa and/or pollen-limited plant taxa that are otherwise under-represented in pollen records and also those that are difficult to identify at species level with pollen. In particular, in stratigraphic sequences in which macrofossils are absent, DNA, when carefully authenticated, may contain an important signal of past flora composition.
Based on these and other previous results from different sediment settings, we conclude that DNA analysis should not be run alone as it is more risky, and results may be biased across settings (the absence of DNA does not necessarily mean the absence of a taxon—it can also be explained by a failure in DNA preservation of a particular species in a particular environment). Rather, we suggest that the best approach to obtain a more complete picture of past vegetation changes is to combine the three proxies considered here (pollen, macrofossils and DNA).
Supplementary Material
Acknowledgements
The authors acknowledge the help and cooperation provided by Eske Willerslev and the laboratory assistance by Kenneth Andersen (Centre for GeoGenetics, Natural History Museum of Denmark, University of Copenhagen). Gold prospectors Antti Kohtamäki and Ami Telilä kindly assisted in the fieldwork in Finnish Lapland. Riitta Kontio is thanked for pollen sample preparation (NF). Dr Dmitry Kaverin and his team are acknowledged for help in coring the NER sequence.
Data accessibility
The pollen, macrofossil and the trnL sequences are available through the Dryad repository (doi:10.5061/dryad.mh981).
Funding statement
This study was supported by the Swedish Research Council (grant 2007-4490) and the Carl Trygger's Foundation (grant 08:303) to L.P.; the Academy of Finland (project 131409) to M.V. and T.R.; the Geological Survey of Finland to I.E.; the Environment Research and Technology Development Fund (S9) of the Ministry of the Environment, Japan, and KAKENHI (nos. 22658046 and 24248025) to Y.S.
Conflict of interests
The authors declare no competing financial interests.
References
- 1.Willerslev E, et al. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795. ( 10.1126/science.1084114) [DOI] [PubMed] [Google Scholar]
- 2.Taberlet P, et al. 2007. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 ( 10.1093/nar/gkl938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jørgensen T, et al. 2012. A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Mol. Ecol. 21, 1989–2003. ( 10.1111/j.1365-294X.2011.05287.x) [DOI] [PubMed] [Google Scholar]
- 4.Willerslev E, et al. 2014. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51. ( 10.1038/nature12921) [DOI] [PubMed] [Google Scholar]
- 5.Pedersen MW, Ginolhac A, Orlando L, Olsen J, Andersen K, Holm J, Funder S, Willerslev E, Kjær KH. 2013. A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quat. Sci. Rev. 75, 161–168. ( 10.1016/j.quascirev.2013.06.006) [DOI] [Google Scholar]
- 6.McLachlan JS, Clark JS, Manos PS. 2005. Molecular indicators of tree migration capacity under rapid climate change. Ecology 86, 2088–2098. ( 10.1890/04-1036) [DOI] [Google Scholar]
- 7.Parducci L, et al. 2013. Molecular- and pollen-based vegetation analysis in lake sediments from central Scandinavia. Mol. Ecol. 22, 3511–3524. ( 10.1111/mec.12298) [DOI] [PubMed] [Google Scholar]
- 8.Birks HH, Birks HJB. 2000. Future uses of pollen analysis must include plant macrofossils. J. Biogeogr. 27, 31–35. ( 10.1046/j.1365-2699.2000.00375.x) [DOI] [Google Scholar]
- 9.Binney HA, et al. 2009. The distribution of late-Quaternary woody taxa in northern Eurasia: evidence from a new macrofossil database. Quat. Sci. Rev. 28, 2445–2464. ( 10.1016/j.quascirev.2009.04.016) [DOI] [Google Scholar]
- 10.Väliranta M, Birks HH, Helmens K, Engels S, Piirainen M. 2009. Early Weichselian interstadial (MIS 5c) summer temperatures were higher than today in northern Fennoscandia. Quat. Sci. Rev. 28, 777–782. ( 10.1016/j.quascirev.2009.01.004) [DOI] [Google Scholar]
- 11.Paus A, Velle G, Berge J. 2011. The Lateglacial and early Holocene vegetation and environment in the Dovre mountains, central Norway, as signalled in two Lateglacial nunatak lakes. Quat. Sci. Rev. 30, 1780–1796. ( 10.1016/j.quascirev.2011.04.010) [DOI] [Google Scholar]
- 12.Anderson-Carpenter L, Mclachlan J, Jackson S, Kuch M, Lumibao C, Poinar H. 2011. Ancient DNA from lake sediments: bridging the gap between paleoecology and genetics. BMC Evol. Biol. 11, 1–15. ( 10.1186/1471-2148-11-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert PDN, Ratnasingham S, de Waard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270, S96–S99. ( 10.1098/rsbl.2003.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parducci L, et al. 2012. Glacial survival of boreal trees in Northern Scandinavia. Science 335, 1083–1086. ( 10.1126/science.1216043) [DOI] [PubMed] [Google Scholar]
- 15.Wood JR, Wilmshurst JM, Wagstaff SJ, Worthy TH, Rawlence NJ, Cooper A. 2012. High-resolution coproecology: using coprolites to reconstruct the habits and habitats of New Zealand's extinct upland moa (Megalapteryx didinus). PLoS ONE 7, e40025 ( 10.1371/journal.pone.0040025.s007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilmshurst JM, Moar NT, Wood JR, Bellingham PJ, Findlater AM, Robinson JJ, Stone C. 2013. Use of pollen and ancient DNA as conservation baselines for offshore islands in New Zealand. Conserv. Biol. ( 10.1111/cobi.12150) [DOI] [PubMed] [Google Scholar]
- 17.Kultti S, Oksanen P, Väliranta M. 2004. Holocene tree line, permafrost, and climate dynamics in the Nenets Region, East European Arctic. Can. J. Earth Sci. 41, 1141–1158. ( 10.1139/e04-058) [DOI] [Google Scholar]
- 18.MacDonald GM, et al. 2000. Holocene treeline history and climate change across northern Eurasia. Quat. Res. 53, 302–311. ( 10.1006/qres.1999.2123) [DOI] [Google Scholar]
- 19.Cremer H, Andreev A, Hubberten HW, Wischer F. 2004. Paleolimnological reconstructions of Holocene environments and climate from Lake Lyadhej-To, Ural Mountains, northern Russia. Arct. Antar. Alp. Res. 36, 147–155. ( 10.1657/1523-0430(2004)036[0147:PROHEA]2.0.CO;2) [DOI] [Google Scholar]
- 20.Väliranta MM. 2006. Long-term changes in aquatic plant species composition in North-eastern European Russia and Finnish Lapland, as evidenced by plant macrofossil analysis. Aquat. Bot. 85, 224–232. ( 10.1016/j.aquabot.2006.05.003) [DOI] [Google Scholar]
- 21.Salonen JS, Seppä H, Väliranta M, Jones VJ, Self A, Heikkilä M, Kultti S, Yang H. 2011. The Holocene thermal maximum and late-Holocene cooling in the tundra of NE European Russia. Quat. Res. 75, 501–511. ( 10.1016/j.yqres.2011.01.007) [DOI] [Google Scholar]
- 22.Jones VJ, et al. 2011. The influence of Holocene tree-line advance and retreat on an Arctic lake ecosystem: a multi-proxy study from Kharinei Lake, North Eastern European Russia. J. Paleolimnol. 46, 123–137. ( 10.1007/s10933-011-9528-7) [DOI] [Google Scholar]
- 23.Salonen VP, Kaakinen A, Kultti S, Miettinen A, Eskola KO, Lunkka JP. 2008. Middle Weichselian glacial event in the central part of the Scandinavian ice sheet recorded in the Hitura pit, Ostrobothnia, Finland. Boreas 37, 38–54. ( 10.1111/j.1502-3885.2007.00009.x) [DOI] [Google Scholar]
- 24.Hättestrand M, Robertsson A. 2010. Weichselian interstadials at Riipiharju, northern Sweden - interpretation of vegetation and climate from fossil and modern pollen records. Boreas 39, 296–311. ( 10.1111/j.1502-3885.2009.00129.x) [DOI] [Google Scholar]
- 25.MacDonald GM, Kremenetski KV, Beilman DW. 2008. Climate change and the northern Russian treeline zone. Phil. Trans. R. Soc. Lond. B 363, 2285–2299. ( 10.1098/rstb.2007.2200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Väliranta M, Sarala P, Eskola T. 2012. Uusia todisteita boreaalisista olosuhteista Veiksel- interstadiaalin aikana. Geologi 64, 1–6. [Google Scholar]
- 27.Helmens KF, Väliranta M, Engels S, Shala S. 2012. Large shifts in vegetation and climate during the Early Weichselian (MIS 5d-c) inferred from multi-proxy evidence at Sokli (northern Finland). Quat. Sci. Rev. 41, 22–38. ( 10.1016/j.quascirev.2012.02.008) [DOI] [Google Scholar]
- 28.Stuiver M, Reimer PJ. 1993. Extended 14C data base and revised Calib 3.0 14C age calibration program. Radiocarbon 35, 215–230. [Google Scholar]
- 29.Reimer PJ, et al. 2013. IntCal13 and MARINE13 radiocarbon age calibration curves 0–50000 years cal BP. Radiocarbon 55, 1869–1887. ( 10.2458/azu_js_rc.55.16947) [DOI] [Google Scholar]
- 30.Heegaard E, Birks HJB, Telford RJ. 2005. Relationships between calibrated ages and depth in stratigraphical sequences: an estimation procedure by mixed-effect regression. Holocene 15, 612–618. ( 10.1191/0959683605hl836rr) [DOI] [Google Scholar]
- 31.The R Development Core Team. 2012. R: a language and environment for statistical computing. See http://www.r-project.org.
- 32.Zabenskie S, Peros M, Gajewski K. 2006. The use of heavy-liquid in the separation of pollen from Arctic lake sediments. Can. Assoc. Palynologists 29, 5–7. [Google Scholar]
- 33.Moore PD, Webb J, Collison ME. 1991. Pollen analysis. Oxford, UK: Blackwell Scientific Publications. [Google Scholar]
- 34.Beug HJ. 2004. Leitfaden der Pollenbestimmung für Mitteleuropa und angrenzende Gebiete. München, Germany: Pfeil. [Google Scholar]
- 35.Bennett KD, Willis KJ. 2001. Pollen. In Tracking environmental change using lake sediments, vol. 3: terrestrial, algal, and siliceous indicators (eds Smol P, Birks HJB, Last WM.), pp. 1–28. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- 36.Bennett KD. 2004. Pollen catalogue of the British Isles See http://chrono.qub.ac.uk/pollen/pc-intro.html.
- 37.Karlsson T. 1997. Förteckning over svenska kärlväxter. The vascular plants of Sweden: a checklist. Sven Bot Tidskrift 91, 241–560. [Google Scholar]
- 38.Willerslev E, Hansen AJ, Poinar HN. 2004. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 19, 141–147. ( 10.1016/j.tree.2003.11.010) [DOI] [PubMed] [Google Scholar]
- 39.Haile J, et al. 2009. Ancient DNA reveals late survival of mammoth and horse in interior Alaska. Proc. Natl Acad. Sci. USA 106, 22 352–22 357. ( 10.1073/pnas.0912510106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Sym. Ser. 41, 1–4. ( 10.1021/bk-1998-0710.ch001) [DOI] [Google Scholar]
- 41.Dabney J, Meyer M, Pääbo S. 2013. Ancient DNA damage. Perspect. Biol. a012567 ( 10.1101/cshperspect.a012567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sønstebø JH, et al. 2010. Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Res. 10, 1009–1018. ( 10.1111/j.1755-0998.2010.02855.x) [DOI] [PubMed] [Google Scholar]
- 43.Hansen AJ, Willerslev E, Wiuf C, Mourier T, Arctander P. 2001. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 18, 262–265. ( 10.1093/oxfordjournals.molbev.a003800) [DOI] [PubMed] [Google Scholar]
- 44.Binladen J, et al. 2005. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics 172, 733–741. ( 10.1534/genetics.105.049718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Väliranta M, Kaakinen A, Kuhry P, Kultti S, Salonen JS, Seppä H. 2011. Scattered late-glacial and early Holocene tree populations as dispersal nuclei for forest development in north-eastern European Russia. J. Biogeogr. 38, 922–932. ( 10.1111/j.1365-2699.2010.02448.x) [DOI] [Google Scholar]
- 46.Väliranta M, Kaakinen A, Kuhry P. 2003. Holocene climate and landscape evolution east of the Pechora Delta, East-European Russian Arctic. Quat. Res. 59, 335–344. ( 10.1016/S0033-5894(03)00041-3) [DOI] [Google Scholar]
- 47.Cooper A, Poinar HN. 2000. Ancient DNA: do it right or not at all. Science 289, 1139 ( 10.1126/science.289.5482.1139b) [DOI] [PubMed] [Google Scholar]
- 48.Gilbert MTP, Bandelt H-J, Hofreiter M, Barnes I. 2005. Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544. ( 10.1016/j.tree.2005.07.005) [DOI] [PubMed] [Google Scholar]
- 49.Oksanen PO, Kuhry P, Alekseeva RN. 2001. Holocene development of the Rogovaya River peat plateau, European Russian Arctic. Holocene 11, 25–40. ( 10.1191/095968301675477157) [DOI] [Google Scholar]
- 50.Andreev AA, Tarasov PE, Ilyashuk BP, Ilyashuk EA, Cremer H, Hermichen WD, Wischer F, Hubberten H-W. 2005. Holocene environmental history recorded in Lake Lyadhej-To sediments, Polar Urals, Russia. Palaeogeogr. Palaeoclim. Palaeoecol. 223, 181–203. ( 10.1016/j.palaeo.2005.04.004) [DOI] [Google Scholar]
- 51.Birks HH, Bjune AE. 2010. Can we detect a west Norwegian tree line from modern samples of plant remains and pollen? Results from the DOORMAT project. Veg. History Archaeobot. 19, 325–340. ( 10.1007/s00334-010-0256-0) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pollen, macrofossil and the trnL sequences are available through the Dryad repository (doi:10.5061/dryad.mh981).


