Abstract
DNA obtained from environmental samples such as sediments, ice or water (environmental DNA, eDNA), represents an important source of information on past and present biodiversity. It has revealed an ancient forest in Greenland, extended by several thousand years the survival dates for mainland woolly mammoth in Alaska, and pushed back the dates for spruce survival in Scandinavian ice-free refugia during the last glaciation. More recently, eDNA was used to uncover the past 50 000 years of vegetation history in the Arctic, revealing massive vegetation turnover at the Pleistocene/Holocene transition, with implications for the extinction of megafauna. Furthermore, eDNA can reflect the biodiversity of extant flora and fauna, both qualitatively and quantitatively, allowing detection of rare species. As such, trace studies of plant and vertebrate DNA in the environment have revolutionized our knowledge of biogeography. However, the approach remains marred by biases related to DNA behaviour in environmental settings, incomplete reference databases and false positive results due to contamination. We provide a review of the field.
Keywords: environmental DNA, ancient, environment, ancient DNA, review
1. Introduction
For over a decade, researchers have exploited the fact that environmental DNA (eDNA) derives not just from microbes, but from a wide range of organisms, including plants and vertebrates. A large proportion of the ancient flora and fauna do not fossilize, but leave extracellular DNA traces in the sediments. In a pioneering 2003 study, sediments from Siberia and New Zealand were found to contain traces of DNA from extinct animals, such as the woolly mammoth and moa birds [1]. The study showed that modern plant DNA could also be recovered from surface soil. The same year, another team reported the retrieval of DNA from the extinct giant ground sloth and other Pleistocene animals from a dry cave in the southwest US [2]. Since then, several studies of both past and present biodiversity have been published using eukaryotic eDNA recovered from a variety of settings including basal ice [3–5] and lake cores [6–10], surface soils [11], cave sediments [12,13], and water from lakes, streams [14–16] and oceans [17,18] (figures 1 and 2). Importantly, studies have revealed that eDNA data and other proxies such as pollen, macrofossils, living mammals and plants seem to complement each other showing wider diversity of species than using the methods separately [9–11,20–22]. Therefore, eDNA should be viewed as a complementary, rather than alternative, approach to assays of more traditional environmental proxies. Here, we discuss the experimental and bioinformatics challenges facing eDNA and provide examples of its uses for addressing biological questions.
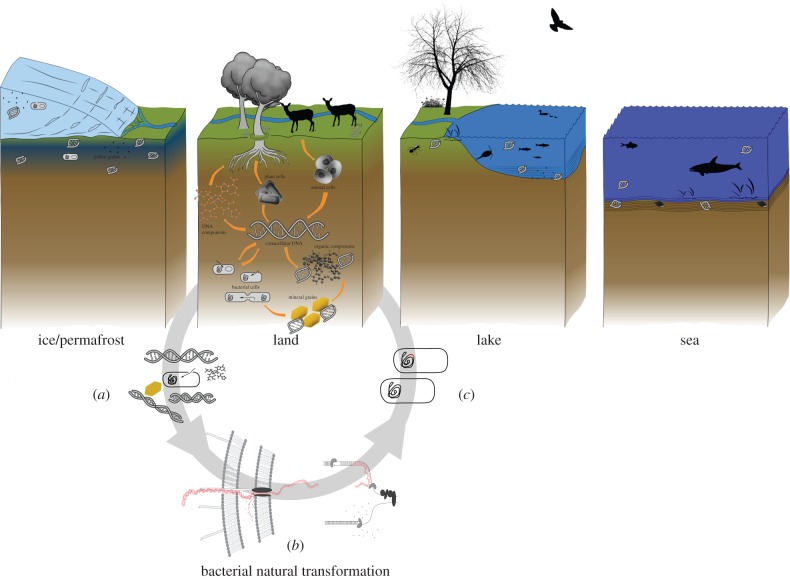
Figure 1.
Environments where eDNA of plants and/or animals have been reported: basal glacier ice, terrestrial sediments, lake, rivers and lake sediments, and ocean water. The eDNA comes mainly from plant fine rootlets, faeces, urine and skin cells. The eDNA can remain in the cells, or be released from the cells in which case it may bind to inorganic particles that protect the DNA from microbial and spontaneous chemical degradation. Extracellular DNA may also be incorporated into the genomes of bacteria (bacterial natural transformation of short and degraded DNA). (a) The last may happen when extracellular DNA meets a bacterium's surface and crosses the outer cell wall via protruding structures named pili. At the inner membrane, one strand of DNA is transported into the cell while the opposite DNA strand is degraded. (b) Once inside the cell, the DNA fragment may encounter the bacterial genome and binding at a single-stranded region during genome replication. (c) When the two new genomes segregate, one of the daughter-cells carries the inserted environmental DNA sequence.
Figure 2.
Geographical distribution of sites where studies have investigated eDNA (adapted from [19]). For references corresponding to numbers, see the electronic supplementary material.
2. Origins and behaviour of environmental DNA
The origins and behaviour of eDNA are still poorly understood. It appears that eDNA can be deposited through skin flakes [23], urine [24], faeces [25,26], eggshells [27], hair [28,29], saliva [30], insect exuviae [31], regurgitation pellets [32], feathers [33], leaves [34,35], root cap cells, in rare cases pollen [9,36], or in living prokaryotes through the secretion of plasmid and chromosomal DNA [37] (figure 1). From bacterial and plant studies, evidence exists that dead cells entering the environment may quickly be lysed with their DNA immediately being released [38]. Upon release into the environment, the DNA molecule has three possible fates.
(a). Metabolism by bacterial and fungal exonucleases
Following its release into the environment, DNA becomes vulnerable to bacterial and fungal DNases, with the former commonly believed to be the primary mechanism for extracellular DNA degradation in the environment [39].
(b). Persistence in the environment
DNA survival can be helped through the binding to environmental compounds such as clay minerals, larger organic molecules and other charged particles, which shields the adsorbed DNA from nuclease activity [40] (figure 1). Binding of nucleases also inhibits their ability to hydrolyse extracellular DNA [39]. For example, clay minerals such as Montmorillonite can absorb more than their own weight in DNA, because of their relatively large negatively charged surface area [41–44]. Furthermore, humic acids, of which some are resistant to decay, also bind DNA molecules due to a negative surface charge, and therefore prolong DNA survival. Similarly, DNA in preserved animal guts and faeces is protected from degradation by absorption to humic acids and other organic molecules. Compared with clays, sand has been found less effective in binding DNA, the primary explanation being its small surface area. However, adsorption to sand is possible and increases with cation concentrations—particularly of divalent cations such as Ca2+ and Mg2+, which are most effective at forming sand–DNA bridges [45].
(c). Natural transformation
Natural transformation is a process through which cells take up extracellular DNA from the surroundings and integrate it into their own genomes [46,47]. Many bacteria are known to be agents for natural transformation, as are some archaea and even a eukaryotic group of micro-invertebrates, the bdelloid rotifers [48–51]. The majority of DNA that microbes take up is quickly degraded and re-metabolized in the cell, but some DNA persists for long enough to recombine with the host genome [52]. Classical natural transformation is efficient with kilobase-long DNA, but recently it has been shown that very short DNA fragments, down to 20 bp long, remain available for integration into the bacterial genome, even when severely damaged (figure 1) [52]. Although the integration depends on similarity between bacteria and source DNA, the authors succeeded in incorporating woolly mammoth mtDNA fragments, albeit after genetically modifying the bacteria to resemble mammoth mtDNA.
In general eDNA, in particular that from ancient samples, is extremely fragmented and chemically modified with abasic sites, deaminated cytosines and cross-links [52–58]. DNA half-life is a complex function of the interplay between the physical, chemical and biological properties of the microenvironment. Turnover time of eDNA in both sea and freshwater was originally thought to be very rapid, just 6.5–25 h [59,60], but more sensitive approaches have shown survival for up to several weeks [16,17,35,61]. By contrast, in soils and sediments, moa DNA from 3000 years (kyr) old dry temperate sediments has been recovered [12], mammoth DNA dating to 30 kyr BP from permafrost sediments has been amplified, as well as 400–600 kyr old plant DNA [1] and approximately 0.5 million year old DNA from glacial basal ice [3] (figure 2).
Most eDNA studies rely on the assumption that the age of the DNA molecule recovered is the same as the age of the sediments in which it is found, but in certain conditions DNA molecules can leach through the strata and contaminate lower layers [12]. With regard to this point, DNA leaching in permanently frozen soil (permafrost) or in sediments recently frozen has not been observed [62,63]. However, in sediments from both temperate and desert environments, leaching has been reported [12,20,64] and must be taken into account as a possible concern [12,64]. In our view, DNA leaching is not the most challenging issue for proper dating of eDNA, rather it is re-deposition of sediments carrying eDNA molecules with them. Therefore, it is crucial for ancient eDNA studies to be supported by good geological profiling, providing evidence of a site's geological stratigraphy and depositional history [65].
3. Experimental design
(a). Sampling and handling of samples for environmental DNA studies
Given the relatively low number of endogenous molecules of DNA from higher organisms in most environmental samples, contamination remains among the greatest experimental challenges to the field. Currently, several strategies for taking eDNA samples exist for aquatic systems [16,61,66,67], lake sediments [10,13,68], permafrost soil [69,70] and ice [1,53,71–74]. The use of trace substances, such as unique plasmid DNA, smeared on exposed surfaces and equipment, represents an efficient means of determining whether contaminants have penetrated inside the sample during sampling, transport, storage or subsequent subsampling [69].
For downstream analyses, samples with ancient DNA must be handled in appropriately designed laboratories divided into a pre- and post-PCR environment to reduce carry-over contamination. For ancient eDNA studies, these should be physically separated, and the former equipped with nightly UV irradiation of surfaces and positive air pressure [75,76]. Bleach and CoPA solution (a copper-bis-(phenanthroline)-sulfate/H2O2 solution, US patent number 5858650) is most efficient when decontaminating surfaces, gloves and equipment [77]. Other DNA decontaminating products such as RNAse away (Molecular Bioproducts) and other detergents are less effective, but in combination with UV-irradiation serve may as a non-corrosive alternative for equipment sensitive to bleach. Carry-over contamination can be limited by wearing gloves, masks and full-body suits [77]. Blank controls are crucial for identifying laboratory contamination, but are not 100% reliable, due to low levels of sporadic contamination and carrier effects [76,78]. Blank controls are likely sufficient for controlling contamination from certain species that are likely showing up only by previously produced amplicons. For other taxa, contaminants can be difficult to distinguish from endogenous DNA. For example, DNA contaminants from various sources are found in reagents [10,21,77–82]. Although most of these are from readily identified domesticated animals or cultivated plants, others such as Salix [83] are not and can be mistaken for genuine environmental diversity. We stress the importance of controls for each new reagent stock and systematically keeping track of these, especially now that the massive throughput of next generation sequencing (NGS) platforms makes it possible to sequence even traces of contamination. For example, commercial PCR primers were recently found to be contaminated with plant DNA (K. Andersen 2013, personal communication). Studies on eDNA using NGS technology have probably overlooked the magnitude of this problem (including our own group). Therefore, recent attempts to compile contamination databases of control sequences are extremely welcome [84].
(b). DNA extraction of environmental samples
The high level of biological complexity in environmental samples makes unbiased extractions a major challenge. The ability to extract the DNA from samples with equal efficiencies seems unlikely, considering the wide range of sample types. Currently, no generic extraction method performs equally well across all environments or taxonomic groups [85–91]. However, numerous commercial and custom extraction protocols have been adjusted to handle different combinations of sample types and organisms. Some of these are generic and have successfully been used for eDNA studies in lakes, ancient sediments and ice [1,4,12,16,17,71,92,93], although a better understanding of extraction bias will benefit the field tremendously.
Inhibition of proteinase, DNA polymerase and DNA ligase activities can preclude eDNA analyses [94]. Several strategies have been developed to identify and overcome this problem: (i) DNA spiking to gauge the presence of inhibitors [95,96], (ii) DNA extract dilution to reduce inhibition, (iii) additional purification (phenol–chloroform, silica-based columns) to remove inhibitors, and (iv) incapacitating the inhibitors by using enzyme facilitators that bind lipids, phenols and other organic inhibitors such as BSA, RSA, Tween20, PEG 400 and Gp32 [94,97].
(c). Generic versus specific primers
Metabarcoding uses generic (or universal) primers, which are designed to target several taxa simultaneously [98–102], in contrast to specific primers, which are designed to amplify only a few selected species. The advantage of using generic primers is the simultaneous amplification of a multitude of taxa and detection of new unexpected taxa. The biggest caveat when using generic primers is that the results might be skewed towards preferential amplification of certain taxa, while others (in particular rare taxa) remain undetected [9,99,101,103]. This problem results from (i) interspecific differences in decaying processes of tissue and DNA, (ii) primer-binding biases due to target sequences not matching equally well to primers [102], (iii) PCR stochasticity, and (iv) inhibition. One disadvantage of specific primers in multispecies surveys is the need for larger volumes of DNA templates, which are often in limited supply in eDNA settings. Therefore, in some cases, generic- and species-specific primers may be used in combination to maximize diversity resolution, as the two approaches may detect non-overlapping taxa [101,104]. Enrichment approaches for specific loci, possibly targeting a range of taxonomic groups simultaneously, might in the future provide a solution to such problems [105,106].
(d). Sequence-to-sample misidentification
To increase overall data output during NGS-based analyses, eDNA can be PCR amplified using unique combinations of 5′-nucleotide-tagged primers, that enable subsequent pooling of amplicons originating from different samples [107]. Originally developed for the FLX platform, subsequent studies explored their use on Illumina platforms—although in this case problems were observed arising from tag recombination during the library amplification steps. This problem has also been observed in non-metabarcoding studies. Specifically, using Illumina sequencing and double-indexing, Kircher et al. [108] reported a significant fraction of sequencing reads with unused combinations of indexes. They identified two major causes: (i) cross-contamination of oligonucleotides carrying different indexes and (ii) chimaera formation in which indexed templates from one library recombine with those from other libraries (‘jumping PCR’) in experiments where multiple sequencing libraries were amplified in bulk. Although unused index-combinations are easily identified, recombination that creates false, but already used index-combinations may introduce significant levels of sample misidentification.
There are several solutions to recognize and/or minimize sequence-to-sample misidentifications: (i) reducing the number of cycles during PCR indexing, (ii) generating a number of PCR replicates of the same sample using different combinations of 5′-nucleotide-tagged primers for each replicate and only keeping sequences consistent across a majority of PCRs (which also reduces sequencing errors), and (iii) using tags that are unique in both ends of the sequence to allow rapid identification of those not used in the study. Even though studies have already looked into the causes and solution to jumping PCR, PCR stochasticity, and PCR-induced artefacts, their respective importance still needs to be tested to optimize how the sequencing output reflects the true diversity present in different environments.
(e). Processing next generation sequencing data and assigning sequences to taxa
Traditional genetic barcodes used in conventional (i.e. non-eDNA) projects exploit DNA barcodes of more than 500 bp in length. Barcodes of such length are inappropriate for eDNA analyses, as the eDNA is often fragmented into less than 150 bp pieces [109]. Therefore, sequence primers targeting short phylogenetically informative regions such as the trnL/rbcL genes [110,111], the 12S rRNA [112,113], 16S rRNA genes [114] and internal transcribed spacers [115,116] have been developed to survey ancient plant, animal, bacterial and fungal diversity. The ecoPrimers software [117] and the PrimerProspector package [118] have proved useful for achieving successful primer design [119–122].
Similar to the challenge of sequencing errors, single base substitutions introduced during PCR, and PCR-derived chimaera formation, can affect the taxonomic identification process. Thus, distinguishing these effects from true biological sequence variation is essential. Different denoising procedures have been developed to do this, initially based around 454/FLX pyrosequencing reads (Life Sciences, Roche), such as PyroNoise [123], Denoiser [124] and Amplicon Noise [125], and for chimaera detection, such as Uchime [126]. Procedures tailored to Illumina platforms, which are more cost-effective per base [127], have also emerged. Caporaso et al. [120] developed a 16S rRNA amplicon sequencing protocol for MiSeq and HiSeq platforms. Paired-end Illumina sequencing of 16S rRNA amplicons was compared to single-end sequencing and was found to increase the detected α-diversity of microbial communities, without affecting the resolution of phylogenetic clustering. A range of additional tools are available to help process NGS data, such as OBITools (http://www.grenoble.prabi.fr/trac/OBITools/) and QIIME [128], which can both handle data from multiple pooled samples.
With regard to taxonomic identification, one of the most popular tools for analysing metagenomic data is MEGAN [129], software that originally used BLAST to infer taxonomic composition. However, BLAST searching does not represent the most appropriate method for metagenomic sequence assignment. This is because alignments are local and not global, and hit similarities provide a measure of the confidence in the local sequence similarity but not of the validity of the assignment per se [130]. Input formats other than BLAST are now compatible with the latest version of the program (MEGAN 5), such as SAM files and QIIME output [131].
Alternative approaches based on phylogenetic placement have been developed, where databases are first screened for orthologues showing significant sequence similarity. Following sequence alignment, Bayesian phylogenetic trees are reconstructed and the query sequence assigned to the highest taxonomic level shared with all members of the smallest supported monophyletic clade to which it belongs. Posterior probability clade support is used as a direct measure of assignment significance [132]. For COI insect and trnL plant sequences, this approach was found to outperform BLAST both in sensitivity and specificity [132]. As the Bayesian framework is computationally intensive and incompatible with the size of NGS datasets, a heuristic approach has been introduced with no apparent loss in sensitivity. This approach is based on neighbour-joining trees and non-parametric bootstrapping for an evaluation of node robustness [132]. We acknowledge the fact that species absent from the database represent an important drawback of this method, as large portions of the biodiversity remain uncharacterized. Using a promising approach based on fuzzy theory and COI sequence data, Zhang et al. [133] have shown that this problem could potentially be addressed during the analyses. Despite this, building a good-quality reference sequence database, properly curated and even including taxonomically validated samples, still represents an essential component of all metabarcoding projects [102].
An important bottleneck observed in previous analyses is the necessity to align query sequences that often number in the millions, against orthologues. Aligning query sequences against a predefined template has provided an efficient solution to this problem. Fast methods based on a diversity of approaches, such as hidden Markov model profiles from the reference alignment, or phylogenetically aware strategies [134], have been proposed [135,136]. The nearest alignment space termination (NAST) procedure [137] is another such approach where the template sequence most similar to the query sequence is first identified using BLAST [138] and then pairwise realigned to the query sequence. Gap spacing originally present in the template alignment is then reintroduced in the pairwise alignment, generating a full global multi-alignment. The NAST procedure is provided with the QIIME software [120,128], which is compatible with Sanger, 454 and Illumina data and performs a full range of analyses for metabarcoding DNA sequences, including operational taxonomic unit (OTU) identification [139,140], α- and β-diversity measurements and clustering methods and UniFrac distances [141]. UniFrac distances are based on the fraction of the total branch length that is shared among samples and reflect how much environments/samples are taxonomically similar. This approach has shown promising results in assessing the microbial taxonomic proximity across environments [142–163] and also in monitoring changes in the human oral microflora following the Neolithic revolution and industrial revolution, in response to major changes in carbohydrate consumption [164]. With the growing availability of environmental metagenomic datasets, SourceTracker [165] appears to be a useful tool that can authenticate DNA profiles, for example, by showing different sources for the samples and their respective negative controls, or by matching samples with their expected tissue source [166].
With ever-reducing sequencing costs, shotgun sequencing now provides an alternative approach to metabarcoding for determining taxonomic profiles. Reads are first aligned to annotated reference genomes or clade-specific [167]/universal [168] markers, and taxon relative abundances can be estimated with appropriate normalization by genome size [169–172]. Such taxonomic profiles are not affected by biases typical of amplicon-based profiles, such as copy-number variation across taxonomic groups [173], target amplification efficiency variability [174] and single marker reliability [175].
The specificity of reference markers for shotgun profiling also limits biases related to evolutionary uninformative conserved regions and horizontal gene transfer [167,172,176,177]. Shotgun profiling is, however, hindered by computational constraints associated with the size of the datasets analysed. With the program MetaPhlAn [167], the speed of read assignment was increased 50-fold compared with commonly employed methods such as PhymmBL [178], BLAST [138], RITA [179] and NBC [180]. The large fraction of taxa present in the environment, but not represented in databases is still problematic, as shown by analyses performed with mOTU [168], which estimated that current databases are only able to detect 43% of species abundance and 58% of richness present in clinical samples of faeces [168].
Shotgun datasets also contain comprehensive and useful information relating to the biological functions used in environmental communities [181]. By using alignment tools such as BLASTX [138], metagenomic reads are aligned to databases of proteins such as NCBInr, KEGG [182], EGGnog [183] or SEED [184], and functional profiles can be analysed in MEGAN [129]. Finally, reference-free alternative approaches based on k-mer counts [185] have also proved to be 860 times faster than BLASTX, with comparable sensitivity and precision, but without loss of accuracy [186].
4. Environmental DNA case studies
(a). Soil, terrestrial sediments and basal ice
Soil and terrestrial sediments represent the most studied eDNA source (figure 2), and recent studies on surface sediments demonstrate that eDNA mirrors the diversity of terrestrial plants [11] and mammals [20] both qualitatively, and to some extent, quantitatively [11,20]. Ancient sediment has revealed the persistence of Late Quaternary megafauna for much longer timespans than their commonly surmised extinction times [19]. This demonstrated the power of eDNA approaches that target short molecular signatures in contrast to palaeontological analyses that require preservation of macrofossils to firmly establish the presence of a given species at a given time period.
Ancient eDNA analyses of permafrost samples distributed across the whole Arctic have provided the largest historical record of vegetation changes over the past 50 kyr [83]. Here, the authors found evidence for a diverse, but rather stable Arctic vegetation dominated by forbs until around the last glacial maximum (LGM), some 20 kyr ago where the diversity declined drastically. As the climate became warmer, a vegetation turnover was detected until the ecosystem was completely dominated by bushes and grass and depleted in forbs. Interestingly, the stomach content and faeces of Arctic megafaunal species revealed a large fraction of forbs in their diet, suggesting that the transition from a forb-dominated to a grass-dominated steppe might have contributed to the massive decline of megafaunal populations after the LGM.
In 1999, the first eDNA study was conducted on ice cores (but on microbial eukaryotes rather than higher organisms) and revealed algae and fungi diversity in the Hans Tausen ice core of northern Greenland [4]. Since then, DNA in basal ice from the DYE-3 ice core of southern Greenland revealed a diverse conifer forest with a full diversity of insects different from those found in Greenland today [3]. By dating this reconstructed environment to beyond the last interglacial period (Eemian 130–115 kyr ago [187]), the authors questioned the common belief at the time, that southern central Greenland was ice-free during the Eemian. Pollen records from a marine sediment core off the south coast of Greenland further supported this claim [188] (figures 1 and 2).
(b). Marine and freshwater
Environmental DNA extracted from contemporary aquatic samples provides a good proxy of the biodiversity in and around the water (figure 2). This was first shown in freshwater ecosystems [14] with the molecular detection of the American bullfrog (Rana catesbeiana) in French wetlands. In subsequent studies, others successfully detected eDNA from invasive and low abundance species, including amphibians [16,67,189–191], fishes [15,16,192–194] and snails [195], but also from endangered amphibians, fishes, mammals and insects [16]. Furthermore, using a quantitative study design, species-specific eDNA concentrations have been found to reflect animal density [16]. The same study also demonstrated that coupling eDNA with high-throughput sequencing can account for entire lake faunas of amphibians and fishes [16], providing cost-effective approaches to monitor biodiversity.
Recently, two studies showed that seawater is also a source of macro-organismal eDNA for detection of whale species [18] and marine fish diversity [17] (figure 2). Importantly, eDNA from fresh and seawater appears to reflect contemporary rather than past diversity, as eDNA decays within a few days or weeks in the water column [16,17,61,196,197].
(c). Lake cores
Lake sediments have traditionally been used for pollen records, but have now been found to contain DNA from fishes [6], mammals [198] and plants [7–10]. This source of information was not only used to infer past human/environment interaction but also addressed a long-lasting controversy in bio-geography: whether spruce survived in Scandinavia ice-free refugia during the last glaciation [8]. Two distinct mtDNA haplogroups were found in present-day Norwegian spruce, of which one is common both in and outside Scandinavia. The other is only known in Scandinavia and could represent the signature of survival in a refugium during the LGM. This was confirmed using eDNA from lake cores in areas shown to have remained ice-free during the LGM, with evidence of spruce DNA including the rare mitochondrial haplogroup.
5. Future of environmental DNA
Among the greatest benefits of eDNA is that it reduces costs and time associated with conventional bio-surveys, such as man-hours, field-training, equipment, permits, safety issues and handling of organisms. At the same time, it provides a means for undertaking large-scale biodiversity comparisons across both time and space. As such, the field of eDNA promises to revolutionize areas of archaeology, ecology and conservation [199]. The next step will be moving from metabarcoding approaches to true metagenomics. With increasing genome data being generated, this should soon be feasible and will allow for better species identifications and quantitative estimates of their abundances in environmental settings. Importantly, however, although the young field of eDNA appears to have a promising future, we emphasize that further basic studies are needed before its potential and limitations are fully explored.
Supplementary Material
Acknowledgements
The authors thank Prof. Kurt H. Kjær for help with figure 1, and Andrea Torti, Mark Lever and Kenneth Andersen for help with the manuscript.
Funding statement
The Danish National Research Foundation supported this work.
References
- 1.Willerslev E. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795. ( 10.1126/science.1084114) [DOI] [PubMed] [Google Scholar]
- 2.Hofreiter M, Mead JI, Martin P, Poinar HN. 2003. Molecular caving. Curr. Biol. 13, R693–R695. ( 10.1016/j.cub.2003.08.039) [DOI] [PubMed] [Google Scholar]
- 3.Willerslev E, et al. 2007. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science 317, 111–114. ( 10.1126/science.1141758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willerslev E, Hansen A, Christensen B, Steffensen JP, Arctander P. 1999. Diversity of Holocene life forms in fossil glacier ice. Proc. Natl Acad. Sci. USA 96, 8017–8021. ( 10.1073/pnas.96.14.8017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould BA, Leon B, Buffen AM, Thompson LG. 2010. Evidence of a high-Andean, mid-Holocene plant community: an ancient DNA analysis of glacially preserved remains. Am. J. Bot. 97, 1579–1584. ( 10.3732/ajb.1000058) [DOI] [PubMed] [Google Scholar]
- 6.Matisoo-Smith E, Roberts K, Welikala N, Tannock G, Chester PI, Feek DT, Flenley JR. 2008. Recovery of DNA and pollen from New Zealand lake sediments. Quat. Int. 184, 139–149. ( 10.1016/j.quaint.2007.09.013) [DOI] [Google Scholar]
- 7.Anderson-Carpenter LL, McLachlan JS, Jackson ST, Kuch M, Lumibao CY, Poinar HN. 2011. Ancient DNA from lake sediments: bridging the gap between paleoecology and genetics. BMC Evol. Biol. 11, 30 ( 10.1186/1471-2148-11-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parducci L, et al. 2012. Glacial survival of boreal trees in northern Scandinavia. Science 335, 1083–1086. ( 10.1126/science.1216043) [DOI] [PubMed] [Google Scholar]
- 9.Parducci L, et al. 2013. Molecular- and pollen-based vegetation analysis in lake sediments from central Scandinavia. Mol. Ecol. 22, 3511–3524. ( 10.1111/mec.12298) [DOI] [PubMed] [Google Scholar]
- 10.Pedersen MW, Ginolhac A, Orlando L, Olsen J, Andersen K, Holm J, Funder S, Willerslev E, Kjær KH. 2013. A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quat. Sci. Rev. 75, 161–168. ( 10.1016/j.quascirev.2013.06.006) [DOI] [Google Scholar]
- 11.Yoccoz NG, et al. 2012. DNA from soil mirrors plant taxonomic and growth form diversity. Mol. Ecol. 21, 3647–3655. ( 10.1111/j.1365-294X.2012.05545.x) [DOI] [PubMed] [Google Scholar]
- 12.Haile JS, et al. 2007. Ancient DNA chronology within sediment deposits: are paleobiological reconstructions possible and is DNA leaching a factor? Mol. Biol. Evol. 24, 982–989. ( 10.1093/molbev/msm016) [DOI] [PubMed] [Google Scholar]
- 13.Haouchar D, Haile J, McDowell MC, Murray DC, White NE, Allcock RJN, Phillips MJ, Prideaux GJ, Bunce M. 2014. Thorough assessment of DNA preservation from fossil bone and sediments excavated from a Late Pleistocene–Holocene cave deposit on Kangaroo Island, South Australia. Quat. Sci. Rev. 84, 56–64. ( 10.1016/j.quascirev.2013.11.007) [DOI] [Google Scholar]
- 14.Ficetola GF, Miaud C, Pompanon F, Taberlet P. 2008. Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. ( 10.1098/rsbl.2008.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jerde CL, Mahon AR, Chadderton WL, Lodge DM. 2011. ‘Sight-unseen’ detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. ( 10.1111/j.1755-263X.2010.00158.x) [DOI] [Google Scholar]
- 16.Thomsen PF, Kielgast J, Iversen LL, Wiuf CC, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E. 2011. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 21, 2565–2573. ( 10.1111/j.1365-294X.2011.05418.x) [DOI] [PubMed] [Google Scholar]
- 17.Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E. 2012. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7, e41732 ( 10.1371/journal.pone.0041732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foote AD, et al. 2012. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7, e41781 ( 10.1371/journal.pone.0041781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haile JS, et al. 2009. Ancient DNA reveals late survival of mammoth and horse in interior Alaska. Proc. Natl Acad. Sci. USA 106, 22 352–22 357. ( 10.1073/pnas.0912510106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen K, et al. 2011. Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 21, 1966–1979. ( 10.1111/j.1365-294X.2011.05261.x) [DOI] [PubMed] [Google Scholar]
- 21.Jørgensen T, et al. 2011. A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Mol. Ecol. 21, 1989–2003. ( 10.1111/j.1365-294X.2011.05287.x) [DOI] [PubMed] [Google Scholar]
- 22.Pawłowska J, Lejzerowicz F, Esling P, Szczucinski W, Zajączkowski M, Pawlowski J. 2014. Ancient DNA sheds new light on the Svalbard foraminiferal fossil record of the last millennium. Geobiology 12, 277–288. ( 10.1111/gbi.12087) [DOI] [PubMed] [Google Scholar]
- 23.Bunce M, Szulkin M, Lerner HRL, Barnes I, Shapiro B, Cooper A, Holdaway RN. 2005. Ancient DNA provides new insights into the evolutionary history of New Zealand's extinct giant eagle. PLoS Biol. 3, e9 ( 10.1371/journal.pbio.0030009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valiere N, Taberlet P. 2000. Urine collected in the field as a source of DNA for species and individual identification. Mol. Ecol. 9, 2150–2152. ( 10.1046/j.1365-294X.2000.11142.x) [DOI] [PubMed] [Google Scholar]
- 25.Poinar HN. 1998. Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281, 402–406. ( 10.1126/science.281.5375.402) [DOI] [PubMed] [Google Scholar]
- 26.Höss M, Kohn M, Pääbo S, Knauer F, Schröder W. 1992. Excrement analysis by PCR. Nature 359, 199 ( 10.1038/359199a0) [DOI] [PubMed] [Google Scholar]
- 27.Strausberger BM, Ashley MV. 2001. Eggs yield nuclear DNA from egg-laying female cowbirds, their embryos and offspring. Conserv. Genet. 2, 385–390. ( 10.1023/A:1012526315617) [DOI] [Google Scholar]
- 28.Higuchi R, von Beroldingen CH, Sensabaugh GF, Erlich HA. 1988. DNA typing from single hairs. Nature 332, 543–546. ( 10.1038/332543a0) [DOI] [PubMed] [Google Scholar]
- 29.Taberlet P, Mattock H, Dubois-Paganon C, Bouvet J. 1993. Sexing free-ranging brown bears Ursus arctos using hairs found in the field. Mol. Ecol. 2, 399–403. ( 10.1111/j.1365-294X.1993.tb00033.x) [DOI] [PubMed] [Google Scholar]
- 30.Nichols RV, Königsson H, Danell K, Spong G. 2012. Browsed twig environmental DNA: diagnostic PCR to identify ungulate species. Mol. Ecol. Resour. 12, 983–989. ( 10.1111/j.1755-0998.2012.03172.x) [DOI] [PubMed] [Google Scholar]
- 31.Hofreiter M, Collins M, Stewart JR. 2012. Ancient biomolecules in Quaternary palaeoecology. Quat. Sci. Rev. 33, 1–13. ( 10.1016/j.quascirev.2011.11.018) [DOI] [Google Scholar]
- 32.Taberlet P, Fumagalli L. 1996. Owl pellets as a source of DNA for genetic studies of small mammals. Mol. Ecol. 5, 301–305. ( 10.1046/j.1365-294X.1996.00084.x) [DOI] [PubMed] [Google Scholar]
- 33.Taberlet P, Bouvet J. 1991. A single plucked feather as a source of DNA for bird genetic-studies. Auk 108, 959–960. [Google Scholar]
- 34.Trevors JT. 1996. Nucleic acids in the environment. Curr. Opin. Biotechnol. 7, 331–336. ( 10.1016/S0958-1669(96)80040-1) [DOI] [PubMed] [Google Scholar]
- 35.Poté J, Ackermann R, Wildi W. 2009. Plant leaf mass loss and DNA release in freshwater sediments. Ecotox. Environ. Safe 72, 1378–1383. ( 10.1016/j.ecoenv.2009.04.010) [DOI] [PubMed] [Google Scholar]
- 36.Levy-Booth DJ, Campbell RG, Gulden RH. 2007. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 39, 2977–2991. ( 10.1016/j.soilbio.2007.06.020) [DOI] [Google Scholar]
- 37.Meier P, Wackernagel W. 2003. Mechanisms of homology-facilitated illegitimate recombination for foreign DNA acquisition in transformable Pseudomonas stutzeri. Mol. Microbiol. 48, 1107–1118. ( 10.1046/j.1365-2958.2003.03498.x) [DOI] [PubMed] [Google Scholar]
- 38.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37–53. ( 10.1051/ebr:2007031) [DOI] [PubMed] [Google Scholar]
- 39.Blum SAE, Lorenz MG, Wackernagel W. 1997. Mechanism of retarded DNA degradation and prokaryotic origin of DNases in nonsterile soils. Syst. Appl. Microbiol. 20, 513–521. ( 10.1016/S0723-2020(97)80021-5) [DOI] [Google Scholar]
- 40.Crecchio C, Stotzky G. 1998. Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 30, 1061–1067. ( 10.1016/S0038-0717(97)00248-4) [DOI] [Google Scholar]
- 41.Khanna M, Stotzky G. 1992. Transformation of Bacillus subtilis by DNA bound on Montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 58, 1930–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietramellara G, Ascher J, Ceccherini MT, Nannipieri P, Wenderoth D. 2007. Adsorption of pure and dirty bacterial DNA on clay minerals and their transformation frequency. Biol. Fertil. Soils 43, 731–739. ( 10.1007/s00374-006-0156-8) [DOI] [Google Scholar]
- 43.Greaves MP, Wilson MJ. 1969. The adsorption of nucleic acids by Montmorillonite. Soil Biol. Biochem. 1, 317–323. ( 10.1016/0038-0717(69)90014-5) [DOI] [Google Scholar]
- 44.Huang YT, Lowe DJ, Churchman GJ, Schipper LA. 2014. Carbon storage and DNA adsorption in allophanic soils and Paleosols. In Soil Carbon (eds McSweeney K, Hartemink AE.), pp. 163–172. Springer International. [Google Scholar]
- 45.Lorenz MG, Wackernagel W. 1987. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl. Environ. Microbiol. 53, 2948–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiological. Rev. 58, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. ( 10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- 48.Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158, 767–778. ( 10.1016/j.resmic.2007.09.004) [DOI] [PubMed] [Google Scholar]
- 49.Vries J, Wackernagel W. 2005. Microbial horizontal gene transfer and the DNA release from transgenic crop plants. Plant Soil 266, 91–104. ( 10.1007/s11104-005-4783-x) [DOI] [Google Scholar]
- 50.Boschetti C, Carr A, Crisp A, Eyres I, Wang-koh Y, Lubzens E, Barraclough TG, Micklem G, Tunnacliffe A. 2012. Biochemical diversification through foreign gene expression in bdelloid rotifers. PLoS Genet. 8, e1003035 ( 10.1371/journal.pgen.1003035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213. ( 10.1126/science.1156407) [DOI] [PubMed] [Google Scholar]
- 52.Overballe-Petersen S, et al. 2013. Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl Acad. Sci. USA 110, 19 860–19 865. ( 10.1073/pnas.1315278110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briggs AW, et al. 2010. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 ( 10.1093/nar/gkp1163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willerslev E, Hansen AJ, Rønn R, Brand TB, Barnes I, Wiuf CC, Gilichinsky D, Mitchell D, Cooper A. 2004. Long-term persistence of bacterial DNA. Curr. Biol. 14, R9–10. ( 10.1016/j.cub.2003.12.012) [DOI] [PubMed] [Google Scholar]
- 55.Deagle BE, Eveson JP, Jarman SN. 2006. Quantification of damage in DNA recovered from highly degraded samples--a case study on DNA in faeces. Front. Zool. 3, 11 ( 10.1186/1742-9994-3-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pietramellara G, Ascher J, Borgogni F. 2009. Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fertil. Soils 43, 731–739. ( 10.1007/s00374-006-0156-8) [DOI] [Google Scholar]
- 57.Allentoft ME, et al. 2012. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B 279, 4724–4733. ( 10.1098/rspb.2012.1745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert MTP, Djurhuus D, Melchior L, Lynnerup N, Worobey M, Wilson AS, Andreasen C, Dissing J. 2007. mtDNA from hair and nail clarifies the genetic relationship of the 15th century Qilakitsoq Inuit mummies. Am. J. Phys. Anthropol. 133, 847–853. ( 10.1002/ajpa.20602) [DOI] [PubMed] [Google Scholar]
- 59.Paul JH, Jeffrey WH, DeFlaun MF. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul JHJ, Jeffrey WHW, David AWA, Deflaun MFM, Cazares LHL. 1989. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl. Environ. Microbiol. 55, 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C. 2011. Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6, e23398 ( 10.1371/journal.pone.0023398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willerslev E, Hansen AJ, Poinar HN. 2004. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 19, 141–147. ( 10.1016/j.tree.2003.11.010) [DOI] [PubMed] [Google Scholar]
- 63.Hebsgaard MB, Gilbert MTP, Arneborg J, Heyn P, Allentoft ME, Bunce M, Munch K, Schweger C, Willerslev E. 2009. ‘The farm beneath the sand’–an archaeological case study on ancient ‘dirt’ DNA. Antiquity 83, 430–444. [Google Scholar]
- 64.Jenkins DL, et al. 2012. Clovis age Western Stemmed projectile points and human coprolites at the Paisley Caves. Science 337, 223–228. ( 10.1126/science.1218443) [DOI] [PubMed] [Google Scholar]
- 65.Arnold LJ, Roberts RG, MacPhee RDE, Willerslev E, Tikhonov AN, Brock F. 2008. Optical dating of perennially frozen deposits associated with preserved ancient plant and animal DNA in north-central Siberia. Quat. Geochronol. 3, 114–136. ( 10.1016/j.quageo.2007.09.002) [DOI] [Google Scholar]
- 66.Young MK, McKelvey KS, Pilgrim KL, Schwartz MK. 2013. DNA barcoding at riverscape scales: assessing biodiversity among fishes of the genus Cottus (Teleostei) in northern Rocky Mountain streams. Mol. Ecol. Resour. 13, 583–595. ( 10.1111/1755-0998.12091) [DOI] [PubMed] [Google Scholar]
- 67.Pilliod DS, Goldberg CS, Arkle RS, Waits LP, Richardson J. 2013. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 70, 1123–1130. ( 10.1139/cjfas-2013-0047) [DOI] [Google Scholar]
- 68.Feek DT, Flenley JR, Chester PI, Welikala N, Matisoo-Smith EA, Tannock GW. 2006. A modified sampler for uncontaminated DNA cores from soft sediments. J. Archaeol. Sci. 33, 573–574. ( 10.1016/j.jas.2005.09.013) [DOI] [Google Scholar]
- 69.Juck DF, Whissell G, Steven B, Pollard W, McKay CP, Greer CW, Whyte LG. 2005. Utilization of fluorescent microspheres and a green fluorescent protein-marked strain for assessment of microbiological contamination of permafrost and ground ice core samples from the Canadian High Arctic. Appl. Environ. Microbiol. 71, 1035–1041. ( 10.1128/AEM.71.2.1035-1041.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lydolph MC, Jacobsen J, Arctander P, Gilbert MTP, Gilichinsky DA, Hansen AJ, Willerslev E, Lange L. 2005. Beringian paleoecology inferred from permafrost-preserved fungal DNA. Appl. Environ. Microbiol. 71, 1012–1017. ( 10.1128/AEM.71.2.1012-1017.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulat SA, et al. 2004. DNA signature of thermophilic bacteria from the aged accretion ice of Lake Vostok, Antarctica: implications for searching for life in extreme icy environments. Int. J. Astrobiol. 3, 1–12. ( 10.1017/S1473550404001879) [DOI] [Google Scholar]
- 72.Rogers SO, Theraisnathan V, Ma LJ, Zhao Y, Zhang G, Shin S-G, Castello JD, Starmer WT. 2004. Comparisons of protocols for decontamination of environmental ice samples for biological and molecular examinations. Appl. Environ. Microbiol. 70, 2540–2544. ( 10.1128/AEM.70.4.2540-2544.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christner BC, Mikucki JA, Foreman CM, Denson J, Priscu JC. 2005. Glacial ice cores: a model system for developing extraterrestrial decontamination protocols. Icarus 174, 572–584. ( 10.1016/j.icarus.2004.10.027) [DOI] [Google Scholar]
- 74.D'Elia T, Veerapaneni R, Rogers SO. 2008. Isolation of microbes from Lake Vostok accretion ice. Appl. Environ. Microbiol. 74, 4962–4965. ( 10.1128/AEM.02501-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Longo MC, Berninger MS, Hartley JL. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93, 125–128. ( 10.1016/0378-1119(90)90145-H) [DOI] [PubMed] [Google Scholar]
- 76.Handt O, et al. 1994. Molecular genetic analyses of the Tyrolean Ice Man. Science 264, 1775–1778. ( 10.1126/science.8209259) [DOI] [PubMed] [Google Scholar]
- 77.Champlot S, Berthelot C, Pruvost M, Bennett EA, Grange T, Geigl E. 2010. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE 5, e13042 ( 10.1371/journal.pone.0013042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malmstrom H, Storå J, Dalén L, Holmlund G, Götherström A. 2005. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol. Biol. Evol. 22, 2040–2047. ( 10.1093/molbev/msi195) [DOI] [PubMed] [Google Scholar]
- 79.Leonard JA, Shanks O, Hofreiter M, Kreuz E, Hodges L, Ream W, Wayne RK, Fleischer RC. 2007. Animal DNA in PCR reagents plagues ancient DNA research. J. Archaeol. Sci. 34, 1361–1366. ( 10.1016/j.jas.2006.10.023) [DOI] [Google Scholar]
- 80.Yao Y-G, Bandelt H-J, Young NS. 2007. External contamination in single cell mtDNA analysis. PLoS ONE 2, e681 ( 10.1371/journal.pone.0000681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boessenkool S, Epp LS, Haile J, Bellemain E, Edwards M, Coissac E, Willerslev E, Brochmann C. 2011. Blocking human contaminant DNA during PCR allows amplification of rare mammal species from sedimentary ancient DNA. Mol. Ecol. 21, 1806–1815. ( 10.1111/j.1365-294X.2011.05306.x) [DOI] [PubMed] [Google Scholar]
- 82.Hofreiter M, Kreuz E, Eriksson J, Schubert G, Hohmann G. 2010. Vertebrate DNA in fecal samples from bonobos and gorillas: evidence for meat consumption or artefact? PLoS ONE 5, e9419 ( 10.1371/journal.pone.0009419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willerslev E, et al. 2015. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51. ( 10.1038/nature12921) [DOI] [PubMed] [Google Scholar]
- 84.Porter TM, Golding GB, King C, Froese DG, Zazula G, Poinar HN. 2013. Amplicon pyrosequencing Late Pleistocene permafrost: the removal of putative contaminant sequences and small-scale reproducibility. Mol. Ecol. Resour. 13, 798–810. ( 10.1111/1755-0998.12124) [DOI] [PubMed] [Google Scholar]
- 85.Miller DN, Bryant JE, Madsen EL, Ghiorse WC. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65, 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67, 2354–2359. ( 10.1128/AEM.67.5.2354-2359.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frostegård A, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65, 5409–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feinstein LM, Sul WJ, Blackwood CB. 2009. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433. ( 10.1128/AEM.00120-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carrigg C, Rice O, Kavanagh S, Collins G, O'Flaherty V. 2007. DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 77, 955–964. ( 10.1007/s00253-007-1219-y) [DOI] [PubMed] [Google Scholar]
- 90.Luna GM, Dell'anno A, Danovaro R. 2006. DNA extraction procedure: a critical issue for bacterial diversity assessment in marine sediments. Environ. Microbiol. 8, 308–320. ( 10.1111/j.1462-2920.2005.00896.x) [DOI] [PubMed] [Google Scholar]
- 91.Terrat S, et al. 2011. Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. J. Microbial. Biotech. 5, 135–141. ( 10.1111/j.1751-7915.2011.00307.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robe P, Nalin R, Capellano C, Vogel TM, Simonet P. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39, 183–190. ( 10.1016/S1164-5563(03)00033-5) [DOI] [Google Scholar]
- 93.Shapiro B, Hofreiter M. 2012. Ancient DNA: methods and protocols. New York, NY: Springer. [Google Scholar]
- 94.Hedman JJ, Rådström PP. 2012. Overcoming inhibition in real-time diagnostic PCR. Methods Mol. Biol. 943, 17–48. ( 10.1007/978-1-60327-353-4_2) [DOI] [PubMed] [Google Scholar]
- 95.Wilson IG. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.King C, Debruyne R, Kuch M, Schwarz C, Poinar H. 2009. A quantitative approach to detect and overcome PCR inhibition in ancient DNA extracts. Biotechnology 47, 941–949. ( 10.2144/000113244) [DOI] [PubMed] [Google Scholar]
- 97.Schwarz C, Debruyne R, Kuch M, McNally E, Schwarcz H, Aubrey AD, Bada J, Poinar H. 2009. New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains. Nucleic Acids Res. 37, 3215–3229. ( 10.1093/nar/gkp159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Phil. Trans. R. Soc. B 360, 1847–1857. ( 10.1098/rstb.2005.1716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taberlet P, et al. 2007. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 ( 10.1093/nar/gkl938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hajibabaei M, Singer G, Hebert P, Hickey DA. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172. ( 10.1016/j.tig.2007.02.001) [DOI] [PubMed] [Google Scholar]
- 101.Jørgensen T, et al. 2012. Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA meta-barcoding. Mol. Ecol. 21, 1980–1988. ( 10.1111/j.1365-294X.2011.05278.x) [DOI] [PubMed] [Google Scholar]
- 102.Taberlet P, et al. 2012. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 21, 1816–1820. ( 10.1111/j.1365-294X.2011.05317.x) [DOI] [PubMed] [Google Scholar]
- 103.Murray D, Bunce M, Cannell BL, Oliver R, Houston J, White NE, Barrero RA, Bellgard MI, Haile J. 2011. DNA-based faecal dietary analysis: a comparison of qPCR and high throughput sequencing approaches. PLoS ONE 6, e25776 ( 10.1371/journal.pone.0025776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cooper A, Stephens J, Ketheesan N, Govan B. 2013. Detection of Coxiella burnetii DNA in wildlife and ticks in northern Queensland, Australia. Vector Borne Zoonotic Dis. 13, 12–16. ( 10.1089/vbz.2011.0853) [DOI] [PubMed] [Google Scholar]
- 105.Hodges E, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527. ( 10.1038/ng.2007.42) [DOI] [PubMed] [Google Scholar]
- 106.Briggs AW, et al. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321. ( 10.1126/science.1174462) [DOI] [PubMed] [Google Scholar]
- 107.Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2, e197 ( 10.1371/journal.pone.0000197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kircher M, Sawyer S, Meyer M. 2011. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 ( 10.1093/nar/gkr771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pääbo S, et al. 2004. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679. ( 10.1146/annurev.genet.37.110801.143214) [DOI] [PubMed] [Google Scholar]
- 110.CBOL Plant Working Group 2009. A DNA barcode for land plants. Proc. Natl Acad. Sci. USA 106, 12 794–12 797. ( 10.1073/pnas.0905845106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Newmaster SG, Fazekas AJ, Ragupathy S. 2006. DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Can. J. Bot. 84, 335–341. ( 10.1139/b06-047) [DOI] [Google Scholar]
- 112.Austin JJ, Arnold EN, Jones CG. 2004. Reconstructing an island radiation using ancient and recent DNA: the extinct and living day geckos (Phelsuma) of the Mascarene islands. Mol. Phylogenet. Evol. 31, 109–122. ( 10.1016/j.ympev.2003.07.011) [DOI] [PubMed] [Google Scholar]
- 113.Beati L, Caceres AG, Lee JA, Munstermann LE. 2004. Systematic relationships among Lutzomyia sand flies (Diptera: Psychodidae) of Peru and Colombia based on the analysis of 12S and 28S ribosomal DNA sequences. Int. J. Parasitol. 34, 225–234. ( 10.1016/j.ijpara.2003.10.012) [DOI] [PubMed] [Google Scholar]
- 114.Soergel DAW, Dey N, Knight R, Brenner SE. 2012. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 6, 1440–1444. ( 10.1038/ismej.2011.208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V. 2005. Land plants and DNA barcodes: short-term and long-term goals. Phil. Trans. R. Soc. B 360, 1889–1895. ( 10.1098/rstb.2005.1720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. 2005. Use of DNA barcodes to identify flowering plants. Proc. Natl Acad. Sci. USA 102, 8369–8374. ( 10.1073/pnas.0503123102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E. 2011. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39, e145 ( 10.1093/nar/gkr732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27, 1159–1161. ( 10.1093/bioinformatics/btr087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917. ( 10.1038/ismej.2010.171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108(Suppl. 1), 4516–4522. ( 10.1073/pnas.1000080107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Epp LS, et al. 2012. New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Mol. Ecol. 21, 1821–1833. ( 10.1111/j.1365-294X.2012.05537.x) [DOI] [PubMed] [Google Scholar]
- 122.Ficetola G, Coissac E, Zundel S, Riaz T, Shehzad W, Bessière J, Taberlet P, Pompanon F. 2010. An in silico approach for the evaluation of DNA barcodes. BMC Genomics 11, 434 ( 10.1186/1471-2164-11-434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6, 639–641. ( 10.1038/nmeth.1361) [DOI] [PubMed] [Google Scholar]
- 124.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7, 668–669. ( 10.1038/nmeth0910-668b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinform. 12, 38 ( 10.1186/1471-2105-12-38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. ( 10.1093/bioinformatics/btr381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shokralla S, Spall JL, Gibson JF, Hajibabaei M. 2012. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 21, 1794–1805. ( 10.1111/j.1365-294X.2012.05538.x) [DOI] [PubMed] [Google Scholar]
- 128.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. ( 10.1101/gr.5969107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Munch K, Boomsma W, Huelsenbeck J, Willerslev E, Nielsen R. 2008. Statistical assignment of DNA sequences using Bayesian phylogenetics. Syst. Biol. 57, 750–757. ( 10.1080/10635150802422316) [DOI] [PubMed] [Google Scholar]
- 131.Huson DH, Weber N. 2013. Microbial community analysis using MEGAN. Amsterdam, The Netherlands: Elsevier. [DOI] [PubMed] [Google Scholar]
- 132.Munch K, Boomsma W, Willerslev E, Nielsen R. 2008. Fast phylogenetic DNA barcoding. Phil. Trans. R. Soc. B 363, 3997–4002. ( 10.1098/rstb.2008.0169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang AB, Muster C, Liang HB, Zhu CD, Crozier R, Wan P, Feng J, Ward RD. 2012. A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding. Mol. Ecol. 21, 1848–1863. ( 10.1111/j.1365-294X.2011.05235.x) [DOI] [PubMed] [Google Scholar]
- 134.Berger SA, Stamatakis A. 2011. Aligning short reads to reference alignments and trees. Bioinformatics 27, 2068–2075. ( 10.1093/bioinformatics/btr320) [DOI] [PubMed] [Google Scholar]
- 135.Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 60, 291–302. ( 10.1093/sysbio/syr010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14, 755–763. ( 10.1093/bioinformatics/14.9.755) [DOI] [PubMed] [Google Scholar]
- 137.DeSantis TZJ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399. ( 10.1093/nar/gkl244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 139.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. ( 10.1093/bioinformatics/btl158) [DOI] [PubMed] [Google Scholar]
- 140.Li W, Jaroszewski L, Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283. ( 10.1093/bioinformatics/17.3.282) [DOI] [PubMed] [Google Scholar]
- 141.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. ( 10.1128/AEM.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aagaard K, et al. 2012. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 7, e36466 ( 10.1371/journal.pone.0036466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bik HM, Halanych KM, Sharma J, Thomas WK. 2012. Dramatic shifts in benthic microbial eukaryote communities following the Deepwater Horizon oil spill. PLoS ONE 7, e38550 ( 10.1371/journal.pone.0038550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bik HM, Sung W, De Ley P, Baldwin JG, Sharma J, Rocha-Olivares A, Thomas WK. 2012. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Mol. Ecol. 21, 1048–1059. ( 10.1111/j.1365-294X.2011.05297.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bokulich NA, Joseph CM, Allen G, Benson AK, Mills DA. 2012. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS ONE 7, e36357 ( 10.1371/journal.pone.0036357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bokulich NA, Bamforth CW, Mills DA. 2012. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS ONE 7, e35507 ( 10.1371/journal.pone.0035507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ceh J, Raina JB, Soo RM, van Keulen M, Bourne DG. 2012. Coral-bacterial communities before and after a coral mass spawning event on Ningaloo Reef. PLoS ONE 7, e36920 ( 10.1371/journal.pone.0036920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, Jefferson KK, Buck GA. 2012. Species-level classification of the vaginal microbiome. BMC Genomics 13(Suppl. 8), S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fierer N, et al. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl Acad. Sci. USA 109, 21 390–21 395. ( 10.1073/pnas.1215210110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. 2011. Microbial biogeography of public restroom surfaces. PLoS ONE 6, e28132 ( 10.1371/journal.pone.0028132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. 2013. Evidence for a persistent microbial seed bank throughout the global ocean. Proc. Natl Acad. Sci. USA 110, 4651–4655. ( 10.1073/pnas.1217767110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hospodsky D, Qian J, Nazaroff WW, Yamamoto N, Bibby K, Rismani-Yazdi H, Peccia J. 2012. Human occupancy as a source of indoor airborne bacteria. PLoS ONE 7, e34867 ( 10.1371/journal.pone.0034867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hou W, et al. 2013. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS ONE 8, e53350 ( 10.1371/journal.pone.0053350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hulcr J, Latimer AM, Henley JB, Rountree NR, Fierer N, Lucky A, Lowman MD, Dunn RR. 2012. A jungle in there: bacteria in belly buttons are highly diverse, but predictable. PLoS ONE 7, e47712 ( 10.1371/journal.pone.0047712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Koberl M, Muller H, Ramadan EM, Berg G. 2011. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 6, e24452 ( 10.1371/journal.pone.0024452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Leff JW, Fierer N. 2013. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 8, e59310 ( 10.1371/journal.pone.0059310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, Banfield JF. 2011. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl Acad. Sci. USA 108, 1128–1133. ( 10.1073/pnas.1010992108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. ( 10.1126/science.1198719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pope PB, et al. 2010. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc. Natl Acad. Sci. USA 107, 14 793–14 798. ( 10.1073/pnas.1005297107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Robeson MS, King AJ, Freeman KR, Birky CWJ, Martin AP, Schmidt SK. 2011. Soil rotifer communities are extremely diverse globally but spatially autocorrelated locally. Proc. Natl Acad. Sci. USA 108, 4406–4410. ( 10.1073/pnas.1012678108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Smith KF, Schmidt V, Rosen GE, Amaral-Zettler L. 2012. Microbial diversity and potential pathogens in ornamental fish aquarium water. PLoS ONE 7, e39971 ( 10.1371/journal.pone.0039971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Verhulst NO, et al. 2011. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE 6, e28991 ( 10.1371/journal.pone.0028991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yatsunenko T, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Adler CJ, et al. 2013. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and industrial revolutions. Nat. Genet. 45, 450–455. ( 10.1038/ng.2536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. ( 10.1038/nmeth.1650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tito RY, et al. 2012. Insights from characterizing extinct human gut microbiomes. PLoS ONE 7, e51146 ( 10.1371/journal.pone.0051146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. ( 10.1038/nmeth.2066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sunagawa S, et al. 2013. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 10, 1196–1199. ( 10.1038/nmeth.2693) [DOI] [PubMed] [Google Scholar]
- 169.Schubert M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomicanalysis using PALEOMIX. Nat. Protoc. 9, 1056–1082. ( 10.1038/nprot.2014.063) [DOI] [PubMed] [Google Scholar]
- 170.Der Sarkissian C, Ermini L, Jónsson H, Alekseev AN, Crubezy E, Shapiro B, Orlando L. 2014. Shotgun microbial profiling of fossil remains. Mol. Ecol. 23, 1780–1798. ( 10.1111/mec.12690) [DOI] [PubMed] [Google Scholar]
- 171.Arumugam M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473, 174–180. ( 10.1038/nature09944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M. 2011. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics 12(Suppl. 2), S4 ( 10.1186/1471-2164-12-S2-S4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29, 181–184. ( 10.1093/nar/29.1.181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4, 642–647. ( 10.1038/ismej.2009.153) [DOI] [PubMed] [Google Scholar]
- 175.Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200 ( 10.1093/nar/gkq873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452. ( 10.1126/science.1147112) [DOI] [PubMed] [Google Scholar]
- 177.von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. 2007. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315, 1126–1130. ( 10.1126/science.1133420) [DOI] [PubMed] [Google Scholar]
- 178.Brady A, Salzberg S. 2011. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nat. Methods 8, 367 ( 10.1038/nmeth0511-367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Parks DH, MacDonald NJ, Beiko RG. 2011. Classifying short genomic fragments from novel lineages using composition and homology. BMC Bioinform. 12, 328 ( 10.1186/1471-2105-12-328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rosen GL, Reichenberger ER, Rosenfeld AM. 2011. NBC: the naive Bayes classification tool webserver for taxonomic classification of metagenomic reads. Bioinformatics 27, 127–129. ( 10.1093/bioinformatics/btq619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tringe SG. 2005. Comparative metagenomics of microbial communities. Science 308, 554–557. ( 10.1126/science.1107851) [DOI] [PubMed] [Google Scholar]
- 182.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. ( 10.1093/nar/gkr988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Powell S, et al. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289. ( 10.1093/nar/gkr1060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. ( 10.1093/nar/gki866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sandberg R. 2001. Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome Res. 11, 1404–1409. ( 10.1101/gr.186401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Edwards RA, Olson R, Disz T, Pusch GD, Vonstein V, Stevens R, Overbeek R. 2012. Real time metagenomics: using k-mers to annotate metagenomes. Bioinformatics 28, 3316–3317. ( 10.1093/bioinformatics/bts599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dahl-Jensen D, et al. 2013. Eemian interglacial reconstructed from a Greenland folded ice core. Nature 493, 489–494. ( 10.1038/nature11789) [DOI] [PubMed] [Google Scholar]
- 188.de Vernal A, Hillaire-Marcel C. 2008. Natural variability of Greenland climate, vegetation, and ice volume during the past million years. Science 320, 1622–1625. ( 10.1126/science.1153929) [DOI] [PubMed] [Google Scholar]
- 189.Goldberg CS, Pilliod DS, Arkle RS, Waits LP. 2011. Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 6, e22746 ( 10.1371/journal.pone.0022746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Olson ZH, Briggler JT, Williams RN. 2012. An eDNA approach to detect eastern hellbenders (Cryptobranchus a. alleganiensis) using samples of water. Wildl. Res. 39, 629 ( 10.1071/WR12114) [DOI] [Google Scholar]
- 191.Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C. 2012. Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 49, 953–959. ( 10.1111/j.1365-2664.2012.02171.x) [DOI] [Google Scholar]
- 192.Minamoto T, Yamanaka H, Takahara T, Honjo MN, Kawabata Z. 2011. Surveillance of fish species composition using environmental DNA. Limnology 13, 193–197. ( 10.1007/s10201-011-0362-4) [DOI] [Google Scholar]
- 193.Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z. 2012. Estimation of fish biomass using environmental DNA. PLoS ONE 7, e35868 ( 10.1371/journal.pone.0035868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Takahara T, Minamoto T, Doi H. 2013. Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS ONE 8, e56584 ( 10.1371/journal.pone.0056584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP. 2013. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshwater Sci. 32, 792–800. ( 10.1899/13-046.1) [DOI] [Google Scholar]
- 196.Alvarez AJ, Yumet GM, Santiago CL, Toranzos GA. 1996. Stability of manipulated plasmid DNA in aquatic environments. Environ. Toxic Water 11, 129–135. () [DOI] [Google Scholar]
- 197.Matsui K, Honjo M, Kawabata Z. 2001. Estimation of the fate of dissolved DNA in thermally stratified lake water from the stability of exogenous plasmid DNA. Aquat. Microb. Ecol. 26, 95–102. ( 10.3354/ame026095) [DOI] [Google Scholar]
- 198.Giguet-Covex C, et al. 2014. Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nature communications 5, 1–7. ( 10.1038/ncomms4211) [DOI] [PubMed] [Google Scholar]
- 199.Kelly RP, et al. 2014. Environmental monitoring. Harnessing DNA to improve environmental management. Science 344, 1455–1456. ( 10.1126/science.1251156) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.