Abstract
This study is part of an ongoing project aiming at determining the ethnogenesis of an eastern Siberian ethnic group, the Yakuts, on the basis of archaeological excavations carried out over a period of 10 years in three regions of Yakutia: Central Yakutia, the Vilyuy River basin and the Verkhoyansk area. In this study, genetic analyses were carried out on skeletal remains from 130 individuals of unknown ancestry dated mainly from the fifteenth to the nineteenth century AD. Kinship studies were conducted using sets of commercially available autosomal and Y-chromosomal short tandem repeats (STRs) along with hypervariable region I sequences of the mitochondrial DNA. An unexpected and intriguing finding of this work was that the uniparental marker systems did not always corroborate results from autosomal DNA analyses; in some cases, false-positive relationships were observed. These discrepancies revealed that 15 autosomal STR loci are not sufficient to discriminate between first degree relatives and more distantly related individuals in our ancient Yakut sample. The Y-STR analyses led to similar conclusions, because the current Y-STR panels provided the limited resolution of the paternal lineages.
Keywords: Yakuts, short tandem repeats, ancient DNA, kinship
1. Introduction
The history of the Yakuts started at the beginning of the second millennium AD, when a group of Turkic-speaking cattle-breeders, pressed by the expansion of Mongolic tribes in the eleventh–thirteenth centuries AD, moved from the south (possibly from the Altai–Sayan and/or Baikal region(s)) to the Lena river basin [1]. During this time, the southern migrants mixed with indigenous populations (probably Mongolian ethnic groups, Tungusic-speaking Evenks and Evens and Yukaghirs), thereby forming the Yakuts. In the following centuries, the Yakuts spread over the vast northeastern region of Siberia. When Russians began to invade their territory in the 1630s, the Yakuts were settled between the Lena, Amga and Aldan rivers (currently Central Yakutia), with pockets of settlement along the Vilyuy River basin and further north in the Verkhoyansk region (figure 1).
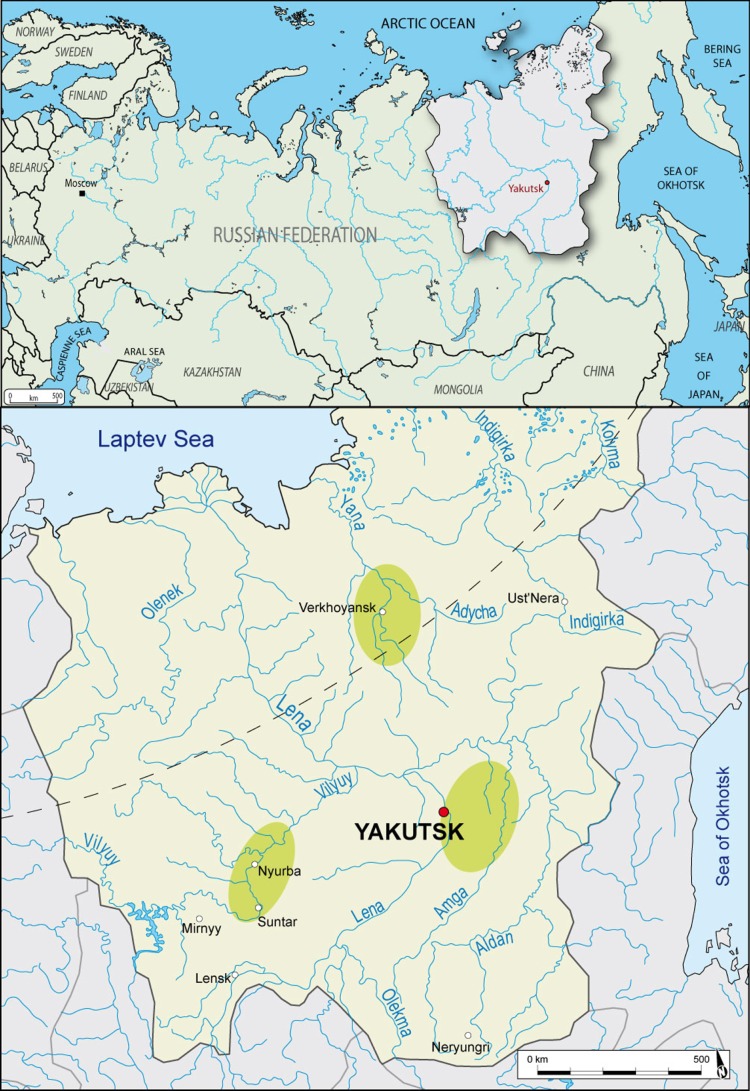
Figure 1.
Localization of Yakutia and map of the archaeological regions of excavations (P. Gérard). (Online version in colour.)
Although many researchers have addressed the issue of the ethnogenesis of the Yakuts, the precise geographical origins of this population as well as the degree of admixture with indigenous populations of Siberia are still unclear. Because the difficulties encountered by molecular biologists might have arisen from the demographic events that occurred in the Yakut population since the Russian colonization, our research group has been involved for 10 years in the study of human remains excavated from Yakut graves dated mainly from the fifteenth to the nineteenth centuries [2–7]. Excavations have been conducted in three regions of Yakutia (Central Yakutia, the Vilyuy River basin and the Verkhoyansk region). In the course of this project, 150 graves were excavated resulting in the recovery of 146 individuals. Genetic data from autosomal, Y-chromosomal and/or mitochondrial DNA were obtained for 130 of them.
In this study, these genetic data were used to examine the biological relationships within and between Yakut burial groups. The main objectives were to: (i) identify members of the same family or clan; (ii) provide insights into the Yakuts' funeral practices; and (iii) determine whether some graves located thousands of kilometres from each other could contain related individuals, in other words, track the migration of individuals and/or families into the different regions of Yakutia. To determine close kinships, a multi-marker strategy was applied. Short tandem repeats (STRs) located on autosomal chromosomes were used to establish individual-specific DNA profiles inherited equally from each parent. STRs present on the non-recombining part of the Y-chromosome (NRY) were used to establish paternal haplotypes allowing the confirmation of paternities deduced from DNA profiling. Sequences in the hypervariable region I (HVI) of the mitochondrial DNA (mtDNA) were finally used to identify maternally related individuals. This combined approach allowed us to not only detect close kinships between individuals from the same or different burial sites, but also some discrepancies between the results obtained from the biparental and uniparental markers. A detailed analysis of this outcome reinforced the view that the Yakut population is unusual in many respects.
2. Material and methods
(a). Ancient human samples
Ancient human individuals were excavated during the years 2002–2012 by the French Archaeological Missions in Eastern Siberia. Most of the burial places were frozen at the time of their discovery allowing for the excellent preservation of the individuals. Genetic data were collected from 130 individuals excavated in Central Yakutia (n = 84), the Vilyuy River basin (n = 23) and the Verhoyansk area (n = 23; figure 1). Dating of the graves was inferred from dendrochronological analyses, archaeological features or radiocarbon 14C dating (electronic supplementary material, table S1). Sex was established from skeletal remains according to the methodology developed by Murail et al. [8]. Age at death was estimated using dental calcification for the children and epiphyseal fusion for the adolescents. Administrative and research work were permitted through the programme of the France–Russia Associated International Laboratory (LIA COSIE number 1029).
(b). DNA extraction
Bones and/or teeth were used for the DNA extractions. To eliminate surface contamination, the outer surface of the bones was abraded to a depth of 2–3 mm with a sanding machine, whereas the teeth were cleaned with bleach, rinsed with ultrapure water and exposed to ultraviolet light for 30 min on each side. Bones or teeth were then powdered in a grinder mill under liquid nitrogen, and DNA was extracted according to protocols previously described [9,10]. Between three and six, extractions were carried out for each individual.
(c). Autosomal short tandem repeat analysis
Autosomal STRs were firstly analysed using the AmpFlSTRs Identifiler (Plus) kit (Life Technologies). Fifteen STRs (D8S1179, D7S820, D3S1358, D13S317, D16S539, D2S1338, D19S433, D5S818, D21S11, CSF1PO, vWA, THO1, TPOX, D18S51 and FGA) and the sex-determining marker amelogenin were amplified in a single PCR. PCRs were performed according to the manufacturer's protocol, except that 32 cycles were used instead of the recommended 28, in a reaction volume of 12.5 µl, thus reducing the volume of the DNA samples. At least two amplifications were performed on each DNA extract. Alleles were assigned according to the International Society for Forensic Genetics guidelines for STR analysis [11].
In some specific cases, the new GlobalFiler kit (Life Technologies) was additionally used. This 24 locus kit allowed the amplification of six additional autosomal STR loci (D1S1656, D12S391, D2S44, D10S1248, D22S1045 and SE33) along with three gender identification markers (amelogenin, DYS391 and a Y-indel). The experimental conditions were those recommended by the manufacturer, except that 32 PCR cycles were used instead of 29. The amplified products were run on a 3500 genetic analyser (Life Technologies) and analysed using the GeneMapper v. 4.1 software (Life Technologies).
(d). Y-chromosomal short tandem repeat and single nucleotide polymorphism analysis
The DNAs of the ancient Yakut males were analysed at 17 Y-chromosomal STR loci (DYS19, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and Y GATA) using the AmpFlSTR Y-Filer kit (Life Technologies) according to the manufacturer's recommendations except that 34 cycles were used instead of 30. DNA from the 29 ancient Yakut males sharing the same 17 Y-chromosomal STR haplotype were genotyped a second time using the PowerPlex Y23 system according to the manufacturer's instructions (Promega) except that 34 cycles were used instead of 30. This kit combines the aforementioned 17 Y-STRs with six additional highly discriminating Y-STR loci (DYS481, DYS533, DYS549, DYS570, DYS576 and DYS643). STR products were run on the 3100 or 3500 genetic analyser (Life Technologies) and analysed using the GeneMapper v. 4.1 software (Life Technologies).
Y-haplogroups were established by single nucleotide polymorphisms (SNPs) typing using a set of 14 Y-chromosomal SNPs (M214, M175, M231, Tat, M128, P43, M242, M346, L54, M3, M120, M130, M217 and M174), the Mass Array platform and the iPLEX Gold technology (Sequenom) as recently described [10]. They were named according to the most recent Y-DNA haplogroup tree 2014 (http://www.isogg.org/tree/).
(e). Mitochondrial DNA analysis
Three hundred and eighty-one base pairs of the HVI region of the mtDNA genome were amplified and sequenced in two overlapping fragments as previously described [12] using the BigDye terminator cycle sequencing kit (Life Technologies). Sequences were read on the 3100 or 3500 genetic analyser (Life Technologies) and analysed using the Sequencher software (Gene Codes Corporation). Haplotypes were assigned to mtDNA haplogroups using a combination of HVI sequence motifs and, in some cases, with SNPs distributed around the coding region of the mtDNA genome, genotyped as previously described [13].
(f). Data analysis
The genetic relationships between individuals were tested by pairwise comparison of the autosomal DNA profiles using the ML-Relate computer program which discriminates between unrelated, half-siblings, full-siblings and parent–offspring pairs [14]. Likelihood ratio (LR) was calculated using the Familias program. A median-joining (MJ) network was constructed with the Network program (Fluxus Technology Ltd) using the N1c1–TatC Y–STR haplotypes. Allele frequencies, observed and expected heterozygosity, Hardy–Weinberg equilibrium and haplotype diversities were calculated using the Arlequin software v. 3.1 [15].
(g). Contamination precautions
Measures were taken to avoid contamination with modern DNA as previously described [7,12,13]. To reduce ancient DNA STR genotyping errors and ensure the reliability of the results, only the amplified products from each sample and locus that were reproducible in at least four different amplification reactions were considered as authentic and included in the consensus DNA profiles reported in the electronic supplementary material, table S2.
3. Results
Of the 146 individual remains analysed by means of the Identifiler kit, 16 DNA samples appeared severely degraded, because no amplifiable product could be obtained (from at least three independent extracts). The remaining extracted samples gave 130 more or less complete autosomal DNA profiles (120 were full DNA profiles, whereas 10 were partial ones with only 6–14 amplified STR loci out of 15). Morphological and molecular typing results for sex determination were in accordance with each other, which was consistent among authentic ancient DNA extracts. Moreover, individual multi-allelic profiles were not mixtures of several individuals' DNA. They were different from each other and from the profiles of all involved in this study.
These Yakut DNA profiles were compared in pairs. To confirm child or (full-/half-)sibling status deduced from this pairwise comparison, Y-haplotypes obtained for the 62 male individuals and mtDNA haplotypes obtained from the 130 ancient Yakuts were also used.
(a). Kinship analyses within multiple graves
Whereas most of the graves excavated in Yakutia over 10 years were individual graves, seven of them were found to contain several individuals. Four were double graves, two were triple graves, and one was a multiple grave containing the remains of five individuals. Kinship investigations therefore focused first on these burials and notably on the grave named Shamanic tree 1 (ST1).
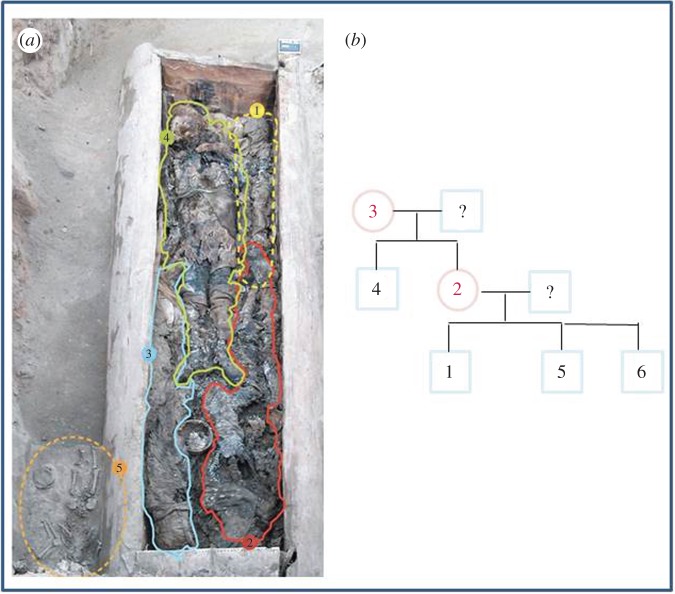
This grave, discovered in Central Yakutia and dated from the early-eighteenth century, was found to contain four mummified individuals along with the skeleton of a young child (found outside the coffin) (figure 2a and the electronic supplementary material, table S1) [16]. Anthropomorphic data revealed that subjects numbered 1 and 5 (ST1–1 and ST1–5) were children of about 5 and 4 years old, respectively; subject 2 (ST1–2) was a female in her 20s; subject 3 (ST1–3) was a female in her 50s and subject 4 (ST1-4) was a male over 30. Autosomal DNA profiling established a mother–offspring relationship between the adult female (ST1–3) and the two young adults (ST1–2 and ST1–4) (LR = 10.819), but also between the female 2 (ST1–2) and the two young children (ST1–1 and ST1–5; LR = 2.79 × 108) suggesting that three generations were present in the same grave (electronic supplementary material, table S2). This result was confirmed by the mtDNA and Y-chromosome analyses which showed that all the individuals belong to the same maternal lineage (electronic supplementary material, table S3) and that the putative brothers (ST1–1 and ST1–5) had the same Y-STR haplotype (electronic supplementary material, table S4). This paternal haplotype was also shared by the adult male (ST1–4), although the latter was not the father, but the maternal uncle of the two children (electronic supplementary material, table S4 and figure 2b). Genetic investigation further revealed that the body of a child about 10 years old (ST2–6) found in a grave (ST2) several metres away from the ST1 grave was that of a third son of female 2 (LR = 1.36 × 1011). From the autosomal DNA profiles of the mother and her three sons, we attempted to reconstruct the DNA profile of the biological father in order to identify him in our ancient Yakut sample. However, no putative father was found among the 130 autosomal STR profiles obtained.
Figure 2.
The Shamanic tree 1 grave. (a) The wooden coffin was found to contain four mummified individuals whereas the skeleton of a young child was found outside. (b) Genealogy of the six Yakut individuals found to belong to the same family. (Online version in colour.)
It was also possible to determine other close familial relationships within the multiple burials. For instance, in the triple grave OTII (excavated in the Vilyuy area and dating from the nineteenth century), one child (approx. 8 years old) was discovered along with two adult females (20–30 years old, individuals A and B; electronic supplementary material, table S1). This child (individual C) was probably the daughter of the female buried head to foot with her (individual B), as revealed by the autosomal DNA profiles (electronic supplementary material, table S2) (LR = 5 × 104). Moreover, both individuals carried the same mtDNA sequence unlike the two adult females who were seemingly not close relatives.
In the same way, the DNA profile of the female skeleton found in a triple grave of the early-nineteen century (excavated in the Verkhoyansk region and named LEP2) shared one allele at each locus with the two male infants (LEP2a and b) found close to her as well as the same mtDNA haplotype (electronic supplementary material, tables S1–S3). These data confirmed a mother–sons relationship (LR = 2.90 × 107). Interestingly, the newborn skeletons were so close in size and bone age that they were thought to be twins. However, because their DNA profiles were not identical, they could not be considered monozygotic twins (electronic supplementary material, table S2).
Nevertheless, not all of the multiple graves contained close relatives. The Tysarastaakh 2 grave (Verkhoyansk region, early-eighteenth century) was found to contain the remains of a child about 5 years old (TYS2-c) and those of an adult female (TYS2-a), who were thought to be a mother and her son. This putative relationship was however disproved by autosomal STR typing (LR = 0), and these individuals also did not share an identical HVI sequence (electronic supplementary material, table S3).
In an eighteenth century grave of the Verkhoyansk region, it was necessary to expand the type of markers tested to determine the exact familial link between two individuals, a male (IE1) and a female (IE2) about 15 years old. Indeed, the genetic analyses performed with the Identifiler kit on these two individuals revealed that they shared a common allele at each of the 15 loci tested along with sharing mtDNA sequences (electronic supplementary material, tables S2 and S3). These results suggest that the two individuals could be a mother and her son (LR = 1.5 × 105). Nevertheless, owing to the young age of these two individuals, we decided to refine the parentage assignment by using other markers. Indels typing revealed a parental exclusion at one locus (the male subject carried an insertion not observed in the female individual) suggesting a brother–sister kinship (LR = 2.69 × 105) rather than a mother–son kinship [17]. This case foreshadowed the difficulties that we would face throughout the genetic analysis of the single graves.
(b). Kinship analyses between graves, sites and regions
We subsequently tested whether individuals buried in separate graves or at different sites or even regions could be close relatives. The pairwise comparison approach notably allowed us to identify a brother–sister kinship between two adults found in separate graves at the Atyyr Meite site in the Verkhoyansk region. This comparison also revealed that these two individuals, a male (AM1) and a female (AM2), might be the children of a male individual (KUR1) excavated on the Kureleekh site, 70 km away from the Atyyr Meite site (LR = 3.6 × 108). Y-STR typing showed moreover that AM1 had the same Y-haplotype as the alleged father KUR1, confirming the paternity between them (electronic supplementary material, table S4). This relationship was also supported by the fact that all three graves were dated from the eighteenth century, making relatedness possible.
Through this approach, other parent–offspring links were revealed. One of those was between a male individual (OulA) and a child about 10 years old (OrtA; LR = 2.95 × 105), both buried in Central Yakutia. Another was between an adult female (MYR1) and a subadult male (KOU; LR = 4.33 × 104), both unearthed in the Verkhoyansk region. Full-sibling relationships were also found. One between the male adult LEP3 and the female adult LEP2 (LR = 6.8 × 103): both graves were 300 m away from each other, and the individuals shared a common mtDNA sequence.
In some cases, however, pairwise comparison revealed discordant results between the biparental and the two uniparental systems used. For instance, the multi-allelic profile obtained from the skeletal remains recovered from the MunU1 grave supported a parent–offspring relationship with the KenEb 2 individual from a distant grave (electronic supplementary material, table S5). Nevertheless, because these two ‘relatives' did not share a Y haplotype (electronic supplementary material, table S4), we had to consider the possibility that the two individuals were not a biological father and his son (as confirmed by the low value of the LR = 7). For other pairs of individuals (OttoA1 and Pokrovsk’ individual (LR = 13); ST1-2 and LEP1 (LR = 98); SeTou and OttoA1 (LR = 44); OTIIA and OttoA1 (LR = 37); KER1 and KUR2 (LR = 3.7 × 103)), the parent–offspring relationship inferred by ML-Relate was excluded by the low values of the LR along with the absence of a common mtDNA sequence and/or by incompatibilities with the dating of the graves.
To further understand these discrepancies, we decided to expand the number of autosomal STR markers analysed and to employ a newly commercialized kit (GlobalFiler, Life Technologies) developed for use with challenging forensic casework samples. This 24 locus STR kit, tested on some of the aforementioned samples, allowed us to exclude false parent–offspring relationships deduced from ML-Relate. As an example, the DNA profiles obtained for ST1–2 and the LEP1 male individual revealed exclusions at two loci (D2S441 and D22S1045), those obtained for SeTou and OttoA1 showed exclusion at the SE33 locus, whereas those established for KER1 and KUR2 showed exclusion at two loci (SE33 and D1S1656; electronic supplementary material, table S2). In each case, this additional genetic information reduced the LR values for a parent–offspring relationship to zero.
This new kit was also used to verify the accuracy of the kinships previously determined, namely the parent–offspring relationships between KUR1 and AM1 and AM2 (LR = 9.5 × 108), between OulA and OrtA (LR = 3.94 × 105), and between MYR1 and KOU (LR = 1.41 × 106). In all of these cases, the 24 STR loci increased the LR values and confirmed previously ascertained kinship relationships.
(c). Reduced diversity of the Y-chromosome versus high diversity of the mitochondrial DNA
The Y-STR analysis, performed on the 62 ancient Yakut male individuals by means of the Y-Filer kit (Life Technologies), revealed low haplotype diversity (0.7726 ± 0.0548). A total of 19 different haplotypes were observed but one of them was carried by almost half of the males tested (29 out of 62; electronic supplementary material, table S4). Almost 79% (15 out of 19) of these Y-haplotypes were found to belong to the N1c1 haplogroup (Tat-C allele). The four others were assigned to haplogroups N1c2b–P43 or C2-M217. The MJ network constructed with the N1c1-Tat haplotypes shows a star-like pattern; most of the haplotypes are grouped around a founder haplotype (the most frequent one) at a distance ranging from one to six mutation steps (electronic supplementary material, figure S1). This pattern, along with the high frequency of the founder haplotype, is indicative of a founder effect [18].
In an attempt to increase the Y-haplotype resolution and the possibility to differentiate between closely and distantly related males, another commercial kit, including six additional STR loci with high gene diversity was used, the PowerPlex Y23 system (Promega). The 29 ancient Yakut males sharing the same 17 Y-STR haplotype were typed with this kit. Despite the addition of the new loci, among which rapidly mutating loci, only four of the 29 male individuals could be distinguished from each other (electronic supplementary material, table S4). The most informative markers for differentiating the Yakut male lineages were DYS643, DYS570 and DYS576, considered to be rapidly mutating Y-STR loci [19]. Thus, it appears from our study that 23 polymorphic Y-STRs are not sufficient to differentiate male lineages in a population with a founder effect.
The results obtained from the mtDNA analysis revealed a higher haplotype diversity of the maternal lineages (0.9559 ± 0.0079). A total of 44 different haplotypes were observed (data not shown). Most of them fall into eastern Eurasian haplogroups (A, B, C, D, G, F, Y and Z) whereas western Eurasian lineages (H, J, T and W) constituted a minority (6.9%). Haplogroups D and C which include 38% and 37% of all lineages, respectively, were prevalent. Whereas D4 was represented by a diverse set of subclades, D5 consisted exclusively of subhaplogroup D5a2. The second dominant haplogroup was C, represented mainly by two major subhaplogroups C4 and C5. These subhaplogroups are present in many Siberian populations, including various Mongol–Turkic populations from south Siberia. However, lineages of subcluster D5 are shared only with present-day Yakuts, Yukaghirs, Evens and Evenks. The influence of these neighbouring populations is therefore clearly visible in the mtDNA lineages of the ancient Yakuts.
4. Discussion
In this study, genealogical relationships were reconstructed using biparental, paternal and maternal genetic systems in a sample of 130 Yakut skeletal remains dating mostly from the fifteenth to the nineteenth centuries and collected as part of a 10-year long field campaign. One of the main strengths of this study was the possibility of combining archaeological, anthropological and molecular data to assess the genetic relationships within this ancient ethnic group. Although both maternal and paternal genetic contributions can be assessed with autosomal markers such as STRs, we wanted to gain a higher power of discrimination by typing STRs of the non-recombining part of the Y chromosome as well as polymorphisms in the HVI region of the mtDNA.
In most cases, the results from the two uniparental marker systems corroborated those from autosomal DNA analyses and enabled us to identify kin groupings in the same grave and reconstitute a partial history of some of the deceased individuals. This multi-marker approach was useful for example in the case of the Shamanic tree 1 grave in which five members of the same family were found. This discovery was unusual, because individual burials were the standard practice in Yakutia [20]. Anthropological and archaeological investigations suggested that these individuals were buried simultaneously, shortly after death, in the wintertime. On the basis of these observations, the hypothesis of a lethal epidemic was considered. Biological samples were taken from the young mother (ST1–2) and subjected to histological investigations. Microscopical examination of pulmonary tissue showed iron inclusions suggestive of the presence of blood after a possible haemorrhagic episode. One of the hypotheses tested was that of a smallpox infection. This possibility was confirmed by PCR amplifications revealing preserved short DNA fragments of the variola virus [16]. We therefore hypothesized that the variola virus found in this mummy might have decimated the entire family and that the child (ST2–6) found in a separate grave might have escaped the epidemic.
This multi-marker strategy also provided evidence against close relatedness between some individuals found in the same burial. Therefore, the ancient Yakuts did not bury individuals together solely on the basis of kinship, but also of social criteria such as clan structure or other unknown criteria such as adoption (possibly the case of the Tysarastaakh 2 grave). Similar results were reported in a previous article showing that the two male individuals found in a double grave at the At-Dabaan site (Central Yakutia, eighteenth century) shared the same maternal lineage, but were not close relatives [2].
Conversely, close familial relationships were revealed for individuals buried in separate graves. In these cases, anthropological and archaeological data supported the accurate determination of the relatedness. Indeed, the dating and location of the graves along with the age at death of the studied individuals, allowed us to establish who was the child and who was the parent. In some instances, these data disproved a kinship suggested by the pairwise comparison of the DNA profiles. As an example, the ML-Relate software suggested a parent–offspring relationship between OttoA1, a woman buried in a nineteenth century grave, and the male individual from the Pokrovsk grave (dated by accelerator mass spectrometry from 2400 to 2200 years BP) [5]. However, these 2000 year distant individuals could not have been closely related.
(a). Inconsistencies in the autosomal short tandem repeat analysis
Despite the use of a commercial kit dedicated to forensic practices, in some cases, false inclusions were inferred by ML-Relate. Autosomal DNA profiles suggested that two individuals could be parent–offspring (one allele shared at each of the 15 amplified locus) when they were not. Full-siblings were also found to share an allele at every locus when they should not have. For half-sibling cases, the risk of false inclusions is more difficult to detect than for parent–offspring or siblings. It is indeed problematic to test relationships in which allele sharing is not required at every locus (e.g. full- or half-siblings and more distant relationships such as cousins, uncle and niece or aunt and nephew, grandfather and granddaughter or grandmother and grandson…). The observed inconsistencies thereby substantially increased the difficulty in establishing with certainty the precise familial relationships among the ancient Yakuts.
In order to accurately identify individual family members, a new commercially available autosomal STRs kit was used. Increasing the number of typed autosomal STR loci to 21 resulted in the complete discrimination of true parent–offspring relatives versus more distant relatives (electronic supplementary material, table S2, part in blue). This result shows that the 15 forensic STR loci were not sufficient to definitively discriminate between pairs of close relatives and unrelated individuals in our ancient human sample. The resulting lack of resolution can be illustrated by a study performed 8 years ago on two Yakut individuals (SS1 and SS2) from the Sytygane Syhe burial site [4]. The autosomal STR analysis suggested that a father–son relationship between SS1 and SS2 was highly probable (LR = 283). Subject SS1 (an adult male) would be the father, and subject SS2 (an immature male) his son. In this previously published study, it was noted that ‘the absence of data for the historical allelic frequencies of the originating population constitutes a bias in determining the parentage relationship’. This result, deduced from the analysis of nine STR loci (using the AmpFlSTR Profiler Plus, Life Technologies) was invalidated by our recent analyses which revealed exclusion at two of the 21 autosomal STR-loci tested and gave an LR value of zero.
To better understand the low resolution of the kits currently used in forensic practices when applied to the Yakut population, we first compared allele frequency distributions observed in our ancient sample with those available in present-day Yakut populations [21,22] as well as in worldwide populations available in the ‘allSTR autosomal database’ (http://allstr.de). No significant difference between modern populations and ancient Yakuts was observed (data not shown). We also tested for heterozygote deficiency and there was no deviation from genetic equilibrium. The p-values from Hardy–Weinberg were not low except for the locus FGA where it was close to 0 (electronic supplementary material, table S6).
(b). Reduced diversity of the Y-chromosome versus high diversity of the mitochondrial DNA
The non-recombining part of the Y chromosome being identical between fathers and sons (except in the cases of mutation), a Y-chromosomal STR analysis was carried out to validate the biological relationships between Yakut males deduced from the autosomal STR analysis. The most surprising result of this Y-chromosomal typing was the inability to differentiate between close and distant patrilineal relatives. Indeed, even though the PowerPlex Y23 kit has been shown to slightly increase the differentiation of both related and unrelated males, the presence of a dominant paternal lineage hampered a fine-scale resolution of the kinship. As an example, in the Shamanic tree 1 family, the maternal uncle (ST1–4) had the same Y-haplotype (defined by 23 STR loci) as the two young children (ST1–1 and ST1–5; electronic supplementary material, table S4) and therefore might have been considered as their father in the absence of autosomal data.
The dominant haplotype belonged to the N1c1–Tat haplogroup like most of the other Y-haplotypes carried by the ancient Yakuts. It appears to be the ancestral haplotype as revealed by the MJ network (electronic supplementary material, figure S1) which shows the other N1c1–Tat haplotypes clustering around it. This star-like expansion, also observed with present-day Yakut populations, has been interpreted as evidence of a dramatic decrease of the effective population size, possibly owing to a strong founder effect [18,23–25]. It is noteworthy that, even though the prevalent ancient haplotype has been transmitted to the present-day Yakuts [26], it has not been found in all populations available for comparison, a fact which may hint at a limited male gene flow from Yakuts to neighbouring Siberian populations.
The observed low level of Y-chromosome diversity in the ancient Yakuts contrasts with the quite high diversity of the maternal lineages. The mtDNA sequences used for identifying maternally related individuals (including mother–son, full-siblings, maternal half-siblings and maternal uncle–nephew) can be classified in two types of haplotypes: high-frequency ones (belonging to haplogroups C and D which occur at similar frequencies and together account for almost 75% of the gene pool) and low-frequency ones (haplogroups A, B, G, F, H, J, T, W or Z) which contribute to the overall high diversity. Although Y-STR typing provides evidence for a founder effect in the Yakut paternal line, this is far less pronounced in the maternal line. The only indication of a founder effect is the high frequency of subhaplogroup D5a2. This subcluster which is dominant not only in the ancient Yakut sample, but also in the present-day Yakut populations might therefore be a founder lineage or result from a bottleneck effect [5,25,27,28]. Its presence among neighbouring Siberian populations (Evens, Evenks or Yukaghirs) could be owing to gene flow with the immigrating Yakuts.
(c). Possible explanations for the discrepancies observed
Considering that the genetic diversity displayed by molecular markers is strongly dependent on the specific demographic history of a population, we tried to understand which events could have led to the low discrimination power of the autosomal DNA profiles along with a large number of indistinguishable Y-STR haplotypes in the ancient Yakuts. According to several authors, the severe limitation of the Y-chromosomal diversity could be owing to the migration to the north of a small group of related Turkic men [5,23,24]. During this expansion along the Lena River, substantial admixture with local women (namely the Yukaghirs- and Tungusic-speaking Evenks and Evens) would have introduced high mtDNA variation into the Yakut founder population and obscured the genetic signature of the recent founder effect based on NRY and STR data [28]. A further explanation for the difference between Y-chromosomal and mtDNA diversity in Yakuts could be linked to specific cultural practices such as patrilocality, strict exogamy and polygyny [29]. In addition, warfare might have contributed to a reduction of the paternal but not the maternal lineages. Nevertheless, the overall reduced autosomal diversity observed also indicates a substantial level of inbreeding possibly owing to a biased reproductive success in men (only a few of them father most of the offspring) or to consanguineous marriages. However, no heterozygote deficiency was observed in our ancient sample.
Therefore, the puzzle is far from being fully resolved. The fact that the Yakut population was severely depopulated by smallpox and tuberculosis epidemics after the Russian colonization could moreover add further complexity to the analysis of the gene pool structure of the Yakuts [30]. The exact story of the Yakuts therefore remains incomplete, and further research is needed to fully elucidate the genetic evolution of the Yakuts.
5. Conclusion
This study is part of a long-term project devoted to the history of the Yakuts. By studying diploid and haploid (mtDNA, NRY) polymorphisms in three groups of ancient Yakuts from Central, Western (the Vilyuy basin) and Northern (the Verkoïansk region) Yakutia, we aimed to further our understanding of their genealogical history. The most conspicuous finding of these analyses, however, was the difficulty in differentiating close and more distant relatives with STR marker sets routinely used in forensic practices. Our study clearly shows that to reduce the risk of adventitious matches and to build a clear picture of biological relationships within ancient human groups in the absence of parental information, it is absolutely essential to combine information from diploid and haploid loci and to increase their number. In addition, because geographical and temporal differences between ancient samples make observations of genetic differentiation difficult to interpret, it is essential to combine archaeological, cultural, biological and historical data. More robust interpretations of ancient DNA data could be gained by a multidisciplinary study. Such investigations are ongoing in our group.
Supplementary Material
Funding statement
This work was supported by the French Archaeological Missions in Oriental Siberia (Ministry of Foreign and European Affairs, France), the North-Eastern Federal University (Yakutsk, Sakha Republic) and the Human Adaptation programme of the French Polar Institute Paul Emile Victor.
References
- 1.Fedorova SA, et al. 2013. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of northeast Eurasia. BMC Evol. Biol. 13, 127 ( 10.1186/1471-2148-13-127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricaut FX, Kolodesnikov S, Keyser-Tracqui C, Alekseev AN, Crubézy E, Ludes B. 2004. Genetic analysis of human remains found in two eighteenth century Yakut graves at At-Dabaan. Int. J. Legal Med. 118, 24–31. ( 10.1007/s00414-003-0411-6) [DOI] [PubMed] [Google Scholar]
- 3.Ricaut FX, Fedoseeva A, Keyser-Tracqui C, Crubézy E, Ludes B. 2005. Ancient DNA analysis of human Neolithic remains found in northeastern Siberia. Am. J. Phys. Anthropol. 126, 458–462. ( 10.1002/ajpa.20257) [DOI] [PubMed] [Google Scholar]
- 4.Ricaut FX, Kolodesnikov S, Keyser-Tracqui C, Alekseev AN, Crubézy E, Ludes B. 2006. Molecular genetic analysis of 400-year-old human remains found in two Yakut burial sites. Am. J. Phys. Anthropol. 129, 55–63. ( 10.1002/ajpa.20195) [DOI] [PubMed] [Google Scholar]
- 5.Amory S, Crubézy E, Keyser C, Alekseev AN, Ludes B. 2006. Early influence of the steppe tribes in the peopling of Siberia. Hum. Biol. 78, 531–549. ( 10.1353/hub.2007.0001) [DOI] [PubMed] [Google Scholar]
- 6.Amory S, Keyser C, Crubézy E, Ludes B. 2007. STR typing of ancient DNA extracted from hair shafts of Siberian mummies. Forensic Sci. Int. 166, 218–229. ( 10.1016/j.forsciint.2006.05.042) [DOI] [PubMed] [Google Scholar]
- 7.Crubézy E, et al. 2010. Human evolution in Siberia: from frozen bodies to ancient DNA. BMC Evol. Biol. 10, 25 ( 10.1186/1471-2148-10-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murail P, Bruzek J, Braga J. 1999. A new approach to sexual diagnosis in past populations: practical adjustments from Van Vark's procedure. Int. J. Osteoarchaeol. 9, 39–53. ( 10.1002/(SICI)1099-1212(199901/02)9:1%3C39::AID-OA458%3E3.0.CO;2-V) [DOI] [Google Scholar]
- 9.Keyser-Tracqui C, Ludes B. 2005. Methods for the study of ancient DNA. Methods Mol. Biol. 297, 253–264. [DOI] [PubMed] [Google Scholar]
- 10.Hollard C, Keyser C, Giscard PH, Tsagaan T, Bayarkhuu N, Bemmann J, Crubezy E, Ludes B. 2014. Strong genetic admixture in the Altai at the Middle Bronze Age revealed by uniparental and ancestry informative markers. Forensic Sci. Int. Genet. 12, 199–207. ( 10.1016/j.fsigen.2014.05.012) [DOI] [PubMed] [Google Scholar]
- 11.Gill P, et al. 2012. DNA commission of the International Society of Forensic Genetics: recommendations on the evaluation of STR typing results that may include drop-out and/or drop-in using probabilistic methods. Forensic Sci. Int. Genet. 6, 679–688. ( 10.1016/j.fsigen.2012.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keyser-Tracqui C, Crubézy E, Ludes B. 2003. Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia. Am. J. Hum. Genet. 73, 247–260. ( 10.1086/377005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keyser C, Bouakaze C, Crubézy E, Nikolaev VG, Montagnon D, Reis T, Ludes B. 2009. Ancient DNA provides new insights into the history of south Siberian Kurgan people. Hum. Genet. 126, 395–410. ( 10.1007/s00439-009-0683-0) [DOI] [PubMed] [Google Scholar]
- 14.Kalinowski ST, Wagner AP, Taper ML. 2006. ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579. ( 10.1111/j.1471-8286.2006.01256.x) [DOI] [Google Scholar]
- 15.Excoffier L, Laval G, Schneider S. 2007. Arlequin (version 3.0): an integrated software package for population genetic data analysis. Evol. Bioinform. Online 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 16.Biagini P, et al. 2012. Variola virus in a 300-year-old Siberian mummy. N. E. J. Med. 367, 2057–2059. ( 10.1056/NEJMc1208124) [DOI] [PubMed] [Google Scholar]
- 17.Hollard C, Mendisco F, Keyser C, Crubézy E, Ludes B. 2011. First application of the investigator DIPplex indels typing kit for the analysis of ancient DNA samples. Forensic Sci. Int. Genet. 3(Supplement Series), e393–e394. ( 10.1016/j.fsigss.2011.09.058) [DOI] [Google Scholar]
- 18.Puzyrev VP, Stepanov VA, Golubenko MV, Puzyrev KV, Maximova NR, Kharkov VN, Spiridonova MG, Nogovitsina AN. 2003. mtDNA and Y-chromosome lineages in the Yakut population. Russ. J. Genet. 39, 816–822. ( 10.1023/A:1024761305958) [DOI] [PubMed] [Google Scholar]
- 19.Thompson JM, et al. 2013. Developmental validation of the PowerPlex® Y23 system: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci. Int. Genet. 7, 240–250. ( 10.1016/j.fsigen.2012.10.013) [DOI] [PubMed] [Google Scholar]
- 20.Crubezy E, Alekeev A. 2007. Chamane: Kyys, jeune fille des Glaces. 167 p. Paris, France: Edts Errances. [Google Scholar]
- 21.Zhivotovsky LA, Akhmetova VL, Fedorova SA, Zhirkova VV, Khusnutdinova EK. 2009. An STR database on the Volga–Ural population. Forensic Sci. Int. Genet. 3, e133–e136. ( 10.1016/j.fsigen.2008.11.001) [DOI] [PubMed] [Google Scholar]
- 22.Zhirkova VV, Fedorova SA, Akhmetova VL, Zhivotovsky LA, Khusnutdinova EK. 2011. Allelic polymorphism of six microsatellite DNA loci in populations of Sakha (Yakutia). Mol. Biol. 45, 221–228. ( 10.1134/S0026893311020221) [DOI] [PubMed] [Google Scholar]
- 23.Pakendorf B, Morar B, Larissa A, Tarskaia LA, Kayser M, Soodyall H, Rodewald A, Stoneking M. 2002. Y-chromosomal evidence for a strong reduction in male population size of Yakuts. Hum. Genet. 110, 211–224. ( 10.1007/s00439-001-0664-4) [DOI] [PubMed] [Google Scholar]
- 24.Kharkov VN, Stepanov VA, Medvedeva OF, Spiridonova MG, Maksimova NR, Nogovitsina AN, Puzyrev VP. 2008. The origin of Yakuts: analysis of the Y-chromosome haplotypes. Mol. Biol. 42, 198–208. ( 10.1134/S0026893308020040) [DOI] [PubMed] [Google Scholar]
- 25.Pakendorf B, Novgorodov IN, Osakovskij VL, Danilova AP, Protod'jakonov AP, Stoneking M. 2006. Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakuts. Hum Genet. 120, 334–353. ( 10.1007/s00439-006-0213-2) [DOI] [PubMed] [Google Scholar]
- 26.Thèves C, Balaresque P, Evdokimova LE, Timofeev IV, Alekseev AN, Sevin A, Crubézy E, Gibert M. 2010. Population genetics of 17 Y-chromosomal STR loci in Yakutia. Forensic Sci. Int. Genet. 4, e129–e320. ( 10.1016/j.fsigen.2010.01.018) [DOI] [PubMed] [Google Scholar]
- 27.Tarskaia LA, Melton P. 2006. Comparative analysis of mitochondrial DNA of Yakuts and other Asian populations. Russ. J. Genet. 42, 1439–1446. ( 10.1134/S102279540612012X) [DOI] [PubMed] [Google Scholar]
- 28.Zlojutro M, Tarskaia LA, Sorensen M, Snodgrass JJ, Leonard WR, Crawford MH. 2009. Coalescent simulations of Yakut mtDNA variation suggest small founding population. Am. J. Phys. Anthropol. 139, 474–482. ( 10.1002/ajpa.21003) [DOI] [PubMed] [Google Scholar]
- 29.Balzer MM. 1994. Yakut . In Encyclopedia of world cultures, Russia and Eurasia/China, vol VI.G.K. (eds Friedrich P, Diamond N.), pp. 404–407. London, UK: Hall and Company. [Google Scholar]
- 30.Dabernat H, et al. 2014. Tuberculosis epidemiology and selection in an autochtonous Siberian population from the 16th–19th century. PLoS ONE 26, e89877 ( 10.1371/journal.pone.0089877) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.