Abstract
When biomolecules attach to engineered nanoparticle (ENP) surfaces, they confer the particles with a new biological identity. Physical format may also radically alter, changing ENP stability and agglomeration state within seconds. In order to measure which biomolecules are associated with early ENP growth, we studied ENPs in conditioned medium from A549 cell culture, using dynamic light scattering (DLS) and linear trap quadrupole electron transfer dissociation mass spectrometry. Two types of 100 nm polystyrene particles (one uncoated and one with an amine functionalized surface) were used to measure the influence of surface type. In identically prepared conditioned medium, agglomeration was visible in all samples after 1 h, but was variable, indicating inter-sample variability in secretion rates and extracellular medium conditions. In samples conditioned for 1 h or more, ENP agglomeration rates varied significantly. Agglomerate size measured by DLS was well correlated with surface sequestered peptide number for uncoated but not for amine coated polystyrene ENPs. Amine-coated ENPs grew much faster and into larger agglomerates associated with fewer sequestered peptides, but including significant sequestered lactose dehydrogenase. We conclude that interference with extracellular peptide balance and oxidoreductase activity via sequestration is worthy of further study, as increased oxidative stress via this new mechanism may be important for cell toxicity.
Keywords: nanoparticles, peptides, corona, agglomerate, LDH, surface
1. Introduction
The agglomeration state of extrinsic particles in biological systems and cell culture media influences their biological fate and impacts [1]. The surface chemistry influences the size distribution, because the bio–nano interface adsorbs surrounding molecules to form a ‘corona’ and affects particle size distribution stability [2]. In vivo, a protein corona forms and becomes a predictor of biological outcomes [3].
Particle size distribution influences clearance efficiency, cellular uptake and translocation in vivo, for atmospheric particulate matter (PM), atmospheric or ultrafine nanoparticles (NPs or UFPs) and engineered nanoparticles (ENPs) [4,5]. When introduced to the human body or other biological systems, a protein/phospholipid corona quickly forms, depending on the route of entry and the molecules encountered. This was first demonstrated using atmospheric particles in human lung lining fluid and later with ENPs in serum [6–8]. Particle surface type strongly influences the corona [9,10]: in serum between 15 and 40% of attached proteins uniquely associated with three surface variants of 100 nm polystyrene ENPs (plain, amine and carboxylate surfaces).
Several studies linked protein corona composition and particle uptake by cells in vitro [3]. Once introduced to cell culture media and biological fluids in vivo, NPs and monodisperse ENPs become polydisperse depending on particle surface characteristics and the conditions such as surfactant and ionic concentration, charge and pH [11]. Surfaces are coated in host molecules to form a ‘corona fingerprint’, and this influences biological outcomes [3,12]. Low concentrations of polymers cause small interfacial force change and alter rheological properties of particles either to promote or reduce aggregation [13,14]. Monopoli et al. [15] showed that corona composition also varies significantly at in vivo protein concentrations of 80% compared with lower in vitro concentrations (cell culture typically contains 10% or less proteins).
Attachment of peptides to the particle surfaces and ENPs is also significant because peptide–ENP constructs generate antibodies against specific peptides [16,17]. Peptides are used as antigens by coupling to carrier proteins so that epitopes are exposed, but ENPs may also anchor peptides to expose or even concentrate epitopes, initializing different or exaggerated biological responses [17]. For example, in the case of IL-8, adsorption to diesel exhaust particles was observed without loss of epitopes [18]. Studies of surfactant proteins have shown that protein sequestration by NPs changes particle uptake by cells and the infectivity of influenza in vitro [19,20]. Cell medium containing heat inactivated serum proteins and complement resulted in lower NP uptake compared with the non-heat inactivated case [4,5]. Such ENP surface sequestration may thereby disrupt the concentration or function of important proteins within the system. Surface modification of monodisperse ENPs in suspension can directly destabilize the size distribution, and previous studies noted agglomeration of NPs prior to any contact with cells [15,21].
In all these examples, the different protein corona compositions probably resulted in dynamic NP agglomeration states, and the cells secreted proteins in response to the presence of the NPs. In no cases were studies to assess this reported: even where characterization was reported, it was always under unrealistic exposure conditions, as cells are constantly secreting proteins [22]. Thus, characterization of NP agglomeration for in vitro studies should consider changes resulting from secreted proteins.
In this study, we hypothesized that exposed polystyrene ENPs would acquire a protein corona from secreted proteins in conditioned medium, without any contact with cells, that this would affect ENP size distributions and that we could identify the peptides responsible. Only conditioned medium (serum-free medium plus cell exudates, minus cell debris) was interacted with ENPs, so that at no time during the experiments were cells exposed to ENPs. The role of opsonization in cellular uptake is addressed in other work, while these experiments isolated the physical chemistry of protein corona formation during particle growth from cellular responses.
2. Material and methods
(a). Preparation of cell cultures
All chemicals were of the highest quality available and obtained from Sigma-Aldrich (Poole, UK) unless otherwise stated. Human A549 epithelial lung carcinoma cells (ECACC no. 86012804) were cultured in T75 (Falcon) culture flasks at 37°C in a humidified chamber, containing 5% carbon dioxide in phenol red-free Dulbecco's modified Eagle's medium (DMEM) supplemented with heat inactivated fetal bovine serum (10% v/v; FBS, Gibco, Life Technologies, UK), l-glutamine (2 mM), streptomycin (100 µg ml−1) and penicillin (100 U ml−1). Cells were subcultured approximately every 3 days at 90–95% confluence.
(b). Preparation of conditioned medium
Conditioned medium was prepared so that ENPs did not contact cells directly in these experiments. In brief, fresh serum-free medium was added to cells for specific time periods, removed, centrifuged, transferred to new cuvettes and ENPs were added for immediate DLS measurements. The detailed methods are explained below.
Cells were seeded into 35 mm dishes at a density of approximately 0.5 × 106 cells per well in complete medium (see §2a) and cultured until 95% confluent (approx. 2–3 days). The cell culture medium was removed and cells were washed carefully with warm phosphate buffered saline (3 × 3 ml) to remove all traces of the serum-containing culture medium. Next, pre-warmed serum-free DMEM (3 ml) was added to each dish and the cells were incubated with the medium for the time periods 0.25, 0.5, 0.75, 1, 6 and 12 h after which the conditioned medium was recovered and mixed with the ENPs. Based on these initial experiments, 1 h of medium incubation with cells was demonstrated to result in sufficient medium conditioning to cause measurable agglomeration in approximately 50% of samples.
In subsequent samples, serum-free medium was incubated with cells for 1 h before being transferred to a 15-ml sterile centrifuge tube. Cellular debris and other suspended material were removed by centrifugation (6000g, 20 min, 4°C, MSE Mistral 3000i) and the supernatant carefully transferred to a clean sterile centrifuge tube and used as soon as possible for dynamic light scattering (DLS) studies.
(c). Size and zeta potential measurements of nanoparticles
Unmodified polystyrene and amine surface treated polystyrene ENPs were purchased (Polyscience, UK) and well characterized using DLS (Zetanano, Malvern Instruments, Malvern, UK). ENPss were nominally 100 nm, as bought, but varied slightly (±20 nm) by surface type. ENPs were suspended in 1 ml nanopure water and unconditioned DMEM in disposable vials (Malvern Instruments, UK). Polydispersity, hydrodynamic diameter and zeta potential were determined. Where polydispersity was more than 0.8 (all samples), the hydrodynamic diameter was identified as the major single peak achieved by fitting a multi-exponential model to the DLS correlation function. The refractive index and other measured characteristics of each ENP have been reported previously [23]. Zeta potential was measured using a Zetamaster (Malvern Instruments, UK).
(d). Dynamic light scattering size measurement of conditioned medium only
Following centrifugation and prior to the addition of ENPs, the conditioned medium alone was analysed by DLS to detect any suspended material. One millilitre of conditioned medium was used as the suspension medium and no ENPs were added before measurement by DLS (as described in §2c).
(e). Dynamic light scattering size measurement of engineered nanoparticles in conditioned medium
Following the addition of the polystyrene ENPs to the conditioned medium, the size distribution was measured at specific intervals after introduction time t = 0. Measurements were made at t ≈ 0.2, 1–2 and 15 h. The change in size distribution over that period indicated the level of agglomeration in the sample, and this agglomeration was due to ENPs interacting with components of the conditioned medium. This experiment was conducted blind, so that the particle types were mixed.
(f). Nanoparticle associated protein analysis
A subset of the samples was taken for proteomics analysis, based on the agglomeration state at t = 1 h. Blinded samples were selected representing a range of ENP growth rates and underwent proteomic analysis. Ten selected samples of ENPs in conditioned medium were immediately taken for analysis of attached protein within the Functional Genomics and Proteomics Facility (University of Birmingham), using a standardized protocol [24]. Briefly, samples were centrifuged to a pellet and washed three times with nanopure water. One hundred microlitres of resuspended sample was taken and 25 µl 10 mM DTT and 100 mM ammonium bicarbonate were added. The sample was heat denatured at 60°C for 15 min, then cooled for 5 min to room temperature. Twenty-five microlitres of 50 mM iodoacetamide was added, then the sample was left for 45 min at room temperature in the dark. Twenty-five microlitres of trypsin was added and samples were incubated at 37°C overnight. Samples were cleaned up on a C18 zip tip and eluted in 50 : 50 acetonitrile/water. Samples were dried and resuspended in 10 µl 0.1% formic acid in water.
Samples were analysed using nano liquid chromatography–mass spectrometry (nanoLC-MS) technique. An UltiMate 3000 high performance liquid chromatograph (HPLC) series (Dionex, Sunnyvale, CA, USA) was used for peptide concentration and separation. Samples were trapped on a µPrecolumn Cartridge, Acclaim PepMap100 C18, 5 µm, 100 Å 300 µm i.d. × 5 mm (Dionex) and separated in Nano SeriesT Standard Columns 75 µm i.d. × 15 cm, packed with C18 PepMap100, 3 µm, 100 Å (Dionex). The gradient used was from 3.2 to 44% solvent B (0.1% formic acid in acetonitrile) for 30 min. Peptides were eluted directly (approx. 300 nl min−1) via a Triversa Nanomate nanospray source (Advion Biosciences, NY, USA) into a linear trap quadrupole (LTQ) Orbitrap Velos electron transfer dissociation (ETD) mass spectrometer (ThermoFisher Scientific, Germany). The data-dependent scanning acquisition was controlled by xcalibur 2.7 software. The mass spectrometer alternated between a full FT-MS scan (m/z 380–1600) and subsequent collision-induced dissociation (CID) MS/MS scans of the 20 most abundant ions. Survey scans were acquired in the Orbitrap with a resolution of 30 000 at m/z 400 and automatic gain control (AGC) 1 × 106. Precursor ions were isolated and subjected to CID in the linear ion trap with AGC 1 × 105. Collision activation for the experiment was performed in the linear trap using helium gas at normalized collision energy to precursor m/z of 35% and activation Q 0.25. The width of the precursor isolation window was 2 m/z and only multiply-charged precursor ions were selected for MS/MS.
The MS and MS/MS scans were searched against the NCBInr database using the Mascot algorithm (Matrix Sciences), a powerful search engine used to identify proteins from primary sequence databases. The experimental mass values were compared with calculated peptide mass or fragment ion mass values, obtained by applying cleavage rules to the entries in a comprehensive primary sequence database (NCBI). By using an appropriate scoring algorithm, the closest match or matches were identified. If the ‘unknown’ protein was present in the sequence database, then the aim was to pull out that precise entry. If the sequence database did not contain the unknown protein, then the aim was to pull out those entries which exhibit the closest homology, often equivalent proteins from related species. Variable modifications were deamidation (N and Q), oxidation (M) and phosphorylation (S, T and Y). The precursor mass tolerance was 5 ppm and the MS/MS mass tolerance was 0.8 Da. Two missed cleavages were allowed and was accepted as a real hit, where the protein contained at least two high confidence peptides.
Control samples (blank samples containing only ENPs) underwent identical analysis.
(g). Protein results analysis
Protein data were analysed blind to avoid bias; samples were identifiable only by numbers. Proteins were considered to be frequently associated with the ENPs if more than six peptides of the protein were found in any of the nine samples analysed, and not present in the untreated control sample. Twenty-five proteins were selected to represent infrequently associated proteins (with only one association in all nine samples).
The frequently associating proteins were extensively characterized in terms of their physicochemical and functional properties. The generic characteristics of the frequently and infrequently associating proteins were compared, using the ProtParam tool from the ExPASy Bioinformatics Portal, to determine whether any specific characteristics (e.g. isoelectric point (pI), hydropathicity and aliphatic index) could be identified which enhanced the propensity of the protein to interact with the polystyrene ENPs. Furthermore, the percentage of four amino acids (asparagine, leucine, serine and valine) previously demonstrated to deposit onto fine airborne particles (PM2.5) [7] was determined to ascertain whether these amino acids played an important functional role in the interactions between proteins and the polystyrene ENPs. Sequence alignment [25] and analysis [26] were then carried out on the frequently associating proteins to identify any conserved sequence motifs present in all eight proteins.
(h). Protein functional analyses
Enrichment analysis for the adsorbed proteins identified by MS was carried out using gene ontology terms. Analysis was performed using Generic Geneontology Term Finder (http://go.princeton.edu/cgi-bin/GOTermFinder) and visualized with Reduce + Visualize Gene Ontology (REVIGO) (http://revigo.irb.hr/) to identify important biological and molecular functions for each protein dataset and grouped dataset (amine-functionalized versus unmodified polystyrene).
3. Results
After centrifugation, no suspended material was detected by DLS in the conditioned medium alone in the size range of the ENPs. In some samples, small sub-10 nm peaks were observed, attributable to unidentified macromolecules.
(a). Nanoparticle agglomeration in conditioned medium
The increases in hydrodynamic diameter of ENPs suspended in nanopure water and conditioned medium were recorded and compared with values for the control ENPs suspended in nanopure water, which exhibited zero growth (0 nm). There was significant variability in growth rates of ENPs in conditioned medium. A change in particle size distribution was typically observed at the first measurement (t = 10 min), with an increase in the hydrodynamic diameter, indicating that a corona formed very quickly. Early rapid agglomeration was predictive of increased agglomerate size at 24 h, and sedimented agglomerates were visible after 24 h in some samples, supporting the DLS observations. The period of medium conditioning with cells (i.e. the contact time between the medium and cells) was a factor in, but not a predictor of, agglomeration.
Table 1 presents the particle size increase observed in 10 blindly selected samples. This subset of samples was selected on the basis of ENP size distribution change only to represent the range observed among more than 30 samples.
Table 1.
Number of associated peptides compared with particle growth.
| sample | no. associated peptides | LDH (%) | particle growth (nm) |
|---|---|---|---|
| control | 0 | 0 | 0 |
| 1 | 144 | 6 | 106 |
| 2 | 284 | 6 | 188 |
| 3 | 331 | 6 | 210 |
| 4 | 224 | 7 | 158 |
| 5a | 70 | 20 | 370 |
| 6a | 21 | 52 | 358 |
| 7 | 61 | 0 | 27 |
| 8 | 67 | 0 | 74 |
| 9 | 131 | 0 | 6 |
aAmine modified.
(b). Peptides and engineered nanoparticle size distribution
A small group of peptides were detected in all samples, including the controls. This indicated either (i) sample contamination during ENP/medium manufacture or sample processing, (ii) contamination of the analytical column or (iii) false positives during the analysis. The low probabilities assigned to the identified proteins were noted: these proteins identified in the control were excluded from further analysis.
Table 1 compares the number of peptides associated with the ENPs and the percentage of these which were identified as LDH by TurboSEQUEST, with the increase in hydrodynamic diameter. Samples, samples 5 and 6 were observed to agglomerate significantly more than the others (p < 0.01) and had fewer proteins and a higher percentage of LDH. They were later identified as the amine-modified samples. Both table 1 and figure 1 show a linear relationship between hydrodynamic diameter and number of peptides detected. The amine-modified samples are not shown in figure 1 because they behaved differently to plain polystyrene. The R2-value for figure 1 is 0.95 (p < 0.01) but 0.84 (p < 0.01) when the outlier is included.
Figure 1.
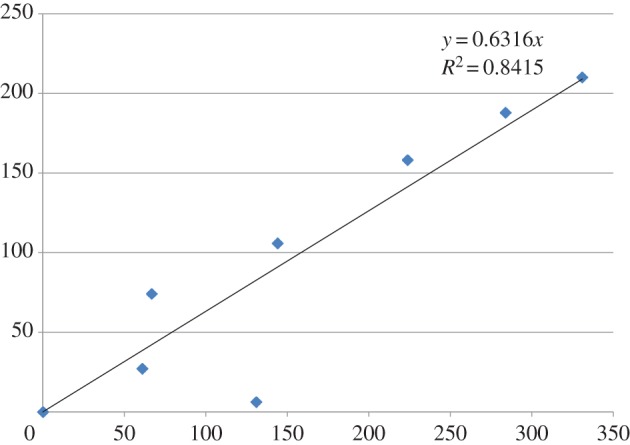
Relationship between nanoparticle hydrodynamic diameter growth in conditioned medium (nm, y-axis) and number of associated peptides (x-axis). (Online version in colour.)
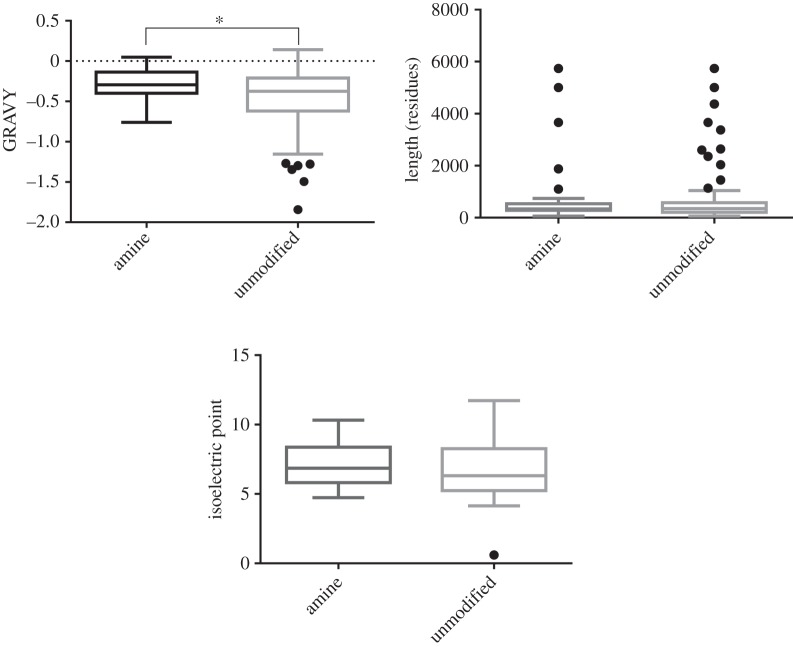
Eight proteins were frequently found to associate with the polystyrene ENPs and their physicochemical properties and functionalities are summarized in table 2. Three of the eight proteins function as oxidoreductase enzymes with NAD+ or NADP+ binding sites. The key properties of these proteins were also investigated and compared to those of 25 proteins that were rarely found to be associated with the polystyrene ENPs. No significant differences were observed in terms of pI, hydropathicity, aliphatic index and amino acid composition (figure 2).
Table 2.
Physicochemical properties and functionalities of the eight proteins found frequently to associate with the polystyrene ENPs.
| protein | total number of peptides associated in the 9 samples | relative molecular mass (kDa) | protein family | cellular process | molecular function | ligands | post translational modifications |
|---|---|---|---|---|---|---|---|
| LDHB_HUMAN (lactose dehydrogenase B) | 52 | 36.62 | LDH/MDH superfamily | glycolysis | oxidoreductase | NAD | acetylation phosphorylation ubiquitination |
| ENOA_HUMAN (alpha enolase) | 42 | 47.14 | enolase family | glycolysis | lyase | DNA, magnesium, metal ions | acetylation phosphorylation |
| AK1BA_HUMAN (aldo/keto reductase family 1 member B10) | 39 | 35.99 | aldo/keto reductase family | reductase | oxidoreductase | NADP | acetylation |
| MUC5B_HUMAN (mucin 5B) | 35 | 590.12 | mucin family | ECM | ECM structural component | n.a. | disulfide bonds glycoproteins |
| LDHA_HUMAN (lactose dehydrogenase A) | 34 | 36.66 | LDH/MDH superfamily | glycolysis | oxidoreductase | NAD | acetylation phosphorylation |
| MUC5A_HUMAN (mucin 5A) | 29 | 526.27 | mucin family | ECM | ECM structural component | n.a. | disulfide bonds phosphorylation glycoproteins |
| CSPG2_HUMAN (chondroitin sulfate proteoglycan core protein 2) | 26 | 372.59 | aggrecan/versican proteoglycan family | cell adhesion/ECM | ECM structural component | calcium, sugar | disulfide bonds phosphorylation glycoproteins |
| ALDOA_HUMAN (fructose bisphosphate aldolase A) | 20 | 39.39 | class I fructose bisphosphate aldolase family | glycolysis | lyase | Schiff base | acetylation phosphorylation |
Figure 2.
Physicochemical characteristics of proteins adsorbed specifically to amine or unmodified particles. The proteins bound to amine or polystyrene particles are similar in terms of chain length (length) and pI; however, proteins bound to amine are more hydrophobic than proteins bound to unmodified particles (Student's t-test p = 0.0224) as shown by their grand average hydropathicity (GRAVY). Box plots show the median value and interquartile range (IQR). Whiskers indicate outliers as 1.5×IQR.
The protein sequences were aligned using UniProt [25] and the amino acid properties of conserved regions were investigated. On the basis of the properties of the polystyrene ENPs employed in this study (negatively charged but considered neutral [not functionally active] and hydrophobic, and those particles having some of the surface charges substituted by amines) regions of conserved positively charged and hydrophobic amino acids were of specific interest. Table 3 summarizes the conserved motifs identified within the eight proteins, noting also the frequency of these motifs in the 25 infrequently associating proteins. None of the infrequently associating proteins contained all the conserved sequence motifs, suggesting that in combination they may play a role in the interaction with the polystyrene ENPs.
Table 3.
Conserved motifs always identified within the eight proteins and present in more than 1000 human proteins, noting also the frequency of these motifs in the 25 infrequently associating proteins. Amino acid sequences within the frequently associating proteins indicated 18 sequence motifs that were common to all eight proteins. (Online version in colour.)
| conserved motif | infrequently associated (%) |
|---|---|
| [RK]XXX[RK] | 96 |
| PXXX[LVI] | 100 |
| G[EK]X[MVI] | 56 |
| VX[LVI]X[KRH] | 76 |
| [LIM]X[NQE][KQR]XG | 16 |
| [ILMV]X[AG][ILMV]X[ILMV] | 64 |
| [KR]X[ILMV]X[ILMV][ST] | 32 |
| [ILMV]X[DE]X[ILMV][ILMV] | 60 |
| [KR][ILMV]XX[ILMV][AG] | 28 |
| [AG]X[ILMV]X[ILMV][ILMV] | 32 |
| [AG]XX[ILMV][AG][ILMV] | 52 |
| [ILMV]X[KR]X[ILMV][AG] | 36 |
| [ILMV][ILMV]X[ILMV][AG] | 72 |
| [ILMV][ILMV]X[AG][ILMV] | 56 |
| [ILMV][ILMV]KXX[AG] | 32 |
| [KR][ILMV]XX[ILMV]G | 12 |
| [DE][DE][ILMV][AG] | 44 |
| [ILMV][ILMV]X[ILMV]G | 40 |
| G[EK]X[MVI] | 56 |
| VX[LVI]X[KRH] | 76 |
| [LIM]X[NQE][KQR]XG | 16 |
| [ILMV]X[AG][ILMV]X[ILMV] | 64 |
| [KR]X[ILMV]X[ILMV][ST] | 32 |
| [ILMV]X[DE]X[ILMV][ILMV] | 60 |
| [KR][ILMV]XX[ILMV][AG] | 28 |
| [AG]X[ILMV]X[ILMV][ILMV] | 32 |
| [AG]XX[ILMV][AG][ILMV] | 52 |
| [ILMV]X[KR]X[ILMV][AG] | 36 |
| [ILMV][ILMV]X[ILMV][AG] | 72 |
| [ILMV][ILMV]X[AG][ILMV] | 56 |
| [ILMV][ILMV]KXX[AG] | 32 |
| [KR][ILMV]XX[ILMV]G | 12 |
| [DE][DE][ILMV][AG] | 44 |
| [ILMV][ILMV]X[ILMV]G | 40 |
Figure 3 shows selected properties of the adsorbed proteins and the roles identified by PANTHER. When the properties of the conserved sequence motifs were considered, hydrophobic and/or positively charged amino acids were conserved, suggesting interactions between the ENP and the protein were predominantly driven by electrostatic forces and hydrophobicity.
Figure 3.
(a) Aminated polystyrene ENP. (b) Unmodified polystyrene ENP. The corona-proteins on both ENP surface types were assigned descriptors based on gene ontology annotations for molecular function using ‘generic gene ontology term finder’ (available online). Descriptors were summarized and grouped by similarity using an online tool (REVIGO) and then simplified to show only the group headings. Tile size represents probability of an annotation appearing within a gene set at a frequency greater than would be expected by chance. There is a larger portfolio of adsorbed proteins on unmodified polystyrene ENPs compared with aminated ENPs which show possible enrichment for proteins involved in the reduction of oxygen.
4. Discussion
(a). Engineered nanoparticle agglomeration in conditioned medium
There was great variability in particle size after a few hours and up to days, despite care being taken to standardize the experimental procedure. Approximately 50% of the 1 h conditioned medium ENPs grew in size (evidence of sample agglomeration), but some only grew a few nanometres compared with the control. As the medium was only conditioned with cells for 1 h, the concentrations of cell secretions were likely low and so variation in ENP growth probably reflects this, as very recently reported elsewhere for gold ENPs [27]. The concentration of polymers in the conditioned medium is therefore predicted to be very variable and this was observable as significant differences in sample surface tension. Variations in the sample meniscus indicate differences in levels of hydrogen bonding between samples. Surface tension differences were observed between plain medium, conditioned medium, centrifuged conditioned medium and conditioned medium containing ENPs.
Another potential source of the variability of the secreted biomolecules in the samples is that in normal culture, cells are distributed across the various phases of the cell cycle, which have quite different protein abundances. Lane et al. [28] demonstrated significant changes in protein abundance for HeLa cells in different stages of the cell cycle and found that a strikingly large proportion of proteins whose abundance changed from G1 to S or from S to G2 were phosphoproteins, consistent with the notion that many changes in protein abundance are controlled by phosphorylation.
This variability in protein content in the conditioned medium is supported by the peptide analysis results. There was a very strong relationship between the numbers of peptides detected in the ENP samples and the growth in hydrodynamic diameter. As the only uncontrolled variable in this experiment was the status of the cells themselves, we must conclude that the status of the cells (population number, health, life stage, etc.) determined these differences. The cell culture protocol was rigorously consistent between each experiment in terms of cell seeding density, length of time in culture and handling of samples, and was identified by a trained cell biologist as identical before medium was added for conditioning. The variation was therefore reduced as far as was practicable.
We centrifuged the conditioned medium to remove particulate and nanoparticulate matter before the introduction of ENPs and peptide identification. As part of this process, we removed material that (i) would normally be present in cell assays and (ii) may be aggregates of proteins/other molecules important in the agglomeration process. Our experiments therefore only reflect a fraction of normal exudates and emissions from cells, which may be significant during cell assays.
No significant differences in the physicochemical properties of frequently and infrequently associating proteins were identified (figure 2). However, there was a significant difference in hydropathicity of proteins attached to the amine versus non-amine particles. While the low number of frequently associating proteins limits this analysis, the data suggested that it was not these fundamental properties that dictated the level of protein–ENP interactions. The amino acid composition of the proteins was also analysed in detail and no significant differences were noted between frequently and infrequently associating proteins.
Comparison of the amino acid sequences within the frequently associating proteins indicated 18 sequence motifs that were common to all eight proteins (table 3). While these motifs were found in a wide range of human proteins, none of the 18 were found in the infrequently associating proteins, suggesting a role for these sequences in associating with the polystyrene ENPs. When the properties of the amino acids within these conserved sequences were considered, the majority of the conserved motifs (94%, n = 17) were noted to contain hydrophobic amino acids (leucine, isoleucine, methionine and valine), two of which (leucine and valine) were previously demonstrated to interact with PM2.5 [7]. We therefore consider that the hydrophobic effect and weak van der Waals forces dominate the interactions between the proteins and polystyrene ENPs. Furthermore, half of the conserved sequences also contained a positively charged amino acid, suggesting some electrostatic interactions between the negatively charged ENPs and the proteins.
The aliphatic index (defined as the relative volume of a protein occupied by aliphatic side chains; alanine, valine, isoleucine and leucine) was demonstrated to be greater in LDHA and LDHB than either the other positively associating proteins or the infrequently associating proteins. This again corresponds with work where the amino acids valine and leucine were demonstrated to associate readily with PM2.5 [7].
The results suggest that ENP agglomeration is driven by LDHA and LDHB as no LDH was found to be associated in samples 7–9 which demonstrated very low aggregation. Aggregation was less in samples 1–4 than 5 and 6; however, no significant differences in the levels of LDH were observed. The reduced aggregation in samples 1–4 may be the result of steric hindrance as these samples had significantly more other proteins associated with them, potentially preventing an interaction between LDHA and LDHB. Alternatively, the increased agglomeration in samples 5 and 6 could result from stronger interactions between LDHA and LDHB and the amine surface in conformations which allow interaction with other LDH monomers or other particles. LDHA and LDHB have several regions of conserved negatively charged amino acid residues which may interact electrostatically with the positively charged amine ENP. Both LDH subunits contain a highly conserved NAD+ binding site which contains the conserved sequence motif VGMACAISILxKxLADELALVD. It is therefore plausible that the amine coated ENP interacts with this motif electrostatically and/or via weak van der Waals forces (between the conserved hydrophobic amino acid residues). The LDH subunits have a much larger binding pocket than the other proteins, which may provide the particle with a favourable site to bind, especially as the binding pocket is lined with hydrophobic residues that would normally be hidden in the centre of a protein. Further work is required to confirm the mechanism of attachment.
Other authors have noted this interaction with LDH [29], and recently a study used circular dichroism to demonstrate the perturbation of the structure of LDH when exposed to a range of nanomaterials [29]. Currently nanotoxicology studies often employ LDH as a simple cytotoxicity marker [10,30,31]. Elevated LDH levels have been detected in in vitro cell studies of NPs [32], but in the literature conclusions about mechanisms and impacts are mixed. If LDH does interact with ENPs as postulated above, toxicology testing of these materials would benefit from use of alternative assays, which could be applied universally to a range of NPs (e.g. MTT).
The association of proteins and the amount/presentation of these proteins on NP surfaces should be of critical importance in determining the in vivo response. This study suggests that LDHA and LDHB have higher affinities for amine coated polystyrene particles, as fewer other proteins are noted to be associated with the plain ENP. This suggests that LDHA and LDHB undergo a slow exchange with the particle, preventing the interaction of other proteins. In vivo, elevated LDH has been associated with poor survival in a number of cancer studies because glycolytic rates in malignant cells are higher than those in their normal tissues of origin (the Warburg effect). It is therefore interesting to note the upregulation of LDH in titanium dioxide NP exposed mice [33].
Three of the eight proteins were identified as oxidoreductase enzymes (table 2) which bind NAD+ or NADP+. Bioinformatics analysis of protein adsorbed specifically to amine particles showed enrichment for oxidoreductase activity of amine-bound particles (figure 3). Unmodified particles had a larger portfolio of adsorbed proteins with less enrichment for any specific group.
We therefore hypothesize that this may be the site where the ENP interacts owing to the conserved hydrophobic and polar residues. The interaction of oxidoreductases with polystyrene ENPs potentially raises an important point in terms of NP-induced production of reactive oxygen species (ROS). The production of ROS and the induction of oxidative stress are considered to be the primary mechanisms by which NPs exert cytotoxicity on cells [11]. The association of these oxidoreductases with NPs could potentially induce oxidative stress in cells and subsequent death. Studies have demonstrated that inhibition of LDHA can induce cell death via oxidative stress mechanisms [34]. Similarly, AK1A is hypothesized to be involved in the detoxification of reactive aldehydes, and therefore sequestration of this protein may enhance the susceptibility of a cell to oxidative stress [35]. Further work is required to establish a greater understanding of the effects of protein–NP complexes.
5. Conclusion
Small differences in conditioned medium (cell culture medium exposed to cells) led to significant alterations in ENP behaviour, such as agglomeration rate. Studying the agglomeration process using DLS and surface-adsorbed proteins using protein sequence analysis, adsorbing peptides at the surface of ENPs were shown to be important in driving size growth. Our results suggest multiple weak interactions via various hydrophobic and charged residues correlated well with ENP size, for uncoated 100 nm polystyrene ENPs. Amine-coated ENPs exhibited more rapid growth than uncoated polystyrene ENPs in conditioned media. Altering the physicochemical properties and surface charge of the ENP increased the binding affinity for LDH and reduced the number of other surface-associated proteins. The expected reduction in oxidoreductase activity has the potential to induce oxidative stress via a peptide-sequestration mediated process. Understanding ENP behaviour in conditioned cell culture medium is crucial for standardized NP toxicity testing in vitro and in vivo, as extracellular biomolecules are emerging as key predictors of the transport, cellular fate and toxicity of ENPs.
Funding statement
The European Centre for Environment and Human Health (part of the University of Exeter Medical School) is part financed by the European Regional Development Fund Programme and the European Social Fund Convergence Programme for Cornwall and the Isles of Scilly (J.T.).
References
- 1.Maiorano G, Sabella S, Sorce B, Brunetti V, Malvindi MA, Cingolani R, Pompa PP. 2010. Effects of cell culture media on the dynamic formation of protein–nanoparticle complexes and influence on the cellular response. ACS Nano 4, 7481–7491. ( 10.1021/nn101557e) [DOI] [PubMed] [Google Scholar]
- 2.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. 2010. What the cell ‘sees’ in bionanoscience. J. Am. Chem. Soc. 132, 5761–5768. ( 10.1021/ja910675v) [DOI] [PubMed] [Google Scholar]
- 3.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DW, Cohen Y, Emili A, Chan WC. 2014. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 8, 2439–2455. ( 10.1021/nn406018q) [DOI] [PubMed] [Google Scholar]
- 4.Lesniak A, Campbell A, Monopoli MP, Lynch I, Salvati A, Dawson KA. 2010. Serum heat inactivation affects protein corona composition and nanoparticle uptake. Biomaterials 31, 9511–9518. ( 10.1016/j.biomaterials.2010.09.049) [DOI] [PubMed] [Google Scholar]
- 5.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. 2012. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano 6, 5845–5857. ( 10.1021/nn300223w) [DOI] [PubMed] [Google Scholar]
- 6.Kendall M, Tetley TD, Wigzell E, Hutton B, Nieuwenhuijsen M, Luckham P. 2002. Lung lining liquid modifies PM2.5 in favour of particle aggregation: a protective mechanism. Am. J. Physiol. Lung 282, L109–L114. [DOI] [PubMed] [Google Scholar]
- 7.Kendall M. 2007. Fine airborne urban particles (PM2.5) sequester lung surfactant and amino acids from human lung lavage. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1053–L1058. ( 10.1152/ajplung.00131.2007) [DOI] [PubMed] [Google Scholar]
- 8.Tenzer S. 2013. Stauber Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 8, 772–781. ( 10.1038/nnano.2013.181) [DOI] [PubMed] [Google Scholar]
- 9.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. 2008. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl Acad. Sci. USA 105, 14 265–14 270. ( 10.1073/pnas.0805135105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clift MJ, et al. 2010. Quantum dot cytotoxicity in vitro: an investigation into the cytotoxic effects of a series of different surface chemistries and their core/shell materials. Nanotoxicology 5, 664–674. ( 10.3109/17435390.2010.534196) [DOI] [PubMed] [Google Scholar]
- 11.Nel A, Xia T, Madler L, Li N. 2006. Toxic potential of materials at the nanolevel. Science 311, 622–627. ( 10.1126/science.1114397) [DOI] [PubMed] [Google Scholar]
- 12.Li R, et al. 2014. Surface interactions with compartmentalized cellular phosphates explain rare earth oxide nanoparticle hazard and provide opportunities for safer design. ACS Nano 8, 1771–1783. ( 10.1021/nn406166n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seagrave JC, Knall C, Mcdonald JD, Mauderley JL. 2004. Diesel particulate material binds and concentrates a proinflammatory cytokine that causes neutrophil migration. Inhal. Toxicol. 16, 93–98. ( 10.1080/08958370490443178) [DOI] [PubMed] [Google Scholar]
- 14.Kendall M, Ding P, Kendall K. 2011. Particle and nanoparticle interactions with fibrinogen: the importance of aggregation in nanotoxicology. Nanotoxicology 5, 55–65. ( 10.3109/17435390.2010.489724) [DOI] [PubMed] [Google Scholar]
- 15.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. 2011. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 133, 2525–2534. ( 10.1021/ja107583h) [DOI] [PubMed] [Google Scholar]
- 16.Lian W, et al. 2004. Ultrasensitive detection of biomolecules with fluorescent dye-doped nanoparticles. Anal. Biochem. 334, 135–144. ( 10.1016/j.ab.2004.08.005) [DOI] [PubMed] [Google Scholar]
- 17.Johnson CJ, Zhukovsky N, Cass AE, Nagy JM. 2008. Proteomics, nanotechnology and molecular diagnostics. Proteomics 8, 715–730. ( 10.1002/pmic.200700665) [DOI] [PubMed] [Google Scholar]
- 18.Seagrave JC. 2008. Mechanisms and implications of air pollution particle associations with chemokines. Toxicol. Appl. Pharmacol. 232, 469–477. ( 10.1016/j.taap.2008.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendall M, et al. 2013. Surfactant protein D (SP-D) alters cellular uptake of particles and nanoparticles. Nanotoxicology 7, 963–973. ( 10.3109/17435390.2012.689880) [DOI] [PubMed] [Google Scholar]
- 20.McKenzie Z, Kendall M, Mackay R-M, Tetley TD, Morgan C, Griffiths M, Clark HW, Madsen J. 2015. Nanoparticles modulate surfactant protein A and D mediated protection against influenza A infection in vitro. Phil. Trans. R. Soc. B 370, 20140049 ( 10.1098/rstb.2014.0049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendall M, Hutton BM, Tetley TD, Nieuwenhuijsen MJ, Wigzell E, Jones F. 2001. Investigation of fine atmospheric particle surfaces and lung lining fluid interactions using XPS. Appl. Surf. Sci. 178, 27–36. ( 10.1016/S0169-4332(01)00248-3) [DOI] [Google Scholar]
- 22.Dowling P, Clynes M. 2011. Conditioned media from cell lines: a complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics 11, 794–804. ( 10.1002/pmic.201000530) [DOI] [PubMed] [Google Scholar]
- 23.Kendall K, Kendall M, Rehfeldt F. 2010. Adhesion of cells, viruses and nanoparticles. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 24.Olsen JV, et al. 2009. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell Proteomic. 8, 2759–2769. ( 10.1074/mcp.M900375-MCP200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, Mcgarvey P, Gasteiger E. 2009. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinform. 10, 136 ( 10.1186/1471-2105-10-136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigoutsos I, Floratos A. 1998. Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics 14, 55–67. ( 10.1093/bioinformatics/14.1.55) [DOI] [PubMed] [Google Scholar]
- 27.Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. 2014. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano 8, 5515–5526. ( 10.1021/nn4061012) [DOI] [PubMed] [Google Scholar]
- 28.Lane KR, et al. 2013. Cell cycle-regulated protein abundance changes in synchronously proliferating HeLa cells include regulation of pre-mRNA splicing proteins. PLoS ONE 8, e58456 ( 10.1371/journal.pone.0058456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barlow PG, Clouter-Baker A, Donaldson K, Maccallum J, Stone V. 2005. Carbon black nanoparticles induce type II epithelial cells to release chemotaxinx for alveolar macrophages. Part. Fibre Toxicol. 2, 11 ( 10.1186/1743-8977-2-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabhu BM, Ali SF, Murdock RC, Hussain SM, Srivatsan M. 2010. Copper nanoparticles exert size and concentration dependent toxicity on somatosensory neurons of rat. Nanotoxicology 4, 150–160. ( 10.3109/17435390903337693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Hitchins VM, Schrand AM, Hussain SM, Goering PL. 2010. Uptake of gold nanoparticles in murine macrophage cells without cytotoxicity or production of pro-inflammatory mediators. Nanotoxicology 5, 284–295. ( 10.3109/17435390.2010.512401) [DOI] [PubMed] [Google Scholar]
- 32.Ruenraroengsak P, et al. 2012. Respiratory epithelial cytotoxicity and membrane damage (holes) caused by amine-modified nanoparticles. Nanotoxicology 6, 94–108. ( 10.3109/17435390.2011.558643) [DOI] [PubMed] [Google Scholar]
- 33.Yu-Mi J, Seul-Ki P, Wan-Jong K, Joo-Hyun H, Mi-Young L. 2011. The effects of TiO nanoparticles on the protein expression in mouse lung. Mol. Cell. Toxicol. 3, 283–289. [Google Scholar]
- 34.Le A, et al. 2010. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl Acad. Sci. USA 107, 2037–2042. ( 10.1073/pnas.0914433107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Y, Penning TM. 2007. Aldo–keto reductases and bioactivation/detoxication. Annu. Rev. Pharmacol. Toxicol. 47, 263–292. ( 10.1146/annurev.pharmtox.47.120505.105337) [DOI] [PubMed] [Google Scholar]