Abstract
Non-Hodgkin’s lymphoma (NHL) is a heterogeneous group of malignancies that originate in lymphatic hematopoietic tissue. Chemotherapy has been used as the main therapy for NHL all the time, and local radiotherapy is also a necessary approach to supplementary treatment. However, resistance of tumor cells to chemo- and radiotherapy often prevent a successful long-term treatment of NHL. MicroRNAs (miRNAs) are a class of approximately 22-nucleotide endogenous non-coding RNAs that play an important regulatory role in gene expression, involving in the process of cell proliferation and differentiation. Alterations of miRNAs have been reported in a variety of human cancers, such as lymphomas, and will critically influence the tumor development and progression. Recently, there is increasing evidence that miRNAs could also influence sensitivity of tumor cells to chemo- and radiotherapy, revealing a crucial role of microRNAs in resistance to anticancer treatment. Therefore, understanding the role of miRNAs in chemo- and radio-resistance of tumor and targeting specific miRNAs will open novel avenues for lymphoma treatment and improve the prognosis of NHL patients. This review outlines the role of miRNAs associated with chemo-and radiotherapy resistance in NHL.
Keywords: MicroRNA, Non-Hodgkin’s lymphoma, chemoresistance, radio-resistance
Introduction
Non-Hodgkin’s lymphoma (NHL) is a heterogeneous group of malignancies that originate in lymphatic hematopoietic tissue. According to different types of lymphoid cells, NHL is further classified into B-cell lymphomas which account for about 90% and T-cell lymphomas which is about 10% [1]. Traditionally, chemotherapy has been used as the main therapy for NHL. According to local mass, regional radiotherapy is also a necessary approach to supplementary treatment [2]. However, resistance of tumor cells to chemo- and radiotherapy often results in the failure of treatment, and a substantial population of patients will eventually relapse. Relapsed lymphomas are refractory to subsequent treatment with the initial chemotherapeutics, moreover, the tumor cells may develop across-resistance to multiple anticancer drugs [3]. Therefore, new targets and more creative methods are required to help overcome the difficulty of and improve the outcome of NHL.
MicroRNAs (miRNAs) are a class of approximately 22-nucleotide endogenous non-coding RNAs, which could be found in almost any section of the DNA. Mature miRNA directs the RISC complex by binding to the 3’ untranslated region (UTR) of their target messenger RNA (mRNA), leading to mRNA degration or protein translation repression. Through controlling gene expression at the post-transcriptional level, miRNAs have been shown to play key regulatory roles in almost all biological process, including cell proliferation, differentiation and apoptosis. They also act as oncogenes or tumor-suppressive genes, involving in tumor development and progression [4-6]. Dysfunctional expression of miRNAs have been reported in several human cancers, including lymphomas [7-9]. More recently, there is increasing evidence that miRNAs could also influence sensitivity of tumor cells to chemo- and radiotherapy, which will lead to the failure of anticancer treatment [10-12]. Therefore, understanding the role of miRNAs in the mechanism of chemo- and radioresistance of tumor and targeting specific miRNAs could help overcome the problem of NHL resistance to anticancer therapy, which will improve the overall response rate and progression-free survival time of NHL patients. This review elaborates the miRNAs that have been reported associated with chemo-and radio-resistance in NHL and other miRNAs expressed in NHL (Tables 1, 2).
Table 1.
MiRNAs that have been reported associated with therapy resistance in NHL
| MiRNAs | Targets | NHL Types |
|---|---|---|
| Downregulation | ||
| miR-15a/16-1 | cyclinD1, MCL-1, BCL-2, BAI2A, RNF41, RASSF5, MKK3, LRIG1 | CLL, MCL, NK/TCL |
| miR-181b | TCL-1, BCL-2, MCL-1, AID, XIAP, FOXP1 | CLL, DLBCL |
| miR-29 | MCL-1, TCL-1, CDK6, IGF1R, DNMT3A, PXND | CLL, DLBCL, MCL, NK/TCL |
| miR-34a | SIRT1, FOXP1 | CLL, DLBCL, MCL |
| Up-regulation | ||
| miR-155 | SOCS1, SMAD5, SHIP1, PIK3R1 | CLL, DLBCL, BL, CTCL, NK/TCL |
| miR-21 | PTEN, PDCD4, CCND2, DPH1 | CLL, DLBCL, CTCL, NK/TCL |
| miR-221/222 | P27 | CLL, DLBCL |
| miR-17-92cluster | PTEN, PHLPP2, BIM, ITIME2F5, T53INP1, TRIM8, IBTB4 | CLL, DLBCL, MCL, BL |
| miR-148b | - | BL |
CLL: chronic lymphocytic leukemia; DLBCL: diffuse large B cell lymphoma; MCL: mantle cell lymphoma; BL: Burkitt’s lymphoma; CTCL: cutaneous T-Cell Lymphoma; NK/TCL NK/T-cell lymphoma.
Table 2.
Other miRNAs expressed in NHL
| MiRNAs | Targets | Functions |
|---|---|---|
| CLL | ||
| miR-106b | ITCH | Inhibit p53-dependent mechanism |
| miR-650 | CDK1, ING4, EBF3 | Inhibit cell cycle progression |
| miR-130a | ATG2B, PICERI | Inhibit cell autophagy and trigger killing |
| miR-9-3 | NF-κB Protein | Inhibit activation of NF-κB |
| DLBCL | ||
| Let-7b | PRDM1 | Overexpression in DLBCL |
| miR-125b | INFAIP3, MAD4 | Up-regulation associated with chemoresistance |
| miR-129-5p | CDK6 | Result in less G arrest and cell death |
| miR-142, 146-5p, 146a, 223, 200c | - | Associated with survival of patients |
| MCL | ||
| miR-101, 26a/b | STMN1 | Decrease expression of STMN1 |
| miR-10a, 20b, 363, 127-3p, 615-3p | - | Associated with survival of patients |
| BL | ||
| miR-155, 98, let-7a, hsa-mir-34b | MYC | Associated with ETF2357 cytotoxic |
| Has-mir-127 | BLIMP1, XBP1 | Block B cell differentiation |
| miR-150 | c-MYB | Reduce cell proliferation |
| CTCL | ||
| miR-122 | - | Up-regulation associated with chemoresistance |
| NK/TCL | ||
| miR-146a | TRAF6 | Enhance cell chemosensitivity |
CLL: chronic lymphocytic leukemia; DLBCL: diffuse large B cell lymphoma; MCL: mantle cell lymphoma; BL: Burkitt’s lymphoma; CTCL: cutaneous T-Cell Lymphoma; NK/TCL NK/T-cell lymphoma.
MicroRNAs associated with chemoresistance in NHL
CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) has been used as the standard treatment for NHL, however, large population of patients undergo relapse, resulting in only about 30% of 3-year overall survival rates [13]. In NHL cells undergoing chemotheraputics, miRNAs can function either as tumor-suppressive genes to inhibit tumor proliferation or oncogenes to promote tumor escape from apoptosis induced by chemotherapy. Expressions of tumor-supressive genes including miR-15a/16-1, miR-34a, miR-181, miR-29, and oncogenes miR-155, miR-21, miR-221/222 as well as other miRNAs in different types of NHL and their crucial roles in resistance to chemotherapy of NHL patints will be elaborated hereinafter.
MicroRNAs in chemoresistance of chronic lymphocytic leukemia (CLL)
MiR-15a/16-1 cluster are demonstrated to be deleted or down-regulated in about 68% of CLL cases, which are located at chromosome 13q14, a 30-kb region of loss in CLL and encodes the DLEU2/miR-15a/16-1 cluster, and that both genes deletion in mice of the region causes proliferation of both human and mouse B cells by modulating the expression of genes controlling cell-cycle progression, leading to CLL [14,15]. Experiments in the New Zealand Black mouse cell line showed cyclin D1, a cell cycle regulator of G1/S transition, was downregulated in protein levels following miR-15a/16-1 addition [16]. In addition, HDAC inhibition induced expression of miR-15a/16 and miR-29, decreasing the levels of the survival protein MCL-1. Mouse model with TCL1-Tg: p53-/- genotype exhibited higher proliferation, higher survival capacity, and more resistance to drug treatment with fludarabine. It was found that p53 deletion resulted in a decrease of miR-15a/16-1, leading to an elevated expression of MCL-1, which was further confirmed in primary leukemia cells from CLL patients with chromosome 17p deletion [17,18]. Expression of miR-15a/16-1 also down-regulated expression of anti-apoptosis protein BCL-2 in CLL, and that enhanced the susceptibility of tumor cells to apoptosis in vitro, loss of mitochondrial function, and activation of cell death [19]. Moreover, genes BAZ2A and RNF41 were found significantly been up-regulated while genes RASSF5, MKK3 and LRIG1 were found significantly down-regulated in CLL patients with down-regulated expression of miR-15a/16-1 [20], however, much work remains to be done to further understand their contributions to CLL.
MiR-34a is a member of the miR-34 family and is regulated by the tumor suppressor p53 [21]. Deletion of 17p was associated with low basal expression of miR-34a and failure to induce expression of miR-34a after DNA damage, and at the same time low expression of miR-34a in CLL was associated with p53 inactivation but also chemo-therapy-refractory disease and apoptosis resistance. Cases with fludarabine -refractory disease had a significantly lower baseline miR-34a expression [22,23]. B-CLL cases carrying the SNP309 G/G polymorphism had significantly lower miR-34a levels even when no other mutations or deletions within the p53 gene were present, indicating microRNA-34a expression correlated with MDM2 SNP309 polymorphism [24]. In addition, DNA-damaging chemotherapeutics, such as etoposide, activate a functional loop linking the main member of the sirtuin family SIRT1 and p53 through the induction of miR-34a, thus identified SIRT1 as part of a p53/miR-34a tumor suppressor network, and nicotinamide enhanced this pathway by negatively regulating SIRT1 [25]. However, details of the underlying mechanisms behind miR-34a pro-apoptotic effect are needed further investigated.
MiR-181b was down-regulated in therapy-refractory CLL cases and its expression levels significantly predicted a good treatment-free survival of CLL patients [26]. MiR-181b and miR-29 were found significantly down-regulated the expression of TCL-1, which was coactivator of the Akt oncoprotein, and high expression of that correlated with aggressive B-CLL phenotype and 11q deletions [27]. MiR-181b targeted the antiapoptotic genes BCL-2 and MCL-1, which were up-modulated in CLL cells of patients with progressive disease. MiR-181b also targeted activation-induced cytidine deaminase (AID), and loss of miR-181b leading to overexpression of AID and genomic instability as well as cancer progression. Furthermore, miR-181a/b inhibiting X-linked inhibitor of apoptosis protein (XIAP), significantly sensitizes CLL cells to fludarabine-induced apoptosis, which might provide a possible therapeutic avenue and a sensitive indicator of the activity of the p53 axis in CLL [28,29]. On the other hand, miR-29 expression is down-regulated in aggressive CLL as compared with indolent CLL. In addition to MCL-1 [17] and TCL-1 [27], CDK6 and DNMT3A are observed down-regulated in miR-29 transgenic mice, however, they are not proved to be tumor suppressors. Interestingly, peroxidasin (PXDN), a p53-responsive gene, whose expression was found be down-regulated in CLL samples compared with normal CD19+ B cells, was suggested to be associated with oncogenic role of miR-29 in B cells [30]. These results may provide candidates for therapeutic agents in CLLs.
MiR-155, miR-21 and miR-221/222 expression levels were found significantly higher in patients with poor prognosis and decreased overall survival or with fludarabine treatment resistance, which validated the prognostic value of the three miRNAs in patients with B-CLL [31-34]. MiR-21 was reported to be involved in down-regulation of phosphatase and tensin homologue (PTEN) in CLL [35], in addition, CCND2 and DPH1, which might act as regulators of cell cycle progression, were also possibly the targets of miR-21 [32]. Furthermore, MDM2 small molecule inhibitor nutlin-3-induced up-regulation of SOCS1 was paralleled and probably mediated by a concomitant down-regulation of miR-155 in nutlin-3 treated primary B-CLL cells, suggesting the existence of the miR-155/SOCS1 axis in the cytotoxic response to nutlin-3 in B-CLL and a potentially important therapeutic target of nutlin-3 in B-CLL[36]. On the other hand, the enforced expression of miR-221/222 in the CLL cell line MEC1 induced a significant decreased expression of p27 protein and exhibited faster progression into the S phase of the cell cycle, indicating a regulatory loop between miR-221/222 and p27 that helped maintaining CLL cells in a resting condition [37]. Although the previous work support miR-155, miR-21 and miR-221/222 as potential therapeutic targets, further study are necessary to be found for the exact mechanism by which they act.
In most recent years, more and more other miRNAs were found to be associated with treatment free survival in patients with CLL or were showed affecting the biologic course of CLL cells. For example, low levels of miR-18, miR-19a and miR-19b as well as high expression of miR-17-5p, miR-92 and miR-223 were showed associated with poor survival in patients with CLL [38,39]. By targeting the ubiquitin ligase Itch, miR106b could initiate a p53-independent mechanism that targets CLL cells [40]. MiR-650 targeted proteins which were important in cell proliferation and survival such as cyclin dependent kinase 1 (CDK1), inhibitor of growth 4 (ING4), and early B-cell factor 3 (EBF3), and inhibited CLL cell cycle progression by regulating p16INK4-mediated pathway [41]. MiR-130a had an effect on cell survival in CLL by targeting ATG2B and DICER1 to inhibit autophagy and trigger Killing of CLL cells [42]. MiR-9-3,which act as a tumor suppressor, was associated with down-regulation of NF-κB protein, and hence might contribute to constitutive activation of NF-κB signaling pathway in CLL [43]. These researches indicate that the expression of these microRNAs in CLL may have an effect on response to chemotherapy of CLL patients, but a more detailed description of these findings is beyond the limits of this review.
MicroRNAs in chemoresistance of diffuse large B cell lymphoma (DLBCL)
As has been described in CLL, miR-34a also has been found to be one of the miRNAs down-regulated in high-grade gastric diffuse large B-cell lymphoma (gDLBLC), which could be directly regulated by myc. Moreover, FOXP1, a hematopoietic oncoprotein overexpressed in gDLBCL, was predicted as a target of miR-34a and (an indirect target) of myc in DLBCL cell lines. Aberrant expression of myc resulted in repression of miR-34a, thus promoted high-grade transformation of B-cell lymphoma by dysregulation of FOXP1. Systemic miR-34a delivery induced apoptosis and abrogated growth of diffuse large B-cell lymphoma in vivo [44,45]. In addition, miR-181a and miR-222 levels were correlated with progression-free survival of DLBCL patients treated with R-CHOP. It was concluded that FOXP1 was also a direct target of miR-181a, and downregulation of FOXP1 expression by miR-181a might at least could in part explain the association between miR-181a and improve survival of DLBCL patients [46]. It might be as well as that miR-181a significantly be decreased NF-κB reporter activity in DLBCL cell lines and also decreased NF-κB reporter activity induced by anti-IgM and TNF-α stimulation by regulating the expression of multiple components of this pathway. MiR-181a expression significantly increased cell death and apoptosis of ABC versus GCB DLBCL, which was associated with a more pronounced G1 phase growth arrest in the ABC DLBCL cells, and miR-181a down-regulation may contribute to the pathogenesis of ABC DLBCL [47]. Interestingly, Epstein-Barr virus encoded latent membrane protein1 (LMP1) transfectants showed that miR-29b was up-regulated among others. Further miR-29b locked nucleic acid (LNA) antisense oligonucleotide transfection into LMP1-expressing cells decreased miR-29b expression and consequently reconstituted oncogene TCL-1, suggesting that LMP1 might negatively regulated TCL-1 through miR-29 up-regulation and presented lymphoma growth antagonistic property [48]. Therefore, replacement therapy of these miRNAs could be considered for the treatment of DLBCL patients with the miRNA-negative, oncogene-overexpressing cases.
In contrast, miR-155, miR-21 and miR-222 were found up-regulated in DLBCL cells. Inositol phosphatase (SHIP1) and PIK3R1 (P85α), two negative regulators of the PI3K-AKT pathway, were both showed down-regulated by miR-155, and consequent diminished SHIP1 expression was further demonstrated to be the result of autocrine stimulation by the pro-inflammatory cytokine tumor necrosis factorα (TNFα). Thus the PI3K-AKT signaling pathway was highly activated in DHL16 cells by overexpression of miR-155, whereas knockdown of miR-155 in OCI-Ly3 cells diminished AKT activity. All above revealed a novel molecular mechanism by which miR-155, through direct targeting and repressing SHIP1 and p85α, activated oncogenetic Akt signaling in DLBCL, resulting in proliferation of tumor cells [49,50]. In addition, miR-155 directly down-regulated the bone morphogenetic protein (BMP)-responsive transcriptional factor SMAD5. And further study in the DLBCL models showed expression of miR-155 blocked TGF-β1-mediated activation of the retinoblastoma protein (RB), reducing the abundance of the inhibitory pRB-E2F1 complex and resulting in G0/G1 transition. Thus miR-155 overexpression lead DLBCL cells resistant to the growth-inhibitory effects of both TGF-β1 and BMPs, through defective induction of p21 and impaired cell cycle arrest, enhancing tumor aggressiveness and facilitating an advantageous survival behavior [51,52].
Tumor suppressor gene phosphatase and tensin homologue (PTEN) and programmed cell death 4 (PDCD4) have been identified as direct targets of miR-21. It was also demonstrated that miR-21 affected the PI3K/AKT pathway through the regulation of PTEN to exert biological effects. Caspase-3 and caspase-9 activity were found significantly increased in miR-21 ASO transfection group compared to the control when cells were exposed to 40 ng/mL CHOP. Meanwhile knockdown of NF-κB decreased miR-21 expression and sensitized the cells to CHOP treatment as well, which indicated that downregulation of miR-21 could significantly increase cell sensitivity to the CHOP regimens and that NF-κB is a key upstream signal of miR-21 [53,54]. As has been mentioned before, expression of miR-222 was proved be also associated with shorter progression-free survival in patients treated with R-CHOP, via inducing downregulation of p27 that might facilitate cell proliferation and survival [37,46]. In short, these miRNAs above could regulate tumor cell proliferation, invasion, and apoptosis. It could be speculated that they have a potential therapeutic application in DLBCL.
More recently, other miRNAs were further found be associated with treatment free survival in patients with DLBCL or were showed affecting the biologic course of DLBCL cells. For instance, high expression levels of miR-142, miR-146-5p, and miR-223 as well as low expression of miR-146a and miR-200c were showed associated with longer relapse or progression-free survival time in DLBCL patients treated with R-CHOP [55-59]. In addition, tumor suppressors miR-127 as well as oncogenes miR-195 and let-7g were found to correlate with event-free and overall survival in DLBCL patients in a multivariate analysis [60]. Let-7, in particular let-7b, was overexpressed in DLBCL relative to normal GCB cells, and overexpression of let-7 was demonstrated contributing to the down-regulation of PRDM1 expression in DLBCL through translation repression [61]. MiR-125b, which were commonly gained and/or overexpressed in DLBCL, enhanced DLBCL aggressiveness via direct down-regulation of TNFAIP3 expression [62]. And miR-129-5p, whose low expression was associated with shorter overall survival in patients treated with R-CHOP, was attempted to speculate that low miR-129-5p lead to up-regulation of CDK6, which resulted in less G1 arrest and cell death [63]. The specific role of these miRNAs in DLBCL is not defined as yet. There remains considerable potential for miRNA targets and functions to be examined in clinical trials.
MicroRNAs in chemoresistance of Mantle cell lymphoma (MCL)
It was the same as in CLL that miR-16-1 regulated the expression of cyclinD1 as well in MCL. Truncated cyclin D1 mRNA expression lead to deletion of miR-16-1 binding sites within the cyclin D1 mRNA 3’-UTR and altered miR-16-1 regulation in MCL, resulting in increased total cyclin D1 mRNA and protein expression, and thus S-phase fraction in MCL cell lines [64]. In addition, in Mino and Jeko-1 cells with oncogenic transcription factor MYC knockdown by small interfering RNAs (siRNAs), there were significantly up-regulated pri-miR-15a/16-1 mRNAs, and it was further confirmed that MYC downregulated the transcription activity of miR-15a/16-1 promoter depended on HDAC3 [65]. The down-regulation of miR-15a/16-1 in MCL has been demonstrated, however, to further understand their role as possible therapeutic targets in MCL, much remains to be done in the future.
It was also shown that MCL patients with down-regulated miR-34a or miR-29 both had short survival and were associated with poor prognosis. Low expression of miR-34a was in cooperation with high expression of the MYC oncogene, and down-regulation of miR-29 lead to frequent overexpression of CDK6, which cooperated with cyclin D1 to further promote cell-cycle progression and the development of MCL [66,67]. In addition, expression of miR-29-c was found inversely correlates with the expression of insulin-like growth factor (IGF-1R) in MCL. Overexpression of miR-29-c down-regulated IGF-R1, inhibiting cell growth and inducing cell cycle arrest in MCL cells, and thus suppressed MCL growth and progression in NOD/SCID mouse xenograft models, which indicated that miR-29-c were implicated in MCL cell cycle control and survival and its down-regulation likely contributed to the tumorigenesis of MCL [68]. However, the role of miR-34a as well as miR-29 in MCL cell survival, growth, and chemoresistance needs more cultural cells and animal models to be further evaluated.
Unlike in CLL and DLBCL, miR-181a presented a different role in MCL. Up-regulation of miR-181a post-transcriptionally down-regulated the proapoptotic protein, BIM, thus enhancing follicular dendritic cell (FDC)-mediated protection against apoptosis in lymphoma cell lines and primary lymphoma cells, leading to FDC-dependent drug resistance [69], which provided us a novel angle of view about chemoresistance mechanism involved in FDC and miRNA.
So far, there have been more miRNAs found to be associated with overall survival and prognosis of patients with MCL. High expression of miR-10a, miR-20b and miR-363 were observed to be associated with a short overall survival and poor prognosis in MCL. MiR-127-3p and miR-615-3p were also proved to be significantly related with overall survival in the MCL training set [70,71], however, more work need to be done to detailed their functions in MCL.
MicroRNAs in chemoresistance of Burkitt’s lymphoma (BL)
Observation of the cytotoxic activity of histone deacetylase inhibitor ITF2357 on BL cell lines found a decrease of the expression of miR-155 and miR-98, which exert oncogenic activity in MYC-associated tumors, and a conversely increase of let-7a and miR-146a, which target the MYC mRNA stabilizer IMP-1 among its tumor suppressive functions, suggesting that cytotoxic activity of ITF2357 was associated to miRNA modulation in BL [72,73]. Let-7a and hsa-mir-34b down-regulates MYC and reverts MYC-induced growth has been demonstrated in Burkitt lymphoma cells and vitro experiments [74,75]. Interestingly, in contrast to CLL and DLBCL, very low levels of BIC (pri-miR-155) and miR-155 are present in BL. Further study showed protein kinase C (PKC) and nuclear factor-κB (NF-κB) were the crucial involvement in the regulation of BIC expression upon B-cell receptor triggering. However, the mechanism was as yet unknown [76].
In addition, there were still miRNAs that showed expression and displayed important role in EBV positive BL. Hsa-miR-127 was demonstrated strongly up-regulated only in EBV-positive BL samples and involved in B-cell differentiation process through posttranscriptional regulation of 2 master regulators of plasma cell differentiation BLIMP1 and XBP1. The overexpression of this miRNA might thus represent a key event in the lymphomagenesis of EBV positive BL, through blocking the B-cell differentiation process [77]. BL cell lines showed an extremely low expression of miR-150. Restored miR-150 expression at physiologic levels in 5 BL cell lines showed the re-expression of miR-150 in Daudi and Raji cells, which were both of EBV-positive germinal center B-cell origin. MiR-150 decreased c-MYB protein levels and reduced proliferation of BL cells, and transduced with miR-150 could be rescued to differentiate toward B-cell terminal stage [78]. Nevertheless, their more functions in BL deserved specific recognition in the future.
MicroRNAs in chemoresistance of Cutaneous T-Cell Lymphoma (CTCL)
Interestingly, other than the mechanisms previous mentioned, miR-181a was found to be down-regulated after Notch activation γ-secretase inhibitors GSI treatment, for which the explanation might be that miR-181a was involved in the protective, pro-survival circuit, and induced cancer cells in response to the pro-apoptotic agents, thus up-regulation of miR-181a should promote apoptosis induced by GSI [79]. But it did not show the exact target of miR-181a in CTCL.
The relationship between miRNAs and JAK/STAT pathway has also been recently elaborated in CTCL. MiR-155 was found as well overexpressed in CTCL patients, and further study showed that aberrant expression of miR-155 and its host gene BIC in CTCL were induced by transcription factor STAT5 that signaled downstream of the IL-2 receptor family, indicating a STAT5/BIC/miR-155 pathway which promoted proliferation of malignant T cells, and therefore would be a potential target for therapy in CTCL [80]. In addition, chromatin immunoprecipitation showed that miR-21 was a direct STAT3 target in Sé zary cells. IL-21 stimulated Sé zary cells resulting in a strong activation of STAT3, and thus subsequent up-regulation expression of miR-21. The results suggested a STAT3/miR-21 pathway which induced anti-apoptosis of Sé zary cells, and consequently might represent a therapeutic target for the treatment of Sé zary Syndrome [81]. However, the targets of miR-155 and miR-21 in CTCL were not shown in above studies.
MiR-122 was found over-expressed in CTCL and decreased the sensitivity to the chemotherapy-induced apoptosis by a signaling circuit involved in the activation of Akt and inhibition of p53. Up-regulation of miR-122 was induced post-transcriptionally by p53, and was thus an amplifier of the antiapoptotic Akt/p53 circuit. It is possible that a pharmacological intervention in this pathway may provide a novel therapy for CTCL [82]. New findings described a novel signaling by cMyc/miR-125b-5p that determined sensitivity to bortezomib in preclinical model of CTCL. MiR-125b-5p increased cellular resistance to proteasome inhibitors through down-regulation of MAD4, overexpression of cMYC induced downregulation of miR-125b-5p and sensitized lymphoma cells to bortezomib, whereas silencing of cMyc was associated with a high miR-125b-5p expression level and tumor growth and thus drug resistance [83]. Information obtained about miRNAs and their targets in CTCL could lead us to develop new therapeutic strategies for this disease, and there remains much to be done to discover more.
MicroRNAs in chemoresistance of NK/T-cell lymphoma (NK/TCL)
MiR-15a was recently revealed that it expressed at a much lower level in nasal NK/T-cell lymphoma (NNKTL) cells and regulated the expression of transcription factor myb and cyclinD1. Reduced miR-15a expression, which correlated with expression of myb and cyclin D1, resulted in poor prognosis of NNKTL patients. Knockdown of LMP1 significantly increased miR-15a expression, suggesting down-regulation of miR-15a might possibly due to LMP1. MiR-15a thus could be a potential target for antitumor therapy and a prognostic marker for NNKTL [84]. In addition, T-lymphoblastic lymphoma/leukemia (T-LBL/ALL) patients with higher expression level of miR-16 had a better overall 1-year survival rate, indicating that miR-16 might be a potential prognostic predictor for T-LBL/ALL [85]. Furthermore, significantly negative correlation of expression between miR-29a and MCL-1 in extranodal NK/T-cell lymphoma (ENKTCL) patients suggested that miR-29a might participate in tumorigenesis and development of ENKTL by targeted-regulating oncogene MCL-1, providing a possible target for ENKTK treatment [86].
On the other hand, reducing expression of miR-21 or miR-155 led to up-regulation of PTEN and PDCD4, or SHIP1, and with down-regulation of pAkt in NK/TCL. Whereas transduction with either miR-21 or miR-155 resulted in down-regulation of PTEN and PDCD4, or SHIP1 and with up-regulation of pAkt, which provided an important new insight into the pathogenesis of NK/TCL and suggested a useful approach to treating NK/TCL by targeting miR-21 or miR-155 [87]. However, due to the limits of current researches, the clinical applicability of miR-21 or miR-155 for NK/TCL still has a long way to go.
Furthermore, miRNA-146a was found down-regulated and might act as a tumor suppressor in NK/TCL. In vitro studies with SNK6 and YT cells showed that miR-146a over-expression inhibited NF-κB pathway by targeted down-regulation of TRAF6, and thus suppressed cell proliferation, induced apoptosis, and enhanced chemosensitivity of NK/TCL [88]. Moreover, By using lentiviral vectors as an alternate method to express miR-101, miR-26a, and miR-26b in NK-YS cells resulted in a significant increase expression of these miRNAs and a corresponding decrease expression of STMN1 [89]. Above findings point out new targets and avenues for the treatment of NK/TCL, more detailed researches need to be done in the further.
Knock-down or re-expression of specific miRNAs using synthetic anti-miRNA oligonucleotides or pre-miRNAs to inhibit oncogenic miRNAs or replace tumor-suppressive miRNAs will correct those specific miRNAs which dys-regulate in gene expression and restore the gene regulatory network and signaling transduction pathways, leading to increased inhibition of lymphoma cell growth and invasion, thus sensitize the NHL cells to chemotherapeutics and improve the NHL resistance to chemotherapy. However, there is still a tough way for discovering more miRNAs as well as their clinical functions and targeted therapeutic applications for NHL.
MicroRNAs associated with radio-resistance in NHL
Radiotherapy is also an important method of NHL anticancer treatment, large amount ofevidence found miRNAs regulate proliferation and apoptosis of lymphoma cells after irradiation, which supports a role for miRNA in regulating key cell signaling pathways that mediates response to radiation-induced lymphoma cellular courses induced by radiation. However, in NHL, in addition to miR-17-92 in MCL and miR-148b in BL cell lines Raji, relatively few miRNAs have been found and studied in detail and the biological relevance of majority remains uncovered. The role of miR-17-92 and miR-148b in resistance to radiotherapy of NHL will be expound then.
MicroRNAs in radio-resistance of MCL
High level expression of miR-17-92 cluster was associated with poorer survival in patients with MCL. MiR-17-92 cluster is consisted of six miRNAs (miR-17-5p, miR-18, miR-19a, miR-19b, miR-20a and miR-92-1) and is transcribed as a polycistronic unit from C13orf25, generally up-regulating in malignant lymphomas with chromosome 13q31-q32 gain/amplification [90]. Overexpression of miR-17-92 cluster has been found significantly promoting cell proliferation, increasing survival cell number, and decreasing cell death in human MCL after different doses of radiation. And it was showed that the protein phosphatase and tensin homolog PTEN and phosphatase PHLPP2 were down-regulated and thus the pAkt activity was enhanced with overexpression of miR-17-92 cluster in MCL cells after irradiation. These findings provide the first direct evidence that overexpression of miR-17-92 cluster increased radio-resistance of human MCL cells, which offers a novel target molecule for improving the radiotherapy of MCL [91]. In addition, miR-17-92 cluster overexpression enhanced cell growth and inhibited chemotherapy induced apoptosis in MCL cell lines via activated the PI3K/AKT pathway by down-regulating multiple proteins such as PTEN, PHLPP2, and the BH3-only pro-apoptotic protein BIM, which were all negative regulator of the PI3K/AKT pathway. Conversely, inhibition of miR-17-92 cluster expression in a xenograft MCL mouse model showed suppressed the PI3K/AKT pathway and inhibited tumor growth. As a consequence, over-expression of miR-17-92 may be associated with poorer survival in MCL patients, which revealed a novel oncogenic pathway in MCL and suggested that targeting the miR-17-92 cluster may provide a new therapeutic method for MCL [92,93]. Existing reports show that the miR-17-92 cluster plays a crucial role in MCL chemo- and radiotherapy resistance, and may be considered as a clinical tool to improve the MCL treatment with current chemotherapeutic regimens.
In addition, miR-17-92 cluster was proved also been up-regulated in other malignant lymphomas including CLL, DLBCL and BL. The enforced expression of miR-17-92 cluster in CLL reduced the expression of the tumor suppressor genes E2F5, TP53INP1, TRIM8 and ZBTB4, and thus protected cells from serum-free-induced apoptosis, which was in part responsible for regulations in the genes expression profile of CLL cells with unmutated IGHV genes that stimulated with the TLR9 agonist CpG [94]. MiR-17-92 cluster was significantly overexpressed in GC-DLBCL, and was a major regulator of B-cell receptor (BCR) pathway components in DLBCL. Inhibition of miR-17-92 cluster blunted the BCR response in DLBCL cell lines by targeting ITIM proteins CD22 and FCGR2B. Stimulation of the BCR response by miR-17-92 cluster also resulted in the enhanced calcium flux and elevated levels of MYC. The MYC-miR-17-92 cluster axis amplified B-cell receptor signaling via inhibition of ITIM proteins, which constituted a novel lymphoma genic feed-forward loop. Overexpression of miR-17-92 cluster in GC-DLBCL in comparison with high-grade Follicular Lymphoma, further supported the oncogenic potential of these miRNAs [95,96]. As a MYC target, modestly increased relative MYC transcript levels with marked relative increases in miR-17-92 cluster were showed in the BL cell lines Raji and Ramos, and the Paediatric Burkitt Lymphoma samples with 13q31 gain also showed higher expression of miR-17, suggest the existence of miR-17-92 cluster overexpression in pBL [97], however, further studies in a larger sample set should be performed to assess the possible relationships between the expression level of miR-17-92 cluster and patient outcomes.
MicroRNAs in radio-resistance of BL
In addition to miR-17-92 cluster, a quantitative real-time polymerase chain reaction (PCR) assay showed the up-modulation of miR-148b after radiation. MiR-148b was located in 12q13, and generally down-regulated in various cancers as a tumor-suppressive gene [98]. It was confirmed that miR-148b could increase the radio-sensitivity of Raji cells and inhibited the proliferation. Further studies showed that miR-148b probably increased the cells radio-sensitivity by promoting radiation-induced cell apoptosis. MiR-148b did not affect the cell cycle profile of post-radiation Raji cells compared with controls, the concrete mechanism involved in the enhanced radiation sensitivity induced by miR-148b need further studies [99]. MiR-148b plays an important role in the response of NHL to ionizing radiation, indicating that miR-148b may be another target associated with sensitivity of NHL cells to radiotherapy.
MiRNA offer potential novel avenues in overcoming the NHL cell resistance to apoptosis induced by radiation. By exploring how the miRNA affect the response of specific lymphoma cells to radiation and intervening the expression of dysregulated miRNAs or their targets we can specifically alter the lymphoma cells sensitivity to radiation. However, which miRNAs and how these miRNAs expression affect the response of lymphoma cells to radiation therapy remains little understanding. Effective agents that successfully improve the lymphoma cells sensitivity to radiation in vivo are lacking, too. Future studies are requested to find more miRNA biomarkers associated with radio-resistance in NHL and to better understand their roles in the lymphoma cells radiobiology.
Future prospective and questions
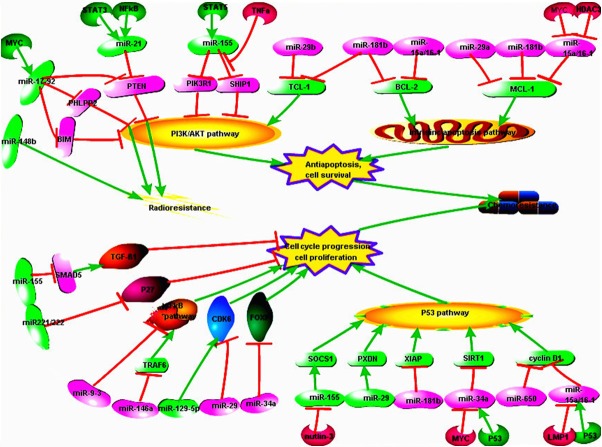
In summary, the mutations and amplifications/deletions as well as altered transcriptional control can lead to dysregulation of miRNAs in NHL, which might play an important role in tumor cell survival, apoptosis and cell cycle progression by targeting several signaling pathways, such as PI3K/AKT pathway, intrinsic pathway, as well as TP53 pathway and so on [100]. Different proteins and pathways targeted by specific miRNAs in different types of NHL with resistant to chemotherapy have been detailed introduced in this article (Figure 1). The exact mechanism about the impact of miRNAs on chemo- and radio-resistance and meaning of them for therapeutic usage provide an emerging strategy for NHL treatment.
Figure 1.
Regulation of miRNAs in NHL. Dysregulation of miRNAs affect different cell signaling pathways by targeting specific genes, leading to cell cycle progression or anti-apoptosis and cell resistance to chemo- and radiotherapy. (Made by Pathway Builder Tool).
However, despite these encouraging researches on the role of miRNAs in sustaining resistance of NHL to chemo- and radiotherapy, many limiting aspects are still have to be addressed. First of all, based on the huge amount of miRNAs in the human genome, it is unlikely that these miRNAs represent an exhaustive list of the miRNAs associated with chemo- and radio-resistance of NHL. Secondly, there is still a considerable lack of elucidating of the precise mechanisms governing miRNAs expression and their influence on targeted genes and cell signaling pathways. Understanding the detailed mechanisms and role of identified miRNAs in chemo- and radio resistance of NHL remains to be a big challenge at the moment. Last but not least, current studies are still at the pre-clinical stage, and the major hurdles including the efficiency and safety of such targeting therapy in clinical patients remain to be resolved. Nevertheless, further extensive researches will lay open the whole number of miRNAs involved in regulating chemo- and radio-resistance of NHL and the way they affect NHL cell sensitivity to therapeutics and radiation. This is an area that is almost certain to flourish as the field matures and it can be expected that in the near future miRNAs and miRNA-targeting oligonucleotides may become promising strategies in overcoming the chemo- and radio-resistance of NHL, and improving the prognosis and life quality of NHL patients.
Acknowledgements
This work was supported by National Natural Science Foundation (No. 81270598; No. 31340009), Natural Science Foundations of Shandong Province (No. Y2007C053, No. 2009ZRB14176, No. ZR2011HQ009 and No. ZR2012HZ003), Technology Development Projects of Shandong Province (No. 2007GG10, 2008GG2NS02018 and No. 2010GSF10250), National Public Health Grand Research Foundation (No. 201202017), Program of Shandong Medical Leading Talent and Taishan Scholar Foundation of Shandong Province.
Disclosure of conflict of interest
None.
References
- 1.Coiffier B. Monoclonal antibody as therapy for malignant lymphomas. C R Biol. 2006;329:241–54. doi: 10.1016/j.crvi.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Campbell BA. The role of radiation therapy in the treatment of stage I-II diffuse large B-cell lymphoma. Curr Hematol Malig Rep. 2013;8:236–42. doi: 10.1007/s11899-013-0170-5. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell SA, Mousavi-Fard S. Non-Hodgkin’s B-cell lymphoma: advances in molecular strategies targeting drug resistance. Exp Biol Med (Maywood) 2013;238:971–90. doi: 10.1177/1535370213498985. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–62. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–41. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 7.Di Lisio L, Martinez N, Montes-Moreno S, Piris-Villaespesa M, Sanchez-Beato M, Piris MA. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood. 2012;120:1782–90. doi: 10.1182/blood-2012-05-402784. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda S, Tagawa H. Dysregulation of microRNAs and their association in the pathogenesis of T-cell lymphoma/leukemias. Int J Hematol. 2014;99:542–52. doi: 10.1007/s12185-014-1535-9. [DOI] [PubMed] [Google Scholar]
- 9.Tagawa H , Ikeda S, Sawada K. Role of microRNA in the pathogenesis of malignant lymphoma. Cancer Sci. 2013;104:801–9. doi: 10.1111/cas.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jardin F, Figeac M. MicroRNAs in lymphoma, from diagnosis to targeted therapy. Curr Opin Oncol. 2013;25:480–6. doi: 10.1097/CCO.0b013e328363def2. [DOI] [PubMed] [Google Scholar]
- 13.Sonneveld P, de Ridder M, van der Lelie H, Nieuwenhuis K, Schouten H, Mulder A, van Reijswoud I, Hop W, Lowenberg B. Comparison of doxorubicin and mitoxantrone in the treatment of elderly patients with advanced diffuse non-Hodgkin’s lymphoma using CHOP versus CNOP chemotherapy. J. Clin. Oncol. 1995;13:2530–9. doi: 10.1200/JCO.1995.13.10.2530. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Salerno E, Scaglione BJ, Coffman FD, Brown BD, Baccarini A, Fernandes H, Marti G, Raveche ES. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol Cancer Ther. 2009;8:2684–92. doi: 10.1158/1535-7163.MCT-09-0127. [DOI] [PubMed] [Google Scholar]
- 17.Sampath D, Liu C, Vasan K, Sulda M, Puduvalli VK, Wierda WG, Keating MJ. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood. 2012;119:1162–72. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Chen G, Feng L, Zhang W, Pelicano H, Wang F, Ogasawara MA, Lu W, Amin HM, Croce CM, Keating MJ, Huang P. Loss of p53 and altered miR15-a/16-1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53(-/-) mouse model and primary human leukemia cells. Leukemia. 2014;28:118–28. doi: 10.1038/leu.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LG, Tang L, Calin G, et al. Expression of MicroRNA (miR) miR-15a/miR-16-1 downregulates expression of BCL-2 protein in chronic lymphocytic leukemia. Blood. 2006;108:792a. [Google Scholar]
- 20.Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL) PLoS One. 2009;4:e7169. doi: 10.1371/journal.pone.0007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 22.Zenz T, Mohr J, Eldering E, Kater AP, Bühler A, Kienle D, Winkler D, Dürig J, van Oers MH, Mertens D, Döhner H, Stilgenbauer S. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113:3801–8. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 23.Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Bühler A, Sarno A, Groner S, Mertens D, Busch R, Hallek M, Döhner H, Stilgenbauer S. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 24.Asslaber D, Pinon JD, Seyfried I, Desch P, Stöcher M, Tinhofer I, Egle A, Merkel O, Greil R. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood. 2010;115:4191–7. doi: 10.1182/blood-2009-07-234823. [DOI] [PubMed] [Google Scholar]
- 25.Audrito V, Vaisitti T, Rossi D, Gottardi D, D’Arena G, Laurenti L, Gaidano G, Malavasi F, Deaglio S. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res. 2011;71:4473–83. doi: 10.1158/0008-5472.CAN-10-4452. [DOI] [PubMed] [Google Scholar]
- 26.Visone R, Veronese A, Balatti V, Croce CM. MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia. Oncotarget. 2012;3:195–202. doi: 10.18632/oncotarget.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 28.Visone R, Veronese A, Rassenti LZ, Balatti V, Pearl DK, Acunzo M, Volinia S, Taccioli C, Kipps TJ, Croce CM. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood. 2011;118:3072–9. doi: 10.1182/blood-2011-01-333484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang YH, Liu P, Hong M, Miao KR, Liu P, Xu W, Li JY. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 30.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T, Croce CM, Pekarsky Y. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107:12210–5. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, Ikuo M, Ghosh AK, Lerner S, Rassenti LZ, Xiao L, Hu J, Reuben JM, Calin S, You MJ, Manning JT, Wierda WG, Estrov Z, O’Brien S, Kipps TJ, Keating MJ, Kay NE, Calin GA. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–9. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, Fabbri M, Lerner S, Barron LL, Rassenti LZ, Jiang L, Xiao L, Hu J, Secchiero P, Zauli G, Volinia S, Negrini M, Wierda W, Kipps TJ, Plunkett W, Coombes KR, Abruzzo LV, Keating MJ, Calin GA. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–52. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moussay E, Palissot V, Vallar L, Poirel HA, Wenner T, El Khoury V, Aouali N, Van Moer K, Leners B, Bernardin F, Muller A, Cornillet-Lefebvre P, Delmer A, Duhem C, Ries F, van Dyck E, Berchem G. Determination of genes and microRNAs involved in the resistance to fludarabine in vivo in chronic lymphocytic leukemia. Mol Cancer. 2010;9:115. doi: 10.1186/1476-4598-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferracin M, Zagatti B, Rizzotto L, Cavazzini F, Veronese A, Ciccone M, Saccenti E, Lupini L, Grilli A, De Angeli C, Negrini M, Cuneo A. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol Cancer. 2010;9:123. doi: 10.1186/1476-4598-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leupin N, Cenni B, Novak U, Hügli B, Graber HU, Tobler A, Fey MF. Disparate expression of the PTEN gene: a novel finding in B-cell chronic lymphocytic leukaemia (B-CLL) Br J Haematol. 2003;121:97–100. doi: 10.1046/j.1365-2141.2003.04227.x. [DOI] [PubMed] [Google Scholar]
- 36.di Iasio MG, Norcio A, Melloni E, Zauli G. SOCS1 is significantly up-regulated in Nutlin-3-treated p53wild-type B chronic lymphocytic leukemia (B-CLL) samples and shows an inverse correlation with miR-155. Invest New Drugs. 2012;30:2403–6. doi: 10.1007/s10637-011-9786-2. [DOI] [PubMed] [Google Scholar]
- 37.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfò L, Rossi C, Ferrari G, Ghia P, Caligaris-Cappio F. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–59. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 38.Culpin RE, Pearce K, Bailey JR, et al. Mature MicroRNAs Mir-18, -19a, -19b, -17-5p and-92 of the Mir-17-92 Cluster Predict for Treatment Free Survival In Patients with Chronic Lymphocytic Leukaemia. Blood. 2010;116:1124–5. [Google Scholar]
- 39.Zhou K, Yi S, Yu Z, Li Z, Wang Y, Zou D, Qi J, Zhao Y, Qiu L. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2012;53:1155–61. doi: 10.3109/10428194.2011.642303. [DOI] [PubMed] [Google Scholar]
- 40.Sampath D, Calin GA, Puduvalli VK, Gopisetty G, Taccioli C, Liu CG, Ewald B, Liu C, Keating MJ, Plunkett W. Specific activation of microRNA106b enables the p73 apoptotic response in chronic lymphocytic leukemia by targeting the ubiquitin ligase Itch for degradation. Blood. 2009;113:3744–53. doi: 10.1182/blood-2008-09-178707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S, Borsky M, Verner J, Doubek M, Brychtova Y, Trbusek M, Hampl A, Mayer J, Pospisilova S. MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood. 2012;119:2110–3. doi: 10.1182/blood-2011-11-394874. [DOI] [PubMed] [Google Scholar]
- 42.Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–72. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 43.Wang LQ, Kwong YL, Kho CS, Wong KF, Wong KY, Ferracin M, Calin GA, Chim CS. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia--implications on constitutive activation of NFkappaB pathway. Mol Cancer. 2013;12:173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craig VJ, Tzankov A, Flori M, Schmid CA, Bader AG, Müller A. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26:2421–4. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 45.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, Schlapbach R, Dirnhofer S, Tzankov A, Müller A. Myc-mediated repression of microRNA-34a promotes highgrade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–36. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH, Gascoyne RD, Tibshirani R, Lossos IS. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–35. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozloski GA, Jiang XY, Bunting KL, Melnick AM, Lossos IS. MiR-181a Is a Master Regulator of the Nuclear Factor-kappa B Signaling Pathway in Diffuse Large B Cell Lymphoma. Blood. 2012:120. [Google Scholar]
- 48.Anastasiadou E, Boccellato F, Vincenti S, Rosato P, Bozzoni I, Frati L, Faggioni A, Presutti C, Trivedi P. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene. 2010;29:1316–28. doi: 10.1038/onc.2009.439. [DOI] [PubMed] [Google Scholar]
- 49.Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–95. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang X, Shen Y, Liu M, Bi C, Jiang C, Iqbal J, McKeithan TW, Chan WC, Ding SJ, Fu K. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am J Pathol. 2012;181:26–33. doi: 10.1016/j.ajpath.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111–6. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang D, Aguiar RC. MicroRNA-155 controls RB phosphorylation in normal and malignant B lymphocytes via the noncanonical TGF-beta1/SMAD5 signaling module. Blood. 2014;123:86–93. doi: 10.1182/blood-2013-07-515254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu L, Song G, Chen L, Nie Z, He B, Pan Y, Xu Y, Li R, Gao T, Cho WC, Wang S. Inhibition of miR-21 induces biological and behavioral alterations in diffuse large B-cell lymphoma. Acta Haematol. 2013;130:87–94. doi: 10.1159/000346441. [DOI] [PubMed] [Google Scholar]
- 54.Bai H, Wei J, Deng C, Yang X, Wang C, Xu R. MicroRNA-21 regulates the sensitivity of diffuse large B-cell lymphoma cells to the CHOP chemotherapy regimen. Int J Hematol. 2013;97:223–31. doi: 10.1007/s12185-012-1256-x. [DOI] [PubMed] [Google Scholar]
- 55.Kwanhian W, Lenze D, Alles J, Motsch N, Barth S, Döll C, Imig J, Hummel M, Tinguely M, Trivedi P, Lulitanond V, Meister G, Renner C, Grässer FA. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1:141–55. doi: 10.1002/cam4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao QW, Li HY, Yao XX, Wang JF. Significance of microRNA-146b-5p in diffuse large B-cell lymphoma and its relationship with risk assessment. Zhonghua Xue Ye Xue Za Zhi. 2012;33:1010–4. [PubMed] [Google Scholar]
- 57.Zhong H, Xu L, Zhong JH, Wang JF. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Exp Ther Med. 2012;3:763–70. doi: 10.3892/etm.2012.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao XX, Wang JF, Wang YH, Gao N. Expression of microRNA-223 and its clinicopathologic correlation in diffuse large B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi. 2012;41:366–70. doi: 10.3760/cma.j.issn.0529-5807.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Berglund M, Hedström G, Amini RM, Enblad G, Thunberg U. High expression of microRNA-200c predicts poor clinical outcome in diffuse large B-cell lymphoma. Oncol Rep. 2013;29:720–4. doi: 10.3892/or.2012.2173. [DOI] [PubMed] [Google Scholar]
- 60.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, Matolcsy A, Bernd HW, Reiniger L, Merz H, Feller AC. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–44. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 61.Nie K, Zhang T, Allawi H, Gomez M, Liu Y, Chadburn A, Wang YL, Knowles DM, Tam W. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: a potential role of the microRNA let-7. Am J Pathol. 2010;177:1470–9. doi: 10.2353/ajpath.2010.091291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci U S A. 2012;109:7865–70. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hedström G, Thunberg U, Berglund M, Simonsson M, Amini RM, Enblad G. Low expression of microRNA-129-5p predicts poor clinical outcome in diffuse large B cell lymphoma (DLBCL) Int J Hematol. 2013;97:465–71. doi: 10.1007/s12185-013-1303-2. [DOI] [PubMed] [Google Scholar]
- 64.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112:822–9. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Chen X, Lin J, Lwin T, Wright G, Moscinski LC, Dalton WS, Seto E, Wright K, Sotomayor E, Tao J. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene. 2012;31:3002–8. doi: 10.1038/onc.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarro A, Clot G, Prieto M, Royo C, Vegliante MC, Amador V, Hartmann E, Salaverria I, Beà S, Martín-Subero JI, Rosenwald A, Ott G, Wiestner A, Wilson WH, Campo E, Hernández L. microRNA expression profiles identify subtypes of mantle cell lymphoma with different clinicobiological characteristics. Clin Cancer Res. 2013;19:3121–9. doi: 10.1158/1078-0432.CCR-12-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, Sotomayor E, Tao J, Cheng JQ. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–9. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin JH, Zhao JJ, Lwin T, et al. MiR-29 MicroRNAs Regulate IGF-1R Expression and Contribute Mantle Cell Lymphoma Growth and Survival. Blood. 2009;114:776. [Google Scholar]
- 69.Lwin T, Lin JH, Choi YS, Zhang X, Moscinski LC, Wright KL, Sotomayor EM, Dalton WS, Tao J. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood. 2010;116:5228–36. doi: 10.1182/blood-2010-03-275925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iqbal J, Shen Y, Liu Y, Liu Y, Fu K, Jaffe ES, Liu C, Liu Z, Lachel CM, Deffenbacher K, Greiner TC, Vose JM, Bhagavathi S, Staudt LM, Rimsza L, Rosenwald A, Ott G, Delabie J, Campo E, Braziel RM, Cook JR, Tubbs RR, Gascoyne RD, Armitage JO, Weisenburger DD, McKeithan TW, Chan WC. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood. 2012;119:4939–48. doi: 10.1182/blood-2011-07-370122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goswami RS, Atenafu EG, Xuan Y, Waldron L, Reis PP, Sun T, Datti A, Xu W, Kuruvilla J, Good DJ, Lai R, Church AJ, Lam WS, Baetz T, Lebrun DP, Sehn LH, Farinha P, Jurisica I, Bailey DJ, Gascoyne RD, Crump M, Kamel-Reid S. MicroRNA signature obtained from the comparison of aggressive with indolent non-Hodgkin lymphomas: potential prognostic value in mantle-cell lymphoma. J. Clin. Oncol. 2013;31:2903–11. doi: 10.1200/JCO.2012.45.3050. [DOI] [PubMed] [Google Scholar]
- 72.Zappasodi R, Di Nicola M, Baldan F, et al. Cytotoxic Activity of Histone Deacetylase Inhibitor ITF2357 on Burkitt’s Lymphoma Cell Lines Is Associated to Micro-RNA Modulation and Transglutaminase 2 Restoration. Blood. 2008;112:565. [Google Scholar]
- 73.Zappasodi R, Cavane A, Iorio MV, et al. MiR-146a up-Regulation Is Associated with Anti-Tumor Activity of Pan-Histone Deacetylase Inhibitor ITF2357 (Givinostat (R)) in Human Burkitt’s Lymphoma. Blood. 2011;118:1174. [Google Scholar]
- 74.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 75.Leucci E, Cocco M, Onnis A, De Falco G, van Cleef P, Bellan C, van Rijk A, Nyagol J, Byakika B, Lazzi S, Tosi P, van Krieken H, Leoncini L. MYC translocation-negative classical Burkitt lymphoma cases: an alternative pathogenetic mechanism involving miRNA deregulation. J Pathol. 2008;216:440–50. doi: 10.1002/path.2410. [DOI] [PubMed] [Google Scholar]
- 76.Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–76. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 77.Leucci E, Onnis A, Cocco M, De Falco G, Imperatore F, Giuseppina A, Costanzo V, Cerino G, Mannucci S, Cantisani R, Nyagol J, Mwanda W, Iriso R, Owang M, Schurfeld K, Bellan C, Lazzi S, Leoncini L. B-cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int J Cancer. 2010;126:1316–26. doi: 10.1002/ijc.24655. [DOI] [PubMed] [Google Scholar]
- 78.Chen S, Wang Z, Dai X, Pan J, Ge J, Han X, Wu Z, Zhou X, Zhao T. Re-expression of microRNA-150 induces EBV-positive Burkitt lymphoma differentiation by modulating c-Myb in vitro. Cancer Sci. 2013;104:826–34. doi: 10.1111/cas.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manfè V, Holst LM, Rosbjerg A, Kamstrup MR, Kaczkowski B, Gniadecki R. Changes in oncomiR expression in CTCL cell lines during apoptosis induced by Notch inhibition. Leukemia Res. 2010;34:235–6. doi: 10.1016/j.leukres.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 80.van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, Willemze R, Tensen CP, Vermeer MH. MicroRNA-21 Expression in CD4+ T Cells Is Regulated by STAT3 and Is Pathologically Involved in Sezary Syndrome. J Invest Dermatol. 2011;131:762–8. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 81.Kopp KL, Ralfkiaer U, Gjerdrum LM, Helvad R, Pedersen IH, Litman T, Jønson L, Hagedorn PH, Krejsgaard T, Gniadecki R, Bonefeld CM, Skov L, Geisler C, Wasik MA, Ralfkiaer E, Ødum N, Woetmann A. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12:1939–47. doi: 10.4161/cc.24987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manfè V, Biskup E, Rosbjerg A, Kamstrup M, Skov AG, Lerche CM, Lauenborg BT, Odum N, Gniadecki R. miR-122 Regulates p53/Akt Signalling and the Chemotherapy-Induced Apoptosis in Cutaneous T-Cell Lymphoma. PLoS One. 2012;7:e29541. doi: 10.1371/journal.pone.0029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manfè V, Biskup E, Willumsgaard A, Skov AG, Palmieri D, Gasparini P, Laganá A, Woetmann A, Ødum N, Croce CM, Gniadecki R. cMyc/miR-125b-5p Signalling Determines Sensitivity to Bortezomib in Preclinical Model of Cutaneous T-Cell Lymphomas. PLoS One. 2013;8:e59390. doi: 10.1371/journal.pone.0059390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Komabayashi Y, Kishibe K, Nagato T, Ueda S, Takahara M, Harabuchi Y. Downregulation of miR-15a due to LMP1 promotes cell proliferation and predicts poor prognosis in nasal NK/T-cell lymphoma. Am J Hematol. 2014;89:25–33. doi: 10.1002/ajh.23570. [DOI] [PubMed] [Google Scholar]
- 85.Xi Y, Li J, Zan L, Wang J, Wang G, Ning Y. Micro-RNA-16 expression in paraffin-embedded specimen correlates with overall survival of T-lymphoblastic lymphoma/leukemia. Hum Pathol. 2013;44:1011–6. doi: 10.1016/j.humpath.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 86.Huang HB, Zhan R, Wu SQ, Xu ZZ, Fan LP. Expression of MCL-1 and microRNA-29a in extranodal NK/T-cell lymphoma tissue. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21:95–8. doi: 10.7534/j.issn.1009-2137.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 87.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–75. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 88.Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, Kim CW. MicroRNA-146a Downregulates NF kappa B Activity via Targeting TRAF6 and Functions as a Tumor Suppressor Having Strong Prognostic Implications in NK/T Cell Lymphoma. Clin Cancer Res. 2011;17:4761–71. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 89.Ng SB, Yan JL, Huang G, Selvarajan V, Tay JL, Lin B, Bi C, Tan J, Kwong YL, Shimizu N, Aozasa K, Chng WJ. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. 2011;118:4919–29. doi: 10.1182/blood-2011-07-364224. [DOI] [PubMed] [Google Scholar]
- 90.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 91.Jiang P, Rao EY, Meng N, Zhao Y, Wang JJ. MicroRNA-17-92 significantly enhances radioresistance in human mantle cell lymphoma cells. Radiat Oncol. 2010;5:100. doi: 10.1186/1748-717X-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rao E, McKeithan T, Jiang CS, et al. The Mir-17 similar to 92 Cluster Enhances Cell Growth and Resistance to Chemotherapy in Mantle Cell Lymphoma by Down-Regulating PTEN, PHLPP2 and BIM. Blood. 2008;112:145. [Google Scholar]
- 93.Rao E, Jiang C, Ji M, Huang X, Iqbal J, Lenz G, Wright G, Staudt LM, Zhao Y, McKeithan TW, Chan WC, Fu K. The miRNA-17 approximately 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26:1064–72. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 94.Bomben R, Gobessi S, Dal Bo M, Volinia S, Marconi D, Tissino E, Benedetti D, Zucchetto A, Rossi D, Gaidano G, Del Poeta G, Laurenti L, Efremov DG, Gattei V. The miR-17 approximately 92 family regulates the response to Toll-like receptor 9 triggering of CLL cells with unmutated IGHV genes. Leukemia. 2012;26:1584–93. doi: 10.1038/leu.2012.44. [DOI] [PubMed] [Google Scholar]
- 95.Fassina A, Marino F, Siri M, Zambello R, Ventura L, Fassan M, Simonato F, Cappellesso R. The miR-17-92 microRNA cluster: a novel diagnostic tool in large B-cell malignancies. Lab Invest. 2012;92:1574–82. doi: 10.1038/labinvest.2012.129. [DOI] [PubMed] [Google Scholar]
- 96.Psathas JN, Doonan PJ, Raman P, Freedman BD, Minn AJ, Thomas-Tikhonenko A. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood. 2013;122:4220–9. doi: 10.1182/blood-2012-12-473090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schiffman JD, Lorimer PD, Rodic V, Jahromi MS, Downie JM, Bayerl MG, Sanmann JN, Althof PA, Sanger WG, Barnette P, Perkins SL, Miles RR. Genome wide copy number analysis of paediatric Burkitt lymphoma using formalin-fixed tissues reveals a subset with gain of chromosome 13q and corresponding miRNA over expression. Br J Haematol. 2011;155:477–86. doi: 10.1111/j.1365-2141.2011.08883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Y, Liu GL, Liu SH, Wang CX, Xu YL, Ying Y, Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin’s Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res. 2012;53:516–25. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Migliore C, Giordano S. Resistance to targeted therapies: a role for microRNAs? Trends Mol Med. 2013;19:633–42. doi: 10.1016/j.molmed.2013.08.002. [DOI] [PubMed] [Google Scholar]