Abstract
Previous studies have shown that the single nucleotide polymorphisms (SNPs) in Methylenetetrahydrofolate reductase (MTHFR) and Glutathione S-transferases (GSTs, including GSTM1, GSTT1) genes play an important role in determining the response of an individual to environmental pathogenesis and significantly relate to incidences of various human tumors, including brain tumors. However, these genes’ polymorphisms on meningioma risk remains poorly understood. The relevant inferences from previous studies are hindered by their limited statistical power and conflicting results. The aim of this meta-analysis is to provide a relatively comprehensive account of the association between these polymorphisms and human meningioma risk. A literature search for eligible studies published before January 1, 2014 was conducted in PubMed, Embase, Web of Science, Cochrane Library, and CNKI databases. Pooled odds ratios (OR) with their corresponding 95% confidence intervals (95% CI) were used to evaluate the strength of the association under a fixed or random effect model according to heterogeneity test results. Heterogeneity and publication bias were evaluated. All statistical analyses were conducted by using the software of STATA 12.0 (STATA Corporation, College Station, TX, USA). For MTHFR C677T (dbSNP: rs1801133) (C T) polymorphism, 9 individual case-control studies from six publications with 1,615 cases and 1,909 controls were obtained. For GSTM1 null polymorphism, there were 4 studies with 417 cases and 1,735 controls. For GSTT1 null polymorphism, there were 4 studies with 405 cases and 1,622 controls. The combined results for the MTHFR C677T show that carriers of the CT genotype may be associated with a higher meningioma risk (OR = 1.20, 95% CI 1.05-1.38, P = 0.009). Stratified analyses show that Caucasians have significantly higher risk if they carry the CT genotype of MTHFR (OR = 1.31, 95% CI 1.05-1.63, P = 0.02). Risk of Caucasians carrying TT + CT genotype is also significantly higher (OR = 1.27, 95% CI 1.02-1.58, P = 0.03). Risk of Caucasians carrying TT genotype is not significantly different compared to control population (OR = 0.96, 95% CI 0.69-1.34, P = 0.82). All of the enrolled studies about GSTM1/GSTT1 are on Caucasians. The pooled ORGSTM1 and ORGSTT1 were not significant in Caucasian population. These results indicate SNPs of MTHFR C677T are related to meningioma risk with ethnic differences. Caucasians carrying CT genotype of MTHFR C677T have significantly higher meningioma susceptibility. SNPs of GSTM1/GSTT1 are not related to meningioma risk.
Keywords: MTHFR, GSTM1, GSTT1, meningioma, gene polymorphism, meta-analysis
Introduction
Meningiomas
Meningioma originates from the arachnoid membrane of the brain and is the second most common primary tumor of the central nervous system (CNS) [1]. In its etiologies, multiple genetic and environmental factors have been implicated. Genetic variants and polymorphisms are the main factors that have been proven or assumed to be involved in meningioma formation. According to the histologic features these tumors are classified into three categories: benign/meningioma (WHO grade I), atypical meningioma (WHO grade II) and anaplastic/malignant meningioma (WHO grade III). Meningioma is a slow-growing benign tumor that accounts for approximately 20% of brain tumors, with an incidence of 2.3/100,000 overall and a 2:1 female-to-male ratio according to literatures [2]. Nowadays Meningioma is a very common brain tumor in the US [3] and also in China [4]. Most meningiomas are benign (WHO Grade I), but up to 20% of meningiomas are assigned to the WHO Grades II (atypical meningioma) and III (anaplastic/malignant meningioma) [5]. Despite largely benign histology, these tumors can lead to serious morbidity owing to their intracranial location [6,7]. The 5-year survival rate of patients with meningiomas could be 81.8% [8]. The etiologies of meningiomas are not well understood. In recent years, multitudinous genes have been identified to be associated with meningiomas [5,9,10]. Evidences from prior epidemiologic studies, although inconsistent, suggest a possible association between the incidence of meningioma and some genes polymorphisms [11,12]. The cryptic genetic background leading to meningiomas includes a great deal of nucleotides mutations, or single nucleotide polymorphisms (SNPs), the latter might be involved in meningiomas development, progression and recurrence. Moreover, more and more genome-wide association studies have suggested genetic polymorphisms as risk factors for brain tumors [13].
MTHFR
Methylenetetrahydrofolate reductase (MTHFR) plays critical roles in the maintenance of DNA normal methylation, de novo synthesis of nucleotide, and DNA repair by modulating the folate metabolism [14,15]. The human MTHFR gene is located on 1p36.3 containing a total of 11 extrons and 10 introns and encoding a protein of 74.6 kDa [14]. The encoded protein of MTHFR can convert 5, 10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is the primary circulating form of folate and is an irreversible conversion [16]. Some mutations in the MTHFR gene, such as C (cytosine) > T (thymidine) at nucleotide 677, leading to a conversion of alanine to valine, are associated with lower folate level when compared to the normal genotype [17]. Thus results in decreased thermal stability and enzyme activity of the encoded protein [18-20]. Some academicians found the MTHFR C677T polymorphism is associated with a number of human malignant tumors, such as cervical cancer [21], hepatocellular carcinoma [22], and breast cancer [23]. However, an other study found there is no association between the MTHFR C677T polymorphism and breast cancer [24]. Because of the complicated molecular mechanisms of tumors and the different genetic backgrounds and environmental exposures, relationship between MTHFR C677T polymorphism and tumors is still controversial and ambiguous.
GSTs
Glutathione S-transferases (GSTs) are dimeric phase II metabolic enzymes that catalyze the reactions between reduced glutathione and toxic. The range of potential GST substrates is very large, including occupational and environmental carcinogens such as solvents, pesticides, and polycyclic aromatic hydrocarbons. Four families of cytosolic soluble GSTs α (GSTA), µ (GSTM), Π (GSTP), and θ (GSTT) are known in humans [25].
GSTM1 is one member of GSTM family. Three alleles of GSTM1 gene locus (GSTM1A, GSTM1B and GSTM1 0) have been found in human populations. GSTM1A and GSTM1B differ by a single amino acid but have similar enzyme activity, whereas GSTM1 0 (null allele) produces no catalyzing activity [26]. GSTM1 null genotype frequencies have been reported to be ethnic differences [27]. A deletion polymorphism was reported also at GSTT1locus (22q12) [28]. The GSTT1 null genotype distribution also has ethnic differences [29,30]. Similar to GSTM1, individuals carrying a homozygous of GSTT1 null genotype encode deficient protein that affects detoxification enzyme activity [26,31].
The knowledge of the role of the GSTs enzymes in detoxification has led to the hypothesis that if an individual’s genotype at GSTs locus encodes a deficient GSTs enzyme it may result in increased risk of tumors. Absent or deficient GSTs enzyme activity may result in increased risk of somatic mutations, subsequently leading to tumor formation [27]. Some epidemiological studies have discovered the association between GSTM1 null genotype and incidences of some tumors, especially oligodendrogliomas [32], myelodysplastic diseases [33], lung cancer [34] and bladder cancers [35]. However, for GSTT1 null polymorphism, no relationship was found with incidence of glioma [36].
The purpose of this study was to examine the effect of folate metabolism gene MTHFR C677T and GSTM1/GSTT1 null polymorphisms on human meningioma risk in total population and the difference in Caucasia and Asian. Although the association of MTHFR C677T and GSTM1/GSTT1 polymorphisms with brain tumor risk has been widely studied, the role of polymorphic variants as risk factors for meningioma has received comparatively little attention. The previous findings of individual case-control studies are paradoxical, possibly due to the small sample sizes and the differences in the source of controls and methods of classification. Thus, we carried out the current meta-analysis by pooling available data from published studies to elucidate the relationship of meningioma incidence risk with the above gene polymorphisms.
Materials and methods
Literature search strategy
A literature search for eligible studies published before January 1, 2014 was conducted among the following electronic databases: PubMed, Embase, Web of Science, Cochrane Library, and CNKI (China National Knowledge Infrastructure) databases. The following key words search strategy was used: (“meningiomas” or “brain tumor”) and (“MTHFR” or “Folate metabolism”) and (“GSTs” or “GSTM1” or “GSTT1” or “Glutathione S-transferases”) and (“genetic polymorphism” or “SNP” or “gene”). The search was done without any limitation on languages and only included studies conducted with human subjects. The reference lists of reviews and retrieved articles were manually screened by two investigators to identify additional potential sources.
Selection criteria
To be included in the analysis, candidate studies had to meet the following criteria: (1) case-control study focused on the relationship between the MTHFR C677T or GSTM1/GSTT1 null polymorphisms and meningioma risk, (2) all patients met the diagnostic criteria for meningioma, (3) sufficient original data for calculating odds ratios (OR) with corresponding 95% confidence intervals (95% CI) was provided. Major reasons for excluding studies were the following: (1) not case-control study, (2) duplicate publications, (3) no usable data reported, (4) missing ethics board approval. This meta-analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [37] with only slight modification to better suit the nature of those studies.
Data extraction
According to the PRISMA guidelines, two independent reviewers checked each full-text report for eligibility and extracted and tabulated the following data from eligible studies: surname of first author, year of publication, country of origin, ethnicity, definition and numbers of cases and controls, genotyping method. Disagreements were resolved through discussion and subsequent consensus between reviewers.
Statistical analysis
To account for the between-study heterogeneity, a statistical test of Cochran’s Q statistic was used. The heterogeneity was indicated by P < 0.05 [38]. Heterogeneity was also assessed through the I2 test, with I2 > 50% indicating significant heterogeneity [39]. When no heterogeneity was found (P > 0.05 or I2 < 50%), a fixed-effect model was used to estimate pooled odds ratios (OR) and their corresponding 95% confidence intervals (CI). Otherwise, a random effects model was applied. The significance of the pooled OR was determined using the Z test. Stratified analyses were performed in ethnicity subgroups (Caucasians, Asians). Sensitivity analyses were performed to assess the stability of pooled results. Each case-control study was omitted in turn to reflect the influence of individual datasets on the pooled results. Begg’s funnel plot and Egger’s linear regression test were used to assess the potential for publication bias [40,41]. All two-tailed P < 0.05 were considered as statistical significance. All analyses were performed using STATA software v12.0 (Stata Corp, College Station, TX, USA).
Results
A total of 126 relevant papers were identified using the prespecified search strategy. In accordance with the inclusion criteria, thirteen case-control studies were included from nine publications [42-50], of which nine were on MTHFR C677T (rs1801133) and four were on GSTM1/GSTT1. A flow chart of the selection process and specific reasons for exclusion from this meta-analysis are shown in Figure 1. A total of 7,703 individual were included in this meta-analysis, of which 2,437 were meningioma patients and 5,266 were controls. The publication years of included studies ranged from 1995 to 2013. The characteristics and methodology of the included studies are summarized in Table 1.
Figure 1.
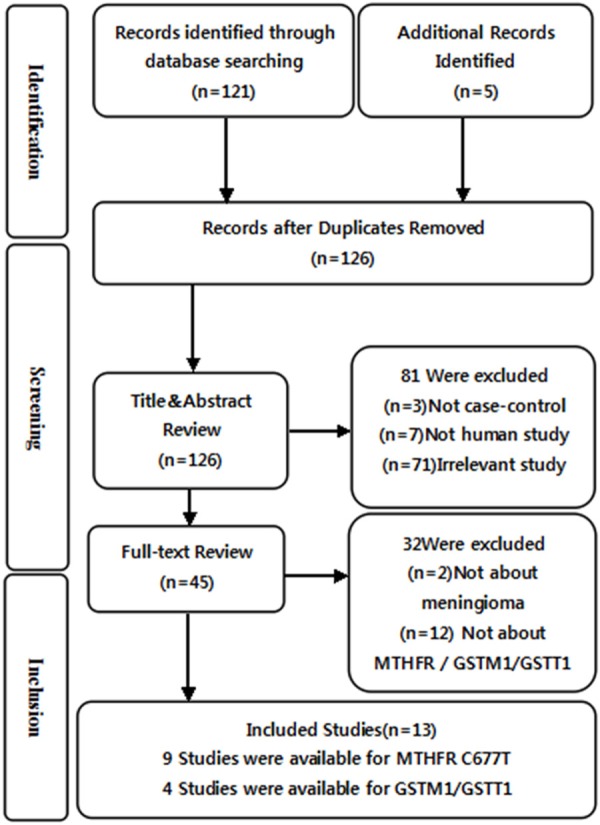
Flow diagram of the selection of studies and specific reasons for exclusion from the present meta-analysis.
Table 1.
Main characteristics of all eligible studies
| Author | Year | Country | Racial descent | Sample size | Control source | Genotyping method | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Case | control | |||||||
| MTHFR C677T (rs1801133) | ||||||||
| Kafadar [42] | 2006 | Turkey | Caucasian | 35 | 98 | HB | PCR-RFLP | |
| Ahn [43] | 2002 | South Korea | Asian | 32 | 254 | HB | PCR-RFLP | |
| Behke FL [44] | 2008 | Finland | Caucasian | 77 | 77 | PB | Illumina | |
| Bethke DM [44] | 2008 | Denmark | Caucasian | 110 | 113 | PB | Illumina | |
| Bethke NO-UK [44] | 2008 | North UK | Caucasian | 174 | 175 | PB | Illumina | |
| Bethke SD [44] | 2008 | Sweden | Caucasian | 149 | 149 | PB | Illumina | |
| Bethke SE-UK [44] | 2008 | Southeast UK | Caucasian | 121 | 123 | PB | Illumina | |
| Li [45] | 2013 | China | Asian | 317 | 320 | HB | PCR-RFLP | |
| Zhang [46] | 2013 | China | Asian | 600 | 600 | PB | PCR-RFLP | |
| GSTM1 Null | ||||||||
| Jose [47] | 1995 | UK | Caucasian | 49 | 577 | HB | PCR-RFLP | |
| Pinarbasi [48] | 2004 | Turkey | Caucasian | 23 | 153 | HB | PCR-RFLP | |
| Roos [49] | 2003 | USA | Caucasian | 169 | 575 | HB | PCR-RFLP | |
| Schwartzbaum [50] | 2007 | European | Caucasian | 176 | 430 | PB | PCR-RFLP | |
| GSTT1 Null | ||||||||
| Jose [47] | 1995 | UK | Caucasian | 47 | 494 | HB | PCR-RFLP | |
| Pinarbasi [48] | 2004 | Turkey | Caucasian | 23 | 153 | HB | PCR-RFLP | |
| Roos [49] | 2003 | USA | Caucasian | 159 | 545 | HB | PCR-RFLP | |
| Schwartzbaum [50] | 2007 | European | Caucasian | 176 | 430 | PB | PCR-RFLP | |
Notes: PCR-RFLP: polymerase chain reaction–restriction fragment length polymorphism; PB: population-based; HB: hospital-based; Behke FL: Finland; Bethke DM: Denmark; Bethke NO-UK: UK-North; Bethke SD: Sweden; Bethke SE-UK: UK-Southeast.
Association between the MATHF C667T polymorphism and meningioma risk
The evaluation of the association between the MTHFR C677T (dbSNP: rs1801133, C>T) polymorphism and meningioma incidence risk is summarized in Table 2. It includes nine case-control studies, with a total of 1,615 meningioma cases and 1,909 controls. The combined results indicate that for total population (Caucasians + Asian) carriers of the CT genotype may be associated with a higher risk of meningioma incidence risk than others genotypes carriers (CT Total: OR = 1.20, 95% CI 1.05-1.38, P = 0.009). Stratified analysis shows that for Caucasians population, not only the CT genotype carriers, but also the sum carriers of CT+TT genotype, have significantly higher meningioma risk when compared to the control population (CT Caucasians: OR = 1.31, 95% CI 1.05-1.63, CT+TT Caucasians: OR = 1.27, 95% CI 1.02-1.58, P = 0.03) (Figures 2, 3 and 4). No statistical significance was obtained for Asian population in this stratified analysis, indicating there are ethnic differences in meningioma susceptibility.
Table 2.
Meta-analysis results for the MTHFR C677T, GSTM1/GSTT1 Polymorphism and meningioma risk
| Genotype | Population | Begg’s test | Egger’s test | |
|---|---|---|---|---|
|
|
||||
| p | p | |||
| MTHFR C677T (rs1801133) | T | Total | > 0.466 | 0.921 |
| Caucasians | > 0.452 | 0.465 | ||
| Asian | 1 | 0.984 | ||
| TT | Total | > 0.602 | 0.886 | |
| Caucasians | > 0.26 | 0.47 | ||
| Asian | 1 | 0.967 | ||
| CT | Total | > 0.602 | 0.738 | |
| Caucasians | > 0.452 | 0.577 | ||
| Asian | 1 | 0.465 | ||
| TT+TC | Total | > 0.348 | 0.978 | |
| Caucasians | > 0.26 | 0.135 | ||
| Asian | 1 | 0.973 | ||
| GSTM1 | Null | Caucasians | > 0.308 | 0.337 |
| Asian | -- | -- | ||
| GSTT1 | Null | Caucasians | > 0.734 | 0.694 |
| Asian | -- | -- | ||
Figure 2.
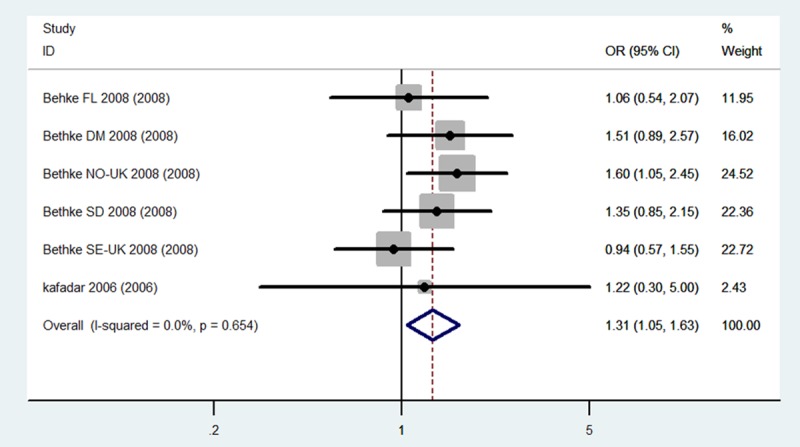
Forest plots for the association between the MTHFR C677T polymorphism and meningioma incidence in CT genotypes among total population.
Figure 3.
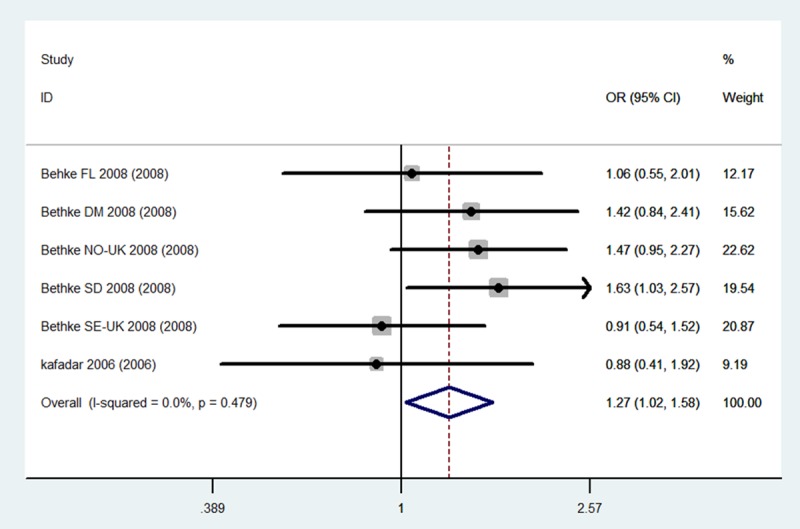
Forest plots for the association between the MTHFR C677T polymorphism and meningioma incidence in CT genotypes among Caucasians.
Figure 4.
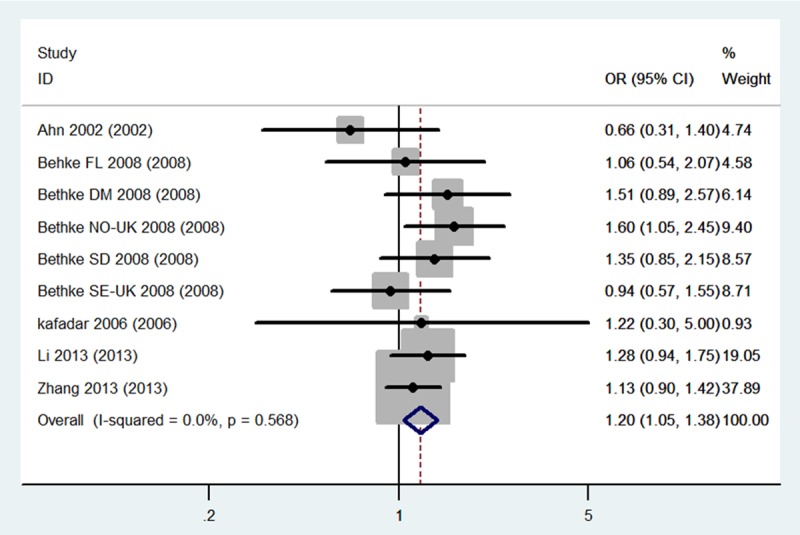
Forest plots for the association between the MTHFR C677T polymorphism and meningioma incidence in CT+TT genotypes among Caucasians.
Association between the GSTM1/GSTT1 null polymorphism and meningioma risk
The evaluation of the association between the GSTM1/GSTT1 null polymorphism and meningioma incidence is summarized in Table 2. For GSTM1 there were four case-control studies, with a total of 417 meningioma cases and 1,735 controls. For GSTT1 genotypes there were also four case-control studies, but with total 405 meningioma cases and 1,622 controls. The random effect model was conducted since heterogeneity obviously existed both in GSTM1 (P = 0.03 and I2 = 66%) (Table 2) and in GSTT1 (P = 0.01 and I2 = 73%) (Table 2). The combined results indicate that Caucasians carriers of both the GSTM1 null genotype and GSTT1 null genotype do not have statistically different risk of meningioma incidence than other allele genotypes carriers (GSTM1: OR = 1.17, 95% CI 0.77-1.79, P = 0.46; GSTT1: OR = 1.59, 95% CI 0.91-2.78, P = 0.1). Unfortunately, all of the studies were conducted for Caucasians and no study derived from Asian was retrieved. Therefore there was no direct evidence showing the GSTM1/GSTT1 null polymorphism association with meningioma incidence in Asian.
Sensitivity analyses and publication bias
Sensitivity analysis was performed to test the influence of each single study to the pooled OR. The pooled OR was not significantly altered when any individual study was deleted for the MTHFR C677T, GSTM1 and GSTT1 analysis. Begg’s test (Figures 5, 6 and 7) and Egger’s test were used to assess the publication bias throughout the studies selected for the meta-analysis. Neither of these tests demonstrated any significant publication bias in studies for the MTHFR, GSTM1 or GSTT1 polymorphisms (Table 3).
Figure 5.
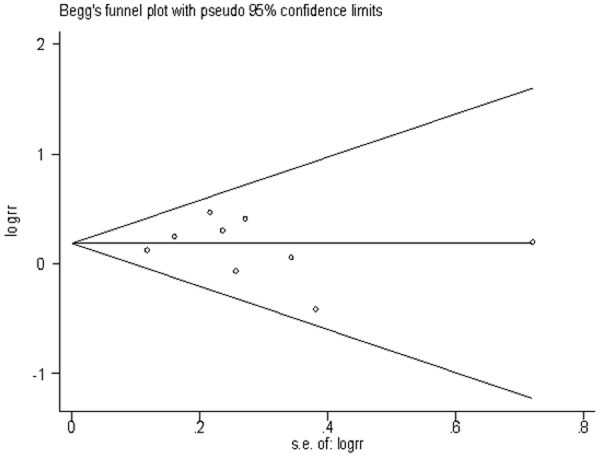
Begg’s funnel plots of publication bias for the association between MTHFR C677T CT genotype and risk of meningioma among total population.
Figure 6.
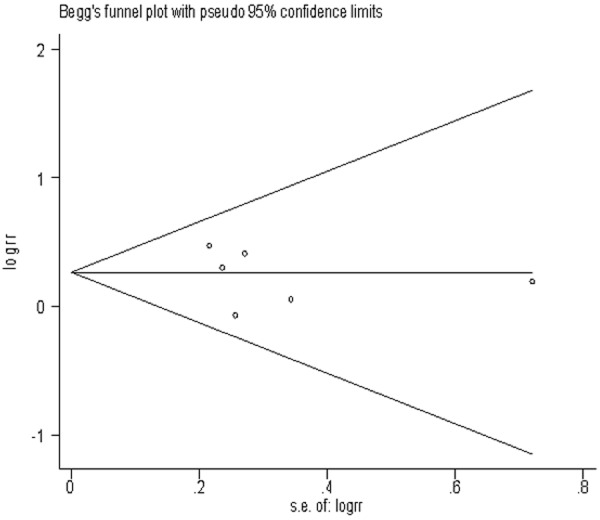
Begg’s funnel plots of publication bias for the association between MTHFR C677T CT genotype and risk of meningioma among Caucasians population.
Figure 7.
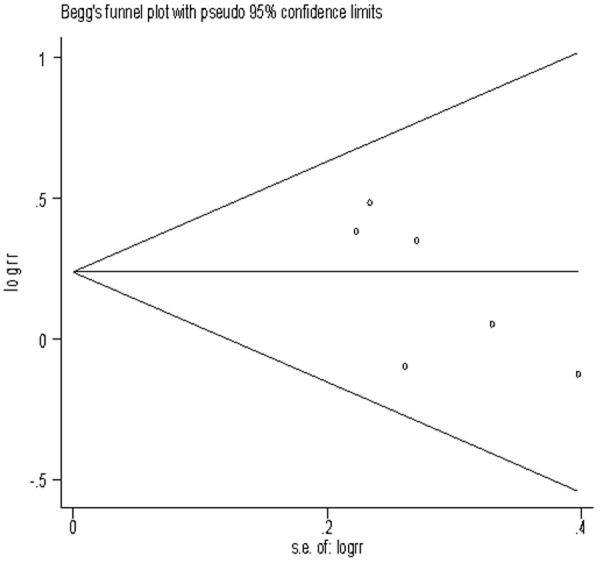
Begg’s funnel plots of publication bias for the association between MTHFR C677T CT+TT genotype and risk of meningioma among Caucasians population.
Table 3.
P values for publication bias tests
| MTHFR | Genotype | Population | N | Test of association | Mode | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| C677T (rs1801133) | CASE | CONTROL | OR | 95% CI | P | χ2 | P | I2 | ||||
| T | Total | 9 | 1101 | 1242 | 1.1 | [0.85, 1.42] | 0.48 | R | 41.5 | 0.00001 | 81% | |
| Caucasians | 6 | 452 | 449 | 1.15 | [0.98, 1.35] | 0.1 | F | 3.75 | 0.59 | 0% | ||
| Asian | 3 | 649 | 793 | 1.06 | [0.93, 1.21] | 0.38 | R | 37.22 | 0.00001 | 95% | ||
| TT | Total | 9 | 192 | 251 | 0.98 | [0.61, 1.57] | 0.94 | R | 31.68 | 0.0001 | 75% | |
| Caucasians | 6 | 77 | 104 | 0.96 | [0.69, 1.34] | 0.82 | F | 2.91 | 0.71 | 0% | ||
| Asian | 3 | 115 | 147 | 0.96 | [0.28, 3.34] | 0.95 | R | 28.77 | 0.00001 | 93% | ||
| CT | Total | 9 | 726 | 771 | 1.2 | [1.05, 1.38] | 0.009 | F | 6.71 | 0.57 | 0% | |
| Caucasians | 6 | 307 | 272 | 1.31 | [1.05, 1.63] | 0.02 | F | 3.3 | 0.65 | 0% | ||
| Asian | 3 | 419 | 499 | 1.14 | [0.95, 1.37] | 0.15 | F | 2.55 | 0.28 | 22% | ||
| TT+TC | Total | 9 | 918 | 1022 | 1.18 | [0.89, 1.57] | 0.26 | R | 28.13 | 0.0004 | 72% | |
| Caucasians | 6 | 384 | 376 | 1.27 | [1.02, 1.58] | 0.03 | F | 4.5 | 0.48 | 0% | ||
| Asian | 3 | 534 | 646 | 1.06 | [0.52, 2.17] | 0.87 | R | 22.86 | 0.0001 | 91% | ||
| GSTM1 | Null | Caucasians | 4 | 230 | 910 | 1.17 | [0.77, 1.79] | 0.46 | R | 8.95 | 0.03 | 66% |
| Asian | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | ||
| GSTT1 | Null | Caucasians | 4 | 92 | 290 | 1.59 | [0.91, 2.78] | 0.1 | R | 10.93 | 0.01 | 73% |
| Asian | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | ||
Discussion
The findings from the current meta-analysis supported that MTHFR C677T variant may exert an increased risk effect on meningioma incidence. From this meta-analysis we demonstrated that carriers of the CT genotype of MTHFR C677T may associate with a higher meningioma risk. The data also show that Caucasians have significantly higher meningioma risk if they carrying the CT genotype, since both Caucasians carriers of the CT genotype and the sum Caucasians carriers of CT+TT genotype have higher meningioma risk according to the stratified analysis. Therefore, we conclude that detecting the MTHFR C677T polymorphism may become a method of forecasting the meningioma susceptibility of human being, especially for Caucasians. In the data no meningioma risk difference was found for Asian in the MTHFR C677T analysis, which may result from the limited sample sizes in the retrieved studies that conducted for Asian. No studies conducted for other ethnicities were retrieved. Therefore, we think the role of MTHFR C677T mutation in meningioma pathogenesis across diverse ethnicities needs further elucidation by more future studies with large sample sizes.
In this study, we present an intensive analysis for associations between GSTM1/GSTT1 variant genotypes and the risk of meningioma incidence. The association between GSTM1 deletion polymorphism (GSTM1 null genotype) and the meningioma risk did not reach statistical significance in our analysis. However, the statistical data (OR = 1.17, 95% CI 0.77-1.79) reflect a trend of positive association. The same, no positive association was confirmed between the meningioma risk and GSTT1 deletion polymorphism (GSTT1 null genotype) but the statistical data (OR = 1.59, 95% CI 0.91-2.78) reflect a positive trend. Although this meta-analysis for the included studies did not found the association between the GSTM1/GSTT1 polymorphisms and the meningioma risk, we believe that amplifying the sample sizes may obtain a different result, since some previous studies have found a positive association between these polymorphisms and types of brain tumors, including brain meningiomas [32,47,49,51,52].
In conclusion, our results indicated MTHFR C677T polymorphism is significantly related to meningioma risk. Even the relationship between the GSTM1/GSTT1 null polymorphisms and meningioma risk cannot be demonstrated by this meta-analysis, we believe it might be positively discovered if relevant studies with large sample sizes be performed in the future. However, we think these conclusions should be received with caution, since the meta-analysis in this study was limited by the quality and quantity of included studies. For one hand, although no statistical significance of publication bias was found in this study, the underlying bias may be produced when only English and Chinese publications were included. On the other hand, it is hard to identify precise gene associations in Asians and other mixed population since the included population was too limited. Further studies should be conducted on a broader scale in order to investigate these gene variants in different ethnicities especially in Asian and to achieve more comprehensively understanding the associations between MTHFR C677T, GSTM1/GSTT1 genes and the risk of meningioma.
Disclosure of conflict of interest
None.
References
- 1.Zang KD. Meningioma: a cytogenetic model of a complex benign human tumor, including data on 394 karyotyped cases. Cytogenet Cell Genet. 2001;93:207–220. doi: 10.1159/000056986. [DOI] [PubMed] [Google Scholar]
- 2.Leone PE, Mendiola M, Alonso J, Paz-y-Miño C, Pestaña A. Implications of a RAD54L polymorphism (2290C/T) in human meningiomas as a risk factor and/or a genetic marker. BMC Cancer. 2003;3:6. doi: 10.1186/1471-2407-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14:v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu J, Little J, Xu T, Zhao X, Guo L, Jia X, Huang G, Bi D, Liu R. Risk factors for meningioma in adults: A case-control study in northeast China. Int J Cancer. 1999;83:299–304. doi: 10.1002/(sici)1097-0215(19991029)83:3<299::aid-ijc2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 5.Semmler A, Simon M, Moskau S, Linnebank M. Polymorphisms of methionine metabolism and susceptibility to meningioma formation. J Neurosurg. 2008;108:999–1004. doi: 10.3171/JNS/2008/108/5/0999. [DOI] [PubMed] [Google Scholar]
- 6.Kousi E, Tsougos I, Fountas K, Theodorou K, Tsolaki E, Fezoulidis I, Kapsalaki E. Distinct peak at 3.8 ppm observed by 3T MR spectroscopy in meningiomas, while nearly absent in high-grade gliomas and cerebral metastases. Mol Med Rep. 2012;5:1011. doi: 10.3892/mmr.2012.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Zhao G, Yang H, Wang D, Yu J, Huang H. Assessment of risk factors for early seizures following surgery for meningiomas using logistic regression analysis. J Int Med Res. 2011;39:1728–1735. doi: 10.1177/147323001103900515. [DOI] [PubMed] [Google Scholar]
- 8.Weber DC, Schneider R, Goitein G, Koch T, Ares C, Geismar JH, Schertler A, Bolsi A, Hug EB. Spot scanning-based proton therapy for intracranial meningioma: long-term results from the Paul Scherrer Institute. Int J Radiat Oncol Biol Phys. 2012;83:865–871. doi: 10.1016/j.ijrobp.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Lomas J, Bello MJ, Arjona D, Alonso ME, MartinezGlez V, Lopez-Marin I, Amiñoso C, de Campos JM, Isla A, Vaquero J. Genetic and epigenetic alteration of the NF2 gene in sporadic meningiomas. Genes Chromosomes Cancer. 2005;42:314–319. doi: 10.1002/gcc.20141. [DOI] [PubMed] [Google Scholar]
- 10.Rajaraman P, Brenner AV, Neta G, Pfeiffer R, Wang SS, Yeager M, Thomas G, Fine HA, Linet MS, Rothman N. Risk of meningioma and common variation in genes related to innate immunity. Cancer Epidemiol Biomarkers Prev. 2010;19:1356–1361. doi: 10.1158/1055-9965.EPI-09-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosking F, Feldman D, Bruchim R, Olver B, Lloyd A, Vijayakrishnan J, Flint-Richter P, Broderick P, Houlston R, Sadetzki S. Search for inherited susceptibility to radiation-associated meningioma by genomewide SNP linkage disequilibrium mapping. Br J Cancer. 2011;104:1049–1054. doi: 10.1038/bjc.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbins SE, Broderick P, Melin B, Feychting M, Johansen C, Andersson U, Brännström T, Schramm J, Olver B, Lloyd A. Common variation at 10p12. 31 near MLLT10 influences meningioma risk. Nat Genet. 2011;43:825–827. doi: 10.1038/ng.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostrom QT, Barnholtz-Sloan JS. Current state of our knowledge on brain tumor epidemiology. Curr Neurol Neurosci Rep. 2011;11:329–335. doi: 10.1007/s11910-011-0189-8. [DOI] [PubMed] [Google Scholar]
- 14.Kutzbach C, Stokstad EL. Mammalian methylenetetrahydrofolate reductase. Partial purification, properties, and inhibition by S-adenosylmethionine. Biochim Biophys Acta. 1971;250:459–477. doi: 10.1016/0005-2744(71)90247-6. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe J, Chrin L. Folate metabolism in filariae: enzymes associated with 5, 10-methylenetetrahydrofolate. J Parasitol. 1980;66:53–58. [PubMed] [Google Scholar]
- 16.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 17.van der Put NM, Gabreëls F, Stevens E, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Liu W, Hao Q, Bao L, Wang K. Folate intake and methylenetetrahydrofolate reductase gene polymorphisms as predictive and prognostic biomarkers for ovarian cancer risk. Int J Mol Sci. 2012;13:4009–4020. doi: 10.3390/ijms13044009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fattal-Valevski A, Bassan H, Korman SH, Lerman-Sagie T, Gutman A, Harel S. Methylenetetrahydrofolate reductase deficiency: importance of early diagnosis. J Child Neurol. 2000;15:539–543. doi: 10.1177/088307380001500808. [DOI] [PubMed] [Google Scholar]
- 20.Schwahn B, Rozen R. Polymorphisms in the methylenetetrahydrofolate reductase gene. Am J Pharmacogenomics. 2001;1:189–201. doi: 10.2165/00129785-200101030-00004. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Zhu J. C677T polymorphism of methylenetetrahydrofolate reductase may contribute to cervical cancer risk in complete over-dominant model. Med Hypotheses. 2013;80:679–683. doi: 10.1016/j.mehy.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Jin F, Qu LS, Shen XZ. Association between the methylenetetrahydrofolate reductase C677T polymorphism and hepatocellular carcinoma risk: a meta-analysis. Diagn Pathol. 2009;4:39–46. doi: 10.1186/1746-1596-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Qiu LX, Wang ZH, Wu XH, Liu XJ, Wang BY, Hu XC. MTHFR C677T polymorphism associated with breast cancer susceptibility: a meta-analysis involving 15,260 cases and 20,411 controls. Breast Cancer Res Treat. 2010;123:549–555. doi: 10.1007/s10549-010-0783-5. [DOI] [PubMed] [Google Scholar]
- 24.Grieu F, Powell B, Beilby J, Iacopetta B. Methylenetetrahydrofolate reductase and thymidylate synthase polymorphisms are not associated with breast cancer risk or phenotype. Anticancer Res. 2004;24:3215–3220. [PubMed] [Google Scholar]
- 25.Hirvonen A. Polymorphisms of xenobiotic-metabolizing enzymes and susceptibility to cancer. Environ Health Perspect. 1999;107:37. doi: 10.1289/ehp.99107s137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidegård J, Vorachek WR, Pero RW, Pearson WR. Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci U S A. 1988;85:7293–7297. doi: 10.1073/pnas.85.19.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 28.Salinas AE, Wong MG. Glutathione S-transferases-a review. Curr Med Chem. 1999;6:279–310. [PubMed] [Google Scholar]
- 29.Nelson HH, Wiencke JK, Christiani DC, Cheng T, Zuo ZF, Schwartz BS, Lee BK, Spitz MR, Wang M, Xu X. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis. 1995;16:1243–1246. doi: 10.1093/carcin/16.5.1243. [DOI] [PubMed] [Google Scholar]
- 30.Lee EJ, Wong JY, Yeoh PN, Gong NH. Glutathione S transferase-theta (GSTT1) genetic polymorphism among Chinese, Malays and Indians in Singapore. Pharmacogenetics. 1995;5:332–334. doi: 10.1097/00008571-199510000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelsey K, Wrensch M, Zuo ZF, Miike R, Wieneke J. A population-based case-control study of the CYP2D6 and GSTTI polymorphisms and malignant brain tumors. Pharmacogenetics. 1997;7:463–468. doi: 10.1097/00008571-199712000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, Sandler D, Taylor J, Bell D, Shore D, Liu E, Bloomfield C, Bloomfield C, Sandler D, Liu E. Increased risk for myelodysplastic syndromes in individuals with glutathione transferase theta 1 (GSTT1) gene defect. Lancet. 1996;347:295–297. doi: 10.1016/s0140-6736(96)90468-7. [DOI] [PubMed] [Google Scholar]
- 34.Pinarbasi H, Silig Y, Cetinkaya O, Seyfikli Z, Pinarbasi E. Strong association between the GSTM1-null genotype and lung cancer in a Turkish population. Cancer Genet Cytogenetics. 2003;146:125–129. doi: 10.1016/s0165-4608(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 35.Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst. 1993;85:1159–1164. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 36.Trizna Z, De Andrade M, Kyritsis AP, Briggs K, Levin VA, Bruner JM, Wei Q, Bondy ML. Genetic polymorphisms in glutathione S-transferase mu and theta, N-acetyltransferase, and CYP1A1 and risk of gliomas. Cancer Epidemiol Biomarkers Prev. 1998;7:553–555. [PubMed] [Google Scholar]
- 37.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2009;151:264–269. [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson D, White IR, Riley RD. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins J, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 40.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 41.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kafadar AM, Yilmaz H, Kafadar D, Ergen A, Zeybek U, Bozkurt N, Kuday C, Isbir T. C677T gene polymorphism of methylenetetrahydrofolate reductase (MTHFR) in meningiomas and high-grade gliomas. Anticancer Res. 2006;26:2445–2449. [PubMed] [Google Scholar]
- 43.Ahn JY, Kim NK, Han JH, Kim JK, Joo JY, Lee KS. Genetic mutation of 5, 10-methylenetetrahydrofolate reductase in the brain neoplasms. Journal of Korean Neurosurgical Society. 2002;32:183–188. [Google Scholar]
- 44.Bethke L, Webb E, Murray A, Schoemaker M, Feychting M, Lönn S, Ahlbom A, Malmer B, Henriksson R, Auvinen A. Functional polymorphisms in folate metabolism genes influence the risk of meningioma and glioma. Cancer Epidemiol Biomarkers Prev. 2008;17:1195–1202. doi: 10.1158/1055-9965.EPI-07-2733. [DOI] [PubMed] [Google Scholar]
- 45.Li R, Wang R, Li Y, Li X, Feng Y, Li Y, Jiang C. Association study on MTHFR polymorphisms and meningioma in northern China. Gene. 2013;516:291–293. doi: 10.1016/j.gene.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Zhou YW, Shi HP, Wang YZ, Li GL, Yu HT, Xie XY. 5, 10-Methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTRR), and methionine synthase reductase (MTR) gene polymorphisms and adult meningioma risk. J Neurooncol. 2013;115:233–239. doi: 10.1007/s11060-013-1218-z. [DOI] [PubMed] [Google Scholar]
- 47.Elexpuru-Camiruaga J, Buxton N, Kandula V, Dias PS, Campbell D, McIntosh J, Broome J, Jones P, Inskip A, Alldersea J. Susceptibility to astrocytoma and meningioma: influence of allelism at glutathione S-transferase (GSTT1 and GSTM1) and cytochrome P-450 (CYP2D6) loci. Cancer Res. 1995;55:4237–4239. [PubMed] [Google Scholar]
- 48.Pinarbasi H, Silig Y, Gurelik M. Genetic polymorphisms of GSTs and their association with primary brain tumor incidence. Cancer Genet Cytogenet. 2005;156:144–149. doi: 10.1016/j.cancergencyto.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 49.De Roos AJ, Rothman N, Inskip PD, Linet MS, Shapiro WR, Selker RG, Fine HA, Black PM, Pittman GS, Bell DA. Genetic polymorphisms in GSTM1, -P1, -T1, and CYP2E1 and the risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2003;12:14–22. [PubMed] [Google Scholar]
- 50.Schwartzbaum JA, Ahlbom A, Lönn S, Warholm M, Rannug A, Auvinen A, Christensen HC, Henriksson R, Johansen C, Lindholm C. An international case-control study of glutathione transferase and functionally related polymorphisms and risk of primary adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2007;16:559–565. doi: 10.1158/1055-9965.EPI-06-0918. [DOI] [PubMed] [Google Scholar]
- 51.Fryer AA, Zhao L, Alldersea J, Pearson W, Strange R. Use of site-directed mutagenesis of allele-specific PCR primers to identify the GSTM1 A, GSTM1 B, GSTM1 A, B and GSTM1 null polymorphisms at the glutathione S-transferase, GSTM1 locus. Biochem J. 1993;295:313–315. doi: 10.1042/bj2950313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ezer R, Alonso M, Pereira E, Kim M, Allen JC, Miller DC, Newcomb EW. Identification of glutathione S-transferase (GST) polymorphisms in brain tumors and association with susceptibility to pediatric astrocytomas. J Neurooncol. 2002;59:123–134. doi: 10.1023/a:1019601305210. [DOI] [PubMed] [Google Scholar]


