Abstract
Cytogenetic abnormalities have emerged as the major novel prognostic factors in multiple myeloma (MM) patients. This meta-analysis comprehensively investigates the association between the cytogenetic abnormalities and survival of MM patients. We searched the PubMed, EMBASE, SCOPUS, and Cochrane databases for articles published until February, 2014. Thirty eligible studies involving 10276 patients were included to examine the association of three chromosomal abnormalities, t (4; 14), del (17p), and Amp (1q21), with survival in MM patients. The main outcome measures were progression-free survival (PFS) and overall survival (OS). Individuals with t (4; 14), del (17p), and Amp (1q21) had low OS and PFS. In a subgroup analysis for therapy regimen, lenalidomide- and bortezomib-based therapies increased the PFS of patients with Amp (1q21) (HR=1.50, 95% CI=0.95-2.36, p=0.084) and t (4; 14) (HR=1.38, 95% CI=0.90-2.11, p=0.143). The presence of del (17p) elicited no significant influence on the prognosis of patients under different therapy regimens. Our meta-analysis provides globally quantifiable confirmation of the adverse prognostic value of t (4; 14), del (17p), and Amp (1q21) in OS and PFS for MM patients. Lenalidomide- and bortezomib-based therapies were partly conducive to improve the prognosis of individuals with t (4; 14). Bortezomib-based therapy can partly improve the PFS of patients with Amp (1q21).
Keywords: Cytogenetic aberrations, multiple myeloma, prognosis, meta-analysis, bortezomib, lenalidomide
Introduction
Multiple myeloma (MM), the second most common hematologic malignancy, is accounting for approximately 1% of all cancer diagnoses. MM is characterized by the expansion and accumulation of clonal plasma cells in the bone marrow (BM), the secretion of monoclonal immunoglobulins, and the presence of osteolytic bone lesions. Although treatment strategies have improved in the last decade, MM remains an incurable disease. Nevertheless, the myeloma patients’ survival time is highly different, ranging from a few months to more than 10 years. This diversity mainly relates to the prognosis of both the tumor and the host. International Staging System is the most widely applied prognostic system in myeloma, which could categorize patients into three groups based on the levels of serum albumin and b2-microglobulin [1]. Notably, these two variables could reflect both patient and tumor factors. While b2-microglobulin serves as a measure of tumor bulk and renal function. Albumin is related to the general state of the patient. This prognostic system is well validated and easily applied; however, it cannot completely explain the heterogeneity of survival time. Therefore, studies on new prognostic factors are necessary to determine the course of the disease, define therapeutic strategies, and predict long-term survival and outcome.
Cytogenetic abnormalities have emerged as the major novel prognostic factors in newly diagnosed MM patients [2-4]. However, previous studies related to the cytogenetic abnormalities is limited because of the small number of analyzable metaphases, the low proliferative activity of plasma cells and the limited extent of BM involvement. Approximately 30% abnormal karyotypes have reported in most large series [5,6]. However, other techniques without obtaining metaphases have obtained genomic aberrations in almost all cases [7-9]. This pitfall has led to the replacement of classical detecting techniques by interphase fluorescence in situ hybridization (FISH) technology, which has the advantage of detection of specific chromosomal changes even in non-cycling interphase cells. FISH technique in myeloma demonstrated a high incidence of chromosomal changes [9,10], suggesting that it can be applied to the assessment of single abnormality with prognostic value [2,11-14].
Some studies assessed the prognostic value of cytogenetic abnormalities detected by FISH in MM patients; however, the association between the genomic abnormalities and clinical outcome of MM patients remains controversial [15-18]. In addition, a few studies have evaluated the prognostic value of novel drugs for cytogenetic aberrations. This meta-analysis was performed to gain complete understanding of the association between the genomic abnormalities and survival outcome of MM patients.
Materials and methods
Search strategy
The PubMed, EMBASE, SCOPUS, and Cochrane databases were searched using the terms “Cytogenetic abnormalities”, or “genomic abnormalities”, or “chromosome abnormalities”, and “multiple myeloma”. Furthermore, we attempted to identify the potentially relevant studies by tracing the reference list of pertinent manuscripts as well as contacting known authors in the articles. No language restrictions were applied. The last search was performed on March 2014.
Study selection
Selected studies must meet the following inclusion criteria: (1) t (4; 14), del (17p), or Amp (1q21) as the exposure factor in MM patients; (2) observational studies only on human beings; (3) hazard ratio (HR) or survival curve for overall survival (OS) or progression-free survival (PFS) [or time to progression (TTP)] with an available or calculable confidence interval (CI) of 95%; and (4) full articles with English language. Accordingly, the following exclusion criteria were applied: (1) studies on the same population or subpopulation, (2) lack of control group, and (3) patients with monoclonal gammopathy of undetermined significance or asymptomatic MM and not MM.
Data extraction and methodological quality appraisal
Two reviewers independently assessed all articles identified by search strategies for relevance and reached a consensus on all items. The following information was obtained from each publication: author names, publication year, population region, country, age of patients, detection techniques, cytogenetic abnormalities, detectable sample size, number of exposed and unexposed cohort, previous therapies, and disease state. The HR and 95% CI were either directly determined from the articles or generated from published Kaplan-Meier curves with the software Engauge Digitizer version 4.1 (free software downloaded from http://sourceforge.net) [19].
Three categories of the Newcastle-Ottawa scale, including selection, comparability, and outcome which contain eight items, were used to assess the quality of selected studies, a maximum of one star can be given for each numbered item for the selection and outcome categories. A maximum of two stars can be given for comparability category [20]. Considering the lack of standard criteria, we defined 0 to 3 stars, 4 to 6 stars, and 7 to 9 stars as low, moderate, and high quality, respectively [21].
Statistical analysis
All statistical analysis was performed using the STATA 11.0 software package and conducted according to PRISMA guidelines [22]. The HR data with a 95% CI for OS and PFS (TTP) were pooled to evaluate the association of t (4; 14), del (17p), or Amp (1q21) with the survival outcome of MM patients. The inter-study heterogeneity was estimate by the Q and I2 statistical tests. I2>50% indicated heterogeneity [23]. If I2 was significant (>50%), the random-effects model was selected. Otherwise, the fixed-effects model was selected. The studies were categorized into therapy, disease state, and assessed quality for the subgroup analyses of the correlation between t (4; 14), del (17p), or Amp (1q21) and patient survival. The Funnel plots of Egger’s linear regression test and Begg’s test was used to assess publication bias. An asymmetrical plot and p<0.05 indicated a statistically significant publication bias.
Results
Characteristics of included studies
The characteristics of the selected trials are summarized in Table 1. The flowchart of selection is shown in Figure 1. The results were published between 2003 and 2013, and had sample sizes ranging from 43 to 1095 participants. Del (17p), Amp (1q21), and t (4; 14) were detected in 19, 16, and 20 of the selected studies, respectively. Twenty-two studies investigated participants with newly diagnosed MM, and eight studies investigated relapsed/refractory MM patients. Among these 30 studies, 5 investigated MM patients who received bortezomib-based therapy, 5 studied MM patients who received lenalidomide-based therapy, and 15 investigated MM patients who received conventional chemotherapy/ACST.
Table 1.
Main characteristics of these studies included in this meta-analysis
| First Author | Year | Country | Study quality | Therapy | Disease status | Sample size | Age (range) | Detection techniques | Cytogenetic abnormalities | Median follow-up months (range) |
|---|---|---|---|---|---|---|---|---|---|---|
| Avet-Loiseau | 2013 | France | 8 stars | CC | newly | 1095 | 72 (66-94) | FISH | t (4; 14), del (17p) | unknown |
| Grzasko | 2013 | Poland | 8 stars | CTD/MPT | newly | 104 | 59 (36-85) | FISH, cIg-FISH | +1q21 | 16.5 (1-53) |
| Avet-Loiseau | 2012 | France | 8 stars | VAD, ASCT | newly | 520 | <66 | FISH | t (4; 14), del (17p), +1q21 | 90.5 |
| Boyd | 2012 | UK | 7 stars | CVAD/CTD, ASCT or MP/CTDa | newly | 1960 | unknown | FISH | del (17p), +1q21 | 44.4 |
| Kiyota | 2012 | Japan | 8 stars | BD | RR | 43 | 63 | FISH G-banding, | t (4; 14), +1q21 | 17 |
| Neben | 2012 | Germany | 7 stars | VAD/PAD | newly | 344 | 57 (25-65) | FISH | t (4; 14), del (17p), +1q21 | 40.9 |
| Nemec | 2012 | Czech | 7 stars | VAD, ASCT | newly | 207 | 57 (33-69) | FISH G-banding, | t (4; 14) | 35.4 (0.4-70.3) |
| Stephens | 2012 | USA | 6 stars | Unknown | newly | 626 | unknown | FISH | del (17p), +1q21 | unknown |
| Chang | 2011 | Canada | 8 stars | Bortezomib | RR | 85 | 59 (32–78) | cIg-FISH | t (4; 14), del (17p), +1q21 | unknown |
| Hose | 2011 | Germany | 6 stars | VAD/TAD/PAD | newly | 554 | 57 (25-77) | FISH | del (17p) | unknown |
| Jiang | 2011 | Canada | 7 stars | VAD, ASCT | newly | 86 | 54 (30-70) | FISH, cIg-FISH | +1q21 | 36.5 |
| Kim | 2011 | Korea | 6 stars | ASCT/Chemotherapy | newly | 102 | 60.7 (35.2-79.8) | FISH, cIg-FISH | t (4; 14), +1q21 | 57.5 (44.8-86.5) |
| Klein | 2011 | Germany | 8 stars | RD | RR | 92 | 65 (29-80) | FISH | t (4; 14), del (17p), +1q21 | 12.1 |
| Shaughnessy | 2011 | USA | 7 stars | VTD | newly | 270 | <75 | FISH | del (17p) | unknown |
| Avet-Loiseau | 2010 | France | 8 stars | BD | newly | 507 | 57 (31-65) | FISH | t (4; 14), del (17p) | 24 |
| Avet-Loiseau | 2010 | France | 7 stars | RD | RR | 207 | 65 (37-89) | FISH | t (4; 14) | unknown |
| Chang | 2010 | Canada | 7 stars | RD | RR | 143 | 62.2 (31.8-80.0) | cIg-FISH | del (17p), +1q21 | 46.9 (5.76-148) |
| Chang | 2010 | Canada | 8 stars | VAD, ASCT | newly | 203 | 55 (31-73) | FISH, cIg-FISH | t (4; 14), del (17p), +1q21 | 36 |
| Dimopoulos | 2010 | Greece | 8 stars | RD/VRD | RR | 99 | unknown | FISH | t (4; 14), del (17p), +1q21 | 11 (0.4-36) |
| Neben | 2010 | Germany | 8 stars | VAD/TAD/PAD, ASCT | newly | 315 | 59 (25-73) | FISH | t (4; 14), del (17p) | unknown |
| Nemec | 2010 | Czech | 7 stars | HDT | newly | 91 | 58 (33-66) | cIg FISH | +1q21 | 39.2 |
| Reece | 2009 | Canada | 7 stars | RD | RR | 130 | 61 (31-84) | FISH | t (4; 14), del (17p) | 41.4 (1.9-139) |
| Schilling | 2008 | Germany | 7 stars | MF, allo-SCT | RR | 101 | 52 (28-68) | FISH | t (4; 14), del (17p) | 33 (3-73) |
| Avet-Loiseau | 2007 | France | 8 stars | VAD/MP/MPT/allo-SCT | newly | 1064 | <66 | FISH | t (4; 14), del (17p) | 41 |
| Gutie´rrez | 2007 | Spain | 7 stars | ASCT | newly | 260 | 60 (39-70) | FISH | t (4; 14) | 34 |
| Moreau | 2007 | France | 7 stars | HDT, ASCT | newly | 100 | 58 (33-65) | FISH | t (4; 14) | 46 |
| Shaughnessy | 2007 | USA | 7 stars | TD | newly | 220 | <75 | FISH | +1q21 | unknown |
| Fonseca | 2006 | USA | 8 stars | HDT | newly | 159 | unknown | FISH | +1q21 | unknown |
| Gertz | 2005 | USA | 7 stars | HDT | newly | 238 | 56 (30-71) | FISH, cIg-FISH | t (4; 14), del (17p) | unknown |
| Fonseca | 2003 | USA | 8 stars | CC | newly | 351 | 63 (35-84) | FISH | t (4; 14), del (17p) | (96-138) |
Newly: newly diagnosed; RR: relapsed/refractory; CC: conventional chemotherapy; VAD: vincristin, adriamycin, and dexamethasone; ASCT: autologous stem cell transplantation; BD: bortezomib plus dexamethasone; RD: Lenalidomide and bortezomib; VRD: lenalidomide, bortezomib and dexamethasone; MP: melphalan, pamidronate; MPT: melphalan, pamidronate plus thalidomide; CVAD: cyclophosphamide, vincristine, doxorubicin and dexamethasone; CTD: cyclophosphamide, thalidomide and dexamethasone; CTDa: attenuated cyclophosphamide, thalidomide and dexamethasone; VTD: Bortezomib, Thalidomide and dexamethasone; TD: Thalidomide and dexamethasone; HDT: high dose therapy; allo-SCT: allogeneic hematopoietic stem cell transplantation.
Figure 1.
Flowchart of selection of studies.
HR for OS
HR was pooled from 29 articles (Figure 2). The pooled HRs of all included studies revealed a statistically significant association between these three chromosome abnormalities and prognosis (Table 2). Del (17p) appeared to be the most significant adverse predictor of prognosis (HR=2.55, 95% CI=2.02-3.20) with the random-effects model. The fixed-effects model was used to estimate the HRs of Amp (1q21) (HR=1.68, 95% CI=1.51-1.88) and t (4; 14) (HR=2.32, 95% CI=2.08-2.59). In a stratified analysis by therapeutic schedule, a decreased risk of HR was found in t (4; 14). Bortezomib-based therapy decreased the risk of OS shortening (HR=1.85, 95% CI=1.28-2.67). An evident trend was found in lenalidomide-based therapy (HR=1.48, 95% CI=1.02-2.16). In a stratified analysis by disease status, newly diagnosed patients with t (4; 14) showed a worse long-term outcome (HR=2.49, 95% CI=2.21-2.80) than relapsed/refractory patients with t (4; 14) (HR=1.44, 95% CI=1.06-1.95). In addition, relapsed/refractory patients with Amp (1q21) had lower OS (HR=2.33, 95% CI=1.55-3.49) than newly diagnosed patients with Amp (1q21) (HR=1.64, 95% CI=1.46-1.84).
Figure 2.
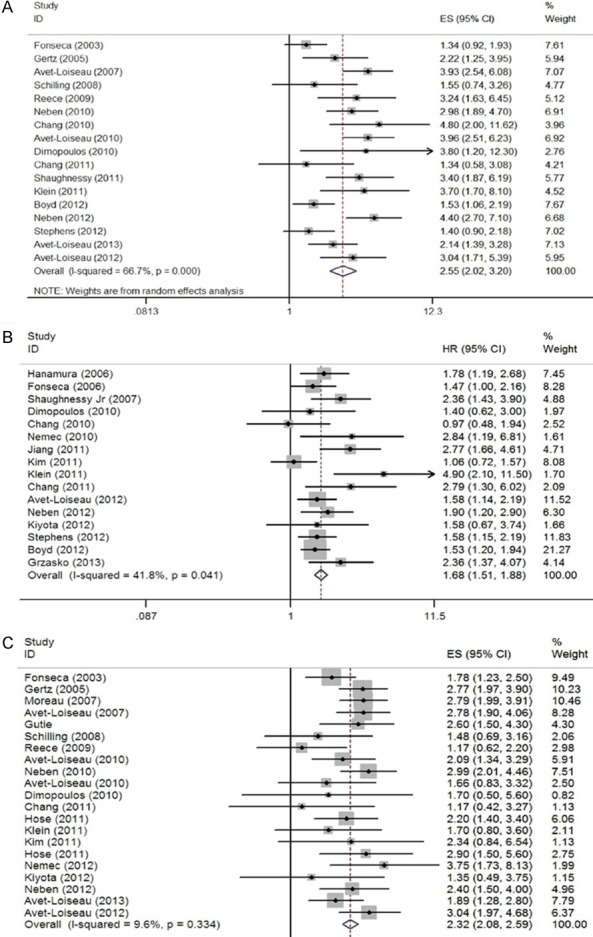
Forest plot of the pooled HRs for overall survival. A: del 17p; B: 1q21+; C: t (4; 14).
Table 2.
Meta-analysis of the cytogenetic abnormalities by FISH
| del 17p | +1q21 | t (4, 14) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| na | HR (95% CI) | P | I2 (%) | na | HR (95% CI) | P | I2 (%) | na | HR (95% CI) | P | I2 (%) | ||
| OS | newly + rel/ref | 17 | 2.55 (2.02-3.20)b | <0.001 | 66.7 | 16 | 1.68 (1.51-1.88) | <0.001 | 41.8 | 21 | 2.32 (2.08-2.59) | <0.001 | 9.6 |
| newly | 12 | 2.59 (1.97-3.39)b | <0.001 | 74.0 | 12 | 1.64 (1.46-1.84) | <0.001 | 37.5 | 14 | 2.49 (2.21-2.80) | <0.001 | 0.0 | |
| rel/ref | 5 | 2.42 (1.69-3.46) | <0.001 | 30.0 | 4 | 2.33 (1.55-3.49) | <0.001 | 45.8 | 7 | 1.44 (1.06-1.95) | 0.02 | 0.0 | |
| PFS | newly + rel/ref | 14 | 2.20 (1.94-2.50) | <0.001 | 43.7 | 13 | 1.50 (1.36-1.66) | <0.001 | 10.1 | 13 | 2.08 (1.82-2.38) | <0.001 | 12.8 |
| newly | 9 | 2.27 (1.85-2.79)b | <0.001 | 51.0 | 8 | 1.51 (1.35-1.68) | <0.001 | 34.8 | 7 | 2.27 (1.95-2.65) | <0.001 | 0.0 | |
| rel/ref | 5 | 2.41 (1.72-3.36) | <0.001 | 38.0 | 5 | 1.45 (1.12-1.87) | 0.004 | 0.0 | 6 | 1.62 (1.25-2.11) | <0.001 | 0.0 | |
| OS | bortezomib | 4 | 3.14 (2.28-4.32) | <0.001 | 41.9 | 2 | 2.17 (1.22-3.84) | 0.008 | 0.0 | 4 | 1.85 (1.28-2.67) | 0.001 | 0.0 |
| lenalidomide | 3 | 3.49 (2.18-5.60) | <0.001 | 0.0 | 2 | 2.59 (0.76-8.84)b | 0.002 | 77.7 | 3 | 1.48 (1.02-2.16) | 0.041 | 0.0 | |
| conventional | 11 | 2.33 (1.75-3.08)b | <0.001 | 70.0 | 12 | 1.64 (1.46-1.84) | <0.001 | 37.5 | 14 | 2.45 (2.17-2.77) | <0.001 | 13.5 | |
| PFS | bortezomib | 3 | 1.98 (1.34-2.91) | 0.001 | 13.0 | 2 | 1.50 (0.95-2.36) | 0.084 | 46.8 | 3 | 1.38 (0.90-2.11) | 0.143 | 0.0 |
| lenalidomide | 4 | 2.79 (1.91-4.08) | <0.001 | 22.2 | 3 | 1.43 (1.05-1.94) | 0.023 | 0.0 | 4 | 1.74 (1.29-2.35) | <0.001 | 0.0 | |
| conventional | 8 | 2.33 (1.80-3.02)b | <0.001 | 61.8 | 8 | 1.51 (1.35-1.68) | <0.001 | 34.8 | 7 | 2.26 (1.91-2.67) | <0.001 | 3.2 | |
Number of comparisons;
Random effects estimate;
newly: newly diagnosed; rel/ref: relapsed or refractory.
HR for PFS
With regard to PFS, the fixed-effect model was used for the meta-analysis of 22 studies (Table 2). The pooled effect sizes of the three abnormalities were in accordance with the OS [del (17p) HR=2.20, 95% CI=1.94-2.50; Amp (1q21) HR=1.50, 95% CI=1.36-1.66; t (4, 14) HR=2.04, 95% CI=1.77-2.35, Figure 3]. The subgroup analysis showed that bortezomib-based therapy can reverse the negative effects of t (4; 14) and Amp (1q21) abnormalities on PFS (HR=1.38, 95% CI=0.90 to 2.11 and HR=1.50, 95% CI=0.95-2.36, respectively). Meanwhile, lenalidomide-based therapy can reduce the risk of survival time shortening. Furthermore, newly diagnosed patients with t (4; 14) showed lower PFS (HR=2.27, 95% CI=1.95-2.65) than relapsed/refractory patients with t (4; 14) (HR=1.62, 95% CI=1.25-2.11). However, no significant difference in PFS was identified between newly diagnosed and relapsed/refractory patients with Amp (1q21).
Figure 3.
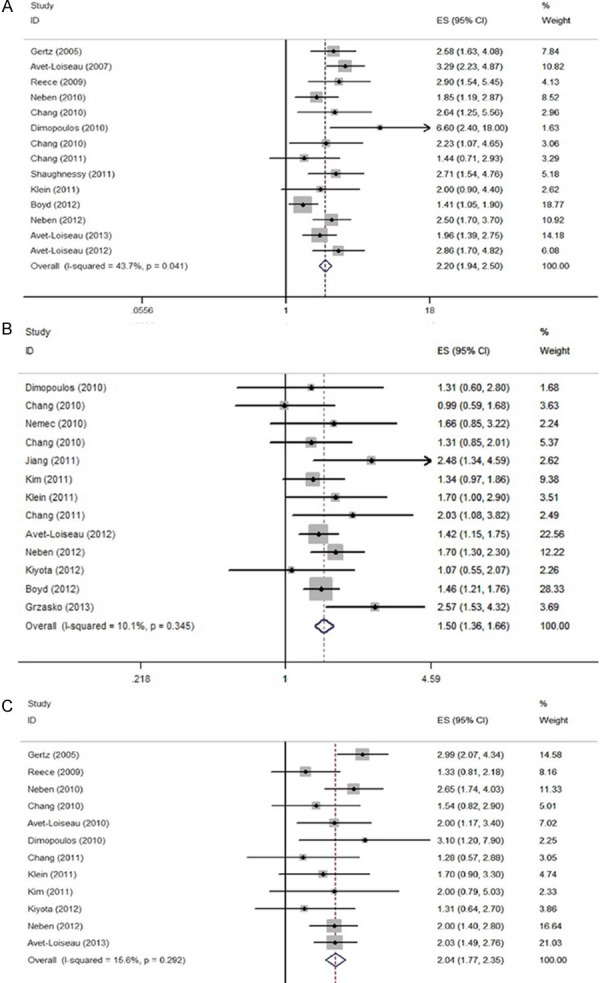
Forest plot of the pooled HRs for progression-free survival. A: del 17p; B: 1q21+; C: t (4; 14).
Evaluation of publication bias
The publication bias (Figure 4) was not statistically prominent among the studies focused on PFS [del (17p), p=0.093; Amp (1q21), p=0.502; t (4; 14), p=0.197] using Begg’s test. The funnel plot from Egger’s test also showed no asymmetry in PFS (p=0.099). In addition, the results for OS from Egger’s test [del (17p), p=0.188; Amp (1q21), p=0.077; t (4; 14), p=0.062) and Begg’s test (the same as Egger’s test) suggested an inconspicuous publication bias.
Figure 4.
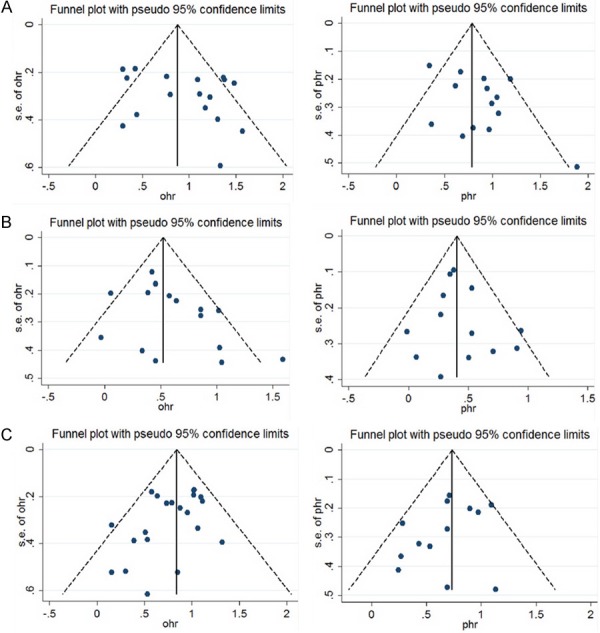
Funnel plots of studies for overall survival and progression-free survival. A: del 17p OS; B: 1q21+ OS; C: t (4; 14) OS; D: del 17p PFS; E: 1q21+ PFS; F: t (4; 14) PFS.
Discussion
This meta-analysis involved 30 independent studies. The association between three chromosome abnormalities and the prognosis of MM was determined. The three abnormalities t (4; 14), del (17p), and Amp (1q21) have been established as valuable predictors of poor outcome in MM patients treated with chemotherapy or stem cell transplantation [24-26]. We also found a strong evidence suggesting that bortezomib- and lenalidomide-based therapies can improve prognosis in patients with t (4; 14) through a subgroup analysis. The current meta-analysis is the first to report these three abnormalities.
FISH, an effective method in prognosis assessment and risk classification for MM [27], can detect specific aberrances in interphase cells and overcome the drawback of the lack of dividing cells required for conventional de-tection of cytogenetics [28]. Recently, FISH was applied to detect several genetic abnormalities, such as del (17p), Amp (1q21), and t (4; 14), which have been related to poor survival. Considering that these aberrations play an important role in MM development, we performed this meta-analysis to comprehensively estimate the significance of these three chromosomal abnormalities.
In MM, the most frequent structural changes are translocations involving the immunoglobulin heavy chain (IgH) switch region on chromosome 14q with various partner genes [29]. These genes include cyclin D1 in t (11; 14) (16%), FGFR3 in t (4; 14) (15%), and C-MAF in t (14; 16) (3% to 5%) [30,31]. Among these 14q32 translocations, t (4; 14) confers an adverse prognosis in patients treated with conventional chemotherapy or autologous stem cell transplant [2,24,32].
Our results indicate that t (4; 14) is significantly associated with poor long-term survival. In addition, this translocation serves a more significant influence in newly diagnosed patients than in relapsed/refractory patients. The translocation leads to the deregulation of the FGFR3 and MMSET genes, which are located at 4p16. FGFR3, a transmembrane receptor tyrosine kinase involved in regulating cell proliferation and differentiation, is overexpressed in many cancers [33]. In MM, FGFR3 translocation results in ectopic expression in plasma cells because it is strongly regulated by 3’IgH enhancers; this finding suggests that this protein exerts an oncogenic function in the pathogenesis of myeloma [30,34]. Another gene dysregulated by t (4; 14), is a multiple myeloma SET domain protein (MMSET) [27] and is an oncogene that contributes to cellular adhesion and clonogenic growth [28]. MMSET and FGFR3 are dysregulated in 100% and approximately 70% of all cases with t (4; 14), respectively [29].
Further stratified analysis suggests that bortezomib- and lenalidomide-based therapies can improve the prognosis of patients with this translocation. With regard to bortezomib-based therapy, the probable mechanism might be related to FGFR3. Anderson [35] et al. have recently demonstrated that MCL-1 downregulation contributes to MM cell apoptosis and confers bortezomib resistance. Moreover, previous studies have suggested that the increased levels of STAT3 and downstream MCL-1 in MM cells are transfected with wild- or mutant-type FGFR3 [36]. Thus, patients who express FGFR3 respond well with bortezomib-based therapy; this finding may be attributed to the enhanced MCL-1 expression caused by FGFR3. Dawson’s study [37] and our meta-analysis obtained consistent results on bortezomib-based therapy for MM. FGFR3 has been considered as a poor prognostic marker. However, Dawson et al. [37] found that patients who express FGFR3 respond equally well and have similar outcomes with bortezomib-based therapy relative to FGFR3-negative patients. This finding suggests that bortezomib-based therapy can overcome the resistance mediated by FGFR3 overexpression. Meanwhile, data concerning the ability of lenalidomide-based therapy to overcome the poor prognostic impact of t (4; 14) are limited. In 130 relapsed/refractory MM patients treated with lenalidomide plus dexamethasone (MM016 trial), those with del (13q) or t (4; 14) had a similar TTP and OS to patients without these abnormalities; this result suggests that lenalidomide-based therapy can partly reverse the prognosis of t (4; 14) [38].
Structural aberrations of chromosome 1 appear in 40% to 48% of all MM cases [6]. The gain of the 1q21 region appears in approximately one third of MM patients [39]. This locus has a pathogenetic function in disease progression; therefore, gains are associated with poor prognosis [40,41]. A previous study in Arkansas showed that patients with either a gain of the 1q21 chromosomal region or with overexpression of the CKS1B gene (located at 1q21) present a poor outcome in the ‘Total Therapy’ prgram [40]. Our meta-analysis obtained results consistent with that in Arkansas in terms of PFS (HR=2.17, p<0.001) and OS (HR=1.68, p<0.001). The probable mechnism might be related to the elevated expression of the cell cycle-related gene CKS1B and strongly correlated with Amp (1q21) [42-44]. CKS1B increases tumor aggressiveness by regulating the ubiquitination and subsequent breakdown of the cyclin-depedent kinase inhibitor p27Kip1, thereby favoring cell proliferation [45,46].
In the subgroup analysis, lenalidomide-based therapy did not show a better effect on prognosis than the conventional therapy. Interestingly, bortezomibbased therapy can partly increase the PFS of patients with Amp (1q21) (p=0.084). However, only a few studies have reported the relation between bortezomib-based therapy and Amp (1q21). A possible explanation for this finding might be attributed to the close relations between Amp (1q21) and high-risk cytogenetic abnormalities, such as t (4; 14). A previous study has determined through multivariate cox proportional hazard analysis that Amp (1q21) is not an independent prognostic marker in MM [39]. This finding suggests that Amp (1q21) is related to other abnormalities. Gains of 1q are frequently observed at late stages of MM. Thus, the locus has a pathogenetic function in disease progression, and gains are associated with poor prognosis [47]. Our results also showed that relapsed/refractory patients with Amp (1q21) had lower survival times than newly diagnosed patients. This result suggests that Amp (1q21) is associated with both disease progression and poor prognosis.
Meanwhile, del (17p) appears in approximately 10% of all MM cases [2,14,24]. This deletion generally involves a major part of the short arm of chromosome 17, leading to the loss of several genes, including the p53 gene at 17p13. The present results indicate that del (17p) is significantly associated with poor OS and PFS for MM and thus serves as an adverse prognostic factor. Furthermore, this deletion is a strong predictive factor for PFS (HR=2.20, p<0.001) and OS (HR=2.55, p<0.001). The poor prognosis might be associated with a loss or disrupted function of the p53 gene, which is common in several human neoplasms, including MM [48]. In addition, p53 deletions are common in MM patients with central nervous system involvement [49] and in patients with plasma cell leukemia who have significantly poor clinical outcomes [50]. Moreover, neither high-dose therapy nor allogeneic transplantation can overcome the extremely poor prognosis of patients with TP53 deletions or mutations [14,51]. In further stratified analysis, no prominent effect on individual prognosis was found in lenalidomide- and bortezomib-based therapies, suggesting that they may not have functions in the cases with del (17p).
The limitations of this meta-analysis should be investigated. First, it is difficult to eliminate heterogeneity by probing into every aspect of confounding factors, such as age, gender, tumor stage, and other genetic aberrations resulting from rare individual patient data, while the meta-regression and subgroup analysis were used to diminish heterogeneity across the articles. Second, the number of published studies was not sufficient, especially for the analyses of the subgroups divided based on the therapeutic regimen. Third, most of the selected studies were from Europe and North America, and only three studies included Asian and South American individuals. Therefore, our results may be applicable only to Europeans and North Americans. Last, the HR was extracted according to an internationally acknowledged methodology. Although this methodology cannot extract data from all studies and cannot accurately measure the absolute HR, it may not cause a significant impact on our study because of our relatively consistent results.
Heterogeneity and publication bias may influence the results of the meta-analysis. In our meta-analysis, no statistically significant heterogeneity existed in the overall or subgroup comparisons. In addition, a significant publication bias from the three chromosome abnormalities was not detected, suggesting the reliability of our results.
This meta-analysis provided a globally quantifiable evaluation of the increased hazards on OS and PFS for MM patients with t (4; 14), del (17p), and del (13q). Interestingly, our results determined that bortezomib-based therapies can improve the prognosis of patients with t (4; 14). Thus, these abnormalities should be considered when determining a patient’s disease stage and therapy. Further research should focus on the gene-specific prediction for survival and gene-targeted treatments.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81372540). The Scientific Research Starting Foundation for Returned Overseas Chellonese Scholars, Ministry of Education (2012); Jiangsu Province’s Medical Elite Program (RC201148); A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions; “333” project of Jiangsu Province (BRA2011217).
Disclosure of conflict of interest
None.
References
- 1.Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, Boccadoro M, Child JA, Avet-Loiseau H, Kyle RA, Lahuerta JJ, Ludwig H, Morgan G, Powles R, Shimizu K, Shustik C, Sonneveld P, Tosi P, Turesson I, Westin J. International staging system for multiple myeloma. J. Clin. Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, Dewald GW, Van Ness B, Van Wier SA, Henderson KJ, Bailey RJ, Greipp PR. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 3.Moreau P, Facon T, Leleu X, Morineau N, Huyghe P, Harousseau JL, Bataille R, Avet-Loiseau H Intergroupe Francophone du Myélome. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002;100:1579–1583. doi: 10.1182/blood-2002-03-0749. [DOI] [PubMed] [Google Scholar]
- 4.Shaughnessy J Jr, Tian E, Sawyer J, McCoy J, Tricot G, Jacobson J, Anaissie E, Zangari M, Fassas A, Muwalla F, Morris C, Barlogie B. Prognostic impact of cytogenetic and interphase fluorescence in situ hybridization-defined chromosome 13 deletion in multiple myeloma: early results of total therapy II. Br J Haematol. 2003;120:44–52. doi: 10.1046/j.1365-2141.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- 5.Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66:380–390. [PubMed] [Google Scholar]
- 6.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 7.Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Singhal S, Mehta J, Siegel D, Shaughnessy J, Barlogie B. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood. 1998;92:4269–4278. [PubMed] [Google Scholar]
- 8.Barlogie B, Alexanian R, Dixon D, Smith L, Smallwood L, Delasalle K. Prognostic implications of tumor cell DNA and RNA content in multiple myeloma. Blood. 1985;66:338–341. [PubMed] [Google Scholar]
- 9.Drach J, Schuster J, Nowotny H, Angerler J, Rosenthal F, Fiegl M. Multiple myeloma: high incidence of chromosomal aneuploidy as detected by interphase fluorescence in situ hybridization. Cancer Res. 1995;55:3854–3859. [PubMed] [Google Scholar]
- 10.Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, Minvielle S, Bataille R Intergroupe Francophone du Myélome. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99:2185–2191. doi: 10.1182/blood.v99.6.2185. [DOI] [PubMed] [Google Scholar]
- 11.Zojer N, Konigsberg R, Ackermann J, Fritz E, Dallinger S, Kromer E, Kaufmann H, Riedl L, Gisslinger H, Schreiber S, Heinz R, Ludwig H, Huber H, Drach J. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000;95:1925–1930. [PubMed] [Google Scholar]
- 12.Facon T, Avet-Loiseau H, Guillerm G, Moreau P, Genevieve F, Zandecki M, Lai JL, Leleu X, Jouet JP, Bauters F, Harousseau JL, Bataille R, Mary JY Intergroupe Francophone du Myélome. Chromosome 13 abnormalities identified by FISH analysis and serum beta2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood. 2001;97:1566–1571. doi: 10.1182/blood.v97.6.1566. [DOI] [PubMed] [Google Scholar]
- 13.Fonseca R, Harrington D, Oken MM, Dewald GW, Bailey RJ, Van Wier SA, Henderson KJ, Blood EA, Rajkumar SV, Kay NE, Van Ness B, Greipp PR. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: an eastern cooperative oncology group study. Cancer Res. 2002;62:715–720. [PubMed] [Google Scholar]
- 14.Chang H, Qi C, Yi QL, Reece D, Stewart AK. p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005;105:358–360. doi: 10.1182/blood-2004-04-1363. [DOI] [PubMed] [Google Scholar]
- 15.Chang H, Trieu Y, Qi X, Jiang NN, Xu W, Reece D. Impact of cytogenetics in patients with relapsed or refractory multiple myeloma treated with bortezomib: Adverse effect of 1q21 gains. Leuk Res. 2011;35:95–98. doi: 10.1016/j.leukres.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Dimopoulos MA, Kastritis E, Christoulas D, Migkou M, Gavriatopoulou M, Gkotzamanidou M, Iakovaki M, Matsouka C, Mparmparoussi D, Roussou M, Efstathiou E, Terpos E. Treatment of patients with relapsed/refractory multiple myeloma with lenalidomide and dexamethasone with or without bortezomib: prospective evaluation of the impact of cytogenetic abnormalities and of previous therapies. Leukemia. 2010;24:1769–1778. doi: 10.1038/leu.2010.175. [DOI] [PubMed] [Google Scholar]
- 17.Chang H, Qi X, Jiang A, Xu W, Young T, Reece D. 1p21 deletions are strongly associated with 1q21 gains and are an independent adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Bone Marrow Transplant. 2010;45:117–121. doi: 10.1038/bmt.2009.107. [DOI] [PubMed] [Google Scholar]
- 18.Avet-Loiseau H, Hulin C, Campion L, Rodon P, Marit G, Attal M, Royer B, Dib M, Voillat L, Bouscary D, Caillot D, Wetterwald M, Pegourie B, Lepeu G, Corront B, Karlin L, Stoppa AM, Fuzibet JG, Delbrel X, Guilhot F, Kolb B, Decaux O, Lamy T, Garderet L, Allangba O, Lifermann F, Anglaret B, Moreau P, Harousseau JL, Facon T. Chromosomal abnormalities are major prognostic factors in elderly patients with multiple myeloma: the intergroupe francophone du myélome experience. J. Clin. Oncol. 2013;31:2806–2809. doi: 10.1200/JCO.2012.46.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, P T. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. Available from: URL: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm.
- 21.Ouyang J, Gou X, Ma Y, Huang Q, Jiang T. Prognostic value of 1p deletion for multiple myeloma: a meta-analysis. Int J Lab Hematol. 2014;36:555–65. doi: 10.1111/ijlh.12189. [DOI] [PubMed] [Google Scholar]
- 22.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62:e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Cochran WG. The comparison of percentages in matched samples. Biometrika. 1950;37:256–266. [PubMed] [Google Scholar]
- 24.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, Leyvraz S, Michallet M, Yakoub-Agha I, Garderet L, Marit G, Michaux L, Voillat L, Renaud M, Grosbois B, Guillerm G, Benboubker L, Monconduit M, Thieblemont C, Casassus P, Caillot D, Stoppa AM, Sotto JJ, Wetterwald M, Dumontet C, Fuzibet JG, Azais I, Dorvaux V, Zandecki M, Bataille R, Minvielle S, Harousseau JL, Facon T, Mathiot C. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 25.Drach J, Ackermann J, Fritz E, Kromer E, Schuster R, Gisslinger H, DeSantis M, Zojer N, Fiegl M, Roka S, Schuster J, Heinz R, Ludwig H, Huber H. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92:802–809. [PubMed] [Google Scholar]
- 26.Klein U, Jauch A, Hielscher T, Hillengass J, Raab MS, Seckinger A, Hose D, Ho AD, Goldschmidt H, Neben K. Chromosomal aberrations +1q21 and del (17p13) predict survival in patients with recurrent multiple myeloma treated with lenalidomide and dexamethasone. Cancer. 2011;117:2136–2144. doi: 10.1002/cncr.25775. [DOI] [PubMed] [Google Scholar]
- 27.Avet-Loiseau H. Role of genetics in prognostication in myeloma. Best Pract Res Clin Haematol. 2007;20:625–635. doi: 10.1016/j.beha.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avet-Loiseau H, Brigaudeau C, Morineau N, Talmant P, Lai JL, Daviet A, Li JY, Praloran V, Rapp MJ, Harousseau JL, Facon T, Bataille R. High incidence of cryptic translocations involving the Ig heavy chain gene in multiple myeloma, as shown by fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999;24:9–15. doi: 10.1002/(sici)1098-2264(199901)24:1<9::aid-gcc2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 30.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J. Clin. Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Avet-Loiseau H, Malard F, Campion L, Magrangeas F, Sebban C, Lioure B, Decaux O, Lamy T, Legros L, Fuzibet JG, Michallet M, Corront B, Lenain P, Hulin C, Mathiot C, Attal M, Facon T, Harousseau JL, Minvielle S, Moreau P Intergroupe Francophone du Myélome. Translocation t (14; 16) and multiple myeloma: is it really an independent prognostic factor? Blood. 2011;117:2009–2011. doi: 10.1182/blood-2010-07-295105. [DOI] [PubMed] [Google Scholar]
- 32.Chang H, Sloan S, Li D, Zhuang L, Yi QL, Chen CI, Reece D, Chun K, Stewart AK. The t (4; 14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. British Journal of Haematology. 2004;125:64–68. doi: 10.1111/j.1365-2141.2004.04867.x. [DOI] [PubMed] [Google Scholar]
- 33.Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NC. Targeting FGFR3 in multiple myeloma: inhibition of t (4; 14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–966. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 34.Ishikawa H, Tsuyama N, Liu S, Abroun S, Li FJ, Otsuyama K, Zheng X, Ma Z, Maki Y, Iqbal MS, Obata M, Kawano MM. Accelerated proliferation of myeloma cells by interleukin-6 cooperating with fibroblast growth factor receptor 3-mediated signals. Oncogene. 2005;24:6328–6332. doi: 10.1038/sj.onc.1208782. [DOI] [PubMed] [Google Scholar]
- 35.Podar K, Gouill SL, Zhang J, Opferman JT, Zorn E, Tai YT, Hideshima T, Amiot M, Chauhan D, Harousseau JL, Anderson KC. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27:721–731. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 36.Guan M, Zhu L, Somlo G, Hughes A, Zhou B, Yen Y. Bortezomib Therapeutic Effect is Associated with Expression of FGFR3 in Multiple Myeloma Cells. Anticancer Res. 2009;29:1–10. [PubMed] [Google Scholar]
- 37.Dawson MA, Opat SS, Taouk Y, Donovan M, Zammit M, Monaghan K, Horvath N, Roberts AW, Prince HM, Hertzberg M, McLean CA, Spencer A. Clinical and immunohistochemical features associated with a response to bortezomib in patients with multiple myeloma. Clin Cancer Res. 2009;15:714–722. doi: 10.1158/1078-0432.CCR-08-1022. [DOI] [PubMed] [Google Scholar]
- 38.Reece D, Song KW, Fu T, Roland B, Chang H, Horsman DE, Mansoor A, Chen C, Masih-Khan E, Trieu Y, Bruyere H, Stewart DA, Bahlis NJ. Influence of cytogenetics in patients with relapsed or refractory multiple myeloma treated with lenalidomide plus dexamethasone: adverse effect of deletion 17p13. Blood. 2009;114:522–525. doi: 10.1182/blood-2008-12-193458. [DOI] [PubMed] [Google Scholar]
- 39.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, Bergsagel PL, Rajkumar SV, Greipp PR, Litzow MR, Price-Troska T, Henderson KJ, Ahmann GJ, Gertz MA. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–2040. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 40.Hanamura I. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grzasko N, Hus M, Pluta A, Jurczyszyn A, Walter-Croneck A, Morawska M, Chocholska S, Hajek R, Dmoszynska A. Additional genetic abnormalities significantly worsen poor prognosis associated with 1q21 amplification in multiple myeloma patients. Hematol Oncol. 2013;31:41–48. doi: 10.1002/hon.2018. [DOI] [PubMed] [Google Scholar]
- 42.Shaughnessy J. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27 Kip1 and an aggressive clinical course in multiple myeloma. Hematology. 2005;10:117–126. doi: 10.1080/10245330512331390140. [DOI] [PubMed] [Google Scholar]
- 43.Shaughnessy JD Jr, Barlogie B. Using genomics to identify high-risk myeloma after autologous stem cell transplantation. Biol Blood Marrow Transplant. 2006;12:77–80. doi: 10.1016/j.bbmt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Pagano M, Benmaamar R. When protein destruction runs amok, malignancy is on the loose. Cancer Cell. 2003;4:251–256. doi: 10.1016/s1535-6108(03)00243-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhan F, Colla S, Wu X, Chen B, Stewart JP, Kuehl WM, Barlogie B, Shaughnessy JD Jr. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood. 2007;109:4995–5001. doi: 10.1182/blood-2006-07-038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H, Jiang N, Jiang H, Saha MN, Qi C, Xu W, Reece D. CKS1B nuclear expression is inversely correlated with p27Kip1 expression and is predictive of an adverse survival in patients with multiple myeloma. Haematologica. 2010;95:1542–1547. doi: 10.3324/haematol.2010.022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, Hollmig K, Zangarri M, Pineda-Roman M, van Rhee F, Cavallo F, Burington B, Crowley J, Tricot G, Barlogie B, Shaughnessy JD Jr. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carson DA, Lois A. Cancer progression and p53. Lancet. 1995;346:1009–1011. doi: 10.1016/s0140-6736(95)91693-8. [DOI] [PubMed] [Google Scholar]
- 49.Chang H, Sloan S, Li D, Keith Stewart A. Multiple myeloma involving central nervous system: high frequency of chromosome 17p13.1 (p53) deletions. Br J Haematol. 2004;127:280–284. doi: 10.1111/j.1365-2141.2004.05199.x. [DOI] [PubMed] [Google Scholar]
- 50.Chang H, Sloan S, Li D, Patterson B. Genomic aberrations in plasma cell leukemia shown by interphase fluorescence in situ hybridization. Cancer Genet Cytogenet. 2005;156:150–153. doi: 10.1016/j.cancergencyto.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Schilling G, Hansen T, Shimoni A, Zabelina T, Perez-Simon JA, Gutierrez NC, Bethge W, Liebisch P, Schwerdtfeger R, Bornhauser M, Otterstetter S, Penas EM, Dierlamm J, Ayuk F, Atanackovic D, Bacher U, Bokemeyer C, Zander A, San Miguel J, Nagler A, Kroger N. Impact of genetic abnormalities on survival after allogeneic hematopoietic stem cell transplantation in multiple myeloma. Leukemia. 2008;22:1250–1255. doi: 10.1038/leu.2008.88. [DOI] [PubMed] [Google Scholar]


