Abstract
Alzheimer’s disease (AD) is characterized by cortical atrophy and disrupted anatomical connectivity, and leads to abnormal interactions between neural systems. Diffusion weighted imaging (DWI) and graph theory can be used to evaluate major brain networks, and detect signs of a breakdown in network connectivity. In a longitudinal study using both DWI and standard MRI, we assessed baseline white matter connectivity patterns in 30 subjects with mild cognitive impairment (MCI; mean age: 71.8+/−7.5 yrs; 18M/12F) from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Using both standard MRI-based cortical parcellations and whole-brain tractography, we computed baseline connectivity maps from which we calculated global “small-world” architecture measures, including mean clustering coefficient (MCC) and characteristic path length (CPL). We evaluated whether these baseline network measures predicted future volumetric brain atrophy in MCI subjects, who are at risk for developing AD, as determined by 3D Jacobian “expansion factor maps” between baseline and 6-month follow-up anatomical scans. This study suggests that DWI-based network measures may be a novel predictor of AD progression.
Keywords: Graph theory, brain networks, white matter, DTI, tractography, ADNI, TBM, small worldness, connectivity
1. Introduction
Alzheimer’s disease (AD), the most common form of dementia, is characterized by memory loss in its early stages, typically followed by a progressive decline in other cognitive domains. People with mild cognitive impairment (MCI) - a transitional stage between normal aging and AD - convert to AD at a rate of about 10–15% per year (Petersen et al., 2001; Bruscoli and Lovestone, 2004). The Alzheimer's Disease Neuroimaging Initiative (ADNI) is one of several major efforts worldwide to identify sensitive biomarkers that may help track or predict brain tissue loss due to AD progression.
AD is marked by pervasive gray matter atrophy, but the brain’s white matter (WM) pathways also progressively decline (Braak and Braak, 1996; Bartzokis, 2011; Braskie et al., 2011; Hua et al., 2013). Recent models of AD suggest that cognitive deficits arise from the progressive disconnection of cortical and subcortical regions, promoted by neuronal loss and white matter injury (Delbeuck et al., 2003; Pievani et al., 2011). Many MRI-based image analysis methods have been used to track structural atrophy of the brain, but diffusion tensor imaging (DTI) is sensitive to microscopic WM injury not always detectable with standard anatomical MRI. DTI may be used to track the highly anisotropic diffusion of water along axons, revealing microstructural WM fiber bundles connecting cortical and subcortical regions and allowing for characterization of the brain’s WM structural network (Hagmann et al., 2008).
Graph theory network topology measures have been used increasingly to analyze brain networks and characterize network organization. “Small-world” network properties have been regarded as typical properties of many kinds of communication networks, and are found in social networks, efficient biological networks, and even in healthy mammalian brain networks (Hilgetag et al., 2000; Achard and Bullmore, 2007; Reijneveld et al., 2007; Iturria-Medina et al., 2008). Networks with a small-world organization can have both functional segregation and specialization of modules and a “low wiring cost’ that supports easy communication across an entire network. Small-world networks are marked by low characteristic path length (CPL) and high mean clustering coefficient (MCC), so they are both integrated and segregated. Studies using various modalities, including cortical thickness analyses, fMRI, and EEG, suggest that AD patients have abnormal small-world architecture in their large-scale structural and functional brain networks, with differences in MCC and CPL that may imply less optimal network topology (Stam et al., 2007; He et al., 2008; Sanz-Arigita et al., 2010; Brown et al., 2011; Toga and Thompson, 2013).
In this study, we assessed 30 ADNI participants showing signs of mild cognitive impairment (MCI). MCI subjects are the target of many clinical trials that aim to slow disease progression, before brain changes are so pervasive that they are irremediable. However, predictors of decline in MCI are sorely needed, as mildly impaired subjects do not usually exhibit drastic changes in most standard biomarkers of AD. Here, we combined DTI with longitudinally acquired standard anatomical MRI (across a 6-month interval) to measure the microstructure and connectivity of white matter tracts, and assess whether variations in the degree and extent of connections might predict future brain decline. We created 68×68 structural connectivity matrices, or graphs, that describe the strength of connections between any pair of brain regions based on baseline structural cortical parcellations and whole-brain tractography. In these graphs, nodes designate brain regions, which are thought of as being connected by edges representing WM fibers. We then used graph theory to describe general properties of the anatomical networks and to characterize connectivity patterns.
Given the recent interest in “small world’ phenomena as a characteristic of biological networks, we examined whether global “small-world architecture’ network measures, MCC and CPL, calculated from baseline connectivity maps were able to predict future volumetric brain atrophy (dynamic tissue loss) over a 6-month follow-up period, as determined by 3D Jacobian “expansion factor maps” of T1-weighted structural scans. That is, we tested whether the intactness of the brain’s anatomical network predicted ongoing brain decline in the future, assessed using the more accepted anatomical MRI methods. In follow-up analyses, we additionally assessed whether several baseline local nodal measures (efficiency, clustering and betweenness) were associated with volumetric brain atrophy. We found that global and nodal network measures may offer a potentially useful biomarker for predicting longitudinal atrophy, at this critical time before the onset of AD.
2. Methods
2.1 Subject information and image acquisition
Data collection for the ADNI2 project (the second phase of ADNI) is still in progress. Here we performed an initial analysis of 30 MCI subjects who had returned for a follow-up evaluation at 6-months (mean age at baseline: 71.8 +/− 7.5 yrs; 18 men / 12 women). We note that in ADNI2 MCI participants include the enrollment of a new early MCI (e-MCI) cohort, with milder episodic memory impairment than the MCI group of ADNI1, now called late MCI (l-MCI) in ADNI2 (Table 1). We additionally analyzed baseline data from 29 cognitively healthy control subjects (CTL) to create a study-specific brain template (mean age at baseline: 73.4+/−5.2 yrs; 15 men/14 women). Detailed inclusion and exclusion criteria are found in the ADNI2 protocol (http://adni-info.org/Scientists/Pdfs/ADNI2_Protocol_FINAL_20100917.pdf).
Table 1.
Demographics and clinical scores for the participants
| e-MCI (n=21) |
l-MCI (n=9) |
p-value for group difference | |
|---|---|---|---|
| e-MCI vs l-MCI | |||
| Age | 71.6 +/− 8.1 | 72.1 +/− 6.6 | 0.87 |
| Sex | 11 M / 10 F | 7 M / 2 F | -- |
| Education (yrs) | 15.8 +/− 2.7 | 16.2 +/− 3.1 | 0.73 |
| MMSE | 27.9 +/− 1.8 | 27.6 +/− 1.7 | 0.63 |
All subjects underwent whole-brain MRI scanning on 3-Tesla GE Medical Systems scanners, on at least one of two occasions (baseline and 6 months). T1-weighted IR-FSPGR (spoiled gradient echo) sequences (256×256 matrix; voxel size = 1.2×1.0×1.0 mm3; TI=400 ms; TR = 6.98 ms; TE = 2.85 ms; flip angle = 11°), were collected as well as diffusion-weighted images (DWI; 35 cm field of view, 128×128 acquired matrix, reconstructed to a 256×256 matrix; voxel size: 2.7×2.7×2.7 mm3; scan time = 9 min; more imaging details may be found at http://adni.loni.usc.edu/wp-content/uploads/2010/05/ADNI2_GE_3T_22.0_T2.pdf). 46 separate images were acquired for each DTI scan: 5 T2-weighted images with no dedicated diffusion sensitization (b0 images) and 41 diffusion-weighted images (b=1000 s/mm2). The DTI protocol for ADNI was chosen after a detailed evaluation of different protocols that could be performed in a reasonably amount time; we reported results of these comparisons previously (Jahanshad et al., 2010; Zhan et al., 2012a). All T1-weighted MR and DWI images were checked visually for quality assurance to exclude scans with excessive motion and/or artifacts after preprocessing corrections; all scans were included.
2.2 Image preprocessing
2.2.1 Preprocessing of baseline and 6-month follow-up anatomical scans
All extra-cerebral tissue was removed from both baseline and 6 month T1-weighted anatomical scans using a number of software packages, primarily ROBEX, a robust automated brain extraction program trained on manually “skull-stripped” MRI data (Iglesias et al., 2011) and FreeSurfer (Fischl et al., 2004). Skull-stripped volumes were visually inspected, and the best one selected and sometimes further manually edited. Anatomical scans subsequently underwent intensity inhomogeneity normalization using the MNI nu_correct tool (www.bic.mni.mcgill.ca/software/). To align data from different subjects into the same 3D coordinate space, each anatomical image was linearly aligned to a standard brain template (the Colin27; Holmes et al., 1998) using FSL flirt (Jenkinson et al., 2002).
2.2.2 Baseline DWI preprocessing
For each subject, all raw DWI volumes were aligned to the average b0 image using the FSL eddy-correct tool (www.fmrib.ox.ac.uk/fsl) to correct for head motion and eddy current distortions. Non-brain tissue was removed from the diffusion-weighted images using the Brain Extraction Tool (BET) from FSL (Smith, 2002). To correct for echo-planar induced (EPI) susceptibility artifacts, which can cause distortions at tissue-fluid interfaces, skull-stripped b0 images were linearly aligned and then elastically registered to their respective baseline T1-weighted structural scans using an inverse consistent registration algorithm with a mutual information cost function (Leow et al., 2007). The resulting linear registration matrices and 3D deformation fields were then applied to the remaining 41 DWI volumes. FA maps were subsequently calculated using FSL dtifit and overlaid on T1 anatomical scans to ensure proper alignment.
2.3 Fiber tractography
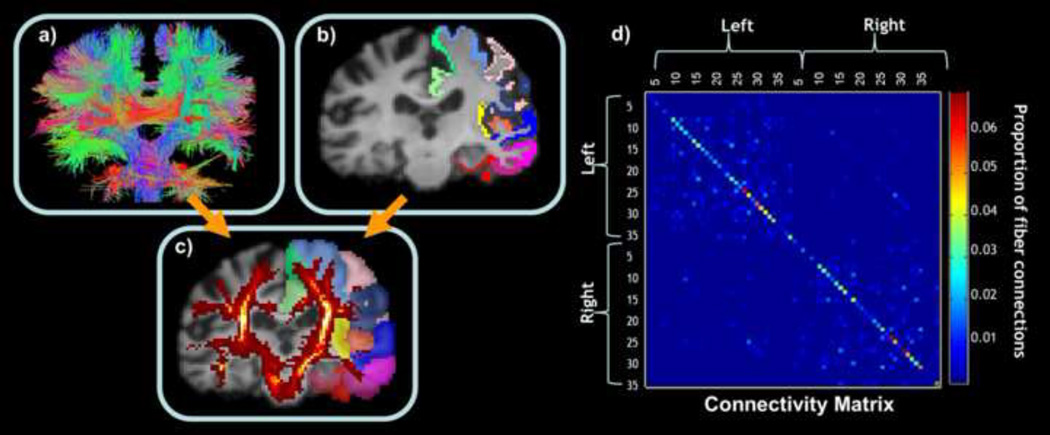
At each voxel, orientation distribution functions (ODFs) were computed using the normalized and dimensionless ODF estimator, derived for Q-ball imaging (QBI) as in (Aganj et al., 2010). The angular resolution of the ADNI data is somewhat limited to avoid long scan times that may tend to increase patient attrition, but the use of an ODF model makes best use of the available angular resolution. Tractography was performed on the linearly aligned sets of DWI volumes by probabilistically seeding voxels with a prior probability based on the FA value. Curves through a seed point receive a score estimating the probability of the existence, computed from the ODFs. We used a voting process provided by the Hough transform to determine the best fitting curves through each point (Figure 1a; Aganj et al., 2011). Elastic deformations obtained from the EPI distortion correction, mapping the average b0 image to the T1-weighted image, were then applied to the resulting tracts’ 3D coordinates. Each subject’s dataset contained approximately 10,000 non-duplicated fibers (3D curves). In prior work, we have determined that this is a sufficient number of fibers to determine most of the common network topology measures accurately (Prasad et al., 2013a). We removed any erroneous fibers traced on the edge of the brain due to high intensity noise. To limit small noisy tracts, we filtered out fibers with less than 10 points.
Figure 1.
(a) EPI-corrected whole-brain tractography calculated from the DWI. (b) Anatomical cortical parcellations in one hemisphere are shown, registered to the same subject’s DWI space. (c) Red fiber density map, where each voxel represents the total number of streamlines that pass through it, overlaid on the dilated labels. (d) Connectivity matrix, in which each colored element represents the proportion of detected fibers connecting each of the colored labels in each hemisphere to each of the other colored labels in (c) – computed as a proportion of the total number of extracted fibers in the brain. This general method was used by us in (Jahanshad et al., 2012a, 2013), to which the reader is referred for further details.
2.4 Automated cortical segmentation
Using FreeSurfer (http://surfer.nmr.mgh.harvard.edu/; Fischl et al., 2004), 34 cortical labels from the Desikan-Killiany atlas (Table 2; Desikan et al., 2006) were automatically extracted in each hemisphere from the raw baseline T1-weighted structural MRI scans. The resulting T1-weighted images and were then aligned to the corrected T1 images, and the linear transformation matrix was applied to the cortical parcellations using nearest neighbor interpolation (to avoid intermixing of labels). This placed the cortical labels in the same space as the tractography, calculated from the DWIs that were elastically registered to the corrected T1 space (Figure 1b). To ensure tracts would intersect cortical labeled boundaries, labels were dilated with an isotropic box kernel of 5×5×5 voxels (Figure 1c; Jahanshad et al., 2011a). Proper alignment of each subject’s cortical parcellations, T1-weighted image, and tractography was verified by visually inspecting the 3 overlaid images.
Table 2.
Index of cortical labels extracted from the anatomical MRI scans by FreeSurfer (Fischl et al., 2004)
| 1 | Banks of the superior temporal sulcus | 19 | Pars orbitalis |
| 2 | Caudal anterior cingulate | 20 | Pars triangularis |
| 3 | Caudal middle frontal | 21 | Peri-calcarine |
| 4 | -N/A- | 22 | Postcentral |
| 5 | Cuneus | 23 | Posterior cingulate |
| 6 | Entorhinal | 24 | Precentral |
| 7 | Fusiform | 25 | Precuneus |
| 8 | Inferior parietal | 26 | Rostral anterior cingulate |
| 9 | Inferior temporal | 27 | Rostral middle frontal |
| 10 | Isthmus of the cingulate | 28 | Superior frontal |
| 11 | Lateral occipital | 29 | Superior parietal |
| 12 | Lateral orbitofrontal | 30 | Superior temporal |
| 13 | Lingual | 31 | Supra-marginal |
| 14 | Medial orbitofrontal | 32 | Frontal pole |
| 15 | Middle temporal | 33 | Temporal pole |
| 16 | Parahippocampal | 34 | Transverse temporal |
| 17 | Paracentral | 35 | Insula |
| 18 | Pars opercularis |
2.5 N×N matrices representing structural connectivity
As in (Jahanshad et al., 2011a), for each subject, a baseline 68×68 (34 right hemisphere labels and 34 left) connectivity matrix was created. Each element described the estimated proportion of the total number of fibers, in that subject, connecting each of the labels to each of the other labels (Figure 1d).
2.6 Graph theory network analyses
We applied the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/) to our weighted baseline connectivity matrices generated above, to compute the measures whose values contribute to small world architecture. In weighted measures, a path between two neighbors with strong connections contributes more than a path between two weakly connected neighbors. Characteristic path length (CPL) is an average measure (across the whole network) of the minimum number of edges necessary to travel from one node to another in the network (i.e., average minimum path length; Watts and Strogatz, 1998). Mean clustering coefficient (MCC) is an average measure (across the whole network) of how many neighbors of a given node are also connected to each other, relative to the total possible number of connections in the network (Onella et al., 2005). Small-worldness, which measures the balance between network differentiation and network integration, is a ratio of the MCC and CPL of a network. As the small worldness measure may falsely report small world topology in highly segregated, but poorly integrated networks (Rubinov and Sporns, 2010), we chose to assess MCC and CPL as joint predictors instead.
In a post hoc analysis, we additionally evaluated several weighted nodal measures to assess the extent to which local connectivity measures can also drive prediction: nodal clustering coefficient (CC), which parallels nodal efficiency (EFF), and nodal betweenness centrality (BTW). The CC measures how many neighbors of a given node are also connected to each other, relative to the total possible connections, while the EFF of a node is the average inverse shortest path length calculated on the neighborhood of a given node. BTW is the fraction of all shortest paths in the network that contain a given node. Nodes with high values of betweenness centrality participate in a large number of shortest paths. The equations to calculate each of these measures can be found in (Rubinov and Sporns, 2010).
2.7 Study specific template creation
A study-specific minimal deformation template (MDT; Gutman et al., 2010) was created using 29 cognitively healthy elderly control (CTL) subjects’ baseline spatially-aligned corrected anatomical volumes. Using a customized template from subjects in the study (rather than a standard atlas or a single optimally chosen subject) can reduce bias in the registrations. The MDT is the template that deviates least from the anatomy of the subjects, and, in some circumstances, it can improve statistical power (Lepore et al., 2007). The MDT was generated by creating an initial affine mean template from all 29 subjects, then registering all the aligned individual scans to that mean using a fluid registration (Leow et al., 2007) while regularizing the Jacobians (Yanovsky et al., 2007). A new mean was created from the registered scans; this process was iterated several times.
2.8 Tensor based morphometry
To quantify 3D patterns of volumetric brain atrophy in MCI, each subject’s 6 month preprocessed T1-weighted scan was elastically registered to its respective corrected baseline T1-weighted scan (Leow et al., 2007). A separate 3D Jacobian map (i.e., volumetric expansion factor map) was created for each subject to characterize the local volume differences between their baseline scan and 6 month scan. To ensure the Jacobians had common anatomical coordinates for statistical analysis, each subject’s respective 3D deformation field - from the elastic registration of the baseline T1-weighted scan to the MDT - was applied to each Jacobian map.
2.9 Statistics
We ran voxel-wise multiple linear regressions, covarying for sex and age, and a partial F test, using baseline MCC and CPL as predictors – both jointly and independently – of the longitudinal volumetric changes. Computing thousands of association tests at a voxel-wise level can introduce a high false positive error rate in neuroimaging studies, if not corrected. To correct for these errors, we used the searchlight method for false discovery rate correction (sFDR; Langers et al., 2007). All statistical maps are thresholded at a corrected p-value to show regression coefficients only in regions that controlled the false discovery rate (q=0.05).
In post hoc analyses, we further ran voxel-wise linear regressions, covarying for sex and age, to detect any associations between baseline CC/EFF and BTW in each of the 68 nodes and the Jacobian maps. To correct for multiple comparisons for each of 68 nodes, we used the sFDR correction at q=0.05/68 or q=0.00074 (Langers et al., 2007).
3. Results
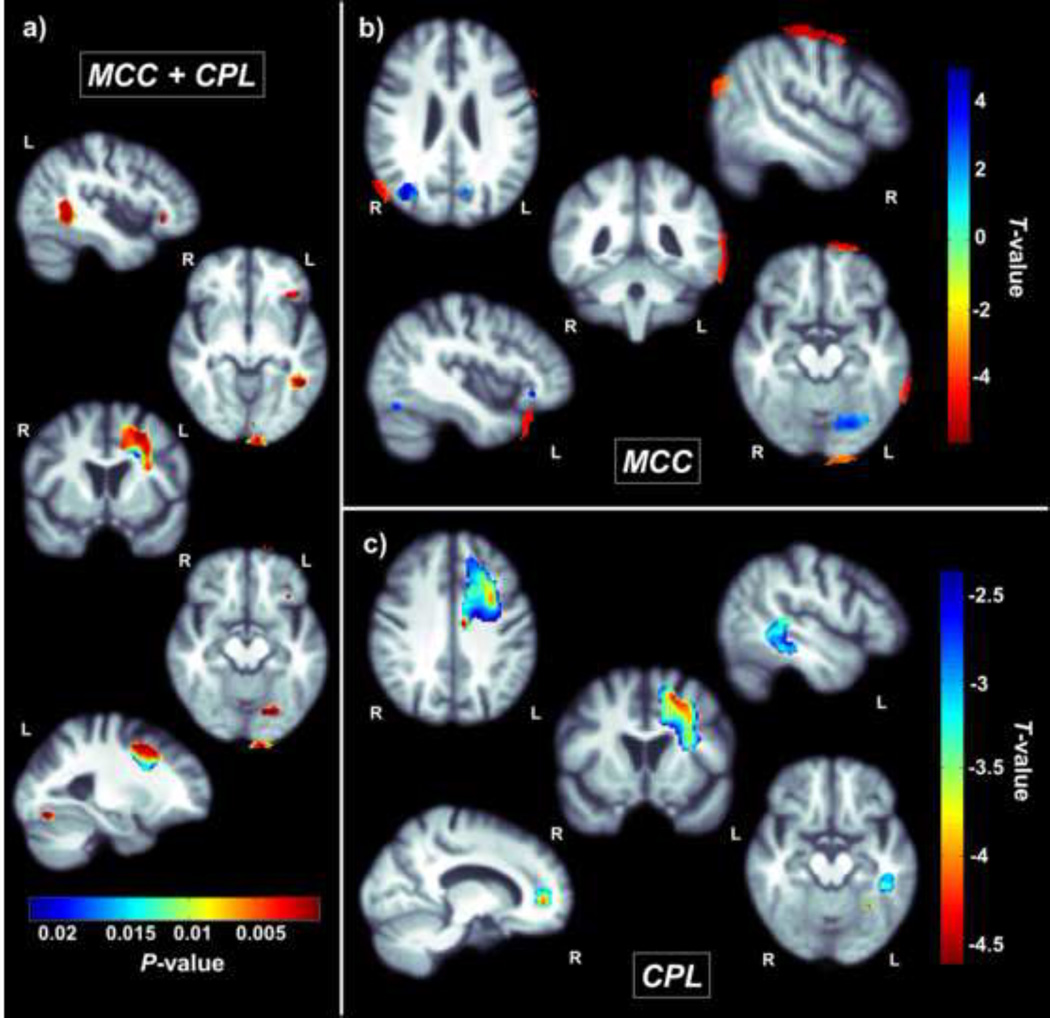
We found a significant association between the baseline global network measures, CPL and MCC, used together as predictors in the same regression model, and 3D volumetric changes over the 6-month follow-up interval (Figure 2a; corrected p<0.05; Langers et al., 2007). Separately, MCC was significantly negatively associated with CSF volume changes surrounding the frontal, parietal, temporal and occipital lobes and positively associated with regional volumetric changes around the right angular gyrus, left posterior orbital gyrus, left precuneus and left fusiform (Figure 2b). CPL was negatively associated with regional volume changes in the right and left anterior corona radiata and left superior corona radiata, as well as the left fusiform and temporal lobe (Figure 2c). This suggests that lower MCC and increased CPL at baseline are associated with decreases in tissue volume and increases in CSF expansion (implying tissue loss) in these regions after 6 months.
Figure 2.
(a) These p-maps show regions where CPL and MCC are joint predictors of volumetric changes on standard anatomical MRI between baseline and a 6-month follow-up scan (corrected p<0.05; Langers et al., 2007) (b) These maps show T-values within regions where only MCC has a significant correlation with volumetric changes (corrected p<0.05; Langers et al., 2007). (c) These maps show T-values within regions where only CPL has a significant negative correlation with volumetric changes (corrected p<0.05; Langers et al., 2007). Lower MCC and higher CPL at baseline are associated with greater volumetric atrophy after 6 months.
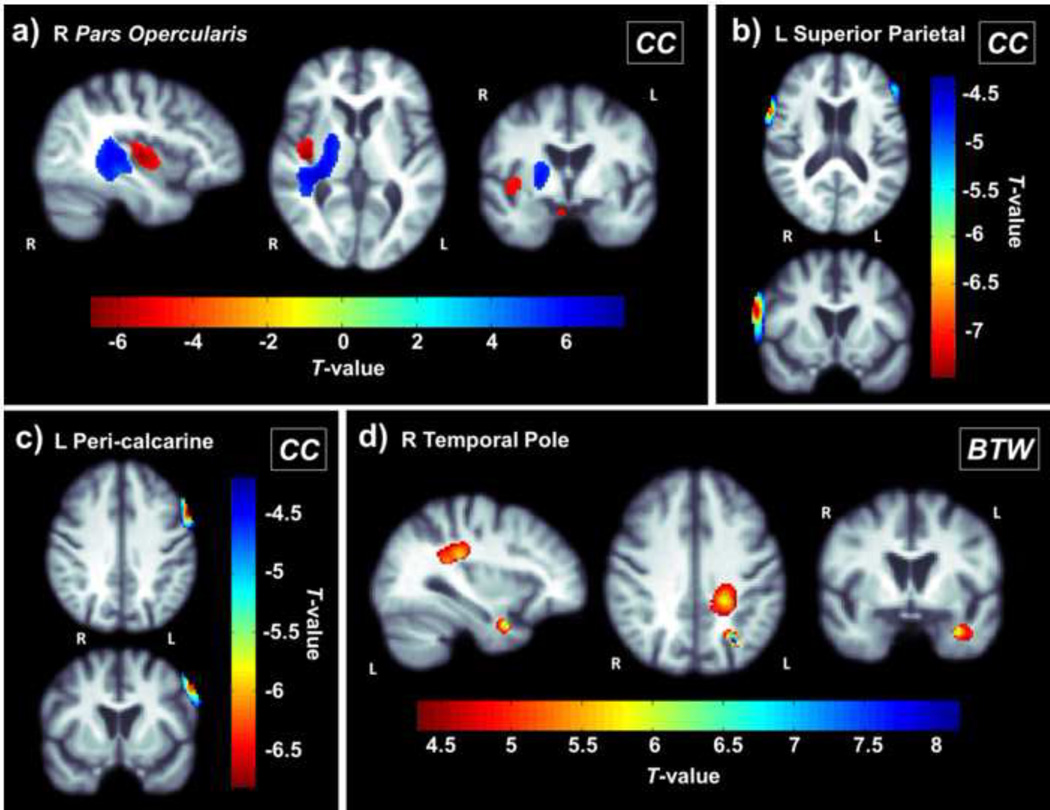
In a post hoc analysis, we also found the right pars opercularis (inferior frontal gyrus) node’s local EFF and CC are significantly positively associated with right internal capsule and temporal lobe and negatively associated with the right insular sulcus/lateral fissure and chiasmatic cistern (Figure 3a; corrected p<0.00074; Langers et al., 2007). The left superior parietal node EFF and CC are significantly negatively associated with CSF volume around the left and right frontal lobe extending towards the right temporal lobe (Figure 3b; corrected p<0.00074; Langers et al., 2007). CC of the left peri-calcarine node was negatively associated with CSF volume around the left frontal lobe (Figure 3c; corrected p<0.00074; Langers et al., 2007). Finally, the right temporal pole BTW was positively associated with the volume of the left temporal lobe, angular gyrus, and posterior corona radiata (Figure 3d; corrected p<0.00074; Langers et al., 2007). Overall, these measures suggest that decreased local CC/EFF and BTW at baseline are associated with atrophy between baseline and a follow-up scan 6 months later.
Figure 3.
Nodal clustering coefficient (CC) in the (a) right pars opercularis (inferior frontal gyrus) (b) left superior parietal node and (c) left peri-calcarine node are significantly associated with 3D patterns of volumetric brain atrophy, implying that increased clustering in these regions is associated with greater future atrophy. These same patterns are associated with efficiency in these nodes, a measure that parallels CC. (d) Right temporal pole betweenness (BTW) is positively associated with volume. These maps show T-values within regions that show a significant association (corrected p<0.00074; Langers et al., 2007).
4. Discussion
There is great interest in predicting which subjects with MCI are likely to decline, as well as in understanding what patterns of organizational decline in the brain may be harbingers of brain tissue loss. Rather than evaluating gross anatomical structures of the brain independently, brain connectivity analyses can evaluate how integrated each region is with others, and thus may be more sensitive to alterations in brain systems as a whole. Several recent studies have suggested that AD progression may involve a loss of small world characteristics in the brain’s structural and functional networks (Stam et al., 2007; He et al., 2008; Sanz-Arigita et al., 2010). This is consistent with theoretical notions that small-world topology may be functionally beneficial and efficient. In this study, we assessed whether abnormalities in small worldness, the balance between network segregation and network integration, at baseline were predictive of volumetric brain decline over a 6-month period.
We found an association between baseline small-world global network measures and volumetric changes in T1-weighted structural scans. Moreover, we found that lower mean clustering (MCC) and higher characteristic path length (CPL) at baseline are associated with greater atrophy. Networks with lower CPL, a measure reflecting speed or ease of functional integration of distributed brain regions, and higher levels of clustering or dense connections within regions across the network (MCC), may indicate a more functionally coherent neural system (Bullmore and Sporns, 2009).
To further investigate which regions or nodes may be driving global MCC associations we assessed nodal clustering (CC), which parallels local nodal efficiency, and found that decreased clustering in the right pars opercularis, left superior parietal node and left peri-calcarine node are significantly associated with patterns of volumetric brain atrophy. To assess which nodes might help facilitate lower global CPL, we evaluated nodal betweenness centrality (BTW), which measures whether a node participates in a large number of shortest paths, facilitating integration between anatomically unconnected regions. We found that lower BTW in the right temporal pole was associated with atrophy.
These regions have been implicated in other DTI network studies. The superior parietal cortex, for example, is known to be affected by AD pathology early on (Jacobs et al., 2012), and is one of few “rich club’ hubs, the set of most highly interconnected nodes, that play a central role in global network integration (van den Heuvel et al., 2012). A DTI study by Lo et al. (2010) also revealed nodal efficiency reductions in several prefrontal areas including the orbital part of the inferior frontal gyrus, and the temporal pole. In a connectivity study involving grey matter volume correlations, the temporal pole, fusiform, cingulate, superior parietal region, and orbital frontal gyrus showed significant changes in the interregional correlations between the normal control and AD groups (Yao et al., 2010).
This study could be extended in several ways. There is a great deal of work in brain connectivity analyses trying to identify subnetworks that are more sensitive to picking up differences in disease. Rather than pick a fixed partition of the cortical surface, other work has attempted to adaptively refine and alter the cortical partition to better sensitize the analysis to group differences in disease (Prasad et al., 2013b). Although such adaptive approaches are elegant, they have the limitation that the cortical connectivity matrices from different studies and cohorts would be quite difficult to compare, as they are not defining connectivity for the same regions of interest. A second line of work has argued that connectivity can be defined in different ways, some of which may be better sensitized to pick up disease-related differences. For instance, some have defined lattice networks where every voxel is considered connected to all its immediate neighbors, and the angular diffusion signal at each voxel is used to define a dense weighted network that is amenable to connectivity analysis (Prasad et al., 2013c; Li et al., 2013). Other approaches use statistical methods to pre-select fibers likely to show associations with disease (Jahanshad et al., 2012b). A third line of work has attempted to threshold the connectivity networks to focus on nodes that have very high connectivity to others, or that might be important hubs or highly connected “centers’ for the network as a whole. This leads to concepts such as network filtrations, k-cores, and rich club coefficients (Dennis et al., 2013), which have begun to be tested for DTI based analysis of connectivity in disease (Daianu et al., 2013a,b,c). When the ADNI2 dataset is much larger, it should be possible to compare many of these methods head-to-head.
How the raw data have been acquired and processed prior to any statistical analysis can have large effects on results as each step is susceptible to sources of error and bias (Jones & Cercignani, 2010). For example, connectivity studies comparing networks derived from 3T and 7T scans have revealed differences between field strengths (Zhan et al., 2012b). Additional limitations may include the limited angular resolution of the ADNI dataset, selected to avoid long scan times that may increase patient attrition. However, the use of an ODF model makes best use of the available angular resolution. The standard single-tensor model is somewhat limited in regions with extensive fiber crossing and mixing, while the ODF model can better resolve multi-fiber trajectories.
TBM voxel-wise analyses assume that a specific voxel location in the brain is identical across all subjects. However, registration accuracy from one subject to another may vary, in particular in aging studies where structures atrophy. Similarly, while tracts were corrected for susceptibility-induced artifacts, remaining distortion could cause misalignment and can lead to spurious results (Jahanshad et al., 2011b).
Different parcellation schemes may also affect graph theory metrics. We used the FreeSurfer Desikan–Killiany atlas (Desikan et al., 2006) for cortical parcellation, which has been widely used for structural connectivity analysis (Honey et al., 2009; Hagmann et al., 2010; Daianu et al., 2013 a,b,c). However, other parcellations are possible and there is still work being done to understand how different parcellation templates and resolutions may influence different kinds of network measures (Hagmann et al., 2010; Zalesky et al., 2010; Bassett et al., 2011, Prasad et al., 2013b).
It appears that the degree of integration and efficiency both across distributed brain regions- CPL and BTW- and locally within regions- MCC, CC- is an important indication of a coherent neural system at baseline, and may be predictive of future decline. These results are preliminary and need to be replicated as ADNI2 progresses and new subjects are scanned. As the longitudinal study progresses, we can later investigate which of these subjects eventually develops AD, and if these early aberrations in connectivity can help to predict a patient’s conversion to AD, future brain tissue loss, and cognitive decline. This study offers evidence that DTI-based network measures may be a novel predictor of AD progression.
Acknowledgments
Algorithm development and image analysis for this study was funded, in part, by grants to PT from the NIBIB (R01 EB008281, R01 EB008432) and by the NIA, NIBIB, NIMH, the National Library of Medicine, and the National Center for Research Resources (AG016570, AG040060, EB01651, MH097268, LM05639, RR019771 to PT). Data collection and sharing for this project was funded by ADNI (NIH Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This research was also supported by NIH grants P30 AG010129 and K01 AG030514 from the National Institute of General Medical Sciences.
Footnotes
Many investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but most of them did not participate in analysis or writing of this report. A complete list of ADNI investigators may be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Disclosure statement
The authors have no potential financial or personal conflicts of interest including relationships with other people or organizations within three years of beginning the work submitted that could inappropriately influence this work. One of the authors, Michael Weiner, receives private funding unrelated to the content of this paper.
References
- Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 2007;3(2):e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aganj I, Lenglet C, Sapiro G, Yacoub E, Ugurbil K, Harel N. Reconstruction of the orientation distribution function in single- and multiple-shell q-ball imaging within constant solid angle. Magn. Reson. Med. 2010;64(2):554–466. doi: 10.1002/mrm.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aganj I, Lenglet C, Jahanshad N, Yacoub E, Thompson PM, Sapiro G. A Hough transform global probabilistic approach to multiple-subject diffusion MRI tractography. Med. Image Anal. 2011;15(4):414–425. doi: 10.1016/j.media.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G. Alzheimer's disease as homeostatic responses to age-related myelin breakdown. Neurobiol. Aging. 2011;32(8):1341–1371. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. NeuroImage. 2011;54(2):1262–1279. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Development of Alzheimer-related neurofibrillary changes in the neocortex inversely recapitulates cortical myelogenesis. Acta. Neuropathol. 1996;92(2):197–201. doi: 10.1007/s004010050508. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Common Alzheimer's disease risk variant within the CLU gene affects white matter microstructure in young adults. J. Neurosci. 2011;31(18):6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KM, Small JW, Bookheimer SY. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proc. Natl. Acad. Sci. USA. 2011;108(51):20760–20765. doi: 10.1073/pnas.1109038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruscoli M, Lovestone S. Is MCI really just early dementia? A systematic review of conversion studies. Int Psychogeriatr. 2004;16(2):129–140. doi: 10.1017/s1041610204000092. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Daianu M, Jahanshad N, Nir TM, Toga AW, Jack CR, Weiner MW, Thompson PM for ADNI. Breakdown of brain connectivity between normal aging and Alzheimer’s disease: a structural k-core network analysis. Brain Connectivity. 2013a;3(4):407–422. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daianu M, Dennis EL, Nir TM, Jahanshad NAW, Jack CR, Weiner MW, Thompson PM for ADNI. Alzheimer’s disease disrupts rich club organization in brain connectivity networks. Int. Symp. Biomed. 2013b doi: 10.1109/ISBI.2013.6556463. Imaging, Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daianu M, Dennis EL, Nir TM, Jahanshad NAW, Jack CR, Weiner MW, Thompson PM for ADNI. Disrupted brain connectivity in Alzheimer’s disease: effects of network thresholding. 2013c Submitted to MICCAI 2013. [Google Scholar]
- Delbeuck X, Van der Linden M, Collette F. Alzheimer’s disease as a disconnection syndrome? Neuropsychol. Rev. 2003;13:79–92. doi: 10.1023/a:1023832305702. [DOI] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, Toga AW, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Wright MJ, Thompson PM. Development of the “rich club” in brain networks from 438 adolescents and adults aged 12 to 30. Int. Symp. Biomed. 2013 doi: 10.1109/ISBI.2013.6556552. Imaging, Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Gutman B, Svarer C, Leow AD, Yanovsky I, Toga AW, Thompson PM. Creating unbiased minimal deformation templates for brain volume registration. OHBM. 2010;2010 [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the Structural Core of Human Cerebral Cortex. PLoS Biol. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of largescale cortical networks in Alzheimer’s disease. J. Neurosci. 2008;28(18):4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, Meuli R, Thiran J-P, Grant PE. White matter maturation reshapes structural connectivity in the late developing human brain. Proc. Natl. Acad. Sci. USA. 2010;107(44):19067–19072. doi: 10.1073/pnas.1009073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of largescale cortical networks in Alzheimer’s disease. J. Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, Burns GA, O'Neill MA, Scannell JW, Young MP. Anatomical connectivity defines the organization of clusters of cortical areas in the macaque and the cat. Phil. Trans. R. Soc. Lond. 2000;355(1393):91–110. doi: 10.1098/rstb.2000.0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Eva AC. Enhancement of MR images using registration for signal averaging. J. Comput. Assist. Tomogr. 1998;22(2):324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. USA. 2009;106(6):2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, Leow AD, Toga AD, Jack AD, Harvey D, Weiner MW, Thompson PM. Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer's disease clinical trials. Neuroimage. 2013;66:648–661. doi: 10.1016/j.neuroimage.2012.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging. 2011;30(9):1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodríguez EJ, Alemán-Gómez Y, Melie-García L. Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory. Neuroimage. 2008;40:1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Jacobs HI, van Boxtel MP, Jolles J, Verhey FR, Uylings HB. Parietal cortex matters in Alzheimer's disease: an overview of structural, functional and metabolic findings. Neurosci. Biobehav. Rev. 2012;36(1):297–309. doi: 10.1016/j.neubiorev.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Jahanshad N, Zhan L, Bernstein MA, Borowski B, Jack CR, Toga AW, Thompson PM. Diffusion tensor imaging in seven minutes: Determining trade-offs between spatial and directional resolution. Proc. IEEE Int. Symp. Biomed. Imaging. 2010;2010:1161–1164. [Google Scholar]
- Jahanshad N, Aganj I, Lenglet C, Joshi A, Jin Y, Barysheva M, McMahon KL, Zubicaray GI, Martin NG, Wright MJ, Toga AW, Sapiro G, Thompson PM. Sex differences in the Human Connectome: 4-Tesla high angular resolution diffusion tensor imaging (HARDI) tractography in 234 young adult twins. Proc. IEEE Int. Symp. Biomed. Imaging. 2011a;2011:939–943. [Google Scholar]
- Jahanshad N, Aganj I, Lenglet C, Sapiro G, Toga AW, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. 4-Tesla high angular resolution diffusion tractography analysis of the human connectome in 234 subjects: Sex differences and EPI distortion effects. ISMRM. 2011b;2011 [Google Scholar]
- Jahanshad N, Valcour VG, Nir TM, Kohannim O, Busovaca E, Nicolas K, Thompson PM. Disrupted brain networks in the aging HIV+ population. Brain Connectivity. 2012a;2(6):335–444. doi: 10.1089/brain.2012.0105-Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Nir TM, Jack CR, Weiner MW, Toga AW, Thompson PM. Boosting power to associate brain connectivity measures and dementia severity using Seemingly Unrelated Regression. MICCAI NIBAD. 2012b;2012 [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR, Saykin AJ, Green RC, Weiner MW, Medland SE, Montgomery GW, Hansell NK, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM ADNI. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady J, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jones DK, Cercignani M. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR in Biomedicine. 2010;23(7):803–820. doi: 10.1002/nbm.1543. [DOI] [PubMed] [Google Scholar]
- Langers DR, Jansen JF, Backes WH. Enhanced signal detection in neuroimaging by means of regional control of the global false discovery rate. NeuroImage. 2007;38:43–56. doi: 10.1016/j.neuroimage.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsk I, Chiang MC, Lee AD, Klunder A, Lu A, Becker J, Davis S, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans. on Medical Imaging. 2007;26(6):822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Lepore N, Brun CC, Pennec X, Chou YY, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Mean template for tensor-based morphometry using deformation tensors. Med. Image Comput. Comput. Assist. Interv. 2007;10(2):826–833. doi: 10.1007/978-3-540-75759-7_100. [DOI] [PubMed] [Google Scholar]
- Li J, Jin Y, Shi Y, Dinov ID, Wang JJ, Toga AW, Thompson PM. Voxelwise spectral diffusional connectivity and its applications to Alzheimer’s disease and intelligence prediction. 2013 doi: 10.1007/978-3-642-40811-3_82. Submitted to MICCAI 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Spulber G, Lehtimäki KK, Könönen M, Hallikainen I, Gröhn H, Kivipelto M, Hallikainen M, Vanninen R, Soininen H. Diffusion tensor imaging and tract-based spatial statistics in Alzheimer's disease and mild cognitive impairment. Neurobiol. Aging. 2011;32(9):1558–1571. doi: 10.1016/j.neurobiolaging.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. J. Neurosci. 2010;30:16876–16885. doi: 10.1523/JNEUROSCI.4136-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D, DeToledo-Morrell L, Urresta F, Gabrieli JD, Moseley M, Fleischman D, Bennett DA, Leurgans S, Turner DA, Stebbins GT. White matter changes in mild cognitive impairment and AD: A diffusion tensor imaging study. Neurobiol. Aging. 2006;27(5):663–672. doi: 10.1016/j.neurobiolaging.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Onnela JP, Saramaki J, Kertesz J, Kaski K. Intensity and coherence of motifs in weighted complex networks. Phys. Rev., E Stat. Nonlinear Soft Matter Phys. 2005;71:065103. doi: 10.1103/PhysRevE.71.065103. [DOI] [PubMed] [Google Scholar]
- Pievani M, de Haan W, Wu T, Seeley WW, Frisoni GB. Functional network disruption in the degenerative dementias. Lancet Neurol. 2011;10(9):829–843. doi: 10.1016/S1474-4422(11)70158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch. Neurol. 2001;58(12):1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Prasad G, Nir TM, Toga AW, Thompson PM ADNI. Tractography density and network measures in Alzheimer's disease. Int. Symp. Biomed. 2013a doi: 10.1109/ISBI.2013.6556569. Imaging, Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, Joshi S, Toga AW, Thompson PM. A dynamical clustering model of brain connectivity inspired by the n-body problem. 2013b doi: 10.1007/978-3-319-02126-3_13. Submitted to MICCAI 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, Joshi S, Nir TM, Toga AW, Thompson PM. Optimizing nodes in brain connectivity analyses using Markov Chain Monte Carlo (MCMC) methods for Alzheimer’s disease classification. 2013c Submitted to MICCAI 2013. [Google Scholar]
- Reijneveld JC, Ponten SC, Berendse HW, Stam CJ. The application of graph theoretical analysis to complex networks in the brain. Clin. Neurophysiol. 2007;118:2317–2331. doi: 10.1016/j.clinph.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage. 2010;3:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SARB, Maris E, Barkhof F, Scheltens P, Stam CJ. Loss of 'small-world' networks in Alzheimer's disease: graph analysis of FMRI resting-state functional connectivity. PLoS One. 2010;5(11):e13788. doi: 10.1371/journal.pone.0013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltems PH. Small-world networks and functional connectivity in Alzheimer's disease. Cereb. Cortex. 2007;17(1):92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Stebbins GT, Murphy CM. Diffusion tensor imaging in Alzheimer's disease and mild cognitive impairment. Behav. Neurol. 2009;21(1):39–49. doi: 10.3233/BEN-2009-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Connectomics sheds new light on Alzheimer’s disease, Biological Psychiatry. 2013;73(5):390–392. doi: 10.1016/j.biopsych.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS, Goni J, Sporns O. High-cost, high-capacity backbone for global brain communication. Proc. Natl. Acad. Sci. USA. 2012;109:11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Yanovsky I, Thompson PM, Osher S, Leow AD. Topology preserving log-unbiased nonlinear image registration: theory and implementation; IEEE Conference on Computer Vision and Pattern Recognition; 2007. pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Zhang Y, Lin L, Zhou Y, Xu C, Jiang T. Abnormal cortical networks in mild cognitive impairment and Alzheimer’s disease. PLoS Comput. Biol. 2010;6:e1001006. doi: 10.1371/journal.pcbi.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yücel M, Pantelis C, Bullmore E. Whole-brain anatomical networks: Does the choice of nodes matter? Neuroimage. 2010;50(3):970–983. doi: 10.1016/j.neuroimage.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Zhan L, Jahanshad N, Ennis DB, Jin Y, Bernstein MA, Borowski BJ, Jack CR, Jr, Toga AW, Leow AD, Thompson PM. Angular versus spatial resolution trade-offs for diffusion imaging under time constraints. Hum. Brain Mapp. 2012a;34(10):2688–2706. doi: 10.1002/hbm.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Jahanshad N, Jin Y, Lenglet C, Mueller BA, Sapiro G, Ugurbil K, Harel N, Toga AW, Lim KO, Thompson PM. Field strength effects on diffusion measures and brain connectivity networks. Brain Connectivity. 2012b;3(1):72–86. doi: 10.1089/brain.2012.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]