Abstract
Brain connectivity is progressively disrupted in Alzheimer’s disease (AD). Here we used a seemingly unrelated regression (SUR) model to enhance the power to identify structural connections related to cognitive scores. We simultaneously solved regression equations with different predictors and used correlated errors among the equations to boost power for associations with brain networks. Connectivity maps were computed to represent the brain’s fiber networks from diffusion-weighted MRI scans of 200 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). We first identified a pattern of brain connections related to clinical decline using standard regressions powered by this large sample size. As AD studies with a large number of DTI scans are rare, it is important to detect effects in smaller samples using simultaneous regression modeling like SUR. Diagnosis of MCI or AD is well known to be associated with ApoE genotype and educational level. In a subsample with no apparent associations using the general linear model, power was boosted with our SUR model--combining genotype, educational level, and clinical diagnosis.
Keywords: Brain connectivity, neuroimaging genetics, HARDI tractography, seemingly unrelated regression (SUR), APOE4, multivariate analysis
1 Introduction
Brain connectivity is progressively disrupted in Alzheimer’s disease (AD). Several new technologies can recover patterns of brain connectivity from scans performed in a clinical setting, such as diffusion-weighted MRI. Connectivity maps are of interest from a neuroscientific point of view but there is also practical interest in whether connectivity measures are useful biomarkers for identifying factors that affect the brain in epidemiological studies or for monitoring brain decline in clinical trials.
Connectivity maps reveal organizational features of the brain not detectable on standard anatomical MRI. There is some interest in whether connectivity measures might help in predicting patient diagnosis or prognosis, either alone or when combined with other biomarkers. Connectivity measures may also provide insight into disease beyond what can be inferred from other imaging measures, or from clinical or cognitive assessments. In particular, diffusion tensor imaging (DTI) and its mathematical extensions (such as high angular diffusion weighted imaging, or q-space imaging) can reveal disease-related changes in white matter integrity (Nir et al., 2013), revealing how various cortical regions are connected to each other. Using diffusion MRI, structural connectivity can be defined in terms of the density or integrity of reconstructed fiber tracts connecting various regions of the brain. Often, cortical regions are identified automatically on T1-weighted structural MRI scans. Based on co-registered diffusion-imaging data, we can then study the trajectories and densities of white matter tracts interconnecting the cortical regions.
Clinical studies of brain connectivity are highly informative and are becoming highly successful. Brain connectivity changes profoundly during development (Hagmann et al., 2008, Hagmann et al., 2010, Dennis et al., 2013) in normal aging (Brown et al., 2011), in elderly people with HIV (Jahanshad et al., 2012), Alzheimer’s disease (Nir et al., 2012, Daianu et al., 2013a) and other neurodegenerative diseases (Toga and Thompson, 2013), and in disorders such as epilepsy (Engel et al., 2013). Such work reveals how diseases disrupt connections and networks, offering insights into their mechanisms and consequences.
Large-scale efforts such as the Alzheimer’s Disease Neuroimaging Initiative, or ADNI, have led to analyses of neuroimaging data in large cohorts of patients. ADNI recently launched a second phase (ADNI-2) of longitudinal data collection to include diffusion-weighted scans, with the goal of studying microstructural integrity and anatomical connectivity (among other measures) in elderly individuals. ADNI-2 is still in its early stages and data are still being collected from AD, MCI, and normal elderly subjects with varying degrees of cognitive impairment. Of the projected 1000 additional subjects in its second phase, ADNI will scan around 300 subjects with DTI. Yet, even in the early stages of data acquisition, connectivity disruptions in AD have been shown using ADNI-DTI (Hasan et al., 2012, Nir et al., 2012, Daianu et al., 2013a, Daianu et al., 2013b, Prasad et al., 2013). DTI can therefore detect changes associated with dementia. Even so, some factors that affect brain-imaging measures require tens of thousands of subjects to detect (Stein et al., 2012, Hibar et al., 2013); efforts are needed to maximize power for discovering factors that predict network decline. Fortunately, combining predictors from all clinical categories can significantly enhance power to predict brain integrity and decline, or in other words, reduce the sample size needed to detect statistical associations (Kohannim et al., 2010, Yuan et al., 2012, Xiang et al., 2013).
To identify clinically relevant changes in the brain’s networks, one typical approach is to fit multiple general linear regression models to identify connections whose integrity is statistically associated with clinical or cognitive scores, or with ratings of dementia severity. Detecting connections whose strength are associated with cognitive decline may help to delineate compromised brain regions and subnetworks. This may focus attention on regions where medication effects may be monitored more specifically. Ideally, one would prefer to analyze a very large cohort of subjects to have enough statistical power to identify all connections associated with changes in cognition. However, as with other measures, the power to relate brain connectivity to clinical parameters is limited by the available sample size. This makes it vital to examine new ways to optimize power to detect clinical associations with images.
In this study, we have two goals. First, we identify a pattern of connections in the brain whose density is associated with clinical decline. To do this, we use a standard regression model where the elements of the connectivity matrix are predicted using widely used cognitive test scores including the global clinical dementia rating (CDR) and Mini-Mental State Examination (MMSE) scores. The classical approach to find brain measures related to disease burden, is to fit a large regression model that includes as many relevant predictors as possible. These predictors may include measures of dementia severity (such as the CDR or MMSE) - or other predictors known to be associated with AD, such as ApoE4 genotype (Reiman et al., 1996), age, and educational level (Stern et al., 1994). In other words, the connectivity matrix is treated as a 2D image and all relevant predictors are fitted to the data at each matrix element, leading to a statistical parametric map of connections that decline in AD.
However, as a second goal, we propose a different and more powerful tactic to pick up connectivity patterns that decline in disease, based on a method known as seemingly unrelated regression (SUR), adapted from econometrics (Zellner, 1962). SUR is quite well known in the financial literature but perhaps not so well known in brain imaging, so we explain it briefly here. In the standard statistical model, we could insert all the predictors we have (MMSE, CDR, educational level, ApoE genotype, etc.) into a single multiple regression equation to predict the values of connectivity matrix elements, C(x,y). If that is done a pattern of connections would be found that relates to clinical decline, so long as there is sufficient power to find an effect. With SUR, we instead write down a set of simultaneous regression equations where each equation in the set does not necessarily have to predict the same outcome measure, i.e., some of the regression equations may predict a different dependent variable, and some predictors may be present or absent in each equation. If the predictors depend on each other statistically, we are then able to use the fact that the errors are correlated among the larger set of equations to solve them more accurately. We essentially use the correlated errors among equations to boost power to find brain connections that decline in AD.
As SUR can be more powerful than a standard regression, we used both SUR and a standard linear regression to identify brain connections related to clinical decline. We hypothesized that SUR would detect associations too weak to detect with the standard model. Our goal was to boost the effect sizes of associations between brain connectivity measures and clinical scores to ‘revive’ significance for tests that would have failed using the standard regression model. The overall goal of our work is to enhance the power to pick up patterns of brain connections that decline in AD. This is particularly useful when the available sample size is limited, but should always be beneficial even in large samples.
2 Methods
2.1 Subject information and image acquisition
Data collection for the second phase of the Alzheimer’s Disease Neuroimaging Initiative (ADNI2) includes diffusion MRI, but, at the time of writing (April 2013), this is still in its early stages. Here we performed an initial cross-sectional analysis of the ADNI DTI data from 200 adults whose DTI scans passed a quality control procedure; the QC process involved checking each scan for cropping or incomplete coverage of the brain, slice or gradient drop-out, stripes or other artifacts, excessive distortion or excessive rotational or translational motion during the scan. Scans that failed QC were excluded. Table 1 shows a summary of relevant demographic information for these 200 participants. Age, sex and educational level (in years) were ascertained for all subjects. Clinical assessments of dementia severity include the Mini-Mental State Exam (Folstein et al., 1975); lower scores denote greater impairment), and the Clinical Dementia Rating (Hughes et al., 1982) (scored as 0, 0.5 or 1; higher scores represent more severe dementia). ApoE4 genotype data to date has been collected for 76 of the 200 individuals who had neuroimaging scans. We conduct a t-test to determine demographic differences between the genotyped group and the full group, as well as between the genotyped group and the non-genotyped group, as seen in Table 2. We note no differences in distribution of sex, age, MMSE score, CDR score or education level between the subgroup and the full group. The genotyped group and the non-genotyped group show nominally significant (p<0.05) differences between MMSE (p=0.015) and education level (p=0.016), however these would not be considered significant when considering all 5 between group comparisons tested (Bonferroni correction would require p < 0.01).
Table 1.
ADNI demographic information: subjects with available DTI scans fall into one of three diagnostic categories, cognitively healthy controls, those with mild cognitive impairment (MCI), and those with Alzheimer’s disease (AD). Apolipoprotein E4 genotype was modeled here in a dominant fashion as 0 or 1, indicating the presence of any epsilon-4 alleles carried by the subject (which confer heightened risk of AD). A dominant model for the genotype was used due to the limited sample size (which limits the number of people with 2 copies of ApoE4).
| Sex | Age | MMSE | Global CDR |
Years of Education (N=198) |
|
|---|---|---|---|---|---|
| Total N=200 | NF= 87, NM=113 | 73.0 (7.5) | 27.2 (2.6) | 0.43 (0.31) | 15.8 (2.8) |
| Genotyped N=76 | NF=30, NM=46 | 74.2 (7.2) | 27.8 (2.0) | 0.41 (0.25) | 16.4 (2.7) |
| APOE4=0 (N=44) | NF=21, NM=23 | 75.8 (7.3) | 27.7 (2.1) | 0.39 (0.27) | 16.1 (2.7) |
| APOE4=1 (N=32) | NF=9, NM=23 | 72.0 (6.7) | 27.8 (1.86) | 0.44 (0.26) | 16.9 (2.7) |
| APOE4 t-test | 0.09 | 0.02 | 0.85 | 0.39 | 0.18 |
Table 2.
Two-sided t-tests were conducted between the genotyped group and the full group for sex, age, MMSE score, CDR score and education level. No significant differences were found. The genotyped group was also compared to the non-genotyped group. While MMSE score and education level are nominally significant at p < 0.05, these would not survive multiple comparisons correction.
| Between group t-test p-values | Sex | Age | MMSE | CDR | Education |
|---|---|---|---|---|---|
| Genotyped (76) vs full group (200) | 0.545 | 0.238 | 0.052 | 0.415 | 0.109 |
| Genotyped (76) vs non-genotyped (124) | 0.369 | 0.082 | 0.015 | 0.253 | 0.016 |
All subjects underwent whole-brain MRI scanning on 3-Tesla GE Medical Systems scanners. T1-weighted SPGR (spoiled gradient echo) sequences (256×256 matrix; voxel size = 1.2×1.0×1.0 mm3; TI = 400 ms; TR = 6.984 ms; TE = 2.848 ms; flip angle=11°) and diffusion-weighted images (DWI; 35 cm field of view; 128×128 acquired matrix, reconstructed to 256×256; voxel size: 2.7×2.7×2.7 mm3; scan time = 9 min) were collected. 46 separate images were acquired for each diffusion MRI scan: 5 T2-weighted images with no diffusion sensitization (b0 images) and 41 diffusion-weighted images (b=1000 s/mm2) chosen to optimize the signal-to-noise ratio in a fixed scan time (Zhan et al., 2012). More details on ADNI protocols may be found at: http://adni-info.org/Scientists/Pdfs/ADNI2_Protocol_FINAL_20100917.pdf and http://adni.loni.usc.edu/wp-content/uploads/2010/05/ADNI2_GE_3T_22.0_T2.pdf.
2.2 Cortical extraction and HARDI tractography
Diffusion-weighted images and structural MRI scans were independently preprocessed and jointly analyzed to map the fiber connections between cortical regions. Non-brain regions were automatically removed from each T1-weighted MRI scan, and from a T2-weighted image from the DWI set using the FSL tool “BET” (http://fsl.fmrib.ox.ac.uk/fsl/). Anatomical scans subsequently underwent intensity inhomogeneity normalization using the MNI “nu_correct” tool (www.bic.mni.mcgill.ca/software/). All T1-weighted images were linearly aligned using FSL (with 6 DOF) to a common space with 1mm isotropic voxels and a 220×220×220 voxel matrix. DWIs were corrected for eddy-current distortions using the FSL tools (http://fsl.fmrib.ox.ac.uk/fsl/). For each subject, the 5 images with no diffusion sensitization were averaged, linearly aligned and resampled to a downsampled version of that same subject’s T1-weighted image (110×110×110, 2×2×2mm). b0 maps were then elastically registered to the T1-weighted scan – which is considered to be a relatively undistorted anatomical reference – to compensate for susceptibility artifacts.
The transformation matrix obtained by linearly aligning the mean b0 image to the T1-weighted volume was applied to each of the 41 gradient directions to properly re-orient the orientation distribution functions (ODFs). We performed high-angular resolution diffusion imaging (HARDI) specific tractography as performed in (Aganj et al., 2011) on the sets of DWI volumes. We note that the diffusion images that ADNI collects are not HARDI, in the sense of having only a modest number of gradients (41), but methods to deal with HARDI data can be used to correctly make use of all the available angular information. We note that this technique exploits the available angular resolution of the data without assuming a tensor model for the diffusion process and is applicable to scans with limited angular resolution (as is the case with ADNI).
Elastic deformations obtained from the EPI distortion correction, mapping the average b0 image to the T1-weighted image, were then applied to the each traced fiber’s 3D coordinates for accurate realignment of the anatomy. Each subject’s dataset contained ~10,000 useable fibers (3D curves) in total, which has shown to be sufficient in a connectivity study of fiber curves traced (Prasad et al, 2013). Fibers artificially traced on the edge of the brain due to imaging artifacts were removed as were fibers shorter than 10mm in length, which were thought to be due to noise in the image.
34 cortical labels per hemisphere, listed in the Desikan-Killiany atlas (Desikan et al., 2006), were automatically extracted from all aligned T1-weighted structural MRI scans using FreeSurfer (version 5.0 http://surfer.nmr.mgh.harvard.edu/) (Fischl et al., 2004). The resulting T1-weighted images and cortical models were aligned to the original T1-weighted input image space and down-sampled using nearest neighbor interpolation to the space of the DWIs (to avoid intermixing of labels). Labels were dilated with an isotropic box kernel of width 5 voxels to ensure tracts would intersect labeled cortical regions.
2.3 Calculating the connectivity matrix
For each subject, a full 68×68 connectivity matrix was created. Each element described the proportion of the total number of extracted fibers in the brain connecting each of the labels, such that the number of fiber counts in each connection is normalized by the overall number of fibers from the full brain tractography; diagonal elements of the matrix describe the total number of fibers passing through a certain cortical region of interest. If more than 50% of subjects had no detected fibers for a specific matrix element, then the connection was considered invalid or insufficiently consistent in the population, and was not included in the analysis. We are more lenient with this threshold here than in studies involving only healthy individuals, in order to include connections that may be affected by the disease.
2.4 General linear regression
The general linear regression model was applied to data from the full sample of 200 subjects to associate cognitive scores from the Mini-Mental State Exam (MMSE) with each element of the connectivity matrix (representing the proportion of white matter fibers connecting the various cortical regions):
| (1) |
The y’s represent the traits being predicted, the α’s represent intercepts, while β’s represent the slopes (regression coefficients) for the fitted regressors, the residual errors (ε) are identically and independently distributed for each individual (1, …, j). yN×N represents the elements of the N×N connectivity matrix. Associations were performed using the ‘lm’ function in the R statistical package. We used the standard false discovery rate procedure (Benjamini and Hochberg, 1995) to correct for multiple statistical tests made across all tested matrix elements (Jahanshad et al., 2013b).
2.5 The seemingly unrelated regressions model
The SUR model (Zellner, 1962) was developed for applications in econometrics. Recently, SUR models have been applied to biomedical applications, e.g., to explain variance in biological traits based on predictors from genome-wide association scans (Saint-Pierre et al., 2011). The SUR model combines regression equations for a number of traits (shown below for two traits, as assessed here) and assumes that the residual errors (ε) are identically and independently distributed for each individual (1, …, j) within traits (labeled 1 and 2 here, for two traits). The model also allows for the residual errors to be correlated for an individual between traits. Different traits may have different sets of predictors (Xi). This means that the sets of predictors for different equations are not identical - Equation 1 can have predictors X1i= [x1i, x2i, x3i, x4i] while Equation 2 can have predictors X2i= [x2i, x4i, x5i, x6i] for an individual i. α represents an intercept, while β represents the slopes (regression coefficients) for the fitted regressors as above.
| (2) |
The idea behind SUR is to fit a number of regression equations at once – not necessarily with the same outcome measure – and use the information on the covariance in the errors from each equation to update the other. Here, the following joint equation was used to boost power to detect associations with cognitive scores (MMSE) on brain measures, when limiting our associations to only those 76 subjects who were genotyped for ApoE4. The two traits examined are (1) element-wise fiber proportions, with the MMSE score as a predictor of interest and age and sex as covariates, and (2) global CDR score (0, 0.5, or 1 generally representing controls, MCI or AD) with ApoE4 genotype and years of formal education as additional predictors:
| (3) |
SUR regressions were carried out using the ‘systemfit’ package in R. We used the false discovery rate procedure (Benjamini and Hochberg, 1995) to correct for multiple statistical tests made across all matrix elements.
2.6 Additional comparisons
In addition to comparing the SUR model to the linear regression model for the entirety of the group (all 200 subjects) to associate MMSE with specific brain connections, we also evaluated a linear regression model of the same association in a subsample consisting of just the 76 genotyped subjects, and an additional linear model including covariates ApoE4 and years of education.
| (4) |
The idea of this test was to confine the analysis to a deliberately small sample, and see if the standard model or SUR could still pick up associations that we know are present when the larger sample is used.
3 Results
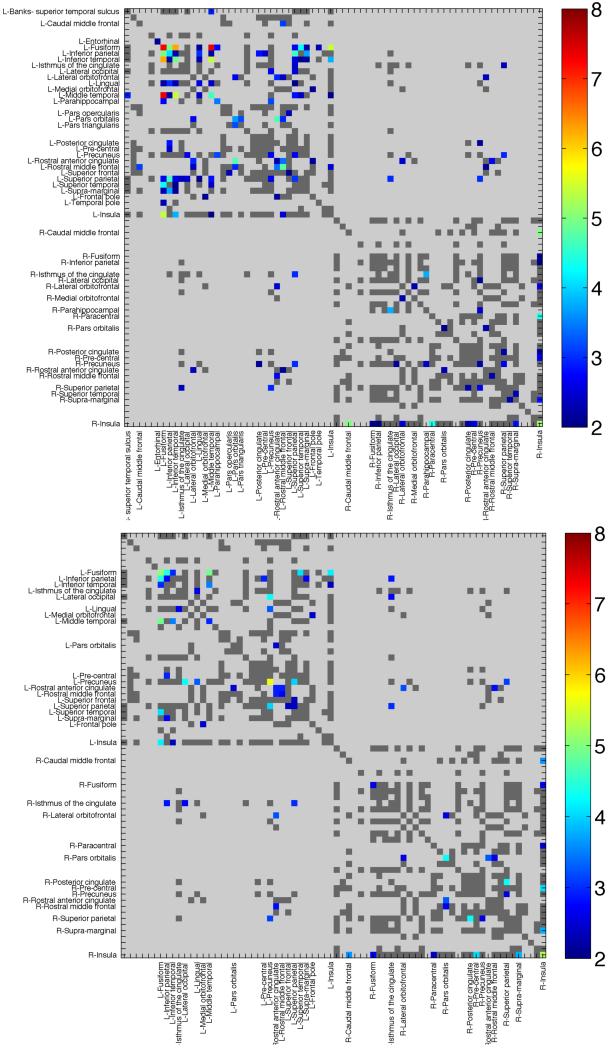
In our full sample of 200 individuals, we found significant and widespread associations of fiber density in the connectivity matrices with MMSE, after adjusting for age and sex. Regions of significance, after correcting for multiple comparisons with an FDR of q=0.05 are shown in Figure 1a. In total, 88 connections were significantly related to MMSE scores (Table 3), after multiple comparisons correction.
Figure 1. Clinical Associations with Structural Connectivity in ADNI.
Connectivity maps show regions of significant association between fiber proportions in individual cortical pathways and (a) MMSE scores, or (b) global CDR measures. Colored elements indicate significant associations. Dark gray regions were considered ‘valid’ (i.e., present in at least 50% of individuals) and were evaluated, but were not found to be significant. Light gray regions were invalid and not assessed. Warmer colors represent lower ‘more significant’ p-values, or higher −log10(p-value). The −log10(p-value) enables us to easily distinguish between the low p-values needed to establish significance after multiple comparisons correction across elements. For example, a p-value of 10−7 has a −log10(p-value) = 7 and can be clearly identified as more significant (red in color) than a p-value of 10−5 or −log10(p-value) =5.
Table 3.
Here we list structural brain network connections significantly associated with MMSE in the full sample (N=200). A connection of one region to itself denotes MMSE is a significant predictor for the fiber density in that particular node. Of the 88 regions, 6 were ‘revived’ (i.e., became significant when they were not before) when using the SUR model, even in a sample 38% of the original size, when power was insufficient for the general linear model to fit. An additional connection not listed (between the right caudal anterior cingulate and the right entorhinal cortex) was also significant with the SUR model. As the power to detect changes is much greater in the larger sample size (Full Model), the p-values for the SUR model are in general slightly higher than those of the full model, or not significant after multiple comparison correction as denoted by ‘–’.
| Connection | Full model significant p-value in a sample of N=200 |
SUR significant p-value in a more challenging sample of N=76 |
|
|---|---|---|---|
| Left Entorhinal | Left Entorhinal | 0.0012 | -- |
| Left Fusiform | Left Fusiform | 0.0008 | -- |
| Left Inferior parietal | Left Fusiform | 0.0075 | -- |
| Left Inferior parietal | Left Inferior parietal | 5.4E-08 | -- |
| Left Inferior temporal | Left Fusiform | 1.1E-05 | -- |
| Left Inferior temporal | Left Inferior parietal | 5.5E-07 | -- |
| Left Inferior temporal | Left Inferior temporal | 0.0069 | -- |
| Left lingual | Left Fusiform | 4.2E-08 | -- |
| Left lingual | Left Inferior parietal | 0.0018 | -- |
| Left lingual | Left Inferior temporal | 0.0021 | -- |
| Left lingual | Left lingual | 5.4E-05 | -- |
| Left Middle temporal | Left Banks of the STS | 0.0060 | -- |
| Left Middle temporal | Left Fusiform | 0.0050 | -- |
| Left Middle temporal | Left Inferior parietal | 3.9E-06 | -- |
| Left Middle temporal | Left Inferior temporal | 8.4E-05 | -- |
| Left Middle temporal | Left lingual | 0.0054 | -- |
| Left Middle temporal | Left Middle temporal | 0.0009 | -- |
| Left Parahippocampal | Left Fusiform | 0.0056 | -- |
| Left Parahippocampal | Left Parahippocampal | 0.0015 | -- |
| Left Pars orbitalis | Lateral orbitofrontal | 0.0038 | 0.0001 |
| Left Pars orbitalis | Left Pars orbitalis | 2.4E-05 | 0.0003 |
| Left Pars triangularis | Left Pars orbitalis | 0.0002 | -- |
| Left Posterior cingulate | Left Inferior parietal | 1.2E-05 | -- |
| Left Pre-central | Left Inferior parietal | 0.0052 | -- |
| Left Precuneus | Left Lateral occipital | 3.9E-06 | -- |
| Left Precuneus | Left Parahippocampal | 0.0028 | -- |
| Left Precuneus | Left Precuneus | 0.0081 | -- |
| Left Rostral anterior cingulate | Lateral orbitofrontal | 0.0080 | 0.0001 |
| Left Rostral anterior cingulate | Left Pars orbitalis | 0.0002 | 0.0002 |
| Left Rostral anterior cingulate | Left Rostral anterior cingulate | 0.0039 | -- |
| Left Rostral middle frontal | Left Caudal middle frontal | 0.0047 | -- |
| Left Rostral middle frontal | Lateral orbitofrontal | 0.0025 | -- |
| Left Rostral middle frontal | Left Pars orbitalis | 0.0034 | 0.0002 |
| Left Rostral middle frontal | Left Rostral anterior cingulate | 0.0022 | -- |
| Left Rostral middle frontal | Left Rostral middle frontal | 0.0011 | -- |
| Left Superior frontal | Left Pars opercularis | 0.0046 | -- |
| Left Superior frontal | Left Superior frontal | 0.0045 | -- |
| Left Superior parietal | Left Fusiform | 0.0031 | -- |
| Left Superior parietal | Left Inferior parietal | 0.0042 | -- |
| Left Superior parietal | Left Inferior temporal | 0.0087 | -- |
| Left Superior parietal | Left Isthmus of the cingulate | 0.0080 | -- |
| Left Superior parietal | Left Lateral occipital | 2.5E-05 | -- |
| Left Superior parietal | Left lingual | 0.0015 | -- |
| Left Superior parietal | Left Middle temporal | 0.0043 | -- |
| Left Superior parietal | Left Precuneus | 0.0092 | -- |
| Left Superior parietal | Left Superior parietal | 0.0004 | -- |
| Left Superior temporal | Left Fusiform | 0.0007 | -- |
| Left Superior temporal | Left Inferior temporal | 0.0049 | -- |
| Left Superior temporal | Left Middle temporal | 0.0002 | -- |
| Left Supra-marginal | Left Fusiform | 0.0006 | -- |
| Left Supra-marginal | Left Inferior parietal | 1.9E-05 | -- |
| Left Supra-marginal | Left Inferior temporal | 0.0011 | -- |
| Left Frontal pole | Left Medial orbitofrontal | 0.0077 | -- |
| Left Frontal pole | Left Rostral anterior cingulate | 0.0022 | -- |
| Left Temporal pole | Left Fusiform | 0.0022 | -- |
| Left Insula | Left Fusiform | 0.0011 | -- |
| Left Insula | Left Inferior temporal | 0.0057 | -- |
| Left Insula | Left Middle temporal | 0.0003 | -- |
| Left Insula | Left Rostral middle frontal | 0.0068 | -- |
| Right Isthmus of the cingulate | Left Superior parietal | 0.0036 | -- |
| Right Lateral orbitofrontal | Left Rostral anterior cingulate | 0.0020 | -- |
| Right Medial orbitofrontal | Right Lateral orbitofrontal | 1.6E-05 | 2.8E-05 |
| Right Parahippocampal | Right Isthmus of the cingulate | 0.0035 | -- |
| Right Pars orbitalis | Right Pars orbitalis | 0.0078 | -- |
| Right Precuneus | Left Posterior cingulate | 0.0082 | -- |
| Right Precuneus | Left Superior parietal | 0.0009 | -- |
| Right Precuneus | Right Lateral occipital | 0.0010 | -- |
| Right Precuneus | Right Parahippocampal | 0.0010 | -- |
| Right Precuneus | Right Precuneus | 1.0E-05 | -- |
| Right Rostral anterior cingulate | Lateral orbitofrontal | 0.0076 | -- |
| Right Rostral anterior cingulate | Left Rostral middle frontal | 0.0084 | -- |
| Right Rostral anterior cingulate | Right Pars orbitalis | 0.0002 | -- |
| Right Rostral middle frontal | Left Rostral anterior cingulate | 0.0075 | -- |
| Right Rostral middle frontal | Right Rostral middle frontal | 0.0058 | -- |
| Right Superior parietal | Left Isthmus of the cingulate | 0.0025 | -- |
| Right Superior parietal | Left Precuneus | 0.0037 | -- |
| Right Superior parietal | Right Posterior cingulate | 4.2E-05 | -- |
| Right Superior parietal | Right Precuneus | 0.0086 | -- |
| Right Supra-marginal | Right Superior temporal | 0.0053 | -- |
| Right Insula | Right Caudal middle frontal | 0.0059 | -- |
| Right Insula | Right Fusiform | 0.0057 | -- |
| Right Insula | Right Inferior parietal | 0.0020 | -- |
| Right Insula | Right Lateral orbitofrontal | 0.0062 | -- |
| Right Insula | Right Paracentral | 0.0025 | -- |
| Right Insula | Right Posterior cingulate | 0.0062 | -- |
| Right Insula | Right Pre-central | 0.0055 | -- |
| Right Insula | Right Supra-marginal | 0.0014 | -- |
| Right Insula | Right Insula | 6.8E-06 | -- |
The global CDR score was also associated with our measures of brain connectivity, as seen in Figure 1b. CDR and MMSE are quite strongly correlated (r=-0.61 in this sample, with a negative sign due to the definitions of which scores are better or worse). Consequently, using both CDR and MMSE in the same linear regression model could lead to a multicollinearity problem. CDR is also associated with differences in the structural network, which supports our hypothesis that an SUR model including both variables may boost power.
In the full available sample of 200 subjects, we tested how the various predictors related to brain connectivity. 38%, or 76 of the 200 individuals were genotyped for ApoE4 in this study. After multiple comparisons correction, we found no significant associations between the 198 subjects with available educational level, or the 76 subjects with ApoE4 information available and brain connectivity. As the second equation used for the SUR model involved only clinical measures, and no imaging measures, we additionally examined whether elements from that equation were significantly associated with each other. On its own (without imaging measures), no significant associations were detected between genotype (ApoE4) or educational level and global CDR, after controlling for age and sex in the subset of 76 subjects, possibly due to the limited sample size. We can therefore conclude that education and genotypes were not directly related predictors of global CDR score or overall brain connectivity within this relatively small sample of individuals.
Next, we fitted a standard linear regression as before, but restricted to the subsample of subjects with ApoE4 data available. In this subgroup of about one third of the full sample, the standard regression approach was not able to predict the matrix elements when the model was fitted in the subsample. Additionally, in this group we found that adding in the additional covariates -- education and genotype -- to the linear model still resulted in no significant associations between the MMSE score with the matrix elements.
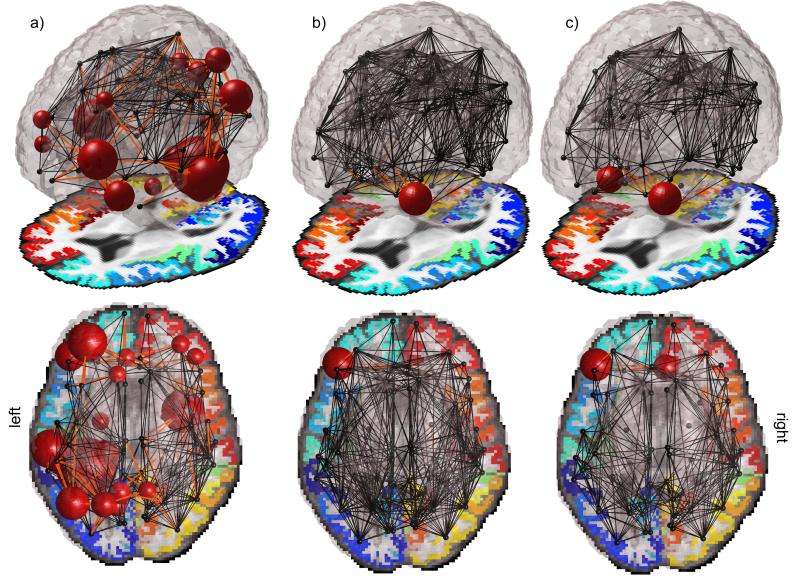
However, when the SUR model was applied to the same subsample (of 76/200 subjects), significant associations between brain connectivity and MMSE were detected, i.e. ‘revived’, in the sense that the standard model could not detect them in the reduced sample, but we know they are true as the standard model could detect them in the full sample. Not all of the same regions were declared significant, but a subsample of connections was implicated when using the SUR model in the much smaller sample than when using a general linear regression in a much larger sample. Seven connections, in total, were found to be significantly associated with MMSE, 6 of which were in the original list; these ‘revived’ connections are marked in Table 3. One connection, between the right caudal anterior cingulate and the right entorhinal cortices was found to associate to MMSE in the SUR model, and this association was not detected in the original linear model, even in the full sample. A visual representation of the full extent of the connections affected and those revived with the SUR model can be seen in Figures 2a and 2b, respectively; it is noteworthy that when filtering connections by those present in 50% of subjects, more connections were filtered out in the larger group. Because of this, we also examined the SUR model only in the valid connections for the full group. This led to similar results, seen in Figure 2c, where the connection between the right caudal anterior cingulate and the right entorhinal cortices was no longer found to be valid, but instead the proportion of fibers intersecting the right medial orbitofrontal cortex was found to be significantly correlated with MMSE again.
Figure 2.
A visual representation of a) the full extent of the connections associated with MMSE, and b) those ‘revived’ with the SUR model are shown. Regions revived with the SUR model in the reduced sample size are the connections between the left pars orbitalis and the left lateral orbitofrontal, left rostral anterior cingulate and the left lateral orbitofrontal, left rostral anterior cingulate and the left pars orbitalis, left rostral middle frontal and the left pars orbitalis, the right medial orbitofrontal and the right lateral orbitofrontal, and also the fiber density within the left pars orbitalis itself. In c), we examined the SUR model restricted to the valid connections for the full group. The connection between the right caudal anterior cingulate and the right entorhinal cortices was no longer found to be valid, but instead the proportion of fibers intersecting the right medial orbitofrontal cortex was again found to be significantly correlated with MMSE.
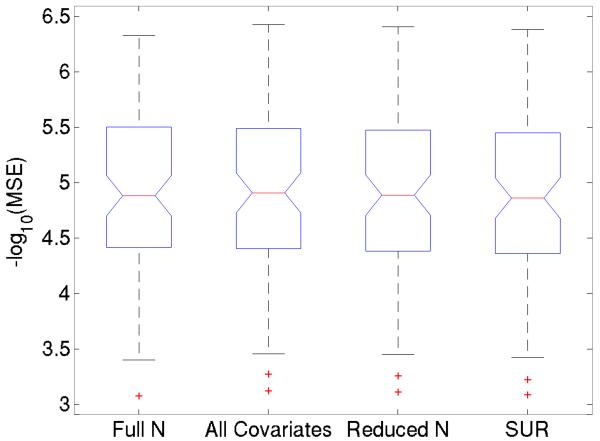
Additionally, for all these association tests of MMSE with network connectivity, we computed the mean-squared error (MSE) of the regressions and found that they were not significantly different between models. Box plots of the −log10 of the error measures on the 88 significant connections found with the entire group are compared for each model in Figure 3.
Figure 3.
For all association tests of MMSE with network connectivity, we found the mean-squared error (MSE) of the regressions; they are not significantly different between models. Box plots of the −log10 of the error measures on the 88 significant connections found with the entire group are compared. From left to right, these represent the linear model using all 200 subjects, the linear model using the 76 subjects with available ApoE data, and also using education level and ApoE4 status as covariates, the linear model using the 76 subjects with only age and sex as covariates, and the SUR model as described in the Methods. No significant differences are seen in the distribution of MSE between the different tests, despite the overall significance in the entire sample, and in the SUR models.
4 Discussion
Here we show, first and foremost, that the density of the brain’s fiber connections is related to clinical assessments of global functioning, and with standard measures of dementia severity, suggesting that connectivity maps may offer a viable image-based biomarker of disease severity. The association of structural connectivity with dementia scores is itself very promising as it can help disentangle the mechanisms by which dementia progresses throughout the brain and by which connections are affected.
Second, we found that our SUR model was more efficient in picking up patterns of brain connections related to clinical decline. In other words, SUR picked up associations in smaller samples of subjects, and found associations not detectable with standard regression. We set up the comparison by finding connections linked to clinical decline in a large sample, and then reducing the sample size to the point where there were too few subjects for standard regression to find the association. With that in mind, we were able to verify whether the associations detected by SUR were reasonable based on the tests in the much larger sample.
If used in conjunction with other standard measures obtained from clinical visits, SUR can enhance the power to detecting brain differences related to clinical decline. By applying SUR to brain connectomes – i.e., connectivity matrices – we developed a novel method to probe the degenerative basis of anatomical brain connectivity in a limited sample size by combining imaging measures with genetic risk assessments and educational history. Using multivariate associations through the ‘seemingly unrelated regression’ approach, we combine two equations of limited power to collectively boost power in mapping associations with brain connectivity. These findings may also help improve efforts in personalizing medicine, in that information on a patient that may seem unrelated to the diagnosis on its own (or simply underpowered) can actually be used to boost the power for other associations.
MMSE and global CDR scores are diffusely associated with fiber density connectivity maps in large enough sample sizes. Most of the MMSE-associated connections were in temporal-parietal regions and the frontal lobe, mainly concentrated in the left hemisphere. These connections shown here to track clinical decline have long been implicated at various points in the trajectory of AD, which usually follows a limbic-to-frontal, temporal and parietal sequence of cortical changes (Braak and Braak, 1991, Thompson et al., 2003, Thompson et al., 2007). Due to the processing of the images, particularly the dilation of the cortical regions, we do not expect this known pattern of cortical thinning to artificially influence our results (i.e., less fibers would not be found simply because regions are thinner, as cortical labels are dilated to ensure inclusion of white matter). Instead, we find that the fibers connecting these regions are reduced in count proportional to all the detected fibers in the brain, offering complementary insight into the structural mechanisms influencing degeneration and cognitive impairment. The strongest MMSE-associated connection was with the fibers intersecting the inferior parietal cortex, a region whose connectivity has been strongly implicated in Alzheimer’s disease (Jacobs et al., 2012).
ApoE4 genotype and poorer educational level have each been consistently associated with risk for AD, across many studies, for example in (Stern et al., 1994). Even so, ADNI2 consists of a relatively highly educated population, and only a fraction of those assessed have been currently genotyped for ApoE4 (others will be genotyped in the future). In our sample, neither genotype nor educational level showed significant associations with the global dementia rating. Although this could be because the sample size to-date is relatively small, we show these factors are insufficient to help predict diagnosis in terms of global CDR scores directly when these factors are entered into the model as linear covariates. Despite this, we can use the knowledge that our sample may be slightly underpowered to pick up the associations and combine two underpowered regression equations. Correlations between the errors of the regression equations, possibly due to the relation of age to ApoE4 status, or sex to education level, end up boosting the power to detect statistical associations at certain connections in the anatomical network. The connections that were revived using the SUR model are mainly focused in the orbitofrontal regions, regions important for connecting frontal monitoring systems to the limbic system (Tekin and Cummings, 2002) and a hub for neurofibrillary tangles in AD (Tekin et al., 2001). Our results here parallel some of the major nodes previously found to associate to AD related changes in network efficiency (Lo et al., 2010).
In the spirit of our work here, there are several other ongoing efforts in statistics to make most efficient use of the available data in ADNI and other studies with large numbers of biomarkers. ADNI has an inevitable pattern of missing data, in that not all subjects were willing or able to have CSF measures or ApoE genotyping, or amyloid PET imaging. By design, only one-third of the sample was scanned with DTI. In a setting where data is missing blockwise, we have previously tested sparse multi-biomarker models that combine across submodels (Yuan et al., 2012). Such methods that transfer or pool information between different problems and models are advantageous in analyzing large datasets of correlated data, and to some extent in all of science.
This work suggests several follow-up studies. Structural connectivity is powerful enough to use as a phenotype to search the genome for genetic variants that affect the brain (Jahanshad et al., 2013b), making it an ideal candidate for also mapping disease related genetic effects. The most dementia-influenced connections are plausible targets to prioritize in the hunt for genes influencing brain integrity and risk for disease (Jahanshad et al., 2013a). Regions where power is boosted in the joint regression model may hold underlying genetic influences, which could potentially be picked up in larger sample sizes. Multivariate models to include more genetic information have shown promise in finding genetic influences on brain structure (Hibar et al., 2011, Kohannim et al., 2013). While the multivariate methods presented here involve including a host of critical patient information with known effects, multivariate methods to include more genetic information may also boost power to detect disease associated changes in the brain. The current study focuses on pinpointing specific connections in the brain that deteriorate as patients decline clinically. Even so, much recent work on brain networks additionally focuses on more general regions or global properties of the connectome as a whole and have been widely successful in identifying developmental or dementia associated changes (Lo et al., 2010, Dennis et al., 2011, Nir et al., 2012, Daianu et al., 2013b). These network properties may be important for identifying brain changes associated with the individual genome; for a review see (Thompson et al., 2013).
In conclusion, models combining a variety of biomarkers may be important for clinical studies, such as ADNI, where several sources of somewhat directly ‘unrelated’ information (clinical, biological, genetic, demographic) are collected all with the goal of identifying biomarkers to better characterize disease.
Acknowledgments
Data collection and sharing for this project was funded by ADNI (NIH Grant U01 AG024904). Investigators within ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. For a complete listing of ADNI investigators, please see http://www.loni.usc.edu/ADNI/Collaboration/ADNI_Manuscript_Citations.pdf. Algorithm development and image analysis for this study was funded by grants to PT from the NIBIB (R01 EB008281, R01 EB008432).
Footnotes
Disclosure statement
ADNI is partially funded by public and private agencies. One of the authors, Michael Weiner, has private funding unrelated to the content of this paper.
References
- Aganj I, Lenglet C, Jahanshad N, Yacoub E, Harel N, Thompson PM, Sapiro G. A Hough transform global probabilistic approach to multiple-subject diffusion MRI tractography. Med Image Anal. 2011;15:414–425. doi: 10.1016/j.media.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, Bookheimer SY. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proc Natl Acad Sci U S A. 2011;108:20760–20765. doi: 10.1073/pnas.1109038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daianu M, Dennis EL, Nir TM, Jahanshad N, Toga AW, Clifford R, Jack J, Weiner MW, Thompson PM. Alzheimer’s disease disrupts rich club organization in brain connectivity networks. 2013a doi: 10.1109/ISBI.2013.6556463. ISBI 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daianu M, Jahanshad N, Nir TM, Toga AW, Clifford R, Jack J, Weiner MW, Thompson PM. Breakdown of Brain Connectivity between Normal Aging and Alzheimer’s Disease: A Structural k-Core Network Analysis Brain Connectivity In submission. 2013b doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, Rudie JD, Brown JA, Johnson K, McMahon KL, de Zubicaray GI, Montgomery G, Martin NG, Wright MJ, Bookheimer SY, Dapretto M, Toga AW, Thompson PM. Altered structural brain connectivity in healthy carriers of the autism risk gene, CNTNAP2. Brain Connectivity. 2011;1:447–459. doi: 10.1089/brain.2011.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, Toga AW, McMahon KL, Zubicaray GId, Martin NG, Hickie I, Wright MJ, Thompson PM. Development of the “Rich Club” in brain networks from 438 adolescents and adults aged 12 to 30. 2013 doi: 10.1109/ISBI.2013.6556552. ISBI 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Engel J, Thompson P, Stern J, Staba R, Bragin A, Mody I. Connectomics and Epilepsy. Current Opinion in Neurology. 2013 doi: 10.1097/WCO.0b013e32835ee5b8. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of psychiatric research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, Meuli R, Thiran JP, Grant PE. White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci U S A. 2010;107:19067–19072. doi: 10.1073/pnas.1009073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M, Lee W, Park B, Han K. In: Connectivity Analysis of Hippocampus in Alzheimer’s Brain Using Probabilistic Tractography - Bio-Inspired Computing and Applications. Huang D-S, et al., editors. Vol. 6840. Heidelberg; Springer Berlin: 2012. pp. 521–528. [Google Scholar]
- Hibar DP, ENIGMA-Consortium . ENIGMA2: Genome-wide scans of subcortical brain volumes in 16,125 subjects from 28 cohorts worldwide. Organization of Human Brain Mapping; Seattle, WA: 2013. +200-co-authors. [Google Scholar]
- Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, Kim S, Pankratz N, Foroud T, Huentelman MJ, Potkin SG, Jack CR, Jr., Weiner MW, Toga AW, Thompson PM. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011;56:1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- Jacobs HI, Van Boxtel MP, Jolles J, Verhey FR, Uylings HB. Parietal cortex matters in Alzheimer’s disease: an overview of structural, functional and metabolic findings. Neuroscience and biobehavioral reviews. 2012;36:297–309. doi: 10.1016/j.neubiorev.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov P, Sprooten E, Mandl RC, Nichols TE, Almassy L, Blangero J, Brouwer RM, Curran JE, Zubicaray GId, Duggirala R, Fox PT, Hong LE, Landman BA, Martin NG, McMahon KL, Medland SE, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Hulshoff Pol HE, Bastin ME, McIntosh AM, Deary IJ, Thompson PM, Glahn DC. Multi-Site Genetic Analysis of Diffusion Images and Voxelwise Heritability Analysis: A Pilot Project of the ENIGMA-DTI Working Group. Neuroimage. 2013a doi: 10.1016/j.neuroimage.2013.04.061. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR, Jr., Saykin AJ, Green RC, Weiner MW, Medland SE, Montgomery GW, Hansell NK, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. ProcNatlAcadSciUSA. 2013b;110:4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Valcour VG, Nir TM, Kohannim O, Busovaca E, Nicolas K, Thompson PM. Disrupted brain networks in the aging HIV+ population. Brain Connectivity. 2012;2:335–344. doi: 10.1089/brain.2012.0105-Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, Jack CR, Jr., Weiner MW, Thompson PM. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiology of aging. 2010;31:1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Rajagopalan P, Hibar DP, Jahanshad N, Grill JD, Apostolova L, Toga AW, J CR, Jr, Weiner MW, Thompson PM. Multilocus genetic profiling to empower drug trials and predict brain atrophy Neuroimage-Clinical In submission. 2013 doi: 10.1016/j.nicl.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:16876–16885. doi: 10.1523/JNEUROSCI.4136-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir T, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM. Small World Network Measures Predict White Matter Degeneration in Patients with Early-Stage Mild Cognitive Impairment. Proceedings / IEEE International Symposium on Biomedical Imaging: from nano to macro IEEE International Symposium on Biomedical Imaging. 2012:1405–1408. doi: 10.1109/ISBI.2012.6235831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, Thompson PM. Effectiveness of Regional DTI Measures in Distinguishing Alzheimer’s Disease, MCI, and Normal Aging. Neuroimage-Clinical In Review. 2013 doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, Nir TM, Toga AW, Thompson PM, ADNI . Tractography density and network measures in Alzheimer’s disease. ISBI; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. The New England journal of medicine. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- Saint-Pierre A, Kaufman JM, Ostertag A, Cohen-Solal M, Boland A, Toye K, Zelenika D, Lathrop M, de Vernejoul MC, Martinez M. Bivariate association analysis in selected samples: application to a GWAS of two bone mineral density phenotypes in males with high or low BMD. Eur J Hum Genet. 2011;19:710–716. doi: 10.1038/ejhg.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga JJ, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O’Brien C, Papmeyer M, Putz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TG, van Tol MJ, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Goring HH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW, Jr., Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Muhleisen TW, Nauck M, Nothen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Renteria ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdes Hernandez MC, Van den Heuvel M, van der Wee NJ, Van Haren NE, Veltman JA, Volzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR, Jr., Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW, Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S, Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJ, Espeseth T, Fernandez G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer-Lindenberg A, Morris DW, Muller-Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Samann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM. Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y, Gurland B, Tatemichi TK, Tang MX, Wilder D, Mayeux R. Influence of education and occupation on the incidence of Alzheimer’s disease. JAMA: the journal of the American Medical Association. 1994;271:1004–1010. [PubMed] [Google Scholar]
- Tekin S, Cummings JL. Frontal-subcortical neuronal circuits and clinical neuropsychiatry: an update. J Psychosom Res. 2002;53:647–654. doi: 10.1016/s0022-3999(02)00428-2. [DOI] [PubMed] [Google Scholar]
- Tekin S, Mega MS, Masterman DM, Chow T, Garakian J, Vinters HV, Cummings JL. Orbitofrontal and anterior cingulate cortex neurofibrillary tangle burden is associated with agitation in Alzheimer disease. Annals of neurology. 2001;49:355–361. [PubMed] [Google Scholar]
- Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. Genetics of the Connectome. Neuroimage In submission. 2013 doi: 10.1016/j.neuroimage.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang MC, Leow AD, Sowell ER, De Zubicaray G, Becker JT, Lopez OL, Aizenstein HJ, Toga AW. Tracking Alzheimer’s disease. Annals of the New York Academy of Sciences. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Connectomics sheds new light on Alzheimer’s disease. BP. 2013;73:390–392. doi: 10.1016/j.biopsych.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Yuan L, Fan W, Wang Y, Thompson PM, Ye J. ADNI. Bi-level multi-souce learning for heterogeneous block-wise missing data. Neuroimage In submision. 2013 doi: 10.1016/j.neuroimage.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. Neuroimage. 2012;61:622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellner A. An Efficient Method of Estimating Seemingly Unrelated Regressions and Tests for Aggregation Bias. J Am Stat Assoc. 1962;57:348–&. [Google Scholar]
- Zhan L, Jahanshad N, Ennis DB, Jin Y, Bernstein MA, Borowski BJ, Jack CR, Jr., Toga AW, Leow AD, Thompson PM. Angular versus spatial resolution trade-offs for diffusion imaging under time constraints. Human Brain Mapping. 2012 doi: 10.1002/hbm.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]