Abstract
An increased flux through glycolysis supports the proliferation of cancer cells by providing additional energy in the form of ATP as well as glucose-derived metabolic intermediates for nucleotide, lipid, and protein biosynthesis. Thus, glycolysis and other metabolic pathways that control cell proliferation may represent valuable targets for therapeutic interventions and diagnostic procedures. In this context, the measurement of glucose uptake and lactate excretion by malignant cells may be useful to detect shifts in glucose catabolism, while determining the activity of rate-limiting glycolytic enzymes can provide insights into points of metabolic regulation. Moreover, metabolomic studies can be used to generate large, integrated datasets to track changes in carbon flux through glycolysis and its collateral anabolic pathways. As discussed here, these approaches can reveal and quantify the metabolic alterations that underlie malignant cell proliferation.
1. INTRODUCTION
Glycolysis is the intracellular biochemical conversion of one molecule of glucose into two molecules of pyruvate with the concurrent generation of two molecules of ATP. Pyruvate is a metabolic intermediate with several potential fates including entrance into the tricarboxylic acid (TCA) cycle within mitochondria to produce NADH and FADH2. These reducing agents subsequently donate electrons to the mitochondrial electron transport chain (ETC), which when fully coupled to the complex V ATP synthase of the mitochondrial inner membrane generates an additional 34 molecules of ATP per glucose. Alternatively, pyruvate can be converted into lactate in the cytosol by lactate dehydrogenase with concurrent regeneration of NAD+ from NADH. Conversion of pyruvate to lactate blocks further ATP production, but the resultant increase in NAD+ drives the first biochemical step in glycolysis (DeBerardinis, Lum, Hatzivassiliou, & Thompson, 2008). An increase in the flow of carbon metabolites through the glycolytic pathway, or glycolytic flux, can increase the rate of ATP production within cells despite being markedly less efficient at generating ATP compared to oxidative phosphorylation (Pfeiffer, Schuster, & Bonhoeffer, 2001).
In addition to generating ATP, glycolysis also supplies biosynthetic intermediates for cell growth and proliferation. For example, glucose-6-phosphate, the first cytosolic product of glucose metabolism, can shunt into the pentose phosphate pathway to drive NADPH generation from NAPD+. NADPH reduces reactive oxygen species produced mainly by respiration to maintain cellular redox balance and to protect the genome from mutations. Carbon flux through the pentose phosphate pathway supplies metabolites for nucleotide biosynthesis that is required for DNA replication and RNA transcription. Another example is 3-phosphoglycerate, a glycolytic metabolite used to synthesize serine, glycine, and cysteine, which in turn supplies one carbon metabolism. Folate and methionine cycles, the components of one carbon metabolism, provide metabolites that support diverse cellular processes including methylation reactions, antioxidant defenses, lipid head group modifications, and nucleotide metabolism (Locasale, 2013).
Warburg (1956) first observed that proliferating tumor cells augment aerobic glycolysis, the conversion of glucose to lactate in the presence of oxygen, in contrast to nonmalignant cells that mainly respire when oxygen is available. This mitochondrial bypass, called the Warburg effect, occurs in rapidly proliferating cells including cancer cells, activated lymphocytes, and pluripotent stem cells. While the Warburg effect is energy inefficient, it is offset by an increased glycolytic flux to provide additional biosynthetic precursors to support rapid cancer cell proliferation (DeBerardinis et al., 2008). This energy compromise supports higher rates of nucleotide synthesis for DNA replication and RNA transcription, phospholipids for membrane production, and amino acids for protein translation to support increased cell division. The Warburg effect has been exploited for clinical diagnostic tests that use positron emission tomography (PET) scanning to identify increased cellular uptake of fluorinated glucose analogs such as 18F-deoxyglucose.
Not all tumors, however, shift to glycolysis for energy production. Some diffuse large B cell lymphomas and glioblastomas remain dependent on oxidative phosphorylation for energy production (Caro et al., 2012; Marin-Valencia et al., 2012). Metabolic enzyme activity is heterogeneous between different tumors even within tumor classes, and glycolytic enzymes can be either increased or decreased in their expression (Hu et al., 2013). Glutamine and fatty acids can also be used by cancers as alternative sources of fuel to make ATP through oxidative phosphorylation (Le et al., 2012; Zaugg et al., 2011).
Although Warburg made his observations over 75 years ago, the detailed mechanisms and consequences of shifting metabolism toward glycolysis are only starting to be revealed. Pyruvate kinase isoformM2 (PKM2), an embryonic splice variant of the glycolytic enzyme pyruvate kinase (PK), is highly expressed in several types of cancer (Christofk, Vander Heiden, Harris, et al., 2008;Lim et al., 2012). PKM2 shows a decreased kinase activity that helps shunt glycolytic intermediates through biosynthetic pathways at the expense of respiration to CO2 (Christofk, Vander Heiden, Harris, et al., 2008; Hitosugi et al., 2012). Phosphorylation of Tyr-105 of PKM2 causes the release of the allosteric activator of PKM2, 1,6-bisphosphate, which decreases its activity (Hitosugi et al., 2012). Anotherglycolytic enzyme, phosphoglycerate dehydrogenase, is amplified in human tumors and directs glycolytic carbon flux into serine biosynthesis instead of continued catabolism to pyruvate (Locasale et al., 2011; Possemato et al., 2011). An increased carbon flux through the serine biosynthesis pathway also supports glycine production, which is used for nucleotide biosynthesis and regulates cell proliferation (Jain et al., 2012).
2. MEASURING GLUCOSE UPTAKE AND LACTATE PRODUCTION
For cells in culture, glycolytic flux can be quantified by measuring glucose uptake and lactate excretion. Glucose uptake into the cell is through glucose transporters (Glut1–Glut4), whereas lactate excretion is through monocarboxylate transporters (MCT1–MCT4) at the cell membrane. A caveat to relatively straightforward measurements of extracellular fluid concentrations of glucose and lactate is that they do not provide information on other possible fates for glucose-derived carbons (Fig. 5.1). Extracellular measurements of glucose and lactate in cell culture media are relatively simple to perform, cell nondestructive, and provide a reasonable estimate of glycolytic flux (Table 5.1).
Figure 5.1. Glucose uptake and lactate excretion approximate glycolytic flux.
Additional metabolic processes that siphon or add metabolic intermediates to the glycolytic pathway remain undefined, which may impact glycolytic flux measurements and complicate data interpretations.
Table 5.1.
Methods to detect glucose uptake and lactate excretion
| Detectable metabolite |
||||||
|---|---|---|---|---|---|---|
| Method | Glucose | Lactate | Equipment | Cell destructive? |
Radioactive? | Advantages and disadvantages |
| Extracellular glucose or lactate kit | Y | Y | Spectrophotometer | N | N | Simple, standard equipment |
| Extracellular bioanalyzer | Y | Y | Bioanalyzer | N | N | Simple |
| ECAR measurement | N | Y | Seahorse XF analyzer | N | N | Coupled with oxygen consumption |
| [3H]-2-DG or [14C]-2-DG uptake |
Y | N | Scintillation counter | Y | Y | Expensive tracers |
| 18FDG uptake | Y | N | Scintillation counter/cyclotron | Y | Y | Short half-life |
| 2-NBDG uptake | Y | N | Flow cytometer or fluorescent microscopy | Y | N | Single cell analysis |
Glucose uptake into cells provides an intracellular estimate of glycolytic flux and can be quantified through detection of entrapped glucose analogs. This approach can quantify glucose uptake at the level of a single cell, but is cell destructive.
2.1. Extracellular glucose and lactate
Commercially available kits and instruments are available to quantify glucose and lactate levels within cell culture media. When using these methods, cell number must be considered as this can greatly affect results. Kit detection methods are usually colorimetric or fluorometric and are compatible with standard lab equipment such as spectrophotometers.
BioProfile Analyzers (Nova Biomedical) or Biochemistry Analyzers (YSI Life Sciences) can measure levels of both glucose and lactate in cell culture media. GlucCell (Cesco BioProducts) can measure only glucose levels in cell culture media. While each commercial method has a different detection protocol, the collection of culture media for analysis is the same. Generally for both extracellular glucose and lactate measurements, cells should be plated at equal densities with the culture media changed 24 h prior to collection. To account for differences in the rates of cell proliferation or death, cells should be counted at the time of media collection for normalization of results to cell number.
2.1.1 Extracellular acidification rate by Seahorse XF analyzer
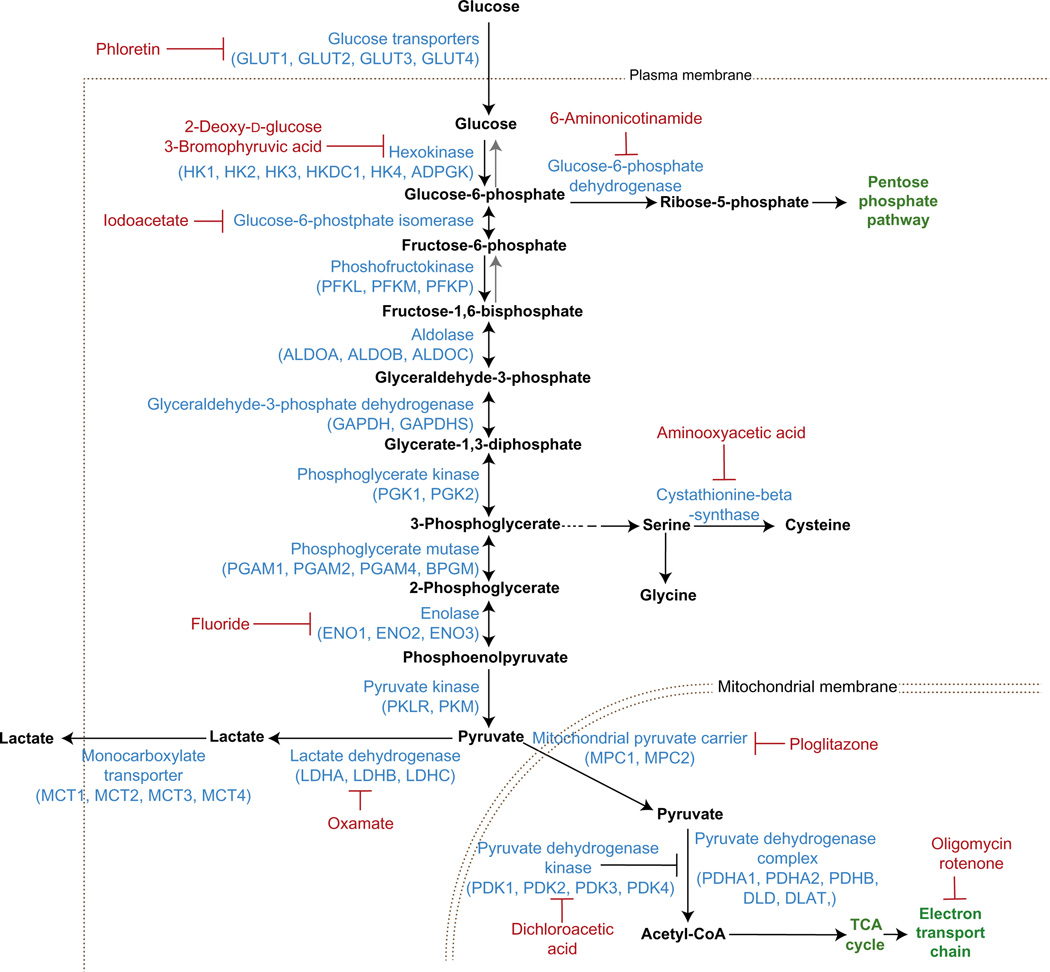
The Seahorse extracellular flux (XF) analyzer (Seahorse Bioscience) is a powerful tool for measuring glycolysis and oxidative phosphorylation (through oxygen consumption) simultaneously in the same cells. Glycolysis is determined through measurements of the extracellular acidification rate (ECAR) of the surrounding media, which is predominately from the excretion of lactic acid per unit time after its conversion from pyruvate (Wu et al., 2007). However, additional metabolic processes in cells, such as CO2 production by the TCA cycle, can change the pH of the media complicating this analysis. Because ECAR is essentially a measurement of pH, a buffering agent, such as sodium bicarbonate, is not included in the assay medium. In addition, bicarbonate and media pH can play a role in regulating glycolysis, which can confound measurements of ECAR. To increase measurement accuracy, chemical inhibitors can be used to determine pH changes that are from lactate excretion versus other sources of media acidification (Fig. 5.2). The Seahorse XF analyzer requires only small cell numbers per assay condition and has injection ports for the addition of up to four inhibitors in each experiment. Available detectors use either a 24-well (XF24) or a 96-well (XF96) assay plate format.
Figure 5.2. Chemical inhibitors of glycolysis.
Inhibitors of glycolytic enzymes are used to quantify metabolite fluxes within the glycolytic pathway. Inhibition of glycolysis with 2-dexoxy-d-glucose (2-DG) is a common tool for measuring glycolysis (e.g., glycolysis stress test).
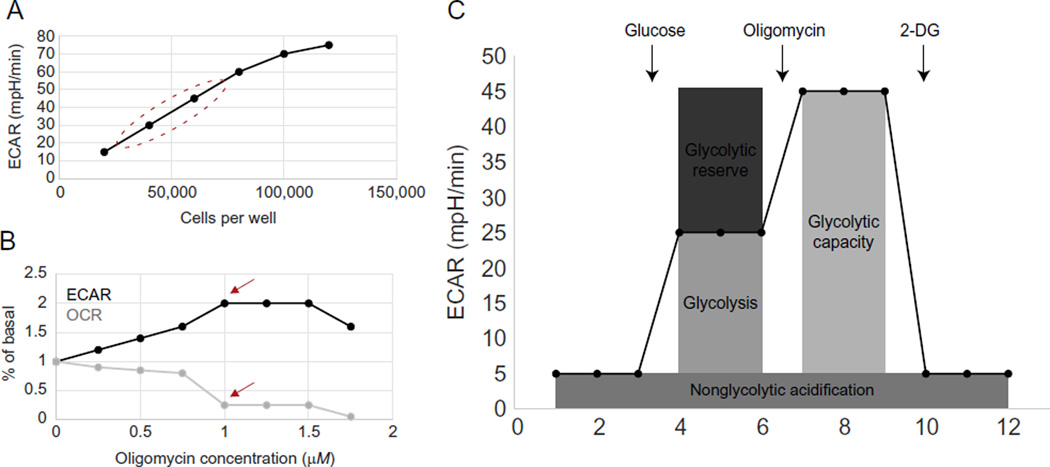
To obtain reproducible data within the linear detection range of the XF analyzer, optimization of the cell number and inhibitor concentration is required. The XF24 analyzer has a two-step cell seeding protocol and generally requires 2 × 104–1 × 105 cells per well. Experimental cell density should be chosen based on three main factors: (1) an ECAR value of at least 20 mpH/min, which is the low end of a potential linear detection range, (2) retained morphology of the plated cells by visual inspection, and (3) a cell density that lies within the linear range of a graph of cell seeding density versus ECAR values (Fig. 5.3A). Metabolic inhibitors used in assays should be titrated to obtain the optimal concentration for maximal response for each cell type without toxicity, as assessed by ECAR and oxygen consumption rate (OCR) measurements (Fig. 5.3B). Loosely adherent and suspension cells can be attached to XF V7 cell culture assay plates by coating these plates with Cell-Tak (BD Bioscience). The example protocol below is for adherent cells used with the XF24 format.
Figure 5.3. ECAR measurements with a Seahorse XF analyzer.
(A) Cell number optimization for XF assays. Circle identifies region with acceptable cell densities. (B, C) Oligomycin optimization for glycolysis stress test. Shown are ECAR (B) and OCR (C) with an oligomycin drug titration. 1 µM is the optimal oligomycin concentration in this case because it stimulates the maximum ECAR (arrow) and inhibits OCR (arrow). (C) Glycolysis, glycolytic capacity, glycolytic reserve, and nonglycolytic acidification can be measured with the glycolysis stress test.
To probe glycolysis, an assay called the glycolysis stress test is often used. In this assay, glucose, oligomycin, and 2-deoxyglucose (2-DG) are inserted through injection ports sequentially while measurements are being made. Glucose is supplied to feed glycolysis, and the difference between ECAR before and after addition of glucose is a measure of the glycolytic rate. Oligomycin inhibits ATP synthase in the ETC, which decreases the ATP/ADP ratio and drives glycolysis. The difference between ECAR before and after oligomycin addition is equal to the glycolytic reserve capacity of cells. 2-DG inhibits glycolysis and therefore provides a baseline ECAR measurement. ECAR after 2-DG addition accounts for the nonglycolytic ECAR of cells (Fig. 5.3C).
2.1.1.1 Protocol for ECAR measurement by Seahorse XF analyzer
Day 1: Seed cells and prepare a sensor cartridge
For adherent cells, wash cells with 1 × PBS, pH 7.4, and add 1 × trypsin until cells begin to visually detach. Add culture media with serum to deactivate trypsin and pipette up and down to create a uniform cell suspension. Count cells (e.g., hemocytometer) and resuspend the total so that the number of desired cells per well is within 100 µl (e.g., for 5 × 104 cells per well, suspend cells at a concentration of 5 × 105 cells per ml). Pipette 100 µl of cell suspension into each well of 24-well V7 plate (Seahorse Bioscience #100777-004). For accurate reproducible measurements, pipette up and down to make sure that cells are uniformly spread throughout the well. One well in each V7 plate row or column should lack cells to be used as a blank control. Put the seeded plate into a 37 °C, 5% CO2 incubator to allow cells to adhere for 1–5 h. Add an additional 150 µl of media and allow cells to grow overnight in a 37 °C, 5% CO2 incubator.
The XF24 and XF96 sensor cartridges (Seahorse Bioscience #100850-001) must be hydrated overnight for consistent results. Add XF calibrant, pH 7.4 (Seahorse Bioscience #100840-000) to each well of a Seahorse 24-well or 96-well plate. Put a sensor cartridge on top of the plate with sensors submerged in calibrant solution. Hydrate at 37 °C without CO2 for up to 72 h. Turn on the Seahorse XF Analyzer to allow the instrument to warm to 37 °C before an assay.
Day 2: Prepare a cell plate for analysis
Warm XF DMEM media (Seahorse Bioscience # 102353-100) to 37 °C in a water bath, add l-glutamine to a final concentration of 2 mM and adjust the pH of the media to 7.4. Remove all but 50–100 µl of media from each well and add 1 ml of 37 °C XF DMEM media, pH 7.4. It is important during this process to always leave a small amount of media in each well so that cells do not dry out and the cell monolayer is not disrupted. Then remove all but 50 µl of media from each well again and add the desired volume of 37 °C XF DMEM media. Place a 24- or 96-well plate in a 37 °C incubator without CO2 for 1 h before running an assay. CO2 will alter the pH of the media and disrupt ECAR measurements.
Inhibitors should be adjusted to the proper concentration in the XF DMEM media. Since inhibitors will be added through injection ports in the cartridge, they should be at a concentration of 10 × higher than the final dilution. Prepare 2 ml stocks of 100 mM glucose, 1 M 2-DG, and the optimized oligomycin concentration chosen by titration. Pipette 61.5 µl of glucose stock solution, 67.5 µl of oligomycin stock solution, and 75 µl of 2-DG stock solution into injection ports A, B, and C, respectively. Make sure to fill all A, B, and C injection ports including blank wells.
The XF24 and XF96 analyzers require the input of a protocol command sequence. Important parameters of this protocol are mix time, wait, and measure time. Cell lines with higher metabolic activity require shorter measurement times than cells with less metabolic activity. A good protocol to start with is a 3-min mix time, 3-min wait, and 3-min measure time. If OCR or ECAR values are too low, a protocol of 2-min mix time, 2-min wait, and 4- or 5-min measure time can improve the data quality.
After running the glycolysis stress test, normalize ECAR data to cell number or protein concentration and plot ECAR against time. Glycolysis, glycolytic capacity, and glycolytic reserve can be calculated with normalized ECAR values using the equations given below (Fig. 5.3C):
2.2. Glucose analog uptake
To determine the glucose uptake rate by cells, a labeled isoform of glucose can be added to the cell culture media and then measured within cells after a given period of time. Two types of glucose analogs are typically used for these studies: radioactive glucose analogs, such as 2-deoxy-d-[1,2-3H]-glucose, 2-deoxy-d-[1-14C]-glucose, or 2-deoxy-2-(18F)-fluoro-d-glucose (18FDG), or fluorescent glucose analogs, such as 2-[N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose (2-NBDG). Measurements of radioactive glucose analog uptake require a scintillation counter, whereas 2-NBDG uptake is usually measured by flow cytometry or fluorescent microscopy. In contrast to extracellular approaches, these techniques require cell harvest and termination of the cell culture.
2.2.1 2-Deoxy-d-[1,2-3H]-glucose or 2-deoxy-d-[1-14C]-glucose
Glucose analog, 2-DG, is brought into cells by a glucose transporter and is phosphorylated within the cytosol by hexokinase (Jenkins, Furler, & Kraegen, 1986). Further metabolism of 2-DG is slow and phosphorylation by hexokinase traps 2-DG within a cell (Hansen, Gulve, & Holloszy, 1994). By using radioactive 2-deoxy-d-[1,2-3H]-glucose or 2-deoxy-d-[1-14C]-glucose, the accumulation of 2-DG can be measured to quantify glucose uptake.
2.2.1.1 Protocol for 2-deoxy-d-[1,2-3H]-glucose or 2-deoxy-d-[1-14C]-glucose uptake
Plate approximately 5 × 104 cells per well in a 24-well plate. Allow time for the cells to attach and spread on the plate and switch to glucose-free or glucose low media. Add 1 µCi 2-deoxy-d-[1,2-3H]-glucose (MP Biomedicals #0127088) or 2-deoxy-d-[1-14C]-glucose (MP Biomedicals #0111012) and incubate for at least 20 min. Wash cells with 1 × PBS, pH 7.4 (Life Technologies #10010-023), and lyse cells with 1% SDS lysis buffer (1% sodium dodecyl sulfate, 10 mM Tris, pH 7.5). Use a scintillation counter to quantify [3H] or [14C] and normalize readings to cell number.
2.2.2 2-Deoxy-2-(18F)-fluoro-d-glucose
18FDG PET scanning has been used in clinical cancer diagnostics since the 1990s, but use of 18FDG for in vitro research protocols has been more limited, likely because of the short half-life of the radioactive fluorine (109.7 min) and limited access to a cyclotron for its manufacture for most research labs. Like other 2-DG analogs, 18FDG is phosphorylated after transport into the cell and cannot be exported (Smith, 2000). 18FDG accumulation occurs in many tumors, or other rapidly dividing cells, due to the Warburg effect, and increased analog trapping contrasts with surrounding normal tissues to provide signal discrimination for in vivo PET imaging. Use of 18FDG for in vitro experiments requires consideration of factors that can influence metabolism including cell density and growth media (Mertens, Mees, Lambert, Van de Wiele, & Goethals, 2012). 18FDG requires instrumentation to measured gamma radiation and increased safety procedures for working with radiation. Because the measurement 18FDG uptake in vitro is uncommon and an alternate method involving fluorescent detection of glucose uptake is available, a detailed work protocol for 18FDG uptake measurements is not provided here.
2.2.3 2-[N-(7-Nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose
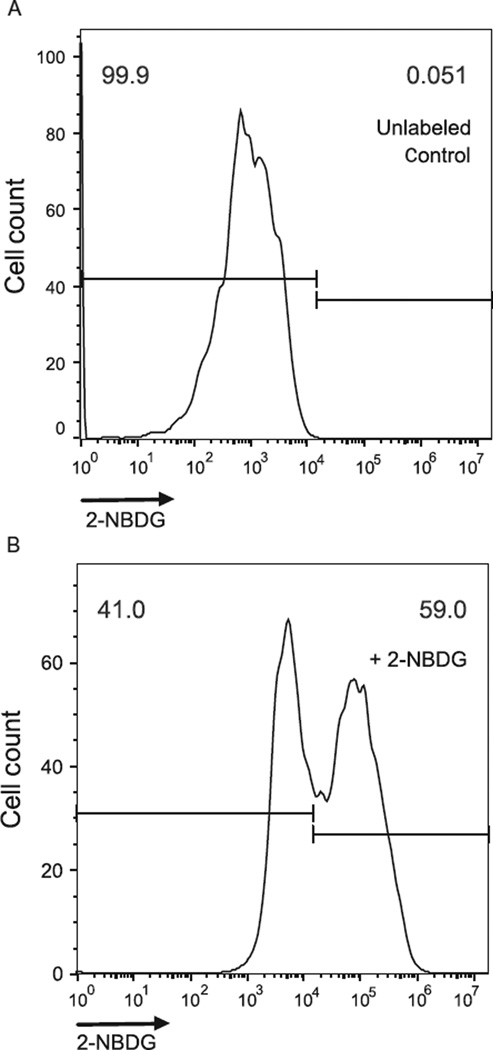
2-NBDG is taken into cells through glucose transporters and phosphorylated by hexokinase. However, 2-NBDG can be dephosphorylated, which can result in efflux from cells, but this analog still provides a good approximation of glucose uptake (O’Neil, Wu, & Mullani, 2005). Because 2-NBDG and glucose are both imported by glucose transporters, unlabeled glucose in the media will impact 2-NBDG uptake (O’Neil et al., 2005; Zou, Wang, & Shen, 2005). Therefore, the use of low-glucose media is optimal when tolerated by cells and available. 2-NBDG can be excited to fluoresce by 465 nm wavelength light and yields a 540-nm wavelength emission, which can be detected using a fluorescence detector channel or filter typically used for green fluorophores. A negative control sample should be included that is not incubated with 2-NBDG to set flow cytometer gating of negative and positive events and compensation when necessary (Fig. 5.4).
Figure 5.4. Measurement of 2-NBDG uptake by flow cytometry.
(A) A negative control that has not been cultured with 2-NBDG. (B) A sample cultured with 2-NBDG.
2.2.3.1 Protocol for 2-NBDG uptake
Approximately 5 × 104 cells should be plated per well. Cell confluence can affect metabolism and should be considered and adjusted appropriately. For each condition to be examined, count and plate cells at equal densities and allow time for cells, if they are adherent, to fully attach to the plate surface or substrate. Wash cells with 1 × PBS, pH 7.4, followed by the addition of culture media (low glucose when possible) supplemented with 10–100 µM 2-NBDG (Life Technologies #N13195), and incubate for 1–12 h at 37 °C with 5% CO2. A negative control should be incubated with the same culture media without adding 2-NBDG. The cell density, concentration of 2-NBDG, and time of incubation can be adjusted for optimal results.
For analysis by flow cytometry, add 1 × trypsin (Life Technologies #25300054), harvest cells, wash twice with ice cold 1 × PBS, pH 7.4, at 4 °C, and keep on ice. Keep the samples on ice to inhibit dephosphorylation and export of 2-NBDG from cells. Analyze the collected cells by flow cytometry within 30 min of harvest. A control sample lacking 2-NBDG should be used to set the flow cytometer compensation and gate parameters for 2-NBDG positive and negative events (Fig. 5.4).
3. MEASURING THE ACTIVITY OF RATE-LIMITING GLYCOLYTIC ENZYMES
While the activity of any single glycolytic enzyme is not a proxy for carbon flux through the entire pathway, specific enzymes limit the rate of glycolysis, and therefore, their activities control the maximum possible flux. Hexokinase, phosphofructokinase, and PK are the main rate-limiting enzymes in glycolysis.
Conversion from NAD+ to NADH or NADP+ to NADPH is often used in enzymatic assays because NADH and NADPH absorb light at 340 nm, while NAD+ and NADP+ lack absorbance at this wavelength (McComb, Bond, Burnett, Keech, & Bowers, 1976). By coupling the enzyme of interest with an NAD+/NADH- or NADP+/NADPH-dependent enzyme, the activity of the enzyme can be measured by the change in absorbance at 340 nm over time. Hexokinase, phosphofructokinase, and PK can be coupled with NAD+/NADH- or NADP+/NADPH-dependent enzymes by using enzymes that react with their products.
3.1. Hexokinase
Hexokinases phosphorylate the 6-hydroxyl of hexose using the γ-PO4 of ATP as a donor group (Wilson, 1995). In humans, hexokinase 4 (HK4) is more specific to glucose than other hexoses and is referred to as glucokinase. An ADP-dependent glucokinase (ADPGK) has been recently discovered in mouse and human genomes, which transfers a phosphoryl group from ADP to glucose (Richter et al., 2012). Hexokinase 1 (HK1), hexokinase 2 (HK2), and hexokinase 3 (HK3) are allosterically inhibited by their product, glucose-6-phosphate, but HK4 is not sensitive to glucose-6-phosphate concentrations (Wilson, 1995). Hexokinase is the first key enzyme in glycolysis because it traps glucose within cells.
High expression ofHK2 is associated with poor prognosis in hepatocellular carcinoma and brain metastasis (Palmieri et al., 2009; Peng, Lai, Pan, Hsiao, & Hsu, 2008). HK2 is required for tumor growth in multiple mouse models of cancer and is a potential target for therapeutics (Patra et al., 2013). Breast cancer cells regulate HK2 through miR-155, which increases expression of HK2 through multiple mechanisms (Jiang et al., 2012). To measure hexokinase activity, glucose is provided as a substrate, and glucose-6-phosphate dehydrogenase (G6PD) further metabolizes glucose-6-phosphate in aNADP+ dependent manner. The absorbance of NADPH at 340 nm will increase as G6PD converts glucose-6-phoshate to fructose-6-phosphate.
3.1.1 Protocol for hexokinase activity assay
Wash cells with 1 × PBS, pH 7.4, and incubate in lysis buffer (50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 1 mM DTT, protease inhibitor cocktail) for 30 min on ice to lyse cells. Prepare reaction buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 0.6 mM ATP (Sigma #A2383), 100 mM glucose (Sigma #G8270), 0.2 mM NADP+ (Sigma #0505), and 0.1 units of glucose-6-phosphate dehydrogenase (Sigma #G4134)) per ml. Add 20 µg of fresh cell lysate to 1 ml of reaction buffer. A negative and positive control should be included without cell lysate and with 0.05 units of hexokinase (Sigma #H5000) added. Mix, incubate at 37 °C, and measure optical absorbance of the reaction at 340 nm with a spectrophotometer every 15 s for 10 min (Yi et al., 2012). Enzyme activity can be represented as the change in absorbance per minute, which should be calculated on a linear portion of the obtained curve.
3.2. Phosphofructokinase
Glucose-6-phosphate can enter the pentose phosphate pathway or continue through the glycolytic pathway. Phosphofructokinase 1 (PFK1) provides the first enzymatic step at which a glucose molecule becomes committed to glycolysis and therefore is subject to regulation (Nelson & Cox, 2008). PFK1 activity depends on the concentrations of AMP, ADP, and ATP with allosteric activation by AMP and ADP and allosteric inhibition by ATP. Additional allosteric inhibition is provided by citrate and activation by fructose 2,6-bisphosphate (Nelson & Cox, 2008).
PFK1 activity has been linked to the Warburg effect through the p53 target protein, TP-53-induced glycolysis and apoptosis regulator (TIGAR). TIGAR inhibits glycolysis and promotes the pentose phosphate pathway by reducing levels of fructose-2,6-bisphosphate, thereby reducing PFK1 activity (Bensaad et al., 2006). Glycosylation of the fructose-2,6-bisphosphate binding site on PFK1 also inhibits glycolytic flux which decreases cell proliferation, thereby reducing cancer aggressiveness (Yi et al., 2012). Measurement of phosphofructokinase activity can be achieved by providing other glycolytic enzymes, aldolase, triosephosphate isomerase, and glyceraldehyde 3-phosphate dehydrogenase, which are NAD+ dependent. An increase of NADH absorbance at 340 nm can be observed as fructose 1,6-bisphosphate is converted to 1,3-bisphosphoglycerate.
3.2.1 Protocol for phosphofructokinase activity assay
Wash cells with 1 × PBS, pH 7.4, and incubate in lysis buffer (50 mM Tris– HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 1 mM DTT, protease inhibitor cocktail) for 30 min on ice to lyse cells. Prepare reaction buffer (50 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 5mM ATP (Sigma #A2383), 0.2 mM NADH (Sigma #N4505), 100 mM KCl, 5 mM Na2HPO4, 5 mM MgCl2, 0.01 AMP (Sigma #01930), 5 mM fructose-6-phosphate (Sigma #F3627), 5 units of triosephosphate isomerase (Sigma #T2507) per ml, 1 unit of aldolase (Sigma #A2714) per ml, and 10 units of glyceraldehyde-3-phosphate dehydrogenase (Sigma #G2267)) per ml. Add 20 µg of fresh cell lysate to 1 ml of reaction buffer. A negative and positive control should be included without cell lysate and with 0.05 units phosphofructokinase (Sigma #F0137) added. Measure optical absorbance of the reaction at 340 nm at room temperature every 15 s for 10 min with a spectrophotometer (Yi et al., 2012). Activity can be represented as the change in absorbance per minute, which should be calculated in a linear portion of the obtained curve.
3.3. Pyruvate kinase
PK catalyzes the final step in glycolysis by transferring a phosphoryl group from phosphoenolpyruvate (PEP) to ADP to form pyruvate and ATP. PK is active as a tetramer protein and is allosterically inhibited by ATP, acetyl-CoA, and long-chain fatty acids (Nelson & Cox, 2008). There are four PK isoforms including PKL, PKR, PKM1, and PKM2. PKL and PKR, the dominant PK isoforms in the liver and red blood cells, undergo allosteric activation by fructose-1,6-bisphosphate (Carbonell, Marco, Felíu, & Sols, 1973). Fructose-1,6-bisphosphate is also an allosteric activator of PKM2, which is a splice variant of the constitutively active PKM1 enzyme isoform (Dombrauckas, Santarsiero, & Mesecar, 2005). The product of PK, pyruvate, can be converted to lactate by lactate dehydrogenase, which is NADH dependent. The activity of PK can then be detected by a loss of absorbance at 340 nm.
3.3.1 Protocol for PK activity assay
Wash cells with 1 × PBS, pH 7.4, and incubate in lysis buffer (50 mM Tris– HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 1 mM DTT, protease inhibitor cocktail) for 30 min on ice to lyse cells. Prepare reaction buffer (50 mM Tris–HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.6 mM ADP (Sigma #A2754), 0.5 mM PEP (Sigma #860077), 0.18 mM NAD+ (Sigma #N1636), 10 µM fructose-1,6-bisphosphate (Sigma #F6803), and 10 units of lactate dehydrogenase (Sigma #59747)) per ml. Add 10 µg of fresh cell lysate to 1 ml of reaction buffer. Measure optical absorbance of the reaction at 340 nm every 15 s for 10 min with a spectrophotometer (Christofk, Vander Heiden, Wu, Asara, & Cantley, 2008; Yi et al., 2012). A positive and a negative control should be run with no cell lysate and 0.05 units of PK (Sigma #P7768) added. Activity can be represented as the change in absorbance per minute, which should be calculated in a linear portion of the obtained curve.
4. METABOLITE MEASUREMENTS AND GLUCOSE TRACING
Unlabeled metabolites or 13C-labeled metabolites can be quantified by mass spectrometry (MS) or nuclear magnetic resonance (NMR) spectrometry. Measurements of unlabeled metabolites provide steady-state data on the levels of all detectable metabolites within the cell. Using metabolite concentrations for extrapolation of flux information is difficult, however, because changes in the steady-state level of a metabolite can be due to increased or decreased flux through any of its associated metabolic pathways. Absolute quantification of an unlabeled metabolite is possible by adding a labeled standard for the metabolite of interest before MS analysis. Metabolite profiling was used to identify decreased serine biosynthesis as a result of PKM2 silencing (Chaneton et al., 2012).
The use of 13C-labeled glucose provides glycolytic flux data by quantifying 13C glucose-derived carbons in downstream metabolites. However, labeled substrates are costly and the label can be diluted by other carbon sources, making measurements challenging for some metabolites. However, detection of 13C-glucose-derived carbons in glycolytic intermediates is robust; making it is a good method for analyzing glycolysis.
While chromatography combined with MS is becoming the most widely used approach for metabolomics, NMR spectroscopy also has advantages for detecting and quantifying metabolites. NMR spectroscopy is quantitative, fast, and reproducible but has a detection limit in the micromolar range (Pan & Raftery, 2007; Want, Cravatt, & Siuzdak, 2005). By contrast, MS is exceedingly sensitive in the nanomolar range, making it useful for complex biosamples. MS analysis, however, requires metabolite separation with gas chromatography (GC) or liquid chromatography (LC), which reduces the speed of data acquisition. GC/MS combines the separation of complex biosamples with the retained sensitivity of MS (Want et al., 2005). However, GC/MS has an additional derivatization step of sample preparation that is required for detecting nonvolatile or large, polar macromolecules (Dietmair, Timmins, Gray, Nielsen, & Krömer, 2010; Want et al., 2005). By contrast, LC/MS detects a broad range of metabolites with simple sample preparation. Following the separation of metabolites by LC, they must be ionized for analysis by MS. Liquid chromatography electrospray ionization mass spectrometry (LC/ESI-MS) combines an automated ionization method, electrospray ionization, with the simplicity of LC and the sensitivity of MS. Therefore, we provide metabolomics protocols that are suitable for use with LC/ESI-MS. Fourier transform ion cyclotron resonance MS (FT-ICR-MS), orbitrap MS, and multipass time of flight MS (multipass TOF-MS) have all be used for metabolomic analysis (Aharoni et al., 2002; Lei, Huhman, & Sumner, 2011). FT-ICR-MS has ultrahigh resolution and mass accuracy below 1 ppm, while orbitrap MS also has high resolving power and mass accuracy between 1 and 5 ppm (Lei et al., 2011).
4.1. Metabolite measurements
Metabolite extraction must be performed to obtain metabolites from cultured cells for LC/ESI-MS analysis. Metabolite extraction generally has two parts: quenching and extraction. Quenching first limits further metabolic activity and removes contaminants, which is then followed by a metabolite extraction step (Sellick, Hansen, Stephens, Goodacre, & Dickson, 2011). Metabolites with a high turnover rate, such as ATP, depend on complete and quick quenching methods for accurate detection. Chemical and physical properties of each metabolite require different methods of quenching and different extraction solutions making it challenging to find one method that works for all metabolites (Dietmair et al., 2010). Glucose-6-phosphate is particularly unstable and requires effective quenching for accurate measurements. Enzymatic assays to measure glucose-6- phosphate can be used to check on the success of quenching and extraction steps. For optimization of extraction, temperature and solvent can be adjusted in both the quenching and extraction step.
4.1.1 Protocol for metabolite extraction of adherent cells for LC/ESI-MS analysis
Approximately 1 × 106 cells are minimally required for detection of metabolites by LC/ESI-MS. Cells can be grown in six-well plates for direct extraction from each well. For normalization to cell number or protein concentration, an extra well of each condition should be plated in parallel for cell counting or protein quantification. Add fresh media to cells 24 h prior to metabolite extraction. Place the six-well plate on ice and wash cells with ice cold 1 × PBS, pH 7.4. Add 400 µl of ice cold HPLC grade methanol (Fisher #A454-4). To maximize quenching speed, methanol can be cooled on dry ice to achieve a temperature as low as −40 °C. Add an equal volume of Milli Q water and gently shake the plate to ensure mixing. Scrape cells off the plate and transfer to a 1.5-ml eppendorf tube. For separation of polar from nonpolar metabolites, add 400 µl of HPLC grade chloroform (Fisher C607-4). Briefly vortex each sample every 5 min for 15–30 min. Transfer the top layer, which is the aqueous phase containing polar molecules, to a new tube. The remaining bottom layer contains nonpolar metabolites, which can also be transferred into a fresh tube. Glycolytic intermediates are polar molecules and should be in the top layer. Dry the contents of both tubes with a vacuum concentrator. Samples can then be stored at −80 °C until analysis. Alternatively, samples can be suspended in 50 µl of initial LC mobile phase solution for immediate LC/ESI-MS analysis. MS measurements should be performed according to the methods described in the instrument documentation.
4.2. Metabolic flux analysis with stable glucose isotopes
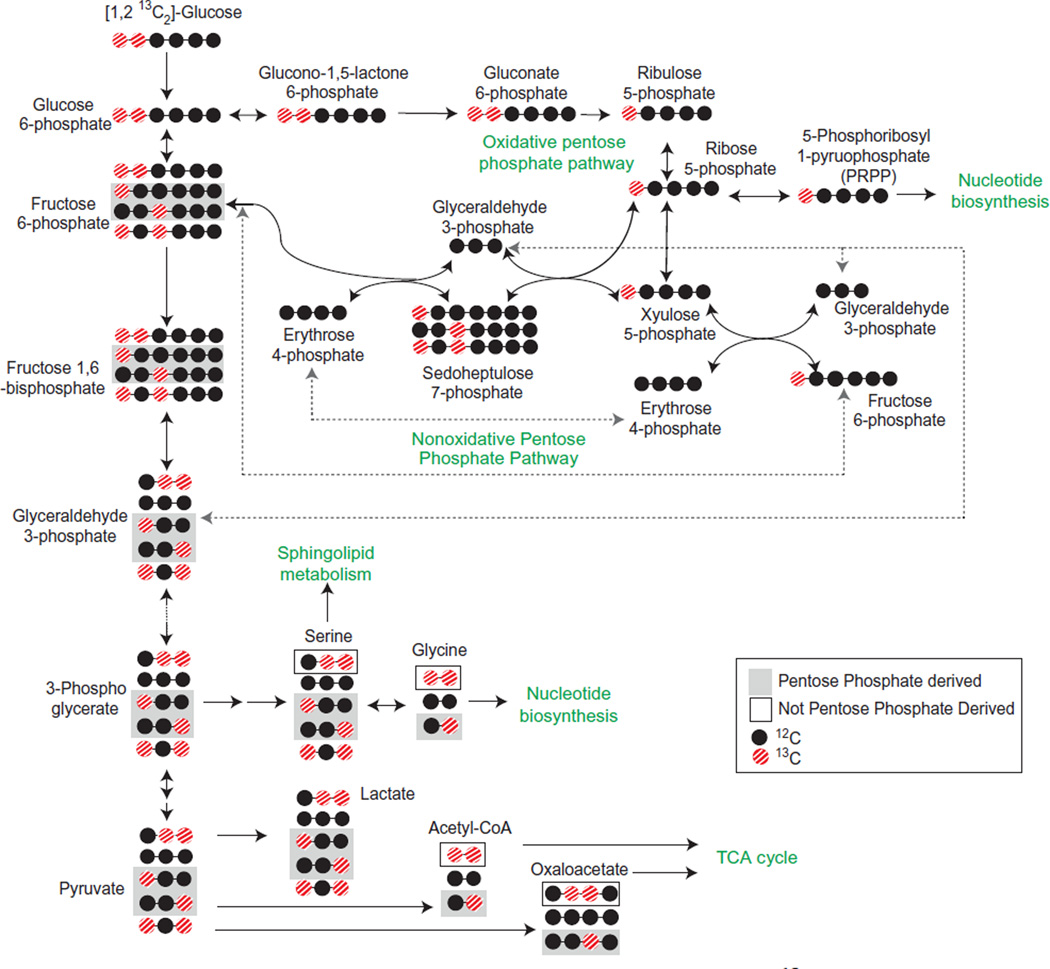
13C can replace any of the six carbons within a glucose molecule. [U-13C6]-glucose is 13C labeled at all six carbons and is often used to trace the glucose contribution to the TCA cycle. [1,2-13C2]-glucose is the best tracer for glycolytic pathway metabolite measurements (Metallo, Walther, & Stephanopoulos, 2009). In addition, carbon flux through the pentose phosphate pathway is not well defined by [U-13C6]-glucose. Instead, [1,2-13C2]-glucose will lose one 13C when it goes through the pentose phosphate pathway, which distinguishes metabolites that have gone through this pathway from those that have gone directly through glycolysis (Fig. 5.5).
Figure 5.5. Glycolytic pathway metabolite flux analysis with [1,2 13C2]-glucose.
Metabolites derived from the pentose phosphate pathway (gray boxes) can be distinguished from metabolites derived directly from glycolysis (black boxes). For simplicity, not all possible permutations of 12C and 13C are provided.
Labeled glucose will compete for uptake into the cell with unlabeled glucose within the media. Therefore, glucose-free or low-glucose media is used for labeled glucose tracing flux experiments. Regular culture media should be replaced with [U-13C6]-glucose (Cambridge Isotope Labs #CLM-1396) or [1,2-13C2]-glucose (Cambridge Isotope Labs #CLM-504) supplemented media 24 h prior to metabolite extraction. The same protocol can be used for extraction of metabolites for metabolic flux analysis as for measurement of unlabeled metabolites. Data analysis, however, is more complex and requires specialized software. Fiatflux, an open source software for use with isotopic tracer experiments, is an enabling tool for data analysis (Zamboni, Fischer, & Sauer, 2005). OpenFLUX and 13CFlux are also software packages available for use for with 13C metabolic flux analysis (Quek, Wittmann, Nielsen, & Krömer, 2009; Weitzel et al., 2013).
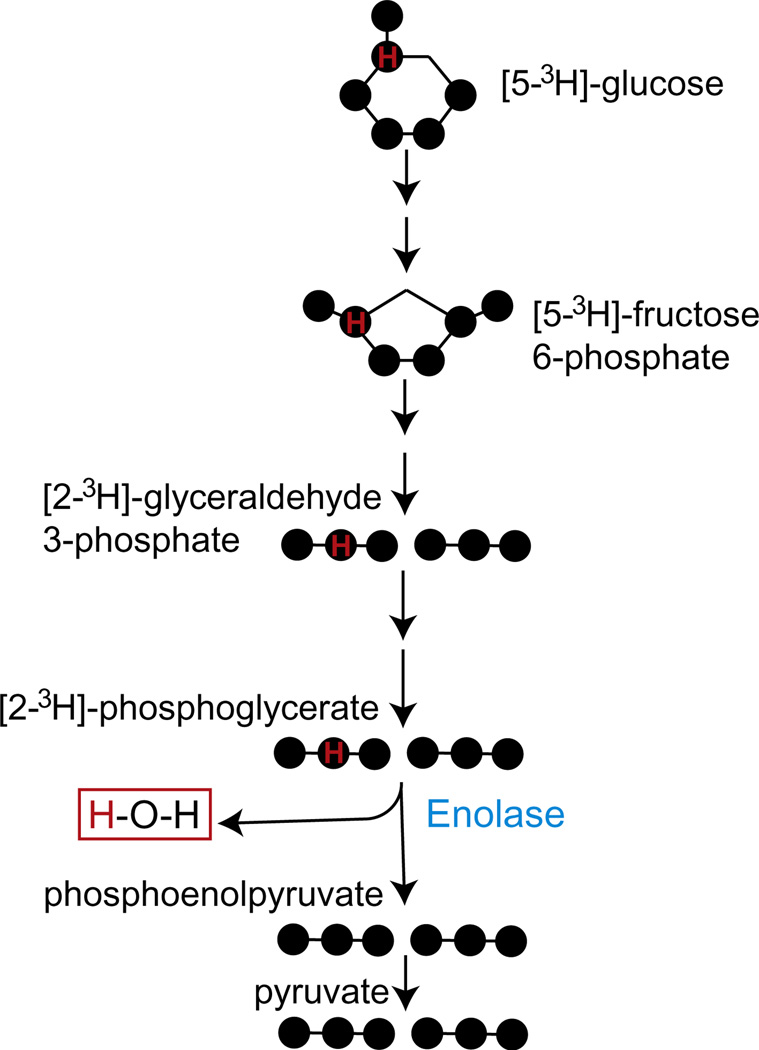
4.3. Release of 3H2O from [5-3H]-glucose
The release of 3H2O from [5-3H]-glucose is a method to quantify glycolytic flux (Neely, Denton, England, & Randle, 1972). A single tritium at the C5 of glucose is removed by a condensation reaction in the ninth step of glycolysis, which is catalyzed by enolase (Fig. 5.6). The resulting 3H2O diffuses freely into the cell culture media and is quantified by scintillation counting. The complete conversion of glucose to pyruvate is not measured by this reaction.
Figure 5.6. Glycolytic flux measurements with [5-13H]-glucose.
A single tritium present on 5C of glucose is released as water in the ninth step of glycolysis catalyzed by enolase.
For detection of 3H2O using a liquid scintillation counter, the [5-3H]-glucose must be removed from the cell supernatant. One method for separation is to allow 3H2O to diffuse into a cold H2O solution while inhibiting the diffusion of [5-3H]-glucose (Vander Heiden et al., 2011). An alternative method is to use ion chromatography to remove the glucose from the cell supernatant (Støttrup et al., 2010).
4.3.1 Protocol for measuring the release of 3H2O from [5-3H]-glucose
Wash 1 × 106 cells with 1 × PBS, pH 7.4, and resuspend in 1 ml Krebs buffer (126 mM NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.2 mM NaH2PO4, 1.2 mM MgCl2, and 2.5 mM CaCl2). Incubate at 37 °C, 5% CO2 for 30 min and then replace with Krebs buffer containing 10 µCi [5-3H]-glucose (Perkin Elmer #NET531005MC) and 10 mM unlabeled glucose. Incubate at 37 °C, 5% CO2 for 1 h. Spin down the sample and remove 500 µl of cell supernatant. Add 500 µl of 0.2 N HCl. Pack a 2-ml Poly-Prep chromatography column (Biorad #731-1550) with 2 ml of AG 1-X8 resin, 200–400 mesh, formate form (Biorad #140-1454). Wash the resin with 20 ml of 1 N NaOH and then with 10 ml of distilled water. Add 1 ml of supernatant solution to the column and collect the sample after glucose removal. Dissolve 3H2O into scintillation solution and quantify by beta-scintillation counting (Støttrup et al., 2010; Vander Heiden et al., 2010).
5. SUMMARY
Each of the discussed methods provides data on the glycolytic flux in mammalian cells, including cancer. Selecting which technique or combinations of techniques are appropriate for a given study depends on equipment availability, budget, and the type of information required. Extracellular glucose and lactate measurements and enzymatic activity assays can be performed with standard laboratory equipment but provide a limited amount of insight. Metabolomics approaches provide data on glycolysis as well as its offshoot pathways but require expensive equipment and laborious data analyses (Table 5.2). All of these methods can be used in combination to obtain a comprehensive assessment of glycolytic flux and downstream metabolic pathways in cancer cells.
Table 5.2.
Glucose tracers used to measure glycolysis and related metabolic pathways
| Detection precision |
|||||
|---|---|---|---|---|---|
| Glucose tracer | Detection method | Glycolysis | PPP | TCA cycle | Price |
| [U-13C6]-glucose | Mass spectrometry | Low | Low | High | $ |
| [1,2-13C2]-glucose | Mass spectrometry | High | High | Ok | $$ |
| [2-13C1]-glucose | Mass spectrometry | High | High | Ok | $$ |
| [5-3H]-glucose | Scintillation counter | High | No | No | $$$ |
PPP, pentose phosphate pathway.
ACKNOWLEDGMENTS
T. T. is supported by a Ruth L. Kirschstein National Research Service Award GM007185. M. A. T. is supported by CIRM grant RB1-01397, UC Discovery Awards Bio07-10663 and 178517, and NIH Grants GM073981, P01GM081621, CA156674, and CA90571.
REFERENCES
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. Omics: A Journal of Integrative Biology. 2002;6(3):217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. http://dx.doi.org/10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Carbonell J, Marco R, Felíu JE, Sols A. Pyruvate kinase. European Journal of Biochemistry. 1973;37(1):148–156. doi: 10.1111/j.1432-1033.1973.tb02969.x. [DOI] [PubMed] [Google Scholar]
- Caro P, Kishan Amar U, Norberg E, Stanley IA, Chapuy B, Ficarro Scott B, et al. Metabolic signatures uncover distinct targets inmolecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22(4):547–560. doi: 10.1016/j.ccr.2012.08.014. http://dx.doi.org/10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. http://dx.doi.org/10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. http://dx.doi.org/10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. http://dx.doi.org/10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. http://dx.doi.org/10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Dietmair S, Timmins NE, Gray PP, Nielsen LK, Krömer JO. Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol. Analytical Biochemistry. 2010;404(2):155–164. doi: 10.1016/j.ab.2010.04.031. http://dx.doi.org/10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. http://dx.doi.org/10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Hansen PA, Gulve EA, Holloszy JO. Suitability of 2-deoxyglucose for in vitro measurement of glucose transport activity in skeletal muscle. Journal of Applied Physiology. 1994;76(2):979–985. doi: 10.1152/jappl.1994.76.2.979. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Elf S, Fan J, Kang H-B, Seo Jae H, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22(5):585–600. doi: 10.1016/j.ccr.2012.09.020. http://dx.doi.org/10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature Biotechnology. 2013;31(6):522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595. http://dx.doi.org/10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins AB, Furler SM, Kraegen EW. 2-Deoxy-D-glucose metabolism in individual tissues of the rat in vivo. International Journal of Biochemistry. 1986;18(4):311–318. doi: 10.1016/0020-711x(86)90036-4. http://dx.doi.org/10.1016/0020-711X(86)90036-4. [DOI] [PubMed] [Google Scholar]
- Jiang S, Zhang L-F, Zhang H-W, Hu S, Lu M-H, Liang S, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. The EMBO Journal. 2012;31(8):1985–1998. doi: 10.1038/emboj.2012.45. http://dx.doi.org/10.1038/emboj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane Andrew N, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metabolism. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. http://dx.doi.org/10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. Journal of Biological Chemistry. 2011;286(29):25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of the M2 isoform of pyruvate kinase is an adverse prognostic factor for signet ring cell gastric cancer. World Journal of Gastroenterology: WJG. 2012;18(30):4037–4043. doi: 10.3748/wjg.v18.i30.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nature Reviews Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics. 2011;43(9):869–874. doi: 10.1038/ng.890. http://dx.doi.org/10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang X-L, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metabolism. 2012;15(6):827–837. doi: 10.1016/j.cmet.2012.05.001. http://dx.doi.org/10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComb RB, Bond LW, Burnett RW, Keech RC, Bowers GN. Determination of the molar absorptivity of NADH. Clinical Chemistry. 1976;22(2):141–150. [PubMed] [Google Scholar]
- Mertens K, Mees G, Lambert B, Van de Wiele C, Goethals I. In vitro 2-deoxy-2-[18F] fluoro-D-glucose uptake: Practical considerations. Cancer Biotherapy and Radiopharmaceuticals. 2012;27(3):183–188. doi: 10.1089/cbr.2011.1125. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Systems Biology for Biotechnological Innovation. 2009;144(3):167–174. doi: 10.1016/j.jbiotec.2009.07.010. http://dx.doi.org/10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely JR, Denton RM, England PJ, Randle PJ. The effects of increased heart work on the tricarboxylate cycle and its interactions with glycolysis in the perfused rat heart. The Biochemical Journal. 1972;128:147–159. doi: 10.1042/bj1280147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger principles of biochemistry. New York: WH Freeman; 2008. [Google Scholar]
- O’Neil RG, Wu L, Mullani N. Uptake of a fluorescent deoxyglucose analog (2-NBDG) in tumor cells. Molecular Imaging and Biology. 2005;7(6):388–392. doi: 10.1007/s11307-005-0011-6. [DOI] [PubMed] [Google Scholar]
- Palmieri D, Fitzgerald D, Shreeve SM, Hua E, Bronder JL, Weil RJ, et al. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Molecular Cancer Research. 2009;7(9):1438–1445. doi: 10.1158/1541-7786.MCR-09-0234. http://dx.doi.org/10.1158/1541-7786.MCR-09-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Analytical and Bioanalytical Chemistry. 2007;387(2):525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- Patra Krushna C, Wang Q, Bhaskar Prashanth T, Miller L, Wang Z, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24(2):213–228. doi: 10.1016/j.ccr.2013.06.014. http://dx.doi.org/10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S-Y, Lai P-L, Pan H-W, Hsiao L-P, Hsu H-C. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncology Reports. 2008;19(4):1045–1053. [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292(5516):504–507. doi: 10.1126/science.1058079. http://dx.doi.org/10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. http://dx.doi.org/10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L-E, Wittmann C, Nielsen L, Krömer J. OpenFLUX: Efficient modelling software for 13C-based metabolic flux analysis. Microbial Cell Factories. 2009;8(1):25. doi: 10.1186/1475-2859-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Richter J, Mehta S, Gribble A, Sutherland-Smith A, Stowell K, et al. Expression and role in glycolysis of human ADP-dependent glucokinase. Molecular and Cellular Biochemistry. 2012;364(1 – 2):131–145. doi: 10.1007/s11010-011-1212-8. http://dx.doi.org/10.1007/s11010-011-1212-8. [DOI] [PubMed] [Google Scholar]
- Sellick CA, Hansen R, Stephens GM, Goodacre R, Dickson AJ. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nature Protocols. 2011;6(8):1241–1249. doi: 10.1038/nprot.2011.366. http://dx.doi.org/10.1038/nprot.2011.366. [DOI] [PubMed] [Google Scholar]
- Smith TA. Mammalian hexokinases and their abnormal expression in cancer. British Journal of Biomedical Science. 2000;57(2):170–178. [PubMed] [Google Scholar]
- Støttrup NB, Løfgren B, Birkler RD, Nielsen JM, Wang L, Caldarone CA, et al. Inhibition of the malate–aspartate shuttle by pre-ischaemic aminooxyacetate loading of the heart induces cardioprotection. Cardiovascular Research. 2010;88(2):257–266. doi: 10.1093/cvr/cvq205. http://dx.doi.org/10.1093/cvr/cvq205. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, et al. Identification of small molecule inhibitors of pyruvate kinase M2. Biochemical Pharmacology. 2010;79(8):1118–1124. doi: 10.1016/j.bcp.2009.12.003. http://dx.doi.org/10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harbor Symposia on Quantitative Biology. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chem Bio Chem. 2005;6(11):1941–1951. doi: 10.1002/cbic.200500151. http://dx.doi.org/10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Weitzel M, Nöh K, Dalman T, Niedenführ S, Stute B, Wiechert W. 13CFLUX2—High-performance software suite for 13C-metabolic flux analysis. Bioinformatics. 2013;29(1):143–145. doi: 10.1093/bioinformatics/bts646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE. Hexokinases. Reviews of physiology, biochemistry and pharmacology. Vol. 126. Berlin, Germany: Springer Berlin – Heidelberg; 1995. pp. 65–198. [DOI] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American Journal of Physiology Cell Physiology. 2007;292(1):C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337(6097):975–980. doi: 10.1126/science.1222278. http://dx.doi.org/10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Fischer E, Sauer U. FiatFlux—A software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics. 2005;6(1):209. doi: 10.1186/1471-2105-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes & Development. 2011;25(10):1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Wang Y, Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. Journal of Biochemical and Biophysical Methods. 2005;64(3):207–215. doi: 10.1016/j.jbbm.2005.08.001. [DOI] [PubMed] [Google Scholar]