Abstract
Mutations in the myocilin gene (MYOC) account for 10% of juvenile open-angle glaucoma cases and 3–4% of adult onset primary open-angle glaucoma cases. It is a secreted glycoprotein found in many ocular and non-ocular tissues and has been linked to elevated intraocular pressure. In human trabecular meshwork (HTM) cells, MYOC expression can be induced by the glucocorticoid dexamethasone (DEX). In this study we examined the role of the calcineurin/NFATc1 (Nuclear Factor of Activated T-cells) pathway in the DEX induction of MYOC in HTM cells. In post-confluent HTM cells treated with either 500 nM DEX or 0.1% ethanol (EtOH; vehicle control) for 0–6 days both protein and mRNA levels of MYOC were increased while DEX was present. The protein and mRNA levels remained elevated for an additional 12 days after the removal of DEX. Only 1 day of DEX treatment was sufficient to trigger a sustained increase in MYOC mRNA that lasted for 4 days after the removal of DEX. Similar to other studies, myocilin protein expression was not seen until the second day of DEX treatment while mRNA increased within one day of DEX indicating that this is a secondary glucocorticoid response. To determine if MYOC gene expression was regulated by calcineurin/NFATc1, HTM cells were pre-treated for 1 h with the calcineurin inhibitors cyclosporin A or INCA-6 prior to the addition of DEX or EtOH for 2 days. NFATc1 siRNA was used to determine if NFATc1 was required for MYOC mRNA expression. Cells were also treated with the ionophone ionomycin to determine if increased cytosolic calcium affected MYOC expression. These studies showed that the DEX induced increase in MYOC mRNA could be inhibited with either CsA or INCA-6 or by transfection with NFATc1 siRNA and that ionomycin was unable to increase MYOC mRNA. Immunofluorescence microscopy was also performed to determine if DEX caused the nuclear translocation of NFATc1. Immunostaining showed that NFATc1 relocated to the nucleus within 15 min of DEX treatment and remained there for up to 2 h. The data suggest that the DEX-induced increase in MYOC expression activates a calcineurin and NFATc1 pathway in a calcium independent mechanism.
Keywords: glucocorticoids, glaucoma, trabecular meshwork, calcineurin, NFATc1, myocilin
1. INTRODUCTION
Glaucoma is a heterogeneous disease characterized by the progressive degeneration of the optic nerve that eventually leads to irreversible blindness. The most common form of glaucoma in the United States is primary open angle glaucoma (POAG), affecting more than 60 million people worldwide (Quigley, 1996). A major risk factor for POAG is increased intraocular pressure (IOP). Although numerous studies indicate that alterations in the conventional outflow pathway are largely responsible for the elevation in IOP, the molecular and cellular mechanisms responsible are still unknown. To date genetic studies have indicated that there are 4 genes linked to adult onset POAG: MYOC (myocilin), WDR36 (WD Repeat Domain 36), OPTN (optineurin), and NTF4 (neutrophin-4) (Fan and Wiggs, 2010; Takamoto and Araie, 2014).
MYOC was one of the first proteins to be linked to glaucoma. It was originally identified because its expression in human trabecular meshwork (HTM) cells can be increased with the glucocorticoid dexamethasone (DEX) (Nguyen et al., 1998; Polansky et al., 1997). Thus, it is thought to play a role in both POAG and steroid-induced glaucoma which clinically mirrors POAG. Mutations in MYOC occur in 10% of juvenile open-angle glaucoma cases and in 3–4% of adult onset POAG cases (Fingert et al., 1999; Fingert et al., 2002; Kwon et al., 2009; Stone et al., 1997). Increasing evidence suggests that mutations in the MYOC gene cause glaucoma through a gain of pathogenic function (Kim et al., 2001; Lam et al., 2000) which prevents MYOC from being secreted from the cell. As a result MYOC accumulates within the endoplasmic reticulum of the cell where it causes endoplasmic reticulum stress, impairing trabecular meshwork cell function and viability (Joe et al., 2003; Wang et al., 2007; Zode et al., 2011).
MYOC is a secreted glycoprotein that is expressed in many structures of the eye, including the trabecular meshwork, iris, ciliary body, sclera, choroid, cornea, lamina cribosa, retina and optic nerve (Adam et al., 1997; Kubota et al., 1997; Ortego et al., 1997; Ricard et al., 2001; Tamm et al., 1999). The function of MYOC is not clear but it may play a role in cell-extracellular matrix interactions (Goldwich et al., 2009; Peters et al., 2005), cell migration (Kwon and Tomarev, 2011) and mitrochondrial function (Sakai et al., 2007). In skeletal muscle, MYOC is part of the dystrophin-associated protein complex by binding α1-syntrophin and may play a role as a regulator of muscle hypertrophy pathways (Joe et al., 2012). Recently, it was shown that MYOC can bind and activate ErbB2/ErbB3 in the sciatic nerve implicating a role for MYOC in myelination in the peripheral nervous system (Kwon et al., 2013).
In addition to DEX, MYOC expression can also be induced in HTM cells with transforming growth factor-β1 (TGF-β1) (Tamm et al., 1999), optineurin (Park et al., 2007), and mechanical stretch (Tamm et al., 1999). The induction of MYOC by both DEX and TGF-β1 is a delayed response, taking days rather than hours to see both MYOC mRNA and protein levels increase (Shepard et al., 2001; Tamm et al., 1999). This delayed response to stimuli is thought to be a secondary response as it requires new protein synthesis of an unidentified factor(s) for induction. Analysis of nucleotides upstream of the MYOC transcription start site support this idea because it failed to identify a functional glucocorticoid response element (Kirstein et al., 2000; Shepard et al., 2001). Recent studies examining how DEX regulates the expression of proteins in the TM show that MYOC is not the only protein up regulated as a result of a secondary glucocorticoid response. The β3 integrin subunit in HTM cells is also up regulated by DEX (Faralli et al., 2013) and this study showed that a calcineurin/NFAT (nuclear factor of activated T-cells) pathway may be involved. Calcineurin is a serine/threonine phosphatase that is modulated by intracellular calcium levels. Upon activation, calcineurin can activate the NFAT family of transcription factors through dephosphorylation (Clipstone and Crabtree, 1992; Emmel et al., 1989). Once dephosphorylated, the NFATs translocate from the cytoplasm to the nucleus where they bind DNA in conjunction with other transcription factors such as AP-1 to induce gene transcription (Chen et al., 1998). There are 5 NFAT family members, NFATc1-5, but only NFATc1-4 are regulated by calcium signaling.
In this study we examined whether the DEX-induced MYOC expression in HTM cells also involved a calcineurin/NFAT pathway. We showed that DEX increased MYOC protein and mRNA levels within 1–2 days and that just one day of DEX treatment was sufficient to induce the increase in mRNA. The increase in MYOC mRNA expression by DEX was dependent on NFATc1 activation by calcineurin, suggesting that NFATc1 may induce the transcription of a gene necessary for MYOC expression in HTM cells. Understanding how MYOC expression is regulated provides potential targets for the treatment of glaucoma.
2. METHODS
2.1 Cell Culture
HTM cells were isolated from cadaver eyes of a 17 (HTM17), 25 (HTM25) and 27-year old (HTM27) donor with no known history of ocular disease in accordance to the tenets of the Declaration of Helsinki. The cells used for each experiment is indicated in the figure legend. The cells were characterized to be HTM cells based on several criteria, as previously described (Alvarado et al., 1982; Filla et al., 2004). These include a cobblestone morphology in stable differentiated postconfluent monolayers, the ability to form cross-linked actin networks (CLANs), the upregulation of MYOC by DEX and the presence of several proteins (fibronectin, collagen IV, laminin, ZO-1, β-catenin and smooth muscle actin) Cells were cultured as previously described in low glucose DME supplemented with 15% FBS (Atlanta Biologicals), 2% L-glutamine, 1% amphotericin B (Thermo Fisher Scientific), 0.05% gentamycin and 1 ng/ml FGF-2 (Peprotech) (Filla et al., 2004). One week after reaching confluency and for all subsequent experiments, serum was reduced to 10% FBS and FGF-2 treatment was stopped. Cells were then treated with either 500 nM DEX or 0.1% ethanol (EtOH; vehicle control). In some experiments, cells were incubated with 1 or 10 μM cyclosporin A (CsA) or 40, 80 or 120 μM INCA-6 (Tocris Bioscience) for 1 h prior to the addition of DEX or EtOH and incubated for 2 days. Fresh CsA, INCA-6, DEX and EtOH were always added after 24 h. Other times 1 or 5 μM ionomycin was added to the media for 2 days. All reagents were obtained from Sigma-Aldrich unless otherwise noted.
2.2 Western Blot Analysis
Western blot analyses were performed as previously described (Faralli et al., 2013) with a few modifications. Briefly, HTM cells were lysed with lysis buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1% NP-40, 0.25% deoxycholate, HALT phosphatase inhibitor cocktail and HALT protease inhibitor cocktail (Thermo Fisher Scientific)). Proteins in the cell lysate (10 μg) were separated on a 10% SDS-PAGE and transferred to Immobilon-FL membrane (Millipore Corp.). The membrane was blocked overnight with 3% bovine serum albumin (BSA) in Tris buffered saline (TBS) containing 0.1% Tween-20 and then incubated with either MYOC monoclonal antibody clone 1.1 (1:1000), or succinate dehydrogenase complex, subunit A (SDHA) monoclonal antibody (Abcam; 1:5000). Antibodies were diluted in 1% BSA/TBS/0.1% Tween-20 and incubated for 1 h at room temperature. The MYOC monoclonal antibody was kindly provided by Dr. Michael Fautsch (Dept. Ophthalmology, Mayo Clinic, Rochester, MN). Membranes were washed and then incubated for 1 h with IRDye 800 conjugated goat anti-mouse or anti-rabbit secondary antibody (Licor; 1:15000) in 1% BSA/TBS/0.1% Tween-20/0.01% SDS. Bound antibody was detected using the Licor Odyssey infrared imaging system using Image Studio Ver. 2.1 (Licor).
2.3 RNA Isolation, Reverse Transcription and Real-Time (RT)-qPCR
Total RNA was isolated using the QIAshredder and RNeasy Plus Mini Kits (Qiagen Inc., Valencia, CA) and RNA concentration was determined using a NanoDrop spectrophotometer. Total RNA (1 μg) was reverse transcribed and RT-qPCR experiments using the synthesized cDNA were performed as previously described (Clark et al., 2013). Primers pairs used were: MYOC forward 5′-GCCCATCTGGCTATCTCAGG-3′ and reverse 5′-CTCAGCGTGAGAGGCTCTCC-3′ and ITGB1 (β1 integrin) forward 5′-GTGGAGAATCCAGAGTGTCCCA-3′ and reverse 5′-GACCACAGTTGTTACGG-3′. Relative quantification of the RT-qPCR data was performed according to Pfaffl (Pfaffl, 2001), using ITGB1 mRNA for normalization. We have determined from previous experiments that ITGB1 mRNA levels are unaffected by the treatments used in this study (Faralli et al., 2013). Statistical comparisons were done using the Student T-test and a p value < 0.05 was considered significant.
To detect NFATc1-4 mRNA expression, cDNA synthesized from untreated HTM cells as described above was used. The cDNA was PCR amplified using 1 U DNA polymerase (Platinum Taq; Invitrogen) with 1.5 mM MgCl2, 125 μM each dNTP and 5 μM gene-specific primer pairs. Primers used were: NFATc1 forward 5′-TGCAAGCCGAATTCTCTGGT-3′ and reverse 5′-CTTTACGGCGACGTCGTTTC-3′ (227 bp fragment), NFATc2 forward 5′-GAGGGGCTGTCAAAGCTCC-3′ and reverse 5′-ACAGTTTTCCCCGTGATTCGG-3′ (162 bp fragment), NFATc3 forward 5′-GCTCGACTTCAAACTCGTCTT-3′ and reverse 5′-GATGCACAATCATCTGGCTCA-3′ (95 bp fragment), and NFATc4 forward 5′-CTTCTCCGATGCCTCTGACG-3′ and reverse 5′-CGGGGCTTGGACCATACAG-3′ (172 bp fragment). The PCR program used was 95°C for 1 min, followed by 35 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec. The products were extended for an additional 5 min at 72°C then separated on a 3% agarose gel and stained with ethidium bromide.
2.4 NFATc1 Knockdown
HTM cells were plated into 6 well plates and grown to confluency as described above. Human NFATc1 siRNA (ON-TARGETplus SMART pool L-003605-00-005) and control siRNA (ON-TARGETplus SMART pool Non-targeting D-001810-10-005) were obtained from Dharmacon/Thermo Fisher Scientific. HTM cells were transfected with 25 nM siRNA using a siRNA transfection reagent (TransIT-TKO reagent; Mirus) according to the manufacturer’s protocol. After 24 h, the media was replaced and DEX or EtOH was added to the appropriate wells and cells were incubated for an additional 48 h. Total RNA was isolated and cDNA was generated as described above. RT-qPCR experiments using the synthesized cDNA were performed as previously described (Clark et al., 2013). The same MYOC and ITGB1 primers were used as described above in addition to NFATc1-specific primers: forward 5′-TGCAAGCCGAATTCTCTGGT-3′ and reverse 5′-CTTTACGGCGACGTCGTTTC-3′. Data were analyzed as described above.
2.5 Immunofluorescence Microscopy
Confluent monolayers of HTM27 cells were treated for 10 days with 500 nM DEX or 0.1% EtOH for 10 days then washed with ice cold PBS and fixed for 15 min in 100% MeOH at −20°C prior to blocking for 1 h with 1% BSA in PBS. Cells were then incubated with a rabbit polyclonal antibody against MYOC followed by Alexa 488 conjugated goat anti-rabbit antibody (Molecular Probes, 1:500). Coverslips were mounted onto slides using Immu-Mount (Shandon) and viewed using an epifluorescence microscope (Axioplan 2; Zeiss) equipped with a digital camera (Axiocam HRm; Zeiss) and image acquisition software (Axiovision ver. 4.5; Zeiss).
For the NFAT localization studies, subconfluent, proliferating HTM cells were plated onto glass coverslips in a 24 well plate at 30,000 cells per well. The next day cells were treated with 500 nM DEX or 0.1% EtOH for 0, 15 min, 30 min, 1 h or 2 h. As a positive control, some cells were treated with 1 μM ionomycin (Sigma-Aldrich) for 1 h. The cells were then washed, fixed and blocked as above and incubated with an NFATc1 monoclonal antibody (Millipore clone 7A6; 1:50) diluted in 0.1% BSA/PBS overnight at 4°C. Cells were then incubated with Alexa 546 conjugated goat anti-mouse antibody (Molecular Probes; 1:500) in 0.1% BSA/PBS for 1 h, mounted and viewed as described above. NFATc1 nuclear localization was quantified by counting the number of cells with nuclei positive for NFATc1 antibody labeling from 15 different fields of view for each condition for each experiment performed (n=3). The percentage of cells with NFATc1 localization versus total number of cells was determined. Data are presented as mean ± S.E.M. Statistical comparisons were done using the Student T-test and a p value < 0.05 was considered significant.
3. RESULTS
3.1 DEX induces MYOC protein and mRNA expression
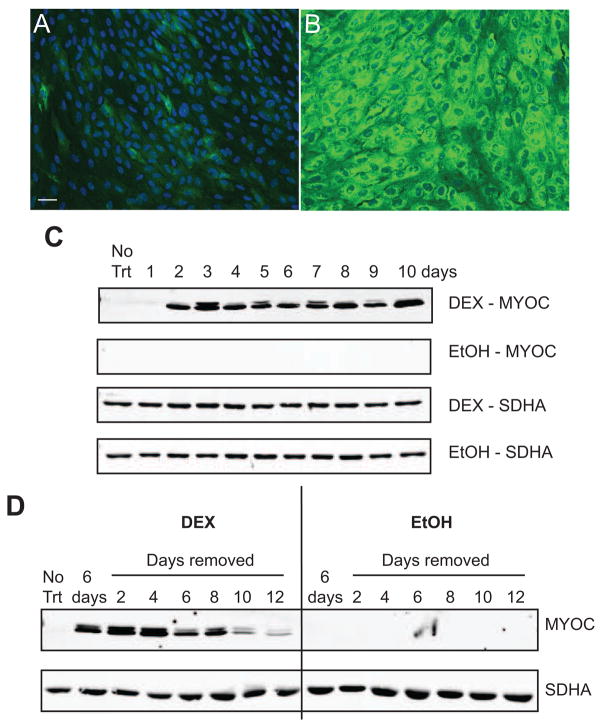
Immunofluorescence microscopy of HTM cells show that there was a low basal level of MYOC expression in EtOH treated cells that increased to 100% expression when cells were treated with DEX (Fig. 1A and B). Similar to other studies using HTM cells (Clark et al., 2001; Shepard et al., 2001), DEX induced the expression of MYOC in a time-dependent manner. Figure 1C shows that protein levels of MYOC could be detected starting 2 days after initiation of DEX treatment and remained high as long as DEX was present (10 days total). MYOC was not detected in the EtOH control lysates, presumably because the level of expression was below the detection level on the Western blots when equal amounts of cell lysate were loaded. Upon removal of DEX after 6 days of treatment, Western blot analysis showed that MYOC expression remained high for 4 days after DEX removal (Fig. 1D) then slowly decreased to near baseline levels 12 days after DEX removal.
Figure 1.
Please change DEX to EtOH and EtOH to DEX so it should read: DEX increases MYOC expression. HTM27 cells were treated with EtOH (A) or DEX (B) for 10 days prior to methanol fixation and labeled with a rabbit polyclonal antibody against MYOC (green) and Hoescht 33342 to label nuclei (blue). Images were taken with the same exposure time. Images of DEX treated cells were deliberately overexposed in order to show the basal levels of MYOC expression. Scale bar = 50 μm. (C) HTM27 cells were treated with DEX or EtOH for 10 days. Cell lysates were collected daily and analyzed for MYOC and SDHA (loading control) expression. (D) HTM27 cells were treated with DEX or EtOH for 6 days followed by media alone for 12 days. Lysate was collected every other day after DEX or EtOH removal and analyzed for the expression of MYOC or SDHA. Blots are representative of 2 biological replicates.
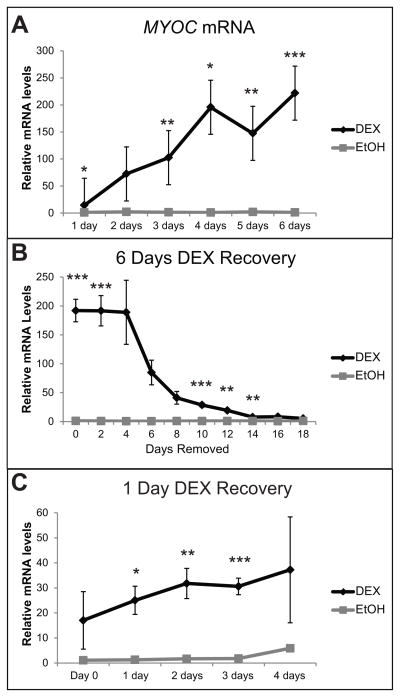
Using RT-qPCR, we found that DEX significantly increased the expression of MYOC mRNA by 13.6 fold (p<0.05) after only 1 day of DEX treatment compared to no treatment (Fig. 2A). The amount of MYOC mRNA continued to increase significantly each day of DEX treatment. After 6 days of DEX treatment there was a 182 fold increase in MYOC mRNA over no treatment, which was significantly higher than the EtOH control (p<0.02). In addition, MYOC mRNA levels remained elevated for up to 14 days after removal of DEX from the medium (Fig. 2B). Interestingly, just one day of DEX exposure was sufficient to induce a prolonged elevation in MYOC mRNA levels in the absence of DEX. As shown in Figure 2C, MYOC mRNA levels increased 17-fold over no treatment after 1 day of DEX treatment (Day 0) and was 30-fold higher than no treatment (p<0.02 compared to EtOH control) 3 days after DEX removal. MYOC mRNA levels remained 6.3 fold higher compared to no treatment 4 days after DEX removal, but this was not significantly different from the EtOH control.
Figure 2.
DEX treatment increases MYOC mRNA. RT-qPCR was performed on HTM27 cells treated with DEX or EtOH. Ct values were corrected for primer efficiency and compared to cells with no treatment as described by Pfaffl (Pfaffl, 2001). Data were then normalized to ITGB1 mRNA (β1 integrin housekeeping gene). (A) Levels of MYOC mRNA in the presence of DEX or EtOH for 1–6 days. (B) Levels of MYOC mRNA in HTM cells treated with DEX or EtOH for 6 days (Day 0) followed by 18 days with no DEX or EtOH. (C) Levels of MYOC mRNA in HTM cells treated with DEX or EtOH for 1 day only (Day 0) followed by 4 days with no DEX or EtOH. n = 3 biological replicates. DEX significantly different from EtOH *p<0.05, **p<0.04, ***p<0.02.
3.2 DEX induction of MYOC involves Calcineurin and NFATc1
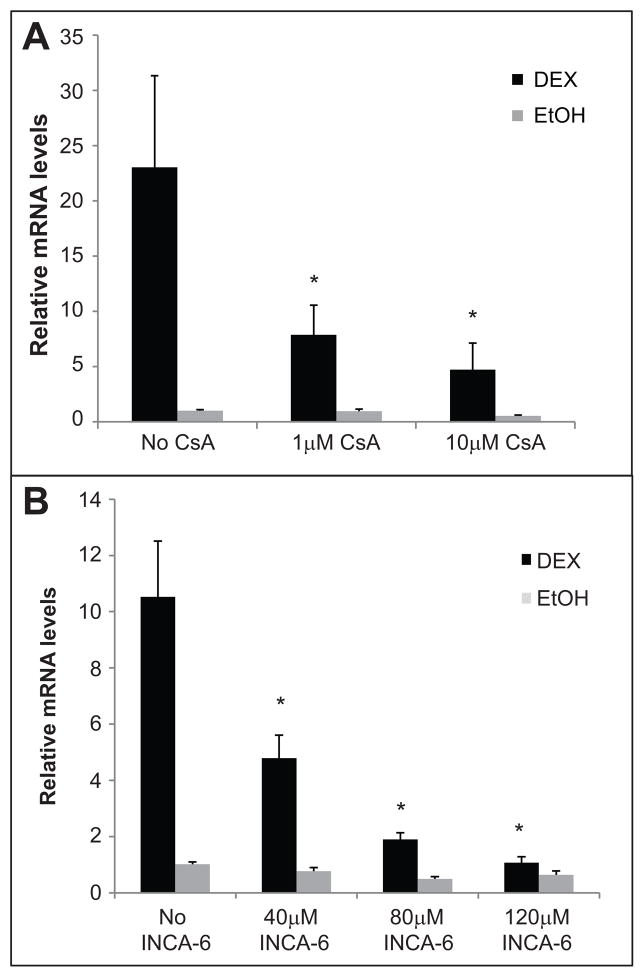
Our previous work showed that, similar to MYOC, ITGB3 (β3 integrin subunit) mRNA expression induced by DEX was also a secondary glucocorticoid response. This increase was inhibited with cyclosporin A (CsA) (Faralli et al., 2013), an immunosuppressant that forms a complex with cyclophilin leading to an inhibition of calcineurin phosphatase activity (Liu et al., 1991). To determine if CsA could also inhibit the DEX-induced increase in MYOC mRNA, we pre-treated HTM cells with 1 or 10 μM CsA for 1 h prior to the addition of DEX or EtOH. For these studies we used HTM cells from three different donors to rule out cell strain variability. Figure 3A shows that CsA significantly reduced the increase in MYOC mRNA caused by DEX treatment (p<0.05). At the highest dose of CsA tested (10 μM), MYOC mRNA expression induced by DEX was significantly reduced by 71% compared to DEX alone (p<0.05; Table 1). We also tested a more selective calcineurin inhibitor called INCA-6. INCA-6 specifically prevents calcineurin from binding NFAT, but does not affect calcineurin activity for other substrates (Roehrl et al., 2004). Figure 3B shows that INCA-6 significantly reduced the increase in MYOC mRNA caused by DEX treatment with all 3 doses tested (40, 80, 120 μM; p<0.05) in all cell strains tested. These data suggested that the DEX induced activation of calcineurin was involved in the upregulation of MYOC.
Figure 3.
CsA and INCA-6 inhibit the DEX-induced expression of MYOC mRNA. RT-qPCR was performed on 3 separate HTM cell lines (HTM17, HTM25 and HTM27). All cell strains had similar results so the data was averaged together. Individual cell strain data is shown in Table 1. Cells were pre-treated for 1 h with 1 or 10 μM CsA (A) or 40, 80 or 120 μM INCA-6 (B) prior to the addition of DEX or EtOH and incubated for 2 days. Fresh CsA, INCA-6, DEX and EtOH was added after 1 day. Ct values were corrected for primer efficiency and compared to cells with no treatment. Data were then normalized to ITGB1 mRNA (β1 integrin housekeeping gene). n = 4 biological replicates. DEX significantly different from DEX + 1 or 10 μM CsA (A) and from DEX + 40, 80 or 120 μM INCA-6 (B) *p<0.05.
Table 1.
Individual Cell Strain Data
| Cell Strain | Average | Avg. % Decrease | |||
|---|---|---|---|---|---|
| HTM27 | HTM25 | HTM17 | |||
| DEX | 100 | 100 | 100 | ||
| DEX + 40μM INCA-6 | 43 | 65 | 35 | 48 ± 9% | 52% |
| DEX + 80μM INCA-6 | 12 | 27 | 29 | 22 ± 5% | 78% |
| DEX + 120μM INCA-6 | 8 | 7 | 22 | 12 ± 5% | 88% |
| DEX + 1μM CsA | 29 | 77 | 87 | 64 ± 18% | 36% |
| DEX + 10μM CsA | 19 | 34 | 34 | 29 ± 5% | 71% |
Table represents the relative amount of MYOC mRNA levels compared to DEX treatment alone (set to 100%) for individual cell strains tested. Figure 3 shows the average of all 3 cell strains.
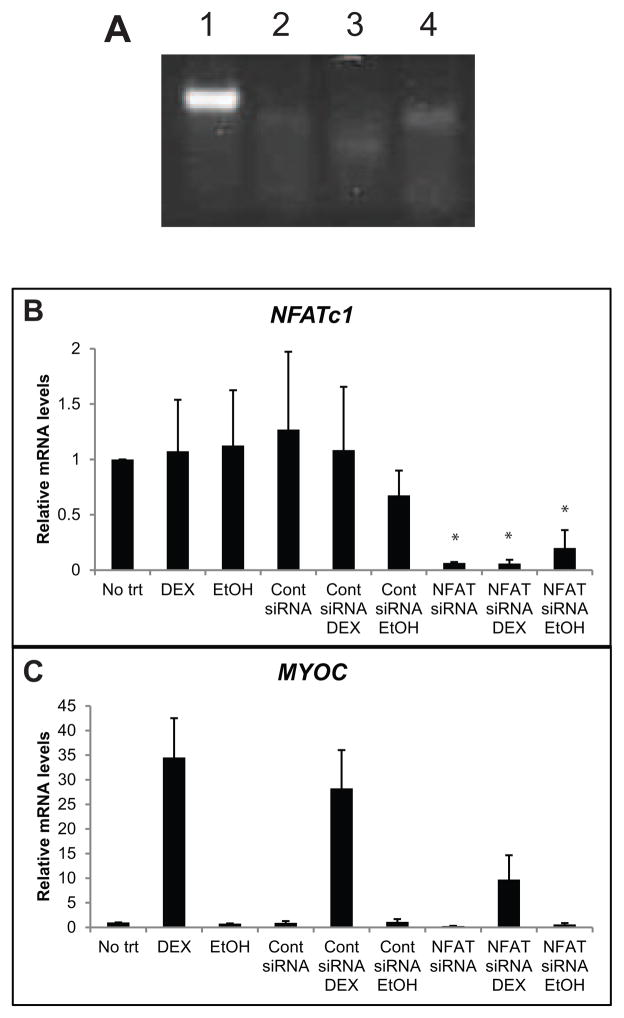
Since, calcineurin is known to activate the NFAT family of transcription factors and cause their relocation to the nucleus, we then investigated whether NFAT was involved in the DEX induced expression of MYOC. To determine which NFATs were expressed in HTM cells we performed PCR analysis using primers specific for NFATc1-4. Figure 4A shows that HTM cells express only NFATc1. To demonstrate that NFATc1 is required for DEX induction of MYOC transcription, we used siRNA directed against NFATc1. As shown in Figure 4B, NFATc1 siRNA decreased NFATc1 mRNA levels by 94% after 72 h (p<0.05), and this decrease was not affected by the presence of DEX or EtOH. This reduction in NFATc1 mRNA levels attenuated the DEX induced increase in MYOC transcription by 72% (Figure 4C), while control siRNA had no effect.
Figure 4.
(A) NFAT expression in HTM27 cells. cDNA was made from HTM cell mRNA. PCR was performed using primers specific for NFATc1 (lane 1), NFATc2 (lane 2), NFATc3 (lane 3) and NFATc4 (lane 4). (B, C) NFATc1 mediates the DEX-induced increase in MYOC mRNA in HTM cells. HTM27 cells were transfected with siRNA directed against NFATc1. 24 h later cells were treated with DEX or EtOH for 48 h. NFATc1 levels (A) or MYOC levels (B) were determined using RT-qPCR 72 h after transfection. Ct values were corrected for primer efficiency and compared to cells with no treatment. Data were then normalized to ITGB1 mRNA. n = 3 biological replicates. Significantly different from no trt, *p<0.05.
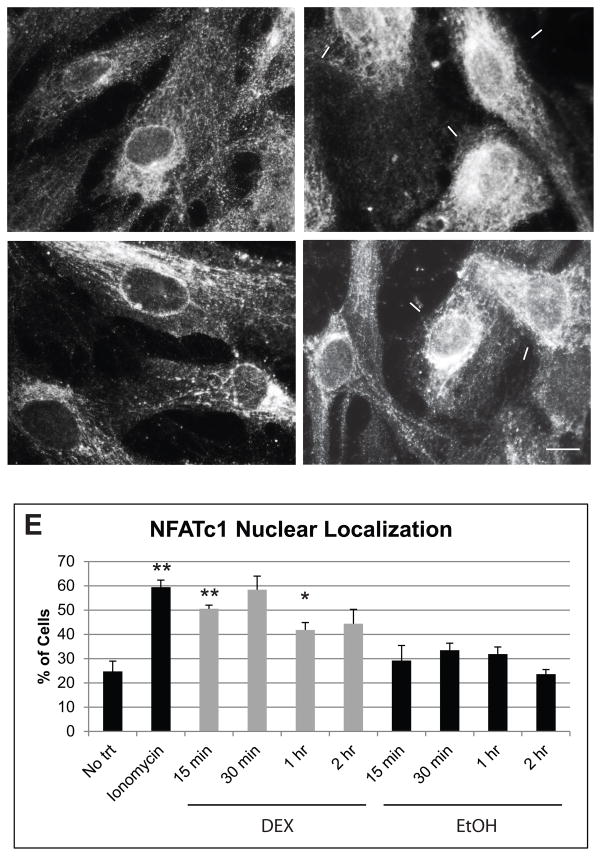
We next performed immunofluorescence microscopy to determine if DEX treatment caused the nuclear translocation of NFATc1 over time. We also treated cells with the Ca2+ ionophore known as ionomycin as a positive control since calcineurin can be activated by an increase in intracellular calcium. Figure 5 shows that NFATc1 was translocated to the nucleus in cells treated with both ionomycin and DEX (Fig. 5B and D respectively), but not in EtOH treated control cells (Fig. 5C). The number of cells with NFATc1 nuclear localization was quantified and shown in Figure 5E. Only 25% of the no treatment control cells showed a nuclear localization of NFATc1. However, if cells were treated with ionomycin, 60% showed a nuclear translocation of NFATc1. We saw a similar increase in cells treated with DEX for 15 min. A 15 min treatment with DEX resulted in 50% of the cells with nuclear labeling for NFATc1, which was significantly different from the no treatment group (p<0.02) and from HTM cells treated with EtOH as a control (p<0.05). At 30 min, 58% of cells treated with DEX showed nuclear localization of NFATc1, which was significantly different from the EtOH control (p<0.02). After 30 min, the number of cells with nuclear localization of NFATc1 following DEX treatment began to decrease, but was still higher than the EtOH controls. Since DEX treatment did not increase NFATc1 translation or transcription as determined by western blotting and RT-qPCR (data not shown), these data together with the siRNA data, suggests that a DEX induced activation of NFATc1 was involved in the increase in MYOC transcription. These also suggest that DEX is utilizing a calcineurin/NFATc1 pathway to upregulate MYOC expression, since calcineurin is the only phosphatase known to activate NFATc1.
Figure 5.
DEX treatment causes the translocation of NFATc1 to the nucleus. HTM27 cells were plated onto coverslips then treated with ionomycin for 1 h or DEX or EtOH for 15 min, 30 min, 1 h or 2 h. Cells were fixed with methanol then incubated overnight with anti-NFATc1 antibody followed by Alexa 546 conjugated secondary antibody. (A) No treatment, (B) 1 h Ionomycin, (C) 15 min EtOH, (D) 15 min DEX. Arrows indicate cells positive for nuclear labeling of NFATc1. Scale bar = 20 μm. (E) The number of cells with NFATc1 nuclear labeling was counted from 15 different fields of view for each treatment group and the percentage of total cells was determined. n = 3 biological replicates. Significantly different from no trt, *p<0.04, **p<0.02.
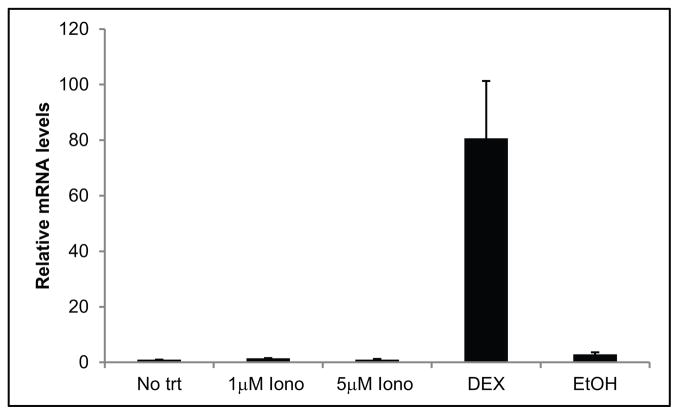
Finally, prior studies by Tumlin et al 1997 (Tumlin et al., 1997) showed that DEX can cause an influx in intracellular calcium through the activation of phospholipase C (PLC) and the generation of inositol triphosphate (IP3) that can activate calcineurin. To determine whether an increase in the cytosolic calcium concentration would be sufficient to activate the calcineurin/NFATc1 pathway and increase MYOC RNA expression, we examined MYOC expression in the presence of ionomycin. These studies showed that ionomycin alone was unable to induce an increase in MYOC mRNA expression (Fig. 6). This suggests that DEX activates the calcineurin/NFATc1 pathway in a calcium independent mechanism.
Figure 6.
Ionomycin does not increase MYOC mRNA. RT-qPCR was performed on HTM27 cells treated with 1 or 5 μM ionomycin or DEX or EtOH for 2 days. Ct values were corrected for primer efficiency and compared to cells with no treatment as described by Pfaffl (Pfaffl, 2001). Data were then normalized to ITGB1 mRNA (β1 integrin housekeeping gene). n = 3 biological replicates.
4. DISCUSSION
In this paper we demonstrate that the DEX-induced increase in MYOC transcription and expression is due to the activation of the calcineurin/NFATc1 pathway. This is supported by the fact that the increase in MYOC expression could be inhibited with the inhibitors CsA and INCA-6 as well as with NFATc1 knockdown using siRNA. A calcineurin/NFATc1 pathway was also recently found to trigger the up regulation of the β3 integrin subunit (Faralli et al., 2013). Since the DEX-induced expression of both proteins is a secondary glucocorticoid response, this suggests that a calcineurin/NFATc1 pathway may be a common mechanism used to regulate a secondary glucocorticoid response in HTM cells and that inhibitors of this pathway may be useful for the treatment or prevention of steroid induced glaucoma.
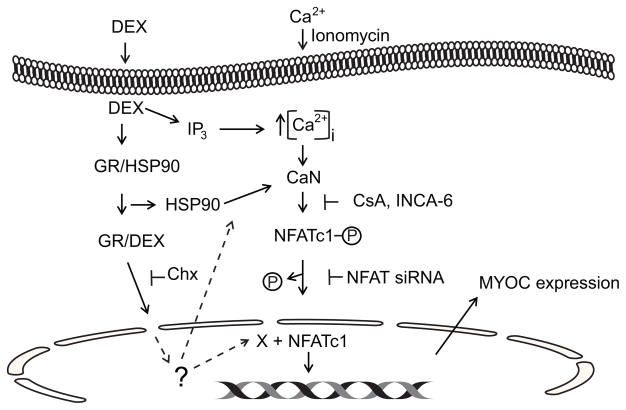
It is still unclear how DEX activates this pathway. The calcineurin/NFATc1 pathway can be activated by several pathways as shown in Figure 7. Since ionomycin failed to induce MYOC expression, a DEX induced increase in cytosolic calcium levels seems to be the unlikely mechanism involved in MYOC expression. Rather, we propose that DEX binding to the glucocorticoid receptor may be responsible for the activation of the calcineurin/NFATc1 pathway. Studies have shown that DEX binding to the glucocorticoid receptor causes a release of heat shock proteins, especially HSP90. HSP90 in turn has been shown to directly activate calcineurin in a calcium independent mechanism (Ranta et al., 2008). Alternatively, the activated glucocorticoid receptor could be inducing the transcription of some factor(s) needed to activate calcineurin. This hypothesis is supported by the fact that the glucocorticoid antagonist RU486 can inhibit the DEX induction of MYOC transcription (Shepard et al., 2001). In addition, there are no known NFAT family member binding sites in the MYOC promoter and the increase in MYOC could be inhibited by cycloheximide indicating that de novo protein synthesis was needed.
Figure 7.
Model of DEX activation of MYOC transcription. Schematic shows possible pathways that DEX can use to activate calcineurin (CaN)/NFATc1. Once activated NFATc1 is dephosphorylated and translocated to the nucleus where it can interact with other factors (X) to control transcription. Dotted lines indicate possible places that the glucocorticoid receptor (GR) could regulate calcineurin/NFATc1 activity. The GR could be promoting the expression of HSP90, cofactor (X) for NFATc1 or other transcription factors need for MYOC expression. Sites where the inhibitors cycloheximide (Chx), cyclosporin A (CsA), and INCA-6 target are indicated.
Together, these data suggest that the glucocorticoid receptor must be inducing the expression of additional factor(s) responsible for the induction of MYOC mRNA within the first 24 h. A plausible factor could be the transcription factor AP-1 which is known to activate gene expression in conjunction with NFATc1 (Chen et al., 1998). Clearly additional studies are needed to determine the mechanisms responsible for the activation of calcineurin/NFATc1.
As in the other studies using HTM cells (Clark et al., 2001; Joe et al., 2011; Shepard et al., 2001), it took up to 24 h for MYOC mRNA to increase and up to 2 days before protein expression could be detected. This increase in MYOC mRNA could be inhibited by cycloheximide indicating that de novo protein synthesis was needed. As previously reported, our studies also showed that the removal of DEX results in a slow decrease in the levels of the MYOC protein and mRNA to control levels. Following a 5 day treatment with DEX, Joe et. al (Joe et al., 2011) showed that about 50% of MYOC mRNA was still present 4 days after removal of DEX. In this study, we saw a similar phenomenon and observed a 50% decrease in MYOC mRNA approximately 6 days after DEX was removed. This prolonged stability of the MYOC mRNA corresponded to the continued expression of the MYOC protein which remained near treated levels for 4 days after DEX removal. This is likely due to the long half-life of MYOC mRNA and not the stability of the protein. MYOC protein levels in TM cells appear to be short-lived and decreased about 50% after 3 h of cycloheximide treatment. (Qiu et al., 2014).
In summary, this is the first study to demonstrate that activation of a calcineurin/NFATc1 pathway is responsible for the DEX-induced up regulation of MYOC and suggests that this may be a common pathway used by DEX to activate protein expression in the TM. Understanding how MYOC expression is regulated by DEX can help us design novel strategies for treating myocilin-related glaucoma.
Highlights.
Myocilin expression induced by dexamethasone is a secondary glucocorticoid response
Calcineurin activity is required for myocilin expression induced by dexamethasone
NFATc1 nuclear localization plays a role in myocilin expression
Acknowledgments
This work was supported by NEI grants EY017006 and EY0020490 (D.M.P.) and a Core grant to the Department of Ophthalmology and Visual Sciences (P30 EY016665).
Abbreviations
- CsA
cyclosporin A
- DEX
dexamethasone
- EtOH
ethanol
- HTM
human trabecular meshwork
- IOP
intraocular pressure
- MYOC
myocilin
- NFAT
Nuclear factor of activated T-cells
- POAG
primary open angle glaucoma
- SDHA
succinate dehydrogenase complex, subunit A
- TGF-β1
transforming growth factor-β1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam MF, Belmouden A, Binisti P, Brezin AP, Valtot F, Bechetoille A, Dascotte JC, Copin B, Gomez L, Chaventre A, Bach JF, Garchon HJ. Recurrent mutations in a single exon encoding the evolutionarily conserved olfactomedin-homology domain of TIGR in familial open-angle glaucoma. Hum Mol Genet. 1997;6:2091–2097. doi: 10.1093/hmg/6.12.2091. [DOI] [PubMed] [Google Scholar]
- Alvarado JA, Wood I, Polansky JR. Human trabecular meshwork cells II. Growth pattern and ultrastructural characteristics. Invest Ophthalmol Vis Sci. 1982;23:464–478. [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Clark AF, Steely HT, Dickerson JE, English-Wright S, Stropki K, McCartney MD, Jacobson N, Shepard AR, Clark JI, Matsushima H, Peskind ER, Leverenz JB, Wilkinson CW, Swiderski RE, Fingert JH, Sheffield VC, Stone EM. Glucocorticoid induction of the glaucoma gene MYOC in human and monkey trabecular meshwork cells and tissues. Invest Ophthalmol Vis Sci. 2001;42:1769–1780. [PubMed] [Google Scholar]
- Clark RW, Nosie AK, Walker T, Faralli JA, Filla MS, Barrett-Wilt G, Peters DM. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Mol Cell Proteomics. 2013;12:194–206. doi: 10.1074/mcp.M112.019745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Emmel EA, Verweij CL, Durand DB, Higgins KM, Lacy E, Crabtree GR. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989;246:1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- Fan BJ, Wiggs JL. Glaucoma: genes, phenotypes, and new directions for therapy. J Clin Invest. 2010;120:3064–3072. doi: 10.1172/JCI43085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli JA, Gagen D, Filla MS, Crotti TN, Peters DM. Dexamethasone increases αvβ3 integrin expression and affinity through a calcineurin/NFAT pathway. Biochim Biophys Acta. 2013;1833:3306–3313. doi: 10.1016/j.bbamcr.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla MS, David G, Weinreb RN, Kaufman PL, Peters DM. Distribution of syndecans 1–4 within the anterior segment of the human eye: expression of a variant syndecan-3 and matrix-associated syndecan-2. Exp Eye Res. 2004;79:61–74. doi: 10.1016/j.exer.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Fingert JH, Heon E, Liebmann JM, Yamamoto T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, Hockey RR, Williams-Lyn D, Trope G, Kitazawa Y, Ritch R, Mackey DA, Alward WL, Sheffield VC, Stone EM. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum Mol Genet. 1999;8:899–905. doi: 10.1093/hmg/8.5.899. [DOI] [PubMed] [Google Scholar]
- Fingert JH, Stone EM, Sheffield VC, Alward WL. Myocilin glaucoma. Surv Ophthalmol. 2002;47:547–561. doi: 10.1016/S0039-6257(02)00353-3. [DOI] [PubMed] [Google Scholar]
- Goldwich A, Scholz M, Tamm ER. Myocilin promotes substrate adhesion, spreading and formation of focal contacts in podocytes and mesangial cells. Histochem Cell Biol. 2009;131:167–180. doi: 10.1007/s00418-008-0518-4. [DOI] [PubMed] [Google Scholar]
- Joe MK, Kee C, Tomarev SI. Myocilin interacts with syntrophins and is member of dystrophin-associated protein complex. J Biol Chem. 2012;287:13216–13227. doi: 10.1074/jbc.M111.224063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe MK, Sohn S, Hur W, Moon Y, Choi YR, Kee C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem Biophys Res Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- Joe MK, Sohn S, Kim TE, Im J, Choi YR, Kee C. Analysis of glucocorticoid-induced MYOC expression in human trabecular meshwork cells. Vision Res. 2011;51:1033–1038. doi: 10.1016/j.visres.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Kim BS, Savinova OV, Reedy MV, Martin J, Lun Y, Gan L, Smith RS, Tomarev SI, John SW, Johnson RL. Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Mol Cell Biol. 2001;21:7707–7713. doi: 10.1128/MCB.21.22.7707-7713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein L, Cvekl A, Chauhan BK, Tamm ER. Regulation of human myocilin/TIGR gene transcription in trabecular meshwork cells and astrocytes: role of upstream stimulatory factor. Genes Cells. 2000;5:661–676. doi: 10.1046/j.1365-2443.2000.00355.x. [DOI] [PubMed] [Google Scholar]
- Kubota R, Noda S, Wang Y, Minoshima S, Asakawa S, Kudoh J, Mashima Y, Oguchi Y, Shimuzu N. A novel myosin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: molecular cloning, tissue expression, and chromosomal mapping. Genomics. 1997;41:360–369. doi: 10.1006/geno.1997.4682. [DOI] [PubMed] [Google Scholar]
- Kwon HS, Johnson TV, Joe MK, Abu-Asab M, Zhang J, Chan CC, Tomarev SI. Myocilin mediates myelination in the peripheral nervous system through ErbB2/3 signaling. J Biol Chem. 2013;288:26357–26371. doi: 10.1074/jbc.M112.446138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Tomarev SI. Myocilin, a glaucoma-associated protein, promotes cell migration through activation of integrin-focal adhesion kinase-serine/threonine kinase signaling pathway. J Cell Physiol. 2011;226:3392–3402. doi: 10.1002/jcp.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. New England Journal of Medicine. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam DS, Leung YF, Chua JK, Baum L, Fan DS, Choy KW, Pang CP. Truncations in the TIGR gene in individuals with and without primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2000;41:1386–1391. [PubMed] [Google Scholar]
- Liu J, Farmer JDJ, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- Nguyen TD, Chen P, Huang WD, Chen H, Johnson D, Polansky JR. Gene structure and properties of TIGR, a olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem. 1998;273:6341–6350. doi: 10.1074/jbc.273.11.6341. [DOI] [PubMed] [Google Scholar]
- Ortego J, Escribano J, Coca-Prados M. Cloning and characterization of subtracted cDNAs from a human ciliary body library encoding TIGR, a protein involved in juvenile open angle glaucoma with homology to myosin and olfactomedin. FEBS Lett. 1997;413:349–353. doi: 10.1016/S0014-5793(97)00934-4. [DOI] [PubMed] [Google Scholar]
- Park BC, Tibudan M, Samaraweera M, Shen X, Yue BY. Interaction between two glaucoma genes, optineurin and myocilin. Genes Cells. 2007;12:969–979. doi: 10.1111/j.1365-2443.2007.01102.x. [DOI] [PubMed] [Google Scholar]
- Peters DM, Herbert K, Biddick B, Peterson JA. Myocilin binding to Hep II domain of fibronectin inhibits cell spreading and incorporation of paxillin into focal adhesions. Exp Cell Res. 2005;303:218–228. doi: 10.1016/j.yexcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JR, Fauss DJ, Chen P, Chen H, Lutjen-Drecoll E, Johnson D, Kurtz RM, Ma ZD, Bloom E, Nguyen TD. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–139. doi: 10.1159/000310780. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Shen X, Shyam R, Yue BYJT, Ying H. Cellular processing of myocilin. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0092845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80:389–393. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta F, Dufer M, Stork B, Wesselborg S, Drews G, Haring HU, Lang F, Ullrich S. Regulation of calcineurin activity in insulin-secreting cells: stimulation by Hsp90 during glucocorticoid-induced apoptosis. Cell Signal. 2008;20:1780–1786. doi: 10.1016/j.cellsig.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Ricard CS, Agapova OA, Salvador-Silva M, Kaufman PL, Hernandez MR. Expression of myocilin/TIGR in normal and glaucomatous primate optic nerves. Exp Eye Res. 2001;73:433–447. doi: 10.1006/exer.2001.1063. [DOI] [PubMed] [Google Scholar]
- Roehrl MHA, Kang S, Aramburu J, Wagner G, Rao A, Hogan PG. Selective inhibition of calcineurin-NFAT signaling by blocking protein-protein interaction with small organic molecules. Proc Natl Acad Sci U S A. 2004;101:7554–7559. doi: 10.1073/pnas.0401835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Shen X, Koha T, Park BC, Noskina Y, Tibudan M, Yue BY. Mitochondrial association of myocilin, product of a glaucoma gene, in human trabecular meshwork cells. J Cell Physiol. 2007;213:775–784. doi: 10.1002/jcp.21147. [DOI] [PubMed] [Google Scholar]
- Shepard AR, Jacobson N, Fingert JH, Stone EM, Sheffield VC, Clark AF. Delayed secondary glucocorticoid responsiveness of MYOC in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2001;42:3173–3181. [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Takamoto M, Araie M. Genetics of primary open angle glaucoma. Jpn J Ophthalmol. 2014;58:1–15. doi: 10.1007/s10384-013-0286-0. [DOI] [PubMed] [Google Scholar]
- Tamm ER, Russell P, Epstein DL, Johnson DH, Piatigorsky J. Modulation of myocilin/TIGR expression in human trabecular meshwork. Invest Ophthalmol Vis Sci. 1999;40:2577–2585. [PubMed] [Google Scholar]
- Tumlin JA, Lea JP, Swanson CE, Smith CL, Edge SS, Someren JS. Aldosterone and dexamethasone stimulate calcineurin activity through a transcription-independent mechanism involving steroid receptor-associated heat shock proteins. J Clin Invest. 1997;99:1217–1223. doi: 10.1172/JCI119278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhuo Y, Liu B, Huang S, Hou F, Ge J. Pro370Leu mutant myocilin disturbs the endoplasm reticulum stress response and mitochondrial membrane potential in human trabecaulr meshwork cells. Mol Vis. 2007;13:618–625. [PMC free article] [PubMed] [Google Scholar]
- Zode GS, Kuehn MH, Nishimura DY, Searby CC, Mohan K, Grozdanic SD, Bugge K, Anderson MG, Clark AF, Stone EM, Sheffield VC. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J Clin Invest. 2011;121:3542–3553. doi: 10.1172/JCI58183. [DOI] [PMC free article] [PubMed] [Google Scholar]