Abstract
Objective
miRNAs are important regulators of gene expression through interaction with the 3′UTR of target mRNAs. The role of miRNAs has been extensively studied in adult human and animal models of heart disease. Hypoplastic left heart syndrome (HLHS) is the most common form of severe congenital heart disease and is an important cause of morbidity and mortality in infants and children. The objective of this work was to analyze the miRNA profile in HLHS patients.
Methods
miRNA profile was determined in the right ventricle (RV) by miRNA array and expression was validated by RT-PCR. Based on bioinformatics analysis, targets were selected and their expression was analyzed by RT-PCR.
Results
In this study we show that the miRNA profile of HLHS is novel with few similarities between pediatric and adult idiopathic dilated cardiomyopathy. Moreover, our analysis identified putative targets for these miRNAs that are known to be important for cardiac development and disease, and that miRNAs and their putative targets are antithetically regulated. We also show that miRNA expression changes with stage of surgery suggesting that volume unloading of the ventricle has important consequences for gene expression.
Conclusions
Our data suggest a unique miRNA profile for HLHS that may be associated with defects in cardiac development and disease.
Introduction
MicroRNAs (miRNA, miRs) are small noncoding ∼22 nucleotide (nt) RNAs capable of modulating the expression of many genes by recognizing a reverse complementary 6 - 8 nucleotide “seed” sequence, most frequently located within 3′ untranslated regions (3′UTR) of mRNAs. The interaction of miRNAs and RNA binding proteins to the 3′UTR can cause translational repression or RNA destabilization. It is estimated that as much as 60% of the genome is subject to their regulation (1).
Over the past few years, several array-based studies have been published detailing changes in miR expression in normal versus failing adult human heart. The results of these studies have been summarized recently (1). miRs that repeatedly surface as dynamically regulated (up or down) in adult heart failure (HF) include: several let-7s, miR-1, miR-133a/b, miR-100, miR-195, miR-199, miR-214, miR-222, miR-23a/b, miR-29a/b, miR-30 family, and miR-320. We have recently shown that pediatric idiopathic dilated cardiomyopathy (IDC) patients display a miRNA profile that is quite different from the adult population suggesting that pediatric IDC is a unique disease process (2).
Here we show that expression of a subset of miRNAs is differentially regulated in the right ventricle (RV) of pediatric patients with hypoplastic left heart syndrome (HLHS), a severe form of congenital heart disease. Furthermore, we show that volume unloading of the RV following surgical palliation normalizes expression of several miRNAs and their putative targets.
Materials and Methods
Tissue Procurement
Human subjects were males and females of all races and ethnic background ≤13 years of age who donated their heart to the institutional review board-approved pediatric heart tissue bank at the University of Colorado. Non-failing (NF) control hearts are obtained from donors whose heart could not be placed for technical reasons. HLHS RV tissue was obtained from explanted hearts of patients at the time of heart transplant. All heart tissue is rapidly flash frozen in the operating room immediately after removal from the subject. A detailed description of all pediatric patients can be found in Table S1. In table S1, RV failure is defined as signs and symptoms of heart failure in the setting of abnormal systolic function on echocardiogram, low cardiac output measured at catheterization [cardiac index below 2.5 L/min/m2] and/or diastolic dysfunction of the RV [end diastolic pressure of the RV >12mmHg] (3). Informed consent was obtained from all patients.
microRNA extraction and array analysis
Total RNA was extracted from the RV of 6 NF and 15 patients with HLHS. The HLHS patients were divided into 2 groups based on their stage of surgical palliation: (1) Stage 3- 5 patients were status post stage 3, the Fontan operation and (2) Stage 1- 10 had no prior surgeries performed or were status post stage 1, the Norwood operation. miRNA extraction was performed using the mirVana™ kit (Ambion) according to manufacturer's recommendation. miRNA expression analysis was performed in house using the TaqMan array human miRNA card (Invitrogen, Inc.) capable of detecting 754 miRNAs.
Array data were analyzed using the Expression Suite Software V1.0 (Invitrogen, Inc). Internal control was determined based on an algorithm generated by the software for changes on baseline miRNA values amongst all samples. In plate A, hsa-miR-361 had the lowest amount of variability among all samples, whereas in plate B hsa-miR-30d* showed the lowest variability in expression.
miRNA and mRNA RT-PCR
Reverse transcription of miRNAs was performed using the miScript Reverse Transcription Kit (Qiagen, Inc) for miRNAs or the iScript Reverse Transcription Kit (Bio-Rad, Inc) for mRNAs according to manufacturer's recommendations and essentially as previously described (2). miRNA expression was normalized to miR-361 and mRNA expression was normalized to 18S. Results obtained for some miRNAs using the miScript kit were confirmed using the TaqMan RT-PCR kit (Invitrogen, Inc). N= 9 NF, 6 stage 3 and 16 stage 1 HLHS patients.
Gene expression primers:
BAZ2A
F 5′ GACGTATTGCTACCCCAGAAG
R 5′ TTGCTTCATCCTCTTCCCAC
FOG2
F 5′ AAAGGCTCAGGTCCCAATG
R 5′ ATGGCCTTCGTAGTTGTACAC
CDK6
F 5′ CCGAAGTCTTGCTCCAGTC
R 5′ GAGTCCAATCACGTCCAAGAT
Sox11
F 5′ GCGAACTTCTCCGACCTG
R 5′ ACCATCAACACCACCATCATC
QKI
F 5′ TGTGGAAGATGCTCAGAACAG
R 5′ TGTAGGTGCCATTCAGAATCG
dHAND
F 5′ GCTACATCGCCTACCTCATG
R 5′ CTGCTCACTGTGCTTTTCAAG
GATA6
F 5′ GCTAGACGTCAGCTTGGAG
R 5′ CTGGAAAGGCTCTGGAGTC
GATA4
F 5′ CTTGCAATGCGGAAAGAGG
R 5′ TGCTGGAGTTGCTGGAAG
Pathways analysis
Pathway analysis was done using the DIANA Lab miRPath v. 2.0 software (http://www.microrna.gr/miRPathv2). Target prediction was done using the DIANA-microT-CDS. p-values for significant pathways are listed in the respective tables. Targets of interest were confirmed by Targetscan.
Statistical analysis
RT-PCR
Statistical analyses were performed using Statview software (SAS Institute, Cary, NC). 2-way Analysis of Variance (ANOVA) was performed on all outcomes. Statistical significance was set a priori at p<0.05 and all data are presented as mean + SEM in the figures.
Array
Analysis of variance (ANOVA) with Benjamini–Hochberg correction for false discovery rate was used to determine differential miRNA expression between groups. Significance was set a q<0.1 (adjusted p-values). Supervised hierarchical clustering was performed on the log scaled array samples that were significantly different by ANOVA (adjusted p-value <.01) using Ward's method. The analysis for miRNA expression was performed using R.
Results
Subject Characteristics
All of the patients included in the no surgery group were listed for primary transplantation (based on parent preference). The primary indication for transplant in all of the stage 1 patients and in 4/6 stage 3 patients was RV failure (as defined in Methods). One of the stage 3 patients had plastic bronchitis in addition to RV failure (with RV failure being the primary indication for transplant) and 2 of the stage 3 patients had protein losing enteropathy, which was their primary indication for transplant. There were 8 patients on milrinone (0.5mcg/kg/min) at the time of transplant. No other inotropes were being used at the time of transplant in this cohort. The time from last surgical palliation procedure to transplant for the stage 1 patients was a mean of 0.7±0.4 years (range 0.4-1.3 years) and for the stage 3 patients was a mean of 3.2±1.9 (range 1.8-6.2 years, p=0.02).
Differential expression of miRNAs in pediatric HLHS patients
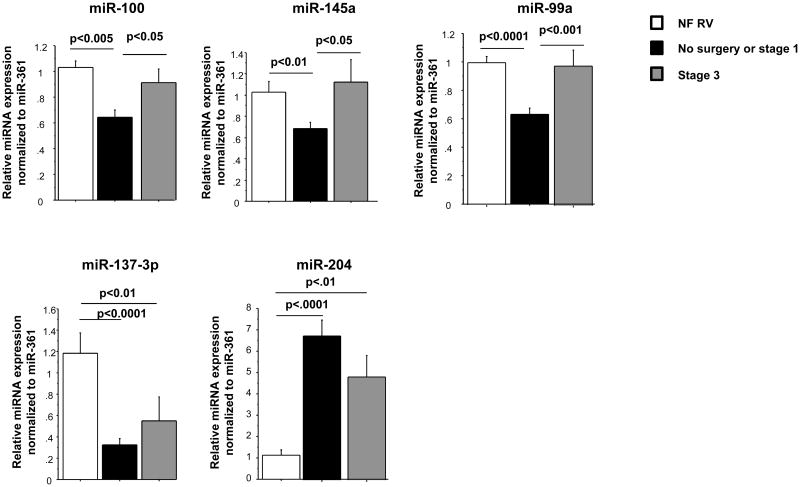
We have previously shown that miRNAs are differentially regulated in pediatric and adult IDC patients (2, 4). Because miRNAs play an important role in the regulation of gene expression and have an effect in cardiovascular diseases, we analyzed their expression in the RV of HLHS patients. HLHS is typified by lack of development of all left sided heart structures (including the left ventricle) and only a single RV is present, which is therefore inherently a volume and pressure overloaded chamber. Management consists of heart transplant in infancy or more commonly a 3 staged surgical palliation approach. There is a serial decrease in the volume load on the single RV across the stages of surgical palliation, but because the RV remains the systemic ventricle the pressure overload persists (5). The increased volume load on the RV of unoperated patients (those awaiting primary transplant) and patients following stage 1 surgery is the same, so these patients were grouped together for the purposes of this study. During stage 3 surgery, all of the systemic venous return is rerouted directly to the lungs resulting in a much lower volume load on the single RV compared to what is present in the unrepaired patient or following stage 1. Failure of the single RV is a common cause of death or need for heart transplant in patients with HLHS, and 1 year survival in these patients is only 68.7% (6). As shown in Table S2, 93 miRNAs were significantly regulated in HLHS hearts with some differences based on surgical stage (qvalue<0.1). Although some of these miRNAs have been shown to be regulated in adults with HF (miR-130a, miR-29b, miR-29c, miR-30 family, miR-208b, miR-499, miR-143 and miR378), most of these miRNAs have not been previously associated with cardiovascular diseases or are antithetically regulated in HLHS patients (miR-145 and miR-100). Figure 1A shows a subset (qvalue<0.025) of miRNAs that are differentially regulated in HLHS patients. Hierarchical clustering grouped the patients into three major distinct clusters (Figure 1B). Non-failing (NF) control patients cluster distinctly compared to those with HLHS. However, distinction between stage 3 cluster and no surgery or stage 1 was not as well defined, suggesting a disease effect. Based on the array data, 6 miRNAs were selected and expression was confirmed by RT-PCR. These miRNAs were selected based on low qvalues (miR-100, miR-145a and miR-137-3p), on being uniquely regulated in pediatric HF (miR-204), or on determining the reproducibility of the array data at higher qvalues (miR-99a). As shown in Figure 1C, changes in expression of miR-204 and miR-137-3p were observed in stage 1 and stage 3. Expression of miR-204 was also increased in pediatric IDC patients and may be an important miRNA specific to pediatric HF irrespective of etiology. Interestingly, expression of miR-100 miR-99 and miR-145 is normalized in stage 3, suggesting that volume unloading of the heart plays an important role in regulating miRNA expression. Although changes in gene expression were virtually identical between array and RT-PCR for miR-204 and -137-3p, some differences were observed in the expression of miR-100, -99a and -145 when both methodologies were compared. Specifically, expression of these miRNAs was significantly down-regulated by RT-PCR and array in stage 1, but expression was normalized in stage 3 by RT-PCR but not by array. It is unclear why these differences were observed but could be due to increased number of samples and repetition in the RT-PCR. Differences in RT-PCR and array methodologies have been observed by us and other groups (2, 7).
Figure 1.
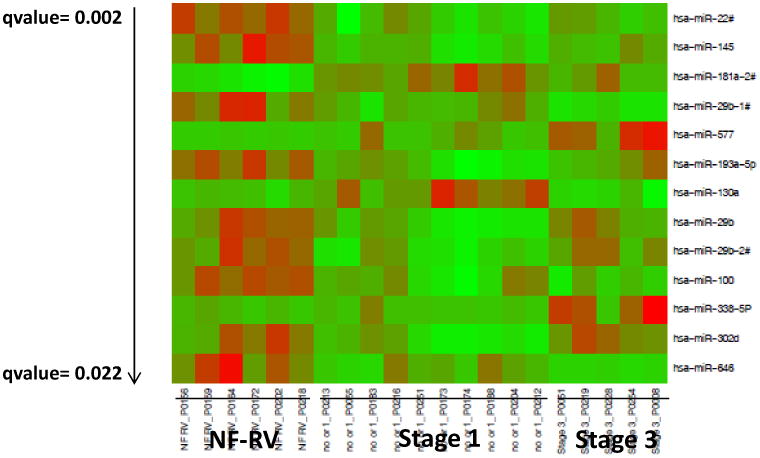
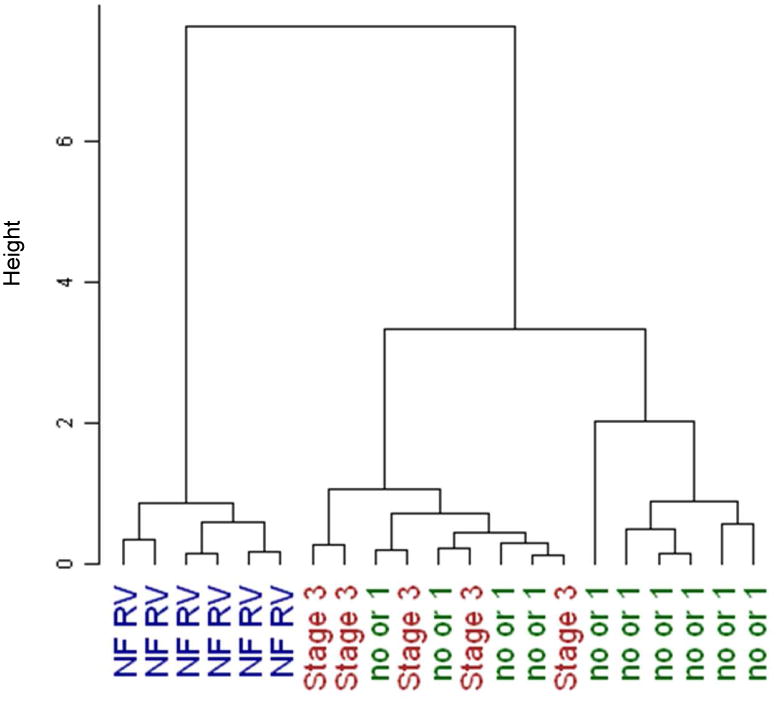
miRNA expression in HLHS patients. (A) Expression profile of miRNAs in non-failing HLHS pediatric patients by miRNA array. Green – down-regulated; Red – up-regulated. Only miRNAs with a qvalue of <0.025 are shown. miRNAs are presented by increasing qvalue. N=6 non-failing right ventricle (NF RV), 10 post-stage 1, 5 post-stage 3 HLHS patients. (B) Supervised hierarchical clustering of array samples based on Ward's method. (C) Relative expression of a subset of miRNAs in non-failing, post-stage 1 and post-stage 3 HLHS patients was confirmed by RT-PCR, as described in the Methods section. N=9 non-failing, 16 post-stage 1, 6 post-stage 3 HLHS patients. Statistically significant expression of miRNAs is shown in the Figure.
Differential expression of putative miRNAs targets in pediatric HLHS patients
To determine the possible role of these miRNAs in cardiovascular diseases, an analysis of possible targets was performed using the Diana Lab software. Pathway analysis was performed for miRNAs that are down-regulated or up-regulated with a qvalue of <0.025. As shown in Table S2 and Figure 1A, 9 miRNAs were down-regulated and 4 miRNAs were up-regulated in HLHS. The pathway analysis results are shown in Table S3 and S4. Among the top 10 most regulated pathways are the PI3K-AKT, cancer-related, MAPK and WNT-signaling pathways. In addition, a pathway analysis of miRNAs whose expression was confirmed by RT-PCR was also performed and is shown in Table S5. Because miR-29b targets collagen genes, the extracellular matrix-receptor interaction pathway is highly represented, and analysis was also performed in the absence of miR-29b (Table S6). A thorough discussion of these pathways and their importance to cardiovascular diseases is detailed in the supplementary material.
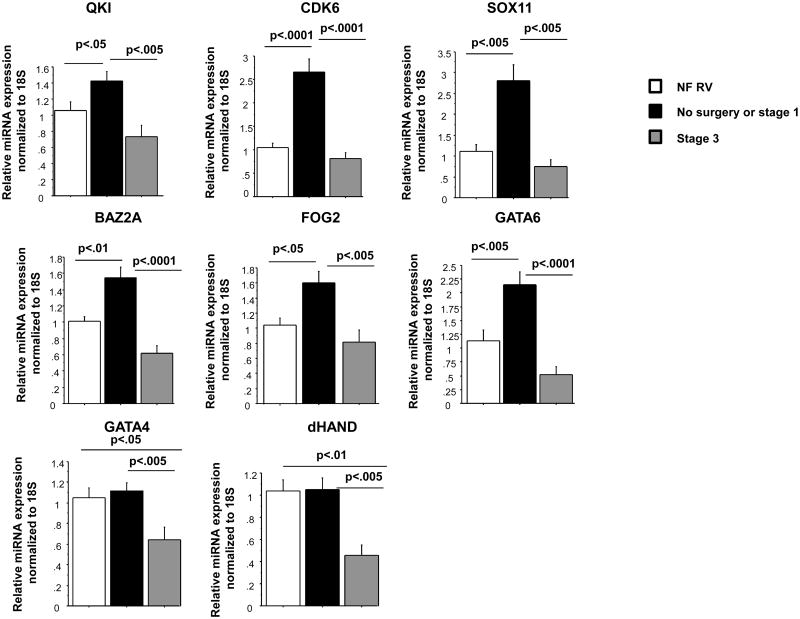
To determine a functional role for miRNAs regulated in HLHS we analyzed expression of potential common targets for RT-PCR-confirmed miRNAs that are down-regulated in stage 1. As shown in Table S7 and Figure 2, there are 5 potential cardiovascular targets that are associated with at least 2 of the 4 down-regulated miRNAs (miR-99a and miR-100 target the same genes). Some of these targets have been confirmed by other studies (Table S7). Expression of QKI, CDK6, SOX11, BAZ2A, FOG2 and GATA6 were increased in no surgery or stage 1 group, but normalized in the stage 3 group. Expression of GATA4 and dHAND was decreased in the stage 3 group. A thorough discussion on the relevance of these targets for HLHS is detailed below.
Figure 2.
Expression analysis of putative miRNA-regulated mRNAs and associated factors. Gene expression was measured by RT-PCR and normalized to 18S. N=9 non-failing right ventricle (NF RV), 16 post-stage 1, 6 post-stage 3 HLHS patients. Statistically significant expression of mRNAs is shown in the Figure.
Discussion
In this study we analyzed the miRNA expression profile in the RV of HLHS patients. Our results suggest that the pediatric HLHS population has a unique miRNA profile, and expression of many miRNAs correlates with stage of surgical palliation and are therefore likely regulated by the volume load on the single RV. However, there are several miRNAs (Table S2) whose expression is similar in children with HLHS regardless of surgical stage. Interestingly several of these miRNAs (bolded in Table S2) were also found to be dynamically regulated by Bernstein et al in the RV of a pressure overloaded mouse model (8). Together these data suggest that while some miRs are modulated by changes in volume load on the RV, other miRs are volume load-independent.
These results indicate that the pathways that may be affected by miRNAs in HLHS are related to cardiac disease and development. We have recently shown that several molecular pathways are differentially regulated in adult and pediatric IDC patients (9), and the results presented here indicate that the pathways regulated in HLHS may be unique to this disease process and may be influenced by miRNAs. Our results show that expression of several genes involved in cardiac disease and development are differentially regulated in HLHS patients. Moreover, because expression of these genes is antithetically regulated compared to the miRNAs that may target them, these miRNAs likely target expression of these genes. Importantly, volume unloading of the heart reverses these changes, indicating that they represent compensatory changes that occur in response to increased volume load of the right ventricle. Collectively, our results suggest that the miRNA profile in cardiac disease is age and disease-specific and that the pediatric HLHS population is unique. These results suggest that pediatric heart disease deserves distinct consideration and challenges the appropriateness of the current paradigm of direct extrapolation of adult heart failure treatment to children.
miRNA-target analysis suggested that expression of several genes important for cardiac disease and development were potential targets for the disregulated miRNAs, and their expression was analyzed by RT-PCR. Of those, Quaking (QKI), Friends of GATA 2 (FOG-2), cyclin-dependent kinase 6 (CDK6) and SRY-box 11 (Sox11) are important for proper heart development (10-12). BAZ2A is an integral component of chromatin re-modeling complexes and is thought to play an important role in chromatin-dependent regulation of transcription (13), and could be involved in transcription deregulation observed in hypertrophy and HF.
QKI is highly expressed during normal cardiac development (10) and protects against ischemia/reperfusion-induced apoptosis in neonatal cardiac myocytes (14). Down-regulation of CDK6 expression, a protein involved in cell-cycle progression, has been implicated in disturbed cardiac chamber formation and trabeculation (11), and phosphorylation of retinoblastoma protein by cyclin D-cdk4/6 is necessary for hypertrophic growth in cardiac myocytes (15). Sox11 null newborn embryos die of cardiac malformation that include ventricular septation defects, double outlet right ventricle and outflow tract malformations (12).
FOG-2 is a GATA co-factor that acts as a scaffold to coordinate interactions between factors (reviewed in (16)). FOG-2 represses GATA function in vitro, and up-regulation of FOG-2 has been observed in human and murine failing hearts (17). Transgenic mouse over-expression of FOG-2 results in depressed cardiac function, activation of the fetal gene program and repression of the SR-Ca2+ ATPase (SERCA) (17), and knockout of FOG-2 results in death due to HF during E12.5-15.5 (16). As shown in Figure 2, FOG-2 expression is increased in volume loaded hearts. Because FOG-2 affects the function of GATA transcriptions factors, and these factors are also involved in proper cardiac development, differentiation (18), cardiac hypertrophy and HF (19), we also looked at expression of GATA4 and GATA6. As shown in Figure 2, GATA6 expression is 1.9 fold higher when compared to NF controls and 4.1 fold higher when compared to volume unloaded hearts. Interestingly, FOG-2 is a putative target for mR-145, which can target GATA6 (20). No differences were observed when GATA4 expression was compared to NF controls but a 1.74 up-regulation was observed when expression was compared to volume unloaded hearts. GATA factors regulate expression of the right ventricle protein dHAND, a protein important for right ventricle formation (21). Moreover, expression of dHAND and GATA4 is increased in pressure overload right ventricular hypertrophy (22). As shown in Figure 2 expression of dHAND is increased in volume loaded hearts when compared to volume unloaded hearts, a pattern similar to that seen for GATA4. We propose that the increase in GATA expression, and consequently dHAND, is a compensatory response to the increased levels of FOG-2. Since all these factors are involved in cardiac hypertrophy and HF, their increased expression may contribute to the pathological hypertrophy and failure of the RV in HLHS. Importantly, expression of these factors is normalized in the volume unloaded heart (stage 3).
There are limitations to this study. Tissue-bank based studies such as this are cross-sectional by nature, so proof of mechanistic associations based on the results presented is not possible. Additionally, tissue-bank studies are limited by the samples available for study. In this study we analyzed miRNA expression based on stage of surgery. Although no surgery and stage 1 groups are younger than stage 3, there was no correlation between miRNA expression and age (determined by regression analysis – not shown). Although the stage 1 patients were closer in time to their last cardiac surgery procedure compared to the stage 3 patients, on average the stage 1 patients were over 6 months removed from their last surgery. Therefore, while it is not possible to know with certainty the role of ischemia-reperfusion or inflammation related to cardiopulmonary bypass in influencing these findings, it seems unlikely that this is a significant factor given the remote timing. In addition, there was no difference in miRNA or gene expression when comparing HLHS with RV failure to those without RV failure or when comparing the no surgery group (listed for primary transplant) and the stage 1 patients (data not shown). Despite these limitations, this study represents the first description of the differences in miRNA expression in the RV of HLHS patients. These findings will be relevant in planning future studies aimed at identifying the mechanisms involved in systemic RV failure and remodeling.
Supplementary Material
Table S1: Pediatric Subject Characteristics and Analyses: M-male, F-female, ACEi – angiotensin converting enzyme inhibitor, PDE – phosphodiesterase, NA – Not Available.
Table S2: List of most significant miRNAs q-value <0.1 Bolded miRNAs were also regulated in a mouse model of pressure overload [5].
Table S3: Pathway analysis of up-regulated miRNAs q-value<0.025
Table S4: Pathway analysis of down-regulated miRNAs q-value<0.025
Table S5: Pathway analysis of genes targeted by miRs-145, - 137, - 99a, -100 and -29b
Table S6: Pathway analysis of genes targeted by miRs-145, - 137, - 99a, -100
Table S7: Putative targets for regulated miRNAs.
Acknowledgments
Sources of Funding: This work was supported by the National Institutes of Health grants: R01 HL107715 (to BLS); American Heart Association 11IRG5070006 (to CCS); Pediatrics Student Research Program at Children's Hospital Colorado (to JS); K12 HD068372 and Addison Scott Memorial Fund and Boedecker Foundation Grant (to SDM)
Disclosures: Carmen Sucharov: Equity in miRagen, Inc., Brian Stauffer: Research support from Forest Laboratories, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Topkara VK, Mann DL. Role of microRNAs in cardiac remodeling and heart failure. Cardiovasc Drugs Ther. 2011 Apr;25(2):171–82. doi: 10.1007/s10557-011-6289-5. [DOI] [PubMed] [Google Scholar]
- 2.Stauffer BL, Russell G, Nunley K, Miyamoto SD, Sucharov CC. miRNA expression in pediatric failing human heart. J Mol Cell Cardiol. 2013 Apr;57:43–6. doi: 10.1016/j.yjmcc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyamoto SD, Stauffer BL, Polk J, Medway A, Friedrich M, Haubold K, et al. Gene expression and beta-adrenergic signaling are altered in hypoplastic left heart syndrome. J Heart Lung Transplant. 2014 Aug;33(8):785–93. doi: 10.1016/j.healun.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008 Aug;45(2):185–92. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellsham-Revell HR, Tibby SM, Bell AJ, Witter T, Simpson J, Beerbaum P, et al. Serial magnetic resonance imaging in hypoplastic left heart syndrome gives valuable insight into ventricular and vascular adaptation. J Am Coll Cardiol. 2013 Feb 5;61(5):561–70. doi: 10.1016/j.jacc.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohye RG, Sleeper LA, Mahony L, Newburger JW, Pearson GD, Lu M, et al. Comparison of shunt types in the Norwood procedure for single-ventricle lesions. N Engl J Med. 2010 May 27;362(21):1980–92. doi: 10.1056/NEJMoa0912461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callari M, Dugo M, Musella V, Marchesi E, Chiorino G, Grand MM, et al. Comparison of microarray platforms for measuring differential microRNA expression in paired normal/cancer colon tissues. PLoS One. 2012;7(9):e45105. doi: 10.1371/journal.pone.0045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy S, Zhao M, Hu DQ, Fajardo G, Hu S, Ghosh Z, et al. Dynamic microRNA expression during the transition from right ventricular hypertrophy to failure. Physiol Genomics. 2012 May 1;44(10):562–75. doi: 10.1152/physiolgenomics.00163.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto SD, Stauffer BL, Nakano S, Sobus R, Nunley K, Nelson P, et al. Beta-adrenergic adaptation in paediatric idiopathic dilated cardiomyopathy. Eur Heart J. 2012 Jul 26; doi: 10.1093/eurheartj/ehs229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justice MJ, Hirschi KK. The role of quaking in mammalian embryonic development. Adv Exp Med Biol. 2010;693:82–92. doi: 10.1007/978-1-4419-7005-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z, Rivkees SA. Rho-associated kinases play an essential role in cardiac morphogenesis and cardiomyocyte proliferation. Dev Dyn. 2003 Jan;226(1):24–32. doi: 10.1002/dvdy.10212. [DOI] [PubMed] [Google Scholar]
- 12.Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004 Aug;24(15):6635–44. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones MH, Hamana N, Nezu J, Shimane M. A novel family of bromodomain genes. Genomics. 2000 Jan 1;63(1):40–5. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Shi X, Liu A, Yang G, Yu F, Zheng Q, et al. RNA binding protein QKI inhibits the ischemia/reperfusion-induced apoptosis in neonatal cardiomyocytes. Cell Physiol Biochem. 2011;28(4):593–602. doi: 10.1159/000335755. [DOI] [PubMed] [Google Scholar]
- 15.Hinrichsen R, Hansen AH, Haunso S, Busk PK. Phosphorylation of pRb by cyclin D kinase is necessary for development of cardiac hypertrophy. Cell Prolif. 2008 Oct;41(5):813–29. doi: 10.1111/j.1365-2184.2008.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fossett N, Schulz RA. Conserved cardiogenic functions of the multitype zinc-finger proteins: U-shaped and FOG-2. Trends Cardiovasc Med. 2001 Jul;11(5):185–90. doi: 10.1016/s1050-1738(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 17.Rouf R, Greytak S, Wooten EC, Wu J, Boltax J, Picard M, et al. Increased FOG-2 in failing myocardium disrupts thyroid hormone-dependent SERCA2 gene transcription. Circ Res. 2008 Aug 29;103(5):493–501. doi: 10.1161/CIRCRESAHA.108.181487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin J, Afouda BA, Hoppler S. Wnt/beta-catenin signalling regulates cardiomyogenesis via GATA transcription factors. J Anat. 2010 Jan;216(1):92–107. doi: 10.1111/j.1469-7580.2009.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Berlo JH, Elrod JW, van den Hoogenhof MM, York AJ, Aronow BJ, Duncan SA, et al. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ Res. 2010 Oct 15;107(8):1032–40. doi: 10.1161/CIRCRESAHA.110.220764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R, Yan G, Zhang Q, Jiang Y, Sun H, Hu Y, et al. miR-145 inhibits isoproterenol-induced cardiomyocyte hypertrophy by targeting the expression and localization of GATA6. FEBS Lett. 2013 Jun 19;587(12):1754–61. doi: 10.1016/j.febslet.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava D, Olson EN. Knowing in your heart what's right. Trends Cell Biol. 1997 Nov;7(11):447–53. doi: 10.1016/S0962-8924(97)01150-1. [DOI] [PubMed] [Google Scholar]
- 22.Bar H, Kreuzer J, Cojoc A, Jahn L. Upregulation of embryonic transcription factors in right ventricular hypertrophy. Basic Res Cardiol. 2003 Sep;98(5):285–94. doi: 10.1007/s00395-003-0410-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Pediatric Subject Characteristics and Analyses: M-male, F-female, ACEi – angiotensin converting enzyme inhibitor, PDE – phosphodiesterase, NA – Not Available.
Table S2: List of most significant miRNAs q-value <0.1 Bolded miRNAs were also regulated in a mouse model of pressure overload [5].
Table S3: Pathway analysis of up-regulated miRNAs q-value<0.025
Table S4: Pathway analysis of down-regulated miRNAs q-value<0.025
Table S5: Pathway analysis of genes targeted by miRs-145, - 137, - 99a, -100 and -29b
Table S6: Pathway analysis of genes targeted by miRs-145, - 137, - 99a, -100
Table S7: Putative targets for regulated miRNAs.