Summary
Teleosts comprise about half of all vertebrate species and exhibit an extraordinary diversity of adult pigment patterns that function in shoaling, camouflage and mate choice and have played important roles in speciation. Here, we review recent studies that have identified several distinct neural crest lineages, with distinct genetic requirements, that give rise to adult pigment cells in fishes. These lineages include post-embryonic, peripheral nerve associated stem cells that generate black melanophores and iridescent iridophores, cells derived directly from embryonic neural crest cells that generate yellow-orange xanthophores, and bipotent stem cells that generate both melanophores and xanthophores. This complexity in adult chromatophore lineages has implications for our understanding of adult traits, melanoma, and the evolutionary diversification of pigment cell lineages and patterns.
Introduction
Studies of vertebrate pigmentation have provided fundamental insights into genetics (Castle and Little, 1910; Wright, 1984a), the cytoskeleton (Fujii, 2000; Kelsh et al., 2009), morphogenesis (Bonaventure et al., 2013; Kelsh et al., 2009), cell lineage diversification (Adameyko et al., 2009; Dorsky et al., 1998; Nitzan et al., 2013a; Thomas and Erickson, 2009), and modes of selection and speciation (Endler, 1980; Fan et al., 2012; Houde, 1997; Houde and Endler, 1990; Price et al., 2008; Seehausen et al., 2008; Wright, 1984b). Whereas an origin of pigment cells in the embryonic neural crest has been known for many years [(Dushane, 1934); reviewed in: (Hall and Hörstadius, 1988; Parichy et al., 2006)], the more specific developmental histories and genetic requirements of pigment cells responsible for adult pigmentation have started to be elucidated only more recently. Here, we review recent advances in understanding lineages of adult pigment cells with an emphasis on recent work in zebrafish Danio rerio (Fig. 1), but also touching upon studies of several other fishes as well as amniotes.
Figure 1.
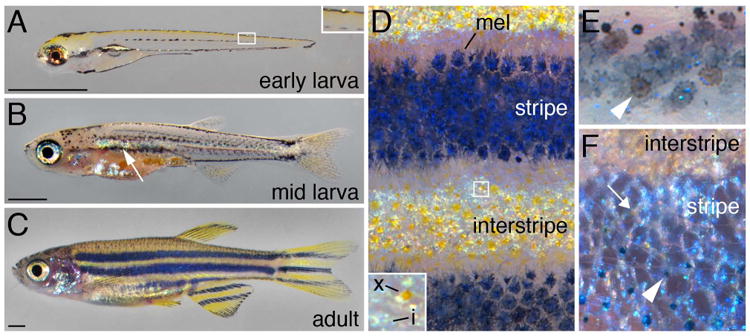
Zebrafish pigment cells and patterns. (A) The EL pigment pattern at 5 dpf. Note stripes of melanophores with a few shiny iridophores and the yellow color imparted by xanthophores. Inset, higher magnification of dorsal melanophore stripe with a single iridescent iridophore; additional EL melanophores occur scattered along the horizontal myoseptum. (B) ∼18 dpf larva developing its adult pattern. Interstripe iridophores have developed (arrow) and adult melanophore stripes are becoming evident. (C) Young adult pattern at ∼2.5 months. Scale bars in A–C, 1 mm. (D) Closeup of adult pattern with dark stripes of melanophores (mel) and iridophores alternating with interstripes of iridescent iridophores and yellow-orange xanthophores. Inset, xanthophore (x) and iridophores (i). (E) In the adult, EL melanophores often have a brownish color (arrowheads), distinguishable from grey-black adult melanophores(Quigley et al., 2004). (F) Different adult iridophore morphologies in the interstripe and stripe. Treatment with epinephrine has contracted melanosomes towards melanophore cell bodies (arrowhead). Very lightly pigment xanthophores are evident at low density as well (arrow).
Pigment cell diversity and stem cells
Adult pigmentation in amniotes depends on the patterned differentiation of melanocytes that contribute melanin to keratinocytes and, ultimately to skin, hair or feathers (Kaelin et al., 2012; Lin et al., 2013). By contrast, teleosts and other ectothermic vertebrates develop several classes of pigment cells, or chromatophores, that retain their pigments intracellularly (Bagnara and Matsumoto, 2006). Overall patterns thus reflect the numbers and arrangements of the chromatophores themselves. Certainly the most studied of these cells is the black melanophore, the melanin-containing cell homologous to the melanocyte of amniotes (and, for this reason, often referred to itself as a melanocyte). Other chromatophores receiving attention recently are yellow-orange xanthophores, having pteridines and carotenoids, iridescent iridophores, with purine-rich stacks of reflecting platelets, and shiny yellow leucophores that contain pteridines and carotenoids as well as reflective crystalline deposits. Nevertheless, the diversity of adult chromatophores is extensive and includes red erythrophores, blue cyanophores and others (Bagnara et al., 2007; Goda and Fujii, 1995; Goda et al., 2013; Khoo et al., 2012; Kimura et al., 2014; Nagao et al., 2014).
A common stem cell origin for different chromatophore classes was suggested by Bagnara et al. (Bagnara et al., 1979) from the observations that cells sometimes contain pigment organelles typical of more than one class, and that organelles themselves sometimes can be mosaic. In this context, “stem cell” referred to a precursor in, or derived from, the neural crest, able to generate multiple differentiated cell types. Yet, stem cells are often defined as being able to self-renew while generating differentiated progeny, and, in this sense, stem cells need not be multipotent. For simplicity in this review, we use the term “stem cell” in reference to latent precursors that normally give rise to adult chromatophores, and note that the degrees to which these cells can self-renew or contribute to multiple chromatophore classes remains often unclear.
Outline of zebrafish pigment pattern development
Adult chromatophore stem cell lineages have been studied most extensively in zebrafish, which exhibit two distinct patterns during their life cycle. Around the time of hatching, the fish has an embryonic/early larval (EL) pattern with stripes of melanophores along the dorsal myotomes and extending over the head, along the ventral myotomes and over the yolk sac, laterally along the horizontal myoseptum, and ventrally under the yolk sac (Kimmel et al., 1995). Iridophores are sparsely distributed within three of these melanophore stripes (and are especially abundant over the swimbladder), whereas xanthophores are widely distributed beneath the epidermis and give an overall yellow cast to the flank (Fig. 1A). This pattern likely provides camouflage while also protecting the nervous system and other tissues from UV damage in shallow water (Arunachalam et al., 2013; Engeszer et al., 2007b; Mueller and Neuhauss, 2014). Development of the EL pattern begins 15–25 hours post-fertilization (hpf) (Raible et al., 1992; Vaglia and Hall, 2000) with the migration of neural crest cells, some of which differentiate as EL melanophores, xanthophores, and iridophores (Dutton et al., 2001; Kelsh et al., 1996; Odenthal et al., 1996; Raible and Eisen, 1994) that organize into the definitive EL pattern by 48 hpf through mechanisms that remain largely, though not entirely (Svetic et al., 2007), unknown.
The zebrafish adult pattern is very different and consists of dark stripes of melanophores with relatively small numbers of iridophores and a few pale xanthophores, alternating with light “interstripes” of abundant iridophores and more heavily pigmented xanthophores (Fig. 1C–F). Distinct classes of iridophores have been identified (Oshima and Kasai, 2002), including elipsoidal cells having small reflecting platelets (S-iridophores) in both stripes and interstripes, and spindle-shaped cells having large reflecting platelets (L-iridophores), associated only with stripe melanophores (Hirata et al., 2003). All of these reside in the hypodermis, between the skin and underlying myotome (Hawkes, 1974a; Hawkes, 1974b; Kirschbaum, 1975; Le Guellec et al., 2004; Rakers et al., 2010), and occupy different strata within this compartment (Hirata et al., 2003; Hirata et al., 2005). Chromatophores are also found intermingled in the dorsal hypodermis, associated with scales in the dermis, and in the fins (Hirata et al., 2005; Tu and Johnson, 2011). The adult pattern functions in shoaling (Engeszer et al., 2007a; Engeszer et al., 2008; Rosenthal and Ryan, 2005) and potentially other behaviors as well (Price et al., 2008).
The beginnings of a transition from EL to adult pattern is evident typically by ∼8 days post-fertilization (dpf)1: by this time a few melanophores have been added to the lateral stripe (Hultman and Johnson, 2010; Milos and Dingle, 1978a; Milos and Dingle, 1978b; Milos et al., 1983); yellow xanthophores, though initially widespread, are no longer apparent; and adult iridophores are evident near the horizontal myoseptum, where the first adult interstripe will develop (Movie 1). Subsequently, lightly pigmented melanophores are found widely over the flank, including where the first two adult stripes will form, dorsal and ventral to the first interstripe (Fig. 1B). Finally, adult interstripe xanthophores appear in association with interstripe iridophores and boundaries between stripes and interstripes become increasingly distinct: melanophores are lost from the interstripe and melanophores within stripes become larger and more stellate (Frohnhofer et al., 2013; Johnson et al., 1995; Maderspacher and Nusslein-Volhard, 2003; Mahalwar et al., 2014; McMenamin et al., 2014; Parichy et al., 2009; Parichy et al., 2000b; Parichy and Turner, 2003b; Patterson et al., 2014; Patterson and Parichy, 2013; Singh et al., 2014; Takahashi and Kondo, 2008). A juvenile pattern forms by ∼28 dpf and, later, new interstripes and stripes are added dorsally and ventrally.
One could posit two very different ways to make an adult pattern: by expanding and rearranging the populations of EL chromatophores, or by replacing EL chromatophores with entirely new populations of adult chromatophores. The predominant strategy used by zebrafish differs between chromatophore classes. In the following sections, we review first the development of embryonic chromatophores that give rise to the EL pattern, and then the development of adult melanophores, iridophores and xanthophores that contribute to the body pattern, as well as chromatophores that form the normal and regenerative patterns of the fins.
Zebrafish mutants with defects in adult pattern hint at a melanophore stem cell
A distinct origin of EL and adult melanophores was first suggested by the observation that lightly pigmented melanophores appear during adult pattern development (Kirschbaum, 1975), implying de novo differentiation from an unpigmented precursor. Additional support for this idea came from several mutants with defects in adult pigmentation. The first of these was sparse (Johnson et al., 1995), shown later to correspond to kita (Parichy et al., 1999), encoding a Kit receptor tyrosine kinase. In kita mutants, there are fewer EL melanophores initially and even these die after several days so that by mid-larval stages melanophores are completely absent (Fig. 2A). By late larval stages, however, new kita-independent melanophores differentiate. Because such cells cannot arise by the proliferation of EL melanophores, which are missing, they must come from unpigmented cells with melanogenic potential; the same precursors might give rise to adult melanophores during normal development. A similar kita mutant phenotype is observed in the zebrafish relative, D. albolineatus, indicating an evolutionary conservation of this kita-independent population of melanophores (Mills et al., 2007).
Figure 2.
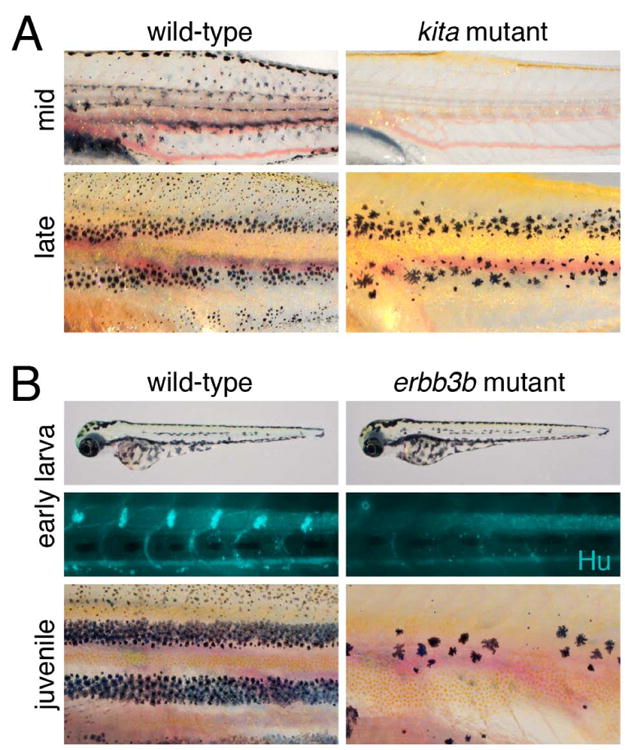
Mutants suggesting a stem cell origin of adult melanophores. (A) kita mutants completely lack melanophores at mid-larval stages but later reacquire stripe (though not dorsal scale) melanophores. (B) erbb3b mutants have normal EL patterns but defects in a ventromedial neural crest migration and lack Hu+ dorsal root ganglia. In the juvenile, most adult melanophores fail to develop; a few large melanophores persist from the EL pattern.
A stem cell origin of adult melanophores was also suggested by the puma (Parichy and Turner, 2003b; Parichy et al., 2003) and picasso (Budi et al., 2008) mutants. In both of these, EL melanophores develop, yet most adult melanophores are missing (Fig. 2B). Despite their similar phenotypes, puma and picasso affect different steps in adult melanophore development. puma corresponds to a mutation in tubulin alpha-8 like-3 (tuba8l3) (Larson et al., 2010), which affects melanophore development autonomously (Parichy et al., 2003). Because this particular allele has a temperature-sensitive phenotype, it was possible to use temperature-shift experiments to define a critical period, corresponding to when adult melanophores normally differentiate, during which time the puma mutation impacted melanophore development (Parichy and Turner, 2003b; Parichy et al., 2003). This suggested that tuba8l3 is involved in the recruitment of melanophores from a latent stem cell.
picasso corresponds to erbb3b, encoding an ErbB3 (EGFR-related) receptor tyrosine kinase (Budi et al., 2008; Lyons et al., 2005). In contrast to puma, pharmacological inhibitors of ErbB activity and transient morpholino knockdown showed that Erbb3b is required for adult melanophore development during embryogenesis, when neural crest cells migrate along a ventromedial migratory pathway between the somites and neural tube, and thus long before these cells actually differentiate (Budi et al., 2008; Dooley et al., 2013). Although erbb3b is expressed by melanophores, genetic mosaic analyses revealed both autonomous and non-autonomous requirements, indicating a role not only within the melanophore lineage but in the tissue environment of these cells as well (Budi et al., 2008). These results suggested that ErbB-dependent precursors to adult melanophores are established during neural crest migration, in parallel with precursors to EL melanophores; later, during the larval-to-adult transformation, these erbb3b-dependent cells are recruited as adult melanophores. Similar adult melanophore defects are observed in mutants for neuregulin1-3, encoding an ErbB3 ligand (Lush and Piotrowski, 2014), and sorbs3, encoding a scaffolding protein that functions in the ErbB pathway (Malmquist et al., 2013). Unlike adult melanophores, most EL melanophores arise independently of this pathway: when ErbB signaling is blocked in embryos, 92% of the normal complement of melanophores develop by early larval stages; most of the “missing” ∼8% would normally populate the lateral stripe after 3 dpf, by which time other EL melanophores have differentiated (Hultman and Johnson, 2010; Milos and Dingle, 1978a).
Both ErbB pathway and tuba8l3 mutants also have defects in the nervous system, with drastically fewer glia and extensive defasciculation of peripheral nerves (Budi et al., 2008; Budi et al., 2011; Larson et al., 2010; Lyons et al., 2005; Parichy et al., 2003). In ErbB-deficient em embryos these phenotypes stem in part from defects in neural crest cell morphogenesis along the ventromedial pathway, with cells either failing to migrate, or migrating but failing to coalesce into dorsal root ganglia (DRG; Fig. 2B) or sympathetic ganglia; gross defects are not apparent for dorsolateral migration, between the epidermis and the somites (Budi et al., 2008; Dooley et al., 2013; Honjo et al., 2008). All of these observations pointed to the possibility that stem cells giving rise to adult melanophores might be established in, and recruited from, medial “extra-hypodermal” regions and perhaps the peripheral nervous system itself (see below).
Larval regenerative melanophores: independent evidence for a melanophore stem cell
Additional indications that zebrafish have a melanophore stem cell come from studies of larval melanophore regeneration. When EL melanophores are ablated by laser or drug treatment at 2–3 dpf, new melanophores arise from proliferative precursors and replace all but the most ventral cells that were lost (Yang and Johnson, 2006; Yang et al., 2004); if the new melanophores are themselves ablated, they too are replaced, suggesting that precursors to regenerative melanophores are self-renewing (Hultman et al., 2009; O'Reilly-Pol and Johnson, 2013; Tryon et al., 2011). In principle, these regenerative melanophores might arise from “left over” precursors to EL melanophores that simply failed to differentiate during embryogenesis. Yet, genetic and cell lineage analyses argue against this idea and instead provide evidence for a stem cell that is distinct from the EL melanophore lineage.
First, regenerative melanophores have requirements different from EL melanophores, but shared with adult melanophores. For example, regeneration fails, just as adult melanophore development fails, when ErbB signaling is blocked genetically, or pharmacologically during neural crest migration (Hultman et al., 2009) (Fig. 3A). Regeneration also fails in homozygous kita null alleles, demonstrating a requirement for Kit signaling (Yang and Johnson, 2006) similar to that exhibited by most adult melanophores (Johnson et al., 1995). Two additional genes required by regenerative but not EL melanophores have been identified by forward genetic screening for melanophore regeneration mutants: glutamine:fructose-6-phosphate aminotransferase 2 (gfpt2) acts within regenerative melanophores to promote terminal differentiation; the RNA helicase gene skiv2l2 promotes the proliferation of regenerative melanophore precursors and is required for regeneration of additional tissues as well (Yang et al., 2007). Regenerative melanophores (Johnson et al., 2011) and adult melanophores (Dooley et al., 2013) also both arise independently of Microphthalmia (Mitf) activity during embryonic stages, when this factor is required for the specification of EL melanophores (Hou and Pavan, 2008; Levy et al., 2006; Lister et al., 1999; Opdecamp et al., 1997).
Figure 3.
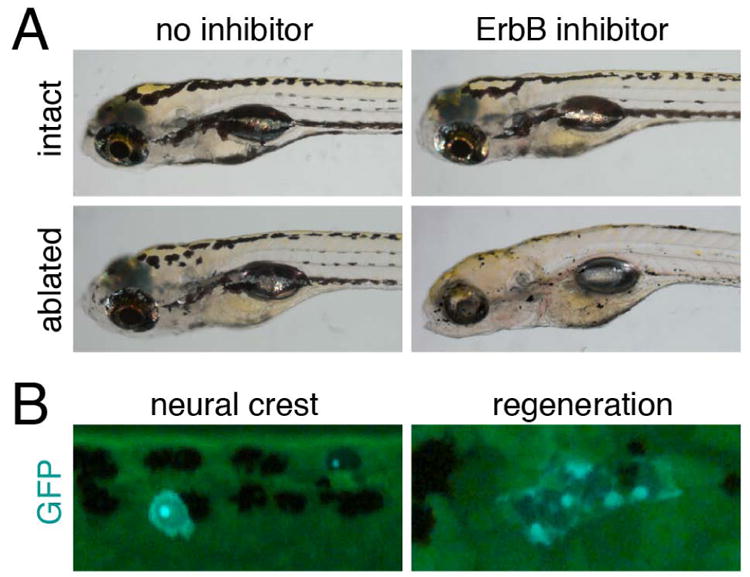
Larval melanophore regeneration requires stem cells. (A) Larvae in which EL melanophores have been ablated and new melanophore regenerated resemble control, unablated larvae. Yet regeneration is impaired when ErbB signaling is inhibited. (B) Transposon-based lineage analyses reveal GFP+ EL melanophores derived directly from neural crest, as well as GFP+ clones of regenerative melanophores. Images courtesy of R. Tryon and S. Johnson.
Second, evidence for distinct lineages of EL melanophores and stem-cell derived regenerative melanophores has come from transposon-based lineage analyses in which individual clones of tyrosinase+ cells were labeled during normal development and in regeneration (Tryon et al., 2011; Tryon and Johnson, 2012) (Fig. 3B). Transposon integration typically occurs by early gastrulation and most tyrosinase+ clones (59%) gave rise to both EL melanophores and regeneration melanophores (Tryon et al., 2011). Nevertheless, some clones, presumably labeled slightly later, gave rise only to EL melanophores (31%) or only to regenerative melanophores (10%). These outcomes suggest a segregation of EL and regenerative fates shortly after the onset of gastrulation and by the time of neural crest dispersal. Further analyses of these data also suggested that 90–190 cells in the early larva are able to generate regenerative melanophores, or, presumably, adult melanophores. This estimate of founding stem cell number is larger than the numbers of progenitors suggested from qualitative observations of patterns in chimeras in mouse and zebrafish (Lin et al., 1992; Mintz, 1967; Mintz, 1971), but not clearly different from observations based on clonal analyses in mouse (Wilkie et al., 2002).
Additional analyses of larval melanophore regeneration have provided further clues to fate segregation and stem cell establishment. For example, applying transposon lineage analyses to fish heterozygous for kita revealed more clones giving rise to EL melanophores and fewer clones giving rise to regenerative melanophores (O'Reilly-Pol and Johnson, 2013). This suggests that Kit signaling promotes adoption of a stem cell fate over EL melanophore fate, and adds to the several roles identified for Kit in melanophore and melanocyte lineages (Besmer et al., 1993; Budi et al., 2011; Hultman et al., 2007; Hultman et al., 2008; Mellgren and Johnson, 2004; Parichy et al., 1999; Rawls and Johnson, 2000; Rawls and Johnson, 2003; Wehrle-Haller, 2003). In a complementary strategy, control and ErbB-inhibited embryos were compared for their responsiveness to transgenically supplied Kit ligand-a (Kitlga) (Hultman et al., 2009). Wild-type embryos overexpressing Kitlga developed extra melanophores derived not from the EL melanophore lineage but instead from the stem cell lineage; surprisingly, ErbB-inhibited embryos overexpressing Kitlga developed even more stem-cell derived melanophores than their wild-type counterparts. This unexpected result suggests that stem cell precursors persist transiently in ErbB-inhibited embryos and remain available for recruitment by Kitlga at these early stages. Thus, ErbB signaling seems to promote the transition of an early precursor, specified for a stem cell fate, to the definitive stem cell itself, which may be more highly regulated, and thus less responsive to Kitlga.
Embryonic origins, niche and genetic requirements of melanophore stem cells
Studies of adult pigment pattern mutants and larval melanophore regeneration indicated the presence of melanophore stem cells but did not identify the cells themselves. This issue was addressed with a combination of molecular markers, fate mapping, and time-lapse imaging in wild-type and mutant backgrounds (Budi et al., 2011). Analyses of reporter expression in a transgenic line, Tg(mitfa:GFP)w47, during early stages of adult pattern formation revealed proliferative extra-hypodermal GFP+ cells, often associated with peripheral nerves (Fig. 4A–C). Although zebrafish mitfa reporters and mitfa transcript can be detected in other chromatophore lineages and in glia (Dooley et al., 2013; Eom et al., 2012; Parichy et al., 2000b) (see below), the melanogenic potential of at least some peripheral nerve associated cells was confirmed by post-embryonic overexpression of Kitlga, which resulted in ectopic melanophore development (Fig. 4D). Consistent with such cells being descendants of an ErbB-dependent stem cell established in the embryo, nerve-associated mitfa:GFP+ cells as well as ectopic Kitlga-induced melanophores were missing in the erbb3b mutant. To test if extra-hypodermal mitfa:GFP+ cells contribute to hypodermal chromatophores, two approaches were used (Fig. 4E,F). First, fate mapping using the vital dye, DiI, showed that labeling of extra-hypodermal cells was followed several days later by the occurence of labeled melanophores in the hypodermis. Second, time-lapse imaging revealed mitfa:GFP+ cells entering the hypodermis from extra-hypodermal locations, after migrating over the dorsal or ventral edges of the myotomes, or via the horizontal or vertical myosepta (Movies 2, 3).
Figure 4.
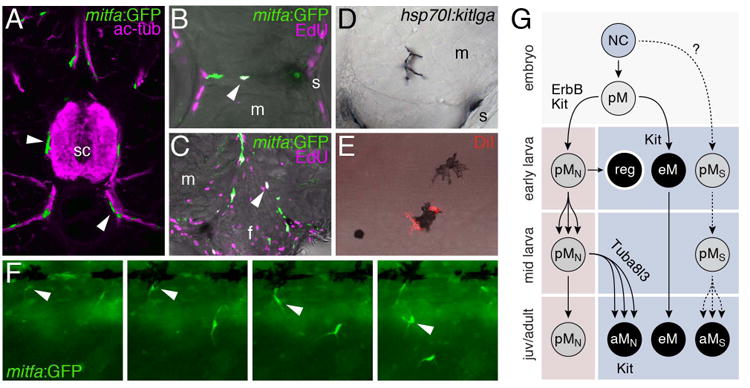
Melanophore stem cells. (A) mitfa:GFP+ cells (green) associated with peripheral nerves marked (ac-tub; magenta) at an early stage of adult pattern development. sc, spinal cord. (B,C) Proliferative mitfa:GFP+ cells in the horizontal myoseptum (B) and near the base of the fin (C) are indicated with white nuclei (arrowheads) in overlays with staining for EdU (magenta). m, myotome; s, skin; f, fin base. (D) Ectopic expression of Kitlga using the heat shock inducible promoter of hsp70l results in melanophore development in the vicinity of peripheral nerves coursing through the myotomes. (E) Injection of DiI in extra-hypodermal regions results in later DiI-labeled melanophores in the hypodermis. (F) Time-lapse frames showing mitfa:GFP+ cells entering the hypodermis (e.g., arrowhead). (G) Model for melanophore lineages showing approximate stages (top to bottom) and later tissue compartments (pink, extra-hypodermal; blue, hypodermal). Progenitors specific for particular chromatophore classes are preceded with a “p”. Neural crest (NC) cells generate a melanophore progenitor (pM) in the embryo that generates ErbB-dependent, peripheral nerve associated melanophore stem cells (pMN) as well as embryonic/early larval melanophores (eM). If eM are ablated, pMN are recruited to the skin as regenerative larval melanophores (reg), but during normal development pMN remain associated with peripheral nerves where they proliferate and ultimately, during pigment pattern metamorphosis, migrate to the hypodermis to generate adult melanophores (aMN). This recruitment fails in tuba8l3 mutants. Some eM persist into the adult though many are lost, particularly from the interstripe. Kit signaling has several roles, promoting establishment of pMN and also the migration, survival, and differentiation of eM and aMN. An additional lineage (pMS) able to generate adult melanophores and possibly other chromatophore classes is hypothetical (see text).
The earliest localization of melanophore stem cells has been examined as well (Dooley et al., 2013). Consistent with previous inferences, mitfa:GFP+ cells were found within the ventromedial neural crest migratory pathway associated with DRG, and were later found more broadly along spinal nerves. By transplanting cells from embryos depleted of EL melanophores to albino hosts, it was further possible to correlate donor, DRG-associated mitfa:GFP+ cells with later patches of melanized melanophores in the adult. This same study identified the sparse-like mutant as allelic to kitlga, and demonstrated that in this background mitfa:GFP+ cells fail to localize at the DRG, consistent with roles for Kit signaling in establishing the melanophore stem cell (O'Reilly-Pol and Johnson, 2013; Yang and Johnson, 2006). Further studies will need to determine whether the peripheral-nerve associated mitfa:GFP+ cells identified at later stages by Budi et al. (2011) correspond to the progeny of early mitfa:GFP+ cells identified by Dooley et al. (2013), or whether additional sources of such cells and melanophores are present.
Whether or not the cells identified so far in zebrafish represent a single population, the existence of peripheral-nerve associated precursors to adult melanophores is reminiscent of peripheral nerve-associated cells that give rise to skin melanocytes in amniotes (Adameyko et al., 2009) and raises the possibility of an evolutionarily ancient origin of this lineage. In chick, these cells express Sox10, an early marker of both melanocyte and glial lineages, and FoxD3—a marker of multipotent neural crest and glia—represses the melanocyte fate; in mouse, nerve-associated melanocyte precursors initially express the promoter of proteolipid protein (PLP), which is active in Schwann cells and their precursors (Adameyko et al., 2009; Nitzan et al., 2013b). In zebrafish, PLP does not mark peripheral glia (Brosamle and Halpern, 2002) but analyses thus far indicate that mitfa:GFP+ cells co-express Sox10 (as would be expected) and also can express Foxd3; interestingly the mitfa:GFP+; Foxd3+ population is especially proliferative (Budi et al., 2011). In principle these multiply labeled cells could represent multipotent precursors adopting a melanophore fate, cells in which an initial melanogenic potential is being repressed, or another lineage entirely. Additional fate mapping should resolve these issues and may provide new insights into the homology of zebrafish and amniote peripheral nerve associated precursors.
Once melanophore stem cells are established in zebrafish, several pathways are required for the recruitment of hypodermal adult melanophores. For example, recruitment fails in tuba8l3 mutants, which exhibit fewer mitfa:GFP+ cells overall; those cells that do reach the hypodermis are more likely than wild-type cells to differentiate, suggesting a compensatory response to their overall depletion (Budi et al., 2011). kita mutants, however, exhibit numerous mitfa:GFP+ cells during adult pattern development, in contrast to the deficiency of these cells at very early stages (Dooley et al., 2013). The later mitfa:GFP+ cells fail to melanize and are more likely to die than those of wild-type, suggesting additional roles for Kit during terminal differentiation (Budi et al., 2011; Mills et al., 2007) as has been inferred in other contexts (Mellgren and Johnson, 2004; Rawls and Johnson, 2000). The few adult melanophores that do differentiate in kita mutants proliferate extensively, likely explaining the late-arising pattern of residual stripes (Fig. 2A) (Budi et al., 2011). Post-embryonic development of mitfa:GFP+ cells or melanophores also requires the gap junction protein Connexin41.8 (Johnson et al., 1995; Maderspacher and Nusslein-Volhard, 2003; Watanabe et al., 2006; Watanabe and Kondo, 2012), the potassium channel Kcnj13 (Haffter et al., 1996; Iwashita et al., 2006; Maderspacher and Nusslein-Volhard, 2003), the cell adhesion molecule Immunoglobulin superfamily 11 (Eom et al., 2012), Endothelin receptor b1a (Budi et al., 2011; Johnson et al., 1995; Parichy et al., 2000a), cell surface scaffolding protein Tetraspanin 3c (Inoue et al., 2014), and Notch signals (Hamada et al., 2014). Finally, additional factors, including Basonuclin-2 (Bnc2) (Lang et al., 2009; Patterson and Parichy, 2013), Colony stimulating factor-1 (Csf1) receptor (Budi et al., 2011; Maderspacher and Nusslein-Volhard, 2003; Parichy et al., 2000b; Parichy and Turner, 2003a; Patterson and Parichy, 2013) and others (Amsterdam et al., 2009; Frohnhofer et al., 2013; Kawakami et al., 2000; Krauss et al., 2013; Lopes et al., 2008; Patterson and Parichy, 2013) act—or are very likely to act—through other cell types to promote adult melanophore development.
Together, these analyses suggest a model for the establishment, maintenance, and recruitment of melanophore stem cells (Fig. 4G) in which early progenitors, that can generate EL melanophores or melanophore stem cells, become restricted for the latter fate and migrate along the ventromedial neural crest migratory pathway, requiring ErbB signaling for the development of DRG at which they localize, in a manner that also requires Kita. These same cells can be recruited as regenerative melanophores when EL melanophores are ablated. During the larval-to-adult transformation, DRG-associated melanophore stem cells begin to proliferate, populate the peripheral nerves more extensively, and they or their progeny migrate to the hypodermis along peripheral nerves and through the myosepta, or after detaching from peripheral nerves and traveling over the edges of the myotomes. Once in the hypodermis, the cells begin to differentiate as melanophores. Meanwhile, some EL melanophores persisting from the early pattern join developing adult stripes, whereas others die or are lost from view as they are covered by iridophores (Budi et al., 2008; Parichy and Turner, 2003b; Patterson and Parichy, 2013; Quigley et al., 2004; Takahashi and Kondo, 2008).
Comparative analyses of adult melanophore development have provided additional insights into EL and stem cell melanophore lineages. Stem cell derived melanophores make the major contribution to adult patterns of zebrafish and most of the zebrafish relatives that have been examined (Mills et al., 2007; Quigley et al., 2005; Quigley et al., 2004). Yet the spotted danio, D. nigrofasciatus, exhibits an evolutionarily derived condition with far fewer stem cell derived melanophores. Instead, stripes that are superficially similar to those of zebrafish arise largely from EL melanophores, which persist and reorganize in the adult. Genetic mosaic analyses demonstrate the species-difference has arisen through evolutionary changes non-autonomous to melanophore lineages (Quigley et al., 2004).
Among more distantly related species, flounders and other flatfishes provide exciting opportunities to examine pigment stem cells in the context of a metamorphosis more dramatic than that of zebrafish. In these fishes a bilaterally symmetric larva—with a bilaterally symmetric pigment pattern—metamorphoses into an asymmetric adult having an upper (ocular) side with a distinct adult pigment pattern, and a lower (blind) side that is relatively unpigmented. In flounder, vital dye labeling and histological analyses suggest that precursors to adult melanophores colonize hypodermis on both upper and lower sides, via the myosepta and also from the bases of the fins (Washio et al., 2013; Watanabe et al., 2008; Yamada et al., 2010). Despite the migration of precursors to both sides, adult melanophores differentiate only on the upper side, a difference that correlates with lower dorsal than ventral levels of a factor known to inhibit melanization, Agouti-signaling protein 1 (Asip1) (Cerda-Reverter et al., 2005; Guillot et al., 2012).
A stem cell origin of adult iridophores
Although melanophores have been most studied, it is iridophores that are the first adult chromatophores to differentiate (Fig. 5A), and these cells play critical roles in establishing the position, and delimiting the width, of subsequently developing melanophore stripes (Frohnhofer et al., 2013; Parichy et al., 2009; Patterson et al., 2014; Patterson and Parichy, 2013; Singh et al., 2014). Given the interplay between iridophore and melanophore patterning it might be anticipated that earlier development of these cells could be coordinated as well. Indeed, mutants with defects in adult melanophore stem cell establishment have iridophore deficiencies (Fig. 5B) and fate mapping has indicated an extra-hypodermal origin of adult iridophores similar to that of melanophores (Budi et al., 2011). Confirming that adult iridophores originate from extra-hypodermal stem cells and providing further insights into this lineage are recent analyses using genetic labeling with ERT2-Cre, driven by the promoter of the very early lineage marker sox10 (Singh et al., 2014). Clones labeled after EL stages that gave rise to adult iridophores originated in the vicinity of the DRG and, interestingly, also generated neurons or glia (Fig. 5C), and small but variable numbers of melanophores in the adult. This suggests an initially multipotent progenitor that becomes further restricted for melanophore vs. iridophore/neural fates (Fig. 5D). In bipotent, directly neural crest derived cells of the embryo, an EL iridophore fate is specified over an EL melanophore fate by the transcription factor Foxd3 (Curran et al., 2010), and it will be interesting to learn whether Foxd3 acts similarly during these post-embryonic stages [and whether such cells might correspond to the multiply labeled cells described by Budi et al. (2011)].
Figure 5.
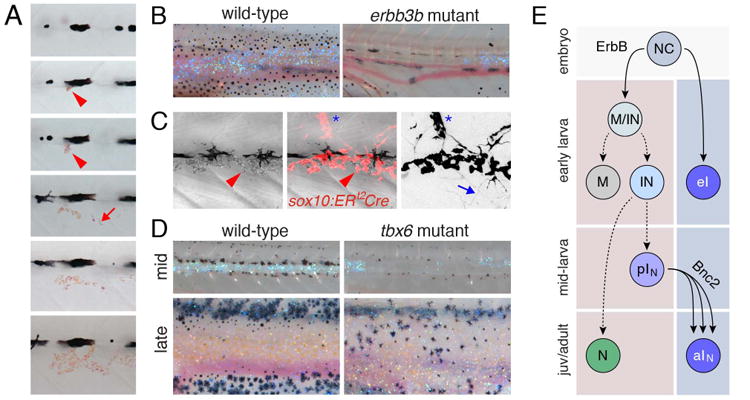
Adult iridophore origins. (A) The first adult iridophores to appear (arrowheads) arise at the horizontal myoseptum and then increase in number (arrow) to form the first interstripe. Shown is a single individual on consecutive days. (B) Iridophores, evident by blue reflectance, are reduced in erbb3b mutants that lack DRG. (C) Genetic labeling at early stages reveals the clonal expansion of iridophores in the hypodermis (arrowhead) and a shared lineage with neurons of the DRG (asterisk); arrow, neuronal arbor (right image inverted to better show neural labeling; courtesy of A. Singh, P. Mahalwar, C. Nüsslein-Volhard). (D) Myosepta provide routes by which iridophore precursors reach the skin as illustrated by defects in iridophore (and melanophore) abundance and patterning in tbx6 mutants. (E) Model for iridophore lineages. NC cells give rise to a few iridophores of the EL pattern (eI) as well as multipotent cells in peripheral nerves (MIN) that can initially generate melanophores, iridophores, neural derivatives but subsequently undergo further restriction (M vs. IN). It is not clear whether M correspond to pMN in Figure 4G, nor whether there exists a progenitor restricted to the iridophore fate alone (pIN), though these seem likely.
An additional factor likely important to adult iridophore specification is Leucocyte tyrosine kinase (Ltk) which acts autonomously to the iridophore lineage (Lopes et al., 2008). ltk is expressed by iridophores and their precursors and in the ltk mutant, shady, EL iridophore specification fails. The several shady alleles also have reduced complements of adult iridophores ranging in severity from complete absence to only a minor deficiency, and it will be interesting to learn precisely when and how Ltk promotes the development of this adult stem cell derived population.
Presumably during or after their specification, adult iridophore precursors migrate to the hypodermis via the horizontal myoseptum (Singh et al., 2014). Mutants with defects in myoseptum development, including choker (Frohnhofer et al., 2013; Svetic et al., 2007) and fused somites (tbx6) (Nikaido et al., 2002; Windner et al., 2012), have corresponding defects in iridophore colonization of the flank as well as later pattern formation (Fig. 5D). Once iridophores have arrived in the hypodermis, the cells proliferate to form dense aggregations at the first interstripe but also spread further dorsally and ventrally, ultimately initiating additional interstripes as well (Patterson et al., 2014; Singh et al., 2014). In mutants for the highly conserved zinc finger protein, Basonuclin-2, the iridophore population fails to expand; adults lack most of their body iridophores as well as melanophores and xanthophores (Lang et al., 2009; Patterson and Parichy, 2013). Although mutant phenotypes identify several additional loci required by adult iridophores (Amsterdam et al., 2009; Haffter et al., 1996; Kawakami et al., 2000; Krauss et al., 2013; Lopes et al., 2008), including genes of the Endothelin signaling pathway (Krauss et al., 2014; Parichy et al., 2000a), the precise molecular mechanisms by which iridophore stem cells are established and recruited to the adult pattern remain to be elucidated.
Origins of adult xanthophores and their relationships to other lineages
EL xanthophores initially over the lateral flank disappear before the adult pattern develops, whereas interstripe xanthophores are the last adult pigment cells on the flank to differentiate (McMenamin et al., 2014; Patterson et al., 2014; Patterson and Parichy, 2013) (Fig. 6A). From results on melanophores and iridophores it might be anticipated that adult xanthophores also arise from stem cells, perhaps associated with peripheral nerves. Yet, two recent studies show this is not the case, and that, instead, many adult xanthophores are derived directly from the EL xanthophore lineage (Mahalwar et al., 2014; McMenamin et al., 2014). In one analysis, early markers or neural crest cells (sox10) or xanthophores [pax7 or colony stimulating factor 1 receptor (see below)] were used with cell transplantation or Cre-recombination to label cells in the hypodermis at early larval stages. These same groups of cells increased in number and differentiated as xanthophores in the adult interstripe and also were present in lower numbers within the melanophore stripes (Mahalwar et al., 2014). In the other analysis (McMenamin et al., 2014), EL xanthophores were shown to give rise directly to adult xanthophores by fate mapping cells that had already differentiated using nuclear-localizing photoconvertible EosFP (nEosFP) driven by the promoter of aldehyde oxidase 3 (aox3), which encodes a pteridine synthesis enzyme (Parichy et al., 2000b; Ziegler et al., 2000): EL xanthophores marked at 4–5 days lost their pigment yet persisted and proliferated, and then many, localizing in the interstripe, re-acquired pigment during adult pattern development (Fig. 6A); others, localizing in the interstripe, failed to do so. This approach also revealed some adult xanthophores that initiated aox3:EosFP expression only after 4–5 days, suggesting they had not yet differentiated by the time of photoconversion; these cells may have been resident initially in the hypodermis as well (and presumably would have been labeled by the earlier lineage markers used by Mahalwar et al. 2014) (Fig. 6B). An aox3 reporter line further showed that cryptic aox3+ cells are widespread during adult pattern development, becoming densely packed and deeply pigmented in the interstripe, but also occurring in the stripes by juvenile stages. Results of both recent studies are thus consistent with earlier observations that lightly pigmented xanthophores occur at low density even within stripes of older, adult fish (Hirata et al., 2003; Parichy and Turner, 2003a). aox3+ cells also are found extra-hypodermally, but are not clearly associated with peripheral nerves. Although there is currently no evidence that extra-hypodermal cells migrate to the hypodermis to differentiate as xanthophores in zebrafish, xanthophore precursors are believed to migrate from extra-hypodermal locations to the skin during flounder metamorphosis (Washio et al., 2013).
Figure 6.
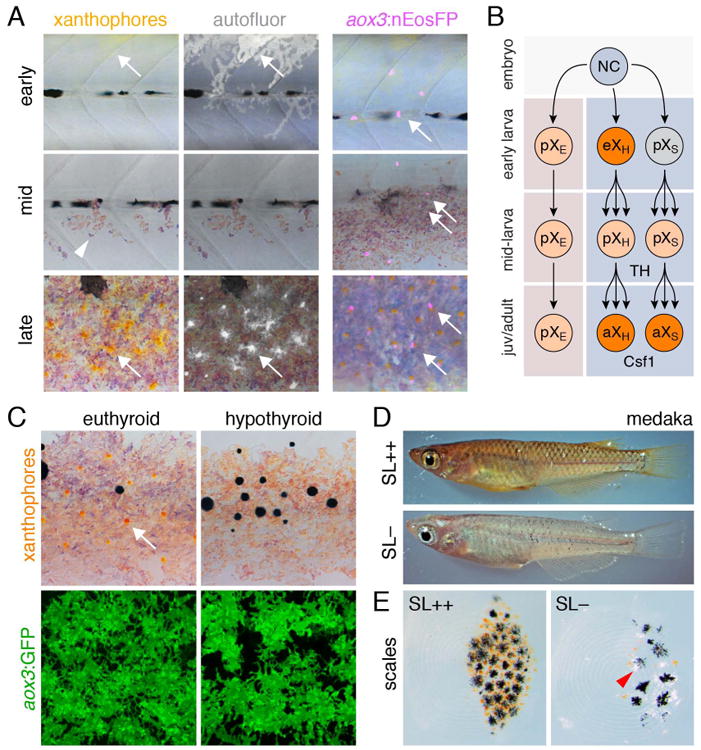
Xanthophore lineage and requirements. (A) Left, Autofluorescent xanthophores (arrows) are present in the early larva, but are not visible on the lateral flank during middle larval stages (though some remain visible along the dorsum; not shown). Arrowhead indicates iridophores. By late larval stages adult xanthophores are visible in the interstripe. Right, EL xanthophores marked with photoconvertible nEosFP (arrow), persist into the adult pattern. Fish are treated with epinephrine, to contract pigment towards cell centers, adjacent to nuclei. (B) In the embryo, NC cells differentiate as early larval xanthophores in the hypodermis (eXH), which then lose their pigment and proliferate, and ultimately re-differentiate as adult xanthophores. Additional aox3+ cells are found extra-hypodermally (pXE) and can differentiate in response to excess TH in zebrafish, or during normal development in other species. Other cells, presumed to arise in the hypodermis (pXS), do not fully differentiate as EL xanthophores, yet do differentiate as adult xanthophores (aXS). (C) In hypothyroid fish, adult xanthophores (arrow) fail to differentiate though aox3:GFP+ precursors remain. (D) In medaka, SL promotes xanthophore development in a ci mutant expressing SL constitively (upper) whereas ci mutants have dramatically fewer xanthophores, evident by reduced yellow coloration, as well as increased numbers of leucophores. Melanophore numbers are not significantly reduced in ci mutants compared to wild-type (Fukamachi et al., 2004). (E) Close-up of scales showing modulation of xanthophore and leucophore (arrowhead) abundances in response to SL. (D and E courtesy S. Fukamachi.).
Several pathways have been studied for roles in promoting adult xanthophore development. First is signaling by Colony stimulating factor-1 (Csf1), the receptor for which, Csf1r, is expressed by xanthophores and their precursors from very early stages and required for their development (Maderspacher and Nusslein-Volhard, 2003; Parichy et al., 2000b; Parichy and Turner, 2003a; Patterson and Parichy, 2013). Csf1 misexpression drives ectopic xanthophore differentiation, and during adult pattern development this factor is expressed by the skin and at high levels by interstripe iridophores, thereby promoting differentiation and probably also increased density of xanthophores in the interstripe (Patterson and Parichy, 2013). In csf1r mutants (pfeffer or panther), xanthophore development mostly fails in the embryo and adult (Haffter et al., 1996; Maderspacher and Nusslein-Volhard, 2003; Parichy et al., 2000b); the absence of EL xanthophores may be directly responsible for the absence of adult xanthophores loss, as would be predicted by aox3:nEosFP fate mapping. Indeed transgenic ablation of EL xanthophores causes adult xanthophore deficiency as well (McMenamin et al., 2014). In a comparative context, signaling through Csf1r is also required for xanthophore development in the guppy Poecilia reticulata (Kottler et al., 2013), whereas increased Csf1 expression has contributed to a marked increase in xanthophore numbers in the pearl danio, D. albolineatus (Patterson et al., 2014).
A second factor required by adult xanthophores is thyroid hormone (TH), known for roles in amphibian and flatfish metamorphosis and for tissue-specific effects in both ectotherms and amniotes (Horn and Heuer, 2009; Laudet, 2011; McMenamin and Parichy, 2013; Shi, 2000; Sirakov et al., 2013). Fish that are deficient for TH, either because they have had their thyroids ablated or because of mutations affecting Thyroid stimulating hormone receptor (Tshr), lack adult xanthophores owing to a failure of terminal differentiation (Fig. 6C) (McMenamin et al., 2014). Conversely, excess TH, either applied directly or resulting from a hyper-activating mutation of Tshr, results in precocious xanthophore differentiation and increased xanthophore proliferation, generating a dramatic increase in xanthophore numbers. Roles for TH in promoting xanthophore development are accompanied by repressive effects on melanophore population expansion; it remains to be determined whether TH effects on xanthophores and melanophores are direct or mediated through other cell types or factors.
Additional clues into adult xanthophore development have come from studies of medaka, Oryzias latipes, which has the same classes of pigment cells as zebrafish but also shiny yellow and white leucophores. An extensive collection of embryonic and adult pigment mutants exists for medaka (Fukamachi et al., 2001; Kelsh et al., 2004; Lamoreux et al., 2005; Tomita, 1975) and these are providing valuable insights complementary to work in zebrafish. For example, the medaka color interfere (ci) mutant exhibits dramatically fewer adult xanthophores and correspondingly more adult leucophores as compared to the wild-type. Positional cloning of ci revealed its correspondence to somatolactin, encoding a pituitary-derived endocrine factor, SL, similar to growth hormone (Fig. 6D) (Fukamachi et al., 2004; Fukamachi et al., 2006; Fukamachi et al., 2009). Interestingly, cobalt variant carp have a similar pigment phenotype associated with reduced SL production (Kaneko et al., 1993).
Consistent with the reciprocal changes in adult xanthophore and leucophore abundance in response to SL, lovely recent analyses of several additional medaka mutants imply a common xanthophore–leucophore progenitor at embryonic stages (Kimura et al., 2014; Nagao et al., 2014). These have identified a critical role for the Pax7a transcription role in development of both cell types, a role consistent with that identified for Pax7 in zebrafish embryonic xanthophore development (Minchin and Hughes, 2008) as well as a genetic association between pax7 allelic variation and a variant xanthophore phenotype in adult cichlids (Roberts et al., 2009). The recent medaka studies further showed that in this species, the Sox5 transcription factor specifies the xanthophore over leucophore fate. A downstream transporter, Slc2a15b, promotes leucophore differentiation, whereas both Slc2a15b and Slc2a11 promote yellow pigmentation in both xanthophores and leucophores. It will be interesting to learn if these same cells persist into the adult, or whether additional stem cells for adult xanthophores and leucophores exist in medaka.
Stem cells contributing to fin pigment pattern and regeneration
The preceding discussion has been limited to pigment cell lineages on the body. Yet, chromatophores on the fins are of considerable interest for at least two reasons. First, some mutants exhibit a decoupling of body and fin patterns, with one developing normally despite defects in the other (Lang et al., 2009; Mellgren and Johnson, 2006; Parichy et al., 2000a) (Fig. 7A,B). This suggests either that body and fin micro-environments differentially regulate chromatophore development, or that body and fins are populated by different chromatophore precursors having distinct genetic requirements. Second, the fin has served as a convenient system for studying regeneration: upon amputation, fin bones (lepidotrichia), vasculature, nerves, skin and pigment pattern are regenerated rapidly, and such cycles of amputation/regeneration can be repeated indefinitely (Goodrich and Nichols, 1931; Morgan, 1900; Tornini and Poss, 2014). Knowing more about the cells that make this possible could provide insights into mechanisms of regeneration and growth control more generally.
Figure 7.
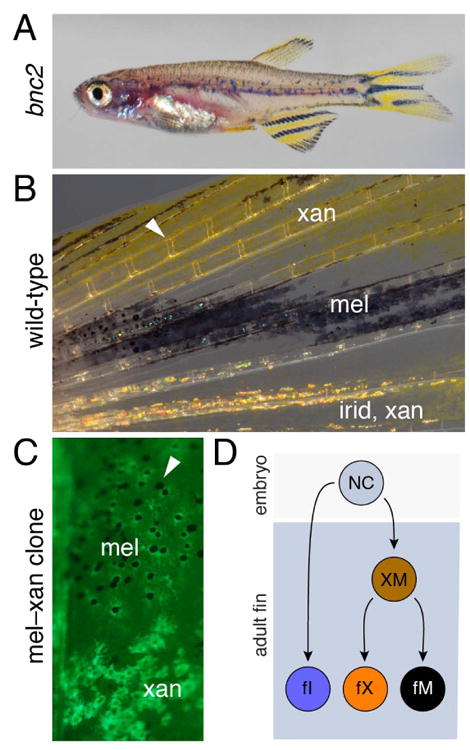
Fin chromatophore patterns and lineages. (A) Despite a severe deficiency in body chromatophores, the bnc2 mutant has normal fin chromatophores and patterns. (B) Detail of fin chromatophores. Arrowhead indicates joint between fin bones. (C) Example of transposon based lineage tracing that resulted in a single GFP+ clone in the anal fin, stretching from proximal (top) to distal (bottom), and contributing to both melanophores and xanthophores. Arrowhead indicates one of several unlabeled melanophores from an additional unmarked clone. (Image courtesy of R. Tryon and S. Johnson.) (D) NC cells giving rise to fin chromatophores are likely to segregate an adult fin iridophore (fI) lineage initially, followed by a bipotent stem cell (XM) able to generate fin xanthophores (fX) and fin melanophores (fM), that also persists into the adult, in which it can generate regenerative xanthophores and melanophores as well (not shown).
During normal development, transposon-based lineage analyses using a tyrosinase reporter showed that developing adult fins are likely to be colonized by fewer than ten precursor cells that give rise to all of the fin melanophores (Tu and Johnson, 2010). Some melanophore clones populated only proximal regions of the fin, whereas others extended from proximally to the distal tip where growth occurs. Interestingly, clones that generated melanophores invariably generated xanthophores, and vice versa (Tu and Johnson, 2010; Tu and Johnson, 2011) (Fig. 7C). This result indicates that founding cells that colonize the fins are not restricted for one or the other fate at the time of labeling, reminiscent of the multiple chromatophore fates sometimes generated by embryonic neural crest cells (Curran et al., 2010; Dutton et al., 2001; Raible and Eisen, 1994) and contrasting with the apparently distinct lineages of body melanophores and xanthophores characterized so far (Budi et al., 2011; Dooley et al., 2013; Mahalwar et al., 2014; McMenamin et al., 2014). The same analyses also revealed only very few clones that also produced iridophores, suggesting an earlier segregation of this fate. Together, these results strongly suggest the existence of one or more populations of stem cells in the fin distinct from those that have been identified so far on the body (Fig. 7D).
During regeneration, melanophores (and xanthophores) rapidly repopulate fin tissue and generate a normal pattern (Goodrich et al., 1954; Goodrich and Nichols, 1931; O'Reilly-Pol and Johnson, 2008), a process that requires the de novo differentiation of melanophores from unpigmented stem cells, rather than proliferation of existing melanophores (Rawls and Johnson, 2000). Moreover, the same clones that give rise to melanophores and xanthophores also give rise to regenerative melanophores and xanthophores (Tu and Johnson, 2010; Tu and Johnson, 2011).
Analyses of kita null and temperature sensitive alleles showed that melanophore regeneration requires Kit signaling, which acts not to establish a fin chromatophore stem cell during embryonic stages but instead promotes the terminal differentiation of regenerating melanophores in the fin itself; in the absence of Kit signaling some melanophores regenerate but they are markedly delayed in their appearance (Rawls and Johnson, 2000; Rawls and Johnson, 2001). These observations led to the classification of fin regenerative melanophores as either primary, kita-dependent cells or secondary, kita-independent cells. Whereas primary melanophores differentiate distally near the front of regenerating tissue, secondary melanophores differentiate more proximally near the plane of amputation. Although melanophore classes having different Kit dependencies might indicate the existence of two distinct populations of stem cells, this hypothesis was excluded by elegant transposon-based lineage analyses using a temperature sensitive kita allele, in which it was possible to challenge cells to regenerate successively at either restrictive or permissive temperature: single clones could regenerate both primary, kita-dependent and secondary, kita-independent regeneration melanophores, implying that a single stem cell relies on Kit signaling for specific cellular processes (Tu and Johnson, 2010). These cells may, for instance, require Kit both for the proliferation and migration towards the distal tip of the regenerate; in the absence of such signaling, the numbers of regenerative melanophores would be reduced and their distribution restricted to the vicinity of the stem cell niche(s).
Even more stem cells?
As the preceding sections show, several distinct lineages of direct developing and stem cell derived chromatophores have now been identified as contributing to the adult pigment pattern of zebrafish. There may be still more lineages. For example, the existence of bipotent melanophore and xanthophore stem cells in the fin (Tu and Johnson, 2010; Tu and Johnson, 2011) suggest such cells might be present on the body as well. If so, these could be the same cells that generate some adult xanthophores inferred from fate mapping (McMenamin et al., 2014). Indeed, extensive regeneration after even repeated laser ablations of adult cells suggest the existence of widespread precursors of still-unknown origin (Nakamasu et al., 2009; Takahashi and Kondo, 2008; Yamaguchi et al., 2007).
A candidate marker for such stem cells or their immediate progeny in adults is mitfa (Lister et al., 1999). Although classically considered a marker of the melanocyte or melanophore lineage, mitfa expression is detectable in cells expressing markers of the xanthophore lineage in the embryo (Parichy et al., 2000b). Later, during development of the adult pattern, mitfa transgenes are typically expressed in the hypodermis by spindle-shaped and highly motile precursors corresponding to melanoblasts, and at lower levels in stellate, slower moving xanthophores and xanthoblasts (Budi et al., 2011; Eom et al., 2012; McMenamin et al., 2014). In the adult, mitfa reporters are expressed in melanophores, xanthophores, and unpigmented cells (Fig. 8A). Although not a substitute for lineage analysis, these expression domains raise the possibility that cells expressing mitfa arise from bipotent or even multipotent stem cells. It will be interesting to learn whether even earlier markers of the neural crest lineage (Luo et al., 2001) identify such stem cells.
Figure 8.
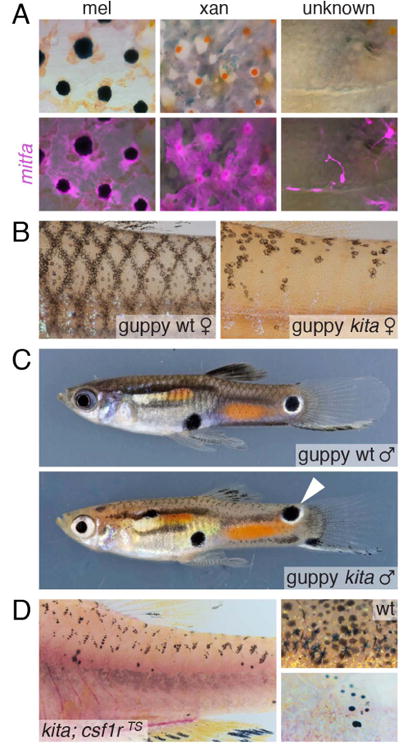
Additional lineages may be present. (A) mitfa transgenes mark adult melanophores, xanthophores and unpigmented cells in the adult. (B) Kita-independent melanophores persist on scales of guppies. (C) Kita-independent melanophore-containing ornaments (e.g., arrowhead) persist in the beautiful patterns of male guppies. (D) In zebrafish, kita-independent melanophores are revealed on scales following csf1r-dependent ablation and regeneration. (Images in B and C courtesy of V. Kottler and C. Dreyer.)
Beyond molecular markers, several phenotypes are consistent with an undocumented source of adult pigment cells as well. As noted above, for example, kita mutants develop adult melanophores despite their defect in establishing nerve-associated melanophore stem cells (Dooley et al., 2013; O'Reilly-Pol and Johnson, 2013) and the complete absence of melanophores at middle larval stages (Johnson et al., 1995; Mills et al., 2007) (Fig. 2A). Similar Kita-independent melanophores are present in adult guppies, and include cells that contribute to the elaborate pattern “ornaments” unique to males (Kottler et al., 2013) (Fig. 8B,C). Moreover, genetic ablations of chromatophores in zebrafish doubly mutant for a null allele of kita and a temperature-sensitive allele of csf1r result in the development of scale xanthophores and melanophores (Budi et al., 2011) (Fig. 8D) even though scale melanophores are not present normally in kita mutants, suggesting that an additional population of stem cells may have been uncovered in this context. Finally, in both tuba8l3 mutants and ErbB-inhibited fish, most adult melanophores are initially lacking but new melanophores eventually develop (Budi et al., 2008; Parichy and Turner, 2003b; Parichy et al., 2003). All of these observations raise the possibility of additional stem cells competent to generate melanophores and perhaps other chromatophore classes.
Conclusions: implications for development, translational biology and evolution
Work so far indicates a surprising diversity of embryonic and adult chromatophore lineages in teleosts, with perhaps more lineages to be discovered. Such analyses add substantially to our understanding of neural crest fates and complement recent studies of other neural crest sub-lineage contributions (or lack thereof) to traits of teleosts and other species (Espinosa-Medina et al., 2014; Mongera and Nusslein-Volhard, 2013; Mongera et al., 2013; Shimada et al., 2013). It will also be interesting to learn how many other adult traits depend substantially on post-embryonic stem cells, as opposed to cells that have already acquired their definitive phenotypes during embryogenesis.
Another area in which studies of adult pigment cell lineages seem poised to make a contribution is our understanding of melanoma onset and progression. Melanoma remains one of the deadliest cancers and its incidence has continued to increase (Balch et al., 2010; Balch et al., 2001; Howlader et al., 2014; Jemal et al., 2009). Several studies of amniotes suggest that the invasive and metastatic potential of melanoma cells is attributable, at least in part, to the re-expression of genes used during early stages of neural crest and melanocyte lineage development (Bailey et al., 2012; Gupta et al., 2005; Kulesa et al., 2013). Despite such realizations, and the clear association of melanoma cells with a neural crest–melanocyte lineage, it remains unclear to what extent melanomas arises from cells at different steps in this lineage (e.g., latent stem cells vs. transit amplifying cells vs. differentiated melanocytes) (Bennett, 2003; Grichnik et al., 2006). Several teleosts have been used for studying melanoma (Meierjohann and Schartl, 2006; Patton et al., 2010; Schartl et al., 2012) and in recent years the zebrafish has become a tractable model, in which melanoma can be induced using the same oncogenic mutation (BRAFV600E) responsible for many human melanomas (Patton et al., 2005). Studies using this system have provided important new insights into genomic alterations in melanoma cells and their metastatic potential, and are aiding in drug discovery (Ceol et al., 2011; Lister et al., 2014; Michailidou et al., 2009; White et al., 2011; Yen et al., 2013). Interestingly, ErbB-inhibited fish remain susceptible to melanoma (Santoriello et al., 2012), adding to the notion that additional stem cell populations may yet be found in fish that contribute to melanoma, and such findings would likely have great translational relevance to human.
In an evolutionary context, studies of teleost pigment cell lineages invite interesting comparisons with other phylogenetic groups. Although the precise origins of adult chromatophores in other ectothermic vertebrates have yet to be elucidated, it seems likely that lineages homologous or analogous to those of teleosts will be found, particularly in amphibians, given the ontogenetic changes in pigment patterns often observed in this group (Niu and Twitty, 1950; Parichy, 1998; Parichy et al., 2006; Stearner, 1946). In a broader phylogenetic context, a potential homology of peripheral nerve associated precursors to teleost melanophores and amniote melanocytes has been cited already, and it is conceivable that teleosts will be found to have adult stem cells homologous to melanocyte stem cells present in hair (Nishimura, 2011; Nishimura et al., 2002; Tanimura et al., 2011) or feather (Lin et al., 2013) follicles as well. Although xanthophores are not found in amniotes, their pigments—pteridines and carotenoids—are found in avian feathers and in the avian iris, and orthologues of a gene required for xanthophore differentiation in medaka (slc2a11b) persist in birds, though they have been lost from mammals, which lack such pigmentation (Kimura et al., 2014). Xanthophore-like cells also have been identified in some species of ascidians (Jeffery, 2006; Jeffery et al., 2008; Jeffery et al., 2004); it will be exciting to learn if these cells depend on a gene regulatory network similar to that of vertebrate xanthophores, and whether or not such a network reflects shared ancestry, or an independent recruitment to such a function in vertebrates and non-vertebrate chordates.
Finally, studies of adult pigment cell lineages also suggest clues to the evolutionary diversification of pigment patterns themselves. Development of adult pigment patterns requires interactions among different pigment cell classes, and changes in these interactions and the timing of pigment cell differentiation are likely to explain some species differences (Kondo and Asai, 1995; Miyazawa et al., 2010; Patterson et al., 2014; Quigley et al., 2005; Watanabe and Kondo, 2012). Diversification of teleost pigment patterns has likely been facilitated by extra genes persisting from whole-genome duplication and the attendant opportunities for sub-functionalization Braasch et al., 2009a; Braasch et al., 2006; Braasch et al., 2009b; Force et al., 1999; Hultman et al., 2007). The existence of multiple chromatophore lineages in teleosts—having different developmental origins and genetic requirements—seems likely to have added another dimension to pattern diversification, in providing additional sources of variation upon which selection can act and additional possibilities for cell-type specialization and complexity. Together, these several factors seem likely to have contributed to the often striking pattern differences across life cycle stages within species (Booth, 1990; Quigley et al., 2004), and stunning pattern diversity across species (Baldwin, 2013; Fan et al., 2012; Greenwood et al., 2012; Kottler et al., 2014; Mills and Patterson, 2008; Seehausen et al., 2008), including the evolution of species-specific ornaments and other pigmentary adaptations (Hughes et al., 2013; Kemp et al., 2009; Kottler et al., 2014; Martin et al., 2014; Santos et al., 2014). All of these possibilities demand further experimental analyses that are likely to be very interesting, whatever their ultimate conclusions might be.
Supplementary Material
Movie 1. Adult pigment pattern development in zebrafish over ∼3 weeks, beginning with the early larval pattern and ending with the juvenile pattern. Animation highlights changes in melanophore pattern and is compiled from repeated daily imaging of a single individual, with frames aligned and rescaled to control for growth (Quigley et al., 2005).
Movie 2. mitfa:GFP+ cells (e.g., arrows) migrating to the hypodermis from the dorsal aspect of the myotome (Budi et al., 2011).
Movie 3. mitfa:GFP+ cells (e.g., arrow) enter the hypodermis via the myoseptum during adult pattern development (Budi et al., 2011).
Acknowledgments
Supported by NIH R01 GM062182 and R01 GM111233 to DMP.
Footnotes
Days post-fertilization are used here for heuristic purposes only. In practice, development rates differ markedly between fish strains and laboratories so age is not an adequate indicator of developmental stage. Normal stages of post-embryonic zebrafish development based on measures of size and developmental milestones have been described and specific stage reporting conventions have been recommended to account for genetic and environmental variation (Parichy et al., 2009).
References
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Muller T, Fritz N, Beljajeva A, Mochii M, Liste I, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–79. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Lai K, Komisarczuk AZ, Becker TS, Bronson RT, Hopkins N, Lees JA. Zebrafish Hagoromo mutants up-regulate fgf8 postembryonically and develop neuroblastoma. Mol Cancer Res. 2009;7:841–50. doi: 10.1158/1541-7786.MCR-08-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam M, Raja M, Vijayakumar C, Malaiammal P, Mayden RL. Natural history of zebrafish (Danio rerio) in India. Zebrafish. 2013;10:1–14. doi: 10.1089/zeb.2012.0803. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Fernandez PJ, Fujii R. On the blue coloration of vertebrates. Pigment Cell Research. 2007;20:14–26. doi: 10.1111/j.1600-0749.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Matsumoto J. Chapter 2. Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In: Nordland JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, Ortonne JP, editors. The Pigmentary System: Physiology and Pathophysiology. New York, New York: Oxford University Press; 2006. [Google Scholar]
- Bagnara JT, Matsumoto J, Ferris W, Frost SK, Turner WA, Jr, Tchen TT, Taylor JD. Common origin of pigment cells. Science. 1979;203:410–5. doi: 10.1126/science.760198. [DOI] [PubMed] [Google Scholar]
- Bailey CM, Morrison JA, Kulesa PM. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment cell & melanoma research. 2012;25:573–83. doi: 10.1111/j.1755-148X.2012.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Ding S, Byrd DR, Cascinelli N, Cochran AJ, Coit DG, Eggermont AM, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J Clin Oncol. 2010;28:2452–9. doi: 10.1200/JCO.2009.27.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, Urist M, Mcmasters KM, Ross MI, Kirkwood JM, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–34. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- Baldwin CC. The phylogenetic significance of colour patterns in marine teleost larvae. Zool J Linn Soc. 2013;168:496–563. doi: 10.1111/zoj.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC. Human melanocyte senescence and melanoma susceptibility genes. Oncogene. 2003;22:3063–9. doi: 10.1038/sj.onc.1206446. [DOI] [PubMed] [Google Scholar]
- Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, Bachvarova RF. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev Suppl. 1993:125–37. [PubMed] [Google Scholar]
- Bonaventure J, Domingues MJ, Larue L. Cellular and molecular mechanisms controlling the migration of melanocytes and melanoma cells. Pigment Cell Melanoma Res. 2013;26:316–25. doi: 10.1111/pcmr.12080. [DOI] [PubMed] [Google Scholar]
- Booth CL. Evolutionary significance of ontogenetic colour change in animals. Biol J Linn Soc. 1990;40:125–163. [Google Scholar]
- Braasch I, Brunet F, Volff JN, Schartl M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol Evol. 2009a;1:479–93. doi: 10.1093/gbe/evp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Braasch I, Volff JN, Schartl M. The endothelin system: evolution of vertebrate-specific ligand-receptor interactions by three rounds of genome duplication. Mol Biol Evol. 2009b;26:783–99. doi: 10.1093/molbev/msp015. [DOI] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008;135:2603–14. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Post-embryonic nerve-associated precursors to adult pigment cells: genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genet. 2011;7:e1002044. doi: 10.1371/journal.pgen.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle WE, Little CC. On a Modified Mendelian Ratio among Yellow Mice. Science. 1910;32:868–70. doi: 10.1126/science.32.833.868. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Reverter JM, Haitina T, Schioth HB, Peter RE. Gene structure of the goldfish agouti-signaling protein: a putative role in the dorsal-ventral pigment pattern of fish. Endocrinology. 2005;146:1597–610. doi: 10.1210/en.2004-1346. [DOI] [PubMed] [Google Scholar]
- Curran K, Lister JA, Kunkel GR, Prendergast A, Parichy DM, Raible DW. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev Biol. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley CM, Mongera A, Walderich B, Nusslein-Volhard C. On the embryonic origin of adult melanophores: the role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development. 2013;140:1003–13. doi: 10.1242/dev.087007. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396:370–3. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- Dushane GP. The Origin of Pigment Cells in Amphibia. Science. 1934;80:620–1. doi: 10.1126/science.80.2087.620-a. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–25. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Endler JA. Natural-Selection on Color Patterns in Poecilia-Reticulata. Evolution. 1980;34:76–91. doi: 10.1111/j.1558-5646.1980.tb04790.x. [DOI] [PubMed] [Google Scholar]
- Engeszer R, Dabarbiano L, Ryan M, Parichy D. Timing and plasticity of shoaling behaviour in the zebrafish, Danio rerio. Animal Behaviour. 2007a;74:1269–1275. doi: 10.1016/j.anbehav.2007.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007b;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- Engeszer RE, Wang G, Ryan MJ, Parichy DM. Sex-specific perceptual spaces for a vertebrate basal social aggregative behavior. Proc Natl Acad Sci U S A. 2008;105:929–33. doi: 10.1073/pnas.0708778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom DS, Inoue S, Patterson LB, Gordon TN, Slingwine R, Kondo S, Watanabe M, Parichy DM. Melanophore migration and survival during zebrafish adult pigment stripe development require the immunoglobulin superfamily adhesion molecule Igsf11. PLoS Genet. 2012;8:e1002899. doi: 10.1371/journal.pgen.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Medina I, Outin E, Picard CA, Chettouh Z, Dymecki S, Consalez GG, Coppola E, Brunet JF. Neurodevelopment. Parasympathetic ganglia derive from Schwann cell precursors. Science. 2014;345:87–90. doi: 10.1126/science.1253286. [DOI] [PubMed] [Google Scholar]
- Fan SH, Elmer KR, Meyer A. Genomics of adaptation and speciation in cichlid fishes: recent advances and analyses in African and Neotropical lineages. Philos Trans R Soc B-Biol Sci. 2012;367:385–394. doi: 10.1098/rstb.2011.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnhofer HG, Krauss J, Maischein HM, Nusslein-Volhard C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development. 2013;140:2997–3007. doi: 10.1242/dev.096719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000;13:300–19. doi: 10.1034/j.1600-0749.2000.130502.x. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Shimada A, Shima A. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat Genet. 2001;28:381–5. doi: 10.1038/ng584. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Sugimoto M, Mitani H, Shima A. Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc Natl Acad Sci U S A. 2004;101:10661–6. doi: 10.1073/pnas.0401278101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamachi S, Wakamatsu Y, Mitani H. Medaka double mutants for color interfere and leucophore free: characterization of the xanthophore-somatolactin relationship using the leucophore free gene. Development Genes and Evolution. 2006;216:152–157. doi: 10.1007/s00427-005-0040-9. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Yada T, Meyer A, Kinoshita M. Effects of constitutive expression of somatolactin alpha on skin pigmentation in medaka. Gene. 2009;442:81–7. doi: 10.1016/j.gene.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Goda M, Fujii R. Blue chromatophores in two species of callionymid fish. Zoological Science. 1995;12:811–813. [Google Scholar]
- Goda M, Fujiyoshi Y, Sugimoto M, Fujii R. Novel Dichromatic Chromatophores in the Integument of the Mandarin Fish Synchiropus splendidus. Biol Bull-Us. 2013;224:14–17. doi: 10.1086/BBLv224n1p14. [DOI] [PubMed] [Google Scholar]
- Goodrich HB, Marzullo CM, Bronson WR. An analysis of the formation of color patterns in two fresh-water fish. J exp Zool. 1954;125:487–505. [Google Scholar]
- Goodrich HB, Nichols R. The development and the regeneration of the color pattern in Brachydanio rerio. J Morphol. 1931;52:513–523. [Google Scholar]
- Greenwood AK, Cech JN, Peichel CL. Molecular and developmental contributions to divergent pigment patterns in marine and freshwater sticklebacks. Evol Dev. 2012;14:351–62. doi: 10.1111/j.1525-142X.2012.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grichnik JM, Burch JA, Schulteis RD, Shan S, Liu J, Darrow TL, Vervaert CE, Seigler HF. Melanoma, a tumor based on a mutant stem cell? J Invest Dermatol. 2006;126:142–53. doi: 10.1038/sj.jid.5700017. [DOI] [PubMed] [Google Scholar]
- Guillot R, Ceinos RM, Cal R, Rotllant J, Cerda-Reverter JM. Transient ectopic overexpression of agouti-signalling protein 1 (asip1) induces pigment anomalies in flatfish. PLoS One. 2012;7:e48526. doi: 10.1371/journal.pone.0048526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Odenthal J, Mullins MC, Lin S, Farrell MJ, Vogelsang E, Haas F, Brand M, Van Eeden FJ, Furutani-Seiki M, et al. Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol. 1996;206:260–76. doi: 10.1007/s004270050051. [DOI] [PubMed] [Google Scholar]
- Hall BK, Hörstadius S. The Neural Crest. New York, New York: Oxford University Press; 1988. [Google Scholar]
- Hamada H, Watanabe M, Lau HE, Nishida T, Hasegawa T, Parichy DM, Kondo S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development. 2014;141:318–24. doi: 10.1242/dev.099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes JW. The structure of fish skin. I. General organization. Cell Tiss Res. 1974a;149:159–172. doi: 10.1007/BF00222270. [DOI] [PubMed] [Google Scholar]
- Hawkes JW. The structure of fish skin. II. The chromatophore unit. Cell Tissue Res. 1974b;149:159–72. doi: 10.1007/BF00222271. [DOI] [PubMed] [Google Scholar]
- Hirata M, Nakamura K, Kanemaru T, Shibata Y, Kondo S. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227:497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- Hirata M, Nakamura K, Kondo S. Pigment cell distributions in different tissues of the zebrafish, with special reference to the striped pigment pattern. Dev Dyn. 2005;234:293–300. doi: 10.1002/dvdy.20513. [DOI] [PubMed] [Google Scholar]
- Honjo Y, Kniss J, Eisen JS. Neuregulin-mediated ErbB3 signaling is required for formation of zebrafish dorsal root ganglion neurons. Development. 2008;135:2615–2625. doi: 10.1242/dev.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Heuer H. Thyroid hormone action during brain development: More questions than answers. Mol Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–76. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Houde AE. Sex, Color, and Mate Choice in Guppies. Princeton, NJ: Princeton University Press; 1997. [Google Scholar]
- Houde AE, Endler JA. Correlated Evolution of Female Mating Preferences and Male Color Patterns in the Guppy Poecilia reticulata. Science. 1990;248:1405–8. doi: 10.1126/science.248.4961.1405. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Koasry CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER Cancer Statistics Review, 1975-2011. Bethesda, MD: National Cancer Institute; 2014. ( http://seer.cancer.gov/csr/1975_2011/, based on November 2013 SEER data submission, posted to the SEER web site, April 2014) [Google Scholar]
- Hughes KA, Houde AE, Price AC, Rodd FH. Mating advantage for rare males in wild guppy populations. Nature. 2013;503:108–10. doi: 10.1038/nature12717. [DOI] [PubMed] [Google Scholar]
- Hultman KA, Bahary N, Zon LI, Johnson SL. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Budi EH, Teasley DC, Gottlieb AY, Parichy DM, Johnson SL. Defects in ErbB-dependent establishment of adult melanocyte stem cells reveal independent origins for embryonic and regeneration melanocytes. PLoS Genetics. 2009;5:e1000544. doi: 10.1371/journal.pgen.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Johnson SL. Differential contribution of direct-developing and stem cell-derived melanocytes to the zebrafish larval pigment pattern. Dev Biol. 2010;337:425–31. doi: 10.1016/j.ydbio.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Scott AW, Johnson SL. Small molecule modifier screen for kit-dependent functions in zebrafish embryonic melanocytes. Zebrafish. 2008;5:279–87. doi: 10.1089/zeb.2008.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kondo S, Parichy DM, Watanabe M. Tetraspanin 3c requirement for pigment cell interactions and boundary formation in zebrafish adult pigment stripes. Pigment Cell Melanoma Res. 2014;27:190–200. doi: 10.1111/pcmr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita M, Watanabe M, Ishii M, Chen T, Johnson SL, Kurachi Y, Okada N, Kondo S. Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: Implications for the regulation of melanosome movement. Plos Genetics. 2006;2:1861–1870. doi: 10.1371/journal.pgen.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR. Ascidian neural crest-like cells: phylogenetic distribution, relationship to larval complexity, and pigment cell fate. J Exp Zool B Mol Dev Evol. 2006;306:470–80. doi: 10.1002/jez.b.21109. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Chiba T, Krajka FR, Deyts C, Satoh N, Joly JS. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: insights into the ancestry and evolution of the neural crest. Dev Biol. 2008;324:152–60. doi: 10.1016/j.ydbio.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–9. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Walker C, Weston JA. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Nguyen AN, Lister JA. mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol. 2011;350:405–13. doi: 10.1016/j.ydbio.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin CB, Xu X, Hong LZ, David VA, Mcgowan KA, Schmidt-Kuntzel A, Roelke ME, Pino J, Pontius J, Cooper GM, et al. Specifying and sustaining pigmentation patterns in domestic and wild cats. Science. 2012;337:1536–41. doi: 10.1126/science.1220893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Kakizawa S, Yada T. Pituitary of “cobalt” variant of the rainbow trout separated from the hypothalamus lacks most pars intermedial and neurohypophysial tissue. Gen Comp Endocrinol. 1993;92:31–40. doi: 10.1006/gcen.1993.1140. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Amsterdam A, Shimoda N, Becker T, Mugg J, Shima A, Hopkins N. Proviral insertions in the zebrafish hagoromo gene, encoding an F-box/WD40-repeat protein, cause stripe pattern anomalies. Curr Biol. 2000;10:463–6. doi: 10.1016/s0960-9822(00)00444-9. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, Haffter P, Odenthal J, Mullins MC, Van Eeden FJ, Furutani-Seiki M, et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–89. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots-A review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009;20:90–104. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, Inoue C, Momoi A, Kondoh H, Furutani-Seiki M, Ozato K, Wakamatsu Y. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;121:841–59. doi: 10.1016/j.mod.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Kemp DJ, Reznick DN, Grether GF, Endler JA. Predicting the direction of ornament evolution in Trinidadian guppies (Poecilia reticulata) Proc Biol Sci. 2009;276:4335–43. doi: 10.1098/rspb.2009.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo G, Lim TM, Phang VPE. Ultrastructure of Erythrophores and Xanthophores of the Siamese Fighting Fish, Betta splendens. Isr J Aquacult-Bamid. 2012;64 [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimura T, Nagao Y, Hashimoto H, Yamamoto-Shiraishi Y, Yamamoto S, Yabe T, Takada S, Kinoshita M, Kuroiwa A, Naruse K. Leucophores are similar to xanthophores in their specification and differentiation processes in medaka. Proc Natl Acad Sci U S A. 2014;111:7343–8. doi: 10.1073/pnas.1311254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum F. Untersuchungen über das Farbmuster der Zebrabarbe Brachydanio rerio (Cyprinidae, Teleostei) Wilhelm Roux's Arch. 1975;177:129–152. doi: 10.1007/BF00848526. [DOI] [PubMed] [Google Scholar]
- Kondo S, Asai R. A reaction-diffusion wave on the skin of the marine angelfish Pomacanthus. Nature. 1995;376:765–8. doi: 10.1038/376765a0. [DOI] [PubMed] [Google Scholar]
- Kottler VA, Fadeev A, Weigel D, Dreyer C. Pigment pattern formation in the guppy, Poecilia reticulata, involves the Kita and Csf1ra receptor tyrosine kinases. Genetics. 2013;194:631–46. doi: 10.1534/genetics.113.151738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler VA, Koch I, Flotenmeyer M, Hashimoto H, Weigel D, Dreyer C. Multiple pigment cell types contribute to the black, blue, and orange ornaments of male guppies (Poecilia reticulata) PLoS One. 2014;9:e85647. doi: 10.1371/journal.pone.0085647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J, Astrinides P, Frohnhofer HG, Walderich B, Nusslein-Volhard C. transparent, a gene affecting stripe formation in Zebrafish, encodes the mitochondrial protein Mpv17 that is required for iridophore survival. Biology open. 2013;2:703–10. doi: 10.1242/bio.20135132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J, Frohnhofer HG, Walderich B, Maischein HM, Weiler C, Irion U, Nusslein-Volhard C. Endothelin signalling in iridophore development and stripe pattern formation of zebrafish. Biology open. 2014;3:503–9. doi: 10.1242/bio.20148441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Morrison JA, Bailey CM. The neural crest and cancer: a developmental spin on melanoma. Cells Tissues Organs. 2013;198:12–21. doi: 10.1159/000348418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoreux LM, Kelsh RN, Wakamatsu Y, Ozato K. Pigment pattern formation in the medaka embryo. Pigment Cell Res. 2005;18:64–73. doi: 10.1111/j.1600-0749.2005.00216.x. [DOI] [PubMed] [Google Scholar]
- Lang MR, Patterson LB, Gordon TN, Johnson SL, Parichy DM. Basonuclin-2 requirements for zebrafish adult pigment pattern development and female fertility. PLoS Genet. 2009;5:e1000744. doi: 10.1371/journal.pgen.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TA, Gordon TN, Lau HE, Parichy DM. Defective adult oligodendrocyte and Schwann cell development, pigment pattern, and craniofacial morphology in puma mutant zebrafish having an alpha tubulin mutation. Dev Biol. 2010;346:296–309. doi: 10.1016/j.ydbio.2010.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V. The origins and evolution of vertebrate metamorphosis. Curr Biol. 2011;21:R726–37. doi: 10.1016/j.cub.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Le Guellec D, Morvan-Dubois G, Sire JY. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio) Int J Dev Biol. 2004;48:217–31. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lin S, Long W, Chen J, Hopkins N. Production of germ-line chimeras in zebrafish by cell transplants from genetically pigmented to albino embryos. Proc Natl Acad Sci U S A. 1992;89:4519–23. doi: 10.1073/pnas.89.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Foley J, Jiang TX, Yeh CY, Wu P, Foley A, Yen CM, Huang YC, Cheng HC, Chen CF, et al. Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science. 2013;340:1442–5. doi: 10.1126/science.1230374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Capper A, Zeng Z, Mathers ME, Richardson J, Paranthaman K, Jackson IJ, Patton EE. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J Invest Dermatol. 2014;134:133–40. doi: 10.1038/jid.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–67. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Lopes SS, Yang X, Muller J, Carney TJ, Mcadow AR, Rauch GJ, Jacoby AS, Hurst LD, Delfino-Machin M, Haffter P, et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169–74. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Lush ME, Piotrowski T. ErbB expressing Schwann cells control lateral line progenitor cells via non-cell-autonomous regulation of Wnt/beta-catenin. eLife. 2014;3:e01832. doi: 10.7554/eLife.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–24. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Maderspacher F, Nusslein-Volhard C. Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development. 2003;130:3447–57. doi: 10.1242/dev.00519. [DOI] [PubMed] [Google Scholar]
- Mahalwar P, Walderich B, Singh AP, Nüsslein-Volhard C. Local reorganization of xanthophores fine-tunes and colors the striped pattern of zebrafish. Science. 2014 doi: 10.1126/science.1254837. [DOI] [PubMed] [Google Scholar]
- Malmquist SJ, Abramsson A, Mcgraw HF, Linbo TH, Raible DW. Modulation of dorsal root ganglion development by ErbB signaling and the scaffold protein Sorbs3. Development. 2013;140:3986–96. doi: 10.1242/dev.084640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RA, Riesch R, Heinen-Kay JL, Langerhans RB. Evolution of male coloration during a post-Pleistocene radiation of Bahamas mosquitofish (Gambusia hubbsi) Evolution. 2014;68:397–411. doi: 10.1111/evo.12277. [DOI] [PubMed] [Google Scholar]
- Mcmenamin SK, Bain EJ, Mccann AE, Patterson LB, Eom DS, Waller ZP, Hamill JC, Kuhlman JA, Eisen JS, Parichy DM. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345:1358–61. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmenamin SK, Parichy DM. Metamorphosis in teleosts. Curr Top Dev Biol. 2013;103:127–65. doi: 10.1016/B978-0-12-385979-2.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierjohann S, Schartl M. From Mendelian to molecular genetics: the Xiphophorus melanoma model. Trends Genet. 2006;22:654–61. doi: 10.1016/j.tig.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. A requirement for kit in embryonic zebrafish melanocyte differentiation is revealed by melanoblast delay. Dev Genes Evol. 2004;214:493–502. doi: 10.1007/s00427-004-0428-y. [DOI] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. pyewacket, a new zebrafish fin pigment pattern mutant. Pigment Cell Res. 2006;19:232–8. doi: 10.1111/j.1600-0749.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- Michailidou C, Jones M, Walker P, Kamarashev J, Kelly A, Hurlstone AF. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis Model Mech. 2009;2:399–411. doi: 10.1242/dmm.001149. [DOI] [PubMed] [Google Scholar]
- Mills MG, Nuckels RJ, Parichy DM. Deconstructing evolution of adult phenotypes: genetic analyses of kit reveal homology and evolutionary novelty during adult pigment pattern development of Danio fishes. Development. 2007;134:1081–90. doi: 10.1242/dev.02799. [DOI] [PubMed] [Google Scholar]
- Mills MG, Patterson LB. Not just black and white: Pigment pattern development and evolution in vertebrates. Semin Cell Dev Biol. 2008;20:72–81. doi: 10.1016/j.semcdb.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milos N, Dingle AD. Dynamics of pigment pattern formation in the zebrafish, Brachydanio rerio. I. Establishment and regulation of the lateral line melanophore stripe during the first eight days of development. J Exp Zool. 1978a;205:205–216. doi: 10.1002/jez.1402270112. [DOI] [PubMed] [Google Scholar]
- Milos N, Dingle AD. Dynamics of pigment pattern formation in the zebrafish, Brachydanio rerio. II. Lability of lateral line stripe formation and regulation of pattern defects. J Exp Zool. 1978b;205:217–224. doi: 10.1002/jez.1402270112. [DOI] [PubMed] [Google Scholar]
- Milos N, Dingle AD, Milos JP. Dynamics of pigment pattern formation in the zebrafish, Brachydanio rerio. III. Effect of anteroposterior location of three-day lateral line melanophores on colonization by the second wave of melanophores. J Exp Zool. 1983;227:81–92. doi: 10.1002/jez.1402270112. [DOI] [PubMed] [Google Scholar]
- Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317:508–22. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B. Gene control of mammalian pigmentary differentiation. I. Clonal origin of melanocytes. Proc Natl Acad Sci U S A. 1967;58:344–51. doi: 10.1073/pnas.58.1.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B. Clonal basis of mammalian differentiation. Symp Soc Exp Biol. 1971;25:345–70. [PubMed] [Google Scholar]
- Miyazawa S, Okamoto M, Kondo S. Blending of animal colour patterns by hybridization. Nat Commun. 2010;1:66. doi: 10.1038/ncomms1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Nusslein-Volhard C. Scales of fish arise from mesoderm. Curr Biol. 2013;23:R338–9. doi: 10.1016/j.cub.2013.02.056. [DOI] [PubMed] [Google Scholar]
- Mongera A, Singh AP, Levesque MP, Chen YY, Konstantinidis P, Nusslein-Volhard C. Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development. 2013;140:916–25. doi: 10.1242/dev.091066. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Regeneration in teleosts. Archiv für Entwickelungsmechanik der Organismen. 1900;10:120–134. [Google Scholar]
- Mueller KP, Neuhauss SC. Sunscreen for fish: co-option of UV light protection for camouflage. PLoS One. 2014;9:e87372. doi: 10.1371/journal.pone.0087372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao Y, Suzuki T, Shimizu A, Kimura T, Seki R, Adachi T, Inoue C, Omae Y, Kamei Y, Hara I, et al. Sox5 functions as a fate switch in medaka pigment cell development. PLoS Genet. 2014;10:e1004246. doi: 10.1371/journal.pgen.1004246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamasu A, Takahashi G, Kanbe A, Kondo S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc Natl Acad Sci U S A. 2009;106:8429–34. doi: 10.1073/pnas.0808622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, Kawakami A, Sawada A, Furutani-Seiki M, Takeda H, Araki K. Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat Genet. 2002;31:195–9. doi: 10.1038/ng899. [DOI] [PubMed] [Google Scholar]
- Nishimura EK. Melanocyte stem cells: a melanocyte reservoir in hair follicles for hair and skin pigmentation. Pigment Cell Melanoma Res. 2011;24:401–10. doi: 10.1111/j.1755-148X.2011.00855.x. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa S. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–60. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- Nitzan E, Krispin S, Pfaltzgraff ER, Klar A, Labosky PA, Kalcheim C. A dynamic code of dorsal neural tube genes regulates the segregation between neurogenic and melanogenic neural crest cells. Development. 2013a;140:2269–79. doi: 10.1242/dev.093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzan E, Pfaltzgraff ER, Labosky PA, Kalcheim C. Neural crest and Schwann cell progenitor-derived melanocytes are two spatially segregated populations similarly regulated by Foxd3. Proc Natl Acad Sci U S A. 2013b doi: 10.1073/pnas.1306287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu MC, Twitty VC. The Origin of Epidermal Melanophores During Metamorphosis in Triturus-torosus. Journal of Experimental Zoology. 1950;113:633–647. [Google Scholar]
- O'reilly-Pol T, Johnson SL. Melanocyte regeneration reveals mechanisms of adult stem cell regulation. Semin Cell Dev Biol. 2008 doi: 10.1016/j.semcdb.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'reilly-Pol T, Johnson SL. Kit signaling is involved in melanocyte stem cell fate decisions in zebrafish embryos. Development. 2013;140:996–1002. doi: 10.1242/dev.088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, Rossnagel K, Haffter P, Kelsh RN, Vogelsang E, Brand M, Van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, et al. Mutations affecting xanthophore pigmentation in the zebrafish, Danio rerio. Development. 1996;123:391–8. doi: 10.1242/dev.123.1.391. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–86. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Oshima N, Kasai A. Iridophores involved in generation of skin color in the zebrafish, Brachydanio rerio. Forma. 2002;17:91–101. [Google Scholar]
- Parichy DM. Experimental analysis of character coupling across a complex life cycle: pigment pattern metamorphosis in the tiger salamander, Ambystoma tigrinum tigrinum. J Morphol. 1998;237:53–67. doi: 10.1002/(SICI)1097-4687(199807)237:1<53::AID-JMOR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Developmental Dynamics. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Mellgren EM, Rawls JF, Lopes SS, Kelsh RN, Johnson SL. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000a;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000b;127:3031–44. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–36. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Reedy MV, Erickson CA. Chapter 5 Regulation of melanoblast migration and differentiation. In: Nordland JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, Ortonne JP, editors. The Pigmentary System: Physiology and Pathophysiology. 2nd. New York, New York: Oxford University Press; 2006. [Google Scholar]
- Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003a;130:817–33. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Turner JM. Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Developmental Biology. 2003b;256:242–257. doi: 10.1016/s0012-1606(03)00015-0. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Turner JM, Parker NB. Essential role for puma in development of postembryonic neural crest-derived cell lineages in zebrafish. Dev Biol. 2003;256:221–41. doi: 10.1016/s0012-1606(03)00016-2. [DOI] [PubMed] [Google Scholar]
- Patterson LB, Bain EJ, Parichy DM. Pigment cell interactions and differential xanthophore recruitment underlying zebrafish stripe reiteration and Danio pattern evolution. Nat Commun. 2014;5:5299. doi: 10.1038/ncomms6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LB, Parichy DM. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet. 2013;9:e1003561. doi: 10.1371/journal.pgen.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Mitchell DL, Nairn RS. Genetic and environmental melanoma models in fish. Pigment Cell Melanoma Res. 2010;23:314–37. doi: 10.1111/j.1755-148X.2010.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–54. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Price AC, Weadick CJ, Shim J, Rodd FH. Pigments, patterns, and fish behavior. Zebrafish. 2008;5:297–307. doi: 10.1089/zeb.2008.0551. [DOI] [PubMed] [Google Scholar]
- Quigley IK, Manuel JL, Roberts RA, Nuckels RJ, Herrington ER, Macdonald EL, Parichy DM. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development. 2005;132:89–104. doi: 10.1242/dev.01547. [DOI] [PubMed] [Google Scholar]
- Quigley IK, Turner JM, Nuckels RJ, Manuel JL, Budi EH, Macdonald EL, Parichy DM. Pigment pattern evolution by differential deployment of neural crest and post-embryonic melanophore lineages in Danio fishes. Development. 2004;131:6053–69. doi: 10.1242/dev.01526. [DOI] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development. 1994;120:495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Rakers S, Gebert M, Uppalapati S, Meyer W, Maderson P, Sell AF, Kruse C, Paus R. ‘Fish matters’: the relevance of fish skin biology to investigative dermatology. Exp Dermatol. 2010 doi: 10.1111/j.1600-0625.2009.01059.x. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Johnson SL. Zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration. Development. 2000;127:3715–24. doi: 10.1242/dev.127.17.3715. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Johnson SL. Requirements for the kit receptor tyrosine kinase during regeneration of zebrafish fin melanocytes. Development. 2001;128:1943–9. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Johnson SL. Temporal and molecular separation of the kit receptor tyrosine kinase's roles in zebrafish melanocyte migration and survival. Dev Biol. 2003;262:152–61. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal GG, Ryan MJ. Assortative preferences for stripes in danios. Animal Behaviour. 2005;70:1063–1066. [Google Scholar]
- Santoriello C, Anelli V, Alghisi E, Mione M. Highly penetrant melanoma in a zebrafish model is independent of ErbB3b signaling. Pigment Cell Melanoma Res. 2012;25:287–9. doi: 10.1111/j.1755-148X.2012.00973.x. [DOI] [PubMed] [Google Scholar]
- Santos ME, Braasch I, Boileau N, Meyer BS, Sauteur L, Bohne A, Belting HG, Affolter M, Salzburger W. The evolution of cichlid fish egg-spots is linked with a cis-regulatory change. Nat Commun. 2014;5:5149. doi: 10.1038/ncomms6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Kneitz S, Wilde B, Wagner T, Henkel CV, Spaink HP, Meierjohann S. Conserved expression signatures between medaka and human pigment cell tumors. PLoS One. 2012;7:e37880. doi: 10.1371/journal.pone.0037880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R, Van Der Sluijs I, Schneider MV, Maan ME, Tachida H, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Shi YB. Amphibian Metamorphosis: From Morphology to Molecular Biology. New York, New York: John Wiley and Sons, Inc.; 2000. [Google Scholar]
- Shimada A, Kawanishi T, Kaneko T, Yoshihara H, Yano T, Inohaya K, Kinoshita M, Kamei Y, Tamura K, Takeda H. Trunk exoskeleton in teleosts is mesodermal in origin. Nat Commun. 2013;4:1639. doi: 10.1038/ncomms2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Schach U, Nusslein-Volhard C. Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat Cell Biol. 2014;16:607–14. doi: 10.1038/ncb2955. [DOI] [PubMed] [Google Scholar]
- Sirakov M, Skah S, Nadjar J, Plateroti M. Thyroid hormone's action on progenitor/stem cell biology: new challenge for a classic hormone? Biochim Biophys Acta. 2013;1830:3917–27. doi: 10.1016/j.bbagen.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Stearner SP. Pigmentation studies in salamanders, with especial reference to the changes at metamorphosis. Physiol Zool. 1946;19:375–404. doi: 10.1086/physzool.19.4.30151927. [DOI] [PubMed] [Google Scholar]
- Svetic V, Hollway GE, Elworthy S, Chipperfield TR, Davison C, Adams RJ, Eisen JS, Ingham PW, Currie PD, Kelsh RN. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development. 2007;134:1011–22. doi: 10.1242/dev.02789. [DOI] [PubMed] [Google Scholar]
- Takahashi G, Kondo S. Melanophores in the stripes of adult zebrafish do not have the nature to gather, but disperse when they have the space to move. Pigment Cell Melanoma Res. 2008;21:677–86. doi: 10.1111/j.1755-148X.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- Tanimura S, Tadokoro Y, Inomata K, Binh NT, Nishie W, Yamazaki S, Nakauchi H, Tanaka Y, Mcmillan JR, Sawamura D, et al. Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell. 2011;8:177–87. doi: 10.1016/j.stem.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–58. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H. Mutant genes in the medaka. In: Yamamoto T, editor. Medaka (Killifish) Biology and Strains, Stock Culture in Biological Field. Tokyo: Keigaku Publishing Company; 1975. pp. 251–272. [Google Scholar]
- Tornini VA, Poss KD. Keeping at arm's length during regeneration. Dev Cell. 2014;29:139–45. doi: 10.1016/j.devcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon RC, Higdon CW, Johnson SL. Lineage relationship of direct-developing melanocytes and melanocyte stem cells in the zebrafish. PLoS ONE. 2011;6:e21010. doi: 10.1371/journal.pone.0021010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon RC, Johnson SL. Clonal and lineage analysis of melanocyte stem cells and their progeny in the zebrafish. Methods Mol Biol. 2012;916:181–95. doi: 10.1007/978-1-61779-980-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Clonal analyses reveal roles of organ founding stem cells, melanocyte stem cells and melanoblasts in establishment, growth and regeneration of the adult zebrafish fin. Development. 2010;137:3931–9. doi: 10.1242/dev.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell. 2011;20:725–32. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglia JL, Hall BK. Patterns of migration and regulation of trunk neural crest cells in zebrafish (Danio rerio) Int J Dev Biol. 2000;44:867–81. [PubMed] [Google Scholar]
- Washio Y, Aritaki M, Fujinami Y, Shimizu D, Yokoi H, Suzuki T. Ocular-Side Lateralization of Adult-Type Chromatophore Precursors: Development of Pigment Asymmetry in Metamorphosing Flounder Larvae. J Exp Zool B Mol Dev Evol. 2013 doi: 10.1002/jez.b.22491. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Washio Y, Fujinami Y, Aritaki M, Uji S, Suzuki T. Adult-type pigment cells, which color the ocular sides of flounders at metamorphosis, localize as precursor cells at the proximal parts of the dorsal and anal fins in early larvae. Dev Growth Differ. 2008;50:731–41. doi: 10.1111/j.1440-169X.2008.01071.x. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Iwashita M, Ishii M, Kurachi Y, Kawakami A, Kondo S, Okada N. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006;7:893–7. doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kondo S. Changing clothes easily: connexin41.8 regulates skin pattern variation. Pigment Cell Melanoma Res. 2012;25:326–30. doi: 10.1111/j.1755-148X.2012.00984.x. [DOI] [PubMed] [Google Scholar]
- Wehrle-Haller B. The role of Kit-ligand in melanocyte development and epidermal homeostasis. Pigment Cell Res. 2003;16:287–96. doi: 10.1034/j.1600-0749.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AL, Jordan SA, Jackson IJ. Neural crest progenitors of the melanocyte lineage: coat colour patterns revisited. Development. 2002;129:3349–57. doi: 10.1242/dev.129.14.3349. [DOI] [PubMed] [Google Scholar]
- Windner SE, Bird NC, Patterson SE, Doris RA, Devoto SH. Fss/Tbx6 is required for central dermomyotome cell fate in zebrafish. Biology open. 2012;1:806–14. doi: 10.1242/bio.20121958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations, Volume 1: Genetic and Biometric Foundations. Chicago IL: University of Chicago Press; 1984a. [Google Scholar]
- Wright S. Evolution and the Genetics of Populations, Volume 3: Experimental Results and Evolutionary Deductions. Chicago, IL: University of Chicago Press; 1984b. [Google Scholar]
- Yamada T, Okauchi M, Araki K. Origin of adult-type pigment cells forming the asymmetric pigment pattern, in Japanese flounder (Paralichthys olivaceus) Dev Dyn. 2010;239:3147–62. doi: 10.1002/dvdy.22440. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Yoshimoto E, Kondo S. Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc Natl Acad Sci U S A. 2007;104:4790–3. doi: 10.1073/pnas.0607790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CT, Hindes AE, Hultman KA, Johnson SL. Mutations in gfpt1 and skiv2l2 cause distinct stage-specific defects in larval melanocyte regeneration in zebrafish. PLoS Genet. 2007;3:e88. doi: 10.1371/journal.pgen.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CT, Johnson SL. Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development. 2006;133:3563–73. doi: 10.1242/dev.02533. [DOI] [PubMed] [Google Scholar]
- Yang CT, Sengelmann RD, Johnson SL. Larval melanocyte regeneration following laser ablation in zebrafish. J Invest Dermatol. 2004;123:924–9. doi: 10.1111/j.0022-202X.2004.23475.x. [DOI] [PubMed] [Google Scholar]
- Yen J, White RM, Wedge DC, Van Loo P, De Ridder J, Capper A, Richardson J, Jones D, Raine K, Watson IR, et al. The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biol. 2013;14:R113. doi: 10.1186/gb-2013-14-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler I, Mcdonaldo T, Hesslinger C, Pelletier I, Boyle P. Development of the pteridine pathway in the zebrafish, Danio rerio. Journal of Biological Chemistry. 2000;275:18926–18932. doi: 10.1074/jbc.M910307199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Adult pigment pattern development in zebrafish over ∼3 weeks, beginning with the early larval pattern and ending with the juvenile pattern. Animation highlights changes in melanophore pattern and is compiled from repeated daily imaging of a single individual, with frames aligned and rescaled to control for growth (Quigley et al., 2005).
Movie 2. mitfa:GFP+ cells (e.g., arrows) migrating to the hypodermis from the dorsal aspect of the myotome (Budi et al., 2011).
Movie 3. mitfa:GFP+ cells (e.g., arrow) enter the hypodermis via the myoseptum during adult pattern development (Budi et al., 2011).


