Abstract
Background
Among the complexities of skeletal muscle differentiation is a temporal distinction in the onset of expression of different lineage-specific genes. The lineage-determining factor MyoD is bound to myogenic genes at the onset of differentiation whether gene activation is immediate or delayed. How temporal regulation of differentiation-specific genes is established remains unclear.
Results
Using embryonic tissue, we addressed the molecular differences in the organization of the myogenin and muscle creatine kinase (MCK) gene promoters by examining regulatory factor binding as a function of both time and spatial organization during somitogenesis. At the myogenin promoter, binding of the homeodomain factor Pbx1 coincided with H3 hyperacetylation and was followed by binding of co-activators that modulate chromatin structure. MyoD and myogenin binding occurred subsequently, demonstrating that Pbx1 facilitates chromatin remodeling and modification prior to myogenic regulatory factor binding. At the same time, the MCK promoter was bound by HDAC2 and MyoD, and activating histone marks were largely absent. The association of HDAC2 and MyoD was confirmed by co-immunoprecipitation, proximity ligation assay (PLA), and sequential ChIP.
Conclusion
MyoD differentially promotes activated and repressed chromatin structures at myogenic genes early after the onset of skeletal muscle differentiation in the developing mouse embryo.
Keywords: myogenesis, somite, gene expression, gene regulation, Pbx, histone deacetylase
INTRODUCTION
Skeletal muscle differentiation is a highly regulated process driven by the MyoD family of basic helix-loop-helix, DNA-binding, lineage determining factors: MyoD, Myf5, Mrf4, and myogenin. These factors have distinct and overlapping roles during embryogenesis and in post-natal skeletal muscle regeneration (reviewed in (Moncaut et al., 2013, Tapscott, 2005)). The activities of these transcription factors are modulated via multiple mechanisms, including heterodimerization with members of a family of E-box proteins, the presence or absence of additional DNA-binding transcription factors at target gene regulatory sequences, and the presence or absence of co-regulatory proteins, many of which directly alter chromatin structure or post-translationally modify histones. Different co-factors positively or negatively influence the transcriptional potential of the MyoD family of regulators. The functional interplay between all of these regulatory molecules has been investigated in great detail, leading to detailed models for the regulation of skeletal muscle-specific genes during differentiation (reviewed in (Berkes and Tapscott, 2005, de la Serna et al., 2006, Moncaut et al., 2013, Puri and Mercola, 2012, Sartorelli and Caretti, 2005, Tapscott, 2005, Guasconi and Puri, 2009)).
Temporal regulation adds an additional level of complexity to the regulatory proteins affecting skeletal muscle differentiation. Detailed time course studies of different tissue culture models for myogenesis revealed multiple clusters of differentiation-specific gene expression profiles that were grouped together based on similar kinetics of gene expression throughout the differentiation process (Bergstrom et al., 2002, Tomczak et al., 2004, Delgado et al., 2003, Moran et al., 2002), but because of differences in experimental models and conditions, we and others have focused our investigations on myogenic genes that are easily distinguished by their times of activation following the onset of differentiation. At its simplest level, there are myogenic genes that are activated early after the onset of differentiation, for example, the gene encoding the myogenin regulatory protein that is required for terminal differentiation, and genes encoding structural and functional components of muscle tissue that are expressed later during differentiation. In particular, studies focusing on MyoD binding to early and late gene promoters revealed that MyoD binding was observed at both early and late gene promoters at early times of differentiation, indicating that MyoD binding was not the determinant for early versus late myogenic gene expression (Blais et al., 2005, Cao et al., 2006, Ohkawa et al., 2006). Subsequent genome-wide profiling of MyoD binding not only supported these findings but revealed that MyoD was bound to many myogenic gene promoters and regulatory sequences even before the onset of differentiation (Cao et al., 2010, Soleimani et al., 2012). Constitutive binding of MyoD at the onset of differentiation raises questions about the functionality of MyoD binding at genes that are being activated in response to MyoD binding and simultaneously at genes that will not be activated until later times. Significant efforts have been made to understand the role of MyoD in gene activation of early genes like myogenin; less information is available about MyoD function at late gene regulatory sequences prior to activation.
Activation of the myogenin gene by MyoD involves cooperation by the homeodomain proteins Pbx and Meis, which form a complex on a subset of myogenic promoters, including the myogenin promoter, prior to expression or activation of MyoD (Berkes et al., 2004, de la Serna et al., 2005, Maves et al., 2007). The presence of Pbx-Meis on the myogenin promoter facilitates both the binding of MyoD to a non-consensus binding site (Berkes et al., 2004, Knoepfler et al., 1999) as well as the interaction of enzymes that serve as coactivators, including acetyltransferases, lysine methyltransferases, the Prmt5 arginine methyltransferase, and the SWI/SNF ATP-dependent chromatin remodeling enzyme (Dacwag et al., 2007, de la Serna et al., 2005, Puri et al., 1997b, Sartorelli et al., 1997, Sartorelli et al., 1999, Simone et al., 2004, Polesskaya et al., 2001b, Puri et al., 1997a, Tao et al., 2011). The presence of MyoD and the chromatin remodeling and modifying enzymes promote chromatin structural changes that increase nuclease accessibility at the myogenin promoter and result in stable binding of MyoD and Mef2 transcription factors to their consensus binding sites (de la Serna et al., 2001a, de la Serna et al., 2005, Gerber et al., 1997, Simone et al., 2004). Inhibition of Pbx expression in zebrafish demonstrated a requirement for Pbx proteins in myogenin expression and in skeletal muscle development (Maves et al., 2007), providing additional physiological relevance to the molecular functions that have been defined for Pbx during skeletal muscle differentiation.
Myogenic genes that will be activated later during differentiation remain repressed at early times of differentiation, despite being bound by MyoD. This suggests that MyoD is present in a repressed or inactive form or that it is associated with co-repressor molecules that prevent its activating function. Previous studies reported that MyoD could be found in association with HDAC1, a class I histone deacetylase, in C2C12 myoblasts and that the association decreased upon differentiation (Mal et al., 2001, Puri et al., 2001). Another report indicated HDAC1 and MyoD were present at the myogenin promoter prior to but not after the onset of C2C12 cell differentiation (Mal and Harter, 2003), but the presence or absence of any HDAC at other genes after differentiation started was not evaluated. More recently, genome wide binding studies identified complexes of Snail and HDAC1/2 at s specific subset of MyoD binding sites in myoblasts, and it was determined that this complex excludes MyoD from these sites. Upon differentiation, the binding of Snail/HDAC complexes was greatly diminished (Soleimani et al., 2012). Subsequently, we used MyoD reprogrammed fibroblasts to demonstrate that MyoD and HDAC2, but not HDAC1, were present at the regulatory sequences of late genes after the onset of differentiation but prior to the time at which these genes became transcriptionally active. MyoD and HDAC2 were displaced by myogenin and co-activating chromatin modifying and remodeling enzymes at the time of gene activation (Ohkawa et al., 2006), suggesting a functional role for HDAC2 in maintaining the silent state of late genes in the immediate timeframe after the induction of differentiation.
Some of the findings about factor interactions with myogenic gene chromatin have been verified in satellite cells and myotubes isolated from neo-natal or adult mouse primary skeletal muscle tissue and/or in mouse whole embryo preparations from E10.5 or later (Dacwag et al., 2007, Ohkawa et al., 2006, Ohkawa et al., 2007). In the case of MyoD and HDAC2, these proteins were observed on late gene regulatory sequences at E10.5. In contrast, at E12.5, when expression of late genes is robust in the somites, MyoD and HDAC2 were no longer observed on late gene regulatory sequences. Instead myogenin and co-activators were observed (Ohkawa et al., 2006), supporting the idea that MyoD and HDAC2 are present on myogenic late gene promoters prior to their activation and that within the same temporal context, MyoD directs different functional outcomes for gene expression at different myogenic genes.
Definitive interpretation of “population-based” experiments, like RT-PCR analysis of myogenic gene expression or ChIP at myogenic genes, in mouse embryonic tissue is difficult because skeletal muscle differentiation is both temporally and spatially controlled. Limb and trunk muscles originate in the somites, which form from the paraxial mesoderm in a sequential, periodic manner, with each new somite forming to the rostral side of the existing somite. In this manner a head to tail continuum of somites forms in the developing embryo. Myogenic gene expression in the somites follows the head to tail continuum of somite development (Pourquie, 2001, Tajbakhsh and Cossu, 1997).
To more precisely understand how regulatory factor binding contributes to the regulation of early and late myogenic genes during embryonic development of limb and trunk skeletal muscle, we analyzed tissue in two ways. Building on our more recent efforts that permitted successful ChIP experiments in E8.5 mouse embryos (Cho et al., 2011), we further refined our approach to assess regulatory factor interactions with chromatin as function of developmental stage instead of time by isolating embryos on the basis of the number of somites. We complemented this strategy with assessment of regulatory factor interactions with myogenic chromatin as a function of spatial organization. E9.5 mouse embryos were dissected to isolate rostral and caudal somite tissues that were positive or negative for myogenin expression, respectively. These methods allowed us to definitively demonstrate that during embryonic development, MyoD is simultaneously associated with co-activators at the myogenin gene and with co-repressors at a late gene and thus is involved in the differential organization of myogenic promoters in a manner that is consistent with the temporal order of myogenic gene expression.
RESULTS
Genetic analyses of mouse and other model organisms have revealed much about mechanisms of transcriptional regulation during skeletal muscle differentiation. More detailed molecular mechanisms involving regulatory factor binding and chromatin modification and remodeling have been elucidated, largely using cell line models for differentiation. Few attempts have been made to directly assess molecular interactions when muscle first begins to differentiate during embryogenesis. For that reason we chose to study the regulation of two representative myogenic genes, myogenin and MCK genes, at the onset of somitogenesis when skeletal muscle differentiation for limb and trunk muscles first begins.
Temporal analysis of myogenin promoter structure during embryogenesis
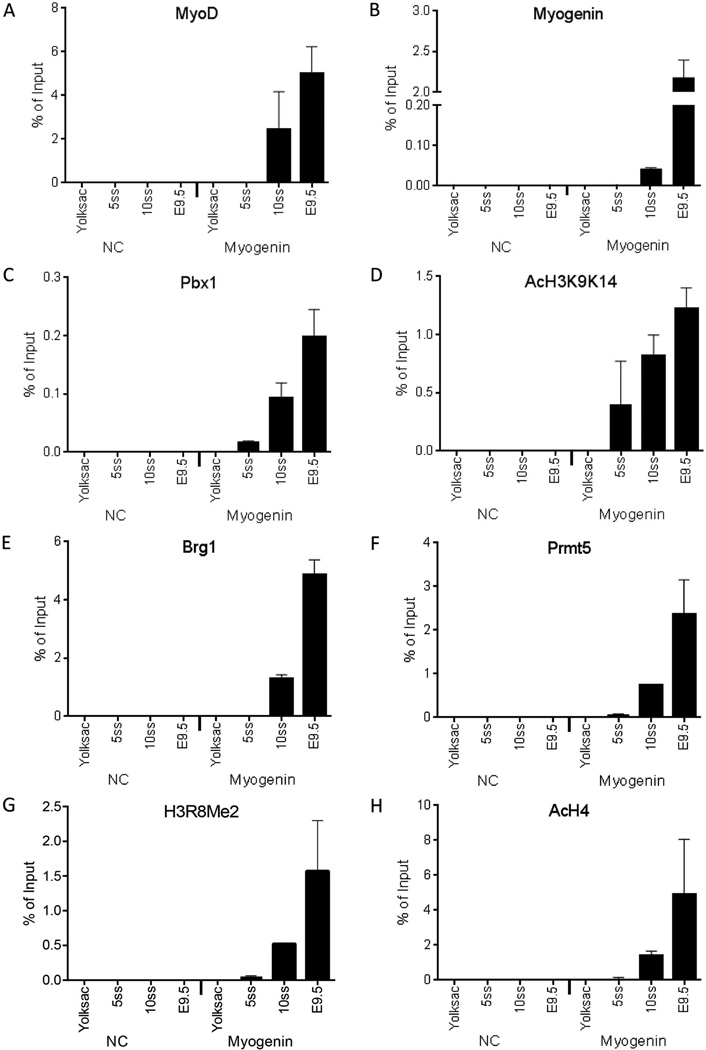
To assess regulatory factor interactions with myogenic promoters, we isolated CD1 mouse embryos at embryonic day 8.5 day (E8.5). Because of variability in developmental stages between littermates and among litters, embryos were staged and pooled according to the number of somites present. Embryos with 5 or 10 somites were used for ChIP experiments where real-time PCR was used as a read-out for precise quantification (Figure 1). Real-time PCR analysis compared amplification of myogenin promoter sequences with negative control (NC) sequences upstream from these loci. E9.5 embryos at 25–40 somites stage, which express abundant levels of myogenin ((Sassoon et al., 1989) and Fig. 3A), were used as a later stage control. ChIP experiments using extracts made from whole embryos were performed as described (Cho et al., 2011).
Figure 1.
Pbx1 and acetylated H3K9K14 are present on the myogenin promoter in embryos containing 5 somites and precede the interaction of the MyoD and myogenin regulators and other co-activators and histone modifications associated with myogenin activation. The binding of (A) MyoD, (B), myogenin, (C) Pbx1, (D) AcH3K9K14, (E) Brg1, (F) Prmt5, (G) H3R8Me2, (H) AcH4 to either the myogenin promoter or to a negative control (NC) sequence in embryos containing 5 or 10 somites (5ss or 10ss), in E9.5 embryos containing 25–40 somites, or in yolk sac was determined by real-time PCR. Data represent the mean of three independent experiments +/− standard deviation. p < 0.05 was obtained for all comparisons between yolk sac values and values for any developmental stage for which signal was obtained as well as for values between different developmental stages, with the following exceptions: MyoD and Pbx1 binding in 10 ss and E9.5 embryos were not statistically different and AcH3K9K14 binding in 5ss, 10ss and E9.5 was not statistically different.
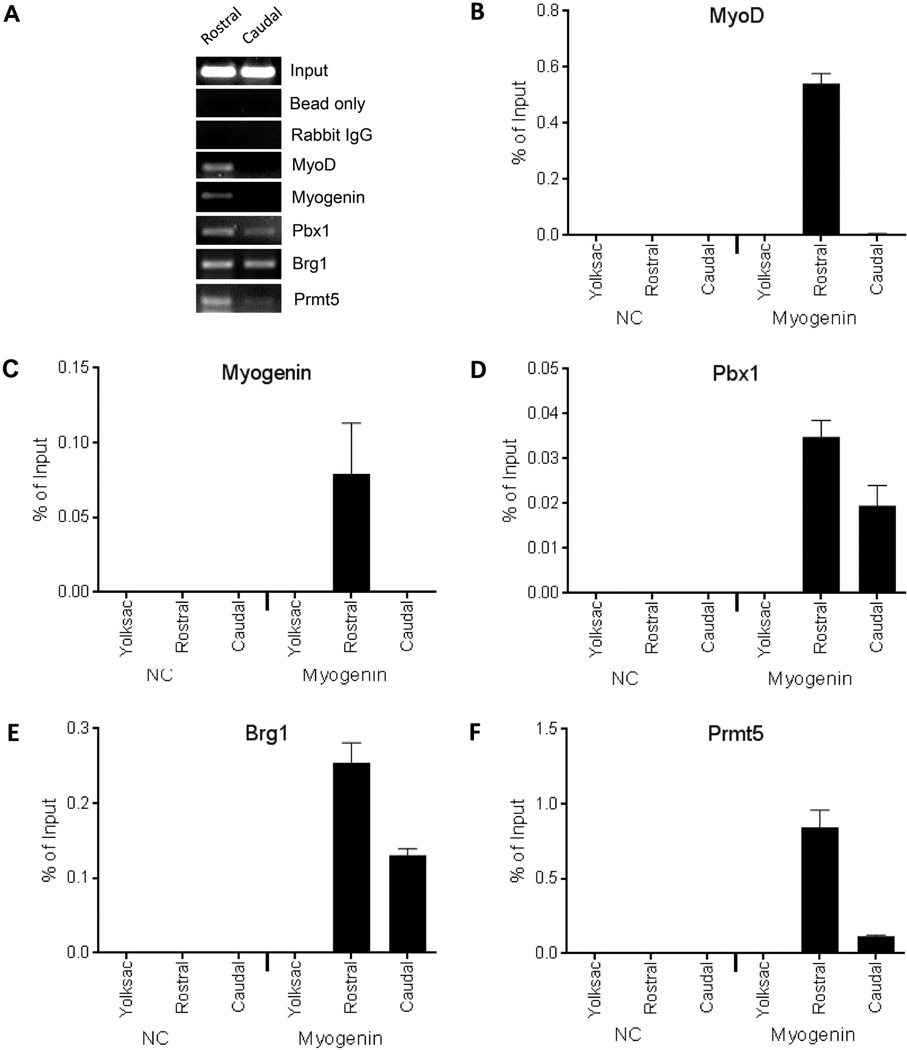
Figure 3.
Pbx1 and the Brg1 and Prmt5 cofactors, but not MyoD or myogenin, are present on the myogenin promoter in caudal somite-containing tissue isolated from E9.5 embryos. (A) Rostral or caudal somite-containing tissue was used for ChIP for the indicated factors and was assayed by semi-quantitative PCR followed by native gel electrophoresis. Real-time PCR was subsequently used to analyze ChIP experiments performed for (B) MyoD, (C) myogenin, (D) Pbx1, (E) Brg1, (F) Prmt5. Binding to either the myogenin promoter or to a negative control (NC) sequence in rostral or caudal somite-containing tissue from E9.5 embryos or in yolk sac was determined. Data represent the mean of three independent experiments +/− standard deviation. p < 0.05 was obtained for all comparisons between yolk sac and rostral tissue values and for all comparisons between rostral and caudal tissue values.
We observed MyoD and myogenin binding at the myogenin promoter in embryos with 10 somites and in E9.5 embryos (Figs. 1A–B). We next examined the binding of Pbx1 at the Myogenin promoter. Pbx1 is a homeodomain protein that has been implicated as a critical regulator of myogenin expression by targeting MyoD and chromatin remodeling enzymes to the myogenin promoter (Berkes et al., 2004, de la Serna et al., 2005, Maves et al., 2007). Pbx1 binding was clearly indicated at the 5 somite stage by real-time PCR analysis (Fig. 1C). This is consistent with the idea that Pbx1 binding precedes interaction of myogenic regulatory factors on the myogenin promoter (Berkes et al., 2004, de la Serna et al., 2005). Diacetylated (K9K14) histone 3 was the only other factor or modified histone detected at the myogenin promoter in 5 somite embryos (Fig. 1D). H3K9 acetylation is the result of the histone acetyl transferases GCN5/PCAF and Tip60. H3K14 acetylation is mediated by GCN5/PCAF, p300/CBP and Myst3 (Jin et al., 2011, Nagy and Tora, 2007, Lee and Workman, 2007). We did not attempt to identify the acetyl transferase responsible for the observed H3 acetylation, but GCN5/PCAF and p300/CBP have been previously implicated by other workers as functioning during skeletal muscle differentiation in general and at the myogenin promoter specifically (Puri et al., 1997b, Sartorelli et al., 1997, Sartorelli et al., 1999, Simone et al., 2004, Polesskaya et al., 2001b, Puri et al., 1997a).
The myogenin promoter in embryos containing 10 somites showed binding by Brg1 (Fig. 1E), an ATPase of the SWI/SNF chromatin-remodeling enzyme that has been shown to mediate chromatin remodeling at the promoter and that is necessary for myogenin expression and subsequent stages of differentiation (de la Serna et al., 2001a, de la Serna et al., 2005, de la Serna et al., 2001b, Simone et al., 2004). Brg1 binding and SWI/SNF enzyme function at the myogenin promoter requires Prmt5 (Dacwag et al., 2007), an arginine methyltransferase that modifies H3R8 (Pal et al., 2004). Analyses of Prmt5 and H3R8 dimethylation at the myogenin promoter indicated that the enzyme and the histone modification could be detected in embryos with 10 somites (Figs. 1F–G). Similarly, hyperacetylated H4 was present at the myogenin promoter in embryos with 10 somites (Fig. 1H). These data clearly indicate that K9 and K14 acetylation of H3 precedes hyperacetylation of H4 during the activation of the myogenin promoter.
These temporal studies of factor binding and modified histone incorporation indicate that the Pbx1 homeodomain factor and the acetyl transferase(s) that mediate H3K9 and K14 acetylation are present and detectable on the myogenin promoter in embryos containing 5 somites. Later during development, when the number of somites has expanded to 10, it is also possible to detect relevant ATP-dependent chromatin remodeling and arginine methyltransferase enzymes, and, by inference, the acetyltransferase(s) that target H4. MyoD and myogenin are also present at the 10 somite stage. Thus we could not distinguish whether myogenic factor binding precedes the binding of chromatin remodeling/modifying enzymes or whether the contributing co-regulatory enzyme binding precedes myogenic regulatory factor binding. Nor could we determine whether binding of all of these factors occurs in a concerted manner. To continue efforts to understand myogenin promoter organization during embryogenesis, we took a different approach where differences in the spatial organization of the developing embryo were used to discriminate between these possibilities.
Myogenin promoter structure examined as a function of spatial organization during embryogenesis
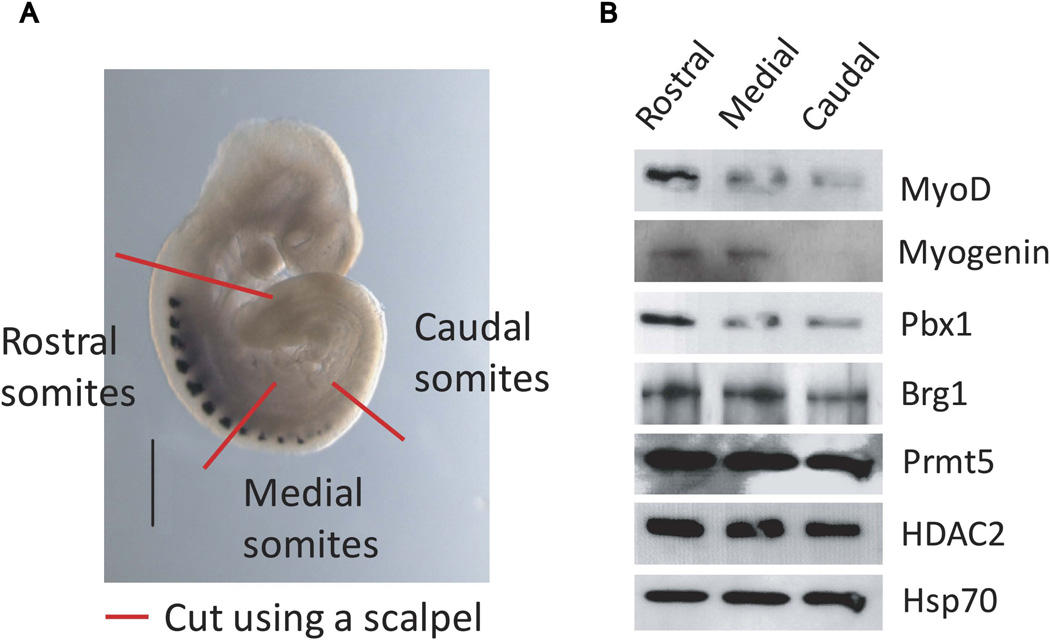
Prior in situ analyses during mouse embryogenesis clearly revealed the spatio-temporal development of the somites and of the skeletal muscle differentiation program (Pourquie, 2001, Tajbakhsh and Cossu, 1997). At E9.5, the somites in the rostral portion, formed at earlier developmental stages, show robust myogenin expression, while the most recently formed somites at the caudal end show no myogenin expression. Somites in the mid-section of the embryo show intermediate levels of myogenin expression, indicative of somatic cells that have recently activated myogenin expression ((Sassoon et al., 1989) and Fig. 3A). We reasoned that if the myogenin expressing somites could be physically separated from the somites that were not yet expressing myogenin, we would have cell populations that could be tested for factor binding and organization of the myogenin promoter. Consequently, we severed the trunk of freshly isolated E9.5 embryos into three sections: a rostral portion containing myogenin-expressing somites, a transitional mid-section with only the anterior somites expressing myogenin, and a caudal section containing no myogenin-expressing somites (Fig. 2A). These tissue sections were treated with pancreatin and trypsin to selectively digest non-somite cells from somites as previously published (Tajbakhsh et al., 1998, Cossu et al., 1996). These somite-enriched cell populations were then analyzed for protein expression by western blot (Fig. 2B) or cross-linked for subsequent ChIP experiments. Western bot analysis confirmed that myogenin was absent from the caudal somite-containing tissue sections (Fig. 2B). MyoD, Brg1, and Pbx1 levels in this region appeared reduced relative to the rostral somite myogenin-positive tissue section, whereas Prmt5 and HDAC2 levels appeared unchanged (Fig. 2B).
Figure 2.
Isolation and characterization of E9.5 somite-enriched tissue. (A) E9.5 embryo hybridized to show myogenin localization showing dissection to divide the trunk region into rostral, medial, or caudal somite-containing tissue. For illustrative purposes, the schematic is overlaid on an image of an E9.5 embryo stained for myogenin protein. (B) Western blot analysis showing the protein levels of the indicated proteins present in extracts from rostral, medial, or caudal somite-containing tissue.
ChIP analysis was restricted to the rostral, myogenin-positive and the caudal, myogenin-negative somite-enriched cell populations. Both semi-quantitative PCR and real-time PCR analyses were performed (Fig. 3). Binding of MyoD and myogenin to the myogenin promoter was observed in the rostral somites but not in the caudal cell population (Figs. 3A–C). In contrast, Pbx1 and the co-activators Brg1 and Prmt5 were detected in both the rostral and caudal somite cell populations (Figs. 3A, 3D–F). These data indicate that in caudal somite-enriched tissue where myogenin expression is not yet detectable, Pbx1 and the chromatin remodeling and arginine modifying co-activating enzymes are present prior to the binding of the myogenic regulatory factors. These data support the whole embryo ChIP analyses (Fig. 1) that showed that Pbx1 was the first transcription factor present on the myogenin promoter and extend those studies by indicating that Pbx1 targets chromatin remodeling and modifying enzymes prior to recruitment of MyoD or myogenin. Together these data provide in vivo evidence indicating how myogenic regulatory factors cooperate with ubiquitous chromatin altering cofactors to promote activation of the myogenin promoter in response to differentiation signaling.
Association of MyoD with HDAC2, a co-repressor of gene expression
Despite the presence and activation of myogenic regulatory factors following signaling to begin the onset of skeletal muscle differentiation, there are temporal differences in the expression of genes activated during the differentiation program. A large number of genes that encode structural and functional components of mature skeletal muscle are expressed subsequent to the expression of myogenin and other “early” genes (Bergstrom et al., 2002, Tomczak et al., 2004, Delgado et al., 2003, Moran et al., 2002). This raises the question of how some myogenic genes are activated in the aftermath of differentiation signaling while others remain silent, despite the myogenic regulator proteins being present.
Prior studies have connected MyoD with class I HDACs prior to skeletal muscle differentiation. Interaction between HDAC1 and MyoD was demonstrated in undifferentiated C2C12 myoblasts (Mal et al., 2001, Puri et al., 2001), and HDAC1 and MyoD could be individually localized to the myogenin promoter by ChIP prior to C2C12 differentiation (Mal and Harter, 2003). On some myogenic genes, a Snail/HDAC1/2 complex binds in the absence of MyoD prior to differentiation (Soleimani et al., 2012). In addition, treatment of C2C12 myoblasts with an HDAC inhibitor prematurely activated MyHC and other late genes and promoted myoblast fusion (Iezzi et al., 2004). Previous work by us demonstrated that MyoD and HDAC2 could be localized to the MCK promoter after the onset of skeletal muscle differentiation in MyoD-reprogrammed fibroblasts with loss of signal coinciding with the onset of MCK gene expression (Ohkawa et al., 2006). Similarly, MyoD and HDAC2 could be localized to the MCK promoter in E10.5 whole embryos, but not in E12.5 limb buds, where MCK was expressed (Ohkawa et al., 2006). We therefore set out to further examine MyoD/HDAC association and function in the context of embryonic development.
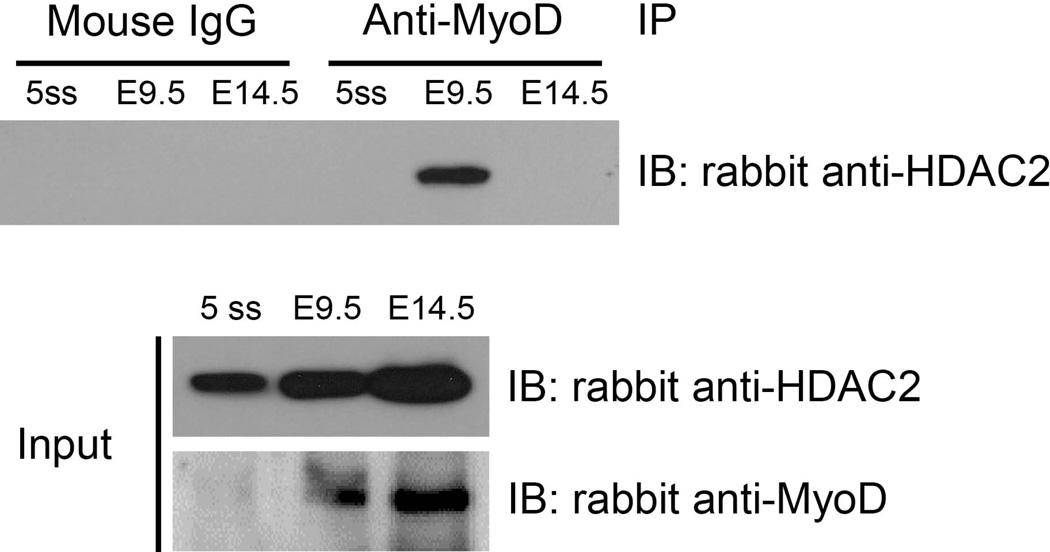
We first performed a co-immunoprecipitation experiment from whole E9.5 embryo extracts, where expression of MCK is limited if expressed at all (Lyons et al., 1991). Extracts from embryos containing 5 somites were used as an early stage control and extracts from E14.5 embryos (head and viscera removed) were used as a control for later time points where skeletal muscle development is more complete and MCK expression is robust (Lyons et al., 1991). Western blots of input material demonstrated the presence of HDAC2 and MyoD at each stage, though the levels of MyoD in 5 somite embryos were barely detectable relative to the levels detected in the later stage tissue samples (Fig. 4). Immunoprecipitation of each sample with a MyoD antibody revealed association with HDAC2 in E9.5 embryos, with no interaction detected at the 5 somite or at the E14.5 stages. We might have expected to detect MyoD-HDAC2 interactions in the 5 somite stage embryos, but perhaps the relative amount of MyoD present was insufficient for efficient pull-down and subsequent detection of the associated HDAC2. We are more confident in interpreting the negative result from the E14.5 embryo tissue, because at this stage, skeletal muscle in the trunk and limbs has formed and we expect a relatively small proportion of the total number of skeletal muscle cells/precursors to be in the initial stages of differentiation. The data indicate an in vivo association between MyoD and HDAC2 at times when skeletal muscle precursors exist but are not yet expressing MCK, thereby providing additional evidence suggesting that this interaction has physiological relevance.
Figure 4.
MyoD and HDAC2 are associated in mouse embryonic tissue. Extracts were made from mouse embryos at the indicated stage and subjected to co-immunoprecipitation experiments as detailed in the Experimental Procedures.
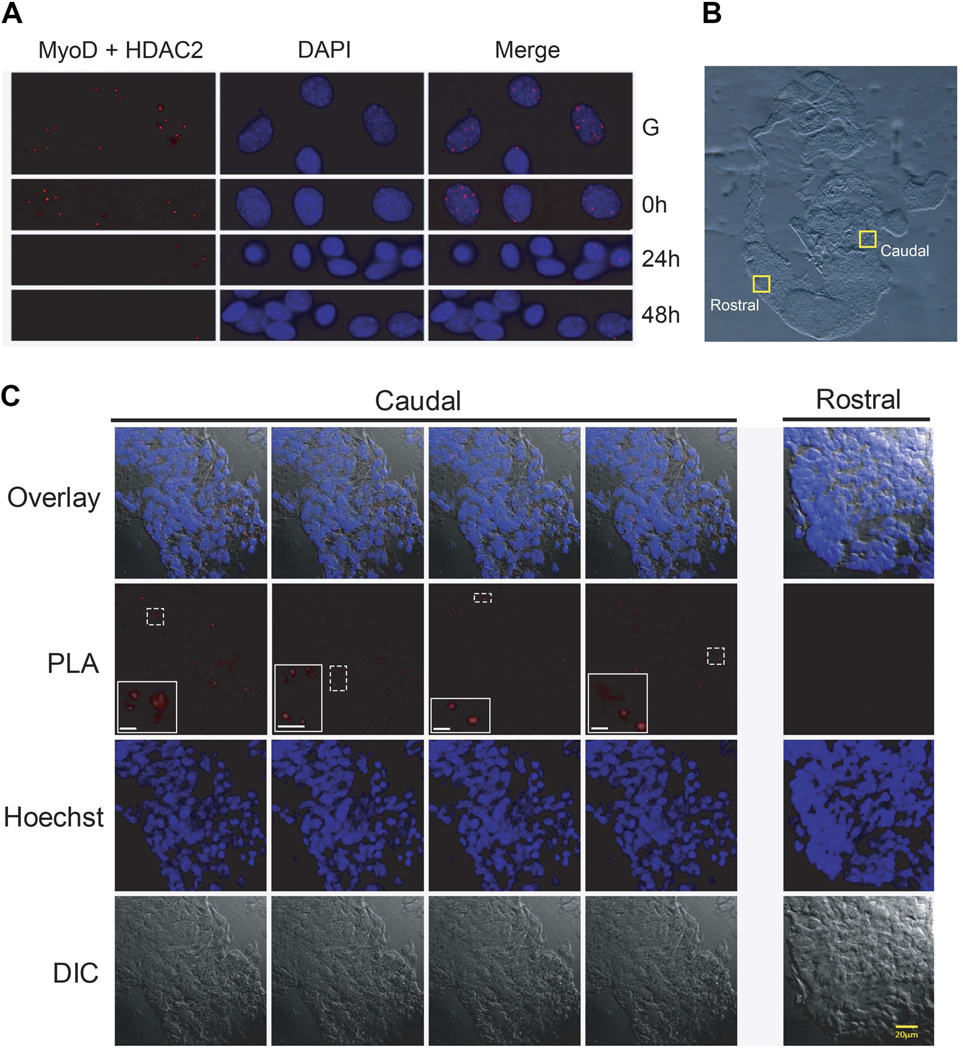
The co-immunoprecipitation experiment is suggestive but provides no spatial information about where in the embryo these interactions occur. We turned to a proximity ligation assay (PLA), which allows identification of co-localized proteins in situ, to document the localization of MyoD-HDAC2 interactions during development. First, however, we used tissue culture cells to verify the interaction and optimize experimental conditions. Analysis of a time course of C2C12 cell differentiation revealed MyoD-HDAC2 interactions in proliferating C2C12 myoblasts and at time 0 of the differentiation process (Fig. 5A), when the cells are approaching confluence and the low-serum differentiation media is introduced. In contrast, few MyoD-HDAC2 interactions were evident at 24 or 72 h post-differentiation (Fig. 5A). The data demonstrate the efficacy of the technique and provide additional support for the conclusion that MyoD-HDAC2 interactions predominantly occur prior to and at the onset of differentiation and are not present at later stages of differentiation or when differentiation is completed.
Figure 5.
MyoD and HDAC2 are associated in C2C12 myoblasts and in embryonic tissue containing caudal, but not rostral somatic tissue. (A) PLA assays were performed on C2C12 cells in growth (G) phase, at the onset of differentiation (0 h), or at 24 or 48 h post-differentiation. DAPI staining and the overlay is shown for each image. (B) Sections of E9.5 mouse embryos were used for PLA assay. The yellow boxes indicate the areas magnified in the following panel. (C) Images from the yellow boxed fields of tissue in the caudal or rostral somite regions in (B) showing the results of the PLA assay, Hoechst staining, the overlay between the PLA and Hoechst staining, or differential interference contrast (DIC). Within the PLA image, the inset images show four different areas, each outlined by a dotted white line. Scale bar in the insets, 5 micrometers. Images presented are representative. The experiment in (A) was performed independently three times. The analysis presented in (B–C) was performed independently on three different embryos.
Serial sections of E9.5 embryos were prepared for the PLA assay. A confocal image of a sample section is presented. Regions from the rostral, myogenin-positive and caudal, myogenin-negative somites that were analyzed are indicated (Fig. 5B). Four representative fields from the myogenin-negative region and one representative field from the myogenin-positive region are presented. PLA signals were observed in all caudal somite fields whereas all rostral fields were completely devoid of PLA signal (Fig. 5C). These data confirm the interaction of MyoD and HDAC2 in the embryo and spatially assign the interactions to somites where the expression of myogenin and of genes expressed at later times of differentiation, such as MCK, has not yet been initiated.
Analysis of factor binding to the MCK promoter during embryogenesis
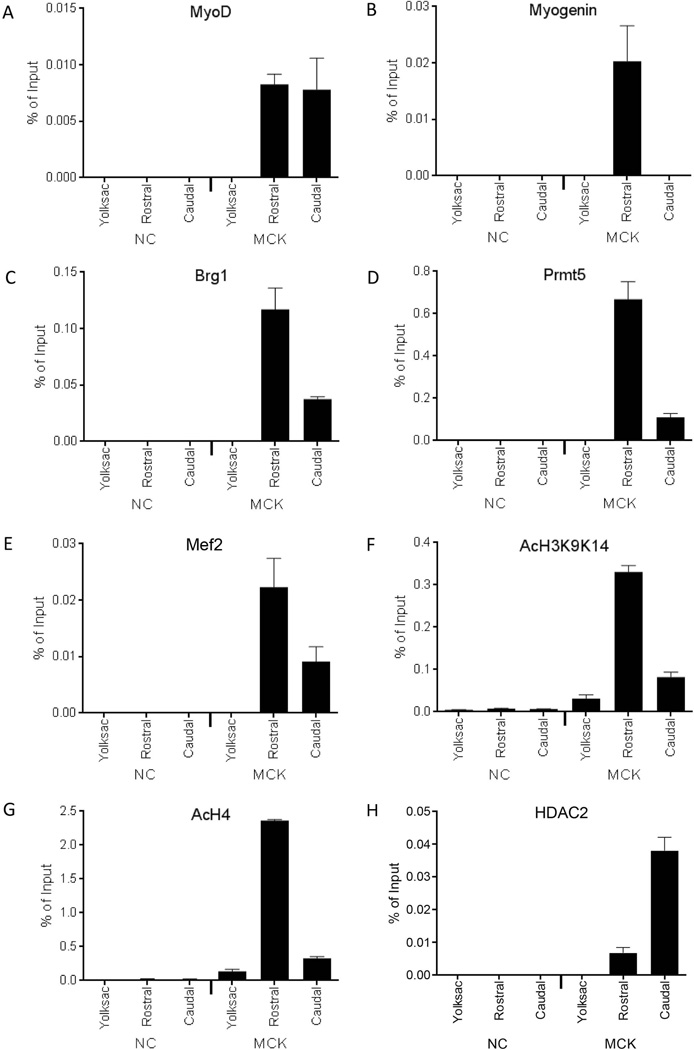
We previously used ChIP to show that MyoD and HDAC2 were individually localized to the MCK promoter in E10.5 embryos but not in E12.5 limb bud or E14.5 limb skeletal muscle tissue (Ohkawa et al., 2006). To investigate the binding of these and other regulators of MCK expression earlier in embryogenesis, we utilized the ChIP approach highlighted in Figure 2. The results indicate that the myogenic regulatory factors MyoD and myogenin were bound to the MCK promoter in the rostral, myogenin-expressing tissue (Figs. 6A–B). As expected, coactivators previously identified at the MCK locus, such as Brg1 and Prmt5 (Figs. 6C–D), the transcription factor Mef2 (Fig. 6E), and modified histones associated with gene activation, diacetylated H3K9K14 and acetylated H4 (Figs. 6F–G), were also enriched at the MCK promoter in the rostral somite-containing tissue. In the caudal, myogenin-negative tissue, the cofactors and modified histones associated with gene activation were present at background levels or at levels reduced relative to that observed in the rostral somite tissue. We note that MyoD was present at the MCK promoter in the myogenin-negative tissue whereas Myogenin was not. Assessment of HDAC2 binding revealed relatively high levels in the caudal somite tissue compared to levels in the rostral somite tissue (Fig. 6H). Collectively the data present a picture suggesting that myogenic regulatory factors, known co-activators, and activating histone marks are present at the MCK promoter in somite tissue where the promoter is active or becoming active. In contrast, the MCK promoter in caudal somite-enriched tissue where MCK gene expression is not yet active is devoid of or has relatively low levels of co-activators and activating histone marks, and is bound by MyoD and HDAC2. Thus both MyoD and HDAC2 can be found in association with the inactive MCK promoter.
Figure 6.
Binding of MyoD, myogenin, co-regulators, and modified histones to the MCK promoter in rostral and caudal somite-containing tissue isolated from E9.5 embryos. Real-time PCR was used to analyze ChIP experiments performed for (A) MyoD, (B) myogenin, (C) Brg1, (D) Prmt5, (E) Mef2, (F) AcH3K9K14, (G) AcH4, (H) HDAC2. Binding to either the MCK promoter or to a negative control (NC) sequence in rostral or caudal somite-containing tissue from E9.5 embryos or in yolk sac was determined. Data represent the mean of three independent experiments +/− standard deviation, except for the HDAC2 experiment, which is the mean of six independent experiments +/− standard deviation. p < 0.05 was obtained for all comparisons between yolk sac and rostral tissue values and for all comparisons other than MyoD and Mef2 between rostral and caudal tissue values.
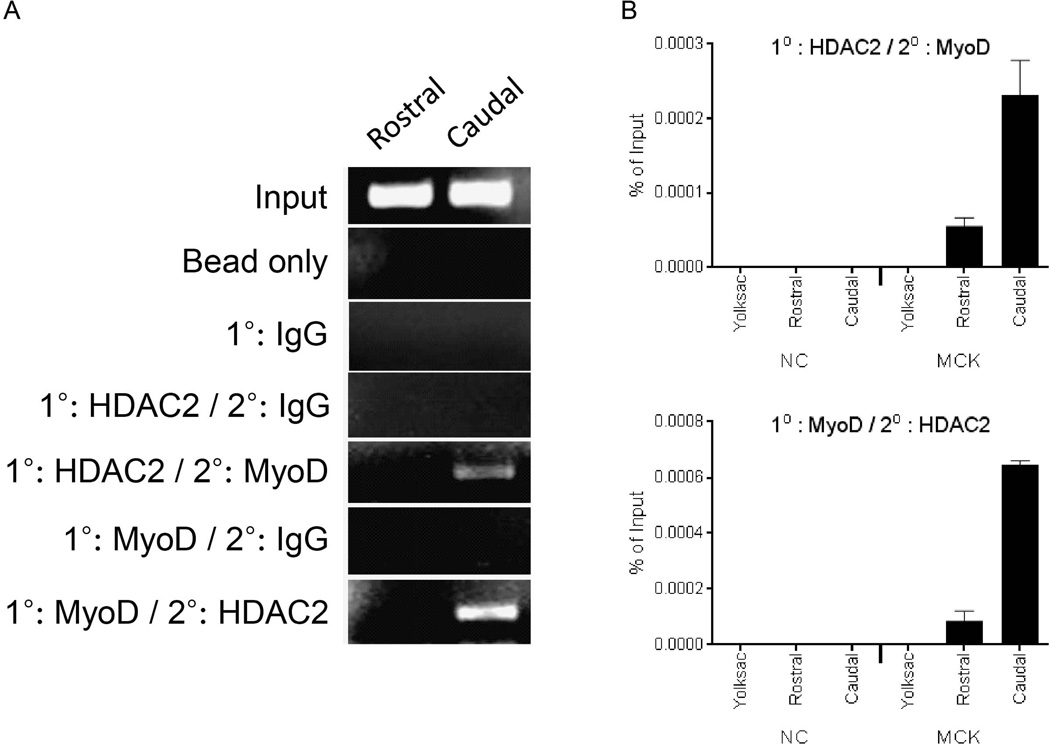
The ChIP experiments presented do not provide evidence that MyoD and HDAC2 are present together at the same time on inactive MCK promoters. To address this question, we performed re-ChIP, also called sequential ChIP, where chromatin from E9.5 rostral or caudal somite-enriched tissue was sequentially immunoprecipitated with MyoD and HDAC2 antibodies, or, conversely, with HDAC2 antibody followed by MyoD antibody. Semi-quantitative PCR results showed that both MyoD and HDAC2 were bound to the MCK promoter in the myogenin-negative caudal tissue but not in the rostral, myogenin-positive tissue (Fig. 7A). Real-time PCR results (Fig. 7B) confirmed the co-localization of MyoD and HDAC2 on the MCK promoter, though these results indicate that the re-ChIP signal was greatly enhanced in the myogenin-negative tissue compared to the signal detected in the myogenin-positive tissue.
Figure 7.
Sequential, or Re-ChIP experiments show co-localization of MyoD and HDAC2 on the MCK promoter in caudal somite-containing tissue. (A) Semi-quantitative PCR analysis of Re-ChIP experiments. The first antibody (1°) and second antibody (2°) used are indicated. (B) Real-time PCR was used to analyze Re-ChIP experiments. Binding to either the MCK promoter or to a negative control (NC) sequence in rostral or caudal somite-containing tissue from E9.5 embryos or in yolk sac was determined. Data represent the mean of three independent experiments +/− standard deviation. p < 0.05 was obtained for all comparisons between yolk sac and rostral tissue values and for all comparisons between rostral and caudal tissue values.
DISCUSSION
During skeletal muscle differentiation, the new program of gene expression that will specify the formation and function of skeletal muscle tissue is initiated. Though the majority of these genes are targets for MyoD and related lineage determining transcription factors, the genes are not expressed with uniform kinetics during development. To address our interest in understanding the molecular basis for temporal regulation of differentiation-specific gene expression, we investigated functional differences between the promoter structure and organization of two representative and well-characterized myogenic genes: the myogenin gene that is expressed early in the differentiation and the gene encoding MCK, which is expressed later in the differentiation process. We performed these analyses in the context of developing mouse embryos, forgoing tissue culture model systems that have traditionally been utilized to identify differentiation-specific changes in chromatin structure and gene regulation. Our efforts demonstrate the feasibility of characterizing gene activation changes in embryonic tissue as a function of both developmental time as well as spatial organization.
Activation of the myogenin promoter
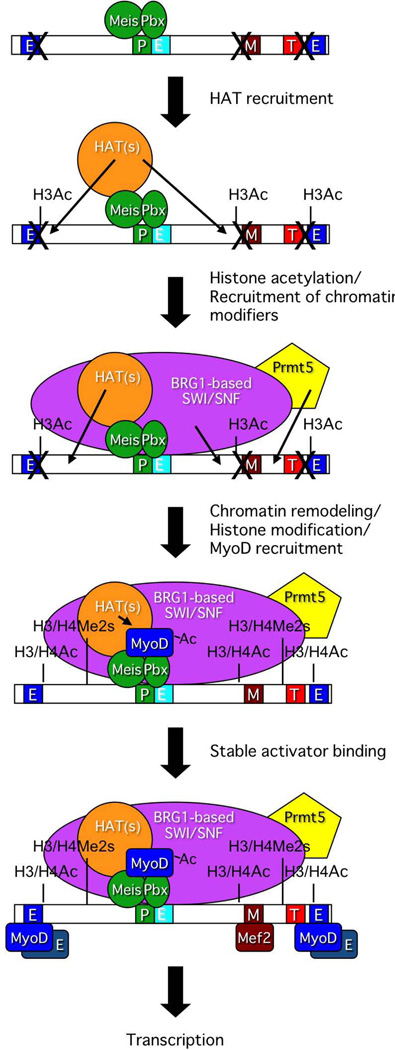
There has been extensive prior characterization of myogenin gene activation. Given our focus on chromatin remodeling enzymes and their interactions with DNA-binding transcription factors and histone modifying enzymes, we monitored the interaction of such factors with the myogenin promoter in the context of embryonic development. Consistent with prior studies using tissue culture cell models for skeletal muscle differentiation, the Pbx-Meis heterodimer, tracked in our assays by the presence of Pbx1, was present on the myogenin promoter in embryos with 5 somites. K9K14 acetylation was also present at the 5 somite stage, but neither MyoD nor myogenin, nor the Brg1 chromatin remodeling enzyme or the histone methyltransferase Prmt5 could be detected. Similarly, the Prmt5 mediated histone mark at H3R8 and acetylated H4 were also not detected (Fig. 1). The data indicate that Pbx initiates association with the myogenin promoter prior to the MyoD lineage determinant and is consistent with prior results and discussions suggesting that Pbx acts as a pioneering factor on the myogenin promoter to initiate the myogenin gene activation process during differentiation (Berkes et al., 2004, Yao et al., 2013). Our temporal study did not further distinguish the order of factor addition to the myogenin promoter, because all of the other factors tested were present on the myogenin promoter by the 10 somite stage of development. However, the spatial analysis (Fig. 3) provided additional novel information. ChIP experiments using the rostral somites, where myogenin was already expressed, showed that MyoD, myogenin, Pbx1, Brg1 and Prmt5 were all present at the myogenin promoter, as would be expected. ChIP experiments using the caudal somites, where the myogenin gene was not yet active, showed the presence of Pbx1, Brg1 and Prmt5 on the myogenin promoter, but not MyoD or myogenin (Fig. 3). The data suggest that Pbx1 is recruiting the chromatin remodeling and arginine methyltransferase enzymes to the promoter prior to recruiting MyoD. This is consistent with co-immunoprecipitation data from tissue culture cell models of differentiation showing association between Pbx1 and Brg1 after the onset of differentiation (de la Serna et al., 2005) and with data showing the physical association of the Brg1 remodeling enzyme with the Prmt5 methyltransferase (Pal et al., 2004, Pal et al., 2003). We therefore propose a refinement of existing models for the step-by-step assembly of regulatory factors and histone modifications that promote activation of the myogenin gene (Fig. 8).
Figure 8.
Proposed model indicating the order of events during activation of the myogenin promoter in mouse embryonic tissue. This schematic diagram is a significantly modified version of the diagram published in Figure 10 of (de la Serna et al., 2005) that was amended with permission from the American Society for Microbiology.
Prior to and at the initial stages of myogenin gene activation, the Pbx-Meis heterodimer marks the chromatin of the myogenin locus (Fig. 8, step 1). The presence of H3K9K14 acetylation coincident with Pbx1 binding at the 5 somite stage (Fig. 1) suggests that one of the initial changes during myogenin activation is the recruitment of H3-specific HATs that will acetylate local chromatin (Fig. 8, step 2). Pbx association with HATs and stimulation of gene expression has been demonstrated by others (Saleh et al., 2000). The exact HAT(s) responsible for H3 acetylation is unclear; both PCAF and p300/CBP co-activate MyoD and acetylate H3, H4, and MyoD (Dilworth et al., 2004, Eckner et al., 1996, Polesskaya and Harel-Bellan, 2001, Polesskaya et al., 2001a, Polesskaya et al., 2001b, Puri et al., 1997a, Puri et al., 1997b, Sartorelli et al., 1997, Sartorelli et al., 1999). Regardless, the evidence presented in Fig. 3 indicates that histone acetylation is followed by association of Brg1, as part of the SWI/SNF chromatin remodeling enzyme, and Prmt5 (Fig. 8, step 3), which in turn would promote histone H3 and H4 symmetric dimethylation, ATP-dependent chromatin remodeling, and H4 acetylation of the myogenin promoter (Fig. 8, step 4). Subsequently, MyoD binding was observed (Fig. 3) In light of the extensive evidence for Pbx1 facilitating MyoD binding to an adjacent site on the myogenin promoter (Berkes et al., 2004, Knoepfler et al., 1999), we incorporated the existing two-step model for MyoD binding in which MyoD first interacts with Pbx/Meis, making protein:protein contacts as well as protein:DNA contacts in the vicinity of the Pbx binding site before transitioning to stably bound interactions with upstream and downstream consensus binding sites and enhanced myogenin transcription ((Berkes et al., 2004, de la Serna et al., 2005); Fig. 8, steps 4–5).
There are two issues to address when considering the model that we present. Genome-wide binding studies in C2C12 cells placed MyoD on the promoter of many myogenic genes prior to differentiation, and the majority of binding sites showed little quantitative difference in occupancy pre- and post-differentiation (Cao et al., 2010). However, a subset of genes, including the myogenin and the MCK genes, showed very low levels of MyoD binding in myoblasts and a substantial increase in occupancy in differentiated cells (Cao et al., 2010). Based on the data presented here, we suggest that either there is no MyoD binding at the myogenin promoter in tissue from embryos containing 5 somites or in somite enriched tissue from the caudal portion of E9.5 embryos or that the levels of binding were below the level of detection in our experiments. A second issue is the absence of information on the MyoD-related factor, Myf5. The absence of MyoD from the myogenin promoter at different times/places during early development does not necessarily reflect the absence of a myogenic regulatory factor. If Myf5 were bound to the myogenin promoter prior to MyoD, then it would remain possible that the chromatin remodeling and arginine methylating enzymes could be recruited by the myogenic factor or by a combination of the myogenic factor and Pbx1. Unfortunately, we were unable to generate reproducible Myf5 ChIP data, despite the existence of prior studies where a commercial Myf5 antibody was successfully used to identify Myf5 binding sites by ChIP (Londhe and Davie, 2011, Parker et al., 2006). Experimental resolution of this question will be a future goal.
Delayed activation of the MCK gene
At the same time that MyoD is involved in the activation of the myogenin gene, the MCK gene remains uninduced. Prior work has implicated an association between MyoD and HDAC1 as well as an association between Snail proteins and HDAC1 and 2 at myogenic promoters prior to the onset of differentiation, but not after (Mal et al., 2001, Puri et al., 2001, Soleimani et al., 2012). Furthermore, HDAC1 can deacetylate acetylated MyoD in vitro as well as act as an inhibitor of differentiation (Mal et al., 2001). Specific examination of the myogenin promoter demonstrated co-localization of MyoD and HDAC1 on the myogenin promoter before differentiation was initiated, but upon differentiation, HDAC1 binding to the promoter was lost, and the PCAF acetyl transferase was bound (Mal and Harter, 2003). Myogenin, however, is expressed soon after the onset of differentiation. Genes expressed at later times following differentiation signaling likely require some additional form of repression, at least temporarily, and this mechanism seems unlikely to involve HDAC1 due to its lack of discernible interaction with MyoD once signaling has occurred. HDAC2, a related but distinct enzyme, has been localized with MyoD on MCK and other “late” gene regulatory sequences in both tissue culture cells and in embryonic tissue from E10.5 mouse embryos (Ohkawa et al., 2006), suggesting co-localization in a manner consistent with keeping these genes off at the early stages of differentiation, but little else is known about a role for HDAC2 in regulating the temporal control of myogenic gene expression during development.
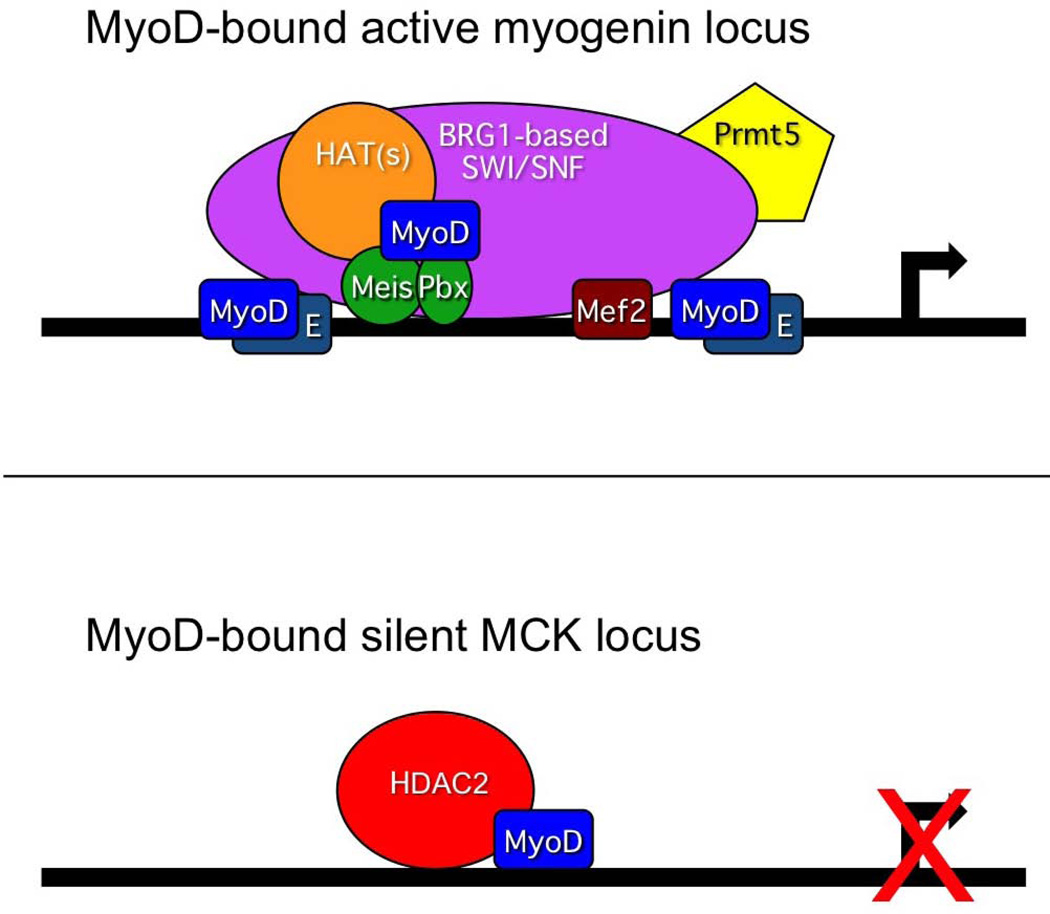
Here, we provide direct evidence of association between endogenous MyoD and HDAC2 in immortalized C2C12 myoblasts prior to and at the onset of differentiation and in situ in the caudal, but not rostral, somites of an E9.5 mouse embryo (Fig. 5). These data indicate a correlation between the association of MyoD and HDAC2 in cells/tissues in which the MCK and other late genes are not expressed. Examination of factor binding in rostral and caudal somite-enriched tissue (Fig. 6) showed that MyoD was present at the MCK promoter in both compartments, despite MCK being active in the rostral tissues but not in the caudal tissue. Binding of co-activators and the presence of activating histone marks were enriched in the rostral tissues and greatly reduced in the caudal tissues. Only HDAC2 showed enrichment in the caudal tissue, suggesting that MyoD is associated with a repressed MCK promoter in the caudal tissues. The re-ChIP experiment (Fig. 7) conclusively demonstrates the co-occupancy of DNA by both factors. Collectively, the data support the idea that MyoD and HDAC2 associate upon differentiation signaling and are present at the MCK promoter in vivo in tissues in which MCK expression has not yet been activated, which suggests that MyoD and HDAC2 act in a repressive manner early in differentiation and contribute to the regulation of temporal control of myogenic gene expression during differentiation. A schematic model illustrating the simultaneous presence of MyoD and identified coactivators at the myogenin locus and MyoD and the associated corepressor HDAC2 at the MCK locus is presented to reflect the differences in the structure of the respective gene regulatory regions after the onset of myogenic differentiation in the somites but prior to the induction of MCK gene expression (Fig. 9). Based on these and prior ChIP results (Ohkawa et al., 2006, Ohkawa et al., 2007), we speculate that the expression of the myogenin gene and subsequent binding of myogenin protein to the MCK locus facilitates the release of MyoD and HDAC2 and leads to activation of MCK expression.
Figure 9.
Proposed model illustrating the simultaneous presence of MyoD with identified coactivators at the myogenin locus state and with the corepressor HDAC2 at the MCK locus after the onset of differentiation in the somites but before activation of the MCK gene.
The complexity of temporal regulation of myogenic gene activation
Finally, the data also reflect the complexity of myogenic gene activation during somite development. Our observations indicate that MyoD is not present at the myogenin promoter in E9.5 rostral somites whereas it is present at the MCK promoter in the same tissues, despite the myogenin gene being activated earlier than the MCK gene. Although we simplify the classification of the temporal expression pattern during myogenesis as “early” and “late”, genome-wide studies of myogenic gene expression and MyoD binding reflect complex temporal regulation, at least in cell line models for differentiation. These studies justify more specific groupings of genes into additional temporal categories (Bergstrom et al., 2002, Tomczak et al., 2004, Delgado et al., 2003, Moran et al., 2002, Blais et al., 2005, Cao et al., 2006, Cao et al., 2010, Soleimani et al., 2012). This additional complexity likely reflects the combination of at least three regulatory mechanisms: regulation of MyoD by post-translational modification, especially by acetylation as discussed above, differences in individual target promoter architecture and cofactor binding, and differences in activation of individual target gene promoters by MyoD and myogenin. Broader studies of myogenic gene activation in the context of somitogenesis may therefore facilitate greater understanding of temporal gene regulation in vivo.
EXPERIMENTAL PROCEDURES
Mice, embryos and cell lines
CD1 mice were housed in the animal care facility at the University of Massachusetts Medical School (Worcester, MA) in accordance with Institutional Animal Care and Use Committee guidelines. E8.5 and E9.5 embryos were isolated and prepared for whole embryo ChIP as described (Cho et al., 2011). Separation and enrichment of rostral, myogenin-positive somites and caudal, myogenin-negative somites from E9.5 embryos was based on previous studies (Cossu et al., 1996, Tajbakhsh et al., 1998) demonstrating that somites could be isolated from surrounding tissue after treatment at 4°C with 0.25% Pancreatin and 0.4% Trypsin in Tyrode’s Solution. C2C12 cells were cultured at 37°C with 5% CO2 in DMEM with 10% fetal bovine serum (FBS), 20 µg/ml gentamicin and 2 mM L-glutamine. Differentiation was induced once cells reached 80% confluence with media supplemented with 2% horse serum instead of 10% FBS.
ChIP assay
E8.5 and E9.5 embryos were isolated and prepared for ChIP using the kits, reagents, and conditions described (Cho et al., 2011). Recovered DNA was analyzed by real-time quantitative PCR in a Bio-Rad iCycler using iQ™ SYBR Green Super Mix (Bio-Rad). Primers used to amplify the myogenin and MCK promoters were described (Cho et al., 2011, Ohkawa et al., 2006). Primers that amplify intergenic sequences 14 kbp upstream of the myogenin gene were used as negative control (NC) regions for all ChIP experiments examining the myogenin promoter. The specific primer sequences are: Myogenin NC forward: 5´- CAG GCA GGA GCA CGG CAG AC -3´; reverse: 5´- ACA CAG CCA GGC GTT CAC TCC-3´. Primers that amplify sequences 11.6 kbp upstream of the MCK gene were used as negative control (NC) regions for all ChIP experiments examining the MCK promoter except Figs. 6E–F, where sequences from the IgH locus were amplified as a negative control using primers previously described (Bergstrom et al., 2002). The specific primer sequences are: MCK NC forward: 5´- CCAGCAGCTCCACACCAGCC -3´; reverse: 5´- GGCCCAGAACGCCTGAACCC -3´. Antibodies: rabbit anti-MyoD (Santa Cruz, sc-304), rabbit anti-Myogenin (Santa Cruz, sc-576), goat anti-Prmt5 (Santa Cruz, sc-22132), rabbit anti-H3R8Me2 (Pal et al., 2004), rabbit anti-AcH4 (Millipore, 06-866), rabbit anti-AcH3K9K14 (Millipore, 06-599), rabbit anti-Brg1 (de La Serna et al., 2000), rabbit anti-Pbx1 (Santa Cruz, sc-889), rabbit anti-HDAC2 (Invitrogen, 51–5100), rabbit anti-Mef2, which recognizes Mef2A, C, and D (Santa Cruz sc-313), normal goat IgG (Santa Cruz, sc-2028), and normal rabbit IgG (Millipore, 12–370). The absence of cross-reactivity between the MyoD and myogenin antibodies was previously published (de la Serna et al., 2005).
Embryo in situ hybridization, lysate preparation and western blotting
The WISH protocol was previously published (Rivera-Perez and Magnuson, 2005). Full-length myogenin cDNA was used as probe. Pelleted embryonic cells were resuspended in 40 µl of RIPA lysis buffer (1% NP 40, 10 mM Tris (pH 7.6), 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate and 2 mM EDTA) with protease inhibitor cocktail (1.25 µl PIC/1ml RIPA buffer). The lysate was incubated on ice for 10 min, with vortexing every 2–3 min. After centrifugation at 4°C at 14000 rpm for 10 min, the supernatant was removed into a new tube. Bradford assay was used for protein quantification based on a standard curve generated from known concentrations of BSA.
25 µg protein were mixed with an equal volume of 2× Laemmli sample buffer (4% SDS, 200 mM DTT, 120 mM Tris-HCl (pH 6.8), 50% glycerol, and 0.02% bromophenol blue) and loaded in lanes of 6–12% SDS-PAGE gels for analysis by immunoblotting. Proteins resolved on the gel were transferred to PVDF membrane (Immobilon-P Transfer Membrane, MILLIPORE) using a current of 100 volts for 1.5 hr. The membranes were blocked with 5% skim milk, 0.05% Tween 20 in 1xPBS for >20 min at RT, then incubated at 4°C overnight with the primary antibody diluted in 5% skim milk in 1X PBS. The next day, the membranes were washed 3 times with 0.05% Tween 20 in 1xPBS for 10 min and subsequently incubated at RT with ECL™ horseradish peroxidase labeled secondary Ab diluted in 5% skim milk for 1.5 hr. The membranes were washed 3 times using 0.05% Tween 20 in 1xPBS for 10 min, developed by enhanced chemiluminescence (Amersham Biosciences) as per the manufacturer’s instructions and exposed to Amersham hyperfilm™ MP (GE Healthcare). Blots were stripped and reprobed. Antibodies used: rabbit anti-MyoD (Santa Cruz, sc-304), rabbit anti-Myogenin (Santa Cruz, sc-576), goat anti-Prmt5 (Santa Cruz, sc-22132), rabbit anti-AcH4 (Milipore, 06-866), rabbit anti-AcH3K9K14 (Milipore, 06-599), rabbit anti-Brg1 (de La Serna et al., 2000), rabbit anti-Pbx1 (Santa Cruz, sc-889), rabbit anti-HDAC2 (Invitrogen, 51–5100), anti-Hsp70 (ABR Affinity Bio Reagents, MA3-006). The absence of cross-reactivity between the MyoD and myogenin antibodies was previously published (de la Serna et al., 2005).
Co-Immunoprecipitation
Freshly dissected embryos were washed 2 times with 1 ml 1xPBS and lysed for 30 min at 4°C in 500 µl of 1% NP-40 lysis buffer (10 mM Tris-HCl pH 7.8, 0.5 mM EDTA, 250 mM NaCl) with protease inhibitor cocktail (8 µl/ml). After centrifugation at 4°C at 14000 rpm for 10 min, 50 µl of supernatants was reserved at 4°C and the rest was pre-cleared at 4°C for 1 hr. on a rotating platform with 500 µl of normal serum (Santa Cruz, sc-2025 or sc-2027) and 80 µl of protein A- or G-sepharose beads that had been washed 3 times with 1% NP-40 lysis buffer prior to use. After centrifugation at 4°C at 2000 rpm for 5 min, the pre-cleared supernatants were incubated on a rotating platform at 4°C overnight with 2–4 µg of the indicated antibody (rabbit anti-HDAC2 (Invitrogen, 51–5100), mouse anti-MyoD (Santa Cruz, sc-32758), or normal mouse IgG (Millipore 12–371)). The following day, the IP samples were mixed with 80 µl of protein A- or G-sepharose beads and incubated at 4°C, with rotation, for 1 hr. After centrifugation at 4°C at 3000 rpm for 2 min, the supernatants were discarded. The beads were washed with 1 ml of 1% NP-40 lysis buffer with protease inhibitor cocktail up to 4 times. Following the final wash, IP samples were resuspended in 60 µl 2× Laemmli sample buffer and boiled for 5 min. For immunoblotting, 10 µl input control was mixed with 10 µl 2× Laemmli sample buffer and all samples were resolved by SDS-PAGE, then visualized using each target antibody.
PLA
Proximity Ligation Assay (PLA) was performed using DuoLink Mouse Plus and Rabbit Minus secondary antibodies and the staining kit from O-Link Biosciences according to the manufacturer’s instructions. Antibodies used were against MyoD (Santa Cruz, sc-32758) and HDAC2 (Invitrogen, 51–5100). Sacrificed E9.5 embryos were flash frozen and sectioned. After completing the PLA on the embryo sections, signals were detected using laser-scanning confocal microscopy (Nikon Eclipse E800) with a 40× phase contrast oil immersion objective (numerical aperture=1.3). Confocal data capture and extraction were performed using manufacturer’s software (Leica). Signals from PLA performed on C2C12 cells were examined using fluorescent microscopy with a green fluorescent filter.
Statistical Analysis
Two-tailed Student t tests were performed to evaluate the statistical significance of quantitated results.
ACKNOWLEDGEMENTS
This work was supported by NIH R01GM56244 and R01GM56244-12S1 to ANI and GM87130 and GM94874 to JARP. We gratefully acknowledge the advice of Professor G. Cossu regarding isolation of somite-enriched tissue. We thank C. Baron for assistance with figure preparation, J. Nickerson for comments on microscopy methods and images, and S. LeBlanc for comments on the manuscript.
REFERENCES
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, Macquarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho OH, Rivera-Perez JA, Imbalzano AN. Chromatin immunoprecipitation assay for tissue-specific genes using early-stage mouse embryos. J Vis Exp. 2011 doi: 10.3791/2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Kelly R, Tajbakhsh S, Di Donna S, Vivarelli E, Buckingham M. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–437. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001a;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Roy K, Carlson KA, Imbalzano AN. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J Biol Chem. 2001b;276:41486–41491. doi: 10.1074/jbc.M107281200. [DOI] [PubMed] [Google Scholar]
- Delgado I, Huang X, Jones S, Zhang L, Hatcher R, Gao B, Zhang P. Dynamic gene expression during the onset of myoblast differentiation in vitro. Genomics. 2003;82:109–121. doi: 10.1016/s0888-7543(03)00104-6. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci U S A. 2004;101:11593–11598. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, Sartorelli V. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 1999;27:3752–3761. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Londhe P, Davie JK. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet Muscle. 2011;1:14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, Muhlebach S, Moser A, Masood R, Paterson BM, Buckingham ME, Perriard JC. Developmental regulation of creatine kinase gene expression by myogenic factors in embryonic mouse and chick skeletal muscle. Development. 1991;113:1017–1029. doi: 10.1242/dev.113.3.1017. [DOI] [PubMed] [Google Scholar]
- Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci U S A. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Moncaut N, Rigby PW, Carvajal JJ. Dial M(RF) for myogenesis. FEBS J. 2013;280:3980–3990. doi: 10.1111/febs.12379. [DOI] [PubMed] [Google Scholar]
- Moran JL, Li Y, Hill AA, Mounts WM, Miller CP. Gene expression changes during mouse skeletal myoblast differentiation revealed by transcriptional profiling. Physiol Genomics. 2002;10:103–111. doi: 10.1152/physiolgenomics.00011.2002. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5457. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007;282:6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MH, Perry RL, Fauteux MC, Berkes CA, Rudnicki MA. MyoD synergizes with the E-protein HEB beta to induce myogenic differentiation. Mol Cell Biol. 2006;26:5771–5783. doi: 10.1128/MCB.02404-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Harel-Bellan A. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J Biol Chem. 2001;276:44502–44503. doi: 10.1074/jbc.M106501200. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol Cell Biol. 2001a;21:5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Fritsch L, Duquet A, Ait-Si-Ali S, Robin P, Vervisch A, Pritchard LL, Cole P, Harel-Bellan A. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 2001b;20:6816–6825. doi: 10.1093/emboj/20.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O. Vertebrate somitogenesis. Annu Rev Cell Dev Biol. 2001;17:311–350. doi: 10.1146/annurev.cellbio.17.1.311. [DOI] [PubMed] [Google Scholar]
- Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997a;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- Puri PL, Mercola M. BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev. 2012;26:2673–2683. doi: 10.1101/gad.207415.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997b;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–371. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Saleh M, Rambaldi I, Yang XJ, Featherstone MS. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol Cell Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, Van Ijcken WF, Grosveld F, Rudnicki MA. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Cossu G. Establishing myogenic identity during somitogenesis. Curr Opin Genet Dev. 1997;7:634–641. doi: 10.1016/s0959-437x(97)80011-1. [DOI] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- Yao Z, Farr GH, 3rd, Tapscott SJ, Maves L. Pbx and Prdm1a transcription factors differentially regulate subsets of the fast skeletal muscle program in zebrafish. Biol Open. 2013;2:546–555. doi: 10.1242/bio.20133921. [DOI] [PMC free article] [PubMed] [Google Scholar]