Abstract
Hair follicle stem cells (HFSCs) and their transit amplifying cell (TAC) progeny sense BMPs at defined stages of the hair cycle to control their proliferation and differentiation. Here, we exploit the distinct spatial and temporal localizations of these cells to selectively ablate BMP signaling in each compartment and examine its functional role. We find that BMP signaling is required for HFSC quiescence and to promote TAC differentiation along different lineages as the hair cycle progresses. We also combine in vivo genome-wide chromatin immunoprecipitation and deep-sequencing, transcriptional profiling, and loss-of-function genetics to define BMP-regulated genes. We show that some pSMAD1/5 targets, like Gata3, function specifically in TAC lineage-progression. Others, like Id1 and Id3, function in both HFSCs and TACs, but in distinct ways. Our study therefore illustrates the complex differential roles that a key signaling pathway can play in regulation of closely-related stem/progenitor cells within the context of their overall niche.
INTRODUCTION
To maintain tissue homeostasis and regeneration, self-renewal and differentiation of stem cells (SCs) must be balanced. The cycling behavior of hair follicle SCs (HFSCs) and their subsequent generation of differentiating progenitor cells offer a unique opportunity to study these processes. HFSCs reside in a two-tiered niche referred to as the bulge and its associated hair germ (HG) just beneath it. During homeostasis, the lower HF below the bulge cycles through bouts of active hair growth (anagen), destruction (catagen) and rest (telogen) (Paus and Cotsarelis, 1999). Telogen can last weeks, and throughout this time, HFSCs are quiescent. However, quiescent HFSCs in the HG are in contact with an underlying dermal papilla (DP) stimulus. Continuous crosstalk leads to a build-up of BMP inhibitory signals and WNT activating signals until the thresholds become sufficient to stimulate a new round of hair growth (Greco et al., 2009).
HG cells begin to proliferate and form a pool of unspecified, short-lived transit amplifying progenitors (TACs), which express sonic hedgehog (SHH)(Hsu et al., 2014). SHH is transiently sensed by bulge HFSCs, which proliferate to self-renew and to form the outer root sheath (ORS) (Hsu et al., 2014; Rompolas et al., 2013) SHH is also sensed by the DP, which elevates FGF7 and BMP inhibitor NOGGIN to sustain proliferation and specification of matrix TACs within the hair bulb (Hsu et al., 2014).
In total, TACs differentiate along seven morphologically and molecularly distinct pathways. At the core of the mature HF is the hair shaft (HS), consisting of an inner pigmented medulla surrounded by a cortex and HS cuticle (K82+). Cortex/cuticle cells exhibit nuclear LEF1 and β-catenin, transcriptional co-activators for WNT target genes encoding HS-specific keratins (AE13+) (DasGupta and Fuchs, 1999; Merrill et al., 2001). Distal to the HS cuticle are the three layers of inner root sheath (IRS). The cuticle and Huxley IRS layers express transcription factor GATA3 and structural protein trichohyalin (AE15+), while the companion layer (CP) between IRS and ORS is marked by keratin 6 (K6), also found in the medulla. Each of their progenitor pools have been lineage traced to the matrix TACs in a spatial distribution that recapitulates this differentiated layering (Fuchs, 2007; Sequeira and Nicolas, 2012).
Accumulating evidence suggests that BMPs (bone morphogenic proteins) play a major role in the regulation of both SCs and their TACs. Drosophila germ line SCs require BMP 2/4 for their maintenance (Xie and Spradling, 1998). By contrast, HF and intestinal SCs use BMP signaling to suppress SC activation (He et al., 2004; Kobielak et al., 2007).
The importance of BMP signaling in the HF has long been recognized. Postnatal inhibition of BMP signaling by ectopic Noggin expression impairs HS formation (Kulessa et al., 2000), and embryonic inhibition of BMP signaling by conditional targeting of Bmpr1a blocks hair lineage specification and/or differentiation (Andl et al., 2004; Kobielak et al., 2003; Ming Kwan et al., 2004; Yuhki et al., 2004). The suppressive effects of inhibiting BMP arise early in the hair lineage, as evidenced by the precocious activation of telogen-phase HFSCs and impaired differentiation that arises when they lack Bmpr1a (Kandyba et al., 2013; Kobielak et al., 2007).
While the consequences of BMP signaling are well-studied, less is known about the molecular mechanisms that underlie how BMP affects HFSC behavior and hair differentiation. Some insights come from Kandyba et al. (2013), who used the keratin 15 (K15) promoter to drive an inducible Cre recombinase and ablate Bmpr1a in telogen-phase HFSCs of the bulge and HG. They identified 16 HFSC/HG mRNAs upregulated by ≥2X, and 80 downregulated mRNAs. Intriguingly, the downregulated genes encoded some inhibitors of HFSC proliferation, such as FGF18, BMP6 and WNT inhibitor DKK3, while upregulated genes included Wnt7a, 7b and Wnt16 (Kandyba et al., 2013). Overall these findings were consistent with prior reports that BMP inhibition a) promotes WNT signaling (Jamora et al., 2003) and b) is a distinguishing feature of the transition of quiescent HFSCs in the HG to an activated state (Greco et al., 2009).
A number of important questions remain. To what extent is this differential expression in mRNAs directly a consequence of changes in pSMAD1/5/8-SMAD4 transcriptional activity? Is BMP activity merely operative in regulating proliferation or does it also influence fate specification and/or differentiation? If the lineage utilizes BMP signaling in different ways, how is this temporally and spatially regulated? In this study, we address these important issues. Using inducible Cre lines, we first analyze the consequences of ablating Bmpr1a selectively in either HFSCs or matrix TACs. Carrying out both RNA-Seq and pSMAD1/5 genome-wide chromatin immunoprecipitation and deep sequencing (ChIP-Seq) analyses on purified HFSCs and TACs, we then identify and validate downstream bona fide pSMAD1/5 targets whose expression is impacted by BMP signaling. Focusing on pSMAD1/5 target genes Gata3 and Ids, we employ a combination of conventional genetics and downstream markers of BMP and other signaling pathways to probe the physiological relevance of these pathways and their effectors in HFSCs, their TAC progeny and their terminal differentiation programs.
RESULTS
BMP Signaling is Temporally Regulated in Both HFSC and TACs
Binding of BMP to their receptors activates an intracellular signaling cascade where SMAD1/5/8 proteins become phosphorylated (activated), translocate to the nucleus and partner with SMAD4 to act as bipartite transcription factors (Massague et al., 2005). In the hair lineage, Smad8 expression is low (Figure S1A), Smad1 and 5 show redundancy, and double knock out mice recapitulate aspects of Bmpr1a cKO mice (Kandyba et al., 2014). Immunoreactivity for nuclear pSMAD1/5 was detected in quiescent HFSCs in early and mid telogen (Figure 1A). This waned as HFs transitioned to anagen. Immunoreactivity remained low through early Ana-IIIa, coincident with the emergence of Shh-expressing TACs and elevated NOGGIN (Hsu et al., 2014; Woo et al., 2012). HFs lacking SHH (K15CrePGR-Shh cKO) failed to downregulate pSMAD1/5 (Figure 1B).
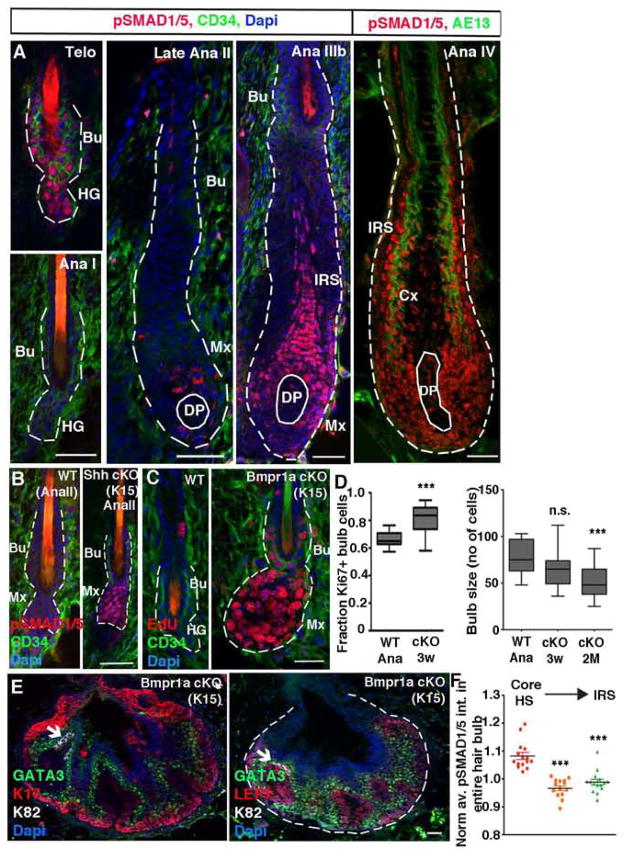
Figure 1. BMP signaling is temporally regulated and required to maintain matrix TACs.
(A) pSMAD1/5 patterns throughout the hair cycle. (B) Early anagen HFSCs fail to downregulate pSMAD1/5 after Shh ablation. (C) Bmpr1a ablation leads to precocious HFSC activation. (D) Quantifications of changes in %Ki67+ hair bulb cells with time following Bmpr1a ablation in HFSCs. (E) Long term loss of BMP signaling leads to HF cysts composed largely of ORS (K17+), early matrix (LEF1+) and IRS-TACs (GATA3+). (F) Within a planar midsection of the hair bulb, more central cortical HS-TACs exhibit stronger nuclear pSMAD1/5 reactivity than more distal GATA3+ TACs. Cre-promoters are given in parentheses. Ana, anagen; Telo, telogen; Bu, bulge; HG, hair germ; DP, dermal papilla; Mx, matrix; Cx, cortex; IRS, inner root sheath. Data are represented as mean±SEM. *** = p<0.001 using Anova. Scale bars = 25 μm.
From early Ana I→IIIb, BMP signaling remained low as activated HFSCs formed the ORS. Signs of pSMAD1/5 immunoreactivity in the bulge resurfaced in Ana-IIIb. At this time, nuclear pSMAD1/5 was also observed in the emerging terminally differentiating IRS(Figure 1A). In maturing Ana-IV HFs, pSMAD1/5 immunolabeling remained high in the terminally differentiating cells particularly within the IRS. These patterns were in agreement with and extended prior developmental studies (Andl et al., 2004), and suggested that BMP signaling may regulate distinct aspects of the HFSC lineage: SC quiescence and terminal differentiation.
Loss of BMP Signaling Affects HF Lineages
When normally quiescent HFSCs are targeted for Bmpr1a loss, they adopt molecular features of activated HFSCs, rapidly progressing to tumor-like cysts (Andl et al., 2004; Kandyba et al., 2013; Kobielak et al., 2003; Kobielak et al., 2007; Ming Kwan et al., 2004; Yuhki et al., 2004). In monitoring the temporal alterations that follow K15-CrePGR-mediated ablation of Bmpr1a in 2nd telogen-phase HFSCs, we observed precocious activation of HFs, accompanied by elevated proliferation within the Bmpr1a-null bulge, HG and early TACs (Figure 1C). These activated Bmpr1a-null HFSCs failed to maintain or return to quiescence.
Early Bmpr1a-null HFs consisted of an upper stalk expressing basal bulge HFSC/ORS markers (SOX9, K17) and an expanding bulb expressing markers of activated HG/early TACs (LEF1, PCAD, Shh, WNT-reporter) (Figure S1B). Cells within Bmpr1a cKO hair bulbs were also hyperproliferative, and this was sustained over time (Figure 1D). As these structures grew, they formed lobular structures whose cells organized in an onion-skin layered fashion analogous to that seen in normal HFs (Figures 1E, S1C–S1E). At this stage, TAC-like cells expressed IRS lineage markers such as GATA3. More centrally in these lobes were weakly LEF1+ cells which spatially corresponded to HS-TACs but which expressed in addition, LHX2, WNT-reporter activity, and other signs of HG/early unspecified TACs. Above these lobes were small numbers of cells expressing markers for ORS, CP, differentiating IRS and cuticle, but not medulla or cortex.
Within 2 months, most Bmpr1a-null HFs had transformed into cysts. Hair bulbs of residual HFs remained proliferative but were noticeably smaller than normal and displayed thinner hair shafts lacking medullary structures; ultimately mice grew bald (Figures S1F and S1G). Thus, loss of BMP signaling appeared to affect TAC lineages to different degrees of severity, with the inner medullary lineage being the most adversely affected. The WT precursors for the HS lineages also appeared to be the most sensitive to BMP signaling, as revealed by their higher pSMAD1/5 intensity measured by quantitative immunofluorescence (Figures 1F and S1H).
Loss of BMP Signaling Expands the IRS Progenitor Pool at the Expense of HS TACs
A priori, the cell populations within K15-CrePGR-derived Bmpr1a-null cysts could merely reflect perturbations arising from a global expansion of HFSCs, which in turn would be expected to expand proliferation within downstream lineages in reverse temporal order of appearance. Subsequent morphological distortions might further alter lineage differentiation. We circumvented these caveats by crossing Bmpr1afl/fl mice to Shh-CreER on the background of the R26YFP reporter line. A benefit of this strategy is that Shh is localized asymmetrically in mature HFs. Since this feature does not affect the onion-skinned symmetry of the differentiating hair layers, it allowed us to examine the consequences of selectively blocking BMP signaling in the Shh+ pocket of TACs, while using the other side of the HF as an internal control. HFs were synchronized by depilation in their 2nd telogen, and then treated with tamoxifen from day 4→10 to induce Cre in full anagen (Figures 2A and 2B).
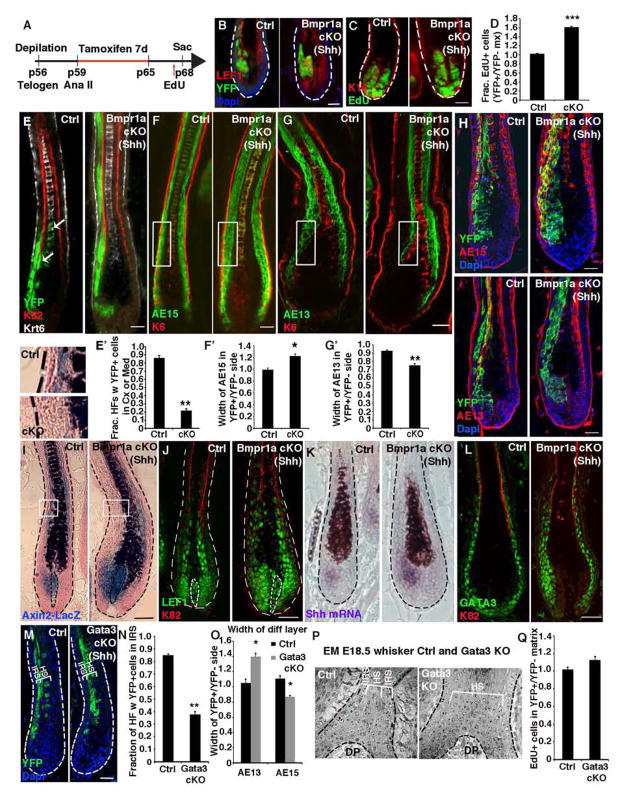
Figure 2. BMP restricts the Shh-expressing IRS progenitor pool.
(A) Experimental timeline for selectively targeting Bmpr1a ablation in Shh-expressing TACs during full-anagen. (B–D) BMPR1a loss results in an expansion of YFP+ cKO TACs and an increase in their proliferation. (E) Lineage tracing of YFP+ TAC progeny. (F and G) Differential distribution of YFP+ progeny reveals an increase in AE15+ IRS and decrease in AE13+ cortex following Bmpr1a ablation in TACs. (H) Bmpr1a-null YFP+ TACs lineage trace to AE15+ IRS but not AE13+ cortex. (I) Axin2-LacZ activity in Shh-expressing TACs, DP and cortex/medulla. Note lack of X-Gal staining (blue) in HS developing from Bmpr1a-targeted side. (J) LEF1+ TACs are expanded upon BMPR1a loss, but LEF1+ cortex is absent. (K) Shh+ IRS TACs are expanded and express elevated Shh in the absence of BMP signaling. (L) GATA3+ cells are expanded following Bmpr1a ablation in Shh+ TACs. (M and N) Lineage tracing of YFP+ progeny following Gata3 ablation in Shh+ TACs. Following loss of GATA3, AE15+ IRS progeny (external to the K82+ HS cuticle) are lost, while AE15+ HS progeny (internal to K82) are expanded. (O) Gata3 ablation in Shh+ TACs leads to an expanded cortical layer (AE13+) and smaller AE15+ IRS. (P) EM of Gata3 straight KO HFs reveals absence of IRS lineage. (Q) Proliferation is unaffected following Gata3 ablation in TACs. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test. Scale bar = 25 μm.
Lineage tracing of control (Shh-CreER; Bmpr1afl/+) HFs revealed that YFP+ TACs contributed predominantly to the three IRS lineages sandwiched between the K6+ CP and K82+ HS cuticle (Figures 2B–2E). YFP+ cells contributed, but less so, to the HS cuticle, K6+ medulla and hair keratin-expressing (AE13+) cortex. By contrast, Bmpr1a-null YFP+ Shh-marked TACs very rarely contributed to HS lineages (Figures 2G–2H).
Quantifications of EdU labeling revealed that YFP+ cells within the Bmpr1a-targeted matrix TAC pool expanded by ~1.6X compared to Ctrl HFs or YFPneg (untargeted) side (Figures 2B–2D, S2A). Thus, autonomous loss of BMP signaling within Shh+ matrix TACs was sufficient to elicit their excessive proliferation independent of changes upstream in the lineage. Moreover, the expansion of this Bmpr1a-null matrix population did not elicit noticeable paracrine effects in untargeted TACs, as confirmed by EdU quantifications of WT, heterozygous and cKO HFs (Figure S2B).
Trichohyalin (AE15+) YFP+ IRS on the Bmpr1a-null side was significantly thickened, while AE13+ cortex was correspondingly thinner (Figures 2F–2H). EdU incorporation verified that BMP signaling was not required for TACs to exit the cell cycle upon terminal differentiation (Figure S2C and S2D). Rather, the imbalance in terminally differentiated layers suggested that a fate skew occurred within the pool of Bmpr1a-null YFP+ TACs, resulting in expansion of IRS-TACs and reduction in HS-TACs.
Impact of Loss of BMP Signaling on WNT Signaling in the Matrix
To evaluate whether the WNT pathway might be affected by loss of BMP signaling, we bred the Axin2-LacZ knockin WNT reporter line to our mice. Analogous to prior results with the TOPGAL reporter for LEF1/β-catenin, the cortex from WT HFs was LacZ+ (Figure 2I). In stark contrast, the cortical zone of the Bmpr1a-null side was selectively LacZneg (see magnified boxed areas). Further reflecting the lack of HS differentiation was the diminished numbers of nuclear LEF1+ terminally differentiating layers on the Bmpr1a-null side just above the bulb (Figure 2J).
By contrast, in the WT matrix, LEF1 is largely cytoplasmic, but upon Bmpr1a-ablation, it was strongly nuclear, suggestive of WNT/β-catenin signaling. Indeed, Wnt10b and Shh were elevated, and Axin2-LacZ and endogenous WNT target genes were activated in this pocket of Bmpr1a-null TACs (Figures 2I–2K; S2E–2G).
GATA3 is Not Suppressed by Bmpr1a Targeting But is Required for TACs to Select the IRS Fate
IRS lineages fail to form when BMP signaling is abrogated during embryogenesis (Andl et al., 2004; Kaufman et al., 2003; Kobielak et al., 2003; Ming Kwan et al., 2004). Thus it was intriguing to find that whether Bmpr1a was ablated in adult HFSCs or the anagen-phase Shh-pocket of TACs, GATA3+ IRS precursors increased while cortical/medulla lineages were diminished (Figures 1E, 2L; S2H).
To ask whether GATA3 is required for the specification of the IRS lineage by adult TACs, we conditionally targeted Gata3 in Shh-CreER, R26YFP mice, as we had done for Bmpr1a. Purified YFP+ targeted TACs lacked GATA3 and showed a reduction in Shh expression (Figures S2I–2K). In contrast to BMPR1a-deficient TACs, TACs lacking GATA3 preferentially contributed to HS lineages (Figures 2M–2O and S2L). The shift in distribution of targeted TACs was accompanied by a reduction of AE15+ cells and a gain in AE13+ and LEF1+ cells on the targeted side of the HF. Electron microscopy of Gata3 straight KO HFs corroborated the lack of IRS and expansion of HS in GATA3’s absence (Figure 2P). Since proliferation of Gata3 cKO TACs remained normal (Figure 2Q), a fate switch must have occurred. Together, these data show that IRS specification by matrix progenitors is dependent upon GATA3, and without it, TACs favor cortical and medullary lineages.
pSMAD1/5 Regulates Transcriptional Networks in Both HFSCs and TACs
To identify the direct transcriptional targets of BMP signaling, we began by using fluorescence activated cell sorting (FACS) to isolate quiescent HFSCs in 2nd telogen (P56) and total matrix TACs in anagen (P28–P32)(Lien et al., 2011) and validated their purity (Figure S3A). We then performed pSMAD1/5/8 antibody ChIP-seq on chromatin isolated from our purified HFSCs and TACs. Our ChIP-seq data were analyzed as established previously [see ENCODE guidelines (Landt et al., 2012)].
After aligning ChIP-seq reads to the mouse genome, we used MACS (Zhang et al., 2008) for peak calling and identified 2008 and 3364 genes that were bound by pSMAD1/5 in HFSCs and TACs, respectively (Figure 3A and Table S1). 60% of pSMAD1/5 peaks found in HFSCs (53% for TACs) were classifieds as enhancer regions (within ±50kb), whereas 31% of HFSCs peaks were in promoters (39% for TACs) (Figure 3B). Interestingly, 76% (1525 of 2008) of pSMAD1/5-bound genes in HFSCs were shared with targets found in TACs, and 78% of these genes had overlapping peaks (Figures 3C and S3B). The overall quality of pSMAD1/5 ChIP data sets were comparable, as judge by normalized read density of peaks in either HFSCs or TACs, or the frequency of binding to non-tissue specific genes (Ramskold et al., 2009) (Figures S3C and S3D).
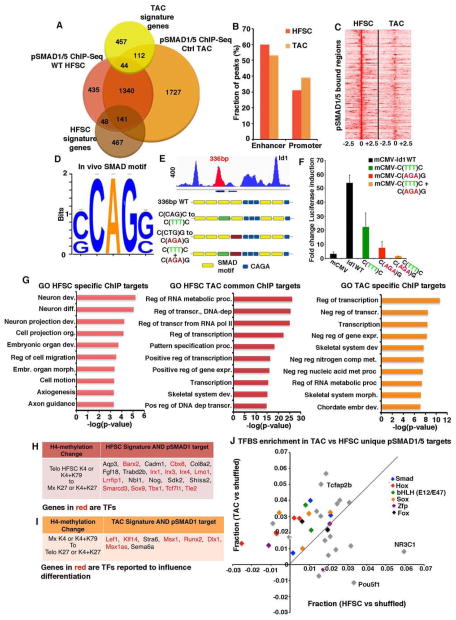
Figure 3. pSMAD1/5 regulates HFSC and TAC transcriptional networks.
(A) Venn diagram of pSMAD1/5 ChIP-seq of HFSC and TAC chromatin, compared to RNA-seq profiling of transcripts ≥2X (p<0.05) changed in the two populations (“signature genes”). (B) SMAD1/5 occupies predominantly enhancer regions. (C) Overlapping pSMAD1/5 peaks for genes bound by pSMAD1/5 in both HFSC and TAC chromatin. (D) Unbiased pSMAD1/5 binding motif analysis. (E) Binding pattern of pSMAD1/5 on the Id1 locus and description of C/G(CAG)G/C mutations introduced. (F) Luciferase reporter assays show decreased BMP sensitivity of mutated Id1 C/G(CAG)G/C enhancer regions. (G) Gene Ontology (GO) analyses of pSMAD1/5 ChIP-seq targets (common or unique) for HFSCs and TACs. (H) Examples of HFSC signature genes bound by pSMAD1/5 and active in HFSCs, but silenced in TACs (I) Examples of TAC signature genes bound by pSMAD1/5 and active in TACs, but silenced in HFSCs. (J) Transcription Factor Binding Site analysis of pSMAD1/5 ChIP targets unique for either HFSCs or TACs reveal enriched binding motifs for putative lineage regulators.
An unbiased de novo-binding motif search of each ChIP data set revealed a (C/G)CAG(G/C) motif (Figure 3D), which was similar, but not identical to, SMAD1/5 motifs reported in various cell types. A previously identified CAGA motif (Dennler et al., 1998) was also enriched within pSMAD1/5 peaks of HFSCs and TACs (Figure S3E). Validating the efficacy of our analyses, we showed that a short Id1 enhancer fragment containing multiple (C/G)CAG(G/C) motifs was effective at inducing Luciferase reporter activity in keratinocytes exposed to BMP-4, and when mutated, reporter activity was diminished (Figures 3E and 3F).
Gene ontology (GO) analysis of pSMAD1/5 targets shared by HFSCs and TACs highlighted transcription factors and transcriptional regulation (Figure 3G and Table S2). Reflective of their stemness and niche environment, HFSC-specific pSMAD1/5 targets showcased genes involved in embryonic tissue development and control of cell migration. By contrast, TAC-specific targets featured both negative and positive regulators of gene expression and metabolism, more reflective of their dynamic but committed state.
A role for BMP signaling in regulating HFSC behavior was further strengthened by the 189 (29%) of telogen HFSC signature genes that were bound by pSMAD1/5 in HFSCs. Even though their expression was downregulated upon TAC specification, many of these genes were still bound by pSMAD1/5 in TACs (p=0.0005), indicating that pSMAD1/5 does not act alone in its governance along the lineage (Figure S3F). That said, consistent with the hierarchical relation between HFSCs and TACs, pSMAD1/5-bound TAC signature genes (26%) overlapped primarily with matrix-unique targets (p=0.0002) (Figure 3A and Table S1).
To gain further insights into how pSMAD1/5-bound genes might be regulated, we next analyzed the chromatin status of pSMAD1/5-bound HFSC and TAC signature genes as they transitioned through the lineage (Lien et al., 2011). Although only a small percentage of HFSC and TAC signature genes were dually marked by H3K4me3:H3K27me3 (poised) chromatin modifications, many of them bound pSMAD1/5 (Figure S3G). Although poised marks may in part reflect heterogeneity within our FACS-purified populations, their increase among pSMAD1/5-bound genes suggested possible relevance of BMP signaling in transitional chromatin states (Bernstein et al., 2006)
Of the 141 HFSC signature genes bound by pSMAD1/5 in both HFSCs and TACs, only two were marked by H3K27me3 alone in HFSCs, while 87% were marked by H3K4me3 ± H3K79me2 (Figure S3H). In TACs, pSMAD1/5-bound HFSC signature genes were downregulated and some acquired H3K27me3 marks. GO analysis revealed that many of the HFSC pSMAD1/5 targets whose expression persisted in TACs showed predicted functions in macromolecular biosynthesis and epithelial development, i.e. sustained throughout the lineage (Figure S3J). By contrast, pSMAD1/5-bound genes active in quiescent HFSCs but silenced in TACs included Tcf7l1, Sox9, Tbx1, and other genes known to regulate chromatin dynamics or key signaling pathways controlling HFSC stemness (Figure 3H). These findings underscore the importance of BMP signaling in general, and pSMAD1/5-bound genes in particular, in governing features of quiescent HFSCs.
Most pSMAD1/5-bound TAC signature genes were in an open chromatin state in TACs but lacked active chromatin marks in HFSCs (Figure S3I). Importantly, this cohort included genes encoding transcription factors LEF1, KLF14, MSX1 and RUNX2 (Figure 3I), which were previously reported to influence hair progenitor cell specification/differentiation (Andl et al., 2004; Hertveldt et al., 2008; Kratochwil et al., 1996). Hence, in both HF stem and progenitor cells, pSMAD1/5 preferentially bound to genes encoding key transcriptional regulators of their identities, even though additional factors appeared to be required to change the status of these genes.
SMADs have been suggested to colocalize with lineage regulators to direct SMAD target specificity. pSMAD1 ChIP targets of TACs were enriched for motifs for HOX, SOX, FOX and bHLH proteins, known to function in early HF differentiation (Figure 3J). pSMAD1 targets of HFSCs were enriched for binding sites of POU and glucocorticoid receptor (NR3C1), previously implicated in HFSC homeostasis. This was suggestive of some lineage state specificity in pSMAD1-bound genes, despite the high degree of overlap.
Loss of BMP Signaling in HFSCs Changes the pSMAD1-Sensitive Transcriptome Towards ORS, HG and TAC Lineages
Previously, 426 probe sets to HFSC signature genes were tested for their sensitivity to BMP signaling: 96 mRNAs were identified, all but 16 of which were downregulated in bulge HFSCs/HG upon Bmpr1a ablation (Kandyba et al., 2013). Based upon our ChIP-seq data of bulge HFSC chromatin, only 29 of these directly bind pSMAD1/5. To gain deeper mechanistic insights into the direct consequences of loss of BMP signaling on HFSCs and their lineages, we therefore performed and qPCR-validated RNA profiling and Illumina sequencing (RNA-Seq) on purified populations of control and Bmpr1a-null HFSCs, and as discussed in the next section, TACs (Figure S4).
By testing 22,000 genes and focusing on purified bulge HFSCs, we expanded the cohort of BMP-sensitive HFSC transcripts. In all, 1938 transcripts were changed by ≥2X in Bmpr1a-null HFSCs relative to their normal counterparts (Figure 4A and Table S3). ~30% of the transcripts upregulated in Bmpr1a-null HFSCs belonged to the WT ORS signature of genes ≥2X upregulated relative to matrix or epidermal progenitors [Figure 4A; (p=0.026)]. Significant overlap was also seen between the signatures of Bmpr1a-null HFSCs and those of WT TACs and HG (Figures 4A and S4C). These data indicate that BMP signaling affects and promotes not only the activated HFSC (HG) signature as hinted previously (Kandyba et al., 2013), but also the signatures of ORS and TAC progenitors.
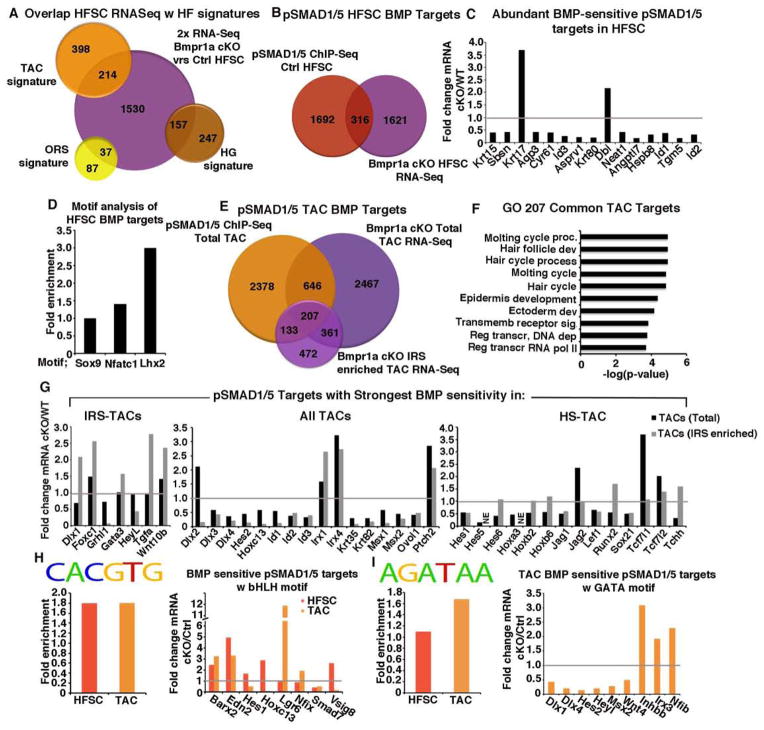
Figure 4. Identification of BMP-sensitive pSMAD1/5-bound targets.
(A) Venn diagram showing overlap between ORS, HG and TAC signatures and the mRNAs ≥2X changed in Bmpr1a cKO versus Ctrl HFSCs. (B) Venn diagram showing overlap between pSMAD1/5-bound genes and mRNAs ≥2X changed in HFSCs. (C) Relative mRNA expression of abundant BMP-sensitive pSMAD1/5 targets in cKO/WT. (D) Motif analysis of HFSC BMP-sensitive pSMAD1/5 targets based on previously published TF motifs (see Supplementary Methods) (E) Venn diagram showing the same analysis as in (B) but for TACs comparing to RNA profiling from IRS enriched or total TACs. (F) Unbiased GO analysis of 207 BMP-sensing pSMAD1/5 targets in TACs show enrichment of HF genes. (G) Differential expression of pSMAD1/5-bound, BMP sensitive targets in either IRS-TACs, HS-TACs or total TACs. (H) Enrichment of bHLH motifs was found in both HFSC and TAC targets, whereas only TAC BMP targets were enriched for GATA motifs.
Gene Ontology (GO) analysis of BMP-sensitive HFSC mRNAs supported roles for BMP signaling in balancing proliferation/self-renewal and keratinocyte identity/differentiation. Thus, mRNAs upregulated in Bmpr1a-null HFSCs encoded many cell cycle and extracellular matrix (ECM) regulators, consistent with their hyperproliferation, while epidermal differentiation mRNAs were downregulated (Table S4). Intriguingly, loss of BMP signaling also elevated Sox9 and Inhbb expression, known to repress epidermal differentiation (Kadaja et al., 2014).
A comparison of our ChIP-Seq and RNA-Seq data revealed 316 HFSC genes that were bound by pSMAD1/5 and differentially expressed upon loss of BMP signaling. This cohort was roughly split in their sensitivity to Bmpr1a-ablation (Figures 4B and S4D), consistent with pSMAD1/5’s impact on both transcriptional activation and repression. GO analysis of these pSMAD1/5-bound, BMP-sensitive targets showed enrichment for ECM, cytoskeleton and developmental processes (Figure S4E). Among BMP-sensitive pSMAD1/5-bound targets that are normally highly transcribed were the upregulated ORS gene Krt17 and the downregulated quiescent HFSC gene Krt15 (Figure 4C). Id1, Id2 and Id3 genes were also among the top pSMAD1/5-bound genes that were highly transcribed in HFSCs, and markedly suppressed upon BMPR1A loss. Id genes are known to be highly sensitive to BMP signaling, and implicated in cell fate determination in some other SCs (Lasorella et al., 2014; Niola et al., 2012). Their function in the HF lineage has been largely unexplored.
Analysis of these 316 HFSC target genes for other transcription factor motifs revealed an appreciable enrichment (3X) for HOX motifs, which overlap with the binding motif for LHX2, a key HFSC transcription factor (Figure 4D) (Folgueras et al., 2013). Motifs for another key HFSC regulator, NFATc1, were also enriched but to a lesser extent (1.4X). These findings highlight the value in determining both the genes bound by pSMAD1/5 and the impact on their expression when BMP signaling is abrogated.
Identifying TAC pSMAD1/5 Target Genes Affected by BMP Signaling
To elucidate how pSMAD1/5-bound genes in TACs are affected by BMP signaling, we began by transcriptionally profiling WT and Bmpr1a-null TACs from both Shh (IRS-enriched) and K15-Cre lineage traced (total matrix) hair bulbs (Figures S4F-S4I). Comparisons of pSMAD1/5 ChIP-Seq targets from total TACs and mRNAs differentially expressed upon loss of BMP signaling in either total or IRS-enriched TACs, yielded 853 and 340 BMP-sensitive, pSMAD1/5-bound target genes, respectively (Figure 4E and Table S5).
By comparing our two RNA-seq datasets, we could designate BMP-sensitive TAC transcripts of the HS lineage as those highly enriched/changed in the total TAC pool over the IRS-TAC enriched pool. We could then refine our IRS-TAC dataset to exclude the HS-TAC mRNAs. The BMP-sensitive transcripts found in both IRS-TACs and HS-TACs constituted our list of BMP-sensitive transcripts common to TACs. We then performed GO analysis on pSMAD1/5-bound targets more prominently regulated by BMPs in IRS-TACs, HS-TACs or equivalently in both populations.
In striking contrast to HFSCs, “common” TAC targets were enriched for genes relevant to the general hair cycle and hair development/differentiation, and most were downregulated upon loss of BMPR1A (Figure 4F and Table S6). Underscoring the efficacy of our classifications, established IRS regulators were among the top pSMAD1/5 targets displaying more prominent BMP sensitivity in IRS-TACs, while our top pSMAD1/5-bound targets with greater sensitivity to BMPs in HS-TACs were enriched for HS-specific genes (Figure 4G).
In light of our earlier sequence motif search for putative factors that might guide pSMAD1/5 to TAC genes versus HFSC genes (Figure 3J), it was particularly notable that Hox and Sox genes were among the top HS-specific TAC targets and Gata3 was featured in IRS-specific TAC targets. Among top targets sensitive to BMPs in all TACs were bHLH genes encoding IDs, Dlx and Msx family of transcriptional regulators. Indeed, when the transcription factor scan was performed on only pSMAD1/5-bound TAC genes showing BMP-sensitivity, bHLH sites (CACGTG) were highly enriched (p=0.00003) (Figure 4H). Intriguingly, bHLH motifs were also enriched in HFSC genes bound by pSMAD1/5 and BMP-sensitive, although different bHLH genes were expressed by HFSCs and TACs. It is interesting to speculate that the BMP-sensitive changes in expression of bHLH family members may in part contribute to this complexity.
By contrast, GATA sites (AGATAA) were enriched preferentially in the pSMAD1/5-bound, BMP-sensitive IRS-TAC enriched genes (p=0.0002) (Figure 4I). Taken together, these data support our genetic evidence that GATA3 and pSMAD1/5 may coordinate lineage determination in the IRS, while pSMAD1/5 and bHLH proteins may function coordinately throughout the lineage. How epigenetic changes impact on these choices and/or further influence pSMAD1/5 occupancy will be interesting to explore in the future.
ID Proteins as Novel Mediators of HFSC Quiescence
Seeking mechanisms that might underlie the activation of proliferation in bulge and HG HFSCs when BMP signaling is blocked, we first focused on Wnts. Previously implicated Wnts (7a,7b,16) (Kandyba and Kobielak, 2014; Kandyba et al., 2013) were preferentially expressed by WT HG versus bulge, and were not markedly affected in Bmpr1a-null bulge HFSCs (Figure S5A). Additionally, even though our ChIP-seq data on WT bulge HFSCs revealed Wnts 7b, 11, 10a and 4 as direct pSMAD1/5 targets, their mRNAs were not appreciably affected in the Bmpr1a cKO bulge (Figures S5A and S5B). These findings supported the view that WNT signaling/BMP inhibition preferentially activates HG proliferation (Hsu et al., 2014).
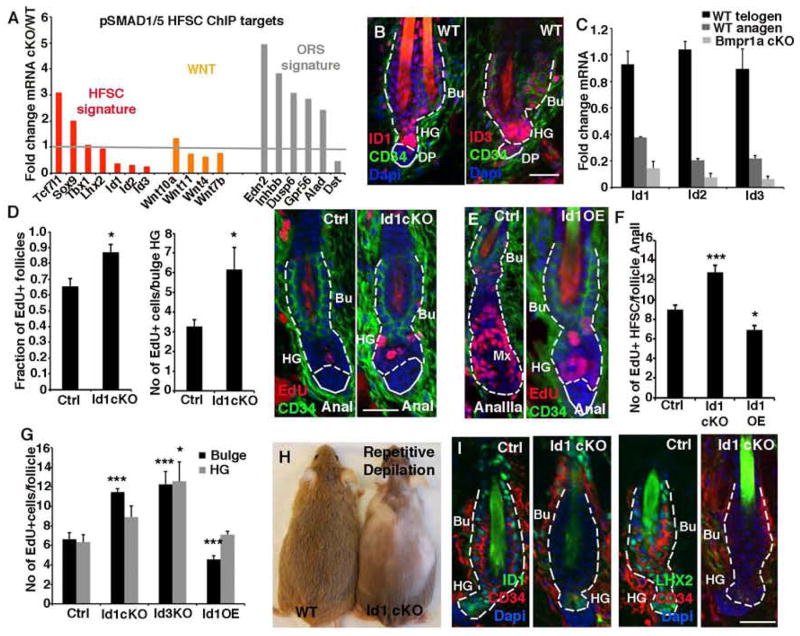
Probing further, we analyzed pSMAD1/5 ChIP targets categorized as either HFSC or ORS signature genes, since the ORS signature was upregulated while most HFSC signature genes were either unaltered or significantly downregulated (Figure 5A). From this list, we were drawn to the Ids since they have been implicated in repressing proliferation (Lasorella et al., 2014). Id1, Id2 and Id3 were each bound by pSMAD1/5 and downregulated upon Bmpr1a targeting in both HFSC and TACs (Figures (Figures 4G, 5A and S5C; Table S3). Anti-pSMAD1/5 ChIP-qPCR on cultured primary mouse keratinocytes (10MK) confirmed our in vivo ChIP-seq, showing a 10–40X enrichment of pSMAD1/5 binding to Id promoters compared to IgG controls (Figures S5C-D). Addition of BMP4 to 10MK for 3hrs resulted in a strong upregulation of Id1-Id3 mRNAs when compared to vehicle-treated cells, and consistent with the antagonistic effects of TGFβ signaling on BMP signaling (Oshimori and Fuchs, 2012), TgfβRII-null 10MK displayed increased Id1-Id3 mRNA expression (Figures S5E and S5F).
Figure 5. ID proteins as novel mediators of HFSC quiescence.
(A) pSMAD1/5 ChIP-seq targets ≥2X differentially expressed upon loss of Bmpr1A in HFSCs. Note that signature genes for quiescent HFSCs are downregulated while those for ORS cells are upregulated in Bmpr1a-null HFSCs. (B) ID1 and ID3 immunostaining in telogen HFs. (C) Verification of Id RNA profiling data. (D) Id1 ablation enhances bulge/HG proliferation at AnaI. (E) Overexpression (OE) of Id1 delays anagen progression. (F) Quantifications of EdU-incorporation in AnaII. (G) ID loss reduces and ID OE enhances proliferation in HFSCs 48 hrs after depilation to induce synchronized activation of hair cycling. (H–I) Id1 cKO mice subjected to repetitive depilation-induced hair cycles eventually fail to regrow hair coat (H) and lose HFSCs. (I). Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test or Anova. Scale bar = 25μm
In WT HFs, ID1, ID2 and ID3 were detected in quiescent (telogen-phase) HFSCs (Figures 5B and S5H). Intensity was strongest in HG, correlating with pSMAD1/5 immunostaining. Id mRNAs were downregulated at telogen→anagen, further correlating with pSMAD1/5 patterns (Figure 5C). qPCR confirmed that Id expression was lost in independently purified Bmpr1a cKO HFSCs.
To elucidate their physiological relevance, we generated K15-CrePGR:R26YFP:Id1fl/fl mice and compared them to Id3 straight null and to tetracycline (Tet)-inducible Id1-overexpressing animals. Targeting efficiency was confirmed at both mRNA and protein levels (Figures S5G and S5H). Although loss of Id1 or Id3 alone was not sufficient to drive HFSCs from telogen→anagen, AnaI Id1 cKO HG progenitors showed enhanced proliferation (Figure 5D). By contrast, when Id1 was overexpressed (OE) in WT AnaI HFs, 3d later they were still in AnaI while their WT counterparts had progressed to AnaIIIa (Figure 5E). Direct comparison of HFSC proliferation in Ana II HFs confirmed that IDs regulated their proliferation (Figure 5F). These findings were further corroborated in vitro with HFSCs purified from these mice (Figures S5I–S5L).
Depilation was used to induce anagen entry of HFs in their 2nd telogen. Within 48h, bulge and HG HFSCs of Id1 and Id3 targeted HFs displayed increased EdU+ labeling while Id1-OE caused significant reductions (Figures 5G, S5M–S5N). This inverse correlation between IDs and proliferation became more pronounced during repetitive rounds of depilation-induced hair cycling, until eventually Id1 cKO animals failed to regrow their hair coat (Figure 5H). Concomitantly, the numbers of cells expressing HFSC markers waned (Figure 5I). These results suggested that ID proteins function to maintain the HFSC pool by restricting their proliferation and wasteful usage.
Roles of BMPs and IDs in Balancing IRS and HS Lineages
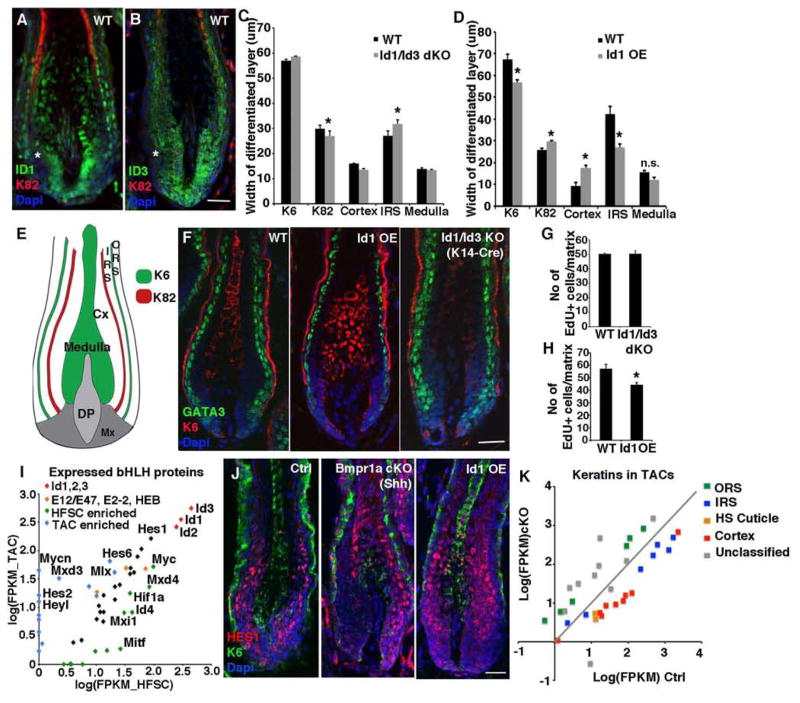
To further explore the importance of Ids as BMP-sensitive pSMAD1/5 targets in HFs, we generated K14Cre-Id1/Id3 double knockout mice (Figure S6A) and compared their anagen HFs to those from Id1-OE HFs. All three IDs were expressed by both IRS and HS lineages (Figures 6A and 6B; S6A and S6B). Intriguingly, however, immunoreactivity was reduced in the matrix pocket where Shh+ IRS-TACs typically reside. The higher ID immunoreactivity in presumptive HS-TACs was also consistent with our earlier findings on pSMAD1/5 levels (Figure 1F).
Figure 6. BMP suppress IRS-progenitors through IDs.
(A and B) ID1 and ID3 immunostaining in TACs and differentiating IRS and cortical progeny. (C and D) Loss of IDs alters the relative contribution of TACs to their differentiated progeny within the HF. (E) Schematic illustration of the differentiated layers of mature HFs. (F) GATA3+ cells in HFs are diminished by Id1 OE and expanded upon Id1/Id3 ablation. (G and H) Quantifications of proliferating (EdU+) TACs in Id1/Id3 dKO HFs (J) or Id1 OE HFs. (I) Expression of bHLH proteins in HFSCs vs TACs show enrichment of HES/HEY proteins in TACs. (J) HES1+ TACs are reduced upon ID loss (Bmpr1a cKO) and expanded with ID OE. (K) Bmpr1a ablation in the total TAC pool leads to their failure to properly express differentiation-specific keratin genes of both IRS and cortical lineages (see also Table S7). Data are represented as mean±SEM. * = p<0.05, Students t-test or Anova. Scale bar = 25μm.
TAC contribution to the various lineages was assessed using lineage-specific K82 and K6 immunolabeling (Figures 6C–6E). Id1/Id3 ablation caused a slight but significant reduction in overall HS width, with correspondingly increased IRS. Conversely, Id1-OE led to enhanced HS and reduced IRS (Figures 6F; S6C–6F). Since the TAC progenitors of these lineages had been targeted, these data best fit a model whereby BMP signaling acts through pSMAD1/5 and Id gene induction to promote cortical progenitors and restrict IRS progenitors. In agreement, Gata3 ablation, which enhanced the cortical TAC pool, also accentuated the pool of TACs expressing ID1 (Figure S6G). Neither GATA3 nor ID loss affected TAC proliferation to the extent seen with BMPR1A loss (Figures 6G and 6H), favoring a role for these downstream targets in lineage choice.
Since bHLH motifs featured prominently in both HFSC and TAC BMP targets, we analyzed their expression in these populations. Strikingly, TACs preferentially expressed Hes/Hey genes, many of which bound pSMAD1/5 and displayed particularly potent BMP sensitivity in HS-TACs (Figures 6I and 4G). HES1 protein also correlated with ID1 levels in Bmpr1a cKO and ID1-OE TACs (Figure 6J).
Finally, even though we uncovered a preference for BMP and ID signaling in specifying HS-TACs, pSMAD1/5 and IDs were clearly present in differentiating IRS cells, and Bmpr1a-null cysts contained GATA3+ IRS progenitors but few AE15+ cells. Exploiting the distinct keratin gene expression patterns in these lineages (Langbein et al., 2010), we analyzed BMP-sensitive patterns of change in 34 keratin mRNAs expressed by TACs in both our RNA profiling datasets. Nearly all of the HS and IRS keratins were downregulated in K15-derived TACs, whereas keratins expressed by the ORS lineage were upregulated (Figure 6K). These findings underscore the impairment of terminal differentiation programs of both lineages in Bmpr1a-null HFs. By contrast, in the Shh-derived TACs, where the HS TACs were lost and the IRS TACs were expanded, the corresponding elevation in IRS genes appeared to be confined to those of the early IRS TAC lineage and not the IRS keratins typical of terminal differentiation (Figure S6H and Table 7). While further studies will be needed, these data are consistent with the view that loss of BMP signaling in the hair lineage acts early, affecting its TACs, while loss of BMP signaling in the IRS lineage acts favorably on the IRS-TACs but acts later in the terminal differentiation of this lineage.
DISCUSSION
Our findings revealed that when BMP signaling is abrogated, quiescent HFSCs proliferate. That said, Bmpr1a-null HFSCs still exhibited a two-tier mechanism of activation, as reflected by the initial sensitivity of the HG in activating Wnt genes (Figure S5A) and rapidly expanding. Additionally and importantly, however, we learned that reduced BMP signaling is not simply a means of activating HFSCs, but rather a prerequisite for generating the GATA3-expressing IRS TACs. Moreover, when Bmpr1a or pSMAD1/5’s downstream effector Id genes were specifically targeted for ablation in TACs, the GATA3+ IRS progenitor pool was expanded and when Gata3 was conditionally ablated in TACs, IRS progenitors were not maintained.
By lineage tracing, we learned that when Bmpr1a is targeted in the Shh-expressing pocket, the few signs of cortical and medulla lineages that arise from this subset of matrix cells are eliminated, while the IRS lineage expands. The functional reliance of HS-TACs on BMP signaling was further supported by the fact that TACs within the WT mid-upper hair bulb just above the Shh-expressing pocket give rise to HS lineages, and these were positive for both pSMAD1/5 and IDs. Given their close proximity to DP, we were at first puzzled by this finding, since BMP inhibitory cues expressed by telogen-phase DP are integral to activating the new hair cycle (Botchkarev et al., 1999; Greco et al., 2009). However, in contrast to the IRS lineage, the HS lineage does not appear until mid-anagen, concomitant with this population of pSMAD1/5, ID+ TACs.
Our ChIP-seq and RNA-seq data added further insight, revealing that pSMAD1/5-bound genes that were upregulated upon BMPR1A loss encode proteins which typify the ORS, HG and IRS-TACs. Consistent with this finding was the abundance of these cells within the K15-CrePGR lineage-traced Bmpr1a-null cysts. Conversely, pSMAD1/5-bound genes encoding factors typical of quiescent HFSCs or HS-TACs tended to be downregulated by BMP signaling loss. These features bolster our conclusions drawn from immunofluorescence and genetic analyses about the differential effects of BMP signaling on these progenitor pools.
The Id genes featured prominently among the cohort of pSMAD1/5 targets that were common between HFSC and TACs and whose expression was profoundly influenced by reductions in BMP signaling. In other cell types, ID loss is often associated with an exit from the cell cycle and differentiation (Lasorella et al., 2014), but similar to B-cell development (Kee et al., 2001) expression of BMPs and ID proteins inversely correlated with HFSC proliferation. Despite its attractive features, however, the paradigm could not account for the fate skew we observed in TACs lacking either BMPR1A or ID1 and ID3. Thus Id1/Id3 dKO HFs were diminished in HS and enhanced in IRS lineages, while Id1 overexpression gave the opposite phenotype. These findings again point to the differences in ways in which not only BMPs, but also their downstream targets, differentially affect progenitors and their lineages within the matrix.
Recent evidence suggests that cell type-specific master regulators may guide the specificity of SMAD binding to their target genes (Trompouki et al., 2011). Exploiting our in vivo pSMAD1/5 ChIP-Seq analyses from both HFSCs and TACs, we identified several master regulatory transcription factors that may differentially direct the occupancy of lineage-specific pSMAD1/5 binding in HFs. We showed genetically that ID proteins promote the HS lineage, and it was both interesting and likely relevant that genes encoding ID-interacting bHLH proteins, particularly of the HES/HEY family (Bai et al., 2007), surfaced among pSMAD1/5-bound HS-TAC genes with higher sensitivity to BMP signaling. When coupled with the established role of Notch signaling in HS differentiation (Pan et al., 2004), our findings suggest a tantalizing crossroads of BMP and Notch signaling through Ids and Hes/Hey genes in orchestrating the early commitment steps of this lineage (Moya et al., 2012). Adding to the intrigue was the enrichment of HOX motifs within the pSMAD1/5 binding regions of the most highly BMP-sensitive HS-TAC genes. This class of transcription factors have been previously found to co-occupy several SMAD-bound target genes in cultured cell lines (Arden, 2004; Walsh and Carroll, 2007) and functionally, they have been shown to be key mediators of HS differentiation (Kulessa et al., 2000).
On the flip-side of TAC specification, we discovered that ablation of Gata3 in Shh/Wnt-expressing TACs resulted in a loss of the IRS progenitor subset and expansion of the LEF1, ID+ TAC subset that forms the HS. Enrichment of GATA sites in IRS-TAC BMP targets as well as pSMAD1/5 occupancy of GATA3 further enhances its prominence in the IRS lineage. That said, we found that many IRS-TAC genes, including Gata3, that bind pSMAD1/5 and are highly sensitive to BMPs are not downregulated when BMP signaling is abrogated. The strong presence of GATA3 IRS-TACs in Bmpr1a-null cysts provided physiological support for this finding. It also unveiled a newfound complexity, since positive regulation of Gata3 must rely upon some additional factor(s), which remains undetermined.
So where does this leave WNTs? While synergistic effects of WNT and BMP signaling have been described, our studies here extend the evidence that in HFs, these pathways are largely antagonistic. Thus, as in the HG, IRS-TACs displayed high Wnt10b and WNT reporter activity. That said, while BMP signaling was also low in this pocket, its lack of symmetry leaves open the possibility that the role of this BMP-low/WNT/SHH-high pocket is for signaling, perhaps to the DP, which in turn could indirectly impact the status of TAC lineage determination.
In HS-TACs, pSMAD1/5 levels were higher and Wnt expression and WNT reporter activity were low. Interestingly, WNT reporter activities were reversed in the differentiated cells of these lineages, as was the intensity of pSMAD1/5 immunolabeling. Finally, pSMAD1/5 targets were not enriched for LEF1/TCF binding motifs, suggesting that in contrast to the hematopoietic system (Trompouki et al., 2011), BMP and WNT signaling regulate temporally and spatially distinct, although interdependent, transcriptional programs.
LEF1 is known to accumulate concomitant with BMP downregulation and WNT upregulation both in developing and postnatal HGs (Botchkarev et al., 1999; Greco et al., 2009; Jamora et al., 2003). Hence it was not surprising to see expansion of LEF1+Shh+ WNT-reporter+ HG and unspecified TAC cells in the cysts that formed soon after Bmpr1a-ablation in HFSCs. However, we also showed that both Lef1 and Wnt genes (see also (Kandyba et al., 2013) are direct pSMAD1/5 targets, and upon BMP loss, Wnt transcripts are elevated while Lef1 transcripts are diminished. These results highlight the paradoxical need for HG cells to start with sufficiently high BMP signaling to allow for sufficient Lef1 transcription, but then diminish BMP signaling to activate Wnt transcription to generate nuclear-LEF1/β-catenin and the unspecified TAC pool.
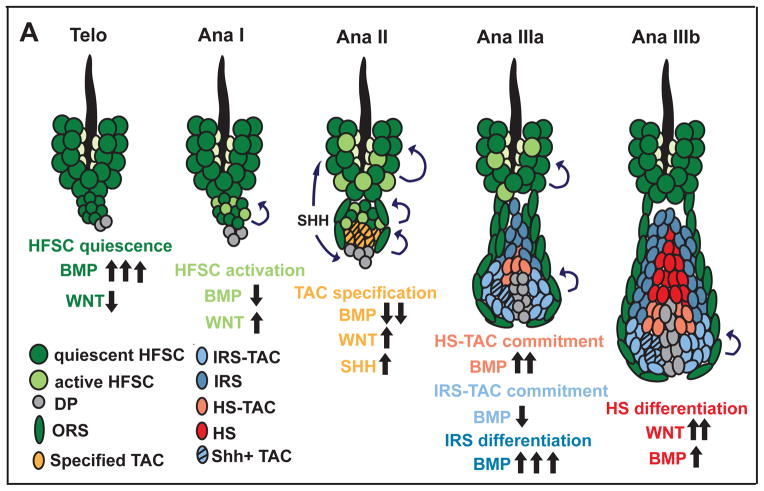
Our findings further revealed that the paradigm repeats itself to progress to the next specification step of the lineage. Reduced BMP signaling and Wnt transcription were features of the Shh+ pocket cells enriched for GATA3+ IRS-TACs, which also accumulated in K15-Cre generated Bmpr1a null cysts. By contrast, HS-TACs displayed higher BMP signaling as evidenced by pSMAD1/5/IDs, and also Lef1 transcripts (diffuse LEF1 protein), and a paucity of Wnt transcripts. Finally to terminally differentiate, each of these specified TACs displayed signs of repeating the paradigm again: differentiating HS cells displayed reduced BMP signaling, stabilized nuclear LEF1/βcatenin protein and activation of WNT reporters. The model in Figure 7 summarizes these results.
Figure 7. Model for how BMP signaling affects the HFSC lineage.
HFSC quiescence is maintained in a BMP high/WNT low environment (Telo). Activation of stem cells in the HG requires combined BMP inhibition and WNT upregulation (AnaI). This leads to the emergence of the TAC pool which expresses SHH, and stimulates bulge HFSCs to self-renew and sustain ORS growth. SHH also stimulates DP to elevate BMP inhibitors and promote TAC expansion. In AnaIIIa, BMP signaling remains relatively low in the lower matrix, which commits to form GATA3+ Shh+IRS-TACs; we posit that SHH signaling in this pocket preferentially affects the adjacent (lower) DP, resulting in higher BMP signaling in the upper-matrix specifying ID+ HS-TACs. Terminally differentiating IRS upregulates BMP signaling; the differentiating cortex is in a complex milieu of DP, IRS and melanocytes, where they receive reduced BMP and elevated WNT signaling. In the differentiating medulla, there are no signs of WNT or BMP signaling, indicating that the two pathways are not always inversely coupled. However, the failure of this lineage to form in the absence of BMPR1A is consistent with a role for BMP signaling in forming their TACs.
Overall, our results on Bmpr1a, Gata3 and Id1/3 loss of function suggest that the intricate balance of HS and IRS TACs arises from temporal and local differences in BMP signaling during the growth phase of the hair cycle. Too much or too little of this pathway perturbs this balance, and depending upon where in the lineage, different fates arise. A final twist to understanding how oscillations in BMP signaling can impact multiple discrete steps in the lineage emerges not only from the intersections between BMP-sensitive pSMAD1/5 target genes encoding ligands for WNTs (Wnts) and Notch (Jaggeds) signaling, but also for downstream effectors of these (Lef1, Hes/Hey, Tcf7l1, Tcf7l2) and other pathways, including Shh (Ptch2) and tyrosine kinases (Tgfα). When coupled with the lineage specific changes in microenvironment of different cell populations along the lineage, the cell stage-specific diversity in pSMAD1/5 targets, and the multiplicity of signaling and transcription factors directly controlled by BMP signaling begins to shed light on how these complex choices can be made by so few signaling pathways. As future functional and chromatin remodeling studies are conducted, remaining mysteries of the complicated lineage specifications of the HF should continue to unfold.
EXPERIMENTAL PROCEDURES
Mice
K14-H2BGFP and K14-Cre mice were generated in the Fuchs lab. Bmprafl/fl;K15-CrePGR;R26YFP have been described (Kandyba et al., 2013). Bmprafl/fl or Gata3fl/fl (Pai et al., 2003) mice were crossed to Shh-CreER and R26YFP reporter mice (JAX). CMV-rtTA:Tre-Id1, Id3 KO and Id1fl/fl mice were a generous gift from the Benezra lab. CD1 mice were from Charles River. Mice were maintained in an AAALAC-approved animal facility and procedures were performed with IACUC-approved protocols.
ChIP-Seq
DNA prepared for ChIP-Seq analysis was prepared as previously described (Lien et al., 2011) using an antibody against pSMAD1/5/8 (Cell Signaling).
RNA Purification and Profiling
For HFSCs, we used Krt15-CrePGR to drive targeting and Rosa26-lox-stop-lox-YFP to aid in purification. Bmpr1afl/fl and Bmpr1afl/+ mice were induced in 2nd telogen and 14d later, SCA1negYFP+CD34+α6high HFSCs were isolated by FACS (Figures S4A–B). For TACs, we used Shh-CreER to drive targeting, Rosa26-lox-stop-lox-YFP to aid in purification and the depilation and induction scheme delineated in Figure 2A to activate the hair cycle synchronously and mark the pocket of Shh-expressing TACs. TACs and early progeny were purified by FACs based upon SCA1negYFP+CD34negα6low expression (Figure S4F). CreER was activated by topical application (1% in EtOH) of tamoxifen. CrePGR was activated using both topical (4% in EtOH) and intraperitoneal injections (1% in corn oil) of RU486. EdU was delivered intraperitoneally (50ug/g)(Invitrogen) and chased for times specified. Total RNAs was purified form FACS-isolated cells sorted into TrizolLS (Invitrogen) using RNeasy Mini Kit (Qiagen) or Direct-Zol RNA Miniprep (Zymo Research). Equal amounts of RNA were reverse-transcribed using VILO (Invitrogen). cDNAs were normalized using primers against HPRT and/or Ppib2. Samples collected for RNA profiling was submitted for Illumina Sequencing.
Luciferase Reporter Assay
336 bp of the Id1 enhancer was amplified from genomic DNA and subcloned into pGL3_Luciferase (Promega) backbone containing a minimal CMV promoter (mCMV). Point mutations were introduced using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). Primary mouse keratinocytes were transfected with the generated pGL3 constructs as well as pGL4 Renilla (Promega). After 24 hours, cells were serum starved for 12 hours, treated with BMP4 for 3 hours and analyzed using a DualGlo Kit (Promega).
Histology and Immunofluorescence
Back skins were embedded in OCT, frozen and cryo-sectioned at 10–14um and fixed for 10 minutes in 4% paraformaldehyde in PBS. Sections were blocked and permeabilized for 1 hour at room temperature in PBS with 0.1% Triton-X, 2.5% normal donkey serum, 2.5% normal goat serum and 0.5% BSA. LacZ staining on OCT sections were performed using standard Xgal protocols. Images were acquired with an Axio Oberver.Z1 epifluorescence microscope equipped with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics), and with an ApoTome.2 (Carl Zeiss) slider that reduces the light scatter in the fluorescent samples, using a Plan-Apochromat 20x/0.8 objective, controlled by Zen software (Carl Zeiss).
Statistics
Data was analyzed and statistics performed (unpaired two-tailed Student’s t-test or Anova) in Prism5 (GraphPad). Significant differences between two groups were noted with asterisks.
Supplementary Material
Supplemental Figure 1 (related to Figure 1) BMP signaling is temporally regulated and required to maintain matrix
(A) Relative quantification of Smad1,5 and 8 by qRT-PCR in both HFSCs and TACs (B) Early cysts are composed predominantly of K17+ cells, whereas only a hand full of cells express the differentiation markers such as AE15. Furthermore, the outer layer of cyst cells are positive for TAC markers LEF1, LHX2 and P-CAD where as the core cells express high levels of SOX9. Early cysts express high levels of the WNT reporter Axin2-LacZ and Shh mRNA. (C–E) Fully developed cysts maintain LHX2 and SHH expression and proliferate readily (EdU) at the epithelial-mesenchymal interface. Differentiation markers K6 and AE15 can be detected in a restricted number of cells, whereas AE13 is absent. (F) Hair shafts formed in the absence of BMPR1a are thinner and lack signs of medulla. (G) Bmpr1a cKO mice grow progressively bald compared to wild type mice. (H) Illustrative images delineating the TAC populations used for quantification of relative pSMAD1/5 intensity in Figure 1F. Scale bars = 25 um.
Supplemental Figure 2 (related to Figure 2) BMP restricts the SHH-expressing IRS progenitor pool
(A) Loss of BMP signaling leads to a unilateral expansion of TAC on the cKO side. (B) Quantification of EdU incorporation of WT, Bmpr1a +/fl and Bmpr1a fl/fl on the Shh-CreER;Rosa26YFP background reveal no effect on proliferation on the YFP- side of the Bmpr1a cKO hair follicle, thus ruling out any paracrine effects from local loss of Bmpr1a. (C and D) The number of proliferating EdU+ cells that also express differentiation markers (AE13 or AE15) is very low and is not significantly altered in the absence of BMP signaling. (E and F) mRNA levels of both Shh and Wnt10b are increased in Shh-Bmpr1a cKO TA progenitors when compared to wild type TAC, correlating to an expansion of the Wnt10b-expressing domain. (G) WNT targets genes are upregulated in the absence of BMP signaling. (H) Analysis of differentiation pattern of K15-Bmpr1a cKO in first anagen after recombination reveals an altered pattern of differentiation with predominantly GATA3+ TAC differentiating into AE15+ IRS at the expense of AE13+ cortex and K6+ medulla. (I) Confirmation of Gata3 targeting in the Shh-CreER mouse. (J) Quantification of the level of Gata3 mRNA in purified YFP+ TACs in control and Gata3 cKO mice. (K) Loss of IRS progenitors in the Gata3 cKO correlates to a reduction in Shh expression and domain (L) Loss of Gata3 results in an expansion of cortical AE13+ and LEF1+ cortical lineages at the expense of AE15+ IRS lineage on the targeted side of the HF. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test or Anova. Scale bar = 25 um
Supplemental Figure 3 (related to Figure 3) pSMAD1/5 regulates transcriptional networks in both HFSCs and TACs
(A) FACS purification strategy and verification of matrix TACs used for ChIP-Seq. Cells were purified using the K14-H2BGFP transgenic mouse line. TACs are GFP low, SCA1 neg, Itga6 low and the identity of the population was verified using TAC and HFSC markers. (B) Common ChIP targets (found in both HFSCs and TACs) are localized within the same genomic region (78% within 1398 bp). (C) The quality of HFSC and TAC ChIP is comparable as judged by either the normalized density reads of all peaks or (D) the fraction of genes bound to non-tissue specific genes. (E) Sequence analysis of CAGA motifs in pSMAD1/5 peaks reveal enrichment of CAG(A/C/G) sequence. (F) pSMAD1/5 targets (either cell type specific or common) that are also HFSC or TAC signature genes analyzed based on expression levels. (G) Global analysis of all HFSC and TAC targets histone modification pattern compared to genome wide distribution (Lien et al., 2011) show enrichment of poised (H3K4me3: H3K27me3) targets in both HF stem and TA progenitor cells. (H) Analysis of histone modification of HFSC signature genes occupied by pSMAD1/5 in both HFSC and TA cells. Most targets bear marks of active transcription (H3K4me3:H3K79me2 or H3K4me3 alone) in telogen, correlating to their high level of expression, but a large fraction of these genes loose their active mark and acquired H3K27me3 in TA progenitors. (I) In comparison H3K4me3:H3K79me2 marks in TAC signature and pSMAD1/5 target genes is reduced in telogen HFSCs. (J) Expression of genes grouped as pSMAD1/5 targets and HFSC signature genes in both HFSC and TAC show that HFSC signature genes are expressed at higher levels in HFSC than TACs. GO analysis of the same group of genes.
Supplemental Figure 4 (related to Figure 4) Identification of bona fide BMP targets
The use of compartment specific Cre lines allows for temporal and spatial regulation of loss of Bmpr1a. (A) K15-CrePGR is active in the CD34+ telogen bulge and the CD34− hair germ, as based on Rosa26YFP reporter activity. YFP+ Bmpr1a +/fl and fl/fl HFSC were purified based on the markers YFP+, SCA1 Neg, CD34+ and Itga6 High. (B) Verification of HFSC RNA-Seq using independently purified K15-Bmpr1a cKO and control HFSCs. (C) Genes differentially expressed in Bmpr1a cKO HFSC and overlapping with signature genes (HG, TAC or ORS) are mostly upregulated in the absence of BMP signaling indicating a preference for ORS, HG and TAC fates. (D) The 316 overlapping BMP sensitive pSMAD1/5 HFSC targets are spit in their sensitivity to BMP signaling. (E) GO analysis of 316 HFSC BMP targets. (F) Shh-CreEr targets a small population of TACs cells correlating to the unilateral expression of endogenous Shh mRNA. SHH+ TACs were purified based on their expression of YFP+, CD34 Neg and Itga6 Low. (G) Verification of Shh+ TAC RNA-Seq using independently purified Shh-Bmpr1a cKO and control TA progenitors.(H) Verification of K15-derived total pool of TAC RNA-Seq. (I) TAC BMP targets are both up and downregulated in the absence of BMP signaling. Scale bar = 25 um
Supplemental Figure 5 (related to Figure 5) ID proteins as novel mediators of HFSC quiescence
(A) Real time qPcr analysis of Wnt16, Wnt7a and Wnt7b from FACS purified K15-Bmpr1a control or cKO HFSC and hair germ. (B) RNA profiling data of published WNT target genes (Lien et al., 2014) in control and Bmpr1a cKO HFSC. (C) pSMAD1/5 binding to Id1, Id2 and Id3 compared to input. Data shown is from wild type HFSC and TAC. (D) Verification of ChIP-Seq targets using ChIP-Pcr in mouse keratinocytes show preferential pSMAD1/5 binding to Ids when compared to IgG. (E) Transient stimulation of keratinocytes with BMP-4 results in up regulation of Id1, Id2 and Id3 mRNAs as compared to control cells. (F) Fold change mRNA in TgfBRII KO mouse keratinocytes compared to wild type. (G) mRNA levels in wild type, K15-Id1 cKO or Id3 KO HFSC. (H) Loss of ID1 and ID3 expression in cKO and KO respectively. ID2 is detected in both HFSC and HG. Overexpression of ID1 results in increased protein levels compared to wild type. (I) Id1 mRNA levels are increased after ID1 overexpression. (J–L) Quantification of colony formation efficiency or colony size of Id1 cKO, Id3 KO or Id1 OE HFSC compared to wild type. (M) Representative image of the number of EdU+ bulge stem cells or hair germ cells 48 hours after depilation of Id1 cKO, Id3 KO or Id1 OE HFSC compared to wild type. (N) ID1 and EdU immunoreactivity in wild type and Id1 OE HFs 48 hours after depilation. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test or Anova. Scale bar = 25 um.
Supplemental Figure 6 (related to Figure 6) BMP suppress IRS-progenitors through ID1 and ID3
(A) Knock out or overexpression confirmation of ID1 and ID3 by immunofluorescence. ID2 is expressed similarly to ID1 and ID3 with the highest expression level in centrally positioned cortical TACs. (B) ID1 is highly expressed by cortical TACs and by differentiating GATA3+ IRS cells during the hair follicle maturation. (C) Loss of ID1 and ID3 results in increased width of the AE15+ IRS lineage, whereas overexpression of ID1 leads to a reduction in IRS. (D) Analysis of ID1 expression in the Shh-Bmpr1a cKO show a specific loss of ID1 correlating to the Shh–expressing pocket and in the forming IRS and cortex on the ablated side of the HF. (E) Analysis of Shh expression in Id1/Id3 cKO (F) and Id1 overexpressing TACs. (G) Loss of GATA3 results in expansion of ID1 expression in comparison to control follicles. (H) Differential expression of keratins in Shh+ TACs in Bmpr1a cKO and WT show preferential loss of cortical keratins whereas IRS specific keratins are largely unaltered. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01 Students t-test or Anova. Scale bar = 25um.
Supplemental Table S1 (related to Figure 3) pSMAD1/5 targets in HFSCs and TACs
Complete list of pSMAD1/5 targets in HFSCs and TACs, as well as pSMAD1/5 targets overlapping with either HFSC or TAC signatures.
GO analysis of genes occupied by pSMAD1/5 in HFSCs, TACs or subdivided into targets unique or common for each population.
RNA-Seq analysis of HFSCs and TACs after loss of BMP signaling using either K15-CrePGR or Shh-CreER to abrogate Bmpr1a.
GO analysis of mRNAs 2x changed after loss of BMP signaling in HFSCs.
List of identified BMP targets in HFSCs and TACs, as well as targets unique or common for IRS- or HS-TACs.
GO analysis of common or IRS/HS-specific TAC targets.
RNA-Seq analysis of keratin expression in the total (IRS+HS) and IRS-enriched TAC populations.
Acknowledgments
We thank RU Genomics Resource Center for Illumina sequencing, S. Mazel, L. Li, S. Semova and S. Tadesse for FACS (RU FACS facility) and the MSKCC Genomics Core Facility for RNA-Sequencing. We thank Fuchs’ lab members: D. Oristian, L. Polak for assistance in mouse research, H.A. Pasolli and C. Kaufman for ultrastructural data, W.-H Lien, M. Kadaja, C.P. Lu for valuable discussions. M.G. was supported by Damon Runyon Cancer Research Foundation and the Wenner-Gren Foundations. E.F. is an HHMI Investigator. This work was supported by grants to E.F. (NIAMS R01-AR31737).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Arden KC. FoxO: linking new signaling pathways. Mol Cell. 2004;14:416–418. doi: 10.1016/s1097-2765(04)00213-8. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, Lindner G, McMahon JA, Peters C, Lauster R, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. Embo J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Hertveldt V, Louryan S, van Reeth T, Dreze P, van Vooren P, Szpirer J, Szpirer C. The development of several organs and appendages is impaired in mice lacking Sp6. Dev Dyn. 2008;237:883–892. doi: 10.1002/dvdy.21355. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Li L, Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaja M, Keyes BE, Lin M, Pasolli HA, Genander M, Polak L, Stokes N, Zheng D, Fuchs E. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 2014;28:328–341. doi: 10.1101/gad.233247.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Hazen VM, Kobielak A, Butler SJ, Kobielak K. Smad1 and 5 but not Smad8 establish stem cell quiescence which is critical to transform the premature hair follicle during morphogenesis toward the postnatal state. Stem Cells. 2014;32:534–547. doi: 10.1002/stem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Kobielak K. Wnt7b is an important intrinsic regulator of hair follicle stem cell homeostasis and hair follicle cycling. Stem Cells. 2014;32:886–901. doi: 10.1002/stem.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Leung Y, Chen YB, Widelitz R, Chuong CM, Kobielak K. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci U S A. 2013;110:1351–1356. doi: 10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat Immunol. 2001;2:242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- Kobielak K, Pasolli HA, Alonso L, Polak L, Fuchs E. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J Cell Biol. 2003;163:609–623. doi: 10.1083/jcb.200309042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci U S A. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Turk G, Hogan BL. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. Embo J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbein L, Yoshida H, Praetzel-Wunder S, Parry DA, Schweizer J. The keratins of the human beard hair medulla: the riddle in the middle. J Invest Dermatol. 2010;130:55–73. doi: 10.1038/jid.2009.192. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Kwan K, Li AG, Wang XJ, Wurst W, Behringer RR. Essential roles of BMPR-IA signaling in differentiation and growth of hair follicles and in skin tumorigenesis. Genesis. 2004;39:10–25. doi: 10.1002/gene.20021. [DOI] [PubMed] [Google Scholar]
- Moya IM, Umans L, Maas E, Pereira PN, Beets K, Francis A, Sents W, Robertson EJ, Mummery CL, Huylebroeck D, et al. Stalk cell phenotype depends on integration of Notch and Smad1/5 signaling cascades. Dev Cell. 2012;22:501–514. doi: 10.1016/j.devcel.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M, Nam HS, Zhuang Y, Benezra R, Di Bernardo D, et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. Paracrine TGF-beta signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, Kopan R. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Paus R, Cotsarelis G. The biology of hair follicles. N Engl J Med. 1999;341:491–497. doi: 10.1056/NEJM199908123410706. [DOI] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira I, Nicolas JF. Redefining the structure of the hair follicle by 3D clonal analysis. Development. 2012;139:3741–3751. doi: 10.1242/dev.081091. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CM, Carroll SB. Collaboration between Smads and a Hox protein in target gene repression. Development. 2007;134:3585–3592. doi: 10.1242/dev.009522. [DOI] [PubMed] [Google Scholar]
- Woo WM, Zhen HH, Oro AE. Shh maintains dermal papilla identity and hair morphogenesis via a Noggin-Shh regulatory loop. Genes Dev. 2012;26:1235–1246. doi: 10.1101/gad.187401.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Yuhki M, Yamada M, Kawano M, Iwasato T, Itohara S, Yoshida H, Ogawa M, Mishina Y. BMPR1A signaling is necessary for hair follicle cycling and hair shaft differentiation in mice. Development. 2004;131:1825–1833. doi: 10.1242/dev.01079. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 (related to Figure 1) BMP signaling is temporally regulated and required to maintain matrix
(A) Relative quantification of Smad1,5 and 8 by qRT-PCR in both HFSCs and TACs (B) Early cysts are composed predominantly of K17+ cells, whereas only a hand full of cells express the differentiation markers such as AE15. Furthermore, the outer layer of cyst cells are positive for TAC markers LEF1, LHX2 and P-CAD where as the core cells express high levels of SOX9. Early cysts express high levels of the WNT reporter Axin2-LacZ and Shh mRNA. (C–E) Fully developed cysts maintain LHX2 and SHH expression and proliferate readily (EdU) at the epithelial-mesenchymal interface. Differentiation markers K6 and AE15 can be detected in a restricted number of cells, whereas AE13 is absent. (F) Hair shafts formed in the absence of BMPR1a are thinner and lack signs of medulla. (G) Bmpr1a cKO mice grow progressively bald compared to wild type mice. (H) Illustrative images delineating the TAC populations used for quantification of relative pSMAD1/5 intensity in Figure 1F. Scale bars = 25 um.
Supplemental Figure 2 (related to Figure 2) BMP restricts the SHH-expressing IRS progenitor pool
(A) Loss of BMP signaling leads to a unilateral expansion of TAC on the cKO side. (B) Quantification of EdU incorporation of WT, Bmpr1a +/fl and Bmpr1a fl/fl on the Shh-CreER;Rosa26YFP background reveal no effect on proliferation on the YFP- side of the Bmpr1a cKO hair follicle, thus ruling out any paracrine effects from local loss of Bmpr1a. (C and D) The number of proliferating EdU+ cells that also express differentiation markers (AE13 or AE15) is very low and is not significantly altered in the absence of BMP signaling. (E and F) mRNA levels of both Shh and Wnt10b are increased in Shh-Bmpr1a cKO TA progenitors when compared to wild type TAC, correlating to an expansion of the Wnt10b-expressing domain. (G) WNT targets genes are upregulated in the absence of BMP signaling. (H) Analysis of differentiation pattern of K15-Bmpr1a cKO in first anagen after recombination reveals an altered pattern of differentiation with predominantly GATA3+ TAC differentiating into AE15+ IRS at the expense of AE13+ cortex and K6+ medulla. (I) Confirmation of Gata3 targeting in the Shh-CreER mouse. (J) Quantification of the level of Gata3 mRNA in purified YFP+ TACs in control and Gata3 cKO mice. (K) Loss of IRS progenitors in the Gata3 cKO correlates to a reduction in Shh expression and domain (L) Loss of Gata3 results in an expansion of cortical AE13+ and LEF1+ cortical lineages at the expense of AE15+ IRS lineage on the targeted side of the HF. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test or Anova. Scale bar = 25 um
Supplemental Figure 3 (related to Figure 3) pSMAD1/5 regulates transcriptional networks in both HFSCs and TACs
(A) FACS purification strategy and verification of matrix TACs used for ChIP-Seq. Cells were purified using the K14-H2BGFP transgenic mouse line. TACs are GFP low, SCA1 neg, Itga6 low and the identity of the population was verified using TAC and HFSC markers. (B) Common ChIP targets (found in both HFSCs and TACs) are localized within the same genomic region (78% within 1398 bp). (C) The quality of HFSC and TAC ChIP is comparable as judged by either the normalized density reads of all peaks or (D) the fraction of genes bound to non-tissue specific genes. (E) Sequence analysis of CAGA motifs in pSMAD1/5 peaks reveal enrichment of CAG(A/C/G) sequence. (F) pSMAD1/5 targets (either cell type specific or common) that are also HFSC or TAC signature genes analyzed based on expression levels. (G) Global analysis of all HFSC and TAC targets histone modification pattern compared to genome wide distribution (Lien et al., 2011) show enrichment of poised (H3K4me3: H3K27me3) targets in both HF stem and TA progenitor cells. (H) Analysis of histone modification of HFSC signature genes occupied by pSMAD1/5 in both HFSC and TA cells. Most targets bear marks of active transcription (H3K4me3:H3K79me2 or H3K4me3 alone) in telogen, correlating to their high level of expression, but a large fraction of these genes loose their active mark and acquired H3K27me3 in TA progenitors. (I) In comparison H3K4me3:H3K79me2 marks in TAC signature and pSMAD1/5 target genes is reduced in telogen HFSCs. (J) Expression of genes grouped as pSMAD1/5 targets and HFSC signature genes in both HFSC and TAC show that HFSC signature genes are expressed at higher levels in HFSC than TACs. GO analysis of the same group of genes.
Supplemental Figure 4 (related to Figure 4) Identification of bona fide BMP targets
The use of compartment specific Cre lines allows for temporal and spatial regulation of loss of Bmpr1a. (A) K15-CrePGR is active in the CD34+ telogen bulge and the CD34− hair germ, as based on Rosa26YFP reporter activity. YFP+ Bmpr1a +/fl and fl/fl HFSC were purified based on the markers YFP+, SCA1 Neg, CD34+ and Itga6 High. (B) Verification of HFSC RNA-Seq using independently purified K15-Bmpr1a cKO and control HFSCs. (C) Genes differentially expressed in Bmpr1a cKO HFSC and overlapping with signature genes (HG, TAC or ORS) are mostly upregulated in the absence of BMP signaling indicating a preference for ORS, HG and TAC fates. (D) The 316 overlapping BMP sensitive pSMAD1/5 HFSC targets are spit in their sensitivity to BMP signaling. (E) GO analysis of 316 HFSC BMP targets. (F) Shh-CreEr targets a small population of TACs cells correlating to the unilateral expression of endogenous Shh mRNA. SHH+ TACs were purified based on their expression of YFP+, CD34 Neg and Itga6 Low. (G) Verification of Shh+ TAC RNA-Seq using independently purified Shh-Bmpr1a cKO and control TA progenitors.(H) Verification of K15-derived total pool of TAC RNA-Seq. (I) TAC BMP targets are both up and downregulated in the absence of BMP signaling. Scale bar = 25 um
Supplemental Figure 5 (related to Figure 5) ID proteins as novel mediators of HFSC quiescence
(A) Real time qPcr analysis of Wnt16, Wnt7a and Wnt7b from FACS purified K15-Bmpr1a control or cKO HFSC and hair germ. (B) RNA profiling data of published WNT target genes (Lien et al., 2014) in control and Bmpr1a cKO HFSC. (C) pSMAD1/5 binding to Id1, Id2 and Id3 compared to input. Data shown is from wild type HFSC and TAC. (D) Verification of ChIP-Seq targets using ChIP-Pcr in mouse keratinocytes show preferential pSMAD1/5 binding to Ids when compared to IgG. (E) Transient stimulation of keratinocytes with BMP-4 results in up regulation of Id1, Id2 and Id3 mRNAs as compared to control cells. (F) Fold change mRNA in TgfBRII KO mouse keratinocytes compared to wild type. (G) mRNA levels in wild type, K15-Id1 cKO or Id3 KO HFSC. (H) Loss of ID1 and ID3 expression in cKO and KO respectively. ID2 is detected in both HFSC and HG. Overexpression of ID1 results in increased protein levels compared to wild type. (I) Id1 mRNA levels are increased after ID1 overexpression. (J–L) Quantification of colony formation efficiency or colony size of Id1 cKO, Id3 KO or Id1 OE HFSC compared to wild type. (M) Representative image of the number of EdU+ bulge stem cells or hair germ cells 48 hours after depilation of Id1 cKO, Id3 KO or Id1 OE HFSC compared to wild type. (N) ID1 and EdU immunoreactivity in wild type and Id1 OE HFs 48 hours after depilation. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01, *** = p<0.001, Students t-test or Anova. Scale bar = 25 um.
Supplemental Figure 6 (related to Figure 6) BMP suppress IRS-progenitors through ID1 and ID3
(A) Knock out or overexpression confirmation of ID1 and ID3 by immunofluorescence. ID2 is expressed similarly to ID1 and ID3 with the highest expression level in centrally positioned cortical TACs. (B) ID1 is highly expressed by cortical TACs and by differentiating GATA3+ IRS cells during the hair follicle maturation. (C) Loss of ID1 and ID3 results in increased width of the AE15+ IRS lineage, whereas overexpression of ID1 leads to a reduction in IRS. (D) Analysis of ID1 expression in the Shh-Bmpr1a cKO show a specific loss of ID1 correlating to the Shh–expressing pocket and in the forming IRS and cortex on the ablated side of the HF. (E) Analysis of Shh expression in Id1/Id3 cKO (F) and Id1 overexpressing TACs. (G) Loss of GATA3 results in expansion of ID1 expression in comparison to control follicles. (H) Differential expression of keratins in Shh+ TACs in Bmpr1a cKO and WT show preferential loss of cortical keratins whereas IRS specific keratins are largely unaltered. Data are represented as mean±SEM. * = p<0.05, ** = p<0.01 Students t-test or Anova. Scale bar = 25um.
Supplemental Table S1 (related to Figure 3) pSMAD1/5 targets in HFSCs and TACs
Complete list of pSMAD1/5 targets in HFSCs and TACs, as well as pSMAD1/5 targets overlapping with either HFSC or TAC signatures.
GO analysis of genes occupied by pSMAD1/5 in HFSCs, TACs or subdivided into targets unique or common for each population.
RNA-Seq analysis of HFSCs and TACs after loss of BMP signaling using either K15-CrePGR or Shh-CreER to abrogate Bmpr1a.
GO analysis of mRNAs 2x changed after loss of BMP signaling in HFSCs.
List of identified BMP targets in HFSCs and TACs, as well as targets unique or common for IRS- or HS-TACs.
GO analysis of common or IRS/HS-specific TAC targets.
RNA-Seq analysis of keratin expression in the total (IRS+HS) and IRS-enriched TAC populations.