Abstract
Mitochondria are small organelles that produce the majority of cellular energy as ATP. Mitochondrial dysfunction has been implicated in the pathogenesis of Parkinson's disease (PD), and rare familial forms of PD provide valuable insight into the pathogenic mechanism underlying mitochondrial impairment, even though the majority of PD cases are sporadic. The regulation of mitochondria is crucial for the maintenance of energy-demanding neuronal functions in the brain. Mitochondrial biogenesis and mitophagic degradation are the major regulatory pathways that preserve optimal mitochondrial content, structure and function. In this mini-review, we provide an overview of the mitochondrial quality control mechanisms, emphasizing regulatory molecules in mitophagy and biogenesis that specifically interact with the protein products of three major recessive familial PD genes, PINK1, Parkin and DJ-1.
Keywords: PD genes, PINK1, Parkin, DJ-1, Mitophagy, Biogenesis
INTRODUCTION
In Parkinson's disease (PD), mitochondrial impairment contributes to the major pathological mechanisms underlying the selective loss of dopaminergic neurons in the substantial nigra (SN) [1]. Mitochondrial homeostasis is crucial for energy production and neuronal survival under stress conditions and is strictly regulated by mitochondrial biogenesis and mitophagy (i.e. mitochondria-selective autophagy) [2]. Mitochondrial impairments have been reported in various samples obtained from human patients with sporadic PD and a complex I deficit has been observed in SN autopsies and the peripheral tissues of patients [3, 4, 5, 6]. In familial PD cases, recessive mutations in PINK1, parkin and DJ-1 have been linked to mitochondrial abnormalities in patients and PD models. For example, mitochondrial impairments are observed in patients carrying PINK1 mutations [7] and PINK1-null mice [8], PINK1-null fruit flies [9, 10] and a PINK1-null dopaminergic neuron model [11]. Parkin-null mice and fruit flies have decreased mitochondrial respiration in the striatum [12] and defective mitochondrial morphology [13], respectively. The accumulation of defective mitochondria is significantly higher in both PINK1-null and DJ-1-null dopaminergic neuron cells [11]; however no significant mitochondrial defects have been previously identified in DJ-1-null mice [14, 15]. These major recessive familial PD genes appear to play important regulatory roles in mitochondrial biogenesis and mitophagy via the direct interactions of their protein products with mitochondrial homeostasis regulators (Table 1). Here we overview the regulatory molecules in mitophagy and biogenesis that specifically interact with the PINK1, Parkin and DJ-1 proteins. Therefore, further insights into the pathogenic mechanism underlying mitochondrial impairment mediated by recessive PD genes may potentially identify neuroprotective drug target(s) to develop mitochondrial therapeutics and alleviate mitochondrial deficits in familial PD.
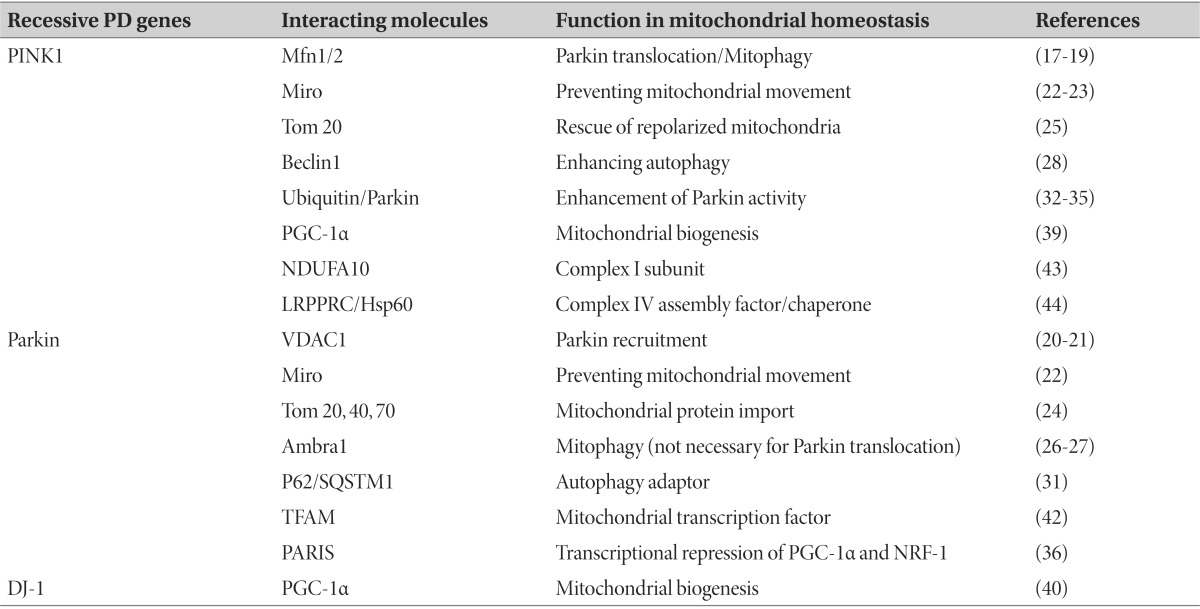
Table 1.
Molecules of mitochondrial homeostasis regulated by recessive PD genes
MOLECULAR INTERACTIONS REGULATING MITOPHAGY
Damaged mitochondria accumulate with age and the accumulation of impaired mitochondria has been implicated in the pathogenesis of PD [1, 2]. Previously, recessive mutations in parkin, an ubiquitin E3 ligase, were identified as a cause of early onset familial PD. Furthermore, Parkin is selectively recruited to damaged mitochondria with low membrane potential in mammalian cells and following recruitment, Parkin mediates the selective elimination of these impaired mitochondria [16]. This finding revealed a link between mitochondrial quality control and the mutated proteins in familial PD, and suggests that a failure to remove impaired mitochondria contributes to PD pathogenesis.
Mitofusin1 and 2 (Mfn1 and Mfn2)
The identity of a factor directing Parkin translocation to damaged mitochondria during mitophagy remained unknown until a phosphorylated form of mitofusin2 (Mfn2) is identified as the receptor for Parkin on damaged cardiac mitochondria [17]. Specifically, Parkin binds to Mfn2, and PINK1 subsequently, phosphorylates Mfn2, which promotes Parkin-mediated ubiquitination. Accordingly, in Mfn2-null cardiac myocytes, the mitochondrial translocation of Parkin and mitophagy are suppressed and functionally impaired mitochondria accumulate in Mfn2-null mouse embryonic fibroblasts (MEFs) and cardiomyocytes [17]. However, previous work employing MEFs from Mfn1/Mfn2 double knockout mice demonstrated normal Parkin translocation to a proton uncoupler carbonyl cyanide p-chlorophenylhydrazone (CCCP)-treated mitochondria [16], and in Drosophila the loss of PINK1 or Parkin increases the level of the profusion factor Mfn. Furthermore, in Drosophila, PINK1 and Parkin mediate the ubiquitination of Mfn1/2 on the outer surface of mitochondria [18, 19], suggesting that Mfn1/2 ubiquitination and/or degradation may provide a mechanism to label terminally damaged mitochondria for mitophagy. In addition, these findings suggest that a tissue-specific mechanism mediates mitophagy.
VDAC1
Voltage-dependent anion channel 1 (VDAC1), a pore-forming protein in the outer mitochondrial membrane, is a target protein of Parkin-mediated Lys 27 poly-ubiquitylation and subsequent mitophagy [20]. Proteomic analyses have shown that that Parkin specifically interacts with VDAC 1, 2, and 3, and that this interaction is disrupted by CCCP treatment [21]. In addition, impaired Parkin translocation to defective mitochondria and mitophagy occurs in the absence of all three VDACs and each VDAC is sufficient to support Parkin recruitment and mitophagy, suggesting that VDACs are functionally redundant [21]. These findings suggest that VDACs serve as mitochondrial docking sites to recruit Parkin to defective mitochondria.
Miro
PINK1 and Parkin regulate mitochondrial distribution and clearance by arresting mitochondrial movement via Miro, a component of the primary motor/adaptor complex that anchors kinesin to the mitochondrial surface [22]. First, to arrest the mitochondria, PINK1 phosphorylates Miro and phosphorylated Miro activates the proteasomal degradation of Miro in a Parkin-dependent manner. Also, the removal of Miro from the mitochondrion detaches kinesin from its surface. Thus, prior to mitochondrial clearance, PINK1 and Parkin may quarantine damaged mitochondria by preventing mitochondrial movement [22]. This observation was further confirmed by proteomics and cellular approaches, demonstrating that upon Parkin translocation to the mitochondria, the ubiquitin-proteasome system (UPS) becomes activated to promote the widespread degradation of outer membrane proteins including Miro 1 and 2 [23], suggesting the importance of remodeling the mitochondrial outer membrane proteome for mitophagy.
Translocase of the outer membrane (TOM) complex
Parkin mediates the proteasome-dependent degradation of outer membrane proteins such as Tom20, Tom40 and Tom70 in depolarized mitochondria [24]. In contrast, the inner membrane and matrix proteins are primarily degraded via mitophagy. In addition, Parkin induces the rupture of the outer membrane of depolarized mitochondria, which is also dependent on proteasomal degradation [24]. Mitophagy is unnecessary for proteasome-dependent degradation of outer membrane proteins as well as outer membrane rupture, suggesting the differential regulatory role of Parkin in the degradation of outer and inner mitochondrial membrane proteins through proteasome- and mitophagy-dependent pathways, respectively [24]. Of interest, PINK1 forms a 700 kDa multimeric complex with the TOM containing Tom70, Tom22 and Tom20 and Tom40, on depolarized mitochondria [25]. Especially, PINK1 directly interacts with Tom20, the main receptor for N-terminal presequences. Although Parkin did not stably interact with the 700 kDa PINK1/TOM complex, PINK1 mediates recruitment of Parkin to induce degradation of TOM complex. Furthermore, Parkin targeted to peroxisomes by PINK1 is sufficient to drive pexophagy, but not on lysosomes [25]. Thus, PINK1 binding to TOM complex may allow rapid reimport of PINK1 to rescue repolarized mitochondria by down-regulation of mitophagy via the PINK1/Parkin pathway [25].
Ambra1
The activating molecule in Beclin 1-regulated autophagy (Ambra1), a Beclin 1-interacting protein, is widely expressed in the adult mouse brain, including midbrain dopaminergic neurons. Furthermore, the endogenous pool of Ambra1 has been shown to be localized at the mitochondria [26]. Ambra1 is a Parkin-interacting protein [27], and mitochondrial depolarization significantly increases the interaction between Parkin and Ambra1; however, Ambra1 has not been found to be ubiquitinated by Parkin. Ambra1 activates class III PI3K during its recruitment to perinuclear clusters of depolarized mitochondria; however, Ambra1 is not necessary for Parkin translocation to depolarized mitochondria but is important for mitophagy [27].
Beclin1
Beclin1 (ATG6), a proautophagic protein, interacts with PINK1 [28]. Beclin1 deficiency also promotes pathogenicity in neurodegenerative diseases, which is supported by the reduced Beclin1 levels in the affected brain regions of patients with Alzheimer's and Huntington's diseases [29, 30], Furthermore, Beclin1 deficiency in cellular and animal models of Alzheimer's and Huntington's diseases promotes the disruption of autophagy, the accumulation of amyloid-β and mutant hungtintin and neuronal cell death [29, 30]. In PD models, PINK1-induced autophagy is also significantly reduced by Beclin1 gene knock down. Specifically, a PINK1(W437X) mutant that has an impaired interaction with Beclin1 cannot enhance autophagy; however a PINK1(G309D) mutant with defective kinase activity still interacts with Beclin1 [28].
p62/SQSTM1
After Parkin translocation to depolarized mitochondria, Parkin mediates ubiquitylation of mitochondria and their transport along microtubules to cluster in the perinuclear region. During Parkin-mediated mitochondrial ubiquitylation, the autophagic adaptor p62/SQSTM1 is recruited to mitochondrial clusters, which is necessary for mitochondrial degradation [20]. However, other studies demonstrated the dispensable role of p62/SQSTM1 in mitophagy. For example, the deletion of p62/SQSTM1 in MEF cells results in the gross loss of mitochondrial perinuclear clustering but does not inhibit mitochondrial clearance [31]. Similarly, mitochondrial-anchored ubiquitin is sufficient to recruit p62/SQSTM1 and promote mitochondrial clustering, but does not promote mitophagy [16]. Thus, p62/SQSTM1 is required for the ubiquitylation-dependent clustering of damaged mitochondria.
Ubiquitin and Parkin
Although the mechanism underlying PINK1-mediated activation of Parkin has remained elusive, Parkin Ser 65 phosphorylation by PINK1 was proposed to be involved in its activation of E3 ligase activity [32, 33]. However, PINK1 still recruits Parkin to mitochondria despite Ser65Ala mutation or individual mutation of all other Ser residues of Parkin, which suggests that another PINK1 substrate might mediate Parkin translocation and activation [34]. Subsequently, PINK1 is also identified as a kinase to phosphorylate ubiquitin (Ub) at Ser 65, a Ser residue that is homologous with the Ser 65 on the Parkin UBL domain [34, 35]. The phosphorylated Ub at Ser 65 acts as a novel Parkin activator. Moreover, PINK1-dependent phosphorylation of both Parkin and Ub is sufficient for full activation of Parkin E3 ligase activity [34, 35].
MOLECULAR INTERACTIONS REGULATING MITOCHONDRIAL BIOGENESIS
Human mitochondrial DNA (mtDNA) encodes 13 proteins that form electron transport chain (ETC) complexes, 22 tRNAs and two rRNAs. The majority of mitochondrial proteins are encoded by nuclear genes; therefore, mitochondrial biogenesis is a complex and sophisticated process, involving both the mitochondrial and nuclear regulation of gene expression. The regulation of mitochondrial biogenesis is achieved by the activation of various transcription factors, such as PGC-1α, TFAM, NRF1 and 2 and ERR-α, β and γ. Mitochondrial biogenesis is also regulated at the posttranscriptional level, involving the TOM complex. Here, we focus on these regulatory molecules of mitochondrial biogenesis, specifically the interactions among PINK1, Parkin and DJ-1.
PARIS
PARIS (a zinc-finger protein ZNF746) is a Parkin-interacting substrate, that accumulates in Parkin knockout models and in human PD patient brains. PARIS mediates the transcriptional repression of peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1α (PGC-1α) and the PGC-1α target gene, NRF-1, by binding to insulin response sequences in the PGC-1α promoter [36]. Moreover, a Parkin conditional knockout in adult mice leads to the progressive loss of dopaminergic neurons due to the accumulation of PARIS. Similarly, the overexpression of PARIS leads to the selective loss of dopaminergic neurons, which is reversed by PGC-1α coexpression [36].
PGC-1α
PGC-1α is characterized as the master regulator of mitochondrial biogenesis by acting as a transcriptional co-activator, that forms heteromeric complexes with transcription factors, such as NRF-1, NRF-2, PPARα, PPARδ, PPARγ, and ERRα [37]. The formation of these complexes results in the regulation of the expression of many nuclear-encoded mitochondrial genes, including the mitochondrial transcription factor A (TFAM) [38]. Of interest, a PGC-1α isoform localizes to the mitochondrial inner membrane and matrix, which colocalizes and interacts with VDAC and PINK1 in brain mitochondria [39]. Moreover, DJ-1 is reported to inhibit the SUMOylation of a transcriptional repressor, PSF, which binds to PGC-1α and suppresses its transcriptional activity [40]. Also, oxidative modification renders DJ-1 unable to inhibit SUMOylation, resulting in attenuated transcriptional synergy between DJ-1 and PGC-1α This attenuation leads to the gradual dysregulation of the SUMO pathway and may cause abnormal mitochondrial gene expression leading to the development of sporadic PD [40].
TFAM
TFAM is a key activator of mitochondrial transcription and is a participant in mitochondrial DNA replication [41]. Parkin overexpression enhances transcription and replication of mitochondrial DNA in proliferating cells, which is attenuated by Parkin gene knock down [42]. In addition, Parkin is associated with TFAM and enhances TFAM-mediated mitochondrial transcription, indicating that Parkin is involved in the regulation of mitochondrial transcription and replication in proliferating cells [42].
MOLECULAR INTERACTIONS REGULATING ELECTRON TRANSPORT CHAINS (ETCs)
Deficiencies in ETC enzymes are a common phenomenon observed in PINK1- and DJ-1-null cellular models. ETCs comprises complex I, II, II, IV and ATP synthase, consisting of approximately 85 subunits and requiring 41 assembly factors, suggesting that these recessive familial PD genes may play regulatory roles in ETC maintenance.
Mitochondrial complex I activity
PINK1-null cells and cells derived from patients with PINK1 mutations display a deficit of mitochondrial complex I activity. When analyzing the phosphoproteome of complex I in the livers and brains of PINK1-null mice, the specific loss of phosphorylation of serine-250 in the complex I subunit NdufA10 occurs [43]. Serine-250 phosphorylation of in NdufA10 is necessary for ubiquinone reduction via complex I activity, therefore, phosphomimetic NdufA10 reverses PINK1 deficits and rescues mitochondrial depolarization in PINK1-null mutant Drosophila. Similarly, complex I deficits are also rescued by phosphomimetic NdufA10 in cells derived from patients with PINK1 mutation [43].
Mitochondrial complex IV activity
In PINK1-null cells, mitochondrial complex IV activity and the levels of specific chaperones, including Hsp60, leucine-rich pentatricopeptide repeat-containing (LRPPRC), and Hsp90, are severely decreased [44]. Interestingly, Hsp60 knockdown results in a decrease in complex IV activity, whereas the antagonistic inhibition of Hsp90 decreases both Hsp60 and complex IV activity. In contrast, the overexpression of the PINK1-interacting factor, LRPPRC, augments complex IV activity by up-regulating Hsp60. A similar recovery of complex IV activity is also induced by the coexpression of Hsp90 and Hsp60, suggesting that PINK1 regulates complex IV activity via interactions with upstream regulators of Hsp60, such as LRPPRC and Hsp90 [44].
Implications for PD
Mitochondrial impairment is considered to be a major pathogenic mechanisms of PD; therefore, specific regulatory molecules involved in mitochondrial homeostasis may be potential drug target(s) for the therapeutic intervention of PD. However, no effective therapy targeting mitochondrial homeostasis is available. Promising drug targets include regulatory molecules that mediate mitochondrial biogenesis and mitophagic degradation, during mitochondrial turnover to protect mitochondrial content, healthy mitochondrial structure and optimal ETC function. In particular, the major recessive familial PD genes, such as PINK1, Parkin and DJ-1, are known to take part in the regulation of mitochondrial biogenesis, mitophagy, and ETC activity via interactions with mitochondrial homeostasis key regulators. Therefore, further insights into the molecular pathways underlying mitochondrial homeostasis involving these three recessive PD genes may help identify an effective drug target for mitochondrial therapeutics in familial PD.
ACKNOWLEDGMENTS
This research was supported in part by a grant (2013-008773) from NRF, the Republic of Korea.
References
- 1.Schapira AH. Mitochondrial dysfunction in Parkinson's disease. Cell Death Differ. 2007;14:1261–1266. doi: 10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- 2.Perier C, Vila M. Mitochondrial biology and Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a009332. doi: 10.1101/cshperspect.a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 4.Bindoff LA, Birch-Machin MA, Cartlidge NE, Parker WD, Jr, Turnbull DM. Respiratory chain abnormalities in skeletal muscle from patients with Parkinson's disease. J Neurol Sci. 1991;104:203–208. doi: 10.1016/0022-510x(91)90311-t. [DOI] [PubMed] [Google Scholar]
- 5.Yoshino H, Nakagawa-Hattori Y, Kondo T, Mizuno Y. Mitochondrial complex I and II activities of lymphocytes and platelets in Parkinson's disease. J Neural Transm Park Dis Dement Sect. 1992;4:27–34. doi: 10.1007/BF02257619. [DOI] [PubMed] [Google Scholar]
- 6.Winkler-Stuck K, Wiedemann FR, Wallesch CW, Kunz WS. Effect of coenzyme Q10 on the mitochondrial function of skin fibroblasts from Parkinson patients. J Neurol Sci. 2004;220:41–48. doi: 10.1016/j.jns.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Hoepken HH, Gispert S, Morales B, Wingerter O, Del Turco D, Mülsch A, Nussbaum RL, Müller K, Dröse S, Brandt U, Deller T, Wirth B, Kudin AP, Kunz WS, Auburger G. Mitochondrial dysfunction, peroxidation damage and changes in glutathione metabolism in PARK6. Neurobiol Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 10.Morais VA, Verstreken P, Roethig A, Smet J, Snellinx A, Vanbrabant M, Haddad D, Frezza C, Mandemakers W, Vogt-Weisenhorn D, Van Coster R, Wurst W, Scorrano L, De Strooper B. Parkinson's disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim JH, Yoon SH, Kim KH, Han JY, Ha JY, Hyun DH, Paek SH, Kang UJ, Zhuang X, Son JH. The antioxidant Trolox helps recovery from the familial Parkinson's disease-specific mitochondrial deficits caused by PINK1- and DJ-1-deficiency in dopaminergic neuronal cells. Mitochondrion. 2011;11:707–715. doi: 10.1016/j.mito.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 13.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg MS, Pisani A, Haburcak M, Vortherms TA, Kitada T, Costa C, Tong Y, Martella G, Tscherter A, Martins A, Bernardi G, Roth BL, Pothos EN, Calabresi P, Shen J. Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial Parkinsonism-linked gene DJ-1. Neuron. 2005;45:489–496. doi: 10.1016/j.neuron.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 15.Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, Pownall S, Wakeham A, You-Ten AJ, Kalia SK, Horne P, Westaway D, Lozano AM, Anisman H, Park DS, Mak TW. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A. 2005;102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziviani E, Whitworth AJ. How could Parkin-mediated ubiquitination of mitofusin promote mitophagy? Autophagy. 2010;6:660–662. doi: 10.4161/auto.6.5.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Vashisht AA, Tchieu J, Wohlschlegel JA, Dreier L. Voltage-dependent anion channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J Biol Chem. 2012;287:40652–40660. doi: 10.1074/jbc.M112.419721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, Torosantucci L, Cassina L, Russo MA, Dallapiccola B, Valente EM, Casari G. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 29.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 31.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, Tanaka K, Matsuda N. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 36.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, Batchu VV, Schubert M, Castellani RJ, Russell JW. A novel PGC-1alpha isoform in brain localizes to mitochondria and associates with PINK1 and VDAC. Biochem Biophys Res Commun. 2013;435:671–677. doi: 10.1016/j.bbrc.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong N, Xu J. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1alpha: regulation by SUMOylation and oxidation. Hum Mol Genet. 2008;17:3357–3367. doi: 10.1093/hmg/ddn230. [DOI] [PubMed] [Google Scholar]
- 41.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 42.Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 43.Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, Aerts L, Overbergh L, Grunewald A, Seibler P, Klein C, Gevaert K, Verstreken P, De Strooper B. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203–207. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 44.Kim KH, Song K, Yoon SH, Shehzad O, Kim YS, Son JH. Rescue of PINK1 protein null-specific mitochondrial complex IV deficits by ginsenoside Re activation of nitric oxide signaling. J Biol Chem. 2012;287:44109–44120. doi: 10.1074/jbc.M112.408146. [DOI] [PMC free article] [PubMed] [Google Scholar]


