Background: Plant flavonoid prenyltransferases (FPTs) transfer prenyl moiety to flavonoid cores and have previously been identified only in Leguminosae.
Results: The newly identified moraceous FPTs, MaIDT, and CtIDT, are distantly related to leguminous FPTs and feature catalytic regioselectivity and promiscuity.
Conclusion: MaIDT and CtIDT evolved independently from leguminous FPTs.
Significance: These findings are valuable for identifying additional evolutionarily different non-Leguminosae FPTs.
Keywords: Flavonoid, Gene Expression, Molecular Evolution, Phylogenetics, Plant Biochemistry, Cudrania tricuspidata, Morus alba, Flavonoid Prenyltransferase, Promiscuity, Regiospecificity
Abstract
Prenylated flavonoids are attractive specialized metabolites with a wide range of biological activities and are distributed in several plant families. The prenylation catalyzed by prenyltransferases represents a Friedel-Crafts alkylation of the flavonoid skeleton in the biosynthesis of natural prenylated flavonoids and contributes to the structural diversity and biological activities of these compounds. To date, all identified plant flavonoid prenyltransferases (FPTs) have been identified in Leguminosae. In the present study two new FPTs, Morus alba isoliquiritigenin 3′-dimethylallyltransferase (MaIDT) and Cudrania tricuspidata isoliquiritigenin 3′-dimethylallyltransferase (CtIDT), were identified from moraceous plants M. alba and C. tricuspidata, respectively. MaIDT and CtIDT shared low levels of homology with the leguminous FPTs. MaIDT and CtIDT are predicted to be membrane-bound proteins with predicted transit peptides, seven transmembrane regions, and conserved functional domains that are similar to other homogentisate prenyltransferases. Recombinant MaIDT and CtIDT were able to regioselectively introduce dimethylallyl diphosphate into the A ring of three flavonoids with different skeleton types (chalcones, isoflavones, and flavones). Phylogenetic analysis revealed that MaIDT and CtIDT are distantly related to their homologs in Leguminosae, which suggests that FPTs in Moraceae and Leguminosae might have evolved independently. MaIDT and CtIDT represent the first two non-Leguminosae FPTs to be identified in plants and could thus lead to the identification of additional evolutionarily varied FPTs in other non-Leguminosae plants and could elucidate the biosyntheses of prenylated flavonoids in various plants. Furthermore, MaIDT and CtIDT might be used for regiospecific prenylation of flavonoids to produce bioactive compounds for potential therapeutic applications due to their high efficiency and catalytic promiscuity.
Introduction
Prenylated flavonoids are an attractive class of plant-specialized metabolites that are primarily distributed in Leguminosae (Fabaceae), Moraceae, Umbelliferae, Guttiferae, Euphorbiaceae, Celastraceae, Compositae, Paulowniaceae, and Zingiberaceae (1–5). These compounds are hybrid molecules that contain structurally divergent flavonoid skeletons and extensively modified prenyl moieties with different chain lengths, and these compounds exhibit a wide range of biological activities, e.g. antioxidative, antitumor, antibacterial, antiviral, and estrogenic activities (1, 3). Notably, the substitution of the flavonoid ring system with prenyl group(s) contributes strongly to these biological activities because the prenyl group(s) increases the lipophilicity of the flavonoids and confers to these molecules a strong affinity to biological membranes that results in greater bioavailability (6).
Due to their diverse chemical structures and impressive biological activities, prenylated flavonoids have been studied in many fields including plant physiology, natural product chemistry, and synthetic chemistry. The prenylation reactions catalyzed by flavonoid prenyltransferases (FPTs)2 have received particular attention and significantly contribute to the structural diversity and further modification of the prenylated flavonoids (7). For example, prenylation is a key step in the biosynthesis of diverse Diels-Alder-type adducts in mulberry trees (8, 9). Consequently, intensive biochemical studies of FPTs have been performed for more than two decades (10, 11). However, the genes coding for these prenyltransferases (PTs) have not been isolated from plants until the recent identification of the first flavonoid-specific PT gene SfN8DT-1 from Sophora flavescens, which is responsible for the specific prenylation of the flavanone naringenin at C-8 (12). Subsequently, further progress was achieved in the molecular biological and biochemical investigations of plant FPTs, and additional relevant genes have been cloned and functionally characterized (Table 1). Moreover, due to their substrate promiscuity in vitro, some soluble types of aromatic PTs from microbes have been demonstrated to be capable of catalyzing the prenylation of flavonoids; e.g. 7-DMATS, a member of DMATS superfamily from Aspergillus fumigatus (18, 19), and some PTs of the CloQ/NphB group from the actinomycetes Streptomyces (20, 21).
TABLE 1.
Identified flavonoid prenyltransferases in plants
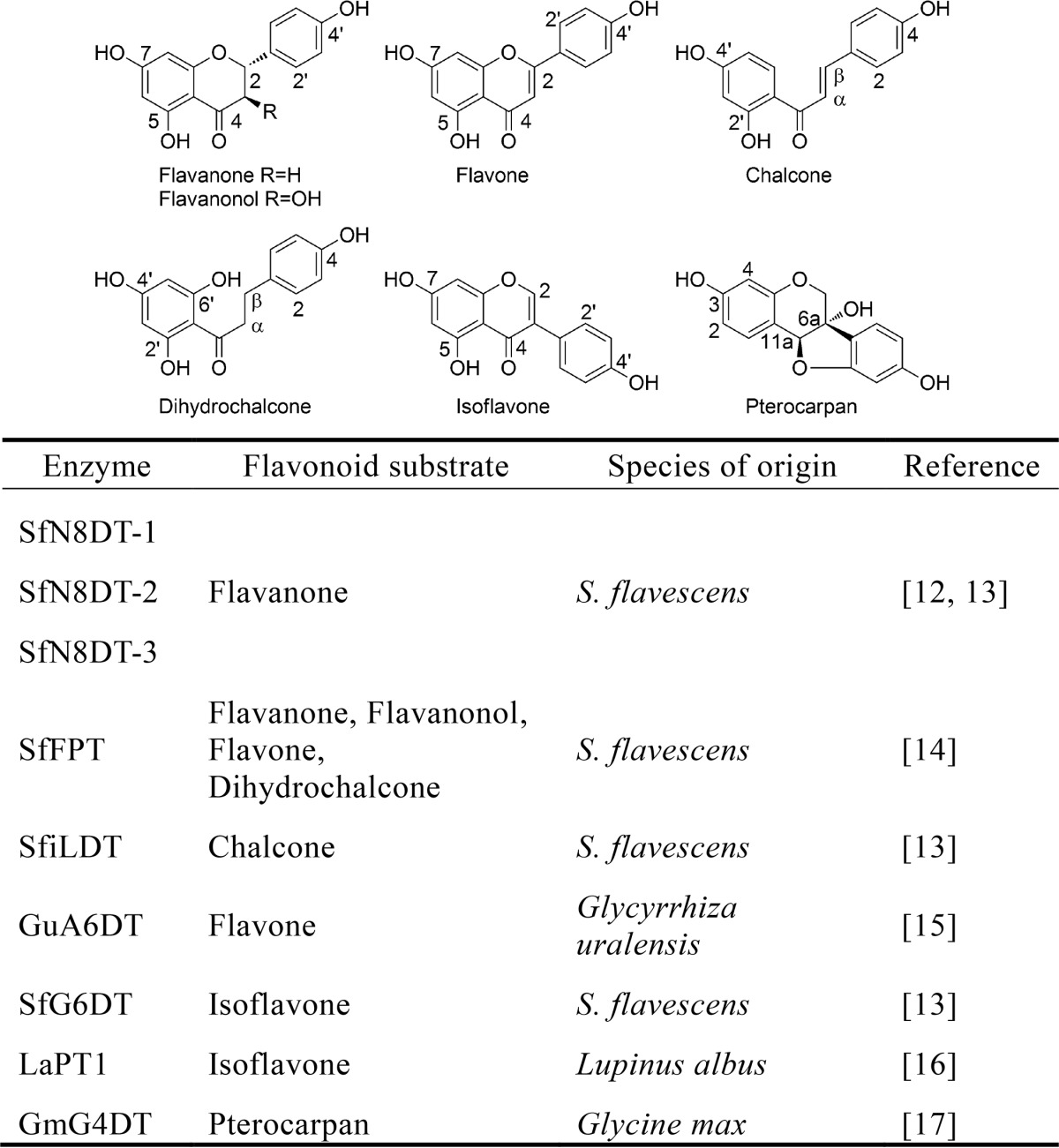
However, to date all of the functionally characterized plant FPTs have been cloned from Leguminosae (Table 1). The genes, phylogeny (in relation to the reported FPTs), and substrate spectra (specificity or promiscuity) of the non-Leguminosae FPTs remain enigmatic. These questions prompted us to search for such enzymes.
Moraceous plants are rich sources of isoprenoid-substituted phenolic compounds and their Diels-Alder-type adducts (22, 23). In our previous work cell suspension cultures of two closely related moraceous trees, Morus alba (mulberry) and Cudrania tricuspidata, were employed to prenylate different types of flavonoids, and the same bioconversions were observed when microsomes of the cell cultures were used (24). Further chemical investigations confirmed that the prenylated flavonoids formed the main chemical compositions of both of the two plant cell suspension cultures and that Diels-Alder-type adducts were present in large amounts in the cell suspension cultures of M. alba (25, 26). These observations indicated that FPTs existed in the cell suspension cultures of each of the two plants. Therefore, in our ongoing work on plant FPTs (14, 15), cell suspension cultures of M. alba and C. tricuspidata were selected as materials from which to isolate non-Leguminosae FPTs.
In this report we describe the characterization of two new FPTs, MaIDT and CtIDT, from the cell suspension cultures of M. alba and C. tricuspidata, respectively. As the first identified non-Leguminosae FPTs, these FPTs share fairly low identities with their homologs in Leguminosae and feature catalytic regioselectivity and promiscuity. These findings not only imply the diversity of FPTs in the plant kingdom but may also illuminate the discoveries of other FPTs in non-Leguminosae plants.
EXPERIMENTAL PROCEDURES
General Experimental Procedures
1H and 13C NMR spectra were recorded on Varian NMR System 600 spectrometers (Varian Inc., Palo Alto, CA). The HPLC-UV/ESI-MS analyses of the enzymatic products were performed as previously described (15) with the exception that the solvent system consisted of a linear gradient from 50% to 100% (v/v) methanol in water with 0.1% formic acid over a period of 30 min followed by an isocratic elution with 100% methanol for 10 min and that the UV detector was set at 350 or 265 nm. For the isolation of the enzymatic products, an approach similar to that previously described was employed (15) with the exception that the semi-preparative reverse-phase HPLC was performed using a linear gradient from 50% to 100% (v/v) methanol in water over a period of 20 min.
Plant Materials
Cell suspension cultures of M. alba were maintained in Murashige and Skoog's medium with 1.0 mg/liter α-naphthalene acetic acid, 0.5 mg/liter 6-benzylaminopurine, and 0.2 mg/liter 2,4-dichlorophenoxy acetic acid. Cell suspension cultures of C. tricuspidata were maintained in 6,7-V medium (58) supplemented with 0.5 mg/liter α-naphthalene acetic acid, 0.2 mg/liter 6-benzylaminopurine, and 0.1 mg/liter 2,4-dichlorophenoxy acetic acid as described previously (24). The cell cultures were subcultured every 15 days.
Chemicals
The prenyl donors dimethylallyl diphosphate (DMAPP), geranyl diphosphate (GPP), farnesyl diphosphate, geranylgeranyl diphosphate, and phytyl diphosphate were chemically synthesized as described previously (27). Chalcones 2-4 were prepared according to the literatures (28, 29). The other tested substrates were purchased from Sigma and BioBioPha (Kunming, Yunnan, China).
Isolation of cDNAs Homologous to AtVTE2–1 from M. alba
Ten-day-old M. alba cell cultures were treated with 0.1 mm methyl jasmonate for 20 h, and total RNA was subsequently extracted with an E.Z.N.A.TM Plant RNA kit (Omega Bio-tek Inc., Doraville, GA) and reverse-transcribed (RT) using a SMARTerTM RACE cDNA Amplification kit (Clontech Inc., Mountain View, CA). The RT products were subjected to rapid amplification of cDNA ends (RACE) according to the manufacturer's protocol. The 3′ ends of MaIDT was obtained using the primer expressed sequence tag (EST)-3′GSP (5′-TGATGCCGATATTGACAGGATAAATAAGCC-3′) that was specific for an EST sequence of the mulberry root (GenBankTM accession number GT734921), which is homologous to AtVTE2–1 (GenBankTM accession number AY089963). The 5′ ends of MaIDT were re-isolated by RT-PCR using the primer MaIDT-5′GSP (5′-AGAGGAGGAGCAGAATAGAAGTGGGCCA-3′). Its full-length clone was acquired by RT-PCR using the gene-specific primer pairs MaIDT-Fw (5′-GAATTCATGGAGCTCTCAATCTCTCACTCT-3′) and MaIDT-Rv (5′-GCGGCCGCTTATATGAAAGGAAATATGACAAACTCCA-3′). In these sequences the restriction sites for subcloning are underlined. The PCR products were cloned into pEASY-Blunt Simple vector (TransGen Biotech Co., Ltd., Beijing, China) for sequencing and then subcloned into pESC-HIS vector (Stratagene Inc., La Jolla, CA), which resulted in the expression construct pESC-HIS-MaIDT.
Isolation of cDNAs Homologous to MaIDT from C. tricuspidata
Ten-day-old C. tricuspidata cell cultures were treated with 0.1 mm methyl jasmonate for 20 h, and total RNA was subsequently extracted with an E.Z.N.A.TM Plant RNA kit and reverse-transcribed as mentioned above. The CtIDT fragment was obtained using the degenerated primer pairs YZ-1 (5′-TAAAYAAGCCKTATYTACCTAT-3′) and YZ-2 (5′-CMAAATGKAAKTCCAAGGG-3′). The complete coding sequence of CtIDT was obtained with 5′- and 3′-RACE with the internal gene-specific primers CtIDT-3′GSP (5′-GAGCTCTCACTTAAGCAAGCATGGTT-3′) and CtIDT-5′GSP (5′-GTGAAGAATTCCGGCCATCAGAGGAT-3′). The full-length CtIDT was acquired using the gene-specific primer pairs CtIDT-Fw (5′-GCGGCCGCATGGCGTTCTCAATCTC-3′) and CtIDT-Rv (5′-ATCGATCTATATAAAGGGAAAAATGACAAATTC-3′). The expression vector pESC-HIS-CtIDT was constructed as described above.
Heterologous Expression in Yeast
The expression vectors pESC-HIS-MaIDT and pESC-HIS-CtIDT were introduced into the yeast strain YPH499. The microsomal fractions of YPH499 expressing pESC-HIS-MaIDT and pESC-HIS-CtIDT were prepared as previously described (14, 15) and resuspended in 100 mm Tris-HCl (pH 9.0). The total protein concentration was determined by the Bradford method (30).
Prenyltransferase Activity
For the quantitative determination of the PT activity, the basic reaction mixture (100 μl) contained 100 mm Tris-HCl (pH 9.0), 10 mm MgCl2, 200 μm prenyl acceptor, 400 μm DMAPP, and 100 μg of protein of the recombinant yeast microsome. The reaction mixtures were incubated at 30 °C, and the reactions were terminated by the addition of 200 μl of methanol. The protein was removed by centrifugation at 14,000 × g for 20 min. The enzymatic products were analyzed by HPLC-UV/ESI-MS under the conditions described above. For the quantitative measurements of the enzyme activity, three parallel assays were carried out routinely, and the values were obtained in duplicate for each assay.
Biochemical Properties of the Recombinant Enzymes
The assays for the determinations of the kinetic parameters (100 μl) of the prenyl acceptors contained 400 μm DMAPP, 100 μg of protein of the recombinant yeast microsome, and prenyl acceptors at final concentrations of 5, 10, 20, 40, 80, 160, and 400 μm. For the determinations of the kinetic parameters of DMAPP, isoliquiritigenin (1) at 400 μm and DMAPP at final concentrations of 5, 10, 20, 40, 80, 160, and 400 μm were used. The incubation time was 30 min. The apparent Km values were calculated from Lineweaver-Burk plots using the Hyper32 software.
To investigate the optimal pH, the enzyme reactions were performed in reaction buffers with pH values in the range of 6.0–8.0 (sodium phosphate buffer), 8.0–10.0 (Tris-HCl buffer), and 10.0–11.0 (CAPS-NaOH buffer) at 30 °C. To assay the optimal reaction temperature, the reaction mixtures were incubated at eight different temperatures that ranged from 4 °C to 50 °C in 100 mm Tris-HCl buffer (pH 9.0). To test the requirement of PT activity for divalent cations, MgCl2, BaCl2, CaCl2, FeCl2, CoCl2, CuCl2, NiCl2, and MnCl2 were individually used with DMAPP and isoliquiritigenin (1) in 100 mm Tris-HCl buffer (pH 9.0) at 30 °C.
Preparative Synthesis of the Enzymatic Products for Structural Elucidation
The assays for the isolation of the enzymatic products (10–15 ml) contained 100 mm Tris-HCl (pH 9.0), 10 mm MgCl2, 1 mm DMAPP, 500 μm prenyl acceptors, and 30 mg of protein of the recombinant yeast microsome. The reaction mixtures were incubated at 30 °C for 16 h and subsequently extracted with ethyl acetate (20 ml × 5). After evaporation of the solvent, the residues were dissolved in methanol and purified by reverse-phase semi-preparative HPLC under the conditions described above. The isolated products were subjected to MS and 1H and 13C NMR spectroscopic analyses, which yielded the following results.
3′-Dimethylallyl isoliquiritigenin (1a): ESI-MS, m/z 325.3 [M+H]+; 1H NMR (600 MHz, acetone-d6): δ 7.83 (1H, d, J = 15.6 Hz, H-β), 7.76 (1H, d, J = 15.6 Hz, H-α), 7.74 (2H, d, J = 8.4 Hz, H-2), 6.93 (1H, d, J = 8.4 Hz, H-3), 6.93 (1H, d, J = 8.4 Hz, H-5), 7.74 (1H, d, J = 8.4 Hz, H-6), 6.53 (1H, d, J = 8.4 Hz, H-5′), 7.98 (1H, d, J = 8.4 Hz, H-6′), 3.37 (2H, d, J = 7.2 Hz, H-1″), 5.28 (1H, t, J = 7.2 Hz, H-2″), 1.78 (1H, s, H-4″), 1.64 (1H, s, H-5″) (supplemental Fig. S1); 13C NMR (150 MHz, acetone-d6): δ 145.0 (C-β), 118.6 (C-α), 193.1 (CO), 127.8 (C-1), 131.9 (C-2), 116.9 (C-3), 161.0 (C-4), 116.9 (C-5), 131.8 (C-6), 116.2 (C-1′), 165.3 (C-2′), 114.5 (C-3′), 162.7 (C-4′), 108.1 (C-5′), 130.4 (C-6′), 22.4 (C-1″), 123.4 (C-2″), 131.6 (C-3″), 18.0 (C-4″), 26.0 (C-5″) (supplemental Fig. S2) (31, 32).
3′-Dimethylallyl-2′,4′-dihydroxychalcone (2a): ESI-MS, m/z 309.1 [M+H]+; 1H NMR (600 MHz, acetone-d6): δ 7.96 (1H, d, J = 15.6 Hz, H-β),7.87 (1H, d, J = 15.6 Hz, H-α), 7.85 (overlapped, 2H; H-2 and H-6), 7.47 (overlapped, 3H; H-3, H-4, and H-5), 6.55 (1H, d, J = 8.4 Hz, H-5′), 8.02 (1H, d, J = 8.4 Hz, H-6′), 3.38 (2H, d, J = 7.2 Hz, H-1″), 5.28 (1H, t, J = 7.2 Hz, H-2″), 1.78 (1H, s, H-4″), 1.65 (1H, s, H-5″) (supplemental Fig. S3); 13C NMR (150 MHz, acetone-d6): δ 144.6 (C-β), 121.9 (C-α), 193.0 (CO), 131.7 (C-1), 129.7 (C-2), 129.9 (C-3), 131.5 (C-4), 129.9 (C-5), 129.7 (C-6), 114.4 (C-1′), 165.4 (C-2′), 116.2 (C-3′), 163.2 (C-4′), 108.3 (C-5′), 130.7 (C-6′), 22.4 (C-1″), 123.3 (C-2″), 131.6 (C-3″), 18.0 (C-4″), 26.0 (C-5″) (supplemental Fig. S4) (33).
3′-Dimethylallyl-2,4,2′,4′-tetrahydroxychalcone (3a): ESI-MS, m/z 340.9 [M+H]+;1H NMR (600 MHz, acetone-d6): δ 8.22 (1H, d, J = 15.6 Hz, H-β),7.79 (1H, d, J = 15.6 Hz, H-α), 6.50 (1H, d, J = 2.4 Hz, H-3), 6.46 (1H, dd, J = 8.4, 2.4 Hz, H-5), 7.69 (1H, d, J = 8.4 Hz, H-6), 6.52 (1H, d, J = 8.4 Hz, H-5′), 7.89 (1H, d, J = 8.4 Hz, H-6′), 3.37 (2H, d, J = 7.2 Hz, H-1″), 5.28 (1H, t, J = 7.2 Hz, H-2″), 1.78 (1H, s, H-4″), 1.64 (1H, s, H-5″) (supplemental Fig. S5); 13C NMR (150 MHz, acetone-d6): δ 140.8 (C-β), 117.6 (C-α), 193.5 (CO), 114.8 (C-1), 159.9 (C-2), 103.7 (C-3), 162.3 (C-4), 109.2 (C-5), 131.8 (C-6), 114.6 (C-1′), 162.5 (C-2′), 115.3 (C-3′), 165.2 (C-4′), 107.9 (C-5′), 130.0 (C-6′), 22.4 (C-1″), 123.4 (C-2″), 131.5 (C-3″), 18.0 (C-4″), 25.9 (C-5″) (supplemental Fig. S6) (34, 35).
3′-Dimethylallylbutein (4a): ESI-MS, m/z 341.0 [M+H]+;1H NMR (600 MHz, acetone-d6): δ 7.76 (1H, d, J = 15.6 Hz, H-β),7.69 (1H, d, J = 15.6 Hz, H-α), 7.34 (1H, d, J = 2.4 Hz, H-2), 6.91 (1H, d, J = 8.4 Hz, H-5), 7.23 (1H, dd, J = 8.4, 2.4 Hz, H-6), 6.53 (1H, d, J = 8.4 Hz, H-5′), 7.97 (1H, d, J = 8.4 Hz, H-6′), 3.37 (2H, d, J = 7.2 Hz, H-1″), 5.27 (1H, t, J = 7.2 Hz, H-2″), 1.77 (1H, s, H-4″), 1.64 (1H, s, H-5″) (supplemental Fig. S7); 13C NMR (150 MHz, acetone-d6): δ 145.4 (C-β), 118.6 (C-α), 193.1 (CO), 128.4 (C-1), 116.0 (C-2), 146.5 (C-3), 149.2 (C-4), 116.5 (C-5), 123.4 (C-6), 114.5 (C-1′), 162.7 (C-2′), 116.2 (C-3′), 165.2 (C-4′), 108.1 (C-5′), 130.3 (C-6′), 22.3 (C-1″), 123.4 (C-2″), 131.5 (C-3″), 18.0 (C-4″), 25.9 (C-5″) (supplemental Fig. S8) (36).
6-Dimethylallylgenistein (7a): ESI-MS, m/z 338.9 [M+H]+; 1H NMR (600 MHz, acetone-d6): δ 8.14 (1H, s, H-2), 6.50 (1H, s, H-8), 7.45 (1H, d, J = 8.4 Hz, H-2′), 6.90 (1H, d, J = 8.4 Hz, H-3′), 6.90 (1H, d, J = 8.4 Hz, H-5′), 7.45 (1H, d, J = 8.4 Hz, H-6′), 3.36 (2H, d, J = 7.2 Hz, H-1″), 5.27 (1H, t, J = 7.2 Hz, H-2″), 1.78 (1H, s, H-4″), 1.64 (1H, s, H-5″) 13.32 (1H, s, 5-OH) (supplemental Fig. S9) (37).
6-Dimethylallyl-2′-hydroxygenistein (8a): ESI-MS, m/z 354.8 [M+H]+; 1H NMR (600 MHz, acetone-d6): δ 8.12 (1H, s, H-2), 6.52 (1H, s, H-8), 6.45 (1H, d, J = 2.4 Hz, H-3′), 6.42 (1H, dd, J = 8.4, 2.4 Hz, H-5′), 7.10 (1H, d, J = 8.4 Hz, H-6′), 3.36 (2H, d, J = 7.2 Hz, H-1″), 5.27 (1H, t, J = 7.2 Hz, H-2″), 1.77 (1H, s, H-4″), 1.64 (1H, s, H-5″) 13.01 (1H, s, 5-OH) (supplemental Fig. S10) (38).
Computer-assisted Sequence Analysis
The sequence identities were obtained via alignment of the amino acid sequences performed with the program BLAST2 SEQUENCES (www.ncbi.nlm.nih.gov). The PT proteins of the plants were aligned using ClustalW (39). From the alignments, a consensus phylogenetic tree was generated by the neighbor-joining method using MEGA6 (40). Bootstrap values are indicated in percentages (only those >70% are presented) on the nodes. The bootstrap values were obtained from 1000 bootstrap replicates. The scale bar corresponds to 0.1 estimated amino acid changes per site. The exon-intron structure diagram was generated using the online Gene Structure Display Server (GSDS) and the exon position and gene length method.
Nucleotide Sequence Accession Number
The nucleotide sequences of MaIDT and CtIDT have been deposited in the GenBankTM database under the accession numbers KM262659 and KM262660, respectively.
RESULTS
Cloning of MaIDT from the Cultured Cells of M. alba
According to previous studies (12–17), plant FPTs from leguminous plants are closely related to each other and cluster together in the phylogenetic tree. Therefore, we initially hypothesized that the FPTs of M. alba would cluster together with the leguminous FPTs and used degenerated primer pairs based on these FPTs to clone the candidate gene(s). Similar cloning strategies proved to be successful in the isolation of SfFPT and GuA6DT from S. flavescens and Glycyrrhiza uralensis in our previous work (14, 15). However, after RT-PCR and RACE, we failed to achieve active PTs. A similar cloning strategy also failed to lead to the identification of active FPT from Epimedium acuminatum (41). Hence, we switched to the Morus EST library for the cloning of these candidate genes. Utilizing the internal sequence of an EST clone that is homologous to the homogentisate PT AtVTE2–1, we succeeded in amplifying the full-length cDNA of MaIDT, and PT activity was observed after the heterologous expression of MaIDT in yeast.
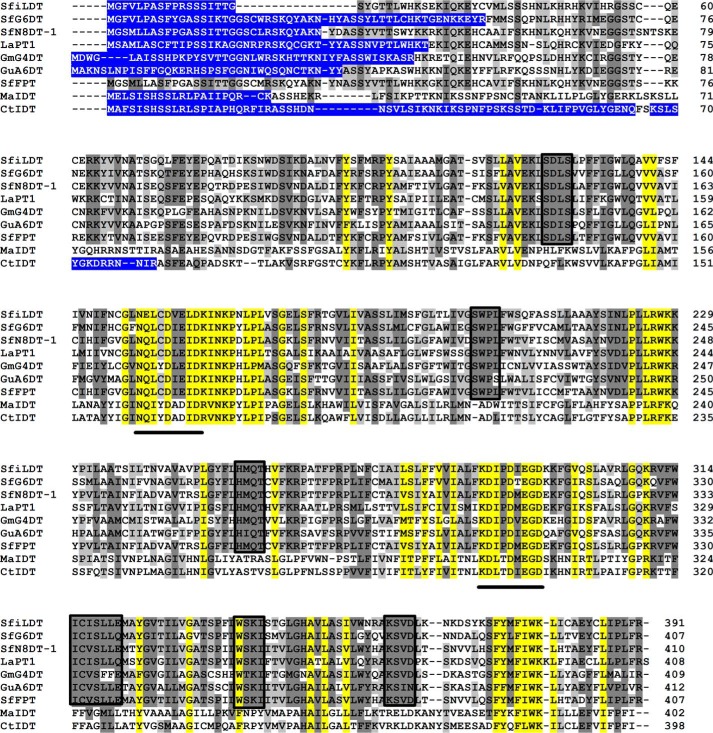
The encoded polypeptide of MaIDT (402 amino acids) had seven putative transmembrane α-helices as predicted by the TMHMM 2.0 program. The polypeptide possessed putative transit peptide sequences for targeting plastids as predicted by Chlorop 1.1, TargetP 1.1, and iPSORT. A multiple alignment of the encoded polypeptide and the reported leguminous FPTs is shown in Fig. 1. The polypeptide possesses a conserved aspartate-rich motif of PTs (N(Q/D)XXDXXXD) in loop 2 and another characteristic sequence that is conserved in the flavonoid/homogentisate PTs in loop 6 (KD(I/L)XDX(E/D)GD). However, several highly conserved sequences, such as SD(L/I)S, SWP(L/S) H(M/I)QT, IC(I/V)SLL(E/Q), and W(S/T)KI were observed in the alignment of the leguminous FPTs and were not apparent in this polypeptide; the functions of these domains are not yet known. Moreover, the polypeptide shared identities of only 24–30% at the amino acid level with the reported leguminous FPTs, although the above data fulfilled the three criteria for selecting FPT candidates that have been adopted in a previous report (12).
FIGURE 1.
Multiple alignment of plant flavonoid prenyltransferases. Shown are: MaIDT (M. alba; KM262659); CtIDT (C. tricuspidata; KM262660); SfFPT (S. flavescens; KC513505); SfN8DT-1 (S. flavescens; AB325579); SfG6DT (S. flavescens; BAK52291); SfiLDT (S. flavescens; AB604223); LaPT1 (L. albus; JN228254); GmG4DT (Glycine max; AB434690); GuA6DT (G. uralensis; KJ123716). The conserved aspartate-rich motifs N(Q/D)XXDXXXD and KD(I/L)XDX(E/D)GD are highlighted with solid underlines. The sequences conserved only in the leguminous FPTs are highlighted with black boxes. The transit peptides have been highlighted in a blue background.
Enzymatic Activity of MaIDT and Identification of the Enzymatic Products
In previous studies mulberry trees and related plants have been found to be rich sources of prenylated flavonoids (22, 23), and prenylation activity has been confirmed in microsomal fractions of mulberry tree cell cultures (42, 43). Accordingly, various flavonoids, including chalcones, flavones, flavanones, and isoflavones have been employed as acceptors to determine enzymatic activities. Of these, isoliquiritigenin (1) is accepted to have the highest efficiency. HPLC-UV/ESI-MS analysis revealed that a product at 24.4 min with a 68-atomic mass unit higher shift in its molecular peak was detected in the incubation mixture with the microsomes from the yeast cells transformed with MaIDT but not with an empty vector (Fig. 2). Product formation was strictly dependent on the presence of MaIDT, DMAPP, Mg2+, and 1. A linear dependence of the product formation on the amount of microsome protein was found up to 200 μg per 100-μl assay and on the reaction time was up to 30 min at 30 °C.
FIGURE 2.
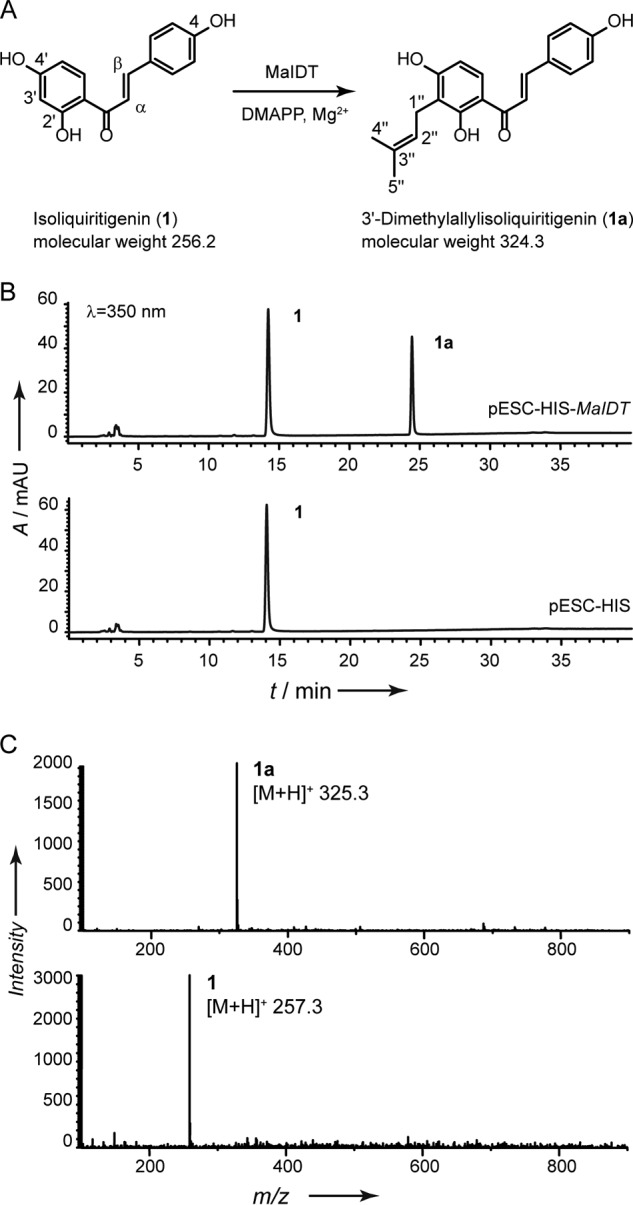
Functional characterization of recombinant MaIDT. A, reaction catalyzed by the recombinant MaIDT. B, incubation containing isoliquiritigenin (1), DMAPP and Mg2+ with microsomal fractions of yeast strain YPH499 harboring either the expression vector pESC-HIS-MaIDT or the empty vector pESC-HIS. mAU, milliabsorbance units. C, MS spectra of the 1a and 1.
To clarify the prenylation properties, the enzymatic product (1a) was subsequently prepared and subjected to NMR analysis. In the 1H NMR spectrum, the presence of the prenyl moiety was suggested by the appropriate signals for two methyls (δH 1.64 and δH 1.78, s, 3H each), a methylene (δH 3.37, d, J = 7.2 Hz, 2H), and a methine group (δH 5.28, t, J = 7.2 Hz, 1H). The location of the prenyl moiety was indicated by the disappearance of the H-3′ signal (1H at δH 6.35) and the change in the H-5′ peak shape, which became an ortho-coupled doublet (δH 6.53, d, J = 8.4 Hz) (supplemental Fig. S1). This observation is supported by a downfield chemical shift of C-3′ from δC 103.7 in 1 to δC 114.5 in 1a (supplemental Fig. S2). The NMR data of 1a were in good agreement with those of 3′-dimethylallyl isoliquiritigenin (31, 32). Therefore, the enzymatic product of MaIDT was unambiguously identified as 3′-dimethylallyl isoliquiritigenin (1a), which indicates that this enzyme regiospecifically transferred a dimethylallyl moiety to 1 at the C-3′ position; thus, this recombinant enzyme was designated as MaIDT (M. alba isoliquiritigenin 3′-dimethylallyltransferase).
Cloning and Identification of CtIDT from Cultured Cells of C. tricuspidata
To increase understanding of the diversity of the FPTs in moraceous plants, a cell suspension culture of C. tricuspidata was chosen for further study. Due to the absence of genetic information about C. tricuspidata, homologous PCR was used to clone candidate gene(s) as previously described (14, 15). Similarly, degenerated primer pairs based on the conserved regions of the leguminous FPTs failed to produce an active FPT. Next, we designed degenerated primer pairs based on the conserved amino acid regions of MaIDT and its closely related genes that are obtained via a BLASTN search of GenBankTM and eventually cloned a full-length cDNA possessing FPT activity (CtIDT). The polypeptide encoded by CtIDT (398 amino acids) fulfilled the three criteria for selecting FPTs (12). This polypeptide shared sequence identities of 66% with MaIDT and only 24–30% with the reported leguminous FPTs. Enzymatic assays containing various flavonoids and HPLC-UV/ESI-MS analyses revealed that CtIDT exhibited a function similar to that of MaIDT (Fig. 3, A and B); thus, this recombinant enzyme was designated as CtIDT (C. tricuspidata isoliquiritigenin 3′-dimethylallyltransferase).
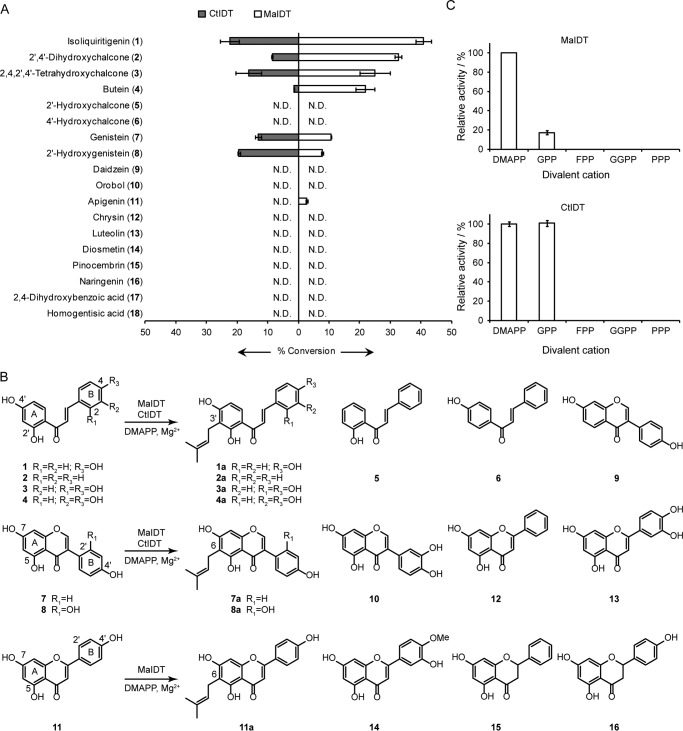
FIGURE 3.
Substrate specificities of the recombinant MaIDT and CtIDT. A, relative enzymatic activities with various flavonoids and simple phenols acting as the prenyl acceptors and DMAPP acting as the prenyl donor. N.D., not determined. B, the reactions catalyzed by the recombinant MaIDT and CtIDT and the chemical structures of the flavonoids used for the substrate specificity analysis. C, relative enzymatic activities with DMAPP, GPP, farnesyl diphosphate (FPP), geranylgeranyl diphosphate (GGPP), and phytyl diphosphate (PPP) as the prenyl donors and 1 as the prenyl acceptor.
Substrate Specificities of MaIDT and CtIDT
To investigate the substrate specificities of the moraceous FPTs, we tested the activities of MaIDT and CtIDT via HPLC-UV/ESI-MS analysis utilizing 18 potential substrates that included various types of flavonoids and structurally simple phenols (Fig. 3, A and B). In addition to isoliquiritigenin (1), MaIDT and CtIDT were also able to catalyze the prenylation of three other chalcones (2′,4′-dihydroxychalcone (2), 2,4,2′,4′-tetrahydroxychalcone (3), and butein (4)) and two isoflavonoids (genistein (7), 2′-hydroxygenistein (8)), and the flavone type substrate apigenin (11) was only accepted by MaIDT. The enzymatic products were prepared from larger scale assays and subjected to MS and NMR spectroscopic analyses with the exception of apigenin (11) (supplemental Fig. S3-S10), the prenylated product of which was identified as 6-dimethylallylapigenin (11a) in a comparison with an authentic standard using a HPLC-UV/ESI-MS analysis (data not shown). The results clearly showed that although the three types of flavonoids could be accepted by MaIDT and CtIDT, one dimethylallyl moiety was regioselectively introduced into the A ring of the skeletons at the same position (C-3′ of the chalcones and C-6 of the isoflavones and flavones).
The relative reactivities of MaIDT and CtIDT toward different flavonoids also provided insight into the “substrate structure-enzyme selectivity relationships.” MaIDT preferred 2′,4′-dihydroxychalcones to isoflavonoids and flavone. The existence of hydroxyls at C-2′,4′ (C-5,7 for isoflavones) of the A ring was crucial for prenylation because the lack of any one of these hydroxyl groups led to no enzymatic product generation (5, 6, and 9). Regarding the B ring, the presence of a hydroxyl group at C-4 position increased the prenylation efficiency, as the conversion rate of 1 by MaIDT was higher than that of 2 (Fig. 3, A and B). In contrast, the hydroxyl group at the C-2/3 (C-2′/3′ for isoflavones) position of B ring reduced the prenylation efficiency as revealed by comparisons of the relative yields of 3 and 4 to 1 and of 8 and 10 to 7 (Fig. 3, A and B).
The prenylation catalyzed by FPTs represents a Friedel-Crafts alkylation of flavonoid skeleton. A carbocation is generated from DMAPP and carries out an electrophilic attack at flavonoid ring, leading to the regio-specific C-C bond formation. Hence, the electron density around attacked carbon at flavonoid ring might play an important role in the course of prenylation. For example, the insufficient electron density around C-6 of daidzein (9) without C-5 hydroxyl group compared with genistein (7) might lead to the non-occurrence of prenylation.
Regarding CtIDT, its substrate specificity was slightly different from that of MaIDT; first, CtIDT accepted isoflavones with higher efficiency than did MaIDT, and this finding is partially in accordance with the fact that 7a has been identified in large amounts in C. tricuspidata cell suspension cultures (26); second, the hydroxyl group at C-2 of chalcones decreased the activity of CtIDT as revealed by comparisons of the relative yields of 3 to 1, whereas the corresponding hydroxyl group at the C-2′ position of the isoflavone increased the activity as revealed by the comparison of the relative yields of 8 to 7.
Other prenyl donors (including GPP, farnesyl diphosphate, geranylgeranyl diphosphate, and phytyl diphosphate) were also tested in the enzymatic assay. The results revealed that GPP could also act as prenyl donor when 1 is used as the acceptor (Fig. 3C).
Biochemical Properties of the Recombinant MaIDT and CtIDT
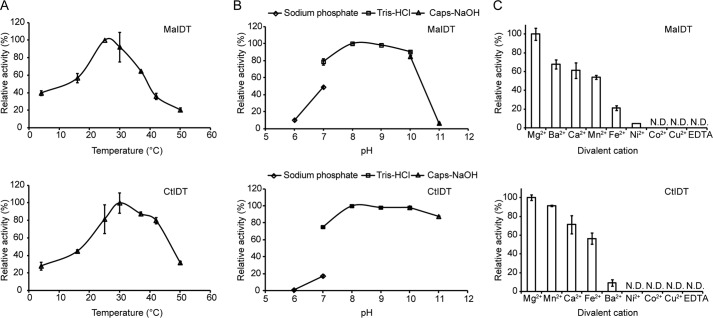
Previous studies have reported that the plant FPT activity is dependent on divalent cations and strongly affected by pH and temperature (10). Further investigations using 1 as an acceptor and DMAPP as a prenyl donor in this study revealed that the highest activities occurred at ∼30 °C and that the activity decreased rapidly at temperatures above 40 °C (Fig. 4A). The analysis of the enzyme activity within the pH range of 6.0 to 11.0 revealed that the optimal pH value was between 8.0 and 9.0 (Fig. 4B). MaIDT activity was observed to decrease in the order of Mg2+ > Ba2+ > Ca2+ > Mn2+ > Fe2+ > Ni2+, and Co2+ and Cu2+ did not lead to the production of 1a; CtIDT activity was found to decrease in the order of Mg2+ > Mn2+ > Ca2+ > Fe2+ > Ba2+, whereas Ni2+, Co2+, and Cu2+ did not lead to the production of 1a (Fig. 4C). No product was detected in the buffer after the addition of EDTA.
FIGURE 4.
Biochemical properties of MaIDT and CtIDT. A, effects of temperature on the enzyme activities of MaIDT and CtIDT. B, pH dependences of MaIDT and CtIDT. C, effects of various divalent metal ions on MaIDT and CtIDT activities.
The apparent Km values of MaIDT and CtIDT for DMAPP as a prenyl donor were calculated as 28.3 and 173.6 μm, respectively. The apparent Km values of MaIDT and CtIDT for 1 were calculated as 23.9 and 109.6 μm, respectively. The apparent Km values of MaIDT and CtIDT for 7 were calculated as 36 and 255.9 μm, respectively. Considering the relatively high Km values of CtIDT for flavonoids, this enzyme might act on other flavonoids (or other aromatic compounds) in C. tricuspidata.
Phylogenetic and Gene Structure Analysis of the Flavonoid Dimethylallyltransferases and Related Enzymes
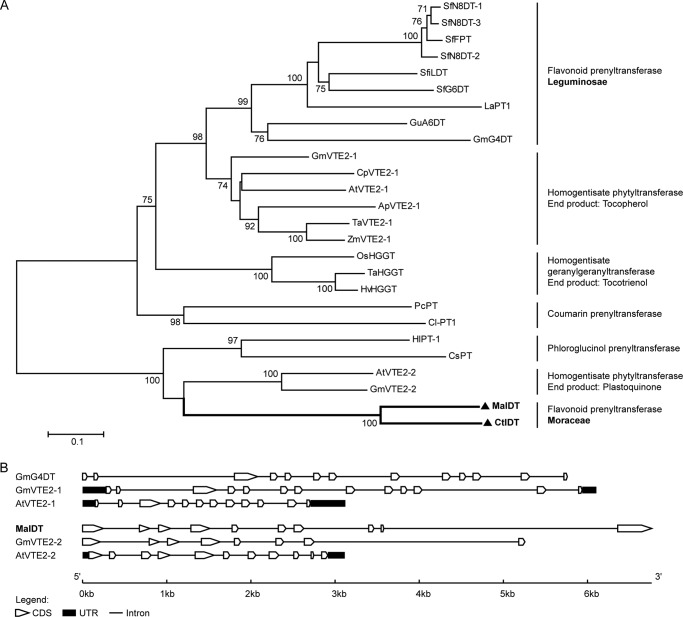
A neighbor-joining phylogenetic tree was constructed to analyze the evolutionary relationships of MaIDT and CtIDT with the other plant aromatic PTs (Fig. 5A). As the first identified FPTs from non-Leguminosae plant species, MaIDT and CtIDT will have particularly significant effects on our understanding of the molecular evolution of plant FPTs. The two moraceous FPTs MaIDT and CtIDT appeared to cluster together to form a new branch with a common origin in the homogentisate phytyltransferases for plastoquinone formation (AtVTE2–2 and GmVTE2–2). This branch is clearly divergent from that of the leguminous FPTs, which were recruited from the vitamin E biosynthetic pathway. Considering the similar substrate spectra and sequence divergence of the leguminous and moraceous FPTs, it can be concluded that they have evolved independently via recruitment from the vitamin E and plastoquinone biosynthetic pathways, respectively. The sequence distance might reflect the taxonomical distance between Moraceae from Leguminosae plants (44). This situation would represent a somewhat subtler example of convergent evolution. Moreover, this type of convergent evolution has been termed “repeated evolution,” which indicates situations in which “a similar function arose not from a completely unrelated sequence but from a homologous, although not orthologous gene” (45, 46). Consequently, the search for new FPTs should not focus solely on the degrees of sequence identity with known enzymes. Furthermore, in evolutionarily distant plants, it is possible that as-yet unidentified FPTs might share low homology with the known FPTs.
FIGURE 5.
Phylogenetic and genome structure analyses of MaIDT and CtIDT. A, the phylogenetic relationship between the MaIDT and CtIDT proteins and the related plant prenyltransferases. The protein sequences were aligned using ClustalW. The neighbor-joining phylogenetic tree was drawn using MEGA6. The bootstrap value was 1000, and the branch lengths represent the relative genetic distances. The abbreviations of the protein sequences and their accession numbers are as follows: MaIDT (M. alba; KM262659); CtIDT (C. tricuspidata; KM262660); SfFPT (S. flavescens; KC513505); SfN8DT-1 (S. flavescens; AB325579); SfN8DT-2 (S. flavescens; AB370330); SfN8DT-3 (S. flavescens; AB604222); SfG6DT (S. flavescens; BAK52291); SfiLDT (S. flavescens; AB604223); LaPT1 (L. albus; JN228254); GmG4DT (G. max; AB434690); GuA6DT (G. uralensis; KJ123716); ZmVTE2–1 (Zea mays; DQ231055); TaVTE2–1 (Triticum aestivum; DQ231056); ApVTE2–1 (Allium porrum; DQ231057); CpVTE2–1 (Cuphea pulcherrima; DQ231058); AtVTE2–1 (Arabidopsis thaliana; AY089963); GmVTE2–1 (G. max; DQ231059); PcPT (Petroselinum crispum; AB825956); Cl-PT1 (Citrus limon; AB813876) OsHGGT (Oryza sativa; AY222862); HvHGGT (Hordeum vulgare; AY222860); TaHGGT (T. aestivum; DQ231056); AtVTE2–2 (A. thaliana; DQ231060); GmVTE2–2 (G. max; DQ231061); HlPT-1 (Humulus lupulus; AB543053); CsPT (Cannabis sativa; see Ref. 51); OsPPT1 (O. sativa; AB263291); AtPPT1 (A. thaliana; AY089963); LePGT1 (Lithospermum erythrorhizon; AB055078); LePGT2 (L. erythrorhizon; AB055079). B, comparisons of the exon-intron structures of MaIDT and the related prenyltransferases. The gene sequence of MaIDT can be acquired from MorusDB with the gene ID Morus014065; the GenBankTM accession numbers of the other gene sequences used have been provided above. CDS, coding sequence; UTR, untranslated regions.
Based on a BLASTP search of the Morus Genome Database (MorusDB) (47, 48), 1 hit with MorusDB accession number Morus014065 was found and shared an identity of 97% with MaIDT, and its genome structure was compared with those of the leguminous FPT GmG4DT and homogentisate PTs involved in vitamin E and plastoquinone biosyntheses, which are available in GenBankTM. The exon-intron structure of MaIDT was quite different from those of the leguminous FPT GmG4DT and the related homogentisate PTs involved in vitamin E biosynthesis, whereas the exon-intron organizations of MaIDT and the homogentisate PTs for plastoquinone biosynthesis were very similar (Fig. 5B). These observations confirm the results of the phylogenetic analysis, which suggested that the leguminous and moraceous FPTs were derived from homogentisate PTs involved in vitamin E and plastoquinone biosynthesis, respectively, and have evolved independently.
DISCUSSION
Prenylation as a Typical Modification of Aromatic Compounds in Moraceous Plants
A large variety of biological activities has been reported for prenylated aromatic compounds, and these natural products are attractive resources in the food and pharmaceutical industries. Aromatic PTs catalyze transfer reactions of prenyl moieties from prenyl diphosphates to diverse aromatic acceptors. Theses PTs are involved in both general and specialized metabolism, play important roles in living organisms, and strongly contribute to the structural diversity and biological activities of aromatic natural products (7, 49).
In moraceous plants, many prenylated aromatic compounds, such as Diels-Alder-type adducts, flavonoids, 2-arylbenzofurans, xanthones, and stilbenes, have been identified, and their biological/pharmacological activities have been extensively studied (9, 22, 23). Of these compounds optically active Diels-Alder-type adducts, which are characteristic mulberry constituents with promising biological activities, are attractive compounds from a biosynthetic point of view, and prenylation is a key biosynthetic step in their formation (8, 9).
Evolution of FPTs
Plant aromatic PTs are divided into two distinct subgroups: one is the 4-hydroxybenzoate PT, which is involved in the biosyntheses of ubiquinone (benzoquinone) and shikonin (naphthoquinone) derivatives; the second subgroup is the homogentisate prenyltransferase (HGPT) family, which is involved in vitamin E and plastoquinone biosyntheses. Thus far, all the identified plant FPTs belong to HGPT family (11–15).
Based on the newly identified moraceous FPTs, the phylogenetic tree of the HGPT gene family was rebuilt. A clear phylogenetic separation of the HGPT genes based on primary metabolism (i.e. those involved in tocopherol, tocotrienol, and plastoquinone biosyntheses) versus specialized metabolism can be observed. The formers phylogenetic group is clustered in a pattern of functional clades because the molecular evolution of this group has been severely constrained by natural selection. However, the latter group exhibits a remarkable flexibility of the HGPT family to evolve enzymes with new substrate specificities. Moreover, several functionally divergent enzymes, such as FPTs, phloroglucinol PTs, and coumarin PTs, have emerged from the primary metabolic HGPTs (50–53). Interestingly, repeated evolution has been observed to exist in the FPTs. The moraceous FPTs MaIDT and CtIDT are grouped in a branch that is clearly different from the branch in which the leguminous FPTs cluster, which implies that FPTs evolved independently in the two plant lineages. Repeated evolution might represent the majority of convergent evolution cases in plant-specialized metabolism, and many such cases have been identified (46); e.g. in each plant lineage the respective linalool synthase is more similar to other terpene synthases, which are responsible for the synthesis of other monoterpenes within that lineage than it is to the linalool synthases from different lineages (54, 55). Furthermore, the phylogeny of FPTs seems to exhibit a pattern of species-specific expansion in Leguminosae and Moraceae. Apparently, the identified moraceous FPTs and leguminous FPTs arose individually by repeated gene duplications and divergence that occurred after the split of the two plant families. However, our understanding of the plant FPTs is currently based on just over 10 genes so far; thus, different FPTs need to be identified to clarify the molecular evolution of plant FPTs.
In some studies the abovementioned species-specific patterns of repeated gene duplications and sequence divergence are referred to as “blooms” (56). In practice, the approach of selecting candidate genes from species-specific blooms has proven successful for the discovery of new genes (56, 57). As FPTs have now been successfully identified in moraceous plants, this approach is likely worthwhile for attempts to clone other FPTs from moraceous plants. Given the great variety of prenylated flavonoids that have been isolated from moraceous plants, a large family of FPTs can be imagined. Moreover, prenylated flavonoids are active constituents of many other non-Leguminosae medicinal plants. As the first successful identification of FPTs from non-Leguminosae plants, this study might inspire the identification and characterization of additional and evolutionarily varied FPTs that are involved in the biosyntheses of pharmacologically active prenylated flavonoids in plants other than those of the Leguminosae and Moraceae families.
In summary, we have characterized the first two non-Leguminosae plant FPTs MaIDT and CtIDT from M. alba and C. tricuspidata, respectively. These FPTs feature catalytic regioselectivity and promiscuity. Multiple alignments, phylogenetic analyses and gene structure analyses revealed that these novel FPTs evolved independently from their counterparts in Leguminosae. These findings provide useful guidance for identifying additional and evolutionarily varied FPTs in non-Leguminosae plants. Furthermore, based on their substrate promiscuities and high catalytic efficiencies, MaIDT and CtIDT might be useful as an environmentally friendly and efficient biological catalysts of the regiospecific prenylation of flavonoids to produce bioactive compounds for potential therapeutic applications.
Supplementary Material
The study was supported by National Natural Science Foundation of China Grant 81273405, Fundamental Research Funds for the Central Universities Grant 2012N06, Program for Doctorate Education Grant 20111106110029, and National Science and Technology Major Project “Key New Drug Creation and Manufacturing,” China Grant 2012X09301002-001-005.

This article contains supplemental Figs. S1–S10.
- FPT
- flavonoid prenyltransferase
- MaIDT
- M. alba isoliquiritigenin 3′-dimethylallyltransferase
- CtIDT
- C. tricuspidata isoliquiritigenin 3′-dimethylallyltransferase
- PT
- prenyltransferase
- HGPT
- homogentisate prenyltransferase
- DMAPP
- dimethylallyl diphosphate
- GPP
- geranyl diphosphate
- ESI
- electrospray mass ionization
- EST
- expressed sequence tag
- RACE
- rapid amplification of cDNA ends
- BLAST
- basic local alignment search tool
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
REFERENCES
- 1. Botta B., Vitali A., Menendez P., Misiti D., Delle Monache G. (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr. Med. Chem. 12, 717–739 [DOI] [PubMed] [Google Scholar]
- 2. Barron D., Ibrahim R. K. (1996) Isoprenylated flavonoids-a survey. Phytochemistry 43, 921–982 [Google Scholar]
- 3. Botta B., Menendez P., Zappia G., de Lima R. A., Torge R., Monachea G. D. (2009) Prenylated isoflavonoids: botanical distribution, structures, biological activities and biotechnological studies. An update (1995–2006). Curr. Med. Chem. 16, 3414–3468 [DOI] [PubMed] [Google Scholar]
- 4. Boland G. M., Donnelly D. M. X. (1998) Isoflavonoids and related compounds. Nat. Prod. Rep. 15, 241–260 [Google Scholar]
- 5. Tahara S., Ibrahim R. K. (1995) Prenylated isoflavonoids-an update. Phytochemistry 38, 1073–1094 [Google Scholar]
- 6. Botta B., Delle Monache G., Menendez P., Boffi A. (2005) Novel prenyltransferase enzymes as a tool for flavonoid prenylation. Trends. Pharmacol. Sci. 26, 606–608 [DOI] [PubMed] [Google Scholar]
- 7. Yazaki K., Sasaki K., Tsurumaru Y. (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70, 1739–1745 [DOI] [PubMed] [Google Scholar]
- 8. Hano Y., Nomura T., Ueda S. (1990) Biosynthesis of optically active Diels-Alder type adducts revealed by an aberrant metabolism of O-methylated precursors in Morus alba cell cultures. J. Chem. Soc. Chem. Commun. 8, 610–613 [Google Scholar]
- 9. Nomura T., Hano Y., Fukai T. (2009) Chemistry and biosynthesis of isoprenylated flavonoids from Japanese mulberry tree. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laflamme P., Khouri H., Gulick P., Ibrahim R. (1993) Enzymatic prenylation of isoflavones in white lupin. Phytochemistry 34, 147–151 [Google Scholar]
- 11. Yamamoto H., Senda M., Inoue K. (2000) Flavanone 8-dimethylallyltransferase in Sophora flavescens cell suspension cultures. Phytochemistry 54, 649–655 [DOI] [PubMed] [Google Scholar]
- 12. Sasaki K., Mito K., Ohara K., Yamamoto H., Yazaki K. (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol. 146, 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sasaki K., Tsurumaru Y., Yamamoto H., Yazaki K. (2011) Molecular characterization of a membrane-bound prenyltransferase specific for isoflavone from Sophora flavescens. J. Biol. Chem. 286, 24125–24134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen R., Liu X., Zou J., Yin Y., Ou B., Li J., Wang R., Xie D., Zhang P., Dai J. (2013) Regio- and stereospecific prenylation of flavonoids by Sophora flavescens prenyltransferase. Adv. Synth. Catal. 355, 1817–1828 [Google Scholar]
- 15. Li J., Chen R., Wang R., Liu X., Xie D., Zou J., Dai J. (2014) GuA6DT, a regiospecific prenyltransferase from Glycyrrhiza uralensis, catalyzes the 6-prenylation of flavones. Chembiochem 15, 1673–1681 [DOI] [PubMed] [Google Scholar]
- 16. Shen G., Huhman D., Lei Z., Snyder J., Sumner L. W., Dixon R. A. (2012) Characterization of an isoflavonoid-specific prenyltransferase from Lupinus albus. Plant Physiol. 159, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akashi T., Sasaki K., Aoki T., Ayabe S., Yazaki K. (2009) Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 149, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kremer A., Li S. M. (2008) Potential of a 7-dimethylallyltryptophan synthase as a tool for production of prenylated indole derivatives. Appl. Microbiol. Biotechnol. 79, 951–961 [DOI] [PubMed] [Google Scholar]
- 19. Yu X., Li S. M. (2011) Prenylation of flavonoids by using a dimethylallyltryptophan synthase, 7-DMATS, from Aspergillus fumigatus. Chembiochem 12, 2280–2283 [DOI] [PubMed] [Google Scholar]
- 20. Kuzuyama T., Noel J. P., Richard S. B. (2005) Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435, 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozaki T., Mishima S., Nishiyama M., Kuzuyama T. (2009) NovQ is a prenyltransferase capable of catalyzing the addition of a dimethylallyl group to both phenylpropanoids and flavonoids. J. Antibiot. 62, 385–392 [DOI] [PubMed] [Google Scholar]
- 22. Nomura T. (1988) Phenolic compounds of the mulberry tree and related plants. In Fortschritte der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, pp. 87–201, Springer, Vienna, Austria: [DOI] [PubMed] [Google Scholar]
- 23. Nomura T., Hano Y. (1994) Isoprenoid-substituted phenolic compounds of moraceous plants. Nat. Prod. Rep. 11, 205–218 [DOI] [PubMed] [Google Scholar]
- 24. Yin Y., Chen R., Zhang D., Qiao L., Li J., Wang R., Liu X., Yang L., Xie D., Zou J., Wang C., Dai J. (2013) Regio-selective prenylation of flavonoids by plant cell suspension cultures of Cudrania tricuspidata and Morus alba. J. Mol. Catal., B Enzym. 89, 28–34 [Google Scholar]
- 25. Tao X. Y., Zhang D. W., Chen R. D., Yin Y. Z., Zou J. H., Xie D., Yang L., Wang C. M., Dai J. G. (2012) Chemical constituents from cell cultures of Morus alba. Zhongguo Zhong Yao Za Zhi 37, 3738–3742 [PubMed] [Google Scholar]
- 26. Yin Y. Z., Wang R. S., Chen R. D., Qiao L. R., Yang L., Wang C. M., Dai J. G. (2012) Chemical constituents from cell suspension cultures of Cudrania tricuspidata. Zhongguo Zhong Yao Za Zhi 37, 3734–3737 [PubMed] [Google Scholar]
- 27. Davisson V. J., Woodside A. B., Poulter C. D. (1985) Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Meth. Enzymol. 110, 130–144 [DOI] [PubMed] [Google Scholar]
- 28. Juvale K., Pape V. F., Wiese M. (2012) Investigation of chalcones and benzochalcones as inhibitors of breast cancer resistance protein. Bioorg. Med. Chem. 20, 346–355 [DOI] [PubMed] [Google Scholar]
- 29. Khatib S., Nerya O., Musa R., Shmuel M., Tamir S., Vaya J. (2005) Chalcones as potent tyrosinase inhibitors: the importance of a 2, 4-substituted resorcinol moiety. Bioorg. Med. Chem. 13, 433–441 [DOI] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Kobayashi M., Noguchi H., Sankawa U. (1985) Formation of chalcones and isoflavones by callus culture of Glycyrrhiza uralensis with different production patterns. Chem. Pharm. Bull. 33, 3811–3816 [Google Scholar]
- 32. Wang H., Yan Z., Lei Y., Sheng K., Yao Q., Lu K., Yu P. (2014) Concise synthesis of prenylated and geranylated chalcone natural products by regiospecific iodination and Suzuki coupling reactions. Tetrahedron Lett. 55, 897–899 [Google Scholar]
- 33. Borges-Argáez R., Peña-Rodríguez L. M., Waterman P. G. (2002) Flavonoids from two Lonchocarpus species of the Yucatan peninsula. Phytochemistry 60, 533–540 [DOI] [PubMed] [Google Scholar]
- 34. Delle Monache G., De Rosa M. C., Scurria R., Vitali A., Cuteri A., Monacelli B., Pasqua G., Botta B. (1995) Comparison between metabolite productions in cell culture and in whole plant of Maclura pomifera. Phytochemistry 39, 575–580 [DOI] [PubMed] [Google Scholar]
- 35. Brandt D. R., Pannone K. M., Romano J. J., Casillas E. G. (2013) The synthetic preparation of naturally-occurring aromatase inhibitors, morachalcone A, isogemichalcone B, and isogemichalcone C. Tetrahedron 69, 9994–10002 [Google Scholar]
- 36. Yin S., Fan C. Q., Wang Y., Dong L., Yue J. M. (2004) Antibacterial prenylflavone derivatives from Psoralea corylifolia, and their structure: activity relationship study. Bioorg. Med. Chem. 12, 4387–4392 [DOI] [PubMed] [Google Scholar]
- 37. Hossain M. M., Kawamura Y., Yamashita K., Tsukayama M. (2006) Microwave-assisted regioselective synthesis of natural 6-prenylpolyhydroxyisoflavones and their hydrates with hypervalent iodine reagents. Tetrahedron 62, 8625–8635 [Google Scholar]
- 38. Tsukayama M., Wada H., Kawamura Y., Yamashita K., Nishiuchi M. (2004) Regioselective synthesis of 6-alkyl- and 6-prenylpolyhydroxyisoflavones and 6-alkylcoumaronochromone derivatives. Chem. Pharm. Bull. 52, 1285–1289 [DOI] [PubMed] [Google Scholar]
- 39. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 40. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Q. Q., Gao J., Zhao P. J., Lu S., Zeng Y. (2011) Cloning and expression analysis of flavonoid prenyltransferase-like genes in Epimedium acuminatum Franch. Plant Physiol. J. 47, 575–580 [Google Scholar]
- 42. Vitali A., Ferrari F., Monache G. D., Bombardelli E., Botta B. (2001) Synthesis and biosynthesis of isocordoin. Planta Med. 67, 475–477 [DOI] [PubMed] [Google Scholar]
- 43. Vitali A., Giardina B., Delle Monache G., Rocca F., Silvestrini A., Tafi A., Botta B. (2004) Chalcone dimethylallyltransferase from Morus nigra cell cultures. Substrate specificity studies. FEBS Lett. 557, 33–38 [DOI] [PubMed] [Google Scholar]
- 44. Zhang S. D., Soltis D. E., Yang Y., Li D. Z., Yi T. S. (2011) Multi-gene analysis provides a well-supported phylogeny of Rosales. Mol. Phylogenet. Evol. 60, 21–28 [DOI] [PubMed] [Google Scholar]
- 45. Pichersky E., Gang D. R. (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 5, 439–445 [DOI] [PubMed] [Google Scholar]
- 46. Pichersky E., Lewinsohn E. (2011) Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566 [DOI] [PubMed] [Google Scholar]
- 47. He N., Zhang C., Qi X., Zhao S., Tao Y., Yang G., Lee T. H., Wang X., Cai Q., Li D., Lu M., Liao S., Luo G., He R., Tan X., Xu Y., Li T., Zhao A., Jia L., Fu Q., Zeng Q., Gao C., Ma B., Liang J., Wang X., Shang J., Song P., Wu H., Fan L., Wang Q., Shuai Q., Zhu J., Wei C., Zhu-Salzman K., Jin D., Wang J., Liu T., Yu M., Tang C., Wang Z., Dai F., Chen J., Liu Y., Zhao S., Lin T., Zhang S., Wang J., Wang J., Yang H., Yang G., Wang J. (2013) Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 4, 2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li T., Qi X., Zeng Q., Xiang Z., He N. (2014) MorusDB: a resource for mulberry genomics and genome biology. Database 10.1093/database/bau054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heide L. (2009) Prenyl transfer to aromatic substrates: genetics and enzymology. Curr. Opin. Chem. Biol. 13, 171–179 [DOI] [PubMed] [Google Scholar]
- 50. Tsurumaru Y., Sasaki K., Miyawaki T., Uto Y., Momma T., Umemoto N., Momose M., Yazaki K. (2012) HlPT-1, a membrane-bound prenyltransferase responsible for the biosynthesis of bitter acids in hops. Biochem. Biophys. Res. Commun. 417, 393–398 [DOI] [PubMed] [Google Scholar]
- 51. Page J. E., Boubakir Z. (June 7, 2012) U. S. Patent 8884100 [P]
- 52. Karamat F., Olry A., Munakata R., Koeduka T., Sugiyama A., Paris C., Hehn A., Bourgaud F., Yazaki K. (2014) A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J. 77, 627–638 [DOI] [PubMed] [Google Scholar]
- 53. Munakata R., Inoue T., Koeduka T., Karamat F., Olry A., Sugiyama A., Takanashi K., Dugrand A., Froelicher Y., Tanaka R., Uto Y., Hori H., Azuma J., Hehn A., Bourgaud F., Yazaki K. (2014) Molecular cloning and characterization of a GDP-specific aromatic prenyltransferase from Citrus limon. Plant Physiol. 166, 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen F., Tholl D., D'Auria J. C., Farooq A., Pichersky E., Gershenzon J. (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crowell A. L., Williams D. C., Davis E. M., Wildung M. R., Croteau R. (2002) Molecular cloning and characterization of a new linalool synthase. Arch. Biochem. Biophys. 405, 112–121 [DOI] [PubMed] [Google Scholar]
- 56. Zerbe P., Hamberger B., Yuen M. M., Chiang A., Sandhu H. K., Madilao L. L., Nguyen A., Hamberger B., Bach S. S., Bohlmann J. (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 162, 1073–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamberger B., Ohnishi T., Hamberger B., Séguin A., Bohlmann J. (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 157, 1677–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Veliky I. A., Martin S. M. (1970) A fermenter for plant cell suspension cultures. Can. J. Microbiol. 16, 223–226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.