Background: Genetic defects in the OSTM1 (osteopetrosis-associated transmembrane protein 1) gene cause autosomal recessive osteopetrosis.
Results: The loss of the transmembrane domain in the OSTM1 gene produces a secreted form of truncated OSTM1 that inhibits osteoclast differentiation and survival.
Conclusion: Extracellular secretion of a truncated OSTM1 is negatively involved in osteoclastogenesis.
Significance: We identified a novel function for the secreted form of truncated OSTM1 in osteoclastogenesis.
Keywords: Bone, Cell Differentiation, Osteoblast, Osteoclast, Osteoporosis, BLIMP1, NFATc1, OSTM1, Osteoclastogenesis, Osteopetrosis
Abstract
Genetic mutations in osteoclastogenic genes are closely associated with osteopetrotic bone diseases. Genetic defects in OSTM1 (osteopetrosis-associated transmembrane protein 1) cause autosomal recessive osteopetrosis in humans. In particular, OSTM1 mutations that exclude the transmembrane domain might lead to the production of a secreted form of truncated OSTM1. However, the precise role of the secreted form of truncated OSTM1 remains unknown. In this study, we analyzed the functional role of truncated OSTM1 in osteoclastogenesis. Here, we showed that a secreted form of truncated OSTM1 binds to the cell surface of osteoclast (OC) precursors and inhibits the formation of multinucleated OCs through the reduction of cell fusion and survival. Truncated OSTM1 significantly inhibited the expression of OC marker genes through the down-regulation of the BLIMP1 (B lymphocyte-induced maturation protein 1)-NFATc1 (nuclear factor of activated T cells c1) axis. Finally, we demonstrated that truncated OSTM1 reduces lipopolysaccharide-induced bone destruction in vivo. Thus, these findings suggest that autosomal recessive osteopetrosis patients with an OSTM1 gene mutation lacking the transmembrane domain produce a secreted form of truncated OSTM1 that inhibits osteoclastogenesis.
Introduction
Osteoclasts (OCs)3 are clinically important cells in bone-related diseases, such as osteopetrosis and osteoporosis; a loss of function of the OCs leads to osteopetrosis, and accelerated bone destruction by OCs results in osteoporosis (1). OCs are multinucleated giant cells derived from monocyte/macrophage lineage precursors through OC differentiation or osteoclastogenesis (2, 3). Macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor κB (RANK) ligand (RANKL), produced by bone-forming osteoblasts (OBs) or immune cells, are key signaling cytokines in osteoclastogenesis (2, 3). M-CSF binds to its receptor c-Fms (M-CSF receptor) to activate downstream signaling pathways, such as Akt and ERK (2, 3). Upon RANKL stimulation, RANK recruits TNF receptor-associated factor 6 (TRAF6), a key adaptor molecule in the RANK-signaling complex, which activates downstream targets of p38, ERK, JNK, Akt, nuclear factor κB (NF-κB), c-Fos, cAMP-response element-binding protein (CREB), and NFATc1 (nuclear factor of activated T cells c1) (2, 3). The final consequence of these signaling pathways is the induction of osteoclastogenic genes, such as TRAP (tartrate-resistant acid phosphatase), cathepsin K, DC-STAMP (dendritic cell-specific transmembrane protein), Atp6v0d2 (V-ATPase subunit d2), Blimp1 (B lymphocyte-induced maturation protein 1), and OSCAR (OC-associated receptor) (2, 3).
The murine Ostm1 (osteopetrosis-associated transmembrane protein 1) gene was first identified in the spontaneous gray-lethal (gl/gl) mouse mutant, which exhibits neurological defects, melanocyte maturation problems, and an osteopetrotic phenotype (4–6). The murine Ostm1 gene encodes a 338-amino acid protein (4). OSTM1 is a type I transmembrane protein containing a signal peptide, a potential transmembrane domain, and a glycosylated extracellular domain with a RING finger domain. Interestingly, a recent report suggests that OSTM1 functions as a β-subunit of ClC-7 to support bone resorption and lysosomal function in OCs (7). Furthermore, the multifunctional roles for OSTM1 as an E3 ubiquitin ligase or a modulator for Wnt/β-catenin signaling have been reported (8, 9).
The gl/gl mutation comprises a genomic deletion encompassing the 5′ region of the Ostm1 gene, including the promoter, the first exon, and a large portion of the first intron, resulting in a null phenotype with an absence of transcription and protein expression (4). In gl/gl mice, an increase in the number of functionally inactive OCs resulting from a defect in cytoskeletal rearrangement during late stage OC maturation has been reported (10, 11). However, a decrease in OC numbers was observed in a human autosomal recessive osteopetrosis (ARO) patient exhibiting a substitution at the donor splice site of intron V, resulting in the production of 261 residues of a truncated OSTM1 protein lacking the transmembrane domain (10, 11). In contrast to the morphology of OCs in gl/gl mice, OCs with an irregularly elongated shape were observed in a histological analysis from an ARO patient (10). In addition, the potential extracellular secretion of this truncated protein has been suggested (7), but the precise role of this secreted mutant protein in ARO patients remains unknown. Hence, it would be interesting to explore the functional relevance of the truncated OSTM1 mutant to OC differentiation or function.
In the present study, we analyzed the functional role of truncated OSTM1, a putative secreted form reflecting a substitution at the donor splice site of intron V of the OSTM1 gene in ARO patients, in osteoclastogenesis. Here we report that the secreted form of truncated OSTM1 mutant is negatively involved in OC differentiation through the down-regulation of the BLIMP1-NFATc1 axis.
EXPERIMENTAL PROCEDURES
Reagents, Cells, Mice, and Plasmids
Antibodies against NFATc1, OSTM1, c-Fos, and TRAF6 and mouse IgG were purchased from Santa Cruz Biotechnology, Inc. Antibodies against FLAG epitope and β-actin were purchased from Sigma-Aldrich. Antibodies against phospho-p65, p65, phospho-p38, p38, phospho-Akt, Akt, phospho-PLC-γ, PLC-γ, phospho-ERK, ERK, phospho-JNK, JNK, phospho-CREB, and CREB were purchased from Cell Signaling Technology (Beverly, MA). The construction of recombinant human soluble RANKL and human M-CSF has been previously described (12). Vitamin D3 (VtD3) and prostaglandin E2 (PGE2) were purchased from Wako Chemical (Osaka, Japan). General chemicals were purchased from Sigma-Aldrich. The Raw264.7 (murine monocytic cell line), NIH3T3 (murine fibroblastic cell line), 293T (human kidney cell line), KMls-8.3.5.1 (murine T-cell hybridoma), S2 (Drosophila cells), MC3T3-E1 (murine osteoblastic cell line), and UAMS32 (murine osteoblastic cell line) cell lines and primary OBs (pOBs) were prepared and maintained as described previously (12–14). The mice were purchased from Daehan Biolink Co. (Umsung, Korea), and the animal study was approved (approval no. CNU-00114) through the Animal Experiment Ethics Committee of Chungnam National University. For the retroviral expression plasmids, the full-length ORF (aa 1–338, pMX-puro-OSTM1) and truncated form (aa 1–268, pMX-puro-OSTM1ΔTM) of the murine Ostm1 gene were amplified through RT-PCR and subcloned into retroviral vector pMX-puro-FLAG (13). Retroviral plasmids expressing the constitutively active form of NFATc1 (caNFATc1), c-Fos, and BLIMP1 were prepared as described previously (12, 15, 16). For protein expression in Drosophila S2 cells, the truncated OSTM1 (aa 35–268, OSTM1ΔTM) was PCR-amplified, fused in frame to the hFc, and subcloned into the pCMV1-FLAG vector (Sigma-Aldrich) harboring the leader sequence tagged with FLAG epitope. The secreted form of OSTM1 (OSTM1ΔTM-hFc) was subcloned into the metal-inducible expression vector pMT/V5-HisA (Invitrogen) through PCR amplification. The following primers were used for the plasmid construction: OSTM1 (sense), 5′-CCC GGA TCC ATG GCT CGG GAC GCG GAG CTG GCG-3′; OSTM1 (antisense), 5′-CCC CTC GAG CTA CTT GTC GTC ATC GTC TTT GTA GTC GGT GGC ATT TTC TTG AAT GT-3′; OSTM1-ΔTM (antisense), 5′-CCC CTC GAG CTA CTT GTC GTC ATC GTC TTT GTA GTC AAT GTT CAT TGC ATC CTC CAC ATC-3′; OSTM1-ΔTM-hFc (sense), 5′-CCC AAG CTT GCG CTC CCC TTC ACC AGC AG-3′; OSTM1-ΔTM-hFc (antisense), 5′-CCC AGA TCT CGT GTC GCT GCA GGT GAC CG-3′.
Osteoclastogenesis
Bone marrow (BM)-derived macrophages (BMMs) and BM-derived OCs (BMOCs) were prepared as described previously (13). Briefly, BM cells were collected from the tibia and femur of 6–8-week-old C57BL/6 male mice and cultured in α-minimal essential medium containing 10% FBS and 5 ng/ml of M-CSF overnight. The non-adherent BM cells were used in subsequent assays. For the differentiation of BMMs, the non-adherent BM cells were seeded at 1 × 105 cells/well in 96-well plates and further cultured in α-minimal essential medium containing 10% FBS and M-CSF (50 ng/ml) for 3 days. For the BMOC formation assay, non-adherent BM cells were seeded at 1 × 105 cells/well in 96-well plates and further cultured with M-CSF (50 ng/ml) and RANKL (200 ng/ml) for 4 days. For the Raw264.7-derived OC-like cell formation assay, Raw264.7 cells were seeded at 2 × 103 cells/well in 96-well plates and further cultured with RANKL (200 ng/ml) for 4 days. For the co-culture assay, non-adherent BM cells (1 × 105 cells/well) were seeded with UAMS32 cells (5 × 103 cells/well) in 96-well plates and cultured with VtD3 (10−8 m) and PGE2 (10−6 m) for 5–6 days. The OC differentiation assay with transgene expression through retroviral infection has been described previously (12). Briefly, retrovirus-transduced BMMs were cultured with M-CSF (150 ng/ml) and puromycin (2 μg/ml) for 2 days. Puromycin-resistant BMMs (1.5 × 104 cells/well) were further cultured with M-CSF (50 ng/ml) and RANKL (200 ng/ml) in 96-well plates for 4 days. The TRAP-positive (TRAP+) multinucleated OCs (TRAP+ MNCs) were analyzed as described below.
Analysis of OCs
OCs were analyzed as described previously (13). Briefly, OCs were fixed with 10% formalin for 10 min and permeabilized in an ethanol/acetone (1:1) mixture for 1 min. For the TRAP solution assay, the OCs were incubated with p-nitrophenyl phosphate substrate for 30 min, and the TRAP activities were analyzed at 405 nm using a microplate reader (Bio-Rad). For the TRAP staining, the OCs used in the TRAP solution assay were further washed twice with PBS and incubated with fast red violet containing naphthol AS-phosphate at room temperature for 30 min. The stained OCs were photographed under a microscope. The TRAP+ MNCs with more than three nuclei were considered as multinucleated OCs. The immunoblotting and Northern blot analyses for OC differentiation were performed as described previously (17). The analyses for gene expression through real-time PCR were performed as described previously (13). β-Actin expression served as an internal control. The primers used for real-time PCR amplification were as follows: cathepsin K (sense), 5′-ACG GAG GCA TTG ACT CTG AAG ATG-3′; cathepsin K (antisense), 5′-GGA AGC ACC AAC GAG AGG AGA AAT-3′, DC-STAMP (sense), 5′-TCC TCC ATG AAC AAA CAG TTC CAA-3′; DC-STAMP (antisense), 5′-AGA CGT GGT TTA GGA ATG CAG CTC-3′, Atp6v0d2 (sense), 5′-TTC AGT TGC TAT CCA GGA CTC GGA-3′; Atp6v0d2 (antisense), 5′-GCA TGT CAT GTA GGT GAG AAA TGT GCT CA-3′, TRAP (sense), 5′-AAA TCA CTC TTC AAG ACC AG-3′; TRAP (antisense), 5′-TTA TTG AAC AGC AGT GAC AG-3′, OSCAR (sense), 5′-TCT GCC CCC TAT GTG CTA TCA-3′; OSCAR (antisense), 5′-AGG AGC CAG AAC CTT CGA AAC-3′, BLIMP1 (sense), 5′-TTC TTG TGT GGT ATT GTC GGG ACT T-3′; BLIMP1 (antisense), 5′-TTG GGG ACA CTC TTT GGG TAG AGT T-3′, OSTM1 (sense), 5′-ACA AGA CTT TGG CCT GCT TTG AGC-3′; OSTM1 (antisense), 5′-C TCB AAG CCC GTT CAG TTT CTG CAT-3′.
Osteoblast Differentiation and Activation
pOB progenitors were isolated from the calvarial bone of newborn mice and cultured in α-minimal essential medium containing 10% FBS, 10 mm β-glycerophosphate, 50 μm ascorbic acid 2-phosphate, and 10 nm dexamethasone as described previously (12, 13). An alkaline phosphatase assay was performed after 3–7 days of culture as described previously (12, 18). An alizarin red S assay was performed after 14–21 days of culture as described previously (12, 18).
Cellular Toxicity Test
For the cellular toxicity test of purified proteins, NIH3T3 cells were cultured with 25 μg/ml of the purified recombinant proteins for 4 days. Subsequently, cell viability was tested using the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan). The CCK-8 activity was measured at 450 nm using a microplate reader.
Cell Fusion and Survival Assay
The cell fusion assay was performed as described previously (13). Briefly, BM cells (2 × 106 cells/ml) were co-cultured on UAMS32 (2 × 105 cells/ml) with VtD3 (10−8 m) and PGE2 (10−6 m) for 6 days to prepare mononuclear TRAP+ OC precursors (preOCs). Subsequently, the preOCs were collected through gentle pipetting. To induce cell-cell fusion, purified preOCs (5 × 105 cells/ml) were seeded in 96-well plates and cultured with M-CSF (50 ng/ml) and RANKL (200 ng/ml) for 24 h in the presence of hFc (25 μg/ml) or OSTM1ΔTM-hFc (25 μg/ml). The OCs formed after cell fusion were analyzed through TRAP staining. The OC survival assay was performed as described previously (17). Briefly, BMOCs were prepared from BM cells after culturing with RANKL (200 ng/ml) and M-CSF (50 ng/ml) for 4 days. To induce the apoptosis of BMOCs, OCs were further cultured without a fresh medium change and cytokine supplementation for 1–3 days. Finally, the surviving OCs were analyzed through TRAP staining.
Secretion Analysis of Truncated OSTM1
NIH3T3 and 293T cells were seeded onto 6-well plates at 1 × 105 cells/well and respectively transfected with 5 μg of pMX-puro-OSTM1 (OSTM1), pMX-puro-OSTM1ΔTM (OSTM1ΔTM), and pMX-puro-FLAG vector using TurboFect transfection reagent (Fermentas). Retrovirus-transduced BMOCs or BMMs expressing the truncated form of OSTM1 (OSTM1ΔTM) were prepared as described above. To collect the secreted proteins, the culture medium was changed to serum-free medium at 16 h after transfection or retroviral infection, and the supernatant was collected 48 h later. The supernatants were precipitated with 10% TCA at 4 °C for 16 h, followed by centrifugation at 1,500 × g for 15 min. The pellets were washed twice with 1 ml of ice-cold acetone and dissolved in loading buffer for SDS-PAGE. The secreted form of OSTM1 was detected through immunoblotting using an anti-FLAG antibody.
Protein Purification
The truncated OSTM1 fused to the hFc domain (OSTM1ΔTM-hFc) was prepared as described previously (19). Briefly, S2-stable cell lines expressing OSTM1ΔTM-hFc were prepared using a Drosophila expression system (Invitrogen), according to the manufacturer's instructions. The hFc fusion protein was purified using a protein A affinity column (Invitrogen), according to the manufacturer's instructions. The purified proteins were subsequently dialyzed in PBS, filtrated through a 0.22-μm membrane, and stored at −70 °C. The hFc protein alone was also generated as a control. The purity of the protein preparation was determined using SDS-PAGE and the Agilent 230 kit assay (Agilent Technologies, Santa Clara, CA). The purified proteins were tested for endotoxin contamination using the limulus amebocyte lysate test kit (Associates of Cape Cod, East Falmouth, MA). To further test endotoxin contamination, the purified proteins were heat-inactivated as described previously (20).
Flow Cytometric Analysis
The cells were seeded at 5 × 103 cells/well onto 24-well plates and cultured for 12 h. The cells were detached through gentle pipetting in cell dissociation solution (Sigma-Aldrich), followed by incubation with purified hFc or OSTM1-ΔTM-hFc (25 μg/ml) for 2 h. The treated cells were subsequently stained with an anti-FLAG antibody (100 ng/ml) at 4 °C for 20 min. After two washing steps with 1 ml of PBS containing 2% FBS, the cells were incubated at 4 °C for 20 min with PE-conjugated goat anti-mouse IgG secondary antibody. The stained cells were analyzed through flow cytometry using a FACSCalibur flow cytometer with CellQuest software (BD Biosciences).
Cell Lines, Culture Supernatant Transfer, Direct Co-culture, and Neutralizing Antibody Treatment
For the construction of stable osteoblastic cell lines, the DNA from the OSTM1 expression plasmid (20 μg) was introduced into UAMS32 cells (5 × 106 cells/ml) through electroporation (600 V and 50 microfarads) using a Gene Pulser Xcell (Bio-Rad). The cells were selected using puromycin (2.5 μg/ml) for 14 days. The puromycin-resistant cells were separated in 96-well plates through serial dilution to isolate individual stable cell lines. The secreted protein was analyzed using TCA precipitation. For the transfer assay, non-adherent BM cells (1 × 105 cells/well) were co-cultured with pOBs (5 × 103 cells/well) in 96-well plates containing VtD3 (10−8 m) and PGE2 (10−6 m). During co-culture, the culture supernatants derived from the UAMS32-stable cell lines expressing OSTM1 or OSTM1ΔTM were transferred daily into the co-culture wells with VtD3 (10−8 m) and PGE2 (10−6 m) for 5–6 days. For the direct co-culture assay, non-adherent BM cells (1 × 105 cells/well) were seeded directly onto UAMS32-stable cell lines (1 × 104 cells/well) expressing OSTM1 or OSTM1ΔTM, instead of pOBs, in 96-well plates and cultured with VtD3 (10−8 m) and PGE2 (10−6 m) for 5–6 days. For the neutralizing antibody treatment assay, polyclonal anti-OSTM1 antibodies (4 μg/ml) were dialyzed and added directly to the wells on the direct co-culture plate for 5–6 days. The human IgG antibody (Sigma-Aldrich) and PBS were used as controls. The OC formation was analyzed using TRAP assays. For the construction of the stable T-cell hybridoma line, the murine T cell hybridoma cell line KMls-8.3.5.1 was infected with retroviral supernatants and selected using puromycin (2.0 μg/ml) for 14 days.
LPS-induced Osteoclastogenesis and Bone Destruction
LPS-induced osteoclastogenesis was performed as described previously (21). Briefly, BMMs (1.5 ×104 cells/ml) were pretreated with RANKL (100 ng/ml) and M-CSF (50 ng/ml) for 1 day. BMMs were further cultured with M-CSF (50 ng/ml) and LPS (10 μg/ml) for 4 days in the presence or absence of hFc or OSTM1-ΔTM-hFc (25 μg/ml). For the LPS-induced bone destruction, a 150-μl dose of LPS (25 mg/kg body weight) was injected into the subcutaneous tissue overlying the periosteum of 7-week-old mouse calvaria. One day later, OSTM1-ΔTM-hFc (2 mg/kg body weight) or PBS control was injected daily for 4 days. Whole calvaria was fixed with 4% paraformaldehyde and stained using the TRAP assay. The bone mineral density unit was measured and calculated using the microcomputed tomography system (GE Healthcare). The calvarial bones were decalcified in 15% EDTA solution at 4 °C for 21 days, followed by dissection and H&E staining. The TRAP+ MNCs were visualized by TRAP staining and counterstained with hematoxylin.
Statistical Analysis
All experiments were performed at least three times. The data represent the means ± S.D. (n = 3/group). Student's t test was used to determine the significance of the differences between the experimental samples.
RESULTS
The Truncated OSTM1 Mutation Is Negatively Involved in Osteoclastogenesis
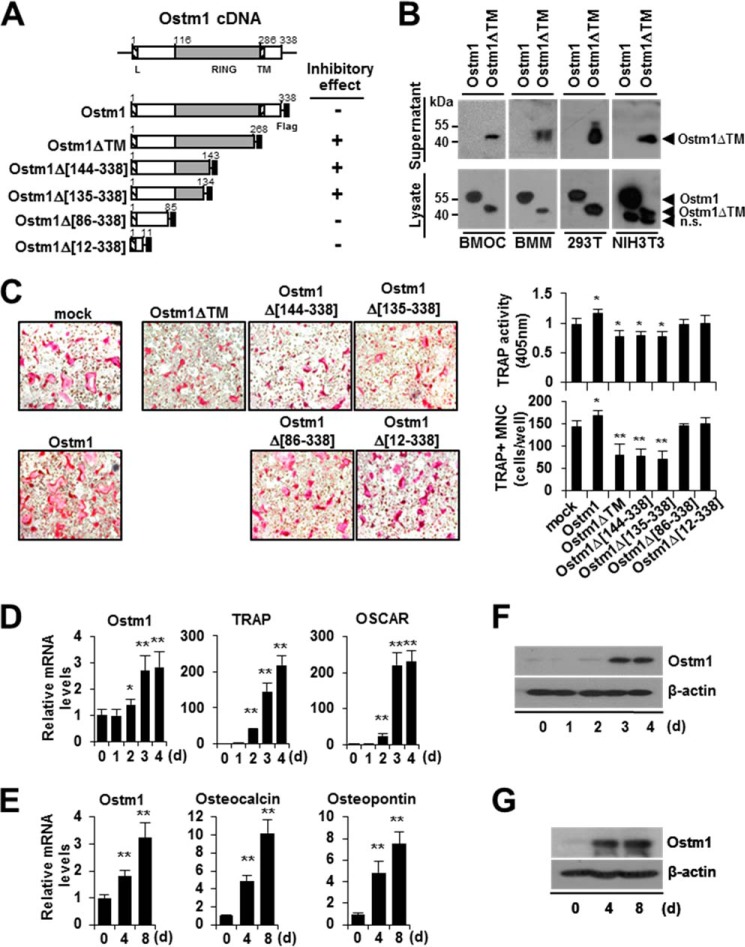
To examine the extracellular secretion of the truncated OSTM1 (OSTM1ΔTM) in BMOCs, we constructed a retroviral plasmid expressing truncated OSTM1 (aa 1–268, which contains three additional residues at the C-terminal region compared with the mutant form created by a substitution at the donor splice site of intron V of the OSTM1 gene in a human ARO patient) lacking the C-terminal 70 amino acid residues that comprise the transmembrane domain and cytoplasmic tail (Fig. 1A). The culture supernatants derived from OSTM1ΔTM-transduced BMOCs were prepared and analyzed by immunoblotting with an anti-FLAG antibody. We observed that a protein band of ∼44 kDa, corresponding to truncated OSTM1, was detected in the culture supernatants of OSTM1ΔTM-transduced BMOCs (Fig. 1B). Similar results were obtained in BMMs and 293T and NIH3T3 cell lines (Fig. 1B). We analyzed whether the expression of OSTM1ΔTM is involved in osteoclastogenesis. BMOC differentiation was reduced (21–45% reduction) compared with the vector control (mock) in both TRAP solution and the TRAP staining assay in response to truncated OSTM1 (OSTM1ΔTM) expression (Fig. 1C). In addition, we observed that the inhibitory function of truncated OSTM1 in OC differentiation was retained in the RING domain of OSTM1 (Fig. 1, A and C). Next, we analyzed whether the expression of Ostm1 is regulated during OC or OB differentiation. The expression of Ostm1 was induced 1.8–3.2-fold during OC or OB differentiation, but the -fold induction was significantly lower than the expression of OC or OB marker genes (Fig. 1, D and E). Similar results were obtained by immunoblotting analysis (Fig. 1, F and G). Hence, these results indicate that the truncated form of OSTM1, secreted from OCs or OBs, is negatively involved in OC differentiation through its extracellular inhibitory function.
FIGURE 1.
Negative effects of the truncated OSTM1 mutant on osteoclastogenesis. A, schematic illustration of the murine Ostm1 cDNA structure and the OSTM1 expression plasmids. The putative RING domain (gray box) and leader/transmembrane sequences (hatched box) are indicated with amino acid residues. Flag, FLAG epitope (black box). The inhibitory effects of OSTM1 deletion mutants in osteoclastogenesis are summarized. B, extracellular secretion of truncated OSTM1 in BMOCs, BMMs, 293T, and NIH3T3 cells. Extracellular secretion of FLAG-tagged OSTM1ΔTM from culture supernatants of cells was detected through immunoblotting with antibodies against the FLAG epitope. n.s., nonspecific band. C, effect of truncated OSTM1 overexpression on OC differentiation. BMMs transduced with retroviral supernatants expressing OSTM1 or OSTM1 truncated mutants were cultured with RANKL and M-CSF. OCs were analyzed and photographed (left; original magnification, ×100) after TRAP staining. The TRAP solution assay was performed (top right panel). The number of TRAP+ MNCs (≥3 nuclei) was counted (bottom right panel). D, analysis of Ostm1 gene expression in BMOCs through real-time PCR. BM cells were differentiated into BMOCs using both M-CSF and RANKL stimulation. β-Actin expression served as an internal control. TRAP, tartrate-resistant acid phosphatase; OSCAR, OC-associated receptor. E, analysis of Ostm1 gene expression in pOBs through real-time PCR. Murine calvaria-derived stromal cells were differentiated into OBs for 8 days. F, immunoblot analysis of Ostm1 gene expression in BMOCs. BMOCs were prepared and analyzed by immunoblotting with specific antibodies. β-Actin was used as the loading control. G, immunoblot analysis of Ostm1 gene expression in pOBs. *, p < 0.05; **, p < 0.01. Error bars, S.D.
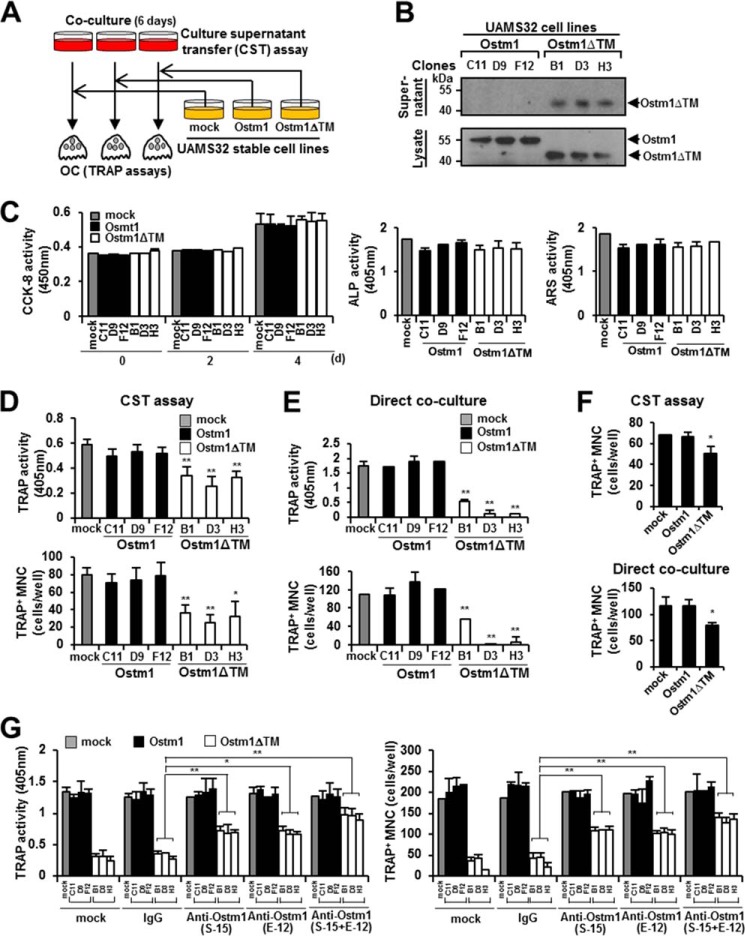
To address this possibility, we designed a strategy to test the functional role of the truncated OSTM1 in osteoclastogenesis (Fig. 2A). We thus generated a stable UAMS32 cell line expressing OSTM1 or OSTM1ΔTM based on a retroviral expression system through puromycin selection. From the puromycin-resistant stable cell line pools, we isolated three individual stable clones. The secreted form of truncated OSTM1 was observed in the individual clones of the UAMS32-OSTM1ΔTM cell line (clones B1, D3, and H3), but not in the UAMS32-OSTM1 cell line (clones C11, D9, and F12) (Fig. 2B). These individual clones were phenotypically normal in osteoblastic functions, such as cell proliferation (CCK-8 assay), OB differentiation (alkaline phosphatase assay), and bone forming activity (alizarin red S assay), compared with the parental UAMS32 cell line (Fig. 2C). Next, we performed a transfer assay using the culture supernatants derived from the UAMS32-stable clones in a co-culture of pOBs and BM cells, as shown in Fig. 2A. We observed that OC differentiation was significantly reduced through the daily transfer of the culture supernatants from the UAMS32-OSTM1ΔTM clones, whereas the transfer of the culture supernatants from UAMS32-OSTM1 clones did not show any effect (Fig. 2D). Furthermore, we observed stronger inhibitory effects in the direct co-culture of UAMS32-OSTM1ΔTM clones and BM cells (Fig. 2E).
FIGURE 2.
Inhibition of OC differentiation, fusion, and survival by the truncated OSTM1 treatment. A, schematic diagram of the culture supernatant transfer assay. B, extracellular secretion of truncated OSTM1 in UAMS32-stable cell lines. C, effects of truncated OSTM1 on OB proliferation, differentiation, and activation. Shown is a cell proliferation assay of the individual cells of UAMS32-stable cell lines expressing OSTM1 or OSTM1ΔTM (left). UAMS32-stable cell lines were cultured with VtD3 (10−8 m) and PGE2 (10−6 m) for 4 days. Cell viability was measured using the CCK-8 assay. Gray box, mock; black box, OSTM1-stable cell lines; open box, OSTM1ΔTM-stable cell lines. The differentiation of OBs was measured using the alkaline phosphatase assay (middle). The formation of mineralized nodules in OBs was measured using the alizarin red S assay (right). D, inhibitory effects of the truncated OSTM1 derived from UAMS32-stable cell lines in OC differentiation using the culture supernatant transfer assay. E, inhibitory effects of the truncated OSTM1 derived from UAMS32-stable cell lines on OC differentiation detected using a direct co-culture assay. F, inhibitory effects in OC differentiation through culture supernatants derived from KMls-8.3.5.1 cell lines expressing OSTM1ΔTM. The top panel shows results for the culture supernatant transfer assay. The bottom panel shows results for the direct co-culture assay. G, rescue experiment using an OSTM1-neutralizing antibody to assess the inhibitory effect of the truncated OSTM1 on osteoclastogenesis. Anti-OSTM1-neutralizing antibodies (clone E-12 or S-15) or antibody mixtures (E-12 + S-15, 1:1 ratio) were added to the direct co-culture plate with non-adherent BM cells and UAMS32-stable cell lines. The human IgG (IgG) antibody (Sigma-Aldrich) and PBS were used as controls. *, p < 0.05; **, p < 0.01. Error bars, S.D.
To exclude cell line-specific effects, we established a stable T-cell hybridoma cell line (KMls-8.3.5.1) expressing OSTM1 or OSTM1ΔTM. Consistent with the results for UAMS32-OSTM1ΔTM clones, we observed significantly reduced OC differentiation in KMls-8.3.5.1-OSTM1ΔTM-stable cell lines in both transfer and direct co-culture assays (Fig. 2F). Furthermore, we conducted an OSTM1-neutralizing antibody treatment assay to exclude the effects of other secreted cellular factors derived from UAMS32-OSTM1ΔTM clones. In the direct co-culture assay, the inhibition of OC differentiation by truncated OSTM1 was significantly rescued after treatment with anti-OSTM1-neutralizing antibodies (clone S-15 or E-12) or antibody mixtures (S-15 and E-12 (1:1 ratio)), whereas treatment with PBS or IgG controls did not produce this effect (Fig. 2G). Hence, these results indicate that osteoclastogenesis is inhibited through the extracellular secretion of truncated OSTM1.
Inhibition of OC Differentiation, Fusion, and Survival through Truncated OSTM1
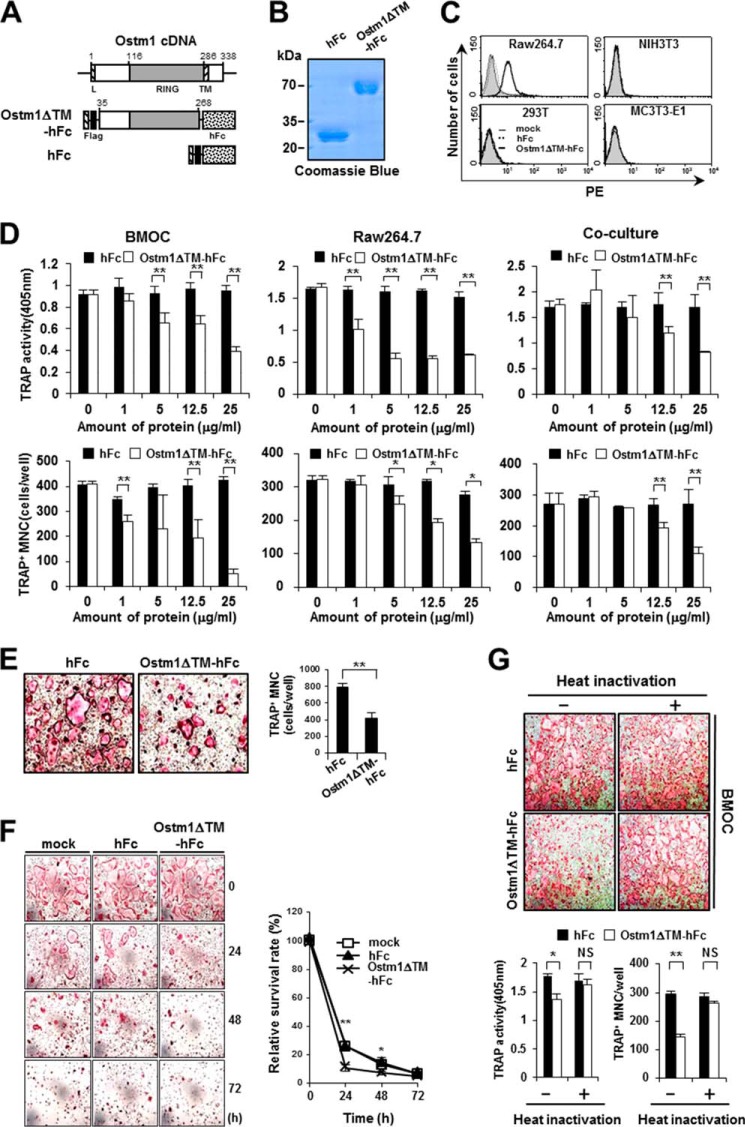
To further address the precise role of truncated OSTM1 in osteoclastogenesis, we purified the truncated OSTM1-hFc fusion protein (OSTM1ΔTM-hFc) or hFc alone from the culture supernatants of S2-stable cell lines grown in spinner cultures (Fig. 3, A and B). The purity of the protein preparations was >98%, as determined by an Agilent 230 kit assay (data not shown). We did not observe any cytotoxicity from the purified proteins in NIH3T3 cells after 4 days of culture (data not shown).
FIGURE 3.
Inhibition of OC differentiation, fusion, and survival by the truncated OSTM1 treatment. A, schematic illustration of the murine Ostm1 cDNA structure and the OSTM1 expression plasmids. Black box, FLAG epitope (Flag); dotted box, Fc fragment of human IgG1 (hFc). B, purification of recombinant OSTM1ΔTM-hFc protein. C, cell surface binding assay with the OSTM1ΔTM-hFc protein. The cell surface binding of OSTM1ΔTM-hFc (thick solid lines) or hFc protein (dotted lines) in various cells was determined through flow cytometry using a mouse anti-FLAG primary antibody and a PE-labeled anti-mouse IgG secondary antibody. Cells treated with the PE-labeled anti-mouse IgG secondary antibody alone were used as a control (mock; gray with thin solid lines). D, inhibitory effects of truncated OSTM1 (OSTM1ΔTM-hFc) on OC differentiation. The number of TRAP+ MNCs (≥3 nuclei) was counted from the BMOC formation assay (left panels), the Raw264.7-derived OC-like cell formation assay (middle panels), and the co-culture assay (right panels). E, OSTM1ΔTM-hFc inhibits the fusion of preOCs. The fusion of TRAP+ mononuclear preOCs was monitored through TRAP staining (left panels; original magnification, ×100). The number of MNCs derived from the fusion of preOCs was counted (right panel). F, OSTM1ΔTM-hFc inhibits the survival of OCs. Apoptosis of BMOCs was induced through the depletion of RANKL and M-CSF for the indicated times. The survival of OCs was monitored through TRAP staining (left panels; original magnification, ×100). TRAP+ MNCs (≥3 nuclei) were counted, and the relative survival rate of OCs after OSTM1ΔTM-hFc treatment was calculated (right). G, effects of heat inactivation of the truncated OSTM1 on OC differentiation. BM cells were differentiated into BMOCs using M-CSF and RANKL in the presence of heat-inactivated proteins (12.5 μg/ml). OCs were analyzed and photographed (top panels; original magnification, ×100) after TRAP staining. The TRAP solution assay was performed (bottom left). The number of TRAP+ MNCs (≥3 nuclei) was counted (bottom right). NS, not significant. *, p < 0.05; **, p < 0.01. Error bars, S.D.
Thus, we examined whether purified OSTM1ΔTM-hFc could bind to the cell surface of preOCs. The surface attachment of purified OSTM1ΔTM-hFc was detected predominantly on Raw264.7 cells but not on NIH3T3, 293T, or MC3T3-E1 cells (Fig. 3C). Raw264.7 cells are well known preOCs that easily differentiate into OC-like cells in response to RANKL stimulation in vitro (3). Hence, we postulate that the attachment of the secreted OSTM1 mutant to the cell surface of preOCs is negatively involved in OC differentiation. Therefore, we determined whether the process of osteoclastogenesis could be inhibited through truncated OSTM1 using three standard in vitro culture methods. In the BMOC formation assay, OSTM1ΔTM-hFc-treated cells showed a marked reduction (26.3–88.2%) in the formation of TRAP+ MNCs in a dose-dependent manner compared with hFc-treated control cells (Fig. 3D, left). Similarly, we observed reduced TRAP+ MNC formation after OSTM1ΔTM-hFc treatment in Raw264.7-derived OC-like cell formation (Fig. 3D, middle) and co-culture assays (Fig. 3D, right). Next, we examined the effect of OSTM1ΔTM-hFc treatment on OC fusion and survival. We observed that the fusion of preOCs was significantly reduced following OSTM1ΔTM-hFc treatment (Fig. 3E). Furthermore, OC survival was also reduced after OSTM1ΔTM-hFc treatment (Fig. 3F). However, the bone-resorbing activities of purified mature OCs were not changed after OSTM1ΔTM-hFc treatment in the presence of M-CSF and RANKL (data not shown).
To exclude the possibility of endotoxin contamination in the purified proteins, we performed a limulus amebocyte lysate assay. The endotoxin contamination in 25 μg of recombinant proteins was below the detectable level of 0.125 endotoxin units/ml. The endotoxin contamination was further examined using the heat inactivation approach (20). When the purified proteins were denatured through heat inactivation, treatment with denatured OSTM1ΔTM-hFc did not inhibit osteoclastogenesis (Fig. 3G), indicating that the inhibition of osteoclastogenesis resulted from OSTM1ΔTM-hFc treatment and not endotoxin contamination during protein preparation. Taken together, these results suggest that truncated OSTM1 is negatively involved in OC differentiation, fusion, and survival in osteoclastogenesis but does not affect the bone-resorbing functions of the already formed mature OCs.
Truncated OSTM1 Inhibits the Expression of OC Markers through the Down-regulation of BLIMP1 and NFATc1
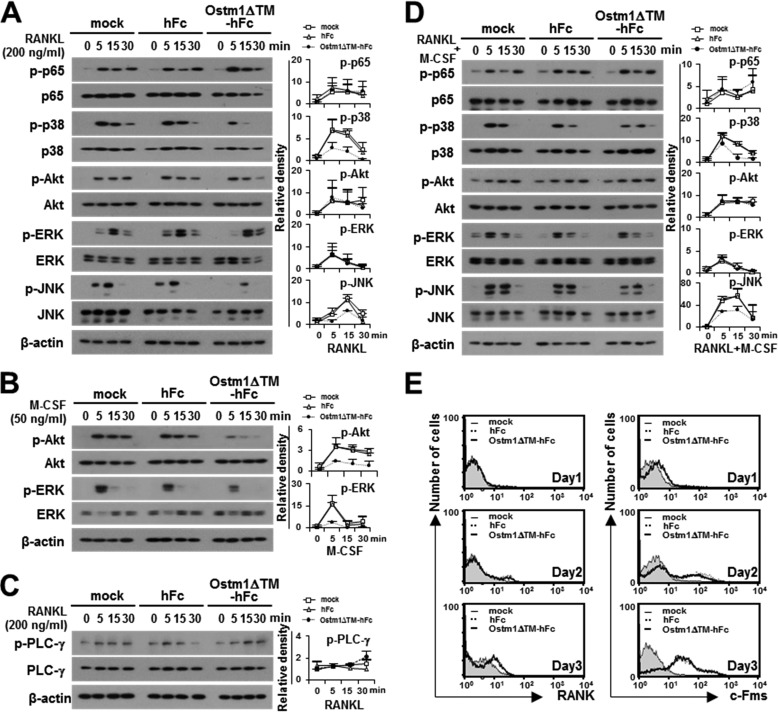
Activation of NF-κB and MAP kinases is crucial for OC differentiation, maturation, and survival (1–3). Thus, we examined whether truncated OSTM1 is involved in the modulation of the RANKL- and M-CSF-mediated signaling pathway in osteoclastogenesis. We observed that both RANKL-induced p38 and JNK activation and M-CSF-induced Akt and ERK activation were significantly reduced in OSTM1ΔTM-hFc-treated BMMs compared with the hFc-treated control cells (Fig. 4, A and B); however, RANKL-mediated costimulatory signaling through the phosphorylation of PLC-γ was not changed in OSTM1ΔTM-hFc-treated BMMs (Fig. 4C). These results indicate that a secreted form of the truncated OSTM1 may lead to distinct negative signals for both M-CSF and RANKL activation. Thus, we tested the effect of OSTM1ΔTM-hFc treatment on the stimulation of both RANKL and M-CSF. The phosphorylation of p38 and JNK by both RANKL and M-CSF stimulation was significantly reduced in OSTM1ΔTM-hFc-treated BMMs compared with the hFc-treated control cells (Fig. 4D). Furthermore, the surface expression of RANK and c-Fms (M-CSF receptor) was not affected after OSTM1ΔTM-hFc treatment (Fig. 4E).
FIGURE 4.
The effect of the truncated OSTM1 on the signaling pathways involved in OC differentiation. A, effect of OSTM1ΔTM-hFc treatment on RANKL-induced signaling. BMMs pretreated with purified proteins (25 μg/ml) for 2 h. The pretreated BMMs were further stimulated with RANKL for the indicated times. The cells were then analyzed through immunoblotting with antibodies recognizing phosphorylated and total p65, p38, Akt, ERK, and JNK. β-Actin was used as the loading control. The relative level of phosphorylated forms was calculated after normalization to total protein input (right). B, effect of OSTM1ΔTM-hFc treatment on M-CSF-induced signaling. The M-CSF-induced signals were analyzed through immunoblotting with antibodies recognizing phosphorylated and total Akt and ERK. The relative level of the phosphorylated form was calculated after normalization to total protein input (right). C, effect of OSTM1ΔTM-hFc treatment on RANKL-induced co-stimulatory signaling. The RANKL-induced co-stimulatory signals were analyzed through immunoblotting with antibodies recognizing phosphorylated and total PLC-γ. The relative level of the phosphorylated form was calculated after normalization to total protein input (right). D, effect of OSTM1ΔTM-hFc treatment on the stimulation of both RANKL and M-CSF. The relative level of the phosphorylated form was calculated after normalization to total protein input (right). E, expression of RANK and c-Fms on the surface of OCs in response to OSTM1ΔTM-hFc treatment. BM cells were differentiated into BMOCs using M-CSF (50 ng/ml) and RANKL (200 ng/ml) for 1–3 days. The cells were stained with antibodies against RANK (left panels) or c-Fms (right panels). Gray with thin solid lines, mock; dotted lines, hFc; thick solid lines, OSTM1ΔTM-hFc. Error bars, S.D.
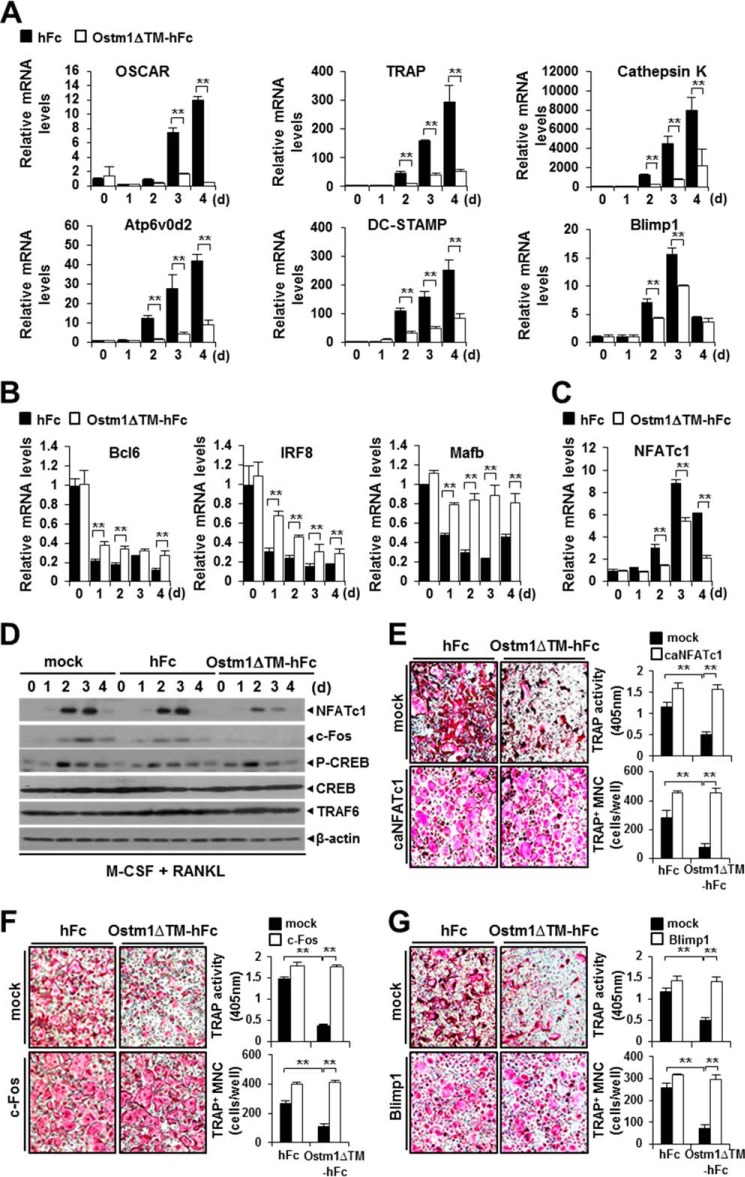
Thus, we analyzed the effect of truncated OSTM1 on the expression of OC marker genes during osteoclastogenesis. OSTM1ΔTM-hFc-treated OCs showed a marked reduction of OC markers, such as OSCAR, TRAP, DC-STAMP, Atp6v0d2, cathepsin K, and Blimp1, compared with hFc-treated control cells (Fig. 5A). BLIMP1 functions as a transcriptional repressor that mediates the repression of anti-osteoclastogenic genes, such as Bcl6 (B-cell lymphoma 6), IRF8 (interferon regulatory factor 8), and MafB (v-Maf musculoaponeurotic fibrosarcoma oncogene family homolog B) (18, 22–24). Thus, we examined the effect of truncated OSTM1 on the expression of anti-osteoclastogenic genes. OSTM1ΔTM-hFc-treated OCs showed relatively higher levels of anti-osteoclastogenic gene expression (Fig. 5B). Consistently, BLIMP1 promoter activity was down-regulated in OSTM1ΔTM-hFc-treated Raw264.7 cells (data not shown).
FIGURE 5.
The truncated form of OSTM1 inhibits the expression of OC markers through the down-regulation of BLIMP1 and NFATc1 expression. A, inhibition of OC marker expression after treatment with OSTM1ΔTM-hFc. Total RNAs were prepared from BMOCs and subjected to real-time PCR analysis. The data were normalized to β-actin. Open boxes indicate treatment of the cultures with OSTM1ΔTM-hFc. hFc was used as a control (black box). B, enhancement of the expression of anti-osteoclastogenic genes after OSTM1ΔTM-hFc treatment. C, inhibition of NFATc1 expression after OSTM1ΔTM-hFc treatment. D, negative effect of NFATc1 and c-Fos expression after OSTM1ΔTM-hFc treatment. BMOCs were prepared and analyzed by immunoblotting with antibodies. TRAF6 was used as the internal control. β-Actin was used as the loading control. E, effects of NFATc1 overexpression. A constitutively active form of NFATc1 (caNFATc1) was transduced into BMMs through retroviral infection. caNFATc1-overexpressed BMMs were further cultured with M-CSF and RANKL after treatment with the purified proteins (25 μg/ml). BMOCs were analyzed through TRAP staining (left panels; original magnification, ×100). The TRAP solution assay was performed (top right panel). The number of TRAP+ MNCs (≥3 nuclei) was counted (bottom right panel). Open boxes, retrovirus-transduced caNFATc1. The empty vector was used as a control (mock; black box). F, effects of c-Fos overexpression. Open boxes, retrovirus-transduced c-Fos. The empty vector was used as a control (mock; black box). G, effects of BLIMP1 overexpression. Open boxes, retrovirus-transduced BLIMP1. The empty vector was used as a control (mock; black box). **, p < 0.01. Error bars, S.D.
NFATc1 is a master transcriptional regulator of osteoclastogenesis, involved in both the induction of OC marker genes and the repression of anti-osteoclastogenic genes (22–24). Interestingly, we observed that the transcription of the NFATc1 gene was significantly reduced (33–59%) after OSTM1ΔTM-hFc treatment compared with the expression of this gene in controls (Fig. 5C). Consistently, the level of NFATc1 protein was also significantly decreased in OSTM1ΔTM-hFc-treated OCs, but the expression of TRAF6 or actin was not affected (Fig. 5D). The NFATc1 promoter activity was also down-regulated in OSTM1ΔTM-hFc-treated Raw264.7 cells (data not shown). NFATc1 induction is regulated by the calmodulin-dependent kinase-CREB activation through Ca2+ signaling, NF-κB, and c-Fos pathways in RANKL-mediated osteoclastogenesis (25–28). We observed that the level of c-Fos protein was significantly decreased in OSTM1ΔTM-hFc-treated OCs, but the phosphorylation of CREB was not affected (Fig. 5D). Furthermore, the inhibition of OC differentiation after OSTM1ΔTM-hFc treatment was completely rescued through NFATc1, c-Fos, or BLIMP1 expression in OSTM1ΔTM-hFc-treated OCs (Fig. 5, E–G). These data indicate that truncated OSTM1 is negatively involved in the regulation of OC marker genes expression through the down-regulation of the NFATc1-BLIMP1 axis in osteoclastogenesis.
Inhibitory Effect of Truncated OSTM1 on LPS-induced OC Formation and Bone Loss
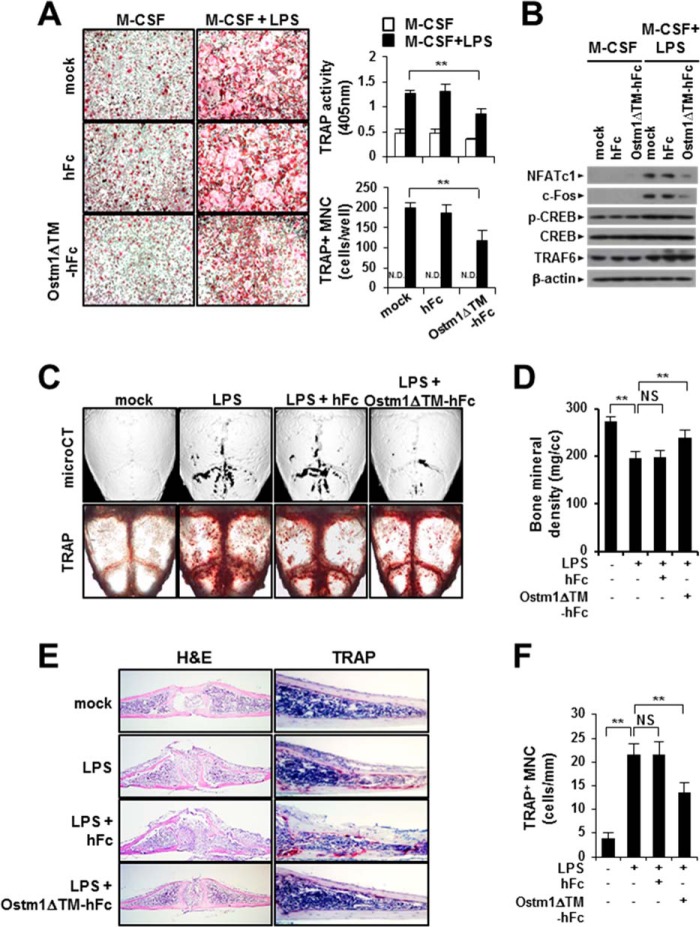
We examined the effect of truncated OSTM1 on OC differentiation under the LPS-induced pathological conditions. To induce OC differentiation through LPS stimulation, BMMs were cultured with M-CSF (30 ng/ml) and LPS (10 μg/ml) for 4 days in the presence or absence of truncated OSTM1. We observed that LPS-induced OC formation was reduced following OSTM1ΔTM-hFc treatment, whereas no inhibition was observed for the hFc control (Fig. 6A). Consistent with these results, NFATc1 and c-Fos protein expression was also significantly decreased in OSTM1ΔTM-hFc-treated OCs, but the phosphorylation of CREB was not affected (Fig. 6B).
FIGURE 6.
The effect of the truncated OSTM1 on LPS-induced OC formation and bone loss. A, effect of OSTM1ΔTM-hFc treatment on LPS-induced OC differentiation. The OCs were differentiated through LPS stimulation for 4 days. LPS-induced OCs were analyzed and photographed (left panels; original magnification, ×100) after TRAP staining. The TRAP solution assay was performed (top right panel). The number of TRAP+ MNCs (≥3 nuclei) was counted (bottom right panel). N.D., not detected. B, down-regulation of NFATc1 expression after OSTM1ΔTM-hFc treatment in LPS-induced OCs. TRAF6 was used as the internal control. β-Actin was used as the loading control. C, effect of OSTM1ΔTM-hFc treatment on LPS-induced bone loss. The TRAP+ OCs on calvaria were visualized after TRAP staining (top panels). Three-dimensional microcomputed tomography images of calvaria are shown in the bottom panels. D, bone mineral density units on calvarial bone were measured through microcomputed tomography analysis. E, effect of OSTM1ΔTM-hFc treatment on LPS-induced OC formation and bone loss. The dissected calvarial tissues were stained through H&E (left panels; original magnification, ×100) or TRAP staining (right panels; original magnification, ×200). F, TRAP+ MNCs on calvaria were counted. NS, not significant. **, p < 0.01. Error bars, S.D.
Next, we determined whether truncated OSTM1 is involved in the modulation of LPS-induced bone destruction in vivo. Bone destruction was induced through the injection of LPS (25 mg/kg body weight) into the subcutaneous tissue overlying the periosteum of mouse calvaria in the presence or absence of truncated OSTM1. We observed that LPS-induced OC formation on mouse calvaria was reduced following OSTM1ΔTM-hFc treatment (Fig. 6C, top panels). Consistent with these results, LPS-induced bone destruction on mouse calvaria was significantly restored after OSTM1ΔTM-hFc treatment (Fig. 6, C (bottom panels) and D). Next, we performed a histological evaluation of mouse calvaria using H&E or TRAP staining. LPS-induced bone erosion was significantly restored after OSTM1ΔTM-hFc treatment (Fig. 6E, left panels). Similarly, TRAP staining revealed reduced TRAP+ MNC formation in the calvarial section following OSTM1ΔTM-hFc treatment (Fig. 6, E (right panels) and F). Hence, these results indicate that truncated OSTM1 reduces LPS-induced bone destruction through the inhibition of OC differentiation in vivo.
DISCUSSION
Genetic mutations in osteoclastogenic genes, such as RANK, RANKL, M-CSF, TRAF6, TCIRG1, CLCN7, and OSTM1, are closely associated with osteopetrotic bone diseases (29). In particular, genetic mutations in the OSTM1 gene lead to the most severe forms of ARO in humans (30). Human OSTM1 contains 334 amino acid residues, encoding a type I transmembrane protein with a putative signal peptide and a potential transmembrane domain (4). Six mutations in the human OSTM1 gene have been implicated in ARO, with an increased risk of early death in clinical reports (4–6,10,30–33). These six human OSTM1 mutations could lead to truncated OSTM1 proteins containing 11, 85, 134, 139, or 261 residues with short unrelated sequences or premature stop codons through frameshift or nonsense mutations (4–6,10,30–33). Although many case studies of human ARO patients have been reported, the functional relevance of these truncated forms of OSTM1 is not clear.
The morphological observation of OCs in bony tissue can be used to evaluate OC differentiation or function. In a recent case study, an ARO patient with a nonsense mutation in the first exon of the OSTM1 gene showed the production of a short 85-residue truncated protein, demonstrating that multinucleated OCs, which are most likely nonfunctional cells, are normally differentiated from precursor cells (31). This phenomenon is similar to the morphological phenotype of OCs resulting from the null mutation in gl/gl mice, showing an increased number of functionally inactive multinucleated OCs (4). However, contrary to these studies, a decrease in OC numbers was observed in another ARO patient with a substitution at the donor splice site of intron V, resulting in the production of a 261-residue truncated protein lacking the putative transmembrane domain (4, 10). In addition, abnormally elongated OCs were observed in this patient, indicating that OC differentiation or maturation might be altered (10). Furthermore, Lange et al. (7) suggested the possibility of extracellular secretion of truncated OSTM1 in ARO patients, although the precise role of the truncated form of OSTM1 remains unknown. Therefore, it is valuable to examine whether the extracellular secretion of truncated OSTM1 is involved in osteoclastogenesis. Interestingly, Chalhoub et al. (4) provided data supporting this possibility in studies involving BAC transgene complementation analyses in gl/gl mice. These authors reported an exceptional BAC 373N3 transgenic clone that failed to rescue the gl/gl phenotype, whereas the other BAC 373N3 transgenic clones showed a fully rescued phenotype (4). This BAC 373N3 transgenic clone has a large genomic deletion beginning in the third intron and extending beyond the 3′ end of the Ostm1 gene, resulting in the production of a 207-residue truncated protein lacking the putative transmembrane domain (4). This mutation is similar to the substitution mutation at the donor splice site of intron V, which results in the production of a 261-residue truncated protein and has been reported in human ARO patients (10). In addition, we observed that truncated OSTM1 reduced OC differentiation (Fig. 3), suggesting a negative role for the secreted form of truncated OSTM1 in osteoclastogenesis. Accordingly, it is reasonable to presume that this truncated OSTM1 is an extracellular secreted mutant form.
OSTM1 is ubiquitously expressed in various tissues, including brain, heart, liver, kidney, and bone (4, 9). However, the primary cell sources of the secreted form of the truncated OSTM1 remain unknown. Interestingly, the gray-lethal mutation leads not only to osteopetrosis but also to coat color defects and neurological damage (5). Furthermore, a recent study demonstrated that a loss of function of the Ostm1 gene results in the deregulation of multiple hematopoietic lineages, including T- and B-cells, in addition to OC lineage cells (34). In rescue experiments in Ostm1-transgenic mice, OSTM1 overexpression in the committed OC lineage under the control of the TRAP promoter was not sufficient to rescue the gl osteopetrotic defect, whereas both gl osteopetrosis and hematopoiesis were fully rescued by the PU.1 promoter in Ostm1-transgenic mice (34). Thus, these data indicate that OSTM1 is involved in intercellular cross-talk in the hematopoietic compartment; however, the complete mode of action is currently unknown. In fact, the involvement of intercellular cross-talk in the hematopoietic lineage cells in gl/gl mice has been previously proposed based on the results of hematopoietic stem cell transplantation experiments (35). Consistent with previous reports, we also observed that osteoclastogenesis is reduced through culture supernatants containing a secreted form of truncated OSTM1 derived from osteoblastic cell lines or T-cell hybridomas (Fig. 2). In addition, we observed that OSTM1 is expressed in bone cells, such as OCs and OBs, in response to differentiation signals (Fig. 1). Thus, these results suggest that a secreted form of the truncated OSTM1 is not only expressed in OC lineage cells but also in various hematopoietic or marrow stromal cells in both humans and mice with ARO.
In conclusion, we identified a novel function for the secreted form of truncated OSTM1 in osteoclastogenesis. Here, we show that a secreted form of truncated OSTM1 is negatively involved in OC differentiation and survival through the down-regulation of BLIMP1 and NFATc1 expression as illustrated in Fig. 7. The formation of multinucleated OCs through cell-cell fusion and OC marker gene expression was significantly reduced through the expression of truncated OSTM1. Moreover, we demonstrated that truncated OSTM1 inhibits osteoclastogenesis via the attenuation of RANKL-induced p38 and JNK activation and M-CSF-induced Akt and ERK activation. Finally, we demonstrated that truncated OSTM1 reduces LPS-induced bone destruction in vivo. Thus, these findings suggest that the loss of the putative transmembrane domain in the OSTM1 gene in ARO patients produces a secreted form of truncated OSTM1 that inhibits osteoclastogenesis.
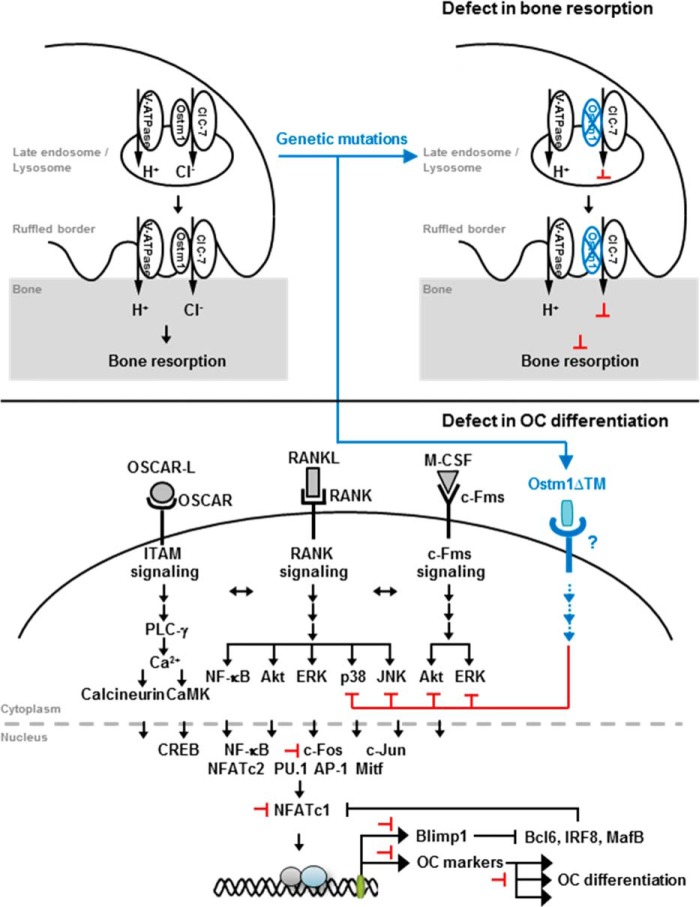
FIGURE 7.
A proposed model for the involvement of truncated OSTM1 in osteoclastogenesis through genetic mutation. The black arrows represent activation signals. The black or black-dashed bar indicates inhibition signals. The effects of genetic mutations in the Ostm1 gene are indicated by changes in color (blue). The proposed inhibition by truncated OSTM1 is shown by the red bars.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by Ministry of Education Grant NRF-2011-0021771, Korea Basic Science Institute Grant D33400, the research fund of Chungnam National University, and Ministry of National Defense Foundation Grant ADD 14-01-06-06.
- OC
- osteoclast
- preOC
- OC precursor
- OB
- osteoblast
- pOB
- primary OB
- RANK
- receptor activator of nuclear factor-κB
- RANKL
- RANK ligand
- M-CSF
- macrophage-colony stimulating factor
- PLC
- phospholipase C
- OSCAR
- OC-associated receptor
- TRAP
- tartrate-resistant acid phosphatase
- CREB
- cAMP-response element-binding protein
- ARO
- autosomal recessive osteopetrosis
- VtD3
- vitamin D3
- PGE2
- prostaglandin E2
- aa
- amino acids
- BM
- bone marrow
- BMM
- BM-derived macrophage
- BMOC
- BM-derived OC
- MNC
- multinucleated OC.
REFERENCES
- 1. Rho J., Takami M., Choi Y. (2004) Osteoimmunology: interactions of the immune and skeletal systems. Mol. Cells 17, 1–9 [PubMed] [Google Scholar]
- 2. Takayanagi H. (2007) Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7, 292–304 [DOI] [PubMed] [Google Scholar]
- 3. Walsh M. C., Kim N., Kadono Y., Rho J., Lee S. Y., Lorenzo J., Choi Y. (2006) Osteoimmunology: interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 24, 33–63 [DOI] [PubMed] [Google Scholar]
- 4. Chalhoub N., Benachenhou N., Rajapurohitam V., Pata M., Ferron M., Frattini A., Villa A., Vacher J. (2003) Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat. Med. 9, 399–406 [DOI] [PubMed] [Google Scholar]
- 5. Pangrazio A., Poliani P. L., Megarbane A., Lefranc G., Lanino E., Di Rocco M., Rucci F., Lucchini F., Ravanini M., Facchetti F., Abinun M., Vezzoni P., Villa A., Frattini A. (2006) Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. J. Bone Miner. Res. 21, 1098–1105 [DOI] [PubMed] [Google Scholar]
- 6. Prinetti A., Rocchetta F., Costantino E., Frattini A., Caldana E., Rucci F., Bettiga A., Poliani P. L., Chigorno V., Sonnino S. (2009) Brain lipid composition in grey-lethal mutant mouse characterized by severe malignant osteopetrosis. Glycoconj. J. 26, 623–633 [DOI] [PubMed] [Google Scholar]
- 7. Lange P. F., Wartosch L., Jentsch T. J., Fuhrmann J. C. (2006) ClC-7 requires Ostm1 as a β-subunit to support bone resorption and lysosomal function. Nature 440, 220–223 [DOI] [PubMed] [Google Scholar]
- 8. Feigin M. E., Malbon C. C. (2008) OSTM1 regulates β-catenin/Lef1 interaction and is required for Wnt/beta-catenin signaling. Cell. Signal. 20, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer T., De Vries L., Meerloo T., Farquhar M. G. (2003) Promotion of G α i3 subunit down-regulation by GIPN, a putative E3 ubiquitin ligase that interacts with RGS-GAIP. Proc. Natl. Acad. Sci. U.S.A. 100, 8270–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quarello P., Forni M., Barberis L., Defilippi C., Campagnoli M. F., Silvestro L., Frattini A., Chalhoub N., Vacher J., Ramenghi U. (2004) Severe malignant osteopetrosis caused by a GL gene mutation. J. Bone Miner. Res. 19, 1194–1199 [DOI] [PubMed] [Google Scholar]
- 11. Rajapurohitam V., Chalhoub N., Benachenhou N., Neff L., Baron R., Vacher J. (2001) The mouse osteopetrotic grey-lethal mutation induces a defect in osteoclast maturation/function. Bone 28, 513–523 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., Choi Y. (2006) v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 13. Yu J., Choi S., Park E. S., Shin B., Yu J., Lee S. H., Takami M., Kang J. S., Meong H., Rho J. (2012) d-chiro-Inositol negatively regulates the formation of multinucleated osteoclasts by down-regulating NFATc1. J. Clin. Immunol. 32, 1360–1371 [DOI] [PubMed] [Google Scholar]
- 14. Rho J., Gong S., Kim N., Choi Y. (2001) TDAG51 is not essential for Fas/CD95 regulation and apoptosis in vivo. Mol. Cell. Biol. 21, 8365–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yasuda T., Kometani K., Takahashi N., Imai Y., Aiba Y., Kurosaki T. (2011) ERKs induce expression of the transcriptional repressor Blimp-1 and subsequent plasma cell differentiation. Sci. Signal. 4, ra25. [DOI] [PubMed] [Google Scholar]
- 16. Kim H., Kim T., Jeong B. C., Cho I. T., Han D., Takegahara N., Negishi-Koga T., Takayanagi H., Lee J. H., Sul J. Y., Prasad V., Lee S. H., Choi Y. (2013) Tmem64 modulates calcium signaling during RANKL-mediated osteoclast differentiation. Cell Metab. 17, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. H., Kim T., Park E. S., Yang S., Jeong D., Choi Y., Rho J. (2008) NHE10, an osteoclast-specific member of the Na+/H+ exchanger family, regulates osteoclast differentiation and survival [corrected]. Biochem. Biophys. Res. Commun. 369, 320–326 [DOI] [PubMed] [Google Scholar]
- 18. Zhao B., Takami M., Yamada A., Wang X., Koga T., Hu X., Tamura T., Ozato K., Choi Y., Ivashkiv L. B., Takayanagi H., Kamijo R. (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim N., Takami M., Rho J., Josien R., Choi Y. (2002) A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J. Exp. Med. 195, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim T., Kim K., Lee S. H., So H. S., Lee J., Kim N., Choi Y. (2009) Identification of LRRc17 as a negative regulator of receptor activator of NF-κB ligand (RANKL)-induced osteoclast differentiation. J. Biol. Chem. 284, 15308–15316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu J., Wang S., Zhang P., Said-Al-Naief N., Michalek S. M., Feng X. (2009) Molecular mechanism of the bifunctional role of lipopolysaccharide in osteoclastogenesis. J. Biol. Chem. 284, 12512–12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyamoto T. (2011) Regulators of osteoclast differentiation and cell-cell fusion. Keio J. Med. 60, 101–105 [DOI] [PubMed] [Google Scholar]
- 23. Miyauchi Y., Ninomiya K., Miyamoto H., Sakamoto A., Iwasaki R., Hoshi H., Miyamoto K., Hao W., Yoshida S., Morioka H., Chiba K., Kato S., Tokuhisa T., Saitou M., Toyama Y., Suda T., Miyamoto T. (2010) The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J. Exp. Med. 207, 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishikawa K., Nakashima T., Hayashi M., Fukunaga T., Kato S., Kodama T., Takahashi S., Calame K., Takayanagi H. (2010) Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakashima T., Hayashi M., Takayanagi H. (2012) New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol. Metab. 23, 582–590 [DOI] [PubMed] [Google Scholar]
- 26. Negishi-Koga T., Takayanagi H. (2009) Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol. Rev. 231, 241–256 [DOI] [PubMed] [Google Scholar]
- 27. Sato K., Suematsu A., Nakashima T., Takemoto-Kimura S., Aoki K., Morishita Y., Asahara H., Ohya K., Yamaguchi A., Takai T., Kodama T., Chatila T. A., Bito H., Takayanagi H. (2006) Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat. Med. 12, 1410–1416 [DOI] [PubMed] [Google Scholar]
- 28. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 29. Del Fattore A., Cappariello A., Teti A. (2008) Genetics, pathogenesis and complications of osteopetrosis. Bone 42, 19–29 [DOI] [PubMed] [Google Scholar]
- 30. Segovia-Silvestre T., Neutzsky-Wulff A. V., Sorensen M. G., Christiansen C., Bollerslev J., Karsdal M. A., Henriksen K. (2009) Advances in osteoclast biology resulting from the study of osteopetrotic mutations. Hum. Genet. 124, 561–577 [DOI] [PubMed] [Google Scholar]
- 31. Maranda B., Chabot G., Décarie J. C., Pata M., Azeddine B., Moreau A., Vacher J. (2008) Clinical and cellular manifestations of OSTM1-related infantile osteopetrosis. J. Bone Miner. Res. 23, 296–300 [DOI] [PubMed] [Google Scholar]
- 32. Ott C. E., Fischer B., Schröter P., Richter R., Gupta N., Verma N., Kabra M., Mundlos S., Rajab A., Neitzel H., Kornak U. (2013) Severe neuronopathic autosomal recessive osteopetrosis due to homozygous deletions affecting OSTM1. Bone 55, 292–297 [DOI] [PubMed] [Google Scholar]
- 33. Ramírez A., Faupel J., Goebel I., Stiller A., Beyer S., Stöckle C., Hasan C., Bode U., Kornak U., Kubisch C. (2004) Identification of a novel mutation in the coding region of the grey-lethal gene OSTM1 in human malignant infantile osteopetrosis. Hum. Mutat. 23, 471–476 [DOI] [PubMed] [Google Scholar]
- 34. Pata M., Héraud C., Vacher J. (2008) OSTM1 bone defect reveals an intercellular hematopoietic crosstalk. J. Biol. Chem. 283, 30522–30530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walker D. G. (1975) Bone resorption restored in osteopetrotic mice by transplants of normal bone marrow and spleen cells. Science 190, 784–785 [DOI] [PubMed] [Google Scholar]