Background: OSR1 modulates ion homeostasis and cell volume in mammal.
Results: The CCT domain and αAL helix of OSR1 act together to suppress its basal activity, while WNKs and MO25 unlock such autoinhibition for full activation.
Conclusion: OSR1 activity is regulated by autoinhibition, WNKs, and MO25.
Significance: This work provides insights into the regulatory mechanisms of OSR1 activation to facilitate functional study.
Keywords: Enzyme Mechanism, Homeostasis, Protein Kinase, Protein Structure, Protein-Protein Interaction, OSR1
Abstract
The oxidative stress-responsive 1 (OSR1) kinase belongs to the mammalian STE20-like kinase family. OSR1 is activated by with no lysine [K] (WNKs) kinases, and then it phosphorylates cation-coupled Cl-cotransporters, regulating ion homeostasis and cell volume in mammalian cells. However, the specific mechanisms of OSR1 activation remains poorly defined, largely due to its extremely low basal activity. Here, we dissect in detail the regulatory mechanisms of OSR1 activation from the aspects of autoinhibition, upstream kinase WNK, and the newly identified master regulator mouse protein-25 (MO25). Based on our structural and biochemical studies, we propose a “double lock” model, accounting for the tight autoinhibition of OSR1, an effect that has to be removed by WNK before MO25 further activates OSR1. Particularly, the conserved C-terminal (CCT) domain and αAL helix act together to strongly suppress OSR1 basal activity. WNKs bind to the CCT and trigger its conformational rearrangement to release the kinase domain of OSR1, allowing for MO25 binding and full activation. Finally, the regulatory mechanisms of OSR1 activation were further corroborated by cellular studies of OSR1-regulated cell volume control through WNK-OSR1 signaling pathway. Collectively, these results provide insights into the OSR1 kinase activation to facilitate further functional study.
Introduction
The mammalian STE20-like kinases were identified as homologues of the yeast Sterile-20 protein kinase, and divided into p21-activated kinase (PAK)4 and germinal center kinase (GCK) families according to their structural differences (1–3). GCKs consist of eight subfamilies, all characterized by the presence of a relatively conserved kinase domain at the N terminus and a diversified C-terminal domain (2). GCKs are involved in multiple cellular processes, including cell growth, apoptosis, polarity, migration, stress responses, and immune regulation (1, 2, 4–6).
Oxidative stress-responsive 1 (OSR1), STE20/SPS1-related proline/alanine-rich kinase (SPAK), and pseudokinase STE20-related adaptor (STRAD) are members of GCK VI subfamily. OSR1 and SPAK share almost 90% sequence identity in the kinase domain. OSR1 comprises of an N-terminal catalytic domain and two regulatory regions, named PF1 [PASK and Fray (Drosophila homolog)] and CCT domains (7). The PF1 domain, adjacent to the C terminus of kinase domain, is likely to regulate OSR1 catalytic activity (8). Besides, a conserved WEW motif is found within the PF1 domain, mediating the association with adaptor protein MO25 (9). The CCT domain is required for recognizing and binding to the RFX[V/I] motif from both OSR1 upstream activators and downstream substrates (10, 11). Recent studies have determined crystal structures of OSR1 kinase domain, as well as the CCT domain in complex with a peptide containing an RFX[V/I] motif derived from WNK4 (12–14). In crystal structure, the OSR1 kinase domain forms a domain-swapped dimer, exchanging each p + 1 loop and αEF helix between dimer-related monomers, highlighting the potential importance of OSR1 homo dimerization (14). Yet the accurate regulatory mechanisms of OSR1 via its PF1 and CCT domains remain unclear.
OSR1 is widely expressed in most tissues, especially in the heart and skeletal muscle, and has been implicated in multiple cellular events (8, 15–18). For example, OSR1 can modulate ion homeostasis in mammalian cells through WNK-OSR1 kinase cascade pathway. During hyperosmotic or hypertonic stress, OSR1 is phosphorylated and activated by WNKs on threonine 185 (Thr-185) in the activation loop (A-loop) and serine 325 (Ser-325) in the PF1 domain, which in turn phosphorylates and activates downstream substrates Na+/Cl− cotransporter (NCC), Na+/K+/2Cl− cotransporter (NKCC) 1 and NKCC2 (11, 19, 20). Mutational analysis indicates that phosphorylation of Thr-185 in OSR1 could significantly enhance its kinase activity, while phosphorylation of Ser-325 only modestly promotes OSR1 kinase activity (19). Moreover, the mammalian target of rapamycin complex 2 (mTORC2), which is activated by phosphatidylinositol-3 kinase (PI3K), can increase OSR1 activity by phosphorylating residue Ser-339 (21, 22). In addition, OSR1 has also been implicated in immune regulation by interacting with protein kinase C-θ (PKCθ) or TNF receptor expressed in lymphoid tissues (RELT) homologues (18, 23).
Recently, the adaptor protein MO25 has been identified as a key regulator that can dramatically enhance the activity of a group of germinal center kinases including OSR1 and SPAK (9). MO25 was initially found to mediate a heterotrimeric complex with the tumor suppressor liver kinase B1 (LKB1) and the catalytically inactive pseudokinase STRAD, and thus activating LKB1 (24–26). Structural analysis of the STRAD-MO25 complex indicates that four conserved sites (A, B, C, and D) in the kinase domain and C-terminal domain of STRAD corporately interact with MO25 (25, 27). Sequence alignment shows a high degree of homology between the four sites of OSR1 and STRAD (9, 28), suggesting OSR1 binds to MO25 in a similar manner.
Here we dissect in detail the regulatory mechanisms of OSR1 kinase activity and propose a “double lock” model, in which two critical structural elements, the αAL helix and the CCT domain, function as two layers of autoinhibition, leading to a remarkably low basal activity that is usually undetectable. In the “locked” state, the αAL helix spatially inserts between the side chains of the conserved lysine and glutamate residues, directly blocking the formation of Lys-Glu ion pair interaction required for an active conformation; while the CCT domain directly interacts with the kinase domain to inhibit both the domain-swapped dimerization, and the recruitment of effector molecule MO25. Structural and biochemical studies indicate that MO25 associates with OSR1 in a similar manner to that observed for STRAD, and that MO25 is required for the full activation of OSR1. During this process, WNKs not only phosphorylate the A-loop of OSR1 to alter the conformation of the catalytic center, but also remove its autoinhibition through direct binding to the CCT domain, thus facilitating the recruitment of MO25 for further activation.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The different constructs of human OSR1, including wild-type or mutant full-length (amino acids 1–527), kinase domain (amino acids 1–293) and kinase domain plus (1–338), MO25 (amino acids 1–341) and WNK1 (amino acids 1–663) were cloned into vector pET28a-HisTEV and expressed by Escherichia coli BL21 (DE3) strain. The OSR1 CCT domain (amino acids 434–527) was cloned into vector pET28a-GSTTEV and expressed by E. coli BL21 (DE3) strain. Protein expression was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside (IPTG) in Terrific Broth medium at 16 °C for 18 h. Bacterial cells were harvested, and proteins were purified at 4 °C by Ni-Sepharose or glutathione-Sepharose. TEV protease was used to cleave His-tagged and GST-tagged proteins to generate untagged proteins. All proteins were further purified by gel filtration and stored in buffer A containing 20 mm Hepes (pH 7.5), 100 mm NaCl, and 1 mm DTT.
In Vitro Kinase Assay
The activities of recombinant OSR1 kinases and related mutants were assayed by autoradiography. Phosphotransferase activity of each reaction was measured in a total assay volume of 40 μl consisting of 50 mm Tris-HCl (pH 7.5), 0.1 mm EGTA, 10 mm magnesium acetate, 1 uCi [γ-32P]ATP, 5 mm MnCl2, and 40 μg of GST-NCC (amino acids 1–100) substrates. The reactions were carried out at 30 °C and were terminated after 30 min by adding loading buffer. Samples were analyzed following electrophoresis and autoradiography. Each experiment was performed at least three times.
Kinetic Assay
Interactions between purified proteins of MO25 (amino acids 1–341) and OSR1 (amino acids 1–338) were detected in vitro by biolayer interferometry experiment (BLI)(Octet Red 96, ForteBio) following a protocol as described previously (29). Biotinylated wild-type OSR1 protein was immobilized on the streptavidin biosensors and incubated with varying concentrations of wild-type or mutant MO25 in 1×kinetics buffer (10 mm Na2HPO4, 2 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, 0.002% Tween 20, 0.1 mg/ml BSA, pH 7.4). The same system was applied to the biotinylated wild-type MO25 with wild-type or mutant OSR1. The BLI experiments comprised 5 steps: 1) Baseline acquisition; 2) Biotinylated proteins loading onto SA biosensor; 3) Second baseline acquisition; 4) Association of interacting protein; 5) Dissociation of interacting protein. Experimental trace about association and dissociation were recorded and analyzed using Octet Data Analysis Software 7.0 (ForteBio).
Pull-down Assay
His-tagged bait proteins were coupled on Ni-Sepharose first, and then mixed with different prey proteins at 4 °C for 2 h in buffer A. The mixtures were washed three times with buffer A containing 20 mm imidazole to avoid the nonspecific binding. The input and output samples were boiled in SDS-PAGE loading buffer and loaded on SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining.
Western Blot
Samples of pull-down assays were boiled and separated electrophoretically on 12% SDS-PAGE gel. Protein bands were transferred onto polyvinylidene fluoride (PVDF) membrane by electroblotting. The membrane was blocked in TBST buffer (25 mm Tris-HCl, pH 7.4, 1.5 m NaCl, 0.5% Tween-20) containing 5% fat-free dry milk for 1 h. The membrane was washed with TBST for three times and incubated with mouse anti-GST antibody overnight. After washing with TBST, the membrane sheets were incubated with peroxidase conjugated anti-mouse antibody for 1 h. The bands were visualized using Thermo ECL detection kit.
Cross-linking
Purified 1 mg/ml wild-type OSR1 or the related fragments was incubated with 0 and 0.005% (v/v) glutaraldehyde (GA) in conjugation buffer (20 mm Hepes pH 7.5, 100 mm NaCl, 1 mm DTT,) at room temperature for 1 h. The reaction was quenched with 50 mm Tris-HCl (pH 8.0) for 30 min. The cross-linked samples were analyzed on an 8% SDS-PAGE followed by CBB staining.
Cell Volume Determination
293T cells were cultured in DMEM supplemented with 10% FBS at 37 °C in 5% CO2. OSR1 and its mutants were then co-transfected into cells with or without MO25 by using Lipofectamine TM 2000 (Invitrogen). After transfection for 24 h, cells were treated with 0.5 m sorbitol using PBS as negative control. Subsequently, the cell culture vessel was placed in a phase-contrast live cell imaging system, the IncucyteTM FLR (Essen Bioscience, Ann Arbor, MI), in a 37 °C incubator supplemented with 5% CO2. Phase-contrast images (750 × 950 μm) were taken by the Incucyte FLR at the indicated time intervals. The approximate number of cells scanned was 200∼600 cells per image. The IncucyteTM FLR was programmed to take 3 images per well of a 96-well plate. It takes 10 min to scan an entire 96-well plate at 3 images per well. Relative confluent was then calculated according the absolute confluent of each well (relative confluent = treatment confluent/negative control confluent). All experiments were repeated at least twice.
RESULTS
The CCT Domain Directly Interacts with the Kinase Domain to Inhibit OSR1 Activation
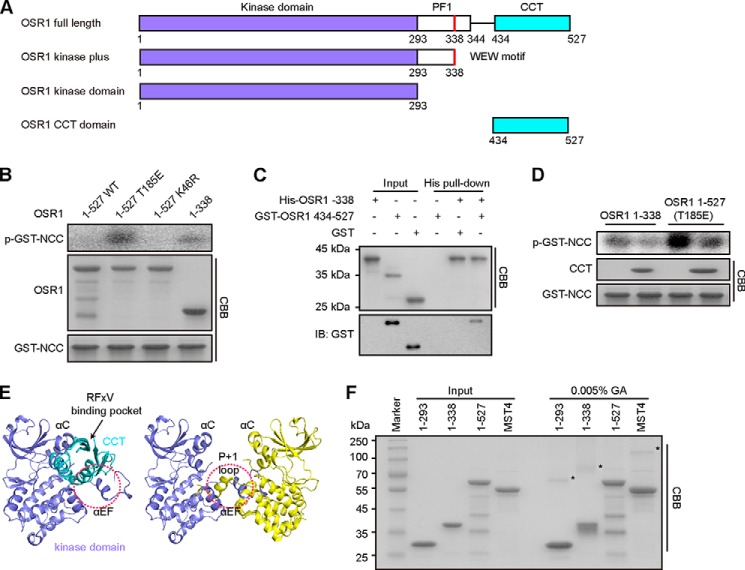
OSR1 comprises a kinase domain, a PF1 domain containing a MO25-binding motif (WEW), and a CCT domain (Fig. 1A). It has been frequently observed that protein kinases contain an autoinhibitory domain outside of the catalytic domain. Removal of the autoinhibitory domain usually results in constitutively activated kinases. To define the mechanisms that regulate the activity of OSR1, we created OSR1 full-length wild-type (WT), T185E mutant (mimicking the phosphorylated active state) (19), K46R mutant (catalytically inactive) and CCT domain-truncated constructs to determine whether it contains an autoinhibitory domain. We first expressed and purified these proteins for in vitro kinase assay. The N-terminal domain of NCC that contains phosphorylation sites of OSR1 was purified as a GST-tagged protein and used as a substrate (11). After incubation with [γ-32P], the results of autoradiography showed that full-length wild-type OSR1 had no detectable activity toward NCC (Fig. 1B). In contrast, the kinase activity of the OSR1 T185E mutant was readily detected, indicating a constitutively activation of OSR1 through mimicking phosphorylation by WNKs. Surprisingly, the truncated fragment (amino acids 1–338), lacking the CCT domain, also exhibited significant kinase activity, even though it contained no phosphorylation-mimicking mutation.
FIGURE 1.
The CCT domain of OSR1 binds to its kinase domain and inhibits OSR1 kinase activity. A, schematic illustration of domain organization for human OSR1. B, catalytic activities of the indicated OSR1 fragments were assayed for determining the ability of OSR1 phosphorylating GST-NCC-(1–100). The relative activity of OSR1 was presented by the phosphorylation levels of GST-NCC, which were quantified by densitometric measurement (ImageJ) (right graph). C, purified recombinant proteins of His-tagged OSR1 kinase plus (1–338) and GST-tagged CCT domain (434–527) were subjected to His pull-down analysis. GST protein was used as a negative control. Pull-down samples were subjected to SDS-PAGE and immunoblotted (IB) with anti-GST antibodies. CBB: Coomassie Brilliant Blue. D, catalytic activities of OSR1-(1–338) and full-length T185E mutant were measured in the presence or absence of the CCT protein by autoradiography. The Coomassie Blue staining shows the total amount of CCT protein and GST-NCC. The relative amounts of phosphorylated-GST-NCC (p-GST-NCC) were quantified by ImageJ (upper graph). E, left panel, docking model of OSR1 CCT domain (cyan, PDB ID: 2V3S) interaction with its kinase domain (slate, PDB ID: 3DAK) by ClusPro 2.0. Right panel, the structure of domain-swapped OSR1 kinase domain (PDB ID: 3DAK). Two molecules are represented in slate and yellow, respectively. The red circles indicate the partially overlapped region. F, cross-linking assay of OSR1 different fragments. Proteins were incubated with 0% and 0.005% (v/v) GA at room temperature for 1 h. The dimers are indicated by asterisks. Full-length MST4 was used as a positive control.
Based on the above results, we speculated that the CCT domain may directly interact with the kinase domain of OSR1 to attain an inactive autoinhibitory conformation, thus resulting in a barely detectable basal activity. To test this possibility, we performed pull-down assay using purified recombinant proteins in vitro. The result showed that His-tagged OSR1 kinase domain plus (amino acids 1–338) could interact with GST-tagged OSR1 CCT domain (amino acids 434–527), but did not interact with the negative control protein GST (Fig. 1C). Furthermore, the activities of OSR1 kinase domain plus and the constitutively active mutant T185E were reduced by addition of the purified CCT domain protein (Fig. 1D). Taken together, these results indicate that the CCT domain can directly interact with the kinase domain of OSR1 to inhibit its activity.
The CCT Domain May Prevent Substrate Binding and Domain-swapped Homodimerization
To further investigate the specific mechanism through which the CCT domain of OSR1 inhibits its kinase activity, we examined the three-dimensional structural information of OSR1. As introduced, the structures of the OSR1 kinase domain, and its CCT domain in complex with a WNK4 peptide have been determined (12–14), but the structure of full-length OSR1 is not yet available. To explore the relative orientation between the CCT domain and the kinase domain of OSR1, we therefore generated a structural model of OSR1 kinase domain in complex with its CCT domain through computational docking (30) by using the resolved crystal structures of OSR1 kinase domain (PDB ID: 3DAK) and CCT domain (PDB ID: 2V3S) (Fig. 1E). In this modeled structure, OSR1 CCT domain was positioned adjacent to the αC helix and A-loop with its RFX[V/I] binding pocket exposed, suggesting that the autoinhibited OSR1 kinase can still bind to upstream WNK kinase and downstream targets. However, the CCT domain would at least partially cover the substrate binding pocket of OSR1 kinase domain, and may particularly contact the αEF helix and p + 1 loop in the C-lobe of OSR1 kinase domain (Fig. 1E). Therefore, the CCT domain of OSR1 may prevent substrate binding to the catalytic center, even though the CCT domain itself can recruit substrate through the RFX[V/I] motif.
Kinase domain-mediated homodimeric conformation could bring the kinase domain together and facilitate trans-autophosphorylation, as in the case we have shown for the mammalian STE20-like kinase 4 (MST4), another member of GCK family (31). In this regard, OSR1 was also observed as a domain-swapped homodimer. However, structural comparison of OSR1 kinase domain alone (PDB ID: 3DAK) and its complex with the CCT domain (modeled in this work) indicated that the region to which the CCT domain binds partially overlaps with that of the kinase domain-mediated homodimeric interface (Fig. 1E). Therefore, we reasoned that the steric hindrance caused by the bound CCT domain would impair the homodimeric interaction of OSR1 kinase domains. To test this hypothesis, we performed cross-linking assay using full-length and truncated OSR1 proteins. MST4 was used as a positive control for this reaction system, since we previously showed that MST4 forms a homodimer (31). As shown in Fig. 1F, the OSR1 kinase domain (amino acids 1–293) and kinase domain plus (amino acids 1–338), both lacking the CCT domain, could form dimers as does full-length MST4. However, 0.005% GA did not induce band shift of full-length OSR1, indicating that the CCT domain plays a negative role in the homodimerization of full-length OSR1. Taken together, these results suggest that the CCT domain acts as a structural lock to suppress OSR1 kinase activity by preventing substrate access and homodimerization of the kinase domain.
The αAL Helix Inhibits OSR1 Kinase Activity through Disruption of the Conserved Lys-Glu Ion Pair
Given the substantially lower basal activity of OSR1 when compared with other kinases such as MST3, MST4, and STK25, we reasoned that there might be certain distinct structural elements in OSR1 kinase domain to which the tight intrinsic autoinhibition may ascribe. Therefore, we performed structural comparison of OSR1 kinase domain with those of MST3, MST4, STK25, as well as the pseudokinase STRADα (Fig. 2A). As expected, the overall structure of OSR1 kinase domain resembles those of MST3, MST4, STK25, and STRADα, featuring a conventional dual lobe architecture. However, unlike MST3, MST4, STK25, and STRADα, the activation loop of OSR1 adopts an inactive conformation through an atypical helix, termed as αAL helix. This structural element is formed by a two-turn helix at the N terminus of the activation loop. Sequence alignment of activation loop including MST3, MST4, STK25, STRADα, and OSR1 indicates that the αAL helix is an extra insertion for OSR1 (Fig. 2A).
FIGURE 2.

Structural element αAL helix of OSR1 inhibits its kinase activity. A, structural comparison and sequence alignment of OSR1 (slate, PDB ID: 3DAK) with MST3 (yellow, PDB ID: 3A7G), MST4 (cyan, PDB ID: 3GGF), STK25 (pink, PDB ID: 2XIK) kinase domains and STRADα (light orange, PDB ID: 3GNI). The αAL helix of OSR1 is colored lime green. Residues lysine and glutamate are shown as sticks. The distances between lysine and glutamate in structures are shown as orange dotted lines. B, αAL helix interacts with αC helix by hydrophobic interaction. ANP and magnesium ion are represented as orange sticks and gray ball, respectively. C, catalytic activities of the purified OSR1 proteins, including OSR1-(1–527) and 6× His-tagged OSR1-(1–527) with αAL deletion, were measured by in vitro kinase assay. GST-NCC was used as a substrate. The CBB staining shows the total amount of kinases and substrates.
Subsequent structural analysis in detail suggests that this αAL helix is important for the autoinhibition of OSR1. The αAL helix is embedded into the groove between N- and C-lobes, which is important for substrate-binding and catalysis (Fig. 2B). Particularly, residues Val-167, Phe-170, and Leu-171 from the αAL helix form hydrophobic interactions with the αC helix, a critical structural element in almost all kinases. Such interactions push the αC helix away from the catalytic core of OSR1, resulting in an inactive conformation. On the other hand, it is generally observed for kinases that a conserved glutamate (Glu-63 in the case of OSR1) from the αC helix would form an ion pair with a conserved lysine (Lys-46 in the case of OSR1) in the VAIK motif when a kinase is activated. However, the potential Lys-46-Glu-63 pairing is disrupted by the inserted αAL helix in OSR1 kinase domain. Instead, residues Lys-46 and Glu-63 form electrostatic interactions with Asp-164 from the DFG motif and Arg-145 from the HRG motif, respectively. Both Asp-164 and Arg-145 are required for catalysis, and thus interactions of these two residues with Lys-46 and Glu-63 may impair their action in catalysis. Although both OSR1 and STRADα belong to the GCK VI subfamily, unlike OSR1, STRADα does not have a helix as the αAL helix of OSR1 to affect the formation of Lys-Glu salt bridge. Next, we compared the length of the Lys-Glu ion pair in several kinases. The distances are 4.0 and 4.2 Å in the active state of MST3 and STK25, respectively. However, the distance becomes 11.9 Å in OSR1, which is comparable with that of MST4 in an inactive state (Fig. 2A). Thus it appears that OSR1 is “structurally inactive” due to the existence of the unique αAL helix.
Next, we performed in vitro kinase assay to verify our structural analysis by detecting the potential inhibitory effect of the αAL helix on OSR1 kinase activity. Two mutants were constructed with the αAL helix deleted on the basis of full-length OSR1 and the kinase domain plus fragment (amino acids 1–338), respectively. As shown in Fig. 2C, either full-length OSR1 or the kinase domain plus fragment, when the αAL helix was removed, exhibited higher kinase activities than their wild-type counterparts. Taken together, these results suggest that the αAL helix acts as another structural lock to inhibit OSR1 kinase activity by interacting with the αC helix and disrupting the conserved Lys-Glu ion pair.
Based on these structural and biochemical studies, we propose a “double lock” model to explain the tight autoinhibition of OSR1. The first lock is the CCT domain of OSR1, which directly associates with its kinase domain to maintain an autoinhibitory conformation by preventing substrate access and the formation of OSR1 homodimer. The second lock is the αAL helix at the N terminus of OSR1 activation loop, which inserts into the catalytic cleft and destroys the Lys-Glu ion pair interaction.
MO25 Binds to OSR1 in a Conserved Manner and Fully Activates OSR1
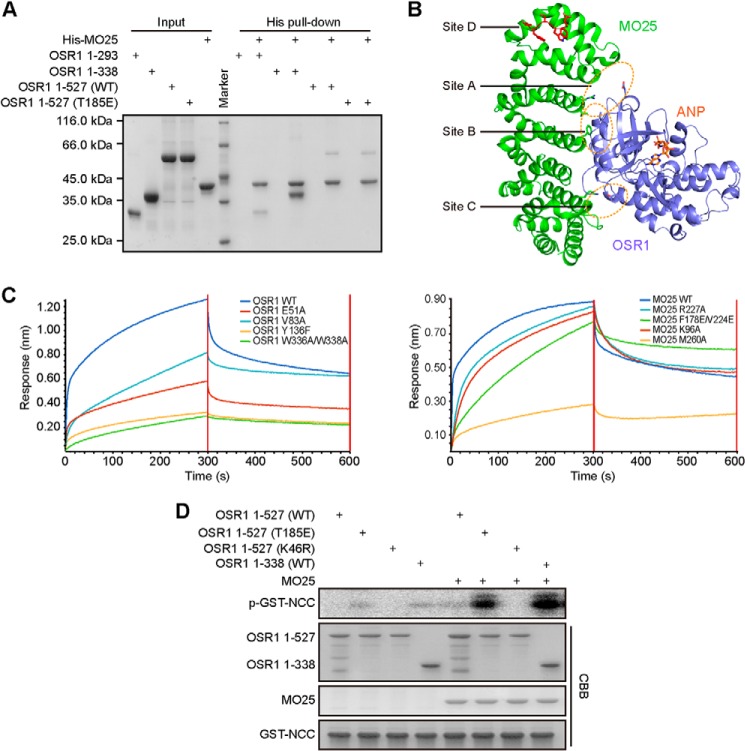
Recent studies have revealed that MO25 can directly bind to and enhance the kinase activity of GCK III (MST3, MST4, and STK25) and GCK VI (OSR1, SPAK, STRAD) kinases (9, 32). To detect the interaction between OSR1 and MO25, we generated several truncation fragments of OSR1 for pull-down assays. OSR1 kinase domain plus (amino acids 1–338), containing the WEW motif (Site D), exhibited the strongest interaction with MO25 (Fig. 3A). In contrast, OSR1 kinase domain (amino acids 1–293) lacking the WEW motif, almost abolished its association with MO25. Surprisingly, full-length OSR1, either wild-type or T185E mutant, showed barely detectable binding to MO25, suggesting an inhibitory role of the CCT domain for MO25 binding.
FIGURE 3.
MO25 interacts with and fully activates OSR1. A, His pull-down to detect the interactions between different constructs of OSR1 and MO25. B, structure of the OSR1-MO25 complex was modeled using the crystal structures of OSR1 kinase domain (slate, PDB ID: 3DAK) and MST4-MO25 complex (PDB ID: 4FZD). MO25 and the kinase domain of OSR1 were colored green and slate, respectively. The Site D of OSR1 and ANP were colored red and orange, respectively. C, interaction between OSR1 kinase domain plus and MO25 was determined by Octet Red 96. The left panel shows the experimental trace obtained from bio-layer interferometry experiments. Wild-type biotinylated MO25 were immobilized on streptavidin biosensors and incubated with mutants of OSR1 proteins. The right panel shows wild-type biotinylated OSR1 incubated with mutants of MO25. D, catalytic activities of OSR1 different fragments were determined in the absence or presence of MO25 using GST-NCC as substrate. The CBB staining shows the total amount of kinase, substrate, and MO25.
Previously, we and others determined the crystal structures of MO25 in complex with different GCK members including MST3, MST4, STK25, as well as the pseudokinase STRADα (25, 28, 31, 33), which defined a unified structural mechanism featuring a MO25-stabilized active conformation of the αC helix and A-loop. The interfaces between MO25 and this group of GCKs are highly conserved within four major sites termed A-D. Since OSR1 also interacts with MO25, we therefore modeled the structure of OSR1-MO25 complex using the crystal structures of OSR1 kinase domain (PDB ID: 3DAK) and MST4-MO25 complex (PDB ID: 4FZD) (Fig. 3B). Subsequently, we performed site-specific mutagenesis to verify this structure model, particularly the four conserved binding sites. To this end, a series of mutants were generated in the contexts of OSR1 kinase domain plus and full-length MO25. Our biolayer interferometry experiments showed that mutations E51A (Site A) and V83A (Site B) of OSR1 partially reduced its binding to MO25 (Fig. 3C). However, single mutation Y136F (Site C), especially double mutation W336A/W338A (Site D) of OSR1, almost abolished OSR1-MO25 interaction. Consistently, substitution of residue Met-260 (Site D) of MO25 with alanine almost completely abrogated its interaction with OSR1. On the other hand, MO25 mutants R227A (Site A), F178E/V224E (Site B), or K96A (Site C) had no significant effect on binding OSR1. Taken together, these results demonstrated that MO25 binds to OSR1 through a similar structural mechanism observed for other MO25-related kinases, and that the WEW motif is critical for the interaction between OSR1 and MO25.
Next we performed in vitro kinase assay using purified recombinant proteins of MO25 and OSR1 (Fig. 3D). Consistent with previous studies (9, 34), our results showed that full-length wild-type OSR1 protein had no detectable activity, even in the presence of MO25. The OSR1 phosphomimetic mutant T185E showed increased activity compared with that of wild-type OSR1, while addition of MO25 further significantly enhanced its activity. Furthermore, we found that OSR1 lacking the CCT domain could be more effectively activated by MO25, again indicating an inhibitory role of the CCT domain during MO25-mediated activation of OSR1.
The CCT-PF1 Region Blocks OSR1 Binding to MO25 While WNK Antagonizes This Effect
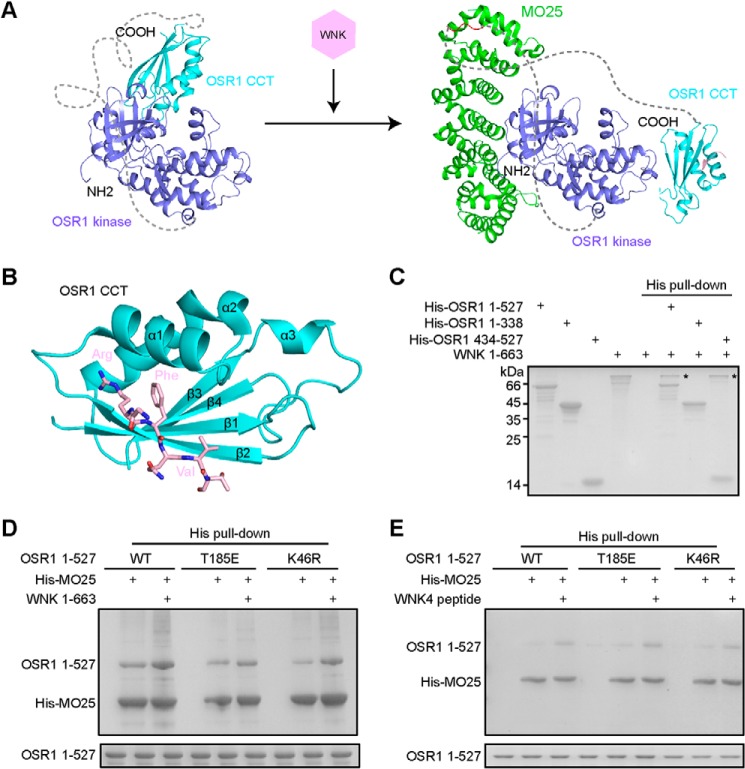
Combining the binding assays for OSR1-MO25 and the kinase assay of OSR1 in the presence or absence of MO25, we hypothesized that the CCT domain, and probably also the immediately preceding PF1 domain can block the interaction between OSR1 and MO25 and therefore inhibit MO25-mediated OSR1 activation. To activate OSR1, the CCT-PF1 region would have to dissociate from the kinase domain where MO25 binds, or at least the conformation of the CCT-PF1 region would have to be altered to allow for direct binding of MO25. Consistent with this notion, full-length OSR1 is unable to bind MO25, but removal of the CCT domain allows for MO25 binding as shown by the truncation mutant (amino acids 1–338). Structurally, MO25 and the CCT domain bind to OSR1 kinase domain at the adjacent regions (Fig. 4A). Besides, a flexible loop between the kinase and CCT domains (amino acids 294–433), whose conformation and orientation relative to the kinase domain would highly depend on whether the CCT domain is attached to or detached from the kinase domain. For example, when the CCT domain is attached to the kinase domain, this PF1 linker may cooperate with the CCT domain to block MO25 binding. We then asked how the CCT/PF1-mediated inhibition is relieved under physiological condition to allow for MO25 binding and thus activation of OSR1. In this regard, it is worth noting that the WNK kinases act as upstream regulators to activate OSR1 through phosphorylation of Thr-185 in the activation loop and Ser-325 in the PF1 domain of OSR1 (19, 34). Moreover, structural study has revealed that WNK kinases interact with OSR1 CCT domain through a unique RFX[V/I] binding motif (12) (Fig. 4B). Therefore, we reasoned that WNKs could be involved in “unlocking” the CCT/PF1-mediated inhibition of MO25 binding to OSR1.
FIGURE 4.
WNKs bind to OSR1 CCT domain, promoting the interaction between OSR1 and MO25. A, structural comparison of OSR1 kinase-CCT domain and OSR1-MO25 complex. The disordered loop (294–433) between OSR1 kinase domain and CCT domain is shown as dotted lines. B, crystal structure of OSR1 CCT domain (cyan) in complex with a WNK4-derived RFXV-containing peptide (pink) (PDB ID: 3DAK). C, purified recombinant proteins of His-tagged OSR1, including full-length (1–527), kinase domain plus (1–338), CCT domain (434–527), and no-tagged WNK1-(1–663) proteins were subjected to His pull-down analysis. D–E, His pull-down assays were performed using purified His-tagged MO25 and untagged OSR1 proteins in the presence or absence of WNK1-(1–663) or WNK4 peptide (GRFQVT). Pull-down samples were subjected to SDS-PAGE. The bottom panel indicates the amount of full-length OSR1 proteins used in each reaction.
To test our hypothesis, we cloned and purified human WNK1 protein (amino acids 1–663). Our pull-down assay showed that WNK1 could interact with both full-length OSR1 and the individual CCT domain (Fig. 4C), consistent with our structural model of the CCT domain bound to the kinase domain showing an exposed RFXV-motif-binding site (Fig. 1E). By contrast, the WNK1 protein did not interact with OSR1 kinase domain plus lacking the CCT domain. Next, we examined the potential effect of WNKs on the interaction between OSR1 and MO25. As shown in Fig. 4, D and E, after incubation with either WNK1 protein or a RFXV-motif-containing peptide (GRFQVT), the interaction between MO25 and OSR1 was significantly enhanced regardless of wild-type, T185E (mimicking phosphorylation by WNKs) or K46R (kinase dead version) mutants. Taken together, these results suggest that in addition to its phosphorylation effect, WNKs can bind to the CCT domain to relieve its inhibition most likely through direct deassociation of the CCT domain from the kinase domain and thus exposing its MO25-binding interface.
OSR1 Regulates Cell Volume Homeostasis in a Manner Dependent on Its Kinase Activity
Previously it has been reported that osmotic stress, such as hyperosmotic stress induced by sorbitol, stimulates the activation of OSR1 (8, 21). The activated OSR1 phosphorylates members of ion cotransporters to maintain cell volume homeostasis. We next assessed the impact of OSR1 kinase activity on the cell volume of HEK 293T cells in the presence or absence of 0.5 m sorbitol. The cell volume reached maximal shrink (∼50%) within half an hour upon stimulation with 0.5 m sorbitol (Fig. 5A). The cells transfected with wild-type OSR1 underwent about 30% volume shrinkage after treatment with 0.5 m sorbitol. Mutation of OSR1 T185E effectively maintained the cell volume within 90%, consistent with the observation that this mutation increases OSR1 activity. The catalytically inactive mutant K46R led to nearly 50% decrease of cell volume. Although the CCT domain truncation increased OSR1 kinase activity in vitro, it failed to maintain cell volume in response to sorbitol-induced hypertonic stress, probably due to that removal of the RFXV motif disables recruitment of both upstream WNK kinases and downstream substrates (Fig. 5B).
FIGURE 5.
The activation of OSR1 promotes cell volume control. A-D, HEK 293T cells were transfected with indicated constructs encoding Flag-tagged OSR1 and/or Myc-tagged MO25. The relative changes of cell size were determined for 3 h at 30-min intervals by Incucyte FLR system. The plus signs indicate cells after treatment with 0.5 m sorbitol. E, model of the regulatory mechanisms of OSR1 activity.
To determine whether MO25 is involved in cell volume control through regulating OSR1 kinase activity, we overexpressed MO25 and OSR1 in HEK 293T cells. As expected, transfection of MO25 prevented cell shrinkage to certain degree (Fig. 5C). Strikingly, cells coexpressing MO25 and wild-type OSR1, especially the T185E mutant, showed no obvious shrink of cell volume upon sorbitol treatment. However, coexpression of wild-type MO25 with OSR1 mutants E51A or V83A that partially disrupted the interaction between MO25 and OSR1, did not maintain cell volume to the level that was observed for wild-type OSR1, indicating a critical role of MO25-induced OSR1 activation (Fig. 5D). Together, these results support the structural mechanism of OSR1 activation by demonstrating that OSR1-regulated cell volume control is dependent on its kinase activity.
DISCUSSION
Regulatory mechanisms of kinase activity are complicated and usually involve various signaling events mediated by upstream activators, phosphatases, adaptor proteins, and even metal ions. OSR1 kinase shows almost undetectable basal activity. Here we demonstrate multiple layers of regulation for OSR1 kinase activity, and propose a “double lock” model to account for its tight autoinhibition. This model features two critical structural elements, the CCT domain and the αAL helix. The CCT domain directly interacts with the kinase domain to not only block substrate access, but also inhibit the domain-swapped homodimerization of the kinase domain and thus its trans-autophosphorylation. More importantly, such intramolecular interaction between the CCT domain and the kinase domain, together with the PF1 region can prevent the kinase domain from binding MO25, a key regulator that is required for full activation of OSR1. On the other hand, OSR1 has a unique αAL helix corresponding to a flexible loop immediately N-terminal the activation loop found in other GCK kinases. This extra helix can interact with the αC helix and disrupt the conserved Lys-Glu ion pair, whose formation is required for kinase activation. Interestingly, such inhibitory helix has also been observed for other low activity kinases such as CDK2, WNK1, and MEK1/2 (35–37), indicative of a general autoinhibitory mechanism. Particularly, WNK1 kinase, which functions as an upstream activator of OSR1, also has relatively low basal activity (36), indicating that the WNK-OSR1 signaling has to be tightly controlled under physiological condition.
MO25 was initially identified as an adaptor protein that forms a heterotrimeric complex with STRAD and LKB1 (24). Lately MO25 was found to also bind to and activate a group of GCKs including OSR1 (9). Our recent structural studies on the regulatory mechanisms of MO25-mediated kinase activation revealed that MO25 alters the conformation of the αC helix of these kinases (28, 31). Mutational study carried out in this work suggests that MO25 may bind to the kinase domain and activate OSR1 in a similar manner, but more dependent on the WXF/W motif. Our structural analysis further suggests that MO25 binding may induce a conformational change of the αAL helix and thus stabilize an active conformation of OSR1. Consistently, a recent study revealed that MO25 binding could help the OSR1 homologous kinase SPAK to attain an active conformation and facilitate its domain-swapped homodimerization (38). However, in resting state of OSR1, the CCT domain binds to the kinase domain and partially covers its MO25-binding site, as shown by the observation that MO25 can better activate the kinase domain than the full-length OSR1. Thus the CCT domain-mediated inhibition of MO25 binding has to be relieved before full activation of OSR1 by MO25.
WNK kinases are upstream activators of OSR1 in the regulation of salt transport and blood pressure. WNKs phosphorylate OSR1 at two conserved sites, residues Thr-185 (A-loop) and Ser-325 (PF1 region) (19). All WNK isoforms contain multiple RFX[V/I] motifs, which recognize and interact with the CCT domain of OSR1 (34, 39). The association between WNKs and the CCT domain of OSR1 is mediated by hydrogen bonds, ionic interactions and hydrophobic packing (12), which is much stronger than that between the kinase and CCT domains of OSR1. Thus, it is likely that WNKs function as an in vivo trigger to relieve the inhibition of OSR1 through binding to the CCT domain and inducing its conformational rearrangement relative to the kinase domain, which would allow for MO25 binding. This notion is supported by our observations that MO25 binding to the full-length OSR1 was dramatically enhanced in the presence of WNK1 or the RFXV-motif-containing peptide. Recently, it has been reported that WNK4 can also interact with MO25 and directly bind to NKCC1 by its CCT-like domain, promoting cotransporter activation in a SPAK/OSR1-independent pathway (40). Mutational analysis indicated that MO25 interacts with WNK kinases in a different manner from its interaction with SPAK/OSR1, which is not contradictory with our results. Moreover, phosphorylation of OSR1 by WNKs is also required for its initial activation since MO25 is unable to activate bacterially expressed OSR1, which was not phosphorylated by WNKs (9).
OSR1 has been implicated in cell volume control. Here, we show that OSR1 regulates cell volume of HEK 293T cells in a manner dependent on its kinase activity. In addition, the CCT domain is also required for such function of OSR1, suggesting the necessity of both kinase activity and substrate recruitment.
Collectively, we propose a multi-step regulatory model for OSR1 kinase activation as shown in Fig. 5E: 1) In the resting state, OSR1 CCT domain associates with the kinase domain, and the αAL helix wedges into the catalytic center, locking OSR1 in an autoinhibited conformation; 2) WNKs bind to the CCT domain of OSR1 and phosphorylate Thr-185 in the A-loop and Ser-325 in the PF1 domain, resulting in an initial activation of OSR1; 3) WNK binding simultaneously alters the intramolecular interactions between the CCT and kinase domains of OSR1, exposing its MO25-binding sites; 4) MO25 binds to and induces conformational changes of OSR1 to stabilize an active conformation for full activation.
Finally, we also noticed that MnCl2 but not MgCl2 can induce SPAK/OSR1 auto-phosphorylation, indicating a preference of divalent cation for these two kinases (39). Our in vitro kinase assay confirmed the selective activation of OSR1 by manganese (data not shown). Exposure to high levels of manganese may cause manganism, a neurological disorder similar to Parkinson disease. The normal concentration of Mn2+ in human adult tissues ranges from 3 to 20 μm and 0.6–1 mm concentration of Mn2+ is sufficient to induce apoptotic cell death in multiple types of cells (41). Moreover, SPAK and OSR1 have important functions in the nervous system, including the developing hypothalamus and neurological disorders (17, 42, 43). Thus it is possible that SPAK and OSR1 may also play a role in manganese-mediated physiology and/or pathology.
This work was supported by the 973 program of the Ministry of Science and Technology of China (2012CB910204), the National Natural Science Foundation of China (31270808, 31300734, 31470736, 31470868), the Science and Technology Commission of Shanghai Municipality (11JC14140000, 13ZR1446400), and the “Cross and Cooperation in Science and Technology Innovation Team” Project of the Chinese Academy of Sciences.
- PAK
- p21-activated kinase
- OSR
- oxidative stress-responsive
- CCT
- conserved C-terminal
- GCK
- germinal center kinase
- SPAK
- STE20/SPS1-related proline/alanine-rich kinase
- STRAD
- pseudokinase STE20-related adaptor
- IPTG
- isopropyl β-d-thiogalactopyranoside
- GA
- glutaraldehyde
- CCT
- conserved C-terminal
- MST
- mammalian STE20-like kinase
- WNK
- with no lysine
- ANP
- adenosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1. Ling P., Lu T. J., Yuan C. J., Lai M. D. (2008) Biosignaling of mammalian Ste20-related kinases. Cell Signal. 20, 1237–1247 [DOI] [PubMed] [Google Scholar]
- 2. Dan I., Watanabe N. M., Kusumi A. (2001) The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230 [DOI] [PubMed] [Google Scholar]
- 3. Record C. J., Chaikuad A., Rellos P., Das S., Pike A. C., Fedorov O., Marsden B. D., Knapp S., Lee W. H. (2010) Structural comparison of human mammalian ste20-like kinases. PloS one 5, e11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delpire E. (2009) The mammalian family of sterile 20p-like protein kinases. Pflugers Archiv : European Journal of Physiology 458, 953–967 [DOI] [PubMed] [Google Scholar]
- 5. Yin H., Shi Z., Jiao S., Chen C., Wang W., Greene M. I., Zhou Z. (2012) Germinal center kinases in immune regulation. Cell Mol. Immunol. 9, 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W., Wang X., Guo T., Li P., Zhao Y., Ji H., Zhang L., Zhou Z. (2014) A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 [DOI] [PubMed] [Google Scholar]
- 7. Leiserson W. M., Harkins E. W., Keshishian H. (2000) Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron 28, 793–806 [DOI] [PubMed] [Google Scholar]
- 8. Chen W., Yazicioglu M., Cobb M. H. (2004) Characterization of OSR1, a member of the mammalian Ste20p/germinal center kinase subfamily. J. Biol. Chem. 279, 11129–11136 [DOI] [PubMed] [Google Scholar]
- 9. Filippi B. M., de los Heros P., Mehellou Y., Navratilova I., Gourlay R., Deak M., Plater L., Toth R., Zeqiraj E., Alessi D. R. (2011) MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J. 30, 1730–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piechotta K., Lu J., Delpire E. (2002) Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1). J. Biol. Chem. 277, 50812–50819 [DOI] [PubMed] [Google Scholar]
- 11. Vitari A. C., Thastrup J., Rafiqi F. H., Deak M., Morrice N. A., Karlsson H. K., Alessi D. R. (2006) Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1. Biochem. J. 397, 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Villa F., Goebel J., Rafiqi F. H., Deak M., Thastrup J., Alessi D. R., van Aalten D. M. (2007) Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep. 8, 839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Villa F., Deak M., Alessi D. R., van Aalten D. M. (2008) Structure of the OSR1 kinase, a hypertension drug target. Proteins 73, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 14. Lee S. J., Cobb M. H., Goldsmith E. J. (2009) Crystal structure of domain-swapped STE20 OSR1 kinase domain. Protein Sci. 18, 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delpire E., Gagnon K. (2006) SPAK and OSR1, key kinases involved in the regulation of chloride transport. Acta Physiologica 187, 103–113 [DOI] [PubMed] [Google Scholar]
- 16. Delpire E., Gagnon K. B. (2008) SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem. J. 409, 321–331 [DOI] [PubMed] [Google Scholar]
- 17. Gagnon K. B., Delpire E. (2012) Molecular physiology of SPAK and OSR1: two Ste20-related protein kinases regulating ion transport. Physiol. Rev. 92, 1577–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Hu J., Vita R., Sun B., Tabata H., Altman A. (2004) SPAK kinase is a substrate and target of PKCtheta in T-cell receptor-induced AP-1 activation pathway. EMBO J. 23, 1112–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vitari A. C., Deak M., Morrice N. A., Alessi D. R. (2005) The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem. J. 391, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richardson C., Sakamoto K., de los Heros P., Deak M., Campbell D. G., Prescott A. R., Alessi D. R. (2011) Regulation of the NKCC2 ion cotransporter by SPAK-OSR1-dependent and -independent pathways. J. Cell Sci. 124, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sengupta S., Lorente-Rodríguez A., Earnest S., Stippec S., Guo X., Trudgian D. C., Mirzaei H., Cobb M. H. (2013) Regulation of OSR1 and the sodium, potassium, two chloride cotransporter by convergent signals. Proc. Natl. Acad. Sci. U.S.A. 110, 18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishida H., Sohara E., Nomura N., Chiga M., Alessi D. R., Rai T., Sasaki S., Uchida S. (2012) Phosphatidylinositol 3-kinase/Akt signaling pathway activates the WNK-OSR1/SPAK-NCC phosphorylation cascade in hyperinsulinemic db/db mice. Hypertension 60, 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cusick J. K., Xu L. G., Bin L. H., Han K. J., Shu H. B. (2006) Identification of RELT homologues that associate with RELT and are phosphorylated by OSR1. Biochem. Biophys. Res. Commun. 340, 535–543 [DOI] [PubMed] [Google Scholar]
- 24. Baas A. F., Boudeau J., Sapkota G. P., Smit L., Medema R., Morrice N. A., Alessi D. R., Clevers H. C. (2003) Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 22, 3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeqiraj E., Filippi B. M., Goldie S., Navratilova I., Boudeau J., Deak M., Alessi D. R., van Aalten D. M. (2009) ATP and MO25alpha regulate the conformational state of the STRADα pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7, e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeqiraj E., Filippi B. M., Deak M., Alessi D. R., van Aalten D. M. (2009) Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milburn C. C., Boudeau J., Deak M., Alessi D. R., van Aalten D. M. (2004) Crystal structure of MO25 alpha in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat. Struct. Mol. Biol. 11, 193–200 [DOI] [PubMed] [Google Scholar]
- 28. Hao Q., Feng M., Shi Z., Li C., Chen M., Wang W., Zhang M., Jiao S., Zhou Z. (2014) Structural insights into regulatory mechanisms of MO25-mediated kinase activation. J. Struct. Biol. 186, 224–233 [DOI] [PubMed] [Google Scholar]
- 29. Ekiert D. C., Friesen R. H., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H. J., Brandenburg B., Vogels R., Brakenhoff J. P., Kompier R., Koldijk M. H., Cornelissen L. A., Poon L. L., Peiris M., Koudstaal W., Wilson I. A., Goudsmit J. (2011) A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 31. Shi Z., Jiao S., Zhang Z., Ma M., Zhang Z., Chen C., Wang K., Wang H., Wang W., Zhang L., Zhao Y., Zhou Z. (2013) Structure of the MST4 in complex with MO25 provides insights into its activation mechanism. Structure 21, 449–461 [DOI] [PubMed] [Google Scholar]
- 32. Gagnon K. B., Rios K., Delpire E. (2011) Functional insights into the activation mechanism of Ste20-related kinases. Cell. Physiol. Biochem. 28, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mehellou Y., Alessi D. R., Macartney T. J., Szklarz M., Knapp S., Elkins J. M. (2013) Structural insights into the activation of MST3 by MO25. Biochem. Biophys. Res. Commun. 431, 604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moriguchi T., Urushiyama S., Hisamoto N., Iemura S., Uchida S., Natsume T., Matsumoto K., Shibuya H. (2005) WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J. Biol. Chem. 280, 42685–42693 [DOI] [PubMed] [Google Scholar]
- 35. De Bondt H. L., Rosenblatt J., Jancarik J., Jones H. D., Morgan D. O., Kim S. H. (1993) Crystal structure of cyclin-dependent kinase 2. Nature 363, 595–602 [DOI] [PubMed] [Google Scholar]
- 36. Min X., Lee B.-H., Cobb M. H., Goldsmith E. J. (2004) Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12, 1303–1311 [DOI] [PubMed] [Google Scholar]
- 37. Ohren J. F., Chen H., Pavlovsky A., Whitehead C., Zhang E., Kuffa P., Yan C., McConnell P., Spessard C., Banotai C., Mueller W. T., Delaney A., Omer C., Sebolt-Leopold J., Dudley D. T., Leung I. K., Flamme C., Warmus J., Kaufman M., Barrett S., Tecle H., Hasemann C. A. (2004) Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 38. Ponce-Coria J., Gagnon K. B., Delpire E. (2012) Calcium-binding protein 39 facilitates molecular interaction between Ste20p proline alanine-rich kinase and oxidative stress response 1 monomers. Am. J. Physiol. Cell Physiol. 303, C1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gagnon K. B., England R., Delpire E. (2006) Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol. Cell. Biol. 26, 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ponce-Coria J., Markadieu N., Austin T. M., Flammang L., Rios K., Welling P. A., Delpire E. (2014) A novel Ste20-related proline/alanine-rich kinase (SPAK)-independent pathway involving calcium-binding protein 39 (Cab39) and serine threonine kinase with no lysine member 4 (WNK4) in the activation of Na-K-Cl cotransporters. J. Biol. Chem. 289, 17680–17688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Latchoumycandane C., Anantharam V., Kitazawa M., Yang Y., Kanthasamy A., Kanthasamy A. G. (2005) Protein kinase Cdelta is a key downstream mediator of manganese-induced apoptosis in dopaminergic neuronal cells. J. Pharmacol. Exp. Ther. 313, 46–55 [DOI] [PubMed] [Google Scholar]
- 42. Grimm P. R., Taneja T. K., Liu J., Coleman R., Chen Y. Y., Delpire E., Wade J. B., Welling P. A. (2012) SPAK isoforms and OSR1 regulate sodium-chloride co-transporters in a nephron-specific manner. J. Biol. Chem. 287, 37673–37690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nugent B. M., Valenzuela C. V., Simons T. J., McCarthy M. M. (2012) Kinases SPAK and OSR1 are upregulated by estradiol and activate NKCC1 in the developing hypothalamus. J. Neurosci. 32, 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]