Background: Assembly of the protein complex associated with Usher syndrome type 2 (USH2) is unclear.
Results: WHRN and PDZD7 heterodimerization and their interactions with USH2A and GPR98 are required for USH2 complex formation.
Conclusion: An USH2 quaternary protein complex may exist in vivo.
Significance: This study provides clues to USH2 pathogenesis and may permit the complex reconstitution for therapeutic intervention.
Keywords: Hair Cell, PDZ Domain, Photoreceptor, Protein Complex, Protein-Protein Interaction, GPR98, PDZD7, USH2A, Usher Syndrome, WHRN
Abstract
Usher syndrome (USH) is the leading genetic cause of combined hearing and vision loss. Among the three USH clinical types, type 2 (USH2) occurs most commonly. USH2A, GPR98, and WHRN are three known causative genes of USH2, whereas PDZD7 is a modifier gene found in USH2 patients. The proteins encoded by these four USH genes have been proposed to form a multiprotein complex, the USH2 complex, due to interactions found among some of these proteins in vitro, their colocalization in vivo, and mutual dependence of some of these proteins for their normal in vivo localizations. However, evidence showing the formation of the USH2 complex is missing, and details on how this complex is formed remain elusive. Here, we systematically investigated interactions among the intracellular regions of the four USH proteins using colocalization, yeast two-hybrid, and pull-down assays. We show that multiple domains of the four USH proteins interact among one another. Importantly, both WHRN and PDZD7 are required for the complex formation with USH2A and GPR98. In this USH2 quaternary complex, WHRN prefers to bind to USH2A, whereas PDZD7 prefers to bind to GPR98. Interaction between WHRN and PDZD7 is the bridge between USH2A and GPR98. Additionally, the USH2 quaternary complex has a variable stoichiometry. These findings suggest that a non-obligate, short term, and dynamic USH2 quaternary protein complex may exist in vivo. Our work provides valuable insight into the physiological role of the USH2 complex in vivo and informs possible reconstruction of the USH2 complex for future therapy.
Introduction
Mutations in USH2A (usherin, MIM *608400), GPR98 (G protein-coupled receptor 98, also known as VLGR1b and MASS1, MIM *602851), WHRN (whirlin, also known as DFNB31, MIM *607928), and PDZD7 (PDZ domain-containing 7, MIM *612971) genes are causal for a spectrum of human diseases, including Usher syndrome type 2 (USH2),2 retinitis pigmentosa without hearing loss, congenital hearing loss without retinitis pigmentosa, and febrile and afebrile seizures (1–8). Among these, USH2 is the most common clinical form of Usher syndrome (USH) and is characterized by combined retinitis pigmentosa and congenital moderate to severe hearing loss. Retinitis pigmentosa is a genetic retinal degenerative disease, manifested in either nonsyndromic or syndromic form (9). Patients with retinitis pigmentosa exhibit early night and peripheral vision loss and late central vision loss, caused by initial rod and subsequent cone photoreceptor cell death. USH2A, GPR98, WHRN, and PDZD7 genes have been found to express in inner ear hair cells and retinal photoreceptors (10–20). In hair cells, proteins encoded by the four genes are colocalized at the ankle link region of the mechanosensitive structure, the hair bundle, during development (10, 11, 16). In photoreceptors, USH2A, GPR98, and WHRN proteins are colocalized at the periciliary membrane complex of the inner segment apex and immediately below the outer segment (21, 22), whereas the localization of PDZD7 remains controversial (1, 16). Knockout or abnormal expression of the Ush2a, Gpr98, Whrn, or Pdzd7 gene causes disorganization and gradual degeneration of hair bundles in mice (10, 11, 16, 21, 22), which leads to reduction of mechanotransduction responses and hearing loss (10, 11, 16). Disruption of Whrn or Ush2a expression in the retina results in slow degeneration (21, 22); vesicles and vacuoles were found to accumulate around the periciliary membrane complex in Whrn mutant photoreceptors (22). Consequently, the four USH genes, USH2A, GPR98, WHRN, and PDZD7, are believed to be important for hair cell development and photoreceptor survival, although each may have relatively different roles in these cellular processes.
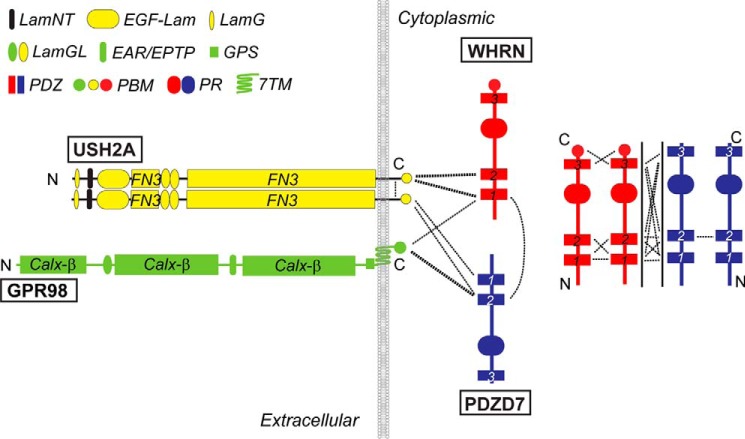
WHRN (Fig. 1) and PDZD7 (Fig. 1) proteins are paralogs sharing 55% similarity in amino acid sequence. Both have several protein-protein interaction domains, including PDZ (PSD95/Dlg1/ZO-1) domains, proline-rich (PR) regions, and harmonin-N like (HNL) domains, suggesting that they are scaffold proteins. USH2A protein is a type I membrane protein with multiple extracellular cell adhesion domains (Fig. 1), and GPR98 protein is a very large adhesion G protein-coupled receptor (GPCR) (23, 24) with multiple tandemly arranged extracellular calcium-binding repeats (Fig. 1). USH2A and GPR98 proteins each have a very short cytoplasmic region carrying a PDZ domain-binding motif (PBM) at their C termini. Biochemical studies demonstrate that the PDZ domains of WHRN and PDZD7 are able to bind the PBMs of USH2A and GPR98 (1, 13, 16, 22, 25). In photoreceptors, USH2A, GPR98, and WHRN proteins show mutual dependence for normal localizations at the periciliary membrane complex, and WHRN is able to recruit USH2A and GPR98 to the periciliary membrane complex (22, 26). In developing cochlear hair cells, some of the USH2A, GPR98, WHRN, and PDZD7 proteins have been demonstrated to be mutually required for normal localizations at the ankle link complex (11, 16). Therefore, USH2A, GPR98, WHRN, and PDZD7 proteins are proposed to form an USH2 protein complex through direct interactions.
FIGURE 1.
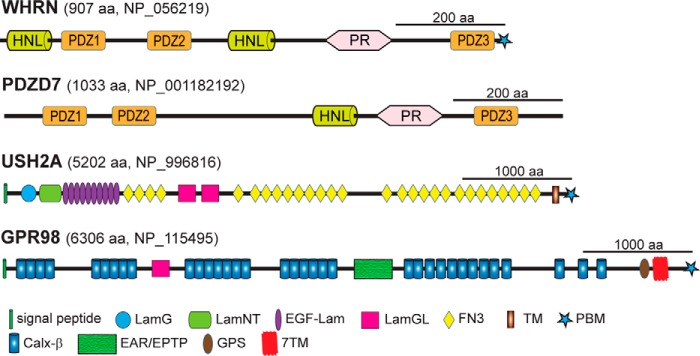
Predicted functional domains of WHRN, PDZD7, USH2A, and GPR98. The longest alternatively spliced isoforms of WHRN, PDZD7, USH2A, and GPR98 are shown. The numbers of amino acids and GenBankTM accession numbers of human sequences are shown at the top of each protein. PR, proline-rich region; LamG, thrombospondin-type laminin G domain; LamNT, N-terminal globular laminin domain; EGF-Lam, laminin EGF-like domain; LamGL, laminin globular-like domain; FN3, fibronectin type III repeat; Calxβ, calx-β motif; EAR/EPTP, epilepsy-associated repeats/epitemptin; GPS, GPCR proteolytic site.
Despite the above findings, no direct evidence has been presented showing the USH2 protein complex formation and its underlying mechanism. Thus, it is unknown how the four USH proteins function together in vivo. Several hurdles exist to addressing these questions in living animals (e.g. the extremely large molecular size of USH2A (5202 aa in humans) and GPR98 (6306 aa in humans) proteins, the existence of transmembrane domains in these two proteins, specific and restricted localizations of the proposed complex on the plasma membrane, and limited amounts of inner ear hair cells and retinal photoreceptors available from animal models. Here, we used a heterologous cell culture expression system to systematically investigate interactions among the known domains of USH2A, GPR98, WHRN, and PDZD7 proteins. Because these interactions are expected to occur inside the cell, we focused on the cytoplasmic regions of USH2A and GPR98 proteins. Based on our findings, we proposed a model for the formation of the USH2 protein complex through direct interactions among its component proteins.
EXPERIMENTAL PROCEDURES
DNA Constructs
DNA constructs were cloned by RT-PCR using the mouse retinal total RNA or subcloned from other constructs containing cDNAs. All cloned constructs were confirmed by DNA sequencing. To generate GFP-, mCherry-, FLAG-, GST-, His-, and HA-tagged proteins, protein cDNAs were cloned in frame with tags into pEGFP-C (Clontech), pmCherry-C1 (modified from pEGFP-C1), p3XFLAG-Myc-CMV-26 (Sigma-Aldrich), pGEX4t-1 (GE Healthcare), pET28a (Novagen, Billerica, MA), and CMV-HA (Clontech) vectors, respectively. GFP-, mCherry-, GAL4 AD-, and GAL4 BD-tagged WHRN FL constructs and GAL4 AD- and GAL4 BD-tagged WHRN PDZ1+2 and WHRN-C constructs were reported previously (22, 27). FLAG-tagged WHRN FL cDNA was subcloned using the GFP- or mCherry-tagged constructs. WHRN PDZ1, PDZ2, PDZ3, PDZ3 PBMΔ, PDZ1+2, and FL PBMΔ cDNAs encoding the protein regions of aa 1–247, 240–469, 710–918, 710–914, 1–469, and 1–914, respectively, (NP_001008791) were cloned from the WHRN FL constructs. GF/AA substitution and β1 deletion in the WHRN PDZ1 and WHRN FL constructs were generated using a site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The GFP-PDZD7 FL construct was described previously (16). PDZD7 PDZ1, PDZ2, PDZ3, PDZ1+2, and FL cDNAs encoding the protein regions of aa 84–64, 209–289, 856–944, 84–289, and 2–1021, respectively (NP_001182194), were cloned from the GFP-PDZD7 FL construct. USH2A (corresponding to aa 5044–5193, NP_067383) and GPR98 (corresponding to aa 6149–6298, NP_473394) cDNAs were inserted into p3XFLAG-Myc-CMV-26, pmCherry-C1, and pEGFP-C1 vectors. FLAG tags were further inserted into the resulting GFP-tagged constructs by PCR. Another USH2A (corresponding to aa 5053–5193, NP_067383) and GPR98 (corresponding to aa 6198–6298, NP_473394) cDNAs were inserted into pGEX4t-1 and pET28a vectors. A third set of USH2A (corresponding to aa 5074–5193, NP_067383) and GPR98 (corresponding to aa 6179–6298, NP_473394) cDNAs were inserted into the pEGFP-C1 vector. The last set of USH2A and GPR98 constructs was used only to measure the molar ratio of components in the USH2 complex and are distinguished from other GFP-tagged USH2A and GPR98 constructs by an asterisk following their names. GAL4 AD-tagged USH2A construct was described previously (22). Full-length mouse vimentin cDNA (corresponding to aa 1–466, NP_035831) was cloned into the pEGFP-C2 vector.
Antibodies
Rabbit and chicken polyclonal antibodies directed against PDZD7, WHRN, and GFP have been reported (16, 22). Another rabbit polyclonal antibody against GFP, a mouse monoclonal antibody against His tag and goat polyclonal antibody against GST were purchased from Abcam (Cambridge, MA). Mouse monoclonal antibodies directed against FLAG tag, HA tag, and BSA were obtained from Sigma-Aldrich. Rabbit polyclonal antibody recognizing mCherry (Clontech) and horseradish peroxidase (HRP)-conjugated AffiniPure secondary antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were also purchased.
Cell Culture and Transfection
HEK293 and COS7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v; HEK293) or 5% (v/v; COS7) fetal bovine serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were transfected with various DNA plasmids using PEI (HEK293, Polysciences, Inc., Warrington, PA), TurboFectTM in vitro transfection reagent (COS7, Fermentas Life Sciences, Glen Burnie, MD), or FuGENE® 6 transfection reagent (COS7, Roche Applied Science) according to the manufacturers' protocols. Cells were harvested for analyses at 24–48 h post-transfection.
FLAG, GST, and His Tag Pull-down Assays
FLAG Pull-down Assays
cDNA constructs of FLAG-tagged proteins and their putative associated proteins were cotransfected into HEK293 cells. After expression of these proteins, cells were homogenized in lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5% (v/v) Triton X-100, 5 mm EDTA, 1× protease inhibitor, and 1 mm DTT). The cell lysates were then cleared by centrifugation at 21,000 × g for 10 min and incubated with anti-FLAG M2-agarose affinity gel (Sigma-Aldrich) for 2 h or overnight with gentle agitation. Agarose beads and their binding proteins were subsequently spun down, washed four times with lysis buffer, and boiled in Laemmli sample buffer for 5 min.
GST Pull-down Assays
GST- and His-tagged proteins were separately expressed in BL21-CodonPlus (DE3)-RIPL cells (Agilent Technologies, Santa Clara, CA) and lysed by sonication and lysozyme treatment in lysis buffer. Wild-type mouse retinas were homogenized in lysis buffer. The Escherichia coli cell lysates containing the GST- and His-tagged proteins and the retinal lysate were mixed and incubated with glutathione-Sepharose beads (GE Healthcare) for 2 h. The Sepharose beads were spun down, washed with lysis buffer three times, and boiled in Laemmli sample buffer for 10 min.
Competitive GST and His Tag Pull-down Assays
GST-USH2A and His-GPR98 cytoplasmic fragments were obtained as described (above). Lysate protein concentrations were determined in Coomassie Blue-stained SDS-polyacrylamide gels by densitometry using BSA (New England Biolabs, Ipswich, MA) as a standard. A GST-USH2A fragment was incubated with the mouse retinal lysate and glutathione-Sepharose beads for 2 h. The beads and their associated proteins were spun down, washed three times with lysis buffer, and incubated with different amounts of His-GPR98 fragment (0 to 6 μg/ml) in lysis buffer for 2 h. Then beads with associated proteins were spun down, and supernatants were mixed with Laemmli sample buffer and boiled for 10 min. The pellets were washed three times with lysis buffer and boiled in Laemmli sample buffer for 10 min. Alternatively, His-GPR98 fragment was first incubated with the mouse retinal lysate and the Ni2+-charged nitrilotriacetic acid-agarose (Novagen, Billerica, MA) for 2 h. The beads and associated proteins were then incubated with the GST-USH2A fragment. The detailed competitive His tag pull-down procedure was similar to the competitive GST pull-down assay. BSA was used in all experiments as a negative control.
All FLAG, GST, and His tag pull-down assays were performed at 4 °C. Inputs and pull-down pellets of these experiments and the supernatants in the competitive pull-down experiments were subjected to standard SDS-PAGE and immunoblotting using appropriate primary antibodies. Signals were developed by sequential incubations with an HRP-conjugated secondary antibody and chemiluminescent substrate and detected using the chemiluminescence mode of a FluorChem Q machine (Proteinsimple, Santa Clara, CA).
Semiquantitative Analysis of Protein Binding Affinities
FLAG pull-down assays were conducted between the two binding partners tagged with FLAG and GFP, respectively. The inputs and pellets were subjected to SDS-PAGE and immunoblotting using anti-FLAG and anti-GFP antibodies. Immunoblot signals were captured under non-saturating conditions; signal intensities of GFP-tagged proteins in the input and pellet lanes and FLAG-tagged proteins in the pellet lanes were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). To normalize differences arising during the transfection and FLAG pull-down steps, the signal intensities of GFP-tagged proteins in the pellets were divided by the signal intensities of their input lanes and the signal intensities of their interacting partners, FLAG-tagged proteins, in the pellet lanes. To reduce variations among different experiments, caused mainly by different exposure times to catch the immunoblot signals, normalized signal intensities of GFP-tagged proteins in the pellets from the interaction between WHRN and USH2A were further normalized by those from the interaction between WHRN and GPR98. The same normalization was also conducted for interactions of PDZD7 with USH2A and GPR98. Student's t tests were conducted to analyze the significance of differences between the interactions of WHRN with USH2A and GPR98 or between the interactions of PDZD7 with USH2A and GPR98.
Stoichiometry of the USH2 Quaternary Protein Complex
Molar ratios were measured in two slightly different USH2 quaternary protein complexes. Besides GFP-tagged WHRN PDZ1 and PDZD7 PDZ2 fragments shared by the two complexes, one complex had the FLAG-GFP-USH2A and GFP-GPR98* cytoplasmic fragments, and the other complex had the FLAG-GFP-GPR98 and GFP-USH2A* cytoplasmic fragments. To measure the molar ratios of the four proteins in complex, HEK293 cells were transfected quadruply with relatively equal amounts of cDNA plasmids of the four protein fragments. Alternatively, HEK293 cells were transfected by the four USH protein fragments individually. The protein expression levels were estimated by signal intensities on the anti-GFP immunoblots. Cell lysates expressing these four proteins were then mixed to generate cell lysates with relatively equal amounts of the four proteins. Standard FLAG pull-down assays were performed using the quadruply transfected or the mixed singly transfected cell lysates, followed by immunoblotting using the anti-GFP antibody. Signal intensities of the three proteins in the FLAG pellet lanes, except for the FLAG-tagged USH2 proteins, were measured using ImageJ software. To calculate the molar amounts of the three protein fragments in the pellet, their signal intensities were divided by their molecular weights. Molar ratios were then calculated by normalizing the molar amounts of the three protein fragments by the molar amount of WHRN fragment in the same pellet for the pull-down experiments using the FLAG-GFP-GPR98 fragment or by the molar amount of the PDZD7 fragment in the same pellet for the pull-down experiments using the FLAG-GFP-USH2A fragment. Substitutions of the FLAG-GFP-USH2A and FLAG-GFP-GPR98 fragments with GFP-USH2A and GFP-GPR98 fragments, respectively, were negative controls in these experiments.
Yeast Two-hybrid Analysis
Yeast AH109 competent cells were made and cotransformations were performed according to the Clontech Yeast Protocols Handbook (PT3024-1). Briefly, three yeast colonies with a diameter of 2–3 mm were picked and grown in 50 ml of YPDA broth at 30 °C with a shaking speed of 260 rpm for 16 h. The resulting yeast broth with an A600 value greater than 1.5 was further transferred to 300 ml of YPDA broth and cultured for another 3 h. After several washes, the competent cells were resuspended in 1× TE, 1× LiAc buffer. For cotransformation, 100 μl of the competent cells were mixed with 0.2 μg of bait and prey plasmid DNAs (0.1 μg each) and 100 μg of carrier DNA. After shaking at 30 °C for 30 min and heat shock at 42 °C for 15 min, the cells were spun down and resuspended in 100 μl of TE buffer. The transformed cells were spread on one DDO (SD/−Leu/−Trp) plate and grown at 30 °C. From the DDO plate, five grown colonies were picked and mixed well in 20 μl of DDO broth. Half of the mixed broth was streaked on a DDO plate, and the other half of the mixed broth was streaked on a QDO (SD/−Leu/−Trp/−Ade/−His) plate with X-α-Gal. Both plates were incubated at 30 °C for 5 days.
RESULTS
USH2A and GPR98 Cytoplasmic Fragments Do Not Interact Directly
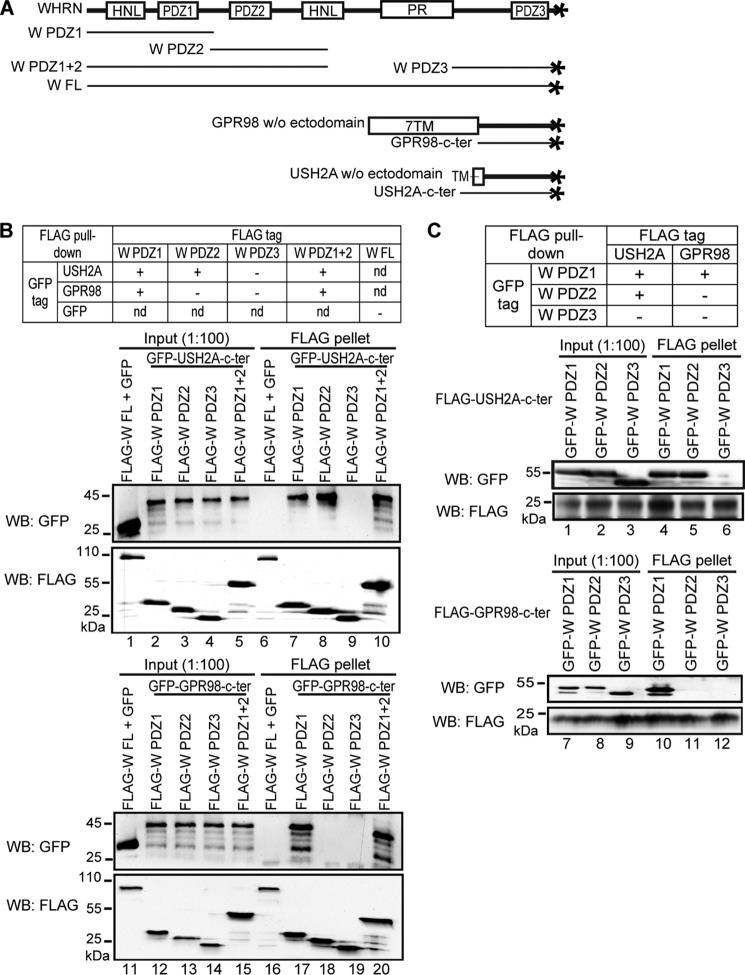
Colocalization of USH2A and GPR98 in photoreceptors and hair cells (22) prompted us to investigate whether these two proteins interacted directly in the proposed USH2 protein complex. Because the USH2 protein complex is presumably present inside the cell, we cloned the entire USH2A and GPR98 cytoplasmic regions and fused them with different tags (Fig. 2A). To determine their interaction, reciprocal FLAG pull-down assays were performed using HEK293 cells double-transfected transiently with differently tagged USH2A and GPR98 cytoplasmic fragments. The ability of FLAG-tagged protein to pull down the other protein would indicate the existence of interaction between these two proteins. We found that USH2A and GPR98 cytoplasmic fragments did not interact directly, although USH2A but not GPR98 cytoplasmic fragment was able to form homodimers (Fig. 2, B and C). Therefore, in order to keep USH2A and GPR98 cytoplasmic fragments in the same protein complex, at least one additional protein that interacts with both USH2A and GPR98 is required.
FIGURE 2.
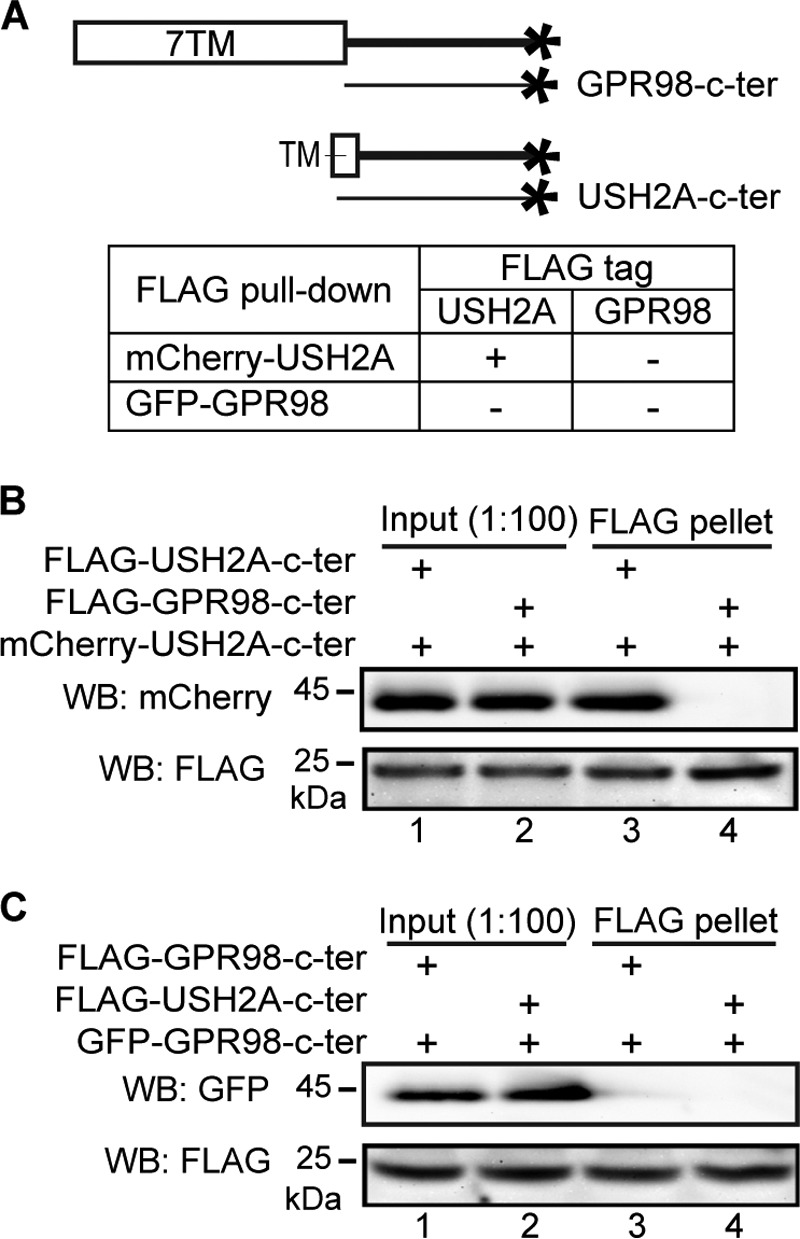
USH2A and GPR98 cytoplasmic fragments do not interact directly, whereas USH2A cytoplasmic fragment interacts with itself. A, diagram showing GPR98 and USH2A fragments used in this experiment and summary of the results. + and −, existence and absence of interactions, respectively. B, FLAG-USH2A (lane 3) but not FLAG-GPR98 (lane 4) cytoplasmic fragment was able to pull down mCherry-USH2A cytoplasmic fragment. C, neither FLAG-GPR98 (lane 3) nor FLAG-USH2A (lane 4) cytoplasmic fragment was able to pull down the GFP-GPR98 cytoplasmic fragment. The anti-FLAG blots in B and C demonstrate success of the FLAG pull-down assays. Lanes 1 and 2 (B and C) are input samples as controls. + (B and C), presence of protein fragments in the reaction.
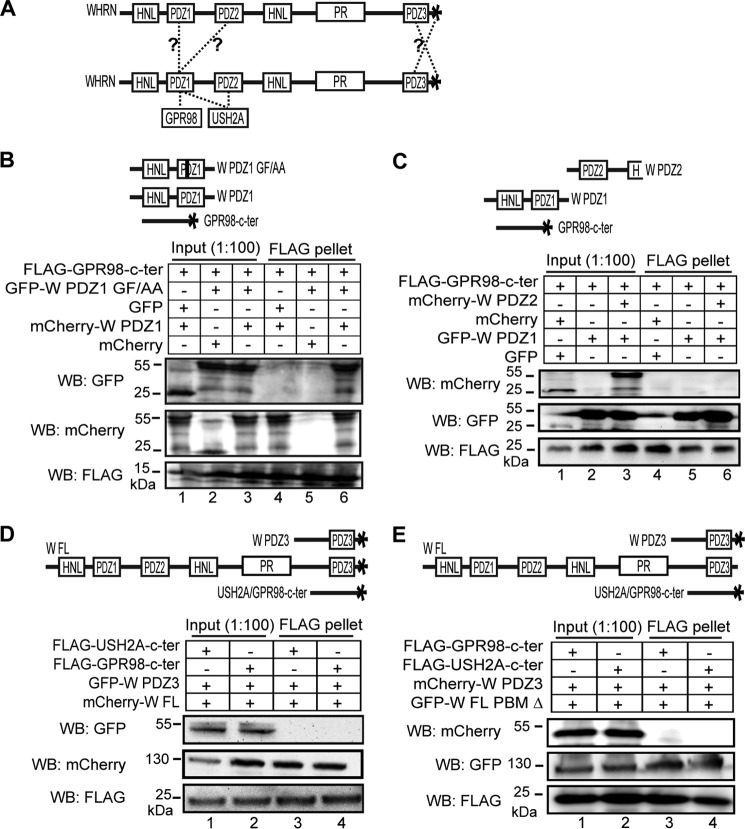
USH2A and GPR98 Interact Differently with WHRN PDZ Domains
USH2A and GPR98 cytoplasmic regions both contain a PBM of the same sequence, Asp-Thr-His-Leu. This PBM of USH2A and GPR98 interacts with WHRN PDZ domains (13, 22, 25). Additionally, WHRN is able to recruit USH2A and GPR98 to their normal locations in photoreceptors (26). Therefore, WHRN may be a candidate link between USH2A and GPR98 in the USH2 protein complex. To examine which WHRN PDZ domains interact with USH2A/GPR98 PBMs, we again performed FLAG pull-down assays. FLAG-WHRN PDZ1, PDZ2, and PDZ1+2 fragments, but not FLAG-WHRN PDZ3 fragment, were able to pull down the GFP-USH2A fragment, whereas the FLAG-WHRN PDZ1 and PDZ1+2 fragments, but not FLAG-WHRN PDZ2 or PDZ3 fragment, were able to pull down the GFP-GPR98 fragment (Fig. 3, A and B). FLAG-WHRN full-length (FL) protein could not pull down GFP, indicating that GFP was not involved in the above interactions. Reciprocally, the FLAG-USH2A fragment was able to pull down WHRN PDZ1 and PDZ2 but not PDZ3 fragments, whereas the FLAG-GPR98 fragment was able to pull down only the WHRN PDZ1 fragment and not the WHRN PDZ2 or PDZ3 fragment (Fig. 3C). Thus, USH2A and GPR98 interact differently with WHRN PDZ domains; the USH2A cytoplasmic fragment interacts with WHRN PDZ1 and PDZ2 domains, whereas the GPR98 cytoplasmic fragment interacts only with WHRN PDZ1 domain. This finding is consistent with a previous report using yeast two-hybrid assays (13).
FIGURE 3.
WHRN PDZ domains interact differently with USH2A and GPR98. A, diagram showing WHRN (W), GPR98, and USH2A fragments used. B, top, summary of the results. Bottom, FLAG-W PDZ1 (lane 7), PDZ2 (lane 8), and PDZ1+2 (lane 10) but not PDZ3 (lane 9) fragments could pull down GFP-USH2A cytoplasmic fragment, whereas FLAG-W PDZ1 (lane 17) and PDZ1+2 (lane 20) but not PDZ2 (lane 18) or PDZ3 (lane 19) fragments could pull down GFP-GPR98 cytoplasmic fragment. FLAG-W FL protein and GFP were used as negative controls (lanes 6 and 16). C, top, summary of the results. Bottom, FLAG-USH2A cytoplasmic fragment could pull down GFP-WHRN PDZ1 (lane 4) and PDZ2 (lane 5) but not PDZ3 (lane 6) fragments, whereas FLAG-GPR98 cytoplasmic fragment could pull down GFP-WHRN PDZ1 (lane 10) but not PDZ2 (lane 11) or PDZ3 (lane 12) fragment. The anti-FLAG blots (B and C) demonstrate the success of the FLAG pull-down assays. Lanes 1–5, 11–15 (B), and lanes 1–3, 7–9 (C) are input samples as controls. +, existence of interactions; −, absence of interactions; nd, not determined.
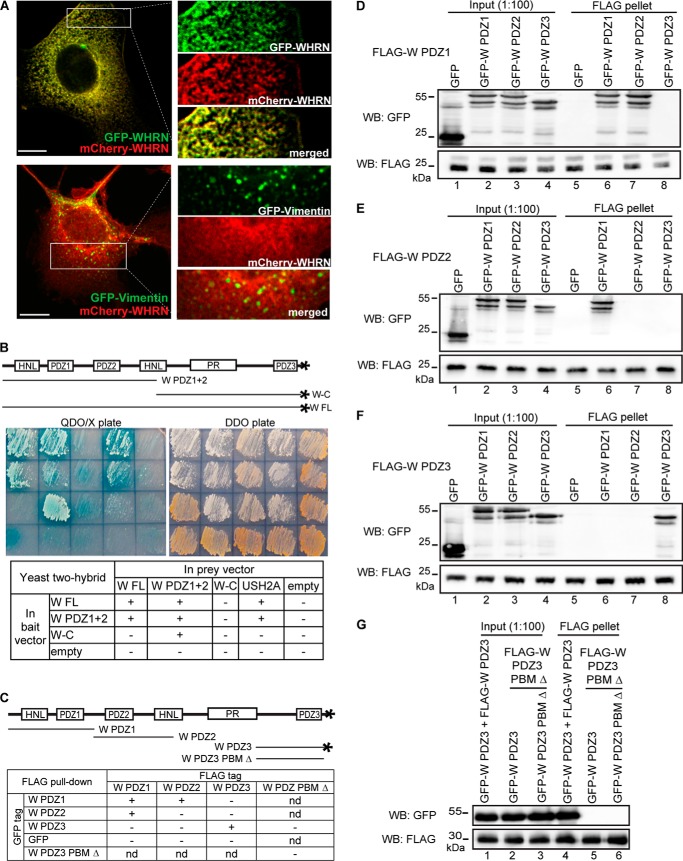
WHRN Forms Homodimers through Its Multiple Domains
WHRN FL and C-terminal half fragments translated in vitro were reported to form dimers (18), suggesting that WHRN dimerization may contribute to the USH2 protein complex formation. To further investigate whether WHRN dimerization occurs in vivo in mammalian cells, we compared the localizations of GFP-tagged and mCherry-tagged WHRN proteins in double-transfected COS7 cells. The two differently tagged WHRN proteins exhibited similar subcellular distributions (Fig. 4A), suggesting colocalization and interaction. As a negative control, mCherry-WHRN did not colocalize with GFP-vimentin, which is an intermediate filament protein and unknown to interact with WHRN. To thoroughly identify WHRN regions responsible for WHRN dimerization, we analyzed the interactions among WHRN FL, N-terminal (WHRN PDZ1+2), and C-terminal (WHRN-C) fragments using yeast two-hybrid analyses. The WHRN PDZ1+2 fragment contains HNL, PDZ1, and PDZ2 domains, and the WHRN-C fragment contains PR, PDZ3, and PBM domains (Fig. 4B). Our results showed that the WHRN PDZ1+2 fragment played an essential role in WHRN dimerization (Fig. 4B), whereas the binding between WHRN PDZ1+2 and WHRN-C fragments might be weak or false due to inconsistent results from two different combinations of bait and prey vectors. We also performed FLAG pull-down assays. WHRN PDZ1 fragment was found to bind to itself as well as to the WHRN PDZ2 fragment (Fig. 4, C and D); WHRN PDZ2 fragment bound only to the WHRN PDZ1 fragment (Fig. 4, C and E); and WHRN PDZ3 fragment bound only to itself (Fig. 4, C and F). Data from yeast two-hybrid analyses and FLAG pull-down assays demonstrate that both WHRN N-terminal and C-terminal regions are involved in WHRN dimerization, and the WHRN PR region may inhibit the dimerization between WHRN C-terminal regions.
FIGURE 4.
WHRN forms homodimers through interactions among its multiple regions. A, GFP-WHRN and mCherry-WHRN colocalize in the cytoplasm when cotransfected in COS7 cells (yellow, top panels). As a negative control, GFP-vimentin and mCherry-WHRN show no colocalization in cotransfected COS7 cells (bottom panels). Signals in white boxes were enlarged and shown (right) in individual and merged channels. Scale bars, 10 μm. B, yeast two-hybrid analysis demonstrates that WHRN (W) FL and W PDZ1+2 fragments form homodimers, whereas the W-C fragment does not. The USH2A cytoplasmic fragment (USH2A) and empty vectors (empty) represent positive and negative controls, respectively. Sample arrangements in the images and table correspond. QDO/X, quadruple dropout medium SD/−Ade/−His/−Leu/−Trp with X-α-Gal to show the existence of interactions. DDO, double dropout medium SD/−Leu/−Trp to show the success of cotransformations. C, diagram of WHRN (W) fragments used in the FLAG pull-down assays (D–G) and summary of the results. D, FLAG-W PDZ1 fragment could pull down GFP-W PDZ1 (lane 6) and PDZ2 (lane 7) fragments but not GFP-W PDZ3 fragment (lane 8) or GFP (lane 5). E, FLAG-W PDZ2 fragment could pull down the GFP-W PDZ1 fragment (lane 6), but not other GFP-W fragments (lanes 7 and 8) or GFP (lane 5). F, FLAG-W PDZ3 fragment could pull down the GFP-W PDZ3 fragment (lane 8) but not other GFP-W fragments (lanes 6 and 7) or GFP (lane 5). G, FLAG-W PDZ3 PBM Δ fragment was unable to pull down the GFP-W PDZ3 fragment (lane 5) or the GFP-W PDZ3 PBM Δ fragment (lane 6). FLAG-W PDZ3 and GFP-W PDZ3 fragments were used as a positive control (lane 4). The anti-FLAG blots (D–G) demonstrate success of FLAG pull-down assays. +, existence of interactions; −, absence of interactions; nd, not determined.
The WHRN PDZ3 fragment used in the above FLAG pull-down experiment has a PDZ domain and a class II PBM, Asn-Val-Met-Leu (28) (Fig. 4C). It was possible that the observed dimerization between WHRN PDZ3 fragments was mediated by interactions between PDZ3 domains and/or between the PDZ3 domain and PBM. To distinguish these possibilities, we generated a mutant WHRN PDZ3 fragment (WHRN PDZ3 PBM Δ), which did not have the PBM (Fig. 4C). FLAG pull-down assays showed that the mutant WHRN PDZ3 fragment pulled down neither the wild-type nor the mutant WHRN PDZ3 fragment (Fig. 4G). Thus, the interaction between the PBM and PDZ3 domain was necessary for WHRN PDZ3 fragment dimerization. The inability of the mutant WHRN PDZ3 fragment to pull down the wild-type WHRN PDZ3 fragment could be explained by unavailability of PBM due to its intramolecular interaction with the PDZ3 domain in the wild-type fragment. In summary, our data demonstrate that WHRN is able to form homodimers through interactions among its three PDZ domains and PBM.
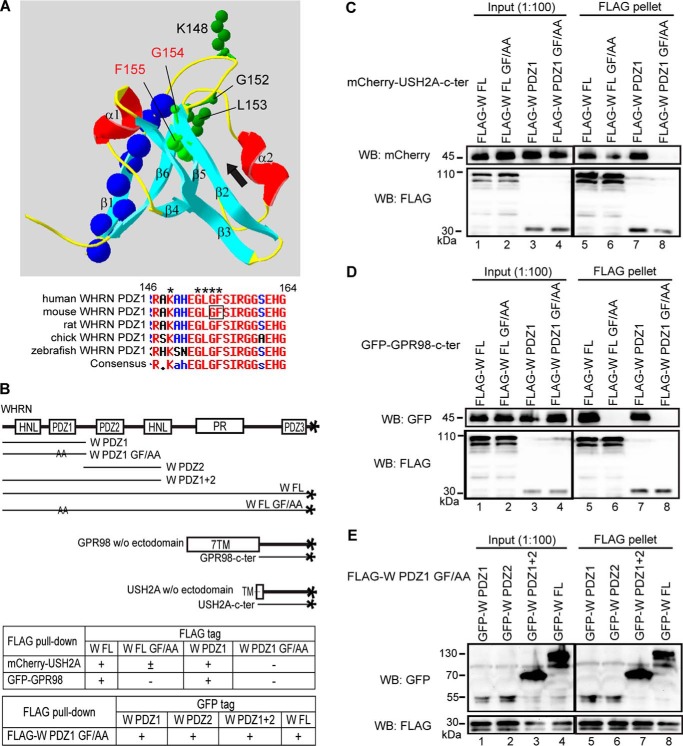
WHRN PDZ1 Domain Independently Dimerizes with WHRN PDZ1/PDZ2 Domain and Interacts with USH2A/GPR98
The ability of WHRN PDZ1 domain to interact with multiple partners, such as itself, WHRN PDZ2, USH2A, and GPR98, prompted us to ask whether the WHRN PDZ1 domain could interact with these partners simultaneously. PDZ domains typically bind to a PBM through their carboxylate-binding loop (28, 29). In WHRN PDZ1 domain, we disrupted the carboxylate-binding loop, Lys-Xaa-Xaa-Xaa-Gly-Leu-Gly-Phe, by substituting key residues Gly154-Phe155 with two alanines (Fig. 5A). FLAG pull-down assays showed that the GF/AA substitution abolished and reduced the binding of WHRN PDZ1 and WHRN FL to the USH2A cytoplasmic fragment, respectively (Fig. 5, B and C). The reduced binding of WHRN FL GF/AA fragment to USH2A was probably due to the remaining WHRN PDZ2 function in this mutant fragment. Similarly, the GF/AA substitution eliminated the bindings of WHRN PDZ1 and FL fragments to the GPR98 cytoplasmic fragment (Fig. 5, B and D). By contrast, the GF/AA substitution did not affect dimerization of the WHRN PDZ1 domain with itself or WHRN PDZ2 domain (Fig. 5, B and E). Thus, the carboxylate-binding loop of the WHRN PDZ1 domain is responsible for interactions with USH2A/GPR98 PBMs but not dimerizations. Further, the inability of the WHRN PDZ1 GF/AA fragment, which includes an intact HNL domain (Fig. 5B), to bind to USH2A or GPR98 cytoplasmic fragment indicates that this HNL domain does not play a role in the binding between WHRN and USH2A or GPR98.
FIGURE 5.
The carboxylate-binding loop of WHRN PDZ1 domain binds to the USH2A/GPR98 PBM. A, three-dimensional structure of human WHRN PDZ1 domain (Protein Data Bank code IUEZ) (top) and sequence alignment of the WHRN PDZ1 carboxylate-binding loop across different species (bottom). From the N to C terminus, WHRN PDZ1 domain has the following β strands and α helixes, β1, β2, β3, α1, β4, β5, α2, and β6. The carboxylate-binding loop, Lys148-Xaa-Xaa-Xaa-Gly152-Leu153-Gly154-Phe155 (green side chains in top panel, asterisks in bottom panel), is located at the N-terminal end of the β2 strand. The PBM-binding groove (black arrow) lies between the β2 strand and α2 helix. The β1 strand is on the opposite side of the carboxylate-binding loop and PBM-binding groove. Residues deleted in the β1 Δ fragments (blue spheres) are labeled. The Gly154-Phe155 residues, replaced by two alanine residues in the GF/AA mutant fragments, are labeled using red type in the top panel and framed in the bottom panel. B, diagram of WHRN (W), GPR98, and USH2A fragments used in the FLAG pull-down assays (C–E) and summary of the results. +, existence of interactions; −, absence of interactions; ±, existence of weak interactions. C, FLAG-W FL GF/AA protein (lane 6) pulled down less mCherry-USH2A cytoplasmic fragment than wild-type FLAG-W FL protein (lane 5). Additionally, FLAG-W PDZ1 GF/AA fragment (lane 8) did not pull down the mCherry-USH2A cytoplasmic fragment, whereas wild-type FLAG-W PDZ1 fragment did (lane 7). D, the FLAG-W FL (lane 6) and PDZ1 (lane 8) GF/AA mutant fragments did not pull down the GFP-GPR98 cytoplasmic fragment, whereas the wild-type FLAG-W FL (lane 5) and PDZ1 (lane 7) fragments did. E, the FLAG-W PDZ1 GF/AA fragment was able to pull down GFP-tagged W PDZ1 (lane 5), PDZ2 (lane 6), PDZ1+2 (lane 7), and FL (lane 8) fragments. The anti-FLAG blots in C–E demonstrate success of the FLAG pull-down assays.
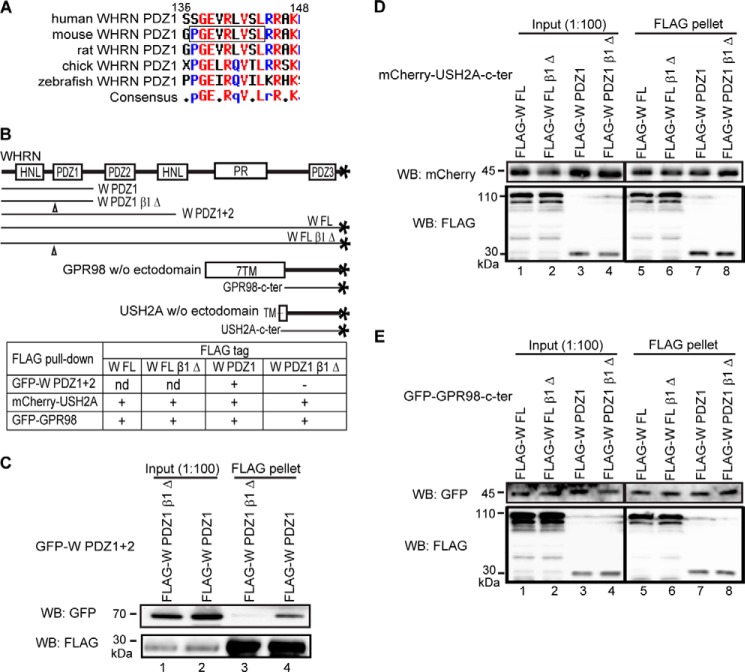
PDZ domains homo- or heterodimerize through their β strands. Among these β strands, β1 serves an important role in several distinct PDZ domain dimerization mechanisms, such as those mediating dimerizations of ZO1 PDZ2, GRIP PDZ6, and Shank1 PDZ domains (30–34). In this study, we deleted the WHRN PDZ1 β1 strand (β1 Δ, Pro-Gly-Glu-Val-Arg-Leu-Val-Ser-Leu 136–144 deletion; Figs. 5A and 6A) and examined the dimerization and PBM-binding abilities of the mutants. β1 strand deletion abolished the ability of the WHRN PDZ1 domain to dimerize with itself and the PDZ2 domain of the WHRN PDZ1+2 fragment (Fig. 6, B and C) but did not affect binding to USH2A or GPR98 PBM (Fig. 6, B, D, and E). Therefore, WHRN PDZ1 domain can interact with partners independently through its carboxylate-binding loop and β1 strand.
FIGURE 6.
WHRN PDZ1 domain homo- and heterodimerizes through its β1 strand. A, sequence alignment of the WHRN PDZ1 β1 strand across different species. The deleted amino acids of the β1 stand are framed here and labeled in Fig. 5A. B, diagram of WHRN (W), GPR98, and USH2A fragments used in the FLAG pull-down assays (C–E) and summary of the results. +, existence of interactions; −, absence of interactions; nd, not determined. C, FLAG-W PDZ1 β1 Δ fragment could not pull down GFP-W PDZ1+2 fragment (lane 3), whereas the wild-type FLAG-W PDZ1 fragment could (lane 4). D and E, deletion of the WHRN PDZ1 β1 strand did not affect bindings of W FL (lanes 5 and 6) and W PDZ1 (lanes 7 and 8) fragments to USH2A (D) or GPR98 (E) cytoplasmic fragment. The anti-FLAG blots in C–E demonstrate success of the FLAG pull-down assays.
WHRN, While Binding to GPR98 or USH2A, Dimerizes Only through Its PDZ1 Domain
We next investigated whether one WHRN protein binding to USH2A/GPR98 could dimerize with another WHRN protein and which WHRN domains are involved in the dimerization (Fig. 7A). We first tested the role of PDZ1 by examining whether the GPR98 cytoplasmic fragment could pull down the mutant WHRN PDZ1 GF/AA fragment in the presence of wild-type WHRN PDZ1 fragment (Fig. 7B). The mutant WHRN PDZ1 GF/AA fragment was unable to bind to GPR98 directly (Fig. 5D). If the wild-type WHRN PDZ1 fragment could dimerize with the mutant WHRN PDZ1 GF/AA fragment and bind to the GPR98 PBM simultaneously, the GPR98 cytoplasmic fragment and mutant WHRN PDZ1 GF/AA fragment could be pulled down together. FLAG pull-down assays using HEK293 cells cotransfected with the three fragments found that the GPR98 cytoplasmic fragment was indeed able to pull down the mutant GFP-WHRN PDZ1 GF/AA fragment, but not GFP, in the presence of the wild-type mCherry-WHRN PDZ1 fragment (Fig. 7B), indicating that WHRN PDZ1 domain can dimerize with itself and bind to GPR98 or USH2A simultaneously.
FIGURE 7.
WHRN, while binding to GPR98 or USH2A, dimerizes only through its PDZ1 domain. A, diagram with the questions to be tested: whether one WHRN protein could bind to GPR98/USH2A and dimerize with another WHRN protein at the same time, and which WHRN regions mediated this dimerization if it occurred. B, FLAG-GPR98 cytoplasmic fragment could pull down GFP-WHRN (W) PDZ1 GF/AA fragment (lane 6) but not GFP (lane 4) in the presence of wild-type mCherry-W PDZ1 fragment. Additionally, the FLAG-GPR98 cytoplasmic fragment could not pull down the GFP-W PDZ1 GF/AA fragment in the presence of mCherry (lane 5). C, FLAG-GPR98 cytoplasmic fragment could not pull down the mCherry-W PDZ2 fragment in the presence of the GFP-W PDZ1 fragment (lane 6), although the FLAG-GPR98 cytoplasmic fragment could pull down the GFP-W PDZ1 fragment (lanes 5 and 6). mCherry and GFP proteins were negative controls (lane 4). Note that, in the anti-GFP blot, weak signals at 55 kDa in lanes 1 and 4 are artifacts caused by sample leaking from other lanes. D, FLAG-USH2A (lane 3) and FLAG-GPR98 (lane 4) cytoplasmic fragments could not pull down GFP-W PDZ3 fragment in the presence of mCherry-W FL protein, although these two FLAG-tagged proteins could pull down the mCherry-W FL protein. E, FLAG-USH2A (lane 4) and FLAG-GPR98 (lane 3) cytoplasmic fragments could not pull down the mCherry-W PDZ3 fragment in the presence of GFP-W FL PBM Δ protein, although these two FLAG-tagged proteins could pull down the GFP-W FL PBM Δ protein. B–E, diagrams of the protein fragments used are shown (top of each panel). The anti-FLAG blots demonstrate the success of the FLAG pull-down assays. + and −, presence and absence of protein fragments in the reaction, respectively.
We then examined whether WHRN PDZ1 domain could bind to USH2A/GPR98 PBMs and form a heterodimer with WHRN PDZ2 domain simultaneously (Fig. 7C). Because USH2A was able to bind to both WHRN PDZ1 and PDZ2 domains (Fig. 3, B and C), which would complicate design and interpretation of our experiment, we decided to use the GPR98 cytoplasmic fragment in this experiment, which bound only to WHRN PDZ1 domain (Fig. 3, B and C). The FLAG-GPR98 cytoplasmic fragment was able to pull down the GFP-WHRN PDZ1 fragment but not the mCherry-WHRN PDZ2 fragment when the three proteins were cotransfected in HEK293 cells (Fig. 7C). Therefore, the GPR98-bound WHRN PDZ1 domain cannot dimerize with the WHRN PDZ2 domain. This result probably holds true when the bindings among WHRN PDZ1 domain, WHRN PDZ2 domain, and USH2A PBM are considered.
To study whether WHRN PDZ3 and PBM participated in WHRN dimerization while WHRN bound to GPR98 or USH2A (Fig. 7D), we transfected WHRN FL and WHRN PDZ3 fragments together with either USH2A or GPR98 cytoplasmic fragment. Interestingly, neither USH2A nor GPR98 could pull down the WHRN PDZ3 fragment in the presence of the WHRN FL protein (Fig. 7D). To exclude the possibility that an intramolecular interaction between PDZ3 domain and PBM of WHRN FL blocked the intermolecular interaction between WHRN FL and WHRN PDZ3 fragments, we repeated the same experiment using WHRN FL PBM Δ protein instead of the wild-type WHRN FL protein and observed the same result (Fig. 7E). Therefore, WHRN FL protein cannot bind the PDZ3 or PBM of another WHRN when its N-terminal region is involved in binding to USH2A or GPR98. Taken together, although the reason is currently unclear, binding of the WHRN N-terminal region with USH2A or GPR98 affects dimerization of WHRN proteins through their PDZ2, PDZ3, and PBM domains. The USH2A/GPR98-bound WHRN can only dimerize with another WHRN through its PDZ1 domain.
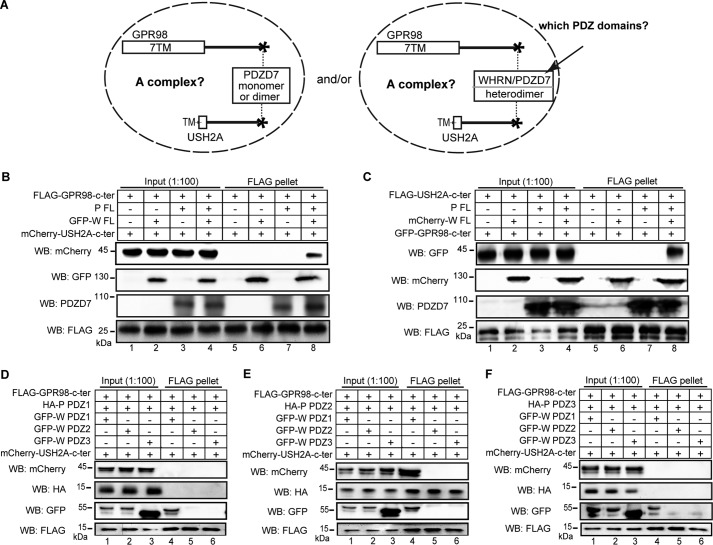
WHRN, USH2A, and GPR98 Are Unable to Form a Complex inside Cells
To test whether WHRN, USH2A, and GPR98 were able to form a complex through binding of the same WHRN protein to both USH2A and GPR98 or through dimerization of USH2A-bound and GPR98-bound WHRN proteins (Fig. 8A), we did triple transfections of HEK293 cells with differently tagged USH2A, GPR98, and WHRN FL fragments. FLAG pull-down assays showed that USH2A and GPR98 fragments were unable to pull down each other in the presence of WHRN FL protein, whereas both USH2A and GPR98 fragments were able to pull down WHRN FL protein, indicating that their interaction domains were functional (lane 6 in Fig. 11, B and C). Therefore, the three proteins could not form a complex. To further verify this finding, we switched to the GST pull-down assay in a cell-free system. We mixed the E. coli cell lysates expressing USH2A and GPR98 cytoplasmic fragments with the mouse retinal lysate containing the endogenous WHRN protein. Similarly, USH2A and GPR98 cytoplasmic fragments could not pull down each other in the presence of the retinal lysate but were able to pull down WHRN from the retinal lysate (Fig. 8B) (data not shown). Additionally, we performed competitive pull-down assays. Increasing amounts of His-tagged GPR98 cytoplasmic fragment, but not BSA, were able to quantitatively remove WHRN from the USH2A-bound pool in the GST pull-down pellet into the supernatant (Fig. 8C). Probably, increasing amounts of GST-USH2A cytoplasmic fragment, but not BSA, were able to quantitatively remove WHRN from the GPR98-bound pool in the His tag pull-down pellet into the supernatant (data not shown). The results from these competitive pull-down assays indicate that the bindings of WHRN to USH2A and GPR98 are mutually exclusive. Therefore, WHRN cannot bind to USH2A and GPR98 simultaneously, and the USH2A-bound and GPR98-bound WHRN proteins are unable to dimerize with each other.
FIGURE 8.
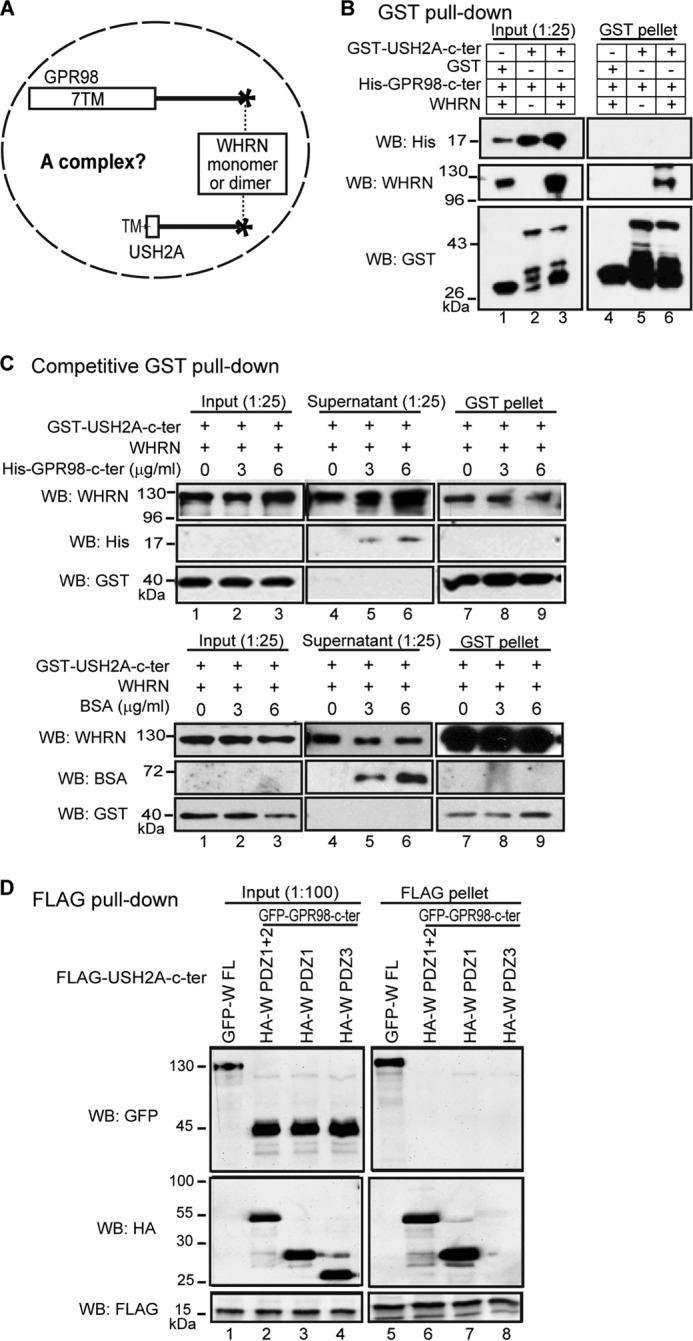
USH2A, GPR98, and WHRN do not form a complex. A, diagram of the question to be tested: whether WHRN, USH2A, and GPR98 can form a complex. B, GST-USH2A cytoplasmic fragment could pull down WHRN but not the His-GPR98 cytoplasmic fragment when the three proteins were mixed (lane 6). Lanes 4 and 5, two different negative controls. C, increasing amounts of His-GPR98 cytoplasmic fragment (top panels) but not BSA protein (bottom panels) competitively removed WHRN from the GST pull-down pellet (lanes 7–9), where WHRN bound to the GST-USH2A cytoplasmic fragment, into the supernatant (lanes 4–6). D, FLAG-USH2A cytoplasmic fragment could not pull down GFP-GPR98 cytoplasmic fragment in the presence of either the HA-WHRN (W) PDZ1+2 (lane 6) or PDZ1 (lane 7) fragment. The HA-W PDZ3 fragment served as a negative control (lane 8). As a positive control, the FLAG-USH2A cytoplasmic fragment could pull down GFP-W FL protein (lane 5). The anti-GST blots in B and C and anti-FLAG blots in D demonstrate the success of the GST and FLAG pull-down assays, respectively. + and −, presence and absence of protein fragments in the reaction, respectively.
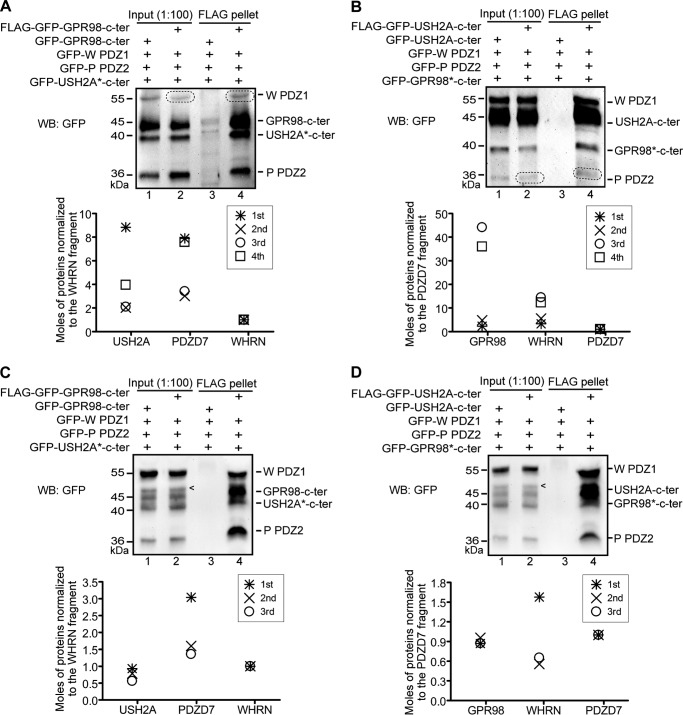
FIGURE 11.
USH2A, GPR98, WHRN, and PDZD7 proteins form a quaternary complex. A, diagram showing the questions to be tested: whether USH2A and GPR98 were able to form a complex with PDZD7 and/or with both PDZD7 and WHRN, and which PDZ domains of WHRN and PDZD7 contributed to the complex formation. B, FLAG-GPR98 cytoplasmic fragment could pull down mCherry-USH2A cytoplasmic fragment only in the presence of both WHRN (W) FL and PDZD7 (P) FL proteins (lane 8) but not in the presence of either W FL (lane 6) or P FL protein (lane 7) alone. Additionally, the FLAG-GPR98 cytoplasmic fragment could not pull down the mCherry-USH2A cytoplasmic fragment directly (lane 5). C, the FLAG-USH2A cytoplasmic fragment could pull down the GFP-GPR98 cytoplasmic fragment only in the presence of both W FL and P FL proteins (lane 8) and not in the presence of either W FL (lane 6) or P FL protein (lane 7) alone. FLAG-USH2A cytoplasmic fragment could not pull down the GFP-GPR98 cytoplasmic fragment directly (lane 5). D–F, FLAG-GPR98 cytoplasmic fragment could pull down the mCherry-USH2A cytoplasmic fragment only in the presence of W PDZ1 and P PDZ2 fragments (lane 4 in E) but not in the presence of any other combinations of W and P fragments (all other lanes in D–F). The anti-FLAG blots (B–F) demonstrate success of the FLAG pull-down assays. + and −, presence and absence of protein fragments in the reaction, respectively.
The WHRN PDZ1 domain was able to dimerize with itself and to interact with GPR98 PBM at the same time (Fig. 7B). Thus, it was possible that the WHRN PDZ1 fragment but not the WHRN FL protein was able to form a complex with GPR98 and USH2A cytoplasmic fragments and that other regions of WHRN FL may interfere with complex formation. To test this, we cotransfected the USH2A and GPR98 cytoplasmic fragments together with WHRN PDZ1 or PDZ1+2 fragment. We found that the FLAG-USH2A cytoplasmic fragment could not pull down the GFP-GPR98 cytoplasmic fragment in the presence of HA-tagged WHRN PDZ1 or WHRN PDZ1+2 fragment (Fig. 8D). Therefore, like the WHRN FL protein, the WHRN PDZ1 fragment could not form a complex with GPR98 and USH2A cytoplasmic fragments.
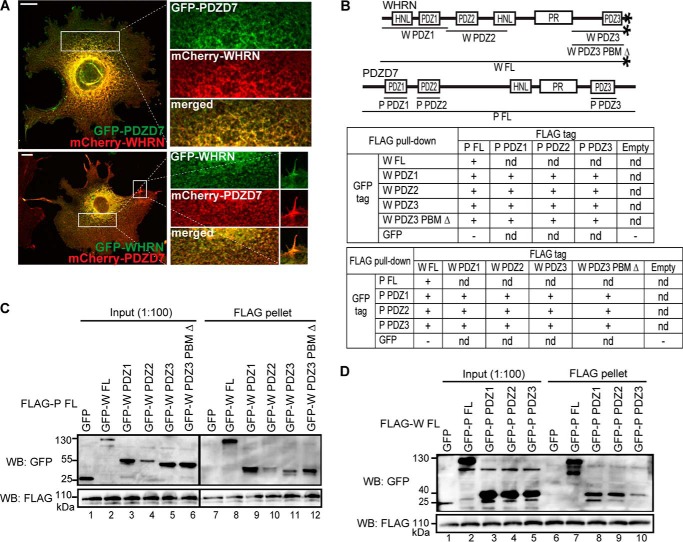
PDZD7 Forms Homodimers through Its PDZ2 Domain and Heterodimers with WHRN through Their Multiple PDZ Domains
Recently discovered in vitro interactions between PDZD7 and WHRN, between PDZD7 PDZ1/PDZ2 and USH2A, and between PDZD7 PDZ2 and GPR98 as well as colocalization of PDZD7 and WHRN in hair cells (1, 16, 19) suggested that PDZD7 might participate in the USH2 protein complex formation. We first investigated whether PDZD7, like its paralog WHRN, could form homodimers. We found that the GFP-tagged and mCherry-tagged PDZD7 proteins colocalized throughout the cytoplasm and filopodia in cotransfected COS7 cells (Fig. 9A). As a negative control, mCherry-PDZD7 protein showed a signal pattern completely different from that of GFP-vimentin, which is not known to interact with PDZD7. This result implied the occurrence of PDZD7 homodimerization in mammalian cells. We further performed FLAG pull-down assays using HEK293 cells cotransfected with various FLAG- and GFP-tagged PDZD7 fragments (Fig. 9B). The PDZD7 FL protein was able to pull down itself and the PDZ2 fragment but not the PDZ1 or PDZ3 fragment (Fig. 9, B and C). Consistently, in reverse FLAG pull-down assays, only interactions between PDZ2 and FL fragments and between PDZ2 fragments themselves were observed (Fig. 9B) (data not shown). Therefore, data from cell culture colocalization and FLAG pull-down experiments demonstrate that PDZD7 is able to form homodimers through the interaction between its PDZ2 domains.
FIGURE 9.
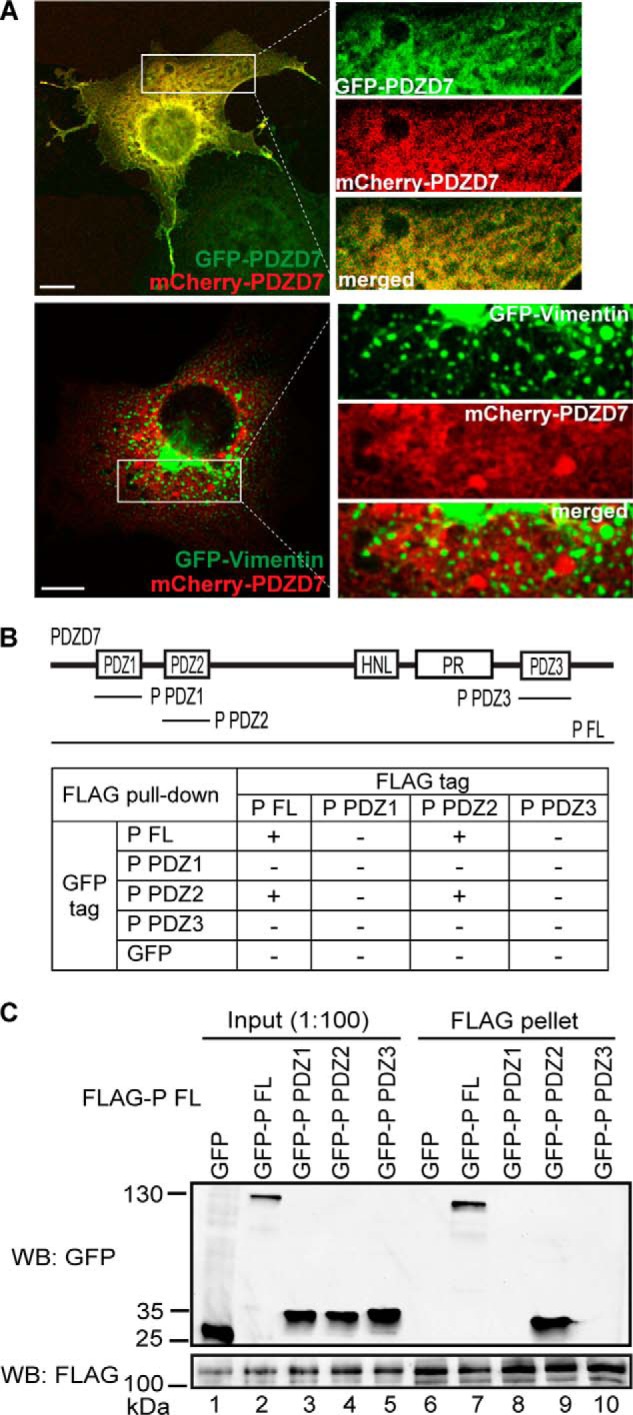
PDZD7 homodimerization is mediated by its PDZ2 domain. A, GFP-PDZD7 and mCherry-PDZD7 proteins colocalized with each other when cotransfected in COS7 cells (top panels). As a negative control, mCherry-PDZD7 and GFP-vimentin proteins did not show similar signal patterns in cotransfected COS7 cells (bottom panels). Signals in white boxes in the images on the left were enlarged and shown (right) in individual and merged channels. Scale bars, 10 μm. B, diagram of PDZD7 (P) fragments used to study PDZD7 homodimerization and summary of results from the FLAG pull-down assays. +, existence of interactions; −, absence of interactions. C, FLAG-P FL protein could pull down GFP-P FL (lane 7) and PDZ2 (lane 9) fragments but not GFP-P PDZ1 (lane 8), GFP-P PDZ3 (lane 10), or GFP (lane 6) fragment. The anti-FLAG blot demonstrates success of the FLAG pull-down assay.
PDZD7 and WHRN heterodimerization, previously suggested by their coimmunoprecipitation (16), was supported here by colocalization of differently tagged PDZD7 and WHRN proteins in COS7 cells (Fig. 10A). To further dissect the PDZD7 and WHRN regions responsible for their heterodimerization, FLAG pull-down assays were exploited. FLAG-PDZD7 FL protein was able to pull down WHRN FL and three individual WHRN PDZ domains (Fig. 10, B and C), and FLAG-WHRN FL protein was able to pull down PDZD7 FL and three individual PDZD7 PDZ domains (Fig. 10, B and D). Consistently, reciprocal FLAG pull-down assays showed that the three WHRN PDZ domains and the three PDZD7 PDZ domains had the ability to bind to each other (Fig. 10B) (data not shown). Additionally, the binding abilities of WHRN PDZ3 and WHRN PDZ3 PBM Δ fragments to the various PDZD7 fragments were similar (Fig. 10B) (data not shown), indicating that it was the WHRN PDZ3 domain, not WHRN PBM, involved in these bindings. Therefore, PDZD7 and WHRN are able to form heterodimers through interactions among their multiple PDZ domains.
FIGURE 10.
Heterodimerization between WHRN and PDZD7 is mediated by their multiple PDZ domains. A, colocalization was observed between GFP-PDZD7 and mCherry-WHRN proteins (top panels) and between GFP-WHRN and mCherry-PDZD7 proteins (bottom panels) when they were cotransfected in COS7 cells. Signals in the cytoplasm (top and bottom images) and filopodia (bottom image), framed in white boxes in the left-hand images, were enlarged and shown on the right in individual and merged channels. Scale bars, 10 μm. B, diagram of WHRN (W) and PDZD7 (P) fragments used in this study and a summary of results from reciprocal FLAG pull-down assays. +, existence of interactions; −, absence of interactions; nd, not determined. C, FLAG-P FL protein could pull down GFP-W FL (lane 8), PDZ1 (lane 9), PDZ2 (lane 10), PDZ3 (lane 11), and PDZ3 PBM Δ (lane 12) fragments but not GFP (lane 7). D, FLAG-W FL protein could pull down GFP-P FL (lane 7), PDZ1 (lane 8), PDZ2 (lane 9), and PDZ3 (lane 10) fragments but not GFP (lane 6). The anti-FLAG blots (C and D) demonstrate the success of the FLAG pull-down assays.
Both PDZD7 and WHRN Are Required for the Formation of a Quaternary Protein Complex with USH2A and GPR98
To explore the role of PDZD7 in USH2 protein complex formation (Fig. 11A), we performed co-transfections in HEK293 cells using variously tagged USH2A cytoplasmic fragment, GPR98 cytoplasmic fragment, WHRN protein, and PDZD7 protein. FLAG-GPR98 cytoplasmic fragment was able to pull down the mCherry-USH2A cytoplasmic fragment only in the presence of both WHRN and PDZD7 proteins and not in the presence of WHRN or PDZD7 protein alone (Fig. 11B). The same result was found using the FLAG-USH2A cytoplasmic fragment to pull down the GFP-GPR98 cytoplasmic fragment (Fig. 11C). Therefore, both WHRN and PDZD7 are required for USH2 protein complex formation. To further dissect WHRN and PDZD7 domains that contribute to complex formation, we performed similar experiments using the three individual PDZ fragments of WHRN and PDZD7. Only the WHRN PDZ1 and PDZD7 PDZ2 domains were required, whereas other PDZ domains of these two proteins were dispensable (Fig. 11, D–F). Therefore, heterodimerization between WHRN PDZ1 domain and PDZD7 PDZ2 domain and interactions of these two PDZ domains with USH2A and GPR98 are essential for the formation of the USH2 quaternary protein complex.
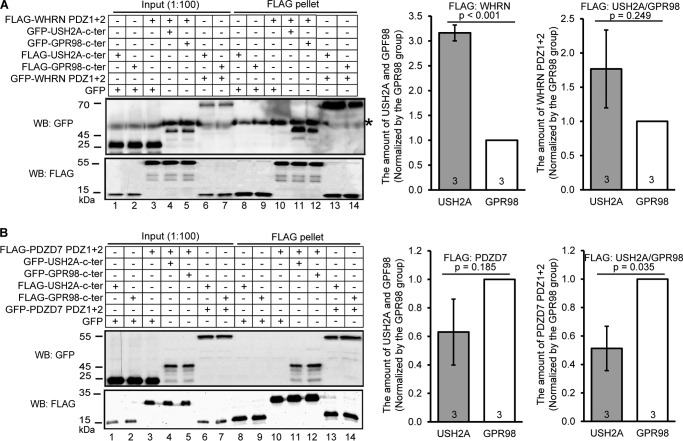
WHRN Binds More Strongly to USH2A than to GPR98, whereas PDZD7 Binds More Strongly to GPR98 than to USH2A
To examine whether WHRN and PDZD7 bound differently to USH2A and GPR98 in the USH2 protein complex, we compared their binding affinities semiquantitatively. FLAG pull-down assays were conducted using HEK293 cells cotransfected with WHRN PDZ1+2 fragment and USH2A or GPR98 cytoplasmic fragment or with PDZD7 PDZ1+2 fragment and USH2A or GPR98 cytoplasmic fragment. It was found that the amount of USH2A fragment pulled down by the FLAG-WHRN PDZ1+2 fragment was significantly more than the amount of GPR98 fragment pulled down by the same WHRN fragment (p < 0.001, Fig. 12A). Reciprocally, the amount of WHRN PDZ1+2 fragment pulled down by the FLAG-USH2A fragment appeared to be more than the amount of the same WHRN fragment pulled down by the FLAG-GPR98 fragment, although this difference was statistically insignificant (p = 0.249; Fig. 12A), which was probably due to the large technical variance inherent in this semiquantitative analysis. On the other hand, the amount of PDZD7 PDZ1+2 fragment pulled down by the FLAG-USH2A fragment was about 50% less than the amount of the same PDZD7 fragment pulled down by the FLAG-GPR98 fragment (p = 0.035; Fig. 12B). Reciprocally, the amount of USH2A fragment pulled down by the FLAG-PDZD7 PDZ1+2 fragment appeared to be about 40% less than the amount of GPR98 fragment pulled down by the same PDZD7 fragment, although this difference was also statistically insignificant (p = 0.185; Fig. 12B). Together, these data suggest that WHRN prefers to bind to USH2A and PDZD7 prefers to bind to GPR98 in the USH2 protein complex in vivo.
FIGURE 12.
WHRN and PDZD7 proteins have different binding affinities to USH2A and GPR98. A, representative immunoblots showing the amount of GFP-USH2A (lane 11) and GFP-GPR98 (lane 12) cytoplasmic fragments pulled down by FLAG-WHRN PDZ1+2 fragment as well as the amount of GFP-WHRN PDZ1+2 fragment pulled down by FLAG-USH2A (lane 13) and FLAG-GPR98 (lane 14) cytoplasmic fragments. GFP served as a negative control (lanes 8–10). Bands labeled by an asterisk are nonspecific. Signal quantification from three independent experiments and p values from Student's t tests are shown (right). B, representative immunoblots showing the amount of GFP-USH2A (lane 11) and GFP-GPR98 (lane 12) cytoplasmic fragments pulled down by FLAG-PDZD7 PDZ1+2 fragment as well as the amount of GFP-PDZD7 PDZ1+2 fragment pulled down by FLAG-USH2A (lane 13) and FLAG-GPR98 (lane 14) cytoplasmic fragments. GFP is a negative control (lanes 8–10). Quantification of signals from three independent experiments and p values from Student's t tests are shown (right). Signals in lanes 11–14 (FLAG blots) and lanes 4–7 (GFP blots) were used to normalize the signals in lanes 11–14 (GFP blots). + and −, presence and absence of protein fragments in the reaction, respectively. Error bars, S.E.
WHRN PBM Has Little Effect on the Interactions between WHRN and USH2A/GPR98
Although WHRN PBM belonged to class II (28), it could interact with WHRN PDZ domains intramolecularly, considering the recent discovery of PDZ domain promiscuity (35, 36). To test whether these potential intramolecular bindings may affect the interactions between WHRN and USH2A/GPR98, we compared the binding of wild-type and mutant WHRN proteins to USH2A or GPR98 cytoplasmic fragment. The mutant WHRN protein (WHRN FL PBM Δ) did not have the PBM. We found no significant difference between the binding of wild-type and mutant WHRN proteins to the USH2A or GPR98 cytoplasmic fragment (data not shown). This result suggests that the potential intramolecular bindings between WHRN PBM and PDZ domains, if they exist, do not affect the binding between WHRN and USH2A or GPR98.
The USH2 Quaternary Protein Complex Has a Variable Molar Ratio of Its Components
To investigate the stoichiometry of components in the USH2 quaternary protein complex, we quadruply transfected HEK293 cells with the USH2A cytoplasmic fragment, GPR98 cytoplasmic fragment, WHRN PDZ1 fragment, and PDZD7 PDZ2 fragment. All protein fragments had a GFP tag. The USH2 protein complex was pulled down by either the USH2A or GPR98 cytoplasmic fragment, which also had a FLAG tag. The molar ratio among the three components except for the FLAG-tagged USH2A or GPR98 fragment in the FLAG pull-down pellet was measured and calculated using signals from anti-GFP immunoblotting. FLAG-tagged USH2A and GPR98 fragments were excluded from the quantification analysis because they might be pulled down excessively in the complex by anti-FLAG-agarose beads. It turned out that the molar ratio among the three protein fragments in the pellet pulled down by either the FLAG-GFP-GPR98 or the FLAG-GFP-USH2A fragment was highly variable from four independent experiments (Fig. 13, A and B). For an unknown reason, we noticed that the amounts of WHRN PDZ1 and PDZD7 PDZ2 fragments in the cell lysate and pull-down pellet were always much smaller than those of the other three proteins in the FLAG-GPR98 and FLAG-USH2A pull-down experiments, respectively (circled bands in Fig. 13, A and B). To avoid the unknown factor leading to significantly uneven expression levels of the four protein fragments, we transfected HEK293 cells separately with the four protein fragments and mixed the singly transfected cell lysates to generate a mixture of approximately equal amounts of the four protein fragments before FLAG pull-down assays. Although molar ratios measured in this way were not as variable as those measured using the quadruply transfected cell lysates, large variations existed among three independent trials (Fig. 13, C and D). The finding that the USH2 quaternary protein complex has variable numbers of USH2A, GPR98, WHRN, and PDZD7 proteins is interpreted as indicating that the cellular complex is non-obligate, dynamic, and heterogeneous.
FIGURE 13.
The USH2 quaternary protein complex has a variable stoichiometry. A and C, representative anti-GFP immunoblots showing GFP-tagged WHRN (W) PDZ1 fragment, USH2A cytoplasmic fragment, and PDZD7 (P) PDZ2 fragment in the pellet pulled down by FLAG-GFP-GPR98 fragment from quadruply transfected cell lysate (lane 4 in A) or the mixed cell lysates individually transfected with each of the four fragments (lane 4 in C). Graphs (bottom) show the calculated moles of USH2A, WHRN PDZ1, and PDZD7 PDZ2 fragments in the FLAG pull-down pellet, normalized by the moles of WHRN PDZ1 fragment in the same pellet, from four (A) or three (C) independent trials. Data from the same experiment (i.e. same FLAG pull-down pellet) are labeled by the same symbols. B and D, representative anti-GFP immunoblots showing GFP-tagged W PDZ1 fragment, GPR98 cytoplasmic fragment, and P PDZ2 fragment in the pellet pulled down by FLAG-GFP-USH2A fragment from their quadruply transfected cell lysate (lane 4 in B) or the mixed cell lysates individually transfected with the four fragments (lane 4 in D). Graphs (bottom) show the calculated moles of GPR98, WHRN PDZ1, and PDZD7 PDZ2 fragments in the FLAG pull-down pellet, normalized by the moles of PDZD7 PDZ2 fragment in the same pellet, from four (B) or three (D) independent trials. Data from the same experiment (i.e. the same FLAG pull-down pellet) are labeled by the same symbols. Substitutions of FLAG-GFP-GPR98 and FLAG-GFP-USH2A fragments with GFP-GPR98 and GFP-USH2A fragments, respectively, are negative controls (lane 3, all panels). Bands circled by dashed lines are the W PDZ1 fragment (A) and P PDZ2 fragment (B). Bands (arrows in C and D) are nonspecific. +, presence of protein fragments in the reaction.
DISCUSSION
We present the first evidence using an in vitro system that USH2A, GPR98, WHRN, and PDZD7 proteins form a quaternary protein complex. Further studies allow us to propose a model explaining how these four USH proteins interact to form this complex (Fig. 14): PDZ1 and PDZ2 domains of WHRN and PDZD7 interact with USH2A PBM, WHRN PDZ1 and PDZD7 PDZ2 domains interact with GPR98 PBM, and the interaction between WHRN PDZ1 and PDZD7 PDZ2 domains is indispensable for linking USH2A and GPR98 cytoplasmic regions in the complex. WHRN prefers to bind to the USH2A cytoplasmic region, whereas PDZD7 prefers to bind to the GPR98 cytoplasmic region. USH2A may exist as oligomers through dimerization of its own cytoplasmic regions. Interactions among the four proteins in the USH2 complex are mainly mediated by PDZ domains, which usually have weak binding affinities in the micromolar range (37). The four USH proteins probably associate and dissociate frequently, which is consistent with our observation that the USH2 protein complex has a variable molar ratio of its four components. Although multiple regions of WHRN and PDZD7 are able to mediate the homo- and heterodimerization of these two proteins, most regions are not required for the USH2 complex formation. They may play a role in regulating the availability of WHRN and PDZD7 to bind to USH2A and GPR98. Because neither WHRN nor PDZD7 alone is able to recruit both USH2A and GPR98 cytoplasmic fragments in the same complex, there are probably no intermediate complexes of the three proteins during the formation of the USH2 quaternary protein complex.
FIGURE 14.
Model for USH2 protein complex formation and in vitro interactions among complex components. Dashed lines indicate the existence of interaction between protein domains they connect. The thickness of dashed lines corresponds to interaction strength. LamNT, N-terminal globular laminin domain; EGF-Lam, laminin EGF-like domain; LamG, thrombospondin-type laminin G domain; LamGL, laminin globular-like domain; EAR/EPTP, epilepsy-associated repeats/epitemptin; GPS, GPCR proteolytic site; PR, proline-rich region; Calxβ, calx-β motif; FN3, fibronectin type III repeat.
In inner ear hair cells, the USH2 protein complex formation can bring the complex components in close proximity, where they act as one functional unit. USH2A could bind to extracellular matrix proteins and/or other transmembrane proteins to facilitate the GPR98-mediated signal transduction. WHRN and PDZD7 may link GPR98 to its intracellular downstream effectors. Because WHRN and PDZD7 may recruit different protein subgroups, it is possible that GPR98 is able to activate functionally synergistic, complementary or opposite intracellular signaling events. Dynamic associations of components in the USH2 protein complex may provide flexibility, thereby allowing rapid on-off switches of signaling. Homo- and heterodimerization of WHRN and PDZD7 may also lead to high level polymerization of these two proteins with the formation of a unique subcellular compartment next to the USH2 protein complex (38–40). In this place, the enrichment of WHRN and PDZD7 could enable their rapid association and dissociation with USH2A and GPR98 and storage of molecules sufficient for GPR98 signaling.
PDZD7 localization could not be determined in mouse photoreceptors in our previous study (16). Additionally, unlike in inner ear hair cells, where PDZD7 is essential for the normal localizations of USH2A, GPR98, and WHRN (16), knockout of Pdzd7 expression in mouse photoreceptors does not affect the localizations of the three USH2 proteins at the periciliary membrane complex (16). These findings suggest that PDZD7 is dispensable for the USH2 complex formation in photoreceptors. Two possibilities may exist. First, another protein, not yet identified, may function at the periciliary membrane complex. This protein may have a domain structure and fulfill a function similar to that of PDZD7. Second, the extracellular regions of USH2A and GPR98 may interact with each other, which leads to formation of the USH2 complex without PDZD7. According to our model (Fig. 14), WHRN may prefer to bind to USH2A in photoreceptors. Interestingly, weak and late onset retinal degeneration has been found in Ush2a−/− and Whrnneo/neo mice (21, 22), whereas no retinal degeneration has been reported in various Gpr98 mutant mice (10, 11, 16, 41–45). Therefore, USH2A and WHRN might have more dominant roles than GPR98 and PDZD7 in photoreceptors. This may explain why, in patients with USH or retinitis pigmentosa, homozygous PDZD7 mutations have not yet been discovered (1, 5), and GPR98 mutations are significantly rarer than USH2A mutations (46).
Previous (13, 22) and current (Fig. 5, B–D) studies demonstrate that WHRN PDZ domains and USH2A/GPR98 PBMs are solely responsible for the interactions between WHRN and USH2A/GPR98 cytoplasmic regions. This could also be true for the interactions between PDZD7 and USH2A/GPR98 cytoplasmic regions. USH2A and GPR98 PBMs have exactly the same amino acid sequence, Asp-Thr-His-Leu. However, USH2A and GPR98 cytoplasmic fragments have different binding specificities and affinities to the PDZ domains of WHRN and PDZD7 (Fig. 14). Recently, it was reported that the amino acids immediately upstream of a PBM could participate in the binding to a PDZ domain (36). From fish, chickens, rodents, monkeys, and humans, 2 and 12 amino acids upstream of the USH2A and GPR98 PBMs are faithfully conserved, respectively. These amino acids in USH2A and GPR98 are completely different. Therefore, the amino acids upstream of USH2A and GPR98 PBMs could be the candidate residues involved in determining the binding specificities and affinities to WHRN and PDZD7 PDZ domains. On the other hand, differences in the carboxylate-binding loop and the PBM-binding groove among the PDZ1 and PDZ2 domains of WHRN and PDZD7 may also contribute to the differential bindings of these PDZ domains to USH2A and GPR98. Among these four PDZ domains, the WHRN PDZ1 and PDZD7 PDZ2 domains have amino acid sequences closest to each other in the carboxylate-binding loop and the PBM-binding groove, whereas, in the same two regions, the amino acid sequence of the PDZD7 PDZ1 domain is least similar to that of the WHRN PDZ1 domain. Further investigation into the mechanism underlying the differential bindings of WHRN and PDZD7 to USH2A and GPR98 is necessary and will provide valuable information regarding the distinct functions of the four USH proteins and the PDZ domain-mediated interactions in multiprotein complexes in general.
In summary, this study demonstrates that interaction between WHRN and PDZD7 is required for formation of the USH2 quaternary protein complex and reveals the dynamic interactions and relative binding affinities of its four component proteins. These findings provide a valuable and plausible model to explain, at a molecular level, how the four USH proteins stay and function together in vivo and why deletions or defects in one of these USH proteins can cause disorganization of the USH2 protein complex and eventually human diseases, such as USH and hearing loss. Further, our findings also suggest that additional proteins interacting with scaffold proteins, WHRN and PDZD7, may contribute unique features to the USH2 protein complex. Our proposed model may facilitate future reconstruction of the USH2 protein complex for therapeutic purposes specifically in photoreceptors and hair cells.
Acknowledgments
We thank Dr. Jeanne M. Frederick for critical reading of the manuscript. We also thank Tihua Zheng and Dr. Li Jiang for purification of the rabbit polyclonal GFP antibody and for sharing the COS7 cell line, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants EY020853 (to J. Y.) and EY014800 (to the Department of Ophthalmology and Visual Sciences, University of Utah). This work was also supported by grants from the Foundation Fighting Blindness (to J. Y.), the E. Matilda Ziegler Foundation for the Blind, Inc. (to J. Y.), Research to Prevent Blindness, Inc. (to J. Y. and the Department of Ophthalmology and Visual Sciences, University of Utah), the Knights Templar Eye Foundation (to Q. C.), the Hearing Health Foundation (to J. Z.), and the National Organization for Hearing Research Foundation (to J. Z.) and a startup package from the Moran Eye Center, University of Utah (to J. Y.).
- USH2
- Usher syndrome type 2
- USH
- Usher syndrome
- WHRN
- whirlin
- USH2A
- usherin
- PBM
- PDZ domain-binding motif
- HNL
- harmonin-N like domain
- GAL4 AD
- activation domain of the GAL4 transcription activator protein
- GAL4 BD
- DNA-binding domain of the GAL4 transcription activator protein
- GF/AA
- the G154A/F155A mutant in the WHRN PDZ1 carboxylate-binding loop
- β1 Δ
- deletion mutant of the WHRN PDZ1 β1 strand
- PBM Δ
- PBM deletion mutant
- GPR98-c-ter
- the GPR98 C-terminal cytoplasmic fragment
- USH2A-c-ter
- the USH2A C-terminal cytoplasmic fragment
- GPCR
- G protein-coupled receptor
- YPDA
- medium blended with yeast extract, peptone, dextrose, and adenine hemisulfate
- DDO
- double dropout medium SD/−Leu/−Trp
- QDO
- quadruple dropout medium SD/−Ade/−His/−Leu/−Trp
- HEK293 cells
- human embryonic kidney 293 cells
- aa
- amino acid(s)
- FL
- full-length
- WB
- Western blot
- TM
- transmembrane domain
- 7TM
- seven-transmembrane domain
- PR
- proline-rich.
REFERENCES
- 1. Ebermann I., Phillips J. B., Liebau M. C., Koenekoop R. K., Schermer B., Lopez I., Schäfer E., Roux A. F., Dafinger C., Bernd A., Zrenner E., Claustres M., Blanco B., Nürnberg G., Nürnberg P., Ruland R., Westerfield M., Benzing T., Bolz H. J. (2010) PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Invest. 120, 1812–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ebermann I., Scholl H. P., Charbel Issa P., Becirovic E., Lamprecht J., Jurklies B., Millán J. M., Aller E., Mitter D., Bolz H. (2007) A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum. Genet. 121, 203–211 [DOI] [PubMed] [Google Scholar]
- 3. Eudy J. D., Weston M. D., Yao S., Hoover D. M., Rehm H. L., Ma-Edmonds M., Yan D., Ahmad I., Cheng J. J., Ayuso C., Cremers C., Davenport S., Moller C., Talmadge C. B., Beisel K. W., Tamayo M., Morton C. C., Swaroop A., Kimberling W. J., Sumegi J. (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280, 1753–1757 [DOI] [PubMed] [Google Scholar]
- 4. Weston M. D., Luijendijk M. W., Humphrey K. D., Möller C., Kimberling W. J. (2004) Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am. J. Hum. Genet. 74, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schneider E., Märker T., Daser A., Frey-Mahn G., Beyer V., Farcas R., Schneider-Ratzke B., Kohlschmidt N., Grossmann B., Bauss K., Napiontek U., Keilmann A., Bartsch O., Zechner U., Wolfrum U., Haaf T. (2009) Homozygous disruption of PDZD7 by reciprocal translocation in a consanguineous family: a new member of the Usher syndrome protein interactome causing congenital hearing impairment. Hum. Mol. Genet. 18, 655–666 [DOI] [PubMed] [Google Scholar]
- 6. Rivolta C., Berson E. L., Dryja T. P. (2002) Paternal uniparental heterodisomy with partial isodisomy of chromosome 1 in a patient with retinitis pigmentosa without hearing loss and a missense mutation in the Usher syndrome type II gene USH2A. Arch. Ophthalmol. 120, 1566–1571 [DOI] [PubMed] [Google Scholar]
- 7. Mburu P., Mustapha M., Varela A., Weil D., El-Amraoui A., Holme R. H., Rump A., Hardisty R. E., Blanchard S., Coimbra R. S., Perfettini I., Parkinson N., Mallon A. M., Glenister P., Rogers M. J., Paige A. J., Moir L., Clay J., Rosenthal A., Liu X. Z., Blanco G., Steel K. P., Petit C., Brown S. D. (2003) Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat. Genet. 34, 421–428 [DOI] [PubMed] [Google Scholar]
- 8. Nakayama J., Fu Y. H., Clark A. M., Nakahara S., Hamano K., Iwasaki N., Matsui A., Arinami T., Ptácek L. J. (2002) A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann. Neurol. 52, 654–657 [DOI] [PubMed] [Google Scholar]
- 9. Hartong D. T., Berson E. L., Dryja T. P. (2006) Retinitis pigmentosa. Lancet 368, 1795–1809 [DOI] [PubMed] [Google Scholar]
- 10. McGee J., Goodyear R. J., McMillan D. R., Stauffer E. A., Holt J. R., Locke K. G., Birch D. G., Legan P. K., White P. C., Walsh E. J., Richardson G. P. (2006) The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J. Neurosci. 26, 6543–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michalski N., Michel V., Bahloul A., Lefèvre G., Barral J., Yagi H., Chardenoux S., Weil D., Martin P., Hardelin J. P., Sato M., Petit C. (2007) Molecular characterization of the ankle-link complex in cochlear hair cells and its role in the hair bundle functioning. J. Neurosci. 27, 6478–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reiners J., van Wijk E., Märker T., Zimmermann U., Jürgens K., te Brinke H., Overlack N., Roepman R., Knipper M., Kremer H., Wolfrum U. (2005) Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum. Mol. Genet. 14, 3933–3943 [DOI] [PubMed] [Google Scholar]
- 13. van Wijk E., van der Zwaag B., Peters T., Zimmermann U., Te Brinke H., Kersten F. F., Märker T., Aller E., Hoefsloot L. H., Cremers C. W., Cremers F. P., Wolfrum U., Knipper M., Roepman R., Kremer H. (2006) The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum. Mol. Genet. 15, 751–765 [DOI] [PubMed] [Google Scholar]
- 14. Zallocchi M., Delimont D., Meehan D. T., Cosgrove D. (2012) Regulated vesicular trafficking of specific PCDH15 and VLGR1 variants in auditory hair cells. J. Neurosci. 32, 13841–13859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zallocchi M., Meehan D. T., Delimont D., Rutledge J., Gratton M. A., Flannery J., Cosgrove D. (2012) Role for a novel Usher protein complex in hair cell synaptic maturation. PLoS One 7, e30573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou J., Zheng T., Ren C., Askew C., Liu X. P., Pan B., Holt J. R., Wang Y., Yang J. (2014) Deletion of PDZD7 disrupts the Usher syndrome type 2 protein complex in cochlear hair cells and causes hearing loss in mice. Hum. Mol. Genet. 23, 2374–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belyantseva I. A., Boger E. T., Naz S., Frolenkov G. I., Sellers J. R., Ahmed Z. M., Griffith A. J., Friedman T. B. (2005) Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat. Cell Biol. 7, 148–156 [DOI] [PubMed] [Google Scholar]
- 18. Delprat B., Michel V., Goodyear R., Yamasaki Y., Michalski N., El-Amraoui A., Perfettini I., Legrain P., Richardson G., Hardelin J. P., Petit C. (2005) Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum. Mol. Genet. 14, 401–410 [DOI] [PubMed] [Google Scholar]
- 19. Grati M., Shin J. B., Weston M. D., Green J., Bhat M. A., Gillespie P. G., Kachar B. (2012) Localization of PDZD7 to the stereocilia ankle-link associates this scaffolding protein with the Usher syndrome protein network. J. Neurosci. 32, 14288–14293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kikkawa Y., Mburu P., Morse S., Kominami R., Townsend S., Brown S. D. (2005) Mutant analysis reveals whirlin as a dynamic organizer in the growing hair cell stereocilium. Hum. Mol. Genet. 14, 391–400 [DOI] [PubMed] [Google Scholar]
- 21. Liu X., Bulgakov O. V., Darrow K. N., Pawlyk B., Adamian M., Liberman M. C., Li T. (2007) Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. U.S.A. 104, 4413–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang J., Liu X., Zhao Y., Adamian M., Pawlyk B., Sun X., McMillan D. R., Liberman M. C., Li T. (2010) Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet. 6, e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liebscher I., Schöneberg T., Prömel S. (2013) Progress in demystification of adhesion G protein-coupled receptors. Biol. Chem. 394, 937–950 [DOI] [PubMed] [Google Scholar]
- 24. Paavola K. J., Hall R. A. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adato A., Lefèvre G., Delprat B., Michel V., Michalski N., Chardenoux S., Weil D., El-Amraoui A., Petit C. (2005) Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum. Mol. Genet. 14, 3921–3932 [DOI] [PubMed] [Google Scholar]
- 26. Zou J., Luo L., Shen Z., Chiodo V. A., Ambati B. K., Hauswirth W. W., Yang J. (2011) Whirlin replacement restores the formation of the USH2 protein complex in whirlin knockout photoreceptors. Invest. Ophthalmol. Vis. Sci. 52, 2343–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L., Zou J., Shen Z., Song E., Yang J. (2012) Whirlin interacts with espin and modulates its actin-regulatory function: an insight into the mechanism of Usher syndrome type II. Hum. Mol. Genet. 21, 692–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sheng M., Sala C. (2001) PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24, 1–29 [DOI] [PubMed] [Google Scholar]
- 29. Ivarsson Y. (2012) Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 586, 2638–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fanning A. S., Lye M. F., Anderson J. M., Lavie A. (2007) Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J. Biol. Chem. 282, 37710–37716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Im Y. J., Lee J. H., Park S. H., Park S. J., Rho S. H., Kang G. B., Kim E., Eom S. H. (2003) Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization. J. Biol. Chem. 278, 48099–48104 [DOI] [PubMed] [Google Scholar]
- 32. Im Y. J., Park S. H., Rho S. H., Lee J. H., Kang G. B., Sheng M., Kim E., Eom S. H. (2003) Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization. J. Biol. Chem. 278, 8501–8507 [DOI] [PubMed] [Google Scholar]
- 33. Gee S. H., Quenneville S., Lombardo C. R., Chabot J. (2000) Single-amino acid substitutions alter the specificity and affinity of PDZ domains for their ligands. Biochemistry 39, 14638–14646 [DOI] [PubMed] [Google Scholar]
- 34. Sugi T., Oyama T., Muto T., Nakanishi S., Morikawa K., Jingami H. (2007) Crystal structures of autoinhibitory PDZ domain of Tamalin: implications for metabotropic glutamate receptor trafficking regulation. EMBO J. 26, 2192–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S. (2008) A specificity map for the PDZ domain family. PLoS Biol. 6, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schreiber G., Keating A. E. (2011) Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 21, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayashi M. K., Tang C., Verpelli C., Narayanan R., Stearns M. H., Xu R. M., Li H., Sala C., Hayashi Y. (2009) The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 137, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu L., Pan L., Zhang C., Zhang M. (2012) Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia. J. Biol. Chem. 287, 33460–33471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson K. R., Zheng Q. Y., Weston M. D., Ptacek L. J., Noben-Trauth K. (2005) The Mass1frings mutation underlies early onset hearing impairment in BUB/BnJ mice, a model for the auditory pathology of Usher syndrome IIC. Genomics 85, 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McMillan D. R., White P. C. (2004) Loss of the transmembrane and cytoplasmic domains of the very large G-protein-coupled receptor-1 (VLGR1 or Mass1) causes audiogenic seizures in mice. Mol. Cell Neurosci. 26, 322–329 [DOI] [PubMed] [Google Scholar]
- 43. Skradski S. L., Clark A. M., Jiang H., White H. S., Fu Y. H., Ptácek L. J. (2001) A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 31, 537–544 [DOI] [PubMed] [Google Scholar]
- 44. Yagi H., Takamura Y., Yoneda T., Konno D., Akagi Y., Yoshida K., Sato M. (2005) Vlgr1 knockout mice show audiogenic seizure susceptibility. J. Neurochem. 92, 191–202 [DOI] [PubMed] [Google Scholar]
- 45. Yagi H., Tokano H., Maeda M., Takabayashi T., Nagano T., Kiyama H., Fujieda S., Kitamura K., Sato M. (2007) Vlgr1 is required for proper stereocilia maturation of cochlear hair cells. Genes Cells 12, 235–250 [DOI] [PubMed] [Google Scholar]
- 46. Millan J. M., Aller E., Jaijo T., Blanco-Kelly F., Gimenez-Pardo A., Ayuso C. (2011) An update on the genetics of Usher syndrome. J. Ophthalmol. 10.1155/2011/417217 [DOI] [PMC free article] [PubMed] [Google Scholar]