Background: Molecular chaperones assist proteins to gain their three-dimensional conformation or triage damaged proteins for degradation.
Results: The chaperone Aha1 prevents aggregation of stressed denatured proteins and favors their ubiquitination.
Conclusion: Aha1 may save the protein folding machinery from overload by misfolded proteins.
Significance: This new function of Aha1 may be crucial to avoid harm to the cell.
Keywords: ATPase, Heat Shock Protein 90 (Hsp90), Molecular Chaperone, Protein Aggregation, Protein Folding, Activator of Hsp90 ATPase (Aha1)
Abstract
Aha1 (activator of Hsp90 ATPase) stimulates the ATPase activity of the molecular chaperone Hsp90 to accelerate the conformational cycle during which client proteins attain their final shape. Thereby, Aha1 promotes effective folding of Hsp90-dependent clients such as steroid receptors and many kinases involved in cellular signaling. In our current study, we find that Aha1 plays a novel, additional role beyond regulating the Hsp90 ATP hydrolysis rate. We propose a new concept suggesting that Aha1 acts as an autonomous chaperone and associates with stress-denatured proteins to prevent them from aggregation similar to the chaperonin GroEL. Our study reveals that an N-terminal sequence of 22 amino acids, present in human but absent from yeast Aha1, is critical for this capability. However, in lieu of fostering their refolding, Aha1 allows ubiquitination of bound clients by the E3 ubiquitin ligase CHIP. Accordingly, Aha1 may promote disposal of folding defective proteins by the cellular protein quality control.
Introduction
The molecular chaperone heat shock protein 90 (Hsp90)2 oversees folding and protein quality control of cellular proteins such as steroid receptors and kinases involved in cellular signaling processes (1). Hsp90 function is dependent on ATP hydrolysis (2, 3) that drives a conformational cycle during which the client protein may fold or is triaged for degradation (4–6). Hsp90 interacts with a multitude of cochaperone proteins that bind to the N-terminal (Hsp90N), middle (Hsp90M), and C-terminal domain (Hsp90C) of the molecular chaperone (4). Among them are Cdc37/p50 and Sba1/p23 that interact with Hsp90N, Aha1 that binds to Hsp90M (7) and to Hsp90N (8), and a cohort of tetratricopeptide repeat (TPR) proteins including Sti/Hop, Tpr1 (9), and the J domain protein Dnajc7 (10) that use the C-terminal EEVD motif of Hsp90 for interaction. It has been shown that some of these interactions are mutually exclusive and even cochaperones that use different places on Hsp90 cannot bind at the same time to the molecular chaperone due to steric exclusion (11, 12). A couple of cochaperone proteins such as Sti1/Hop, Cdc37/p50, Sba1/p23, and Aha1 regulate the ATPase activity of the molecular chaperone and may thereby adjust the speed of the Hsp90 conformational cycle to the demands of a particular client protein (4, 13). Using purified yeast proteins it was shown that Aha1 stimulates the intrinsic ATPase activity of Hsp90 about one order of magnitude (14, 15), while Cdc37/p50, Sti1/Hop and Sba1/p23 have an inhibiting effect. For efficient activity the N-terminal domain of Aha1 (Aha1N) and the C-terminal domain of Aha1 associate with the Hsp90 dimer in an asymmetric fashion, contacting Hsp90M and the catalytic Hsp90N domain (8, 16). Furthermore, the association between Hsp90 and the cochaperones Aha1 and Cdc37/p50 that regulate its ATP hydrolysis rate is controlled by phosphorylation of the molecular chaperone at distinct tyrosine residues during the chaperone cycle (17).
Several studies indicate that Aha1 together with Hsp90 promotes optimal activation of client proteins (11, 14, 15, 18). Although Aha1 is not an essential protein, deletion in yeast cells dramatically reduces activity of the ectopically expressed Hsp90 dependent model kinase v-Src. In mammalian cells Aha1 supports the activity of steroid receptors and kinases such as Raf, suggesting that Aha1-dependent Hsp90 ATPase stimulation plays a critical role in the folding process. On the other hand, Aha1 has adverse effects in the case of the difficult to fold Hsp90 client CFTR (cystic fibrosis transmembrane conductance regulator) and the disease causing CFTRΔF508 mutant. Aha1 promotes degradation of CFTR, a process that could be rescued by down-regulation of Aha 1 protein levels (19).
The two somewhat conflicting observations prompted us to analyze the interaction between Hsp90 and Aha1 in a series of tissues. We consistently find the majority of Aha1 independent of Hsp90 as a self-contained protein suggesting that Aha1 may serve additional functions beyond the stimulation of Hsp90 ATPase activity. Next, we tested chaperone activity of Aha1 and found that this cochaperone prevents aggregation of denatured proteins. Surprisingly, rather than attempting to refold bound clients, Aha1 holds them for ubiquitination by the cellular protein quality control.
EXPERIMENTAL PROCEDURES
Expression Constructs and Purification of Recombinant Proteins
Human hHsp90 and yeast yHsp90, human hAha1, Δ22hAha1, hAha1C, hAha1N, and yeast yAha1 constructs in the pProExHTa vector were expressed in Escherichia coli BL21(DE3) pLsyS cells after induction with 0.2 mm IPTG at 18 or 25 °C for 5 - 16 h (15, 20). Proteins were purified on nickel-nitriloacetate chelate affinity resin followed by gel filtration on a Sephacryl S-400 column (GE Healthcare) and ResourceQ (GE Healthcare) ion exchange chromatography on an ÄktaPurifier system (GE Healthcare). His6 tags could be removed by treatment with TEV (tobacco etch virus) protease. Hop was prepared as reported previously (20). GroEL was expressed at 34 °C from a plasmid in E. coli and purified by subsequent ion exchange chromatography on DE-52 cellulose and gel filtration on a Sephacryl S-300 (GE Healthcare) column. Firefly luciferase with C-terminal Myc and His6 tags (Luc-MycHis) was expressed at 30 °C from a pET3a vector (Novagen) in E. coli strain B834 (Novagen). The protein was purified on nickel-nitriloacetate chelate affinity resin followed by ion exchange chromatography on a MonoQ (GE Healthcare) column. Human CHIP was expressed in E. coli from a pProExHTa vector and Hsp40 from a modified pET23a plasmid (21) and both proteins purified by Nickel-nitriloacetate chelate affinity chromatography. Hsc70 was purified from bovine liver by subsequent steps of DE-52 cellulose chromatography, ATP-agarose affinity chromatography and hydroxyapatite chromatography using Bio-Gel HTP (Bio-Rad).
Gel Permeation Chromatography Analysis of Tissue and Cell Lysates
Heart, brain, and liver tissue from rhesus macaque (German primate Center, Göttingen), murine lung, and HEK293 culture cells were homogenized on ice in 25 mm Tris pH 7.4, 50 mm KCl, 1 mm DTT, 1 mm EDTA, 1 mm EGTA, 1 mm NaF supplemented with HALT protease inhibitor mixture (Pierce) using a Potter homogenizer. Following centrifugation, supernatant proteins were separated on a Superose 12 HR 10/30 column equilibrated in the same buffer using an ÄktaPurifier system (22). Fractions containing 500 μl were collected and analyzed by SDS-PAGE on 12% gels. To detect proteins by Western blotting, antibodies ab56721 (Abcam) specific for Aha1, SRA-1500 (Stressgen Biotechnologies) specific for Hop and the rabbit polyclonal antibody R699 raised against the highly conserved peptide NKTKPIWTRNPDDI deduced from the human Hsp90α/β sequences were used. Purified Hsp90, Aha1, and Hop were run as standards and visualized by Coomassie staining.
Prevention of Rhodanese Aggregation Assay
Prevention of rhodanese aggregation was performed essentially as described (23). Briefly, 100 μm bovine liver rhodanese (Sigma) denatured in 6 m guanidinium HCl, 30 mm Tris pH 7.4, 1 mm DTT was rapidly diluted to 0.5 μm into 30 mm Tris pH 7.4, 50 mm KCl containing the chaperonin GroEL at rhodanese:GroEL (14-mer) ratios of 1:0.125, 1:0.25, 1:0.5, and 1:1 (corresponding to 50, 100, 200 and 400 μg/ml of GroEL per 16.7 μg/ml rhodanese) or in buffer alone to allow maximal aggregation. To test whether Aha1 can suppress protein aggregation, we tested rhodanese aggregation in buffer containing the same amounts of yeast yAha1, human hAha1, or the truncated fragments Δ22hAha1, hAha1C, and hAha1N. As control, we diluted rhodanese into buffer supplemented with matching amounts of bovine serum albumin (BSA).
Measurement of Hsp90 ATPase Activity
Hsp90 ATPase activity was measured essentially as described by a coupled enzymatic system (3, 20, 24). Briefly, ADP produced by the molecular chaperone is re-phosphorylated to ATP by pyruvate kinase (PK) that converts phosphoenolpyruvate to pyruvate. Pyruvate is reduced to lactate by lactate dehydrogenase (LDH) that oxidizes NADH to NAD+ and this reaction can be monitored by the decrease in absorption at 340 nm. Since PK and LDH catalyzed reactions are fast compared with Hsp90 dependent ATP hydrolysis, the ATPase rate is directly coupled to NADH consumption and to the decrease of absorption at 340 nm. ATPase experiments were performed at 37 °C in 40 mm Hepes-KOH buffer pH 7.4, 2 mm MgCl2 and contained 2 μm of yHsp90 or hHsp90, 20 μm of yAha1, hAha1, or Δ22hAha1 where indicated, 0.1 μg/μl of PK and LDH (Roche), 1 mm ATP, 2 mm phosphoenol pyruvate, 0.1 mm NADH, and 1% DMSO. Parallel assays received 100 μm Radicicol in 1% DMSO to inhibit the Hsp90 ATPase and measure background activity. Thus, subtraction of two corresponding experiments in the presence or absence of Radicicol allows determining the specific ATP hydrolysis rate of Hsp90.
Refolding of Denatured Firefly Luciferase
Firefly luciferase (Sigma) was denatured at 10 μm in 6 m guanidinium HCl, 30 mm Tris pH 7.4, 1 mm DTT and rapidly diluted to 0.1 μm into refolding buffer (10 mm MOPS, pH 7.4, 50 mm KCl, 5 mm MgCl2; 1 mm ATP) that contained either 1 μm Hsc70; 2 μm Hsp40 and 5% rabbit reticulocyte lysate or 5% rabbit reticulocyte lysate only as control and incubated for 1 h at 30 °C for refolding, essentially as described (25, 26). To test the effect on luciferase refolding, hAha1 was added in increasing amounts from 2 to 8 μm, as indicated, together with Hsc70 and Hsp40. BSA served as a control. Alternatively, denatured firefly luciferase was refolded in buffer containing 20% RRL only. To test the effect on luciferase refolding, hAha1 was added in increasing amounts from 4 to 16 μm, as indicated. BSA served as a control. Samples were withdrawn after 5, 15, 30, 45, and 60 min as indicated, added to luciferase assay reagent (Promega) and refolding measured by means of luminescence using an EnSpire Multimode Reader (PerkinElmer).
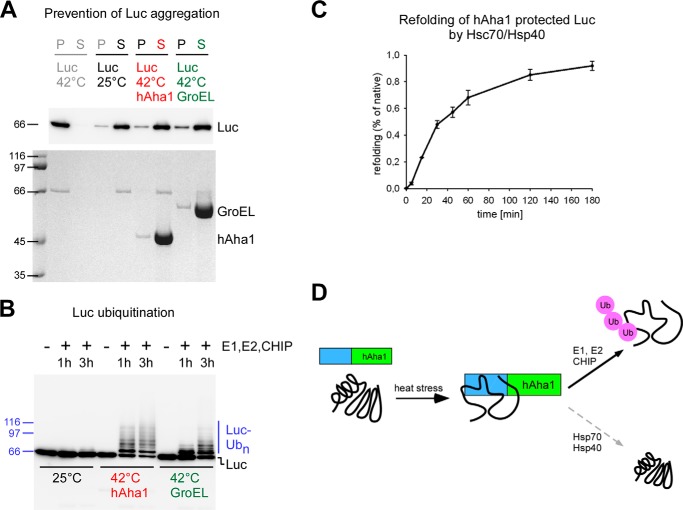
Prevention of Luciferase Aggregation and Ubiquitination Assay
Purified Luciferase with a C-terminal MycHis6 tag (Luc-MycHis) (50 μg/ml) was incubated for 5 min in 50 mm Tris, pH 7.4, 5 mm MgCl2 alone or in the presence of 650 μg/ml hAha1, yAha1, Δ22hAha1, or GroEL at 25 °C. Thereafter, the samples were thermally stressed at 42 °C for 10 min and subsequently spun for 5 min in a microcentrifuge at full speed at 4 °C to separate pellet and supernatant. Native luciferase kept in buffer at 25 °C was used as a control. The samples were analyzed by 10% SDS-PAGE and Western blotting with an anti Myc antibody detecting Luc-MycHis6. Aliquots of the supernatants were added to an ubiquitination mixture in the same buffer as above supplemented with 0.1 μm E1 (UBE1L, Enzo Life Sciences), 1 μm E2 (UbcH5a, Enzo Life Sciences), 5 μm of freshly prepared CHIP (an E3 ubiquitin ligase), 100 μm ubiquitin (Enzo Enzo Life Sciences), 2 mm ATP and incubated for the time indicated at 30 °C. Samples were analyzed by Western blotting with the Myc-specific antibody 9E10 (from hybridoma cells supernatant). The appearance of a ladder of additional bands in 8 kDa increments above the basal signal indicates ubiquitination of the client protein Luc-MycHis.
In Vivo Effect of hAha1 on Heat-shocked Luciferase
Firefly luciferase in a pcDNA3.1 vector was cotransfected into HEK293 cells together with hAha1 or Δ22hAha1 in a pCMV vector adding a Myc-tag to the protein or together with Silencer Select Pre-designed siRNA specific for hAha1 (ID s20802, Ambion Invitrogen). hAha1 levels were analyzed by blotting with the monoclonal antibody ab56721 (Abcam). Cells were grown nearly to confluence and then heat shocked at 43 °C for 30 min. Duplicates kept at 37 °C served as controls. Protein concentrations in the cell lysates were measured by the Bradford protein assay (Bio-Rad), and specific luciferase activity was determined after analysis using the luciferase assay as described above.
RESULTS
The Majority of Aha1 Exists Independent of Hsp90
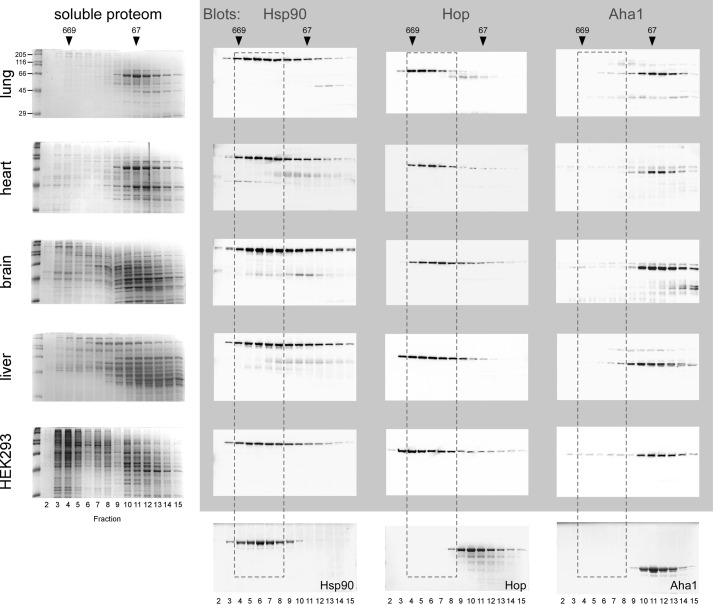
In vitro experiments using cochaperone proteins that compete for binding to Hsp90 have contributed valuable biochemical data to understand the regulation of the Hsp90 reaction cycle involving sequential cochaperone action (12). However, the situation in the “real” environment of a cell or a tissue is complex due to expression of the whole set of tissue specific cochaperones at their particular levels and together with client proteins. In a cell or tissue protein levels and affinities of all potential Hsp90 binders determine to which extent a particular cochaperone may be found together with Hsp90. Thus, analysis of such a complex ensemble offers an opportunity to challenge our current view of the Hsp90 system and may reveal functions hidden until now. Accordingly, we separated lung, heart, brain, liver tissue, and a lysate from HEK293 cells by gel filtration chromatography and identified Hsp90 and the cochaperones Aha1 and Hop by Western blotting using purified proteins for comparison (Fig. 1). In contrast to Hop, the majority of Aha1 migrates at a position outside of Hsp90 complexes, consistently in all five samples (Fig. 1). This suggests that much of the Aha1 protein may be available to serve functions in addition to stimulating the chaperone's ATPase activity.
FIGURE 1.
The majority of Aha1 is independent of high molecular weight Hsp90 complexes. Soluble lysate proteins from lung, heart, brain, liver tissue, and HEK293 cells were separated by gel filtration chromatography, fractions analyzed by SDS-PAGE gels and Hsp90, Hop, and Aha1 identified by Western blotting as indicated by gray shading. The five Coomassie-stained gels on the left show the soluble proteome of the respective tissues and the three gels on the bottom purified Hsp90, Hop, and Aha1 proteins as standards for comparison. The position of the bulk of purified Hsp90 that migrates as a dimer according to the purified Hsp90 standard is labeled by dashed lines, encompassing fractions 4–8. The dashes indicate that Hop in lysates is shifted toward higher molecular weight complexes together with Hsp90 as compared with the Hop standard. In contrast, most of Aha1 stays put, indicating the absence of complex formation. Marker proteins to calibrate the gel filtration column are shown on top (thyroglobulin, 669 kDa; BSA, 66 kDa). Molecular weights of markers proteins on SDS-PAGE gels are indicated.
Aha1 Has Autonomous Chaperone Activity and Prevents Aggregation of the Denatured Client Protein Rhodanese
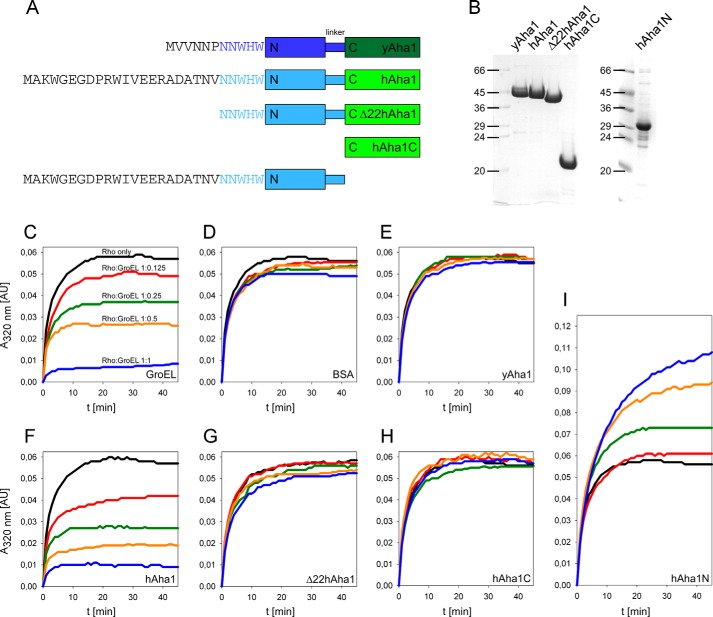
A hallmark of chaperone activity is the ability to shield misfolding denatured proteins from aggregation and protect the cell from adverse effects. To test the hypothesis that Aha1 possesses autonomous chaperone activity, we tested its ability to prevent aggregation of a client protein in comparison to GroEL. The well characterized chaperonin GroEL suppresses precipitation of denatured rhodanese in contrast to bovine serum albumin (BSA) as a negative control (23). We generated several constructs of hAha1 and yeast Aha1 (yAha1) and purified the proteins to homogeneity (Fig. 2, A and B). When identical amounts of GroEL and hAha1 were used, both proteins prevented rhodanese aggregation to the same extent in a concentration-dependent manner compared with the BSA control (Fig. 2, C, D, and F), suggesting that hAha1 possesses indeed autonomous chaperone activity. By contrast, yAha1 that markedly differs from hAha1 with regard to its amino acid composition N-terminal to the conserved NNWHW motif, does not prevent rhodanese aggregation (Fig. 2E). This led us to the assumption that the 22 amino acids present in hAha1 but absent from yAha1 convey the chaperone activity. Thus, we trimmed this fragment from hAha1 to generate Δ22hAha1 (Fig. 2, A and B) and found that this deletion abolished chaperone activity almost completely (Fig. 2G). Furthermore, cleaving off the complete N-terminal domain of hAha1 to generate hAha1C (Fig. 2, A and B) produced a likewise inactive protein (Fig. 2H) and confirmed this result. Addition of denatured rhodanese to hAha1N led to a further increase of the A320 nm signal (Fig. 2I), suggesting that hAha1N can bind to rhodanese but cannot keep the formed complex in solution, so that both proteins aggregate together. In conclusion, the 22 amino acid N-terminal part is necessary but not sufficient for chaperone activity, and the whole hAha1 protein is required to keep denatured rhodanese in solution.
FIGURE 2.
hAha1 prevents aggregation of denatured rhodanese. A, schematic view of yAha1 and hAha1 proteins used in the prevention of aggregation assay. The N-terminal sequences of yeast and human Aha1 are shown. B, Coomassie gel of purified yeast and human Aha1 proteins. C, prevention of rhodanese (Rho) aggregation. Denatured rhodanese was rapidly diluted to 0.5 μm into buffer alone or in buffer containing increasing amounts of GroEL at rhodanese:GroEL (14-mer) molar ratios of 1:0.125, 1:0.25, 1:0.5, and 1:1. D, Rhodanese was diluted into buffer alone or in buffer containing increasing amounts of BSA, (E) yAha1, (F) hAha1, (G) Δ22hAha1, (H) hAha1C, and (I) hAha1N as described under “Experimental Procedures.” Absorbance was recorded continuously every 2 s at 320 nm (A320 nm) for 45 min. All curves are the means of three independent measurements.
Aha1 Protects Firefly Luciferase from Heat-induced Denaturation in Vitro and in Vivo
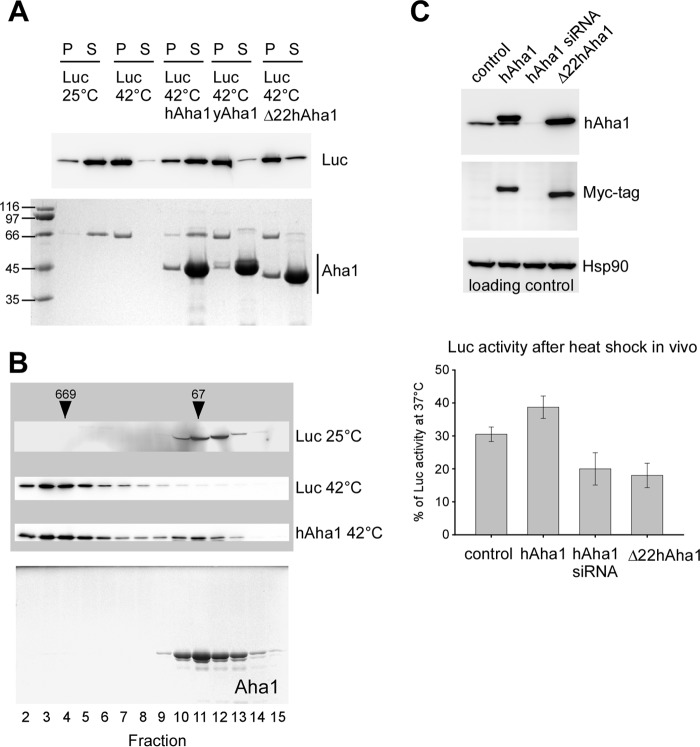
To substantiate the role of Aha1 as a chaperone we tested the effect of hAha1, yAha1 and Δ22hAha1 upon heat treatment of luciferase. Luciferase stays soluble at 25 °C but precipitates after incubation at 42 °C and can be detected in the pellet (Fig. 3A). When the enzyme was heat shocked in the presence of hAha1, the majority of luciferase stayed in solution while yAha1 and Δ22hAha1 showed not such an effect (Fig. 3A), confirming the results obtained with rhodanese. We further analyzed the complex formed between hAha1 and luciferase after heat shock using gel filtration chromatography. This treatment led to binding of luciferase to hAha1 and formation of a soluble high molecular weight complex (Fig. 3B) proving that hAha1 acts as an autonomous chaperone independent of Hsp90 that prevents precipitation of the otherwise aggregation prone client protein.
FIGURE 3.
hAha1 binds to heat-denatured luciferase. A, luciferase (Luc) remains in the supernatant (S) at 25 °C but precipitates in the pellet (P) at 42 °C in the absence of a chaperone. hAha1 but not yAha1 or Δ22hAha1 prevents aggregation of temperature-sensitive luciferase at 42 °C. hAha1, yAha1, and Δ22hAha1 are shown on the Coomassie-stained SDS-PAGE gel (bottom); the distribution of luciferase between supernatant and pellet is shown on a corresponding Western blot (top). Molecular weights are indicated by marker proteins. B, luciferase (Luc) was exposed to 42 °C for heat shock in the presence of purified hAha1 (see “Experimental Procedures”), subsequently analyzed by gel filtration chromatography and proteins detected by Western blotting as indicated by gray shading. Fractions and marker proteins (thyroglobulin, 669 kDa; BSA, 66 kDa) are indicated. Heat shock at 42 °C results in a shift of luciferase to higher molecular weight fractions together with hAha1 compared with luciferase kept at 25 °C and to hAha1 alone. The Coomassie gel showing hAha1 alone was taken from Fig. 1. C, in vivo effect of hAha1 on luciferase in heat-shocked HEK293 cells. Overexpression of hAha1 or Δ22hAha1 and down-regulation of endogenous hAha1 by siRNA is indicated by Western blotting with an antibody specific to hAha1. Blotting with the Myc tag antibody identifies hAha1 and Δ22hAha1 overexpressed from a pCMV vector. The Hsp90 blot serves as a loading control. hAha1 overexpression protects luciferase activity after heat shock compared with control cells, to cells in which hAha1 was silenced and to cells overexpressing Δ22hAha1. Measurements of luciferase activity were performed in triplicate, and standard deviation is indicated by error bars.
To address the physiological role of hAha1, we expressed luciferase in HEK293 cells together with hAha1, Δ22hAha1 or in cells in which endogenous hAha1 had been silenced by siRNA (Fig. 3C). After heat shock, luciferase activity was higher in hAha1 and lower in siRNA treated cells compared with the empty vector control (Fig. 3C). This outcome suggests that the holdase activity of hAha1 can protect heat stressed luciferase in vivo. A similar protective effect on luciferase activity following heat shock has been observed for the cochaperone CHIP (27). Overexpression of Δ22hAha1 resulted in luciferase activity similar to siRNA cells. This negative effect may be due to the ability of Aha1 to form dimers in vivo (28). Accordingly, Δ22hAha1 may form mixed dimers with endogenous hAha1 that appear inactive in binding to denatured proteins.
The N-terminal 22 Amino Acids of hAha1 Are Dispensable for Stimulating Hsp90 ATPase Activity
It has been shown that ATP binding and hydrolysis is necessary for the in vivo function of Hsp90 in yeast (2, 3) and many biochemical properties of the Hsp90-Aha1 complex have been studied mainly using yeast proteins. Human Hsp90 (hHsp90) shows a ∼10-fold lower intrinsic ATPase rate than yeast Hsp90 (yHsp90) but there are differing reports to which extent this activity can be stimulated by Aha1 (16, 29). Therefore, we purified yHsp90 and hHsp90 side by side following exactly the same procedure (see “Experimental Procedures”) and determined their ATPase activities (Fig. 4). As expected, the basic activities of yHsp90 and hHsp90 differed by ∼one order of magnitude (Fig. 4). To stimulate intrinsic ATP hydrolysis, we added Aha1 to 10-fold excess, which has been shown to be saturating (14). While yHsp90 could be stimulated ∼8-fold, hAha1 stimulated hHsp90 only ∼4-fold under these conditions (Fig. 4), suggesting the idea that Aha1 may have lost importance as Hsp90 ATPase activator during evolution from yeast to mammals. Next, we tested Δ22hAha1 for Hsp90ATPase stimulation. Clipping off the N-terminal 22 amino acids did not alleviate the stimulating effect of Δ22hAha1 on Hsp90 ATPase activity (Fig. 4), supporting the view that this part has an exclusive role in the prevention of client protein aggregation.
FIGURE 4.
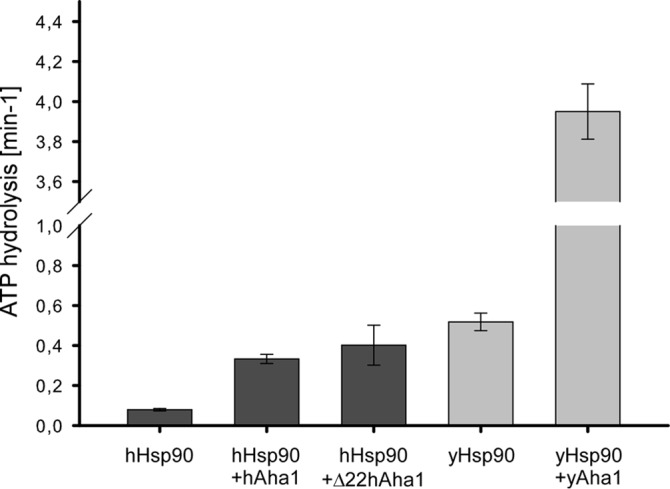
The N-terminal 22 amino acids of hAha1 are dispensable for stimulating Hsp90's ATPase activity. The ATPase activity of hHsp90 (dark gray) and yHsp90 (light gray) was measured with and without hAha1, Δ22hAha1 or yAha1 using a coupled enzymatic system described in “Experimental Procedures.” The N-terminal 22 amino acids of hAha1 that are necessary for prevention of rhodanese aggregation are dispensable for hHsp90 ATPase stimulation. All measurements were performed in triplicate, and standard deviation is indicated by error bars.
hAha1 Prevents Hsc70/Hsp40-dependent Refolding of Denatured Firefly Luciferase
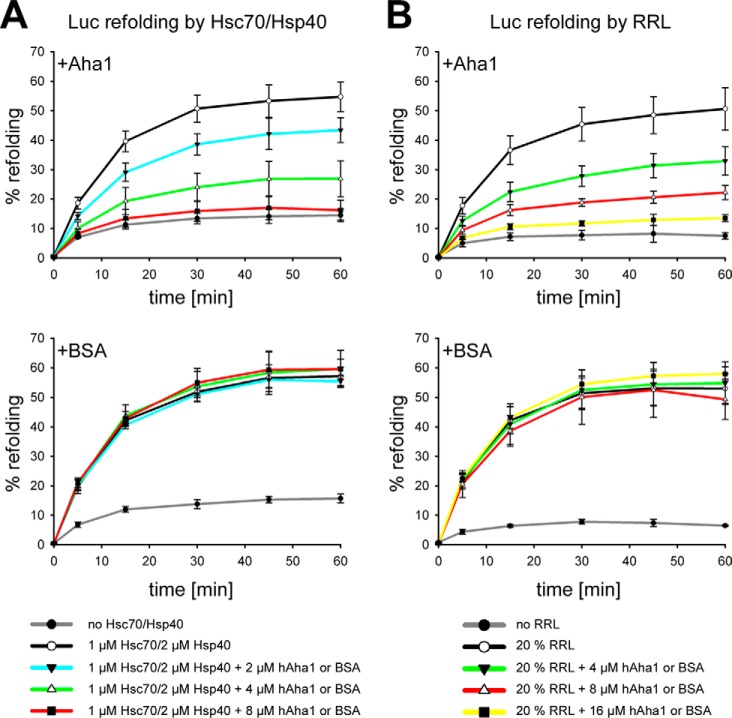
Heat and chemically denatured luciferase can be refolded efficiently by Hsc70 and its cochaperone Hsp40 when supplemented with a minimal amount of rabbit reticulocyte lysate (RRL) or alternatively in higher concentrated RRL in an energy-dependent fashion (25, 26). Accordingly, these test systems allow analyzing candidate proteins for their ability to either participate in or to block reactivation of denatured luciferase. Accordingly, we tested hAha1 for its ability to assist in Hsc70/Hsp40-dependent refolding of denatured firefly luciferase (Fig. 5A). hAha1 exhibited inhibition of Hsc70/Hsp40-dependent luciferase refolding in a concentration-dependent manner in contrast to the BSA added as a control protein (Fig. 5A). Similar results were obtained when we added hAha1 to 20% RRL as refolding system that contained no additional Hsc70/Hsp40 (Fig. 5B). Obviously, hAha1 does not collaborate with the Hsc70/Hsp40 system in refolding but sequesters the bound luciferase protein. This poses the question how hAha1 may get rid of the bound denatured client protein.
FIGURE 5.
hAha1 prevents refolding of denatured luciferase. A, refolding of denatured luciferase in 1 μm Hsc70/2 μm Hsp40 is inhibited by increasing amounts of hAha1 but not affected by BSA. B, refolding of denatured luciferase in 20% RRL. hAha1 prevents refolding of denatured luciferase in contrast to BSA. Refolding is indicated as percentage of native luciferase. All measurements were performed in triplicate, and standard deviation is indicated by error bars.
hAha1 Holds Aggregation Prone Firefly Luciferase Competent for Ubiquitination by the Cellular Protein Quality Control
Since hAha1 does not collaborate with the Hsc70/Hsp40 system in refolding of denatured firefly luciferase, we asked how bound client proteins could be further processed or disposed from hAha1. To this purpose firefly luciferase was either kept at 25 °C or thermally denatured at 42 °C in the presence of hAha1, GroEL as a control, or without addition of another protein (Fig. 6A). GroEL can hold denatured luciferase but not refold it because the client protein does not fit into the size-limited cage of chaperonin (30, 31). After this treatment the reaction mixtures were separated in pellet and supernatant and blotted for the presence of luciferase protein (Fig. 6A). Luciferase stays in solution at 25 °C but precipitates at 42 °C in the absence of a chaperone (Fig. 6A). hAha1 and GroEL prevent aggregation of heat-denatured luciferase (Fig. 6A), and the majority of the protein binds to the chaperone and remains soluble in the supernatant. This is consistent with our finding that luciferase forms a soluble higher molecular weight complex together with hAha1 (Fig. 3). Next, supernatants containing soluble firefly luciferase kept at 25 °C or incubated together with hAha1 or GroEL at 42 °C were supplemented with components of the ubiquitination machinery including E1, E2 enzymes, and the E3 ubiquitin ligase CHIP (32) (Fig. 6B). CHIP has autonomous chaperone activity to recognize polypeptides and can ubiquitinate nonnative luciferase independently of Hsp70 (27). The majority of hAha1-bound denatured luciferase was ubiquitinated, as can be noticed by the appearance of a characteristic ubiquitin ladder added to the protein (Fig. 6B). In contrast, native luciferase that was kept at 25 °C received no ubiquitin addition (Fig. 6B). To test whether luciferase bound to the holdase hAha1 is also competent for reactivation we added the supernatant to the Hsc70/Hsp40 containing refolding system. Only ∼1% refolding to the original activity prior to heat denaturation could be achieved by this procedure (Fig. 6C) pointing to the view that hAha1 does not collaborate efficiently with the Hsc70/Hsp40 refolding system. Since hAha1 is lacking ATPase activity, this might cause it to prevent release of bound client proteins to the Hsc70/Hsp40 system for refolding.
FIGURE 6.
hAha1 prevents aggregation of heat-denatured luciferase and allows client protein ubiquitination by the E3 ubiquitin ligase CHIP. A, hAha1 prevents aggregation of temperature sensitive luciferase (Luc) at 42 °C similar to the chaperonin GroEL. Luciferase remains in the supernatant (S) at 25 °C but precipitates in the pellet (P) at 42 °C in the absence of a chaperone. GroEL and hAha1 are shown on the Coomassie stained SDS-PAGE gel (bottom); the distribution of luciferase between supernatant and pellet is shown on a corresponding Western blot (top). Molecular weights are indicated by marker proteins. B, majority of hAha1 protected luciferase undergoes ubiquitination by the E3 ubiquitin ligase CHIP. Molecular weights are indicated by marker proteins. C, only a minor portion of hAha1-protected luciferase can be reactivated by Hsc70/Hsp40. D, model summarizing the role of the heat stress inducible hAha1 as an autonomous chaperone. hAha1 prevents stress denatured proteins from aggregation and holds them competent for ubiquitination by the E3 ubiquitin ligase CHIP but only to a minor extent for refolding by the Hsc70/Hsp40 chaperone system.
DISCUSSION
In this study we report a novel function of hAha1 beyond its established activity to stimulate the ATPase activity of the molecular chaperone Hsp90. We reveal that hAha1 is an autonomous chaperone that can bind to denatured proteins such as luciferase and rhodanese and prevent them from aggregation (Fig. 6D). However, rather than promoting refolding, hAha1 seems to dispose most of its bound clientele through allowing ubiquitination by the CHIP-dependent cellular protein quality control machinery (Fig. 6D), a prelude to proteolytic degradation of denatured proteins. This pathway may play an important role during cellular stress conditions, such as heat shock, that induce overexpression of Aha1 (14).
Mollapour et al. (33) have reported that diminished Hsp90/Aha1 interaction associated with reduced ATPase stimulation has little effect on the activation of some client proteins, pointing to an additional role of Aha1 beyond Hsp90 ATPase activation. In line with this suggestion is our finding that the majority of Aha1 in mammalian cells and tissues is self-contained and independent of Hsp90 (Fig. 1). Furthermore, hHsp90 has a much lower intrinsic ATP hydrolysis rate than yHsp90 and shows less stimulation by hAha1 than yeast Hsp90 by yAha1 (Fig. 4). This proposes a change of Aha1's main focus from Hsp90 ATPase activator to stand-alone chaperone, although such a transition may be flexible and not exclusive. The finding that an N-terminal part in hAha1 that is lacking from yAha1 is necessary for client binding (Fig. 2E) may support the view that such an altered role has emerged during evolution.
It has been shown that overexpression of Aha1 destabilized wild-type CFTR (cystic fibrosis transmembrane conductance regulator) protein and the folding-deficient mutant CFTRΔF508 and promoted ER associated degradation (19). The Aha1-dependent activation of Hsp90's ATPase rate accelerates the conformational cycle of the molecular chaperone Hsp90 during which client proteins fold. This could reduce the required time of interaction between CFTR and Hsp90 and eventually result in release of the client protein and subsequent degradation (19). This explanation relies on Aha1 function as an activator of the Hsp90 ATPase. In addition to this reasoning, our results provide an alternative avenue to understand the role of Aha1 in this process. Since hAha1 has chaperone activity, it might directly recognize and bind to clients that are not properly folded and triage them for ubiquitination by the cellular protein quality control. In conclusion, the chaperone activity of hAha1 may play an important role under stress conditions that challenge the cell with an overflow of denatured proteins or when proteins are irreversibly misfolded. Under such conditions, Aha1 may promote disposal of damaged proteins by ubiquitin-dependent degradation, rather than attempting their refolding, to protect the cellular protein folding machineries from overload and avert harm from the cell.
Acknowledgments
We thank Ulrich Hartl for GroEL and Luc-MycHis6 expression constructs, and Wolfgang Linke for continuous backup.
This work was supported by the German Science Association (DFG Grant OB 163/1-1, to W. O.) and by Ruhr-University Bochum FoRUM Grant F698 (to W. O.).
- Hsp90
- heat shock protein 90
- Aha1
- activator of Hsp90 ATPase
- Luc
- luciferase
- RRL
- rabbit reticulocyte lysate
- Rho
- rhodanese.
REFERENCES
- 1. Taipale M., Jarosz D. F., Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 2. Obermann W. M., Sondermann H., Russo A. A., Pavletich N. P., Hartl F. U. (1998) In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 143, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1998) ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17, 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pearl L. H., Prodromou C. (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 5. Pearl L. H., Prodromou C., Workman P. (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 410, 439–453 [DOI] [PubMed] [Google Scholar]
- 6. Li J., Buchner J. (2013) Structure, function and regulation of the hsp90 machinery. Biomed. J. 36, 106–117 [DOI] [PubMed] [Google Scholar]
- 7. Meyer P., Prodromou C., Liao C., Hu B., Mark Roe S., Vaughan C. K., Vlasic I., Panaretou B., Piper P. W., Pearl L. H. (2004) Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Retzlaff M., Hagn F., Mitschke L., Hessling M., Gugel F., Kessler H., Richter K., Buchner J. (2010) Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol. Cell 37, 344–354 [DOI] [PubMed] [Google Scholar]
- 9. Lotz G. P., Brychzy A., Heinz S., Obermann W. M. (2008) A novel HSP90 chaperone complex regulates intracellular vesicle transport. J. Cell Sci. 121, 717–723 [DOI] [PubMed] [Google Scholar]
- 10. Brychzy A., Rein T., Winklhofer K. F., Hartl F. U., Young J. C., Obermann W. M. J. (2003) Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 22, 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harst A., Lin H., Obermann W. M. J. (2005) Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem. J. 387, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J., Richter K., Reinstein J., Buchner J. (2013) Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat. Struct. Mol. Biol. 20, 326–331 [DOI] [PubMed] [Google Scholar]
- 13. Siligardi G., Panaretou B., Meyer P., Singh S., Woolfson D. N., Piper P. W., Pearl L. H., Prodromou C. (2002) Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277, 20151–20159 [DOI] [PubMed] [Google Scholar]
- 14. Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., Singh S., Millson S. H., Clarke P. A., Naaby-Hansen S., Stein R., Cramer R., Mollapour M., Workman P., Piper P. W., Pearl L. H., Prodromou C. (2002) Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 15. Lotz G. P., Lin H., Harst A., Obermann W. M. J. (2003) Aha1 Binds to the Middle Domain of Hsp90, Contributes to Client Protein Activation, and Stimulates the ATPase Activity of the Molecular Chaperone. J. Biol. Chem. 278, 17228–17235 [DOI] [PubMed] [Google Scholar]
- 16. Koulov A. V., Lapointe P., Lu B., Razvi A., Coppinger J., Dong M. Q., Matteson J., Laister R., Arrowsmith C., Yates J. R., 3rd, Balch W. E. (2010) Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol. Biol. Cell 21, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu W., Mollapour M., Prodromou C., Wang S., Scroggins B. T., Palchick Z., Beebe K., Siderius M., Lee M. J., Couvillon A., Trepel J. B., Miyata Y., Matts R., Neckers L. (2012) Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol. Cell 47, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes J. L., Sharp S. Y., Hobbs S., Workman P. (2008) Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Research 68, 1188–1197 [DOI] [PubMed] [Google Scholar]
- 19. Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 20. Tripathi V., Obermann W. M. (2013) A primate specific extra domain in the molecular chaperone Hsp90. PLoS One 8, e71856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obermann W. M., Gautel M., Weber K., Fürst D. O. (1997) Molecular structure of the sarcomeric M band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 16, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hawle P., Siepmann M., Harst A., Siderius M., Reusch H. P., Obermann W. M. (2006) The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol. Cell Biol. 26, 8385–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. (1991) Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate [see comments]. Nature 352, 36–42 [DOI] [PubMed] [Google Scholar]
- 24. Ali J. A., Jackson A. P., Howells A. J., Maxwell A. (1993) The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32, 2717–2724 [DOI] [PubMed] [Google Scholar]
- 25. Minami Y., Höhfeld J., Ohtsuka K., Hartl F. U. (1996) Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J. Biol. Chem. 271, 19617–19624 [DOI] [PubMed] [Google Scholar]
- 26. Nimmesgern E., Hartl F. U. (1993) ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 331, 25–30 [DOI] [PubMed] [Google Scholar]
- 27. Rosser M. F., Washburn E., Muchowski P. J., Patterson C., Cyr D. M. (2007) Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 282, 22267–22277 [DOI] [PubMed] [Google Scholar]
- 28. Berg M., Michalowski A., Palzer S., Rupp S., Sohn K. (2014) An in vivo photo-cross-linking approach reveals a homodimerization domain of Aha1 in S. cerevisiae. PLoS One 9, e89436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter K., Soroka J., Skalniak L., Leskovar A., Hessling M., Reinstein J., Buchner J. (2008) Conserved conformational changes in the ATPase cycle of human Hsp90. J. Biol. Chem. 283, 17757–17765 [DOI] [PubMed] [Google Scholar]
- 30. Frydman J., Nimmesgern E., Erdjument-Bromage H., Wall J. S., Tempst P., Hartl F.-U. (1992) Function in protein folding of TRiC, a cytosolic ring-complex containing TCP1 and structurally related subunits. EMBO J. 11, 4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchberger A., Schröder H., Hesterkamp T., Schönfeld H. J., Bukau B. (1996) Substrate shuttling between the DnaK and GroEL systems indicates a chaperone network promoting protein folding. J. Mol. Biol. 261, 328–333 [DOI] [PubMed] [Google Scholar]
- 32. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 33. Mollapour M., Bourboulia D., Beebe K., Woodford M. R., Polier S., Hoang A., Chelluri R., Li Y., Guo A., Lee M. J., Fotooh-Abadi E., Khan S., Prince T., Miyajima N., Yoshida S., Tsutsumi S., Xu W., Panaretou B., Stetler-Stevenson W. G., Bratslavsky G., Trepel J. B., Prodromou C., Neckers L. (2014) Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol. Cell 53, 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]