Background: The physical basis for polymerase fidelity remains unclear.
Results: Molecular dynamics simulations, NMR, and pre-steady-state kinetics of a mutator polymerase reveal conformational states of the active site that serve as fidelity checkpoints.
Conclusion: Protein dynamics, largely concealed by x-ray crystallography, govern incorporation fidelity.
Significance: Strategies have been inspired for engineering (anti)mutator polymerases and attenuated viruses.
Keywords: Plus-stranded RNA Virus, Poliovirus, RNA Polymerase, RNA Virus, Viral Replication, Lethal Mutagenesis, Polymerase Fidelity, Population Genetics
Abstract
RNA viruses encoding high- or low-fidelity RNA-dependent RNA polymerases (RdRp) are attenuated. The ability to predict residues of the RdRp required for faithful incorporation of nucleotides represents an essential step in any pipeline intended to exploit perturbed fidelity as the basis for rational design of vaccine candidates. We used x-ray crystallography, molecular dynamics simulations, NMR spectroscopy, and pre-steady-state kinetics to compare a mutator (H273R) RdRp from poliovirus to the wild-type (WT) enzyme. We show that the nucleotide-binding site toggles between the nucleotide binding-occluded and nucleotide binding-competent states. The conformational dynamics between these states were enhanced by binding to primed template RNA. For the WT, the occluded conformation was favored; for H273R, the competent conformation was favored. The resonance for Met-187 in our NMR spectra reported on the ability of the enzyme to check the correctness of the bound nucleotide. Kinetic experiments were consistent with the conformational dynamics contributing to the established pre-incorporation conformational change and fidelity checkpoint. For H273R, residues comprising the active site spent more time in the catalytically competent conformation and were more positively correlated than the WT. We propose that by linking the equilibrium between the binding-occluded and binding-competent conformations of the nucleotide-binding pocket and other active-site dynamics to the correctness of the bound nucleotide, faithful nucleotide incorporation is achieved. These studies underscore the need to apply multiple biophysical and biochemical approaches to the elucidation of the physical basis for polymerase fidelity.
Introduction
The list of (re)emerging viruses continues to expand, and with this expansion comes an increased risk of the evolution of a highly virulent, highly transmissible strain and a corresponding pandemic. It is cost-prohibitive to treat every (re)emerging agent as the etiologic agent of the next pandemic. However, when that pandemic comes, a rapid response with antiviral therapies and vaccines will be absolutely essential. With this circumstance in mind, the viral RNA-dependent RNA polymerase (RdRp)5 is a very attractive target for both the design of broad spectrum, antiviral therapies and mechanism-based strategies for viral attenuation and vaccine development (1, 2).
It is becoming increasingly clear that the ability of a virus population to be sustained in the presence of antiviral responses of the host and other bottlenecking events requires a genetically diverse virus population. In the presence of this genetic diversity, selection can act to evolve the most robust population for that place and time. For RNA viruses, an important source of this genetic diversity is the nucleotide misincorporation frequency of the RdRp (3). For example, in poliovirus one or two errors are introduced into the viral genome during each replication cycle. Interestingly, in the coronaviruses (e.g. severe acute respiratory syndrome virus), a proofreading exonuclease exists that can remove mistakes made by the RdRp (4). It is possible that both enzymatic (e.g. deamination) and oxidative (e.g. 8-oxo-G) modifications of RNA also contribute to genetic diversity (5).
Numerous examples exist in the literature providing some evidence for sensitivity of viral populations to mutagens like the antiviral agent ribavirin. Resistance to this drug yields viruses harboring mutations in the RdRp that decrease genetic diversity while simultaneously decreasing virus fitness (6–10). Active-site mutagenesis of the RdRp or the proofreading exonuclease can lead to increased genetic diversity but nevertheless decreases virus fitness (11–14). Collectively, observations such as these lead to the suggestion that an optimal genetic diversity exists for maximal fitness. When tested, viruses exhibiting perturbed genetic diversity are attenuated and serve as vaccine strains (15).
The ability to rationally design RdRp derivatives with increased or decreased nucleotide incorporation fidelity would be of great practical value. The RdRp from poliovirus (PV) is an ideal model system for elucidating physical mechanisms governing nucleotide incorporation fidelity because of the substantial tools that can be applied to this system. Of particular importance to the question of the mechanistic basis of fidelity are the following: pre-steady-state kinetics (16, 17), crystallography (18, 19), NMR (20, 21), molecular dynamics (MD) simulations (22–25), as well as the existence of both low (H273R)- and high-fidelity (G64S) RdRp derivatives (5, 6, 8).
Several observations suggest that nucleotide incorporation fidelity of PV RdRp is governed by the conformational dynamics of the active site. First, residues implicated in nucleotide incorporation fidelity are remote from the active site, suggesting allosteric control of the conserved structural motifs in the active site involved in nucleotide binding and/or the nucleotidyl transfer reaction (Fig. 1A). It is now well established that dynamics and allostery are inextricably linked (26). Second, a conformational change step (Fig. 1B, ERnNTP ⇄ *ERnNTP) observed kinetically has been implicated as the step affected in the high fidelity G64S RdRp (6). The structure of G64S RdRp is essentially identical to the wild-type enzyme (27). Finally, structural studies show that a conformational rearrangement of conserved residues is required for stable binding of an incoming nucleotide (Fig. 1C) (18, 19).
FIGURE 1.

Nucleotide addition by PV RdRp. A, PV RdRp is a prototypical RdRp with fingers, palm, and thumb subdomains. Shown is the WT PV RdRp-RNA complex (PDB code 3OL6) (19). The conserved structural motifs are essential for faithful, efficient, nucleotide addition and are color-coded as follows: motif A, dark green; motif B, tan; motif C, dark cyan; motif D, blue; motif E, golden yellow; motif F, red; and motif G, light green. Gly-64 and His-273 are residues remote from the active site that increase or decrease PV RdRp fidelity, respectively (5, 6, 8). B, nucleotide addition has been shown to require at least five kinetically resolvable steps: step 1, binding of nucleotide to the RdRp-RNA complex; step 2, conformational change to produce a catalytically competent state; step 3, chemistry; step 4, conformational change to permit translocation; step 5, pyrophosphate release (16). Step 2 is a fidelity checkpoint, the physical basis of which is only partially understood (17). C, indicated is the current understanding of nucleotide addition of PV RdRp gleaned from crystal structures (PDB codes 1RA6, 3OL6, and 3OL7) (18, 19). Shown are the conserved residues of conserved structural motifs, colored by motif as indicated for A, that feature prominently in the studies reported herein. In the free enzyme, the conformation of the residues shown will not support binding of RNA or NTP. The binary complex shows substantial rearrangement of most residues; however, NTP binding will still be inhibited by the steric clashes with Asp-238. The closest structure to the catalytic state solved is for a complex after nucleotide incorporation. Here it is clear that every residue shown has had to undergo a conformational change relative to the binary complex for nucleotide addition to occur. Filling in the gaps in going from binary complex to post-incorporation complex is the focus of this study.
Here we exploited the PV H273R and G64S RdRp to interrogate the physical basis for nucleotide incorporation fidelity. Our data are consistent with a model in which equilibrium between two conformational states of the active site that interconvert on the nanosecond timescale are linked to the fidelity checkpoint that occurs on the millisecond timescale. Perturbation of this equilibrium alters nucleotide incorporation fidelity, as shown for the PV RdRp derivatives studied here. Importantly, the active-site amino acid residues contributing to the two conformational states are highly conserved, leading us to propose these interactions as a starting point for the development of engineered RdRp with altered fidelity that can be extrapolated to other RNA virus systems.
MATERIALS AND METHODS
Construction of H273R Expression Vector
Mutation encoding substitution of His-273 by arginine was introduced into the WT PV RdRp using overlap extension PCR and subcloning into the expression plasmid pET26-Ub as described previously (5, 28).
Expression and Purification of Proteins
Expression and purification of WT and H273R proteins were performed as described previously (5, 28) using the ubiquitin fusion system. The Escherichia coli BL21(DE3)/pCG1 cells containing either pET26-Ub-WT or pET26-Ub-H273R fusion plasmid were grown at 30 °C overnight in 100 ml of NZCYM medium supplemented with kanamycin at 25 μg/ml (K25), chloramphenicol at 20 μg/ml (C20), and dextrose at 0.1%. The starting overnight culture was used to inoculate 2 liters NZCYM medium supplemented with K25/C20; cells were grown at 37 °C to an OD600 ≈ 1.0. The cells were chilled to 25 °C and induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside (IPTG). Cell growth continued for an additional 4 h at 25 °C before harvesting. Cell pellets were washed once with buffer containing 10 mm Tris, pH 8.0, and 1 mm EDTA and stored at −80 °C. Frozen cell pellets were thawed on ice, suspended in lysis buffer (100 mm potassium phosphate, pH 8.0, 0.5 mm EDTA, 20% glycerol, 1 mm dithiothreitol (DTT), 60 μm ZnCl2, 2.8 μg/ml pepstatin A, and 2.0 μg/ml leupeptin), and disrupted by passage through a French pressure cell at 20,000 p.s.i. Phenylmethanesulfonyl fluoride (PMSF) and Nonidet P-40 were added immediately after lysis to final concentrations of 1.0 mm and 0.1%, respectively. To precipitate nucleic acid, polyethyleneimine was slowly added to the cell lysate at a concentration of 0.025%. The lysate was stirred for 30 min at 4 °C and then centrifuged at 25,000 rpm. Ammonium sulfate fractionation was done by slowly adding ammonium sulfate at 40% saturation to the clear solution from the polyethyleneimine precipitation step. The fraction of proteins containing RdRp was pelleted by centrifugation at 25,000 rpm for 30 min at 4 °C. The ammonium sulfate pellet was suspended in buffer A (50 mm Tris, pH 8.0, 20% glycerol, 1 mm DTT, 0.1% Nonidet P-40, and 60 μm ZnCl2) and diluted to 50 mm NaCl. The protein sample was loaded onto a phosphocellulose column (Whatman, P-11) that was pre-equilibrated with buffer A containing 50 mm NaCl (∼1 ml bed volume/20 mg of total protein was used) at a flow rate of 1.0 ml/min. The column was washed to baseline with the equilibrating buffer and eluted with a linear gradient from 50 to 350 mm NaCl in buffer A. Fractions containing the protein (WT or H273R), checked by SDS-PAGE, were pooled and diluted to a final concentration of 50 mm NaCl and were further fractionated on an S-Sepharose column followed by a Q-Sepharose column (equilibrated, washed, and eluted in the same way as the phosphocellulose column). Protein fractions eluted from the Q-Sepharose column were diluted to adjust the salt concentration to 50 mm NaCl and finally concentrated by passing over a Q-Sepharose column equilibrated with buffer B (50 mm HEPES, pH 7.5, 20% glycerol, 1 mm DTT, 0.1% Nonidet P-40, and 60 μm ZnCl2) containing 50 mm NaCl. The protein was stripped from the Q-Sepharose column using 500 mm NaCl in buffer B. The concentrated protein (∼10 mg/ml) was aliquoted and stored at −80 °C.
Pulse-Chase and Pulse-Quench Experiments
Pulse-chase and pulse-quench experiments were performed in the reaction buffer (50 mm HEPES, pH 7.5, 10 mm 2-mercaptoethanol, 60 μm ZnCl2, and 5 mm MnCl2) at 30 °C using a rapid chemical quench-flow instrument (KinTek Corp., Austin, TX). The enzyme solution (4 μm), WT or H273R, was incubated with a symmetrical self-complementary 10-nt RNA substrate containing uracil as the templating base for the first incorporation (S/S-U, 20 μm) (GE Healthcare) and rapidly mixed with [α-32P]ATP (100 μm) (MB Biomedicals), and then reactions at various time points were either pulse-quenched or pulse-chased by adding a large excess of ATP (20 mm) (Sigma) to proceed for an additional 30 s. Reactions were quenched with HCl to a final concentration of 1.2 n followed by immediate neutralization using 3.0 m KOH. Samples were quantitated via sequencing gel analysis. Kinetic parameters were obtained by simulation of the data using KinTek Explorer 2.03 (KinTek Corp.). All rate constants were determined experimentally, except where noted. The agreement between experimental data and kinetic simulations was determined by visual inspection.
Stopped-flow Nucleotide Incorporation Assays
Stopped-flow pre-steady-state nucleotide incorporation assays were performed using a model SF-2001 stopped-flow apparatus (KinTek Corp.) equipped with a water bath. The incorporation of the correct nucleotide (UTP) and nucleotide with an incorrect sugar (2′-dUTP) was achieved by using the symmetric RNA duplex S/S-UA substrate (Eurofins Genomics, Huntsville, AL) with the sequence 5′-GpyrCAUGGGCCCA-3′), where pyrC or pyrrolo-C at the +1 templating position is a fluorescent analog of the cytidine nucleoside that retains its Watson-Crick base-pairing capacity with G. For incorporation of the correct nucleotide (UTP) next to the site of misincorporation, we used the symmetric RNA duplex S/S-UG substrate (Eurofins Genomics) with the sequence 5′-GpyrCAUGGGCCCG-3′), containing pyrC at the +1 templating position and a G-U mismatch at the 3′-end. All reactions were conducted at 30 °C in the reaction buffer (50 mm Hepes, pH 7.5, 10 mm 2-mercaptoethanol, 5 mm MgCl2, and 60 μm ZnCl2). The reaction conditions for PV RdRp, WT or H273R, were optimized. For WT assays, the enzyme (0.5 μm) was incubated with pyrC-labeled S/S-UA primed template (0.25 μm) or pyrC-labeled S/S-UG primed template (1.5 μm) in the reaction buffer for 3 min and then mixed rapidly with UTP or 2′-dUTP solutions of different concentrations. For H273R, similar assays were performed using H273R enzyme (2 μm) and S/S-UA (1 μm) or S/S-UG (2.5 μm). The excitation wavelength was 350 nm, and fluorescence emission was monitored using a 440-nm cut-on filter (model HW440lp, Chroma Technology Corp., Rockingham, VT). For each experiment, at least four fluorescent traces were averaged. Relative fluorescence was plotted as a function of time and fit to a single exponential equation (Equation 1), yielding an observed rate constant, kobs,
where A is the amplitude and C is the end point. Values for kobs were plotted as a function of nucleotide concentration and fit to a hyperbolic equation (Equation 2), providing estimates for kpol, the maximal rate constant for nucleotide incorporation, and Kd,app, the apparent dissociation constant for nucleotide.
Solvent Deuterium Kinetic Isotope Effect (SDKIE)
Experiments were performed essentially as described above for the stopped-flow nucleotide incorporation assays, except enzyme, substrates, and buffers were prepared in 100% D2O. The pD was used instead of pH for solutions in D2O and was adjusted according to the relationship (pD = pH + 0.4). The SDKIE was calculated as the quotient kpol (H2O)/kpol (D2O).
Crystallization, Data Collection, and Determination of H273R Structure
The H273R construct used for crystallization contained two mutations (L446D and R455D) in the thumb, introduced to disrupt interface I of PV RdRp (27). Prior to crystallization, the H273R protein, purified as described above, was further fractionated on a HiLoad 26/60 Superdex 200 gel filtration column, equilibrated with buffer containing 5 mm Tris, pH 7.5, 200 mm NaCl, 0.1 mm EDTA, and 2 mm DTT as a polishing step. Crystals of H273R were grown using vapor diffusion at 20 °C by mixing equal volumes of the protein (10 mg/ml) and the crystallization reservoir containing 2 m sodium acetate and 0.1 m sodium cacodylate, pH 6.8. For data collection, crystals were soaked briefly in the crystallization solution, adjusted to contain 20% glycerol as a cryoprotectant, prior to flash-cooling in a nitrogen stream and use in the diffraction experiment. Diffraction data were collected at the 19-ID beamline at the Advanced Photon Source (Argonne, IL). The data were integrated, merged, and scaled using DENZO and SCALEPACK (29). The structure was determined by molecular replacement in PHASER (CCP4) (30) using the structure of WT PV RdRp (PDB 1RA6) (18) as the search model. The model was built using Coot (31, 32) and refined with REFMAC5 (33, 34).
Molecular Dynamics Simulations
System Setup
The WT PV RdRp and the G64S mutant in their free forms were investigated by all-atom MD simulations (25 ns) in our previous study (23). Thus, simulations for the two systems described here (termed WT and G64S, respectively) started from the end of the previous 25-ns trajectories and extended to a total length of 150 ns. For the H273R mutant, the solved crystal structure (PDB code 4R0E) was used to prepare the initial model. The two surface residues Asp-446 and Asp-455 introduced to help in crystallization were mutated back to the WT residues Leu-446 and Arg-455, respectively. It should be mentioned that the two surface mutations, L446D and R455D, have no impact on the observed dynamics of the WT protein.6 The starting coordinates for the WT binary complex were obtained from the crystal structure (PDB code 3OL6) (19). The primer-template RNA of the crystal structure was trimmed to include only nucleotides that are within 5 Å distance from protein atoms. The initial simulated structure (termed WT-RNA) included protein residues 1–461 (chain A), a 13-mer RNA template strand (chain B), a 9-mer RNA primer strand (chain C), and 153 structural waters. Also, the two surface residues, Asp-446 and Asp-455, reported in the crystal structure were mutated back to the WT residues. For the binary complexes of the mutants (G64S and H273R), because there is no available crystal structures these complexes were constructed based on the WT-RNA complex by single residue replacement, leaving other protein residues and RNA base sequences unchanged. The steric clashes generated in the two mutant systems (G64S-RNA and H273R-RNA) were removed by subsequent energy minimization and equilibration.
MD Simulations Protocol
MD simulations were carried out using the program AMBER12 (35, 36) with AMBER99bsc0 (37), a version of AMBER99SB (38) with improved parameters for nucleic acid. In all simulations, an integration time step of 1 fs was used. Periodic boundary conditions using cutoff radii of 9 Å (in simulations of the free forms) and 12 Å (in simulations of the RNA-bound forms) were applied in calculating nonbonded interactions. Electrostatic interactions were treated using the particle mesh Ewald method (39, 40). The SHAKE algorithm (41) was employed to constrain all bond lengths involving hydrogen atoms. All MD simulations were performed in explicit water (TIP3P model) (42), imposing a minimal distance of 20 Å between the edge of the solvent box and any protein atom. The simulations were conducted following a scheme that was essentially similar to our previously published work (23). Specifically, the coordinates of the simulated systems were first relaxed to remove any steric clashes between the atoms of proteins, RNA, waters, and ions in multiple steps using SANDER; each system was subjected to two cycles of energy minimization followed by short constrained dynamics (100 ps) under conditions of constant pressure and temperature (NPT). Subsequently, the systems were energy-minimized by applying convergence criteria for the energy gradient DRMS (the root-mean-square of the Cartesian elements of the gradient) of 0.1 kcal/mol-Å. Next, each system was slowly heated to 300 K over a period of 150 ps under conditions of constant volume and temperature (NVT) and applying a Berendsen thermostat (43). This was followed by a 200-ps NVT dynamics before switching to NPT dynamics for another 150 ps, applying the Berendsen method with temperature and pressure coupling constants of 1 ps (weak coupling). The NPT dynamics was continued for the remainder of the MD simulations utilizing the parallel version of PMEMD in AMBER12. Analyses were performed using PTRAJ and CPPTRAJ programs (44) of the AMBER package on MD trajectories, which were generated from snapshots retained every 1 ps. All MD simulations were carried out on 128 Intel Xeon X5675 Six-Core processors managed by the High Performance Computing Group at The Pennsylvania State University and on 128 Cray XT3 processors (TeraGrid resources; transitioned to XSEDE in 2011) managed by the Pittsburgh Supercomputing Center.
Cluster Analysis
To identify distinct conformations generated over the last 100 ns of MD simulations, we performed the cluster analysis. The refinement “means” algorithm (45) in PTRAJ was used to cluster the structures extracted at 20-ps intervals across the trajectories. Initially, the number of clusters was determined by carrying out the cluster analysis using the average-linkage (45) algorithm in PTRAJ.
Principal Component Analysis (PCA)
PCA (46, 47) was carried out to separate major motions from irrelevant noise sampled during MD simulations over the last 75 ns by diagonalizing the variance/covariance matrix (S) of the positional deviations of Cα atoms with respect to the average structure. The elements of this matrix are given by Equation 3,
where Sij is the element of the covariance (cov) matrix, S; ri represents the coordinates x, y, and z of the Cαi atom; rj represents the coordinates x, y, and z of the Cαj atom; 〈ri〉 represents the coordinates x, y, and z of the Cαi atom in the average structure; 〈rj〉 represents the coordinates x, y, and z of the Cαj atom in the average structure; and t denotes the time average over the MD trajectory.
Diagonalization of the matrix S produced an orthogonal set of eigenvectors (or modes) that describe the directions of maximum variation in the observed conformational space sampled during simulations. The variance around an average structure represented atomic displacements or motion. In this analysis, our focus was to identify the regions that exhibited changes in the dynamics of the H273R-RNA complex relative to the WT-RNA complex. To do so, for each Cα atom, contributions to the total variance along the top 10 modes (contained ∼50% of the total variance) were summed and normalized to the average of the least contributing residues (5% of total residues). The PCA sum corresponding to WT-RNA was subtracted from that of H273R-RNA to obtain the ΔPCA values for each residue. The ΔPCA values were mapped onto the structure as shown in Fig. 9.
FIGURE 9.
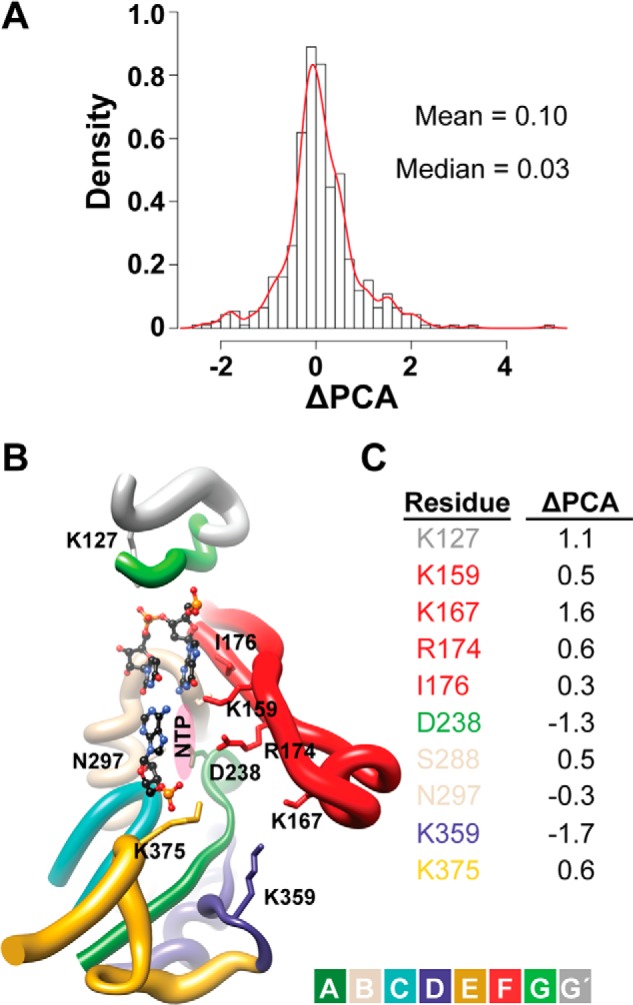
Backbone of functional residues contribute more to the overall motions observed for the H273R RdRp-RNA complex relative to WT. A, we performed a PCA for both the WT and H273R RdRp-RNA complexes. To identify regions that exhibited changes in the overall dynamics of H273R RdRp-RNA complex relative to WT, the sum of the top 10 principal components for WT were substracted from the sum calculated for H273R, yielding a ΔPCA value. The frequency (density) of each observed ΔPCA value was plotted. A ΔPCA value of zero represents no change. Most residues did not exhibit a change; however, as indicated by the tails, outliers exist. B, the ΔPCA values were plotted on the structure of the RdRp-RNA complex. The active site is shown. Colors represent the conserved structural motifs. Motif G′ is defined here for the first time and represents the residues adjacent to canonical motif G that exhibit dynamics equivalent to motif G. The radii of the tubes indicate the magnitude of the ΔPCA values on a scale from largest radius (2.4) to smallest radius (−2.4). C, the ΔPCA values of residues shown in B are indicated explicitly. Colors represent the conserved structural motifs.
Dynamic Cross-correlation Map (DCCM)
Information about correlated motions sampled during MD simulations was obtained from the DCCM analysis (48). In this analysis, the cross-correlation of the atomic displacements of Cα atoms was examined across the last 75 ns of the MD simulations. For the displacement vectors Δri and Δrj of atoms i and j, respectively, the cross-correlation is given by Equation 4,
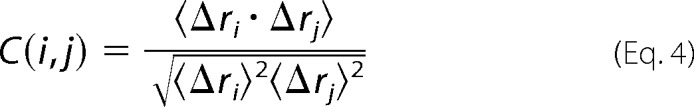 |
Matrix C, for which the elements cij are given by the above equation, was calculated for Cα atoms in PTRAJ. The calculated matrix was visualized and analyzed using MATLAB 2012b (MathWorks, Natick, MA). For completely correlated motions c(i,j) = 1, and for completely anti-correlated motions c(i,j) = −1. Complete correlation indicates that the motions have the same phase as well as the same period. Deviations from 1 (or −1) imply either that the motions of i and j are less correlated (or anti-correlated) or that they deviate from motion along a straight line.
Residence Time Analysis
This analysis was performed using Cα atoms and snapshots extracted from the last 50 ns of the MD trajectories. In this analysis, we viewed the trajectory of Cα atoms as a collection of successive local cages. For a given Cα atom, the residence time was the average time that the atom spent in a cage. To identify a cage, the atomic coordinates over a 3-ps window were grouped, and a running average of the center of mass was calculated. A cage was identified when the distance between three consecutive centers of mass varied less than 5%. The cage identified was characterized by its center of mass and radius; the latter was defined as the one that circumscribed 80% of the cage positions. We considered that an atom left its cage when it spent more than 6 consecutive ps outside of it. This analysis brings new insights to the dynamics of the relatively less mobile atoms, which exhibit longer residence time compared with the more mobile atoms. Further information and details of this analysis will be presented in a separate publication.
Protein NMR
NMR samples were prepared as described previously (20, 49). All NMR experiments were done at 293 K following previously published procedures (20, 49) using a Bruker Advance III 600 MHz spectrometer equipped with a 5-mm inverse detection triple resonance (1H/13C/15N) single axis gradient TCI cryoprobe.
Figure Preparation
All structures in Figs. 1–14 were prepared with CHIMERA 1.9 (50).
FIGURE 2.

Crystal structure of H273R RdRp. A, structural model of H273R RdRp (green) is superimposed onto the structure of WT RdRp (gray), indicating very little difference between these structures (r.m.s.d. ∼0.33 Å). The site of mutation is indicated. B, the 2Fo − Fc electron density map contoured at the 1.4 σ level is shown at the site of mutation. The larger Arg side chain is accommodated without any steric issues.
FIGURE 3.

Analysis of the r.m.s.d. for trajectories of MD simulations. To determine the duration of time to use for analysis, the r.m.s.d. for the backbone atoms of the palm subdomain of the free (WT, G64S, and H273R) and binary complexes (WT-RNA, G64S-RNA, and H273R-RNA) were plotted (black line). In addition, the r.m.s.d. values for the sugar-phosphate backbone of the primer RNA (cyan) and template RNA (blue) of the RNA substrate were plotted. The proteins exhibited no dramatic fluctuations after 50 ns; therefore, the last 100 ns of the trajectories were used for analysis. Over this range of time, the amplitude of motion of the primed template was larger but regular.
FIGURE 4.
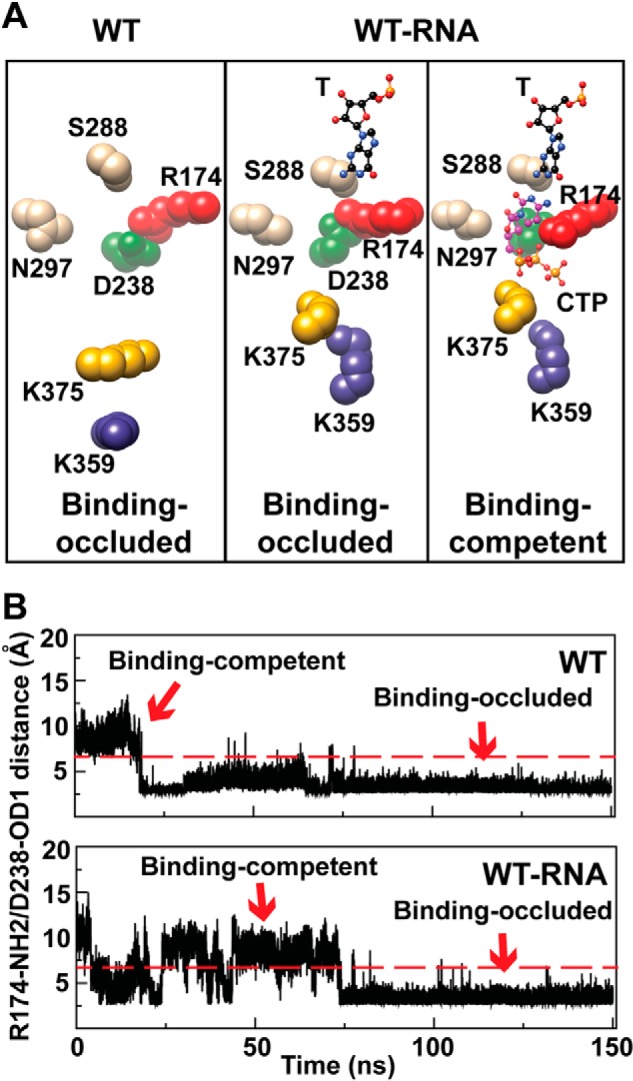
RNA binding to WT RdRp induces switching between two conformational states of the NTP-binding site. A, cluster analysis of the last 100 ns of the MD trajectories of the free and binary complex was performed as described under “Materials and Methods”. Shown is the NTP-binding site. For WT one conformation predominated that occluded the NTP-binding site (WT, Binding-occluded). Binding of RNA caused the appearance of two clusters. One cluster would clearly occlude the NTP-binding site (WT-RNA, Binding-occluded), but the other should be competent for NTP binding, as the interaction between Arg-174 and Asp-238 was disrupted (WT-RNA, Binding-competent). To highlight how the observed changes impact NTP binding, CTP has been included in the illustration of the binding-competent state. Residues shown and colors are as indicated in the legend for Fig. 1A. B, the distance between Arg-174-NH2 and Asp-238-OD1 atoms was used as a probe of the binding-occluded (<7 Å) or binding-competent (>7 Å) state. Shown here is a plot of the distance during the simulation for the free enzyme (WT) and binary complex (WT-RNA). The binding-occluded conformation was present 88% of the time for WT and 60% of the time for WT-RNA.
FIGURE 5.
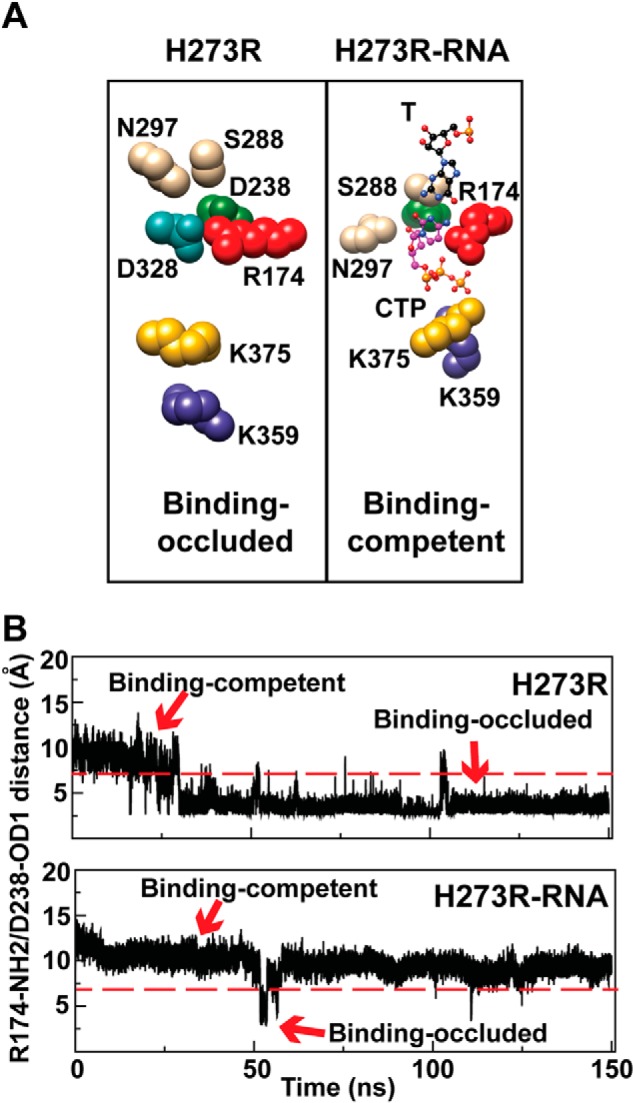
NTP binding-competent state is preferred for the H273R RdRp-RNA complex. A, cluster analysis of the last 100 ns of the MD trajectories of the free and binary complex was performed as described under “Materials and Methods.” Shown is the NTP-binding site. For the free enzyme, the NTP binding-occluded conformation predominated (H273R, Binding-occluded). For the binary complex, the NTP binding-competent conformation predominated (H273R-RNA, Binding-competent). CTP has been included in the illustration for reference. Residues shown and colors are as indicated in the legend to Fig. 1. B, the distance between Arg174-NH2 and Asp238-OD1 atoms was used as a probe of the binding-occluded (<7 Å) or binding-competent (>7 Å) state. Shown here is a plot of the distance during the simulation for the free enzyme (H273R) and binary complex (H273R-RNA). The binding-occluded conformation was present 70% of the time for H273R; the binding-competent conformation was present 98% of the time for H273R-RNA.
FIGURE 6.
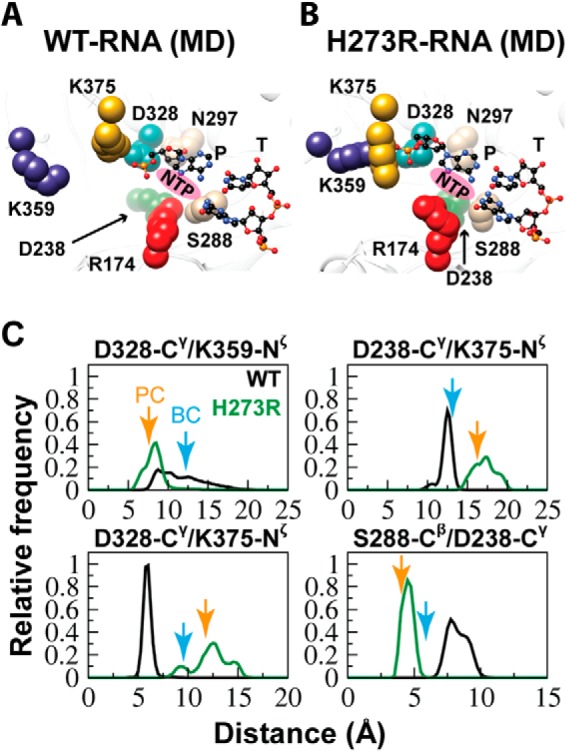
H273R RdRp-RNA complex resembles the post-incorporation complex. A, the average structure for the active site of WT RdRp-RNA complex observed during the MD simulation is shown using the same orientation and color scheme described in the legend for Fig. 1. The NTP-binding site is indicated by the magenta oval. B, same as in A but for the H273R RdRp-RNA complex. C, each panel shows the distribution of interatomic distances for the indicated pair during the MD simulation. The WT RdRp-RNA complex is abbreviated as WT and shown in black; the H273R RdRp-RNA complex is abbreviated as H273R and shown in green. The interatomic distance measured in the crystal structure of the binary complex (BC) is indicated by the cyan arrow, and that in the crystal structure of the post-incorporation complex (PC) is indicated by the orange arrow.
FIGURE 7.
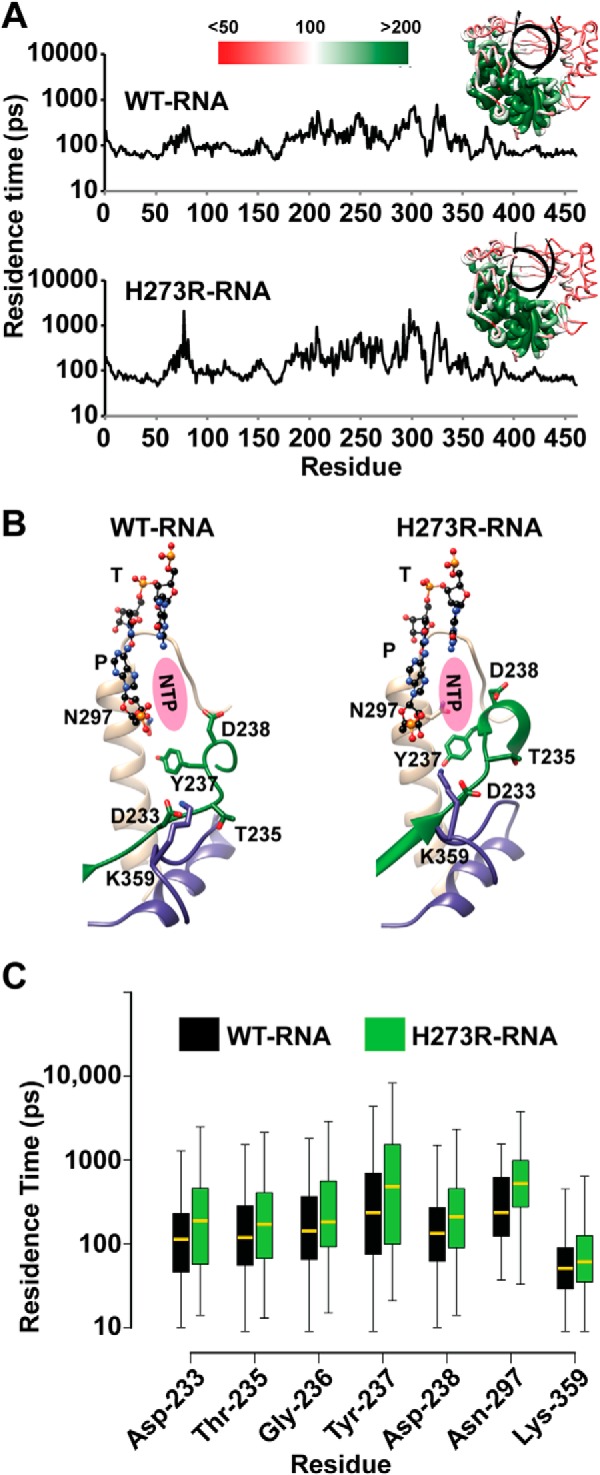
Key active-site residues exhibit reduced flexibility in the H273R RdRp-RNA complex. A, to evaluate increased inflexibility of residues in the H273R RdRp-RNA complex relative to the WT complex, we calculated a parameter that we term “residence time.” Details are provided under “Materials and Methods.” Briefly, the duration of time each Cα atom spends in a predetermined, residue-specific volume is calculated. Inflexible residues will have the longest residence time. Shown is a plot of the average residence time during the MD simulation for each residue of the WT (above) and H273R (below) RdRp-RNA complexes. The insets show the residence times plotted on the structure using the indicated red-to-green scale. The palm subdomain exhibited the least flexibility for both enzymes. B, the plots shown in A were compared for differences. Shown are active-site residues that exhibited reduced flexibility in the H273R RdRp-RNA complex relative to the WT complex. Note that the conformations of these residues also differ with the conformations for the mutator polymerase appearing preorganized for catalysis. C, we compared the plots shown in A using a Wilcoxon rank-sum test. Shown is a box plot of the residence time for the two complexes. The lower and upper ends of the box indicate the cutoff for 25 and 75% of the data, respectively; the lower and upper whiskers extending from the box indicate the minimum and maximum values of the data, respectively. The median residence time is indicated by the yellow line in the box. The p values for the differences, from left to right, were: 0.0009, 0.0409, 0.0456, 0.0549, 0.0004, 0.0200, and 0.00003.
FIGURE 8.
Magnitude of positive correlation increased for functional residues of the H273R RdRp-RNA complex. We calculated the cross-correlation of the dynamics for all Cα atoms of both the WT and H273R RdRp-RNA complexes as described under “Materials and Methods.” Instead of plotting data for all 461 Cα atoms, we only plotted residues with known functional significance, as indicated by the labels of the bracketed residues. Correlation scores range from +1 (blue, positive correlation) to −1 (brown, negative correlation). The red outline indicates residues of the H273R RdRp-RNA complex that exhibit higher values of positive correlation relative to WT. for simplicity of presentation, the analogous pairs on the opposite side of the diagonal were not highlighted.
FIGURE 10.
Side chains of residues controlling interactions with nascent base pair exhibit increased flexibility in the H273R RdRp-RNA complex relative to WT. A, residues exhibiting high ΔPCA values in Fig. 9 interact with the nascent base pair. These residues are shown in the context of the templating base for the average structure of WT and H273R RdRp-RNA complexes observed during the simulation. Colors are again as described in the legend for Fig. 1. B, to evaluate the conformational flexibility of side chains, dihedral angles were evaluated for every residue over the entire duration of the simulation. The dihedral angles for lysine are indicated; the arrow indicates the “plus” direction. A polar coordinate system from −180° to +180° was used to plot each dihedral angle. The example shown here is for dihedral angles χ1 − χ3 of Lys-159. C, cluster analysis was performed for each dihedral angle of each residue of interest. In all cases, no more than three clusters (conformations) were observed. Here the distribution of each conformation is plotted for each dihedral angle of each residue for both the WT and H273R RdRp-RNA complexes. The increased conformational flexibility of the side chains of the H273R RdRp-RNA complex is readily seen.
FIGURE 11.
Analysis of G64S RdRp and its complex with RNA. A, the distance between Arg174-NH2 and Asp238-OD1 atoms was used as a probe of the binding-occluded (<7 Å) or binding-competent (> 7Å) state. Shown here is a plot of the distance during the simulation for the free enzyme (G64S) and binary complex (G64S-RNA). The binding-competent conformation was present 95% of the time for G64S and 71% of the time for G64S-RNA. B, the average structure for the active site of the G64S RdRp-RNA complex observed during the MD simulation is shown using the same orientation and color scheme described in the legend for Fig. 1. The NTP-binding site is indicated by the magenta oval. Although NTP binding will not be occluded by the Arg-174-Asp-238 interaction, this structure represents neither of the complexes described in Fig. 6, A and B. C, the residence time experiment described in Fig. 7 was performed for the G64S RdRp-RNA complex. Shown is a box plot of the residence time for the WT and G64S RdRp-RNA complexes. The lower and upper ends of the box indicate the cutoff for 25 and 75% of the data, respectively; the lower and upper whiskers extending from the box indicate the minimum and maximum values of the data, respectively. The median residence time is indicated by the yellow line in the box. The Wilcoxon rank-sum test gave p values for the differences that were greater than 0.05 for Asp-233 and Asn-297, suggesting no difference between these two complexes to the level observed between WT and H273R RdRp-RNA complexes shown in Fig. 7. D, shown is the DCCM for G64S RdRp-RNA complex as described in the legend for Fig. 8. Correlation scores range from +1 (blue, positive correlation) to −1 (brown, negative correlation). The red outline indicates residues of the G64S RdRp-RNA complex that differ from WT. For simplicity of presentation, the analogous pairs on the opposite side of the diagonal were not highlighted. In this case, the G64S complex showed negative correlations of many residues that were positively correlated for the WT complex.
FIGURE 12.
Met-187 resonance in RdRp-RNA-NTP complex distinguishes the high- and low-fidelity enzyme. A, shown is the Met-187 resonance from the 1H-13C HSQC spectra for WT, H273R, and G64S RdRp in the absence and presence of RNA substrate (binary complex) and after adding UTP (correct nucleotide) to form the ternary complex. In WT RdRp, each complex exhibits a unique Met-187 resonance. In H273R RdRp, a unique Met-187 resonance was not observed for the H273R RdRp-RNA-UTP complex. The G64S RdRp derivative and its complexes behave like the WT. B, as in A, the Met-187 resonance is shown in complexes with UTP (correct), CTP (incorrect base), and 2′-dUTP (incorrect sugar). For WT RdRp, both an incorrect base and an incorrect sugar are “sensed” in the ternary complex, as indicated by the change in the Met-187 resonance. For H723R RdRp, Met-187 conformations correlating to proofreading of the base pair and sugar configuration are not achieved. C, dial plots for the χ1 − χ3 dihedrals of Met-187 side chain are shown. Consistent with the NMR data, conformational sampling of Met-187 is observed in the MD simulation for WT RdRp-RNA but not for H273 RdRp-RNA. D, T362I RdRp was reported previously to exhibit a loss of sugar selection (21). Here we show the Met-187 resonance for the T362I-RNA complex. This Met-187 responds to binding of the correct nucleotide (UTP) and a nucleotide with an incorrect base (CTP) but not a nucleotide with an incorrect sugar configuration (2′-dUTP). The environment of Met-187 in T362I RdRp appears to be substantially different from WT when bound to a nucleotide with an incorrect base. E, average structures of the WT RdRp-RNA complex and H273R RdRp-RNA complex from MD simulations were used to illustrate the Met-187 environment, defined as residues within a radius of 5 Å of Met-187. The nascent base pair is shown, as modeled from the post-incorporation complex (PDB code 3OL7). In the WT RdRp-RNA complex, Met-187 interacts with Phe-296, and the motif-B loop, which contains Ser-288, is not in a catalytically competent conformation. The vicinity of this loop to the nascent base pair likely links changes in Met-187 to both RNA and nucleotide binding. In H273R, Met-187 has a different side chain conformation and now interacts with Met-286, an interaction that occurs because of the substantial change in conformation of motif B. Differences between the complexes are also observed at Leu-268 and Tyr-264.
FIGURE 13.
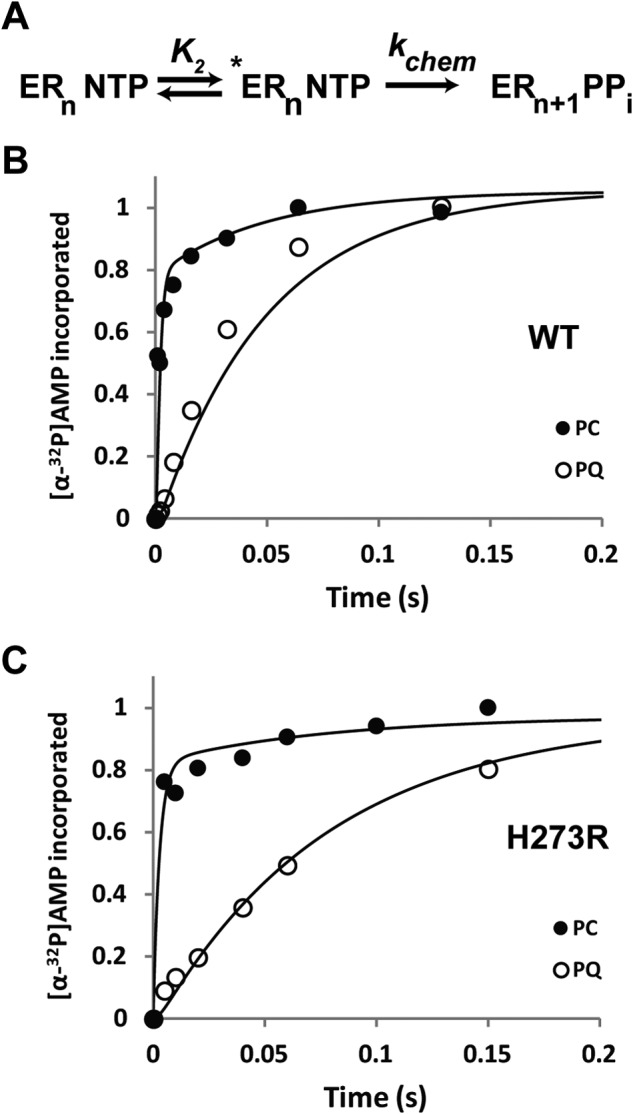
Monitoring fidelity checkpoints, a kinetic approach. A, shown is the portion of the kinetic mechanism for the single nucleotide addition cycle of PV RdRp that provides the first kinetic checkpoints: K2 and kchem. B, an isotope-trapping experiment was performed for WT RdRp-RNA complex and kinetic simulation used to obtain estimates for K2 and kchem. The RdRp-RNA complex (ERn) is pulsed with α-32P-labeled ATP to form the ternary complex (ERnATP) and initiate the reaction. Half of the reaction is quenched as a function of time, the so-called pulse-quench experiment. The other half of the reaction is chased by the addition of unlabeled ATP, thus the pulse-chase experiment. In this case, any remaining *ERnATP will be converted to ERn+1PPi; however, the remaining *ERnATP will depend on both K2 and kchem. Therefore, in the chase experiment, “extra” product will appear that can be accounted for by the fraction that was protected. The data are plotted, and the line represents the mechanism described in A, with K2 = 3.0 and kchem = 30 s−1. C, an isotope-trapping experiment was performed for the H273R RdRp-RNA complex, and kinetic simulation was used to obtain estimates for K2 and kchem. The data are plotted, and the line represents the mechanism described in A, with K2 = 6.0 and kchem = 15 s−1. These data are consistent with H273R making the conformational change step more facile.
FIGURE 14.
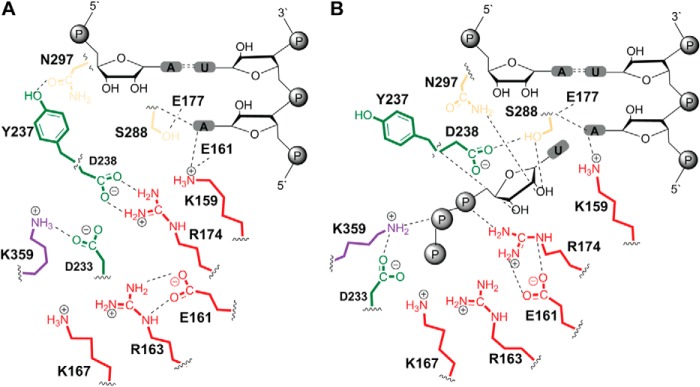
Conserved residues contributing to the transition from the NTP binding-occluded state to the NTP binding-competent state. The NTP binding-occluded state (A) and NTP binding-competent state (B) are shown. Residues of conserved structural motifs A (green), B (tan), D (blue), and F (red) that interact directly or indirectly with bound nucleotide participate in the occluded-to-competent transition. The stability of the nucleotide binding-competent state should be proportional to the number of interactions between the enzyme and nucleotide, providing a possible explanation for the fidelity of nucleotide selection.
Structure Deposition
The atomic coordinates and structure factors have been deposited in the PDB under ID code 4R0E.
RESULTS
PV H273R RdRp Is an Error-prone Polymerase
We recently described a PV mutant that exhibits a mutator phenotype in cells and that is attributable to a change of His-273 of the RdRp to Arg (H273R) (5). In that study, we showed that the GMP misincorporation frequency was ∼3-fold higher than observed for the wild-type (WT) enzyme. To determine whether the error-prone activity of the H273R RdRp was limited to base mispairing, we evaluated the ability of this enzyme to utilize a nucleotide with an incorrect sugar configuration (Table 1, 2′-dUTP) and the ability of this enzyme to extend a mispaired primer terminus (Table 1, S/S-UG RNA). To do this, pre-steady-state kinetic methods were employed to determine the maximal rate constant for single nucleotide incorporation (kpol) and the apparent dissociation constant for nucleotide (Kd,app). As shown previously (5), the catalytic efficiency (kpol/Kd,app) of the H273R RdRp was 10-fold higher than the WT (Table 1, compare line 7 with line 1). In both experiments, H273R RdRp appeared more promiscuous than the WT, even after taking into account the differences in catalytic efficiency.
TABLE 1.
Kinetic parameters for UTP and 2′-dUTP incorporation by WT and H273R enzymes in protonic and deuteronic solvents
Values are rounded to 1 significant figure. Standard errors range from 10 to 20% of the measured values.
| No. | Enzyme | RNA | NTP | Solvent | kpol | Kd,app | kpol/Kd,app | D2O effecta |
|---|---|---|---|---|---|---|---|---|
| s−1 | μm | μm−1s−1 | ||||||
| 1 | WT | S/S-UA | UTP | H2O | 200 | 400 | 5 × 10−1 | 3 |
| 2 | S/S-UA | UTP | D2O | 70 | 60 | 1 | ||
| 3 | S/S-UA | 2′-dUTP | H2O | 8 | 1000 | 8 × 10−3 | 1 | |
| 4 | S/S-UA | 2′-dUTP | D2O | 7 | 600 | 1 × 10−2 | ||
| 5 | S/S-UG | UTP | H2O | 10 | 1000 | 1 × 10−2 | 1 | |
| 6 | S/S-UG | UTP | D2O | 10 | 500 | 2 × 10−2 | ||
| 7 | H273R | S/S-UA | UTP | H2O | 200 | 40 | 5 | 4 |
| 8 | S/S-UA | UTP | D2O | 60 | 20 | 3 | ||
| 9 | S/S-UA | 2′-dUTP | H2O | 40 | 300 | 1 × 10−1 | 2 | |
| 10 | S/S-UA | 2′-dUTP | D2O | 20 | 100 | 2 × 10−1 | ||
| 11 | S/S-UG | UTP | H2O | 40 | 200 | 2 × 10−1 | 3 | |
| 12 | S/S-UG | UTP | D2O | 10 | 70 | 1 × 10−1 |
a The D2O effect was calculated as the ratio of kpol(H2O)/kpol(D2O).
Crystal Structure of H273R RdRp Reveals No Significant Differences Relative to WT
Solution of the x-ray crystal structure of the high fidelity PV G64S RdRp was unable to show a significant difference relative to WT, and the altered fidelity was attributed to differences related to dynamics (23). Although we did not expect substantial structural perturbations with this enzyme, we pursued the x-ray crystal structure of the H273R RdRp to make sure that we did not miss any significant structural difference that could explain the biochemical and biological differences observed for the H273R mutant. Data collection and refinement statistics are shown in Table 2. Superposition of the Cα atoms of H273R and the WT RdRp structures showed a root-mean-square deviation (r.m.s.d.) of 0.33 Å, which was essentially no change (Fig. 2A). Most of the changes were localized to the site of the H273R substitution; these changes accommodated the arginine side chain very well (Fig. 2B).
TABLE 2.
Data collection and refinement statistics for the H273R structure
| Space group | P65 |
| Unit Cell (Å) | a = b = 126.44, c = 113.24 |
| Resolution limits (Å) | 20–3.0 (3.11–3.0)a |
| Total observations | 11,7019 |
| Unique reflections | 19,450 |
| Redundancy | 5.7 (5.6)a |
| Completeness (%) | 99.8 (99.9)a |
| Rmerge (%)b | 6.7 (31.3)a |
| Rcrystc | 18.9 |
| Rfreec | 21.3 |
| No. of protein atoms | 3702 |
| No. of water molecules | 26 |
| r.m.s.d. bond length (Å) | 0.008 |
| r.m.s.d. bond angle (°) | 1.48 |
a Values in parentheses are for the highest resolution shell.
b Rmerge = Σ|Ih − 〈Ih〉|/ΣIh, over all h where Ih is the intensity of reflection h.
c Rcryst and Rfree = Σ‖Fo| − |Fc‖/Σ |Fo|, where Fo and Fc are the observed and calculated amplitudes, respectively. Rfree was calculated using the 5% of data excluded from the refinement.
MD Reveal a Nucleotide Binding-occluded State and Nucleotide Binding-competent State That Interconvert on the Nanosecond Timescale
MD Design
Previously, we performed 25-ns all-atom and 1 μs coarse-grained MD simulations of four picornaviral RdRp (23, 24). These studies revealed conserved and correlated motions of the conserved structural/functional motifs comprising the RdRp active site. Analysis of the high fidelity G64S RdRp revealed a substantial change in these motions, which we interpreted as the basis for the change in fidelity relative to WT (23). In those studies, only the free enzymes were analyzed. In this study, we performed a 150-ns all-atom MD simulation for WT, H273R, and G64S RdRp in the absence and presence of primed template RNA. The primer was 9 nt in length, and the template was 13 nt in length (see “Materials and Methods”). The RdRp-RNA complexes were derived from the structure of the WT RdRp-RNA complex (PDB code 3OL6) (19) (see Materials and Methods”). Analysis of the r.m.s.d. of the backbone of the stable core of the protein (palm subdomain) revealed that all trajectories had reached stability by 50 ns (Fig. 3). We used only the last 100 ns of the trajectory for data analysis. The average r.m.s.d. value decreased when RNA was bound (compare each enzyme in the absence and presence of RNA shown in Fig. 3), consistent with RNA binding reducing the overall flexibility of the protein. The sugar-phosphate backbone of the primer, which is in a stable duplex with the template and has substantial interactions with the enzyme (19), was least flexible (Fig. 3). In contrast, the template was much more flexible (Fig. 3); most of this flexibility derived from the n + 2 to n + 4 regions of template that interact with the fingers subdomain away from the active site (19).
Identification of an Equilibrium between the Nucleotide Binding-occluded and Nucleotide Binding-competent States
A comparison of the crystal structures of the WT RdRp-RNA complex (binary in Fig. 1C) to the post-incorporation complex (Fig. 1C) clearly revealed the need for reorganization of the nucleotide-binding site to occur to permit nucleotide binding (19). The side chains of two residues conserved in essentially all RdRp are at the center of this reorganization: Arg-174 (motif F) and Asp-238 (motif A) (Fig. 1C). We performed a cluster analysis of these and surrounding residues using the structures of the conformations observed over the last 100 ns of the simulation for WT RdRp and the WT RdRp-RNA complex. The binding-occluded conformation was the only state observed for WT RdRp (Fig. 4A). In contrast, both a binding-occluded state and a binding-competent state were observed once RNA was bound (Fig. 4A). To obtain a kinetic perspective of the interconversion of these states, we plotted the distance between the Arg-174 guanidinium moiety and Asp-238 carboxylate moiety over the entire trajectory for WT RdRp and WT RdRp-RNA complex (Fig. 4B). The WT RdRp started in a binding-competent conformation during equilibration but sampled the binding-occluded conformation early in the simulation and remained in that conformation for the remainder of the trajectory (Fig. 4B). The presence of RNA caused the sampling of the two states to occur more often, with the population of each state essentially equal (Fig. 4B).
Preorganization of the H273R RdRp Active Site as a Contributor to Its Error-prone Activity
Nucleotide Binding-competent State Favored by H273R RdRp-RNA Complex
We performed a cluster analysis over the last 100 ns of the simulation for the H273R RdRp and H273R RdRp-RNA complex. In the absence of RNA, the nucleotide binding-occluded state was favored by H273R RdRp. The average structure of this state was similar to that observed for WT. However, there was a reorientation of the Asp-238 side chain that would be expected to further antagonize nucleotide binding (Fig. 5A) because of steric interference and the movements of ribose-binding determinants, Ser-288 and Asn-297 (19). The presence of RNA shifted nearly all of the observed conformations to those that would be classified as binding-competent (Fig. 5A). The interconversion of states was again monitored by following the distance between Arg-174 and Asp-238 as a function of time (Fig. 5B). The H273R RdRp started in the binding-competent state, but the binding-occluded state predominated (∼70% of the whole simulation time) once was sampled (Fig. 5B). In contrast, the H273R RdRp-RNA complex existed most of the time (∼98% of the whole simulation time) in the binding-competent state (Fig. 5B).
H273R RdRp-RNA Complex Resembles the Post-incorporation Complex Observed Crystallographically
We also performed a comparison of the average structures of the RdRp-RNA complexes for WT and H273R observed by MD (Fig. 6, A and B). In addition to changes in the positions of Arg-174 and Asp-238, substantial differences in the positions of other side chains were also evident (compare Asp-328, Ser-288, Asn-297, Lys-359, and Lys-375 in Fig. 6A with the corresponding residues in Fig. 6B). Interestingly, these very same residues showed the largest positional differences when the x-ray crystal structures for the binary complex and post-incorporation complex were compared (Fig. 1C) (19). The post-incorporation complex is the structure most representative of the catalytic state for all of the PV RdRp structures solved to date (19). The nucleotide was incorporated in the crystal but translocation did not occur. The binary complex could be distinguished from the post-incorporation complex by comparing the distances between several side chains, as indicated in Fig. 6C. We determined the distance between the indicated residues (the distances between Cγ of Asp-328 and Nζ of Lys-359 and Lys-375 and between Cγ of Asp-238, Nζ of Lys-375, and Cβ of Ser-288) at 1-ps intervals over the last 75 ns of the simulation for both the WT and H273R RdRp-RNA complexes. The relative frequency of each distance was plotted for each complex (Fig. 6C). In all cases, the inter-side chain distances observed for the H273R RdRp-RNA complex were closest to those observed in the post-incorporation complex (Fig. 6C). For the WT RdRp-RNA complex, this complex was closest to the binary complex, although the level of coincidence was not as observed for the H273R RdRp-RNA complex (Fig. 6C).
Key Active-site Residues of H273R RdRp-RNA Complex Exhibit Reduced Backbone Dynamics
Our previous MD studies of picornaviral RdRp emphasized changes in dynamics occurring for residues, structural elements, and/or domains exhibiting the most motion (23, 24). To obtain information about differences in regions of RdRp exhibiting limited flexibility, we performed a “residence time” analysis. This analysis provides a kinetic perspective of the motions that is hidden in other analyses, such as a B-factor analysis. The technical details of this new approach are provided under “Materials and Methods.” Essentially, we created a sphere that circumscribes 80% of the conformational space sampled by an atom. Next, we determined the length of time that the atom spends in that sphere. Greater time indicates lower flexibility. A major advantage of this approach is that rigid residues are not present in the baseline but appear as peaks, permitting these residues to be identified easily by visual inspection. The data for the WT and H273R RdRp-RNA complexes are shown in Fig. 7A. The median residence time was ∼100 ps. We plotted the residence time for each Cα atom on the enzyme structure using a scale that ranged from half of the median value to twice the median value (see insets in Fig. 7A). The palm subdomain was the least flexible.
We used the nonparametric Wilcoxon rank-sum test (51) in R (52) to determine whether statistically significant differences existed between the WT and H273R RdRp-RNA complexes. This analysis identified 109 residues exhibiting statistically significant differences (p < 0.05) between the complexes. Of these, only 11 residues became more flexible (shorter residence time) in the H273R RdRp-RNA complex. We focused our attention on residues of the palm known to contribute to nucleotide binding and/or catalysis. These are shown in Fig. 7B and include residues located in conserved structural motifs A, B, and D. All of these residues exhibited longer residence times in the H273R RdRp-RNA complex than observed for the WT complex (Fig. 7C).
Correlated Motions of Functionally Important Residues Are Intensified in the H273R RdRp-RNA Complex
In our first MD study of PV RdRp, we created a DCCM, which showed that the strongest correlated motions, both positive and negative, were observed for functional motifs (23). We repeated this analysis for the RdRp-RNA complexes (Fig. 8). To facilitate interpretation of the data, we again used a scale from −1 (negatively correlated) to +1 (positively correlated) but restricted our attention to functional residues, as indicated in Fig. 8. Residues that were positively correlated in the WT RdRp-RNA complex were also positively correlated in the H273R RdRp-RNA complex, but the intensity of the correlation was not always the same. The most striking difference between the WT and H273R RdRp-RNA complexes was that the H273R RdRp-RNA complex exhibited a substantial increase in the number and intensity of the positively correlated residues (highlighted in Fig. 8). For example, Thr-235, Gly-236, Tyr-237, and Asp-238 are all more positively correlated with Arg-163, Lys-167, and Arg-174 (Fig. 8, compare H273R-RNA with WT-RNA). All of these residues interact with the nucleotide substrate. A similar trend existed for the positive correlation between residues interacting with the templating base (Thr-114, Ser-115, Lys-159, Leu-175, Ile-176, Ala-178, and Ser-179) and residues interacting with the nucleotide substrate (Thr-235, Gly-236, Tyr-237, and Asp-238) (Fig. 8). Furthermore, residues of motif A (Asp-233 to Asp-238) appeared more positively correlated with each other in H273R RdRp-RNA relative to WT RdRp-RNA (Fig. 8).
Flexibility of the H273R RdRp Nascent Base Pair-binding Pocket as a Contributor to Its Error-prone Activity
Principal Component Analysis Reveals Larger Amplitudes in the Backbone Motions of Motifs F and G/G′ for the H273R RdRp-RNA Complex
The overall dynamics of a protein can be considered as the sum of a distribution of atomic fluctuations around an average position of that atom. The variance of an atom, representing its motion, can be used to calculate principal components (also known as modes). The number of modes is equivalent to the number of atoms evaluated multiplied by three. Each principal component is associated with an eigenvalue that defines its magnitude of motion and thus permits each principal component to be ranked.
We performed a PCA (46, 47) for the Cα atoms of the WT and H273R RdRp-RNA complexes to obtain information on the major motions observed during the simulations and the major contributors to these motions. As observed for the RdRp alone (23), conserved structural motifs (A, D, E, F, and G/G′) exhibited the largest amplitude of motions. To compare the WT and H273R RdRp-RNA complexes, we used the top 10 modes (accounting for ∼50% of the total variance contained in the last 75 ns of the simulations) to calculate the relative displacement for each residue in the protein. A relative displacement was calculated by normalizing to the average displacement observed for the least flexible 5% of residues. We calculated a ΔPCA value by subtracting the relative PCA value for each residue of the WT RdRp-RNA complex from that of the corresponding residue in the H273R RdRp-RNA complex. A histogram of the data is shown in Fig. 9A. A ΔPCA value of zero means that no change was observed between the two complexes. The ΔPCA mean was 0.1, and the ΔPCA median was 0.03. Therefore, most residues changed very little between the two complexes.
We plotted the ΔPCA values on the structure such that the magnitude of the motion was indicated by varying radii of the tube representation of the structure (Fig. 9B). In general, the conserved structural motifs were more dynamic for the H273R RdRp-RNA complex than that observed for the WT (Fig. 9B). The exceptions were motifs A and D, consistent with the residence time analysis (Fig. 7). This observation led to further scrutiny of the changes observed in the H273R RdRp-RNA complex. Residues in conserved structural motifs exhibiting differences with an absolute value greater than 0.3 are listed in Fig. 9C and are indicated explicitly in the structural model shown in Fig. 9B. All of the residues shown interact with the triphosphate or ribose of the nucleotide substrate and/or the nascent base pair. Seven of the 10 listed active-site residues demonstrated positive ΔPCA values. We concluded that enzyme determinants for substrate binding are substantially more flexible in the H273R RdRp-RNA complex relative to the WT complex.
Dihedral Angle Analysis Reveals Enhanced Flexibility of Side Chains of Motifs F and G/G′ for the H273R RdRp-RNA Complex
The enhanced flexibility of the Cα backbone of motifs F and G/G′ of the mutator polymerase complex was particularly intriguing to us. The average structure of the WT RdRp-RNA complex observed during the simulation represents the nucleotide binding-occluded state, whereas that of the H273R RdRp-RNA complex represents the nucleotide binding-competent state (Fig. 10A). As shown in Fig. 10A, in going from the occluded to the competent state there is a rearrangement of motif F (Tyr-157, Lys-159, Arg-174, and Ile-176) and motif G/G′ (Lys-127). In the occluded state, motif F protrudes into the nucleotide-binding pocket and prevents the templating base from adopting a conformation competent for base-pairing. These impediments to binding are lost in the competent state.
Flexibility of the backbone of motifs F and G/G′ could clearly impact the equilibrium between the states. If specificity is imparted by side chain interactions, then conformational flexibility of the side chains might also be expected to impact specificity. To obtain information on side chain conformations sampled during simulations, we performed analysis of the side chain dihedrals. We used dial plots to present the conformational space sampled by the dihedrals (−180° to +180°) at 1-ps intervals. The analysis is illustrated for the side chain of Lys-159 (Fig. 10B). The lysine side chain has four dihedral angles: χ1, χ2, χ3, and χ4 (Fig. 10B). The position of each angle can be defined by a polar coordinate and plotted accordingly (Fig. 10B). In the example shown, it is readily apparent that angles χ1− χ3 of the H273R RdRp-RNA complex sampled unique conformations relative to the WT complex (Fig. 10B). We used k-means algorithm in MATLAB R2102b (MathWorks) to cluster the observed conformations. Our input specified the existence of no more than five clusters. In general, the conformational space sampled by the dihedrals was trimodal (Fig. 10C), clustering around ±180°, +60°, and −60° as is visually apparent (Fig. 10B). The cluster analysis provided a quantitative analysis of the conformational flexibility of key side chains of motifs F and G/G′ (Fig. 10A) and a quantitative comparison of the impact of the H273R substitution on the distribution of states. In all cases, the side chain flexibility of the indicated residues was increased in the mutator polymerase complex relative to WT (Fig. 10C).
The High Fidelity G64S RdRp
The very first PV RdRp derivative exhibiting perturbed incorporation fidelity contained the G64S substitution but conferred a high fidelity (anti-mutator) phenotype (6, 8). In a previous study, we performed a MD analysis of the unbound G64S RdRp (23). In that study we observed changes between the dynamics of that G64S and WT RdRp that were on par with that observed between WT PV RdRp and WT foot-and-mouth disease virus RdRp (23). The PV and foot and mouth disease virus enzymes share only 30% sequence identity (23). In that study, the relative rigidity of the active site of G64S PV RdRp compared with WT RdRp was suggested to play a role in the enhanced fidelity of G64S mutant. However, we were not able to hone in on a more detailed mechanism for the physical basis of RdRp incorporation fidelity. We have extended the analytics reported above to the G64S RdRp and RdRp-RNA complex.
In the absence of RNA, G64S RdRp favors the nucleotide binding-competent conformation, as defined by the distance between Arg-174 and Asp-238, across the entire simulation (Fig. 11A). For G64S RdRp-RNA, the binding-competent conformation was also observed over the last 100 ns of the MD trajectory. However, the addition of RNA induces sampling of the nucleotide binding-occluded conformation, which was observed during the first 50 ns of the simulation (∼30% of the total length of simulation) (Fig. 11A). The average structure observed for the G64S RdRp-RNA complex (Fig. 11B) was unique relative to both the WT and H273R RdRp-RNA complexes (Fig. 6, A and B, respectively). Because the average structure was produced using the last 75 ns of the trajectory, the observed state should be primarily “binding-competent” based on the Arg-174 to Asp-238 distance (Fig. 11A). However, the key active-site residues, overall, did not adopt a conformation approximating a catalytically competent state as observed for the H273R RdRp-RNA complex (compare Fig. 11B with Fig. 6B), and there was no hint of stable preorganization of any active-site residues relative to WT PV RdRp-RNA as observed for the H273R RdRp-RNA complex (Fig. 6C).
The residence time analysis revealed that residues of motif A (residues 235–238) in the G64S RdRp-RNA complex, in contrast to the H273R RdRp-RNA complex, have shorter residence times than that of WT RdRp-RNA (p < 0.05), see Fig. 7C. Lys-359 exhibited a longer residence time in G64S RdRp-RNA compared with WT, whereas the differences for Asp-233 and Asn-297 were not significant (p > 0.05) compared with what observed between H273R RdRp-RNA and WT (Fig. 7C). Most astonishing, however, was the observation that many residues that exhibited a positive correlation in the WT and H273R RdRp-RNA complexes (Fig. 8) exhibited a negative correlation in the G64S RdRp-RNA complex (Fig. 11D).
Experiments Support Loss of a Fidelity Checkpoint for H273R RdRp-RNA Complex
States of the RdRp Evaluated by Using NMR
We have shown that [methyl-13C]methionine labeling of the PV RdRp permits NMR to be used to distinguish the different states of the RdRp: free, bound to RNA (binary complex), and bound to RNA and nucleotide (ternary complex) (20). The resonance for Met-187 senses transitions of the WT enzyme from the free protein to the binary complex and to the ternary complex, giving rise to three non-overlapping resonances (Fig. 12A, WT). When this experiment was performed for H273R RdRp, free enzyme could be distinguished from the binary complex, but the addition of nucleotide failed to cause a detectable change in the Met-187 resonance (Fig. 12A, H273R). The G64S RdRp appeared identical to the WT (Fig. 12A, G64S). We suggest that the perturbation of the Met-187 resonance observed in the presence of nucleotide represents a state competent for “proofreading” the bound nucleotide, a state that is not achieved in the H273R RdRp.
For WT RdRp-RNA complex, the Met-187 resonance was also diagnostic for the correctness of the bound nucleotide (Fig. 12B, WT). The Met-187 resonances were different for a correct nucleotide (Fig. 12B, UTP, WT), a nucleotide with an incorrect base pair (Fig. 12B, CTP, WT), and a nucleotide with an incorrect sugar configuration (Fig. 12B, 2′-dUTP, WT). For the H273R RdRp-RNA complex, the Met-187 resonance was the same regardless of the correctness of the bound nucleotide (Fig. 12B, H273R). The lost conformational sampling of Met-187 between WT and H273R was also observed in our MD simulations (Fig. 12C). The dial plot shows substantial conformational sampling of the Met-187 side chain for WT but essentially none for H273R (Fig. 12C).
We reported previously a PV RdRp derivative with reduced stringency of selection against nucleotides with an incorrect sugar configuration (21). This derivative changed Thr-362 of motif D to Ile (T362I RdRp) (21). Here we show that for the T362I RdRp-RNA complex, the Met-187 resonance senses nucleotide binding (Fig. 12D, left panel). Binding of a nucleotide with an incorrect base pair leads to a Met-187 resonance at a new position relative to that observed for WT (Fig. 12D, CTP in right panel). However, binding of a nucleotide with an incorrect sugar configuration produces a Met-187 resonance that is coincident with that of a correct nucleotide (Fig. 12D, right panel, 2′-dUTP). Collectively, these data suggest that Met-187 can be used to predict the ability of a derivative to achieve a conformation capable of proofreading the bound nucleotide and the types of nucleotide for which specificity will be reduced. Evaluation of the Met-187 environment for the WT and H273R RdRp-RNA complexes reveals differences that are in large part caused by a conformational change of the motif-B loop, residues 286–291 (Fig. 12E).
Solvent Deuterium Isotope Effect
The kinetic mechanism for PV RdRp includes a conformational change step preceding nucleotidyl transfer (Fig. 1B, step 2) (16) that is used as a fidelity checkpoint. The equilibrium constant for this step for WT is 0.6, imposing a barrier for correct nucleotide incorporation that is partially rate-limiting for nucleotide addition and that becomes even more daunting for incorrect nucleotide incorporation. Because two protons transfer in the transition state for nucleotide addition, an SDKIE is observed for the WT (53). Therefore, we can use the SDKIE to determine the extent to which this conformational change step contributes to the rate constant for nucleotide addition (kpol) (16). If the conformational change step becomes slower (more rate-limiting) than the WT, then the SDKIE should be smaller than observed for the WT. A faster conformational change step (less rate-limiting) will yield a SDKIE larger than that observed for WT. We measured the SDKIE for H273R RdRp during correct nucleotide incorporation, incorporation of nucleotide with an incorrect sugar configuration, and extension of a mispaired primer terminus (Table 1). In all cases, the SDKIE observed for H273R RdRp was always higher than that observed for WT (Table 1). We conclude that the conformational change step is no longer effective as a fidelity checkpoint because of an increase in the equilibrium constant for this step.
Isotope Trapping
Unfortunately, we were unable to convert the value for the change in the SDKIE to a value for the change in equilibrium constant. To get a better idea of the magnitude of the change in the equilibrium constant for this step, we used an isotope-trapping experiment (16). In this experiment, [α-32P]NTP was pulsed into the reaction. The reaction was then either quenched or chased by the addition of an excess of unlabeled NTP. The equilibrium constant for the conformational change step (K2) would determine any difference in the observed kinetics of [α-32P]NMP incorporation. Kinetic simulation to the minimal mechanism shown in Fig. 13A could then be used to approximate the value for K2. In the case of PV RdRp, this experiment becomes very clean if the Mn2+ is used as the divalent cation where chemistry is completely rate-limiting (17). We performed this experiment herein for both the WT (Fig. 13B) and H273R (Fig. 13C). These data are consistent with at least a 2-fold increase in K2 for H273R RdRp. Clearly, this is a lower limit, as essentially all of the pulsed nucleotide is chased into product as fast as it can be measured using the chemical quench-flow device (Fig. 13C).
DISCUSSION
Accurate replication and transcription of the genomes of all organisms are essential for life. As a result, a major objective of polymerase enzymology for decades has been the elucidation of the physical basis for the fidelity of nucleotide addition (54–57). For most organisms, high fidelity replication is beneficial. This is not the case for positive-strand RNA viruses and likely other types of RNA viruses as well (3). It turns out that genetic diversity within a viral population is essential for the virus to evade extinction (58). Importantly, an optimal genetic diversity exists that is required for maximal viral fitness (59). Therefore, the fidelity of nucleotide addition by viral RdRp is a determinant of viral virulence and, as such, represents a target for antiviral therapy and viral attenuation (i.e. live virus vaccine candidate) (2, 60). To harness the full therapeutic and prophylactic potential of RdRp fidelity, an understanding of the physical/structural basis for RdRp fidelity is required.
In all polymerase systems for which a complete kinetic mechanism of nucleotide addition exists, a conformational change after nucleotide binding contributes substantially to the fidelity of nucleotide addition (54–56). The viral RdRp is no different (Fig. 1B) (16). In most polymerases, this conformational change has been related to structural transitions of the fingers and thumb subdomains from an open state to a closed state (55). Because the fingers and thumb subdomains of the RdRp are made inflexible by the presence of fingertip structural elements unique to the RdRp, the conformational change step used for fidelity in this class of polymerases cannot involve structural transitions of the fingers and thumb subdomains from an open state to a closed state (Fig. 1A). Structures of the PV RdRp alone and in complex with RNA before or after nucleotide addition reveal conformational changes of residues of conserved structural motifs (Fig. 1C) (19, 61). Whether these conformational changes contribute to the fidelity of nucleotide addition is not known.
One approach that our laboratory has taken in studying the conformational dynamics of the PV RdRp that might contribute to the fidelity of nucleotide addition is MD simulations (23, 24). We were motivated by the suggestions of others that motions sampled by enzymes are not random and represent those used to channel the enzyme through its reaction coordinate (62) and that motions on the ps-ns timescale may be linked to those on the μs-ms timescale (63). One way to look at this difference in timescales is to consider the much slower macroscopic rate of the biochemical reaction as being determined by averaging over a large number of trajectories similar to those sampled on the ns timescale during MD simulations (23, 64). The energy of this large number of trajectories is distributed according to the Boltzmann law. At temperatures often used to carry out biochemical reactions, only a very small fraction of the trajectories can channel the enzyme through its reaction coordinate. Thus, to detect a measurable amount of the progress of a biochemical reaction averaging over a μs-ms timescale is required. This latter timescale is clearly of relevance for conformational changes that control the fidelity of nucleotide addition by PV RdRp and other polymerases (16, 65). An approach other than x-ray crystallography is clearly warranted, as the crystal structure of the high fidelity PV G64S RdRp could not explain the functional change (27). This conclusion is the same for the low-fidelity H273R RdRp described herein (Fig. 2).
Our previous MD simulations of the PV RdRp in the absence of RNA and nucleotide substrate showed that the conserved structural motifs contributed most to the motions observed (23). These motions were correlated (positively or negatively) in a manner that was relevant to function. Observations made for PV RdRp were reproduced with three other picornaviral RdRp, engendering confidence in both the approach and the findings (23). In this current study, we extended the simulation time of the free PV RdRp, and one of the most unexpected observations was that the nucleotide-binding pocket remained in an occluded state, with Arg-174 and Asp-238 interacting quite stably (Fig. 4). We have extended our goal in this study to interrogate the PV RdRp-RNA binary complex. In general, the RdRp-RNA complex was less flexible than the free enzyme (Fig. 3). This observation was expected, as ligand binding often leads to reduced protein flexibility. However, the presence of RNA increased the flexibility of the active site. For example, the nucleotide-binding pocket now sampled two conformations instead of one; one of these conformations was competent for nucleotide binding (Fig. 4). This unexpected observation is quite interesting and provides meaningful insights. Just as the RdRp alone samples conformations that will permit binding to RNA (23), the RdRp-RNA complex samples conformations that will permit nucleotide binding and catalysis. The observed dynamics of the enzyme and its RNA complex thereof are consistent with the reaction coordinate traversed along the nucleotide incorporation reaction.
Two extremes were observed for the nucleotide-binding and catalytic sites of the WT RdRp-RNA complex. A schematic of these states is presented in Fig. 14. Essentially all of the residues of conserved structural motifs with side chains that interact with some portion of the incoming nucleotide substrate are also found to interact in the absence of nucleotide (Fig. 14, compare panels A and B). In the nucleotide binding-competent state, the RdRp interacts with all functional groups on the nucleotide: triphosphate, ribose hydroxyls, and nascent base pair. In the absence of a 2′-OH, for example, Ser-288 and Asn-297 might remain in the nucleotide binding-occluded conformation, leading to expulsion of this nucleotide with an incorrect sugar configuration. Myriad similar examples can be gleaned from Fig. 14, leading us to propose that the thermodynamic stability of intermediate states relative to the nucleotide binding-occluded state may contribute to nucleotide specificity/selection.
In addition to the two states of the nucleotide-binding pocket, two states of the primer- and template-binding pockets were noted. Residues that interact with primer (Tyr-326, Gly-327, Asp-328, Asp-329, Leu-374, and Lys-375) are all positively correlated (Fig. 8). Residues 326–329 are part of the conserved structural motif C. These residues form a β-turn that can toggle between an extended conformation and a bent conformation, with only this latter conformation supporting catalytically competent alignment of the primer. Residues that interact with a template/nascent base pair (Lys-127, Tyr-157, Lys-159, Arg-174, and Ile-176) transition from a loose to a tight interaction (Fig. 10A). As mentioned above, these motions are also positively coordinated (Fig. 8). Collectively, our data reveal a coordinated sampling of two states of all active-site elements that may contribute to nucleotide selection and the catalytic efficiency of the RdRp.
We propose that the equilibrium between the nucleotide binding-occluded and nucleotide binding-competent states observed on the nanosecond timescale is related to the conformational change step linked to fidelity observed on the millisecond timescale (16). Two observations are consistent with this possibility. First, in the MD simulation of the H273R RdRp-RNA complex, the binding-competent state is favored (Fig. 5B). Second, the H273R RdRp-RNA complex transits the fidelity checkpoint on the millisecond timescale, measured biochemically much more easily than the WT (Fig. 13 and D2O effect in Table 1). This preorganization of the H273R RdRp-RNA complex in a conformation that has already bypassed the fidelity checkpoint is also supported by the fact that the average structure of the H273R RdRp-RNA complex observed during the simulation is most reminiscent of the RdRp-RNA complex that was observed crystallographically immediately following nucleotide incorporation (Fig. 6) (19). In addition, results from the NMR (Fig. 12) suggest that the H273R ternary complex bypassed the fidelity checkpoint that exists in the WT.
If our proposal is correct, then nucleotide incorporation fidelity can be tuned by altering the stability of the interactions shown in Fig. 14, consequently shifting the equilibrium between the binding-occluded and binding-competent states. Because all of these residues are highly conserved in the RdRp of positive-strand RNA viruses in mammals, these interactions may represent new polymerase mechanism-based strategies for viral attenuation. Our data suggest that favoring the nucleotide binding-competent state will produce a mutator RdRp similar to the H273R mutant. However, a complete disruption of the interactions, especially of residues participating in multivalent interactions, will not represent the best strategy. Several years ago, our laboratory changed Asp-238 and Asn-297 to Ala. Both substitutions perturbed the fidelity of the RdRp but debilitated the corresponding mutant virus to an extent that would preclude any practical use of the mutant viruses (66). Learning how to exploit this occluded-to-competent transition to engineer RdRp derivatives of practical value clearly has merit.
In addition to perturbation of the occluded-to-competent transition, enhanced flexibility of the active site may also contribute to the relaxed specificity observed for H273R RdRp. Because of the overall conservation in shape of an incoming nucleotide of the correct sugar configuration base-paired via Watson-Crick hydrogen bonds to the templating nucleotide, a relatively rigid active site should promote high fidelity. A mispaired base pair would be either too big or too small for a binding pocket evolved to bind to a canonical base pair. The ability to accommodate the noncanonical features of a base pair should promote promiscuity during nucleotide selection. This was demonstrated in our simulations, where active-site residues belonging to motifs F and G as well as residues of the loop of motif B and motif E were more flexible in the H273R RdRp-RNA complex than the WT (Fig. 9). Particularly striking was the conformational flexibility of the nascent base pair-binding pocket (Fig. 10).
This study also highlights the likelihood that the conformational change step observed kinetically, which is written as a single step, actually comprises many steps. The transition from occluded to competent involves the breaking and making of numerous hydrogen bonds and van der Waal interactions (Fig. 14). The simulation provides no evidence that this transition occurs in a concerted fashion (supplemental Movie S1). Without the loss of the Arg-174/Asp-238 interaction, nucleotide cannot bind (supplemental Movie S2). Interactions of the triphosphate with Arg-174 and other residues of motif F likely contribute to the first step in nucleotide binding, which is completely blind to the correctness of the bound nucleotide. What happens last is the closure of motif D to permit Lys-359 to donate a proton to the pyrophosphate leaving group (53). The simulation would suggest that stable interaction of Lys-359 with the pyrophosphate will release its interaction with Asp-233, thus permitting this residue to contribute to formation of the binding site of the divalent cation essential for catalysis (supplemental Movie S3). Binding of this “catalytic” metal ion (also known as metal A) is thought to occur immediately prior to catalysis (54).
We propose that at least two additional structural changes of functional significance exist between the formation of the nucleotide binding-competent state and the closure of motif D. One structural change is reported on by Met-187 in our NMR experiments (Fig. 12). When the correct nucleotide binds to WT RdRp-RNA complex, the Met-187 resonance shifts (Fig. 12A). This perturbation does not occur for the incorrect nucleotide (Fig. 12B, CTP). Only when the Met-187 resonance shifts to the position observed for binding of the correct nucleotide is it possible to observe movement of the Met-354 resonance, which reports on motif D (49). The environment of Met-187 in the H273R RdRp-RNA complex is different than that observed in WT and is no longer responsive to nucleotide binding (Fig. 12, A and B), consistent with the loss of a fidelity checkpoint in the H273R enzyme. Notably, MD simulations showed side chain dihedrals of Met-187 sampling different conformations in the WT RdRp-RNA complex but not in the mutator complex (Fig. 12C), in agreement with the observed Met-187 resonances in the NMR experiment (Fig. 12, A and B).
The Met-187 environment is governed, in part, by the conformation of the motif-B loop (Fig. 12E, see residues 286–291). A comparison of the average structures of the WT and H273R RdRp-RNA complexes obtained from the MD simulations shows that the side chain of Met-286 located in the motif-B loop moves from a site remote from the Met-187 side chain (Fig. 12E, see panel WT-RNA) to a site nearby the Met-187 to form a hydrophobic interaction (Fig. 12E, see panel H273R-RNA; also see supplemental Movie S4). Unfortunately, the Met-286 resonance was not observed in the NMR experiment, potentially because of exchange broadening. The conformational change of the motif-B loop facilitates the interaction of Ser-288 with nucleotide, as observed in the post-incorporation complex structure (Fig. 1C). Stabilizing this binding-competent conformation of Ser-288 in the WT RdRp-RNA complex may require the presence of a correct sugar configuration and possibly also correct base-pairing. In the H273R RdRp-RNA complex, this conformation is partially achieved in the absence of nucleotide, again consistent with this derivative bypassing a fidelity checkpoint.
We are not the first to attribute functional importance to conformational changes of the motif-B loop. Peersen and Sholders (61) observed a variety of conformations of this loop in structures of the free PV RdRp solved using crystals produced at a variety of ionic strengths. The conformations were defined as “in” or “out” based on Cys-290 and “up” or “down” based on Ser-288 (61). The down orientation of Ser-288 is consistent with our binding-competent conformation. PV RdRp derivatives with substitutions in motif B that stabilized the down conformation were more efficient than those that stabilized the up conformation (61). Interestingly, the C290V RdRp was 3-fold more active than WT, and we have shown that this derivative exhibits a mutator phenotype in vitro.7 Another important aspect of the Peersen study is the observation that reduced flexibility of the motif-B loop impedes translocation (61). In the context of our study, we would suggest that translocation resets the motif-B loop for another cycle of faithful nucleotide addition. Whether motif-B dynamics are essential for translocation remains unclear, as an enzyme stabilized in the down conformation might also be expected to exhibit a translocation defect, but this defect was not observed (61). Verdaguer and colleagues (67) have suggested that the motif-B loop might represent a druggable target for the development of allosteric inhibitors of RdRp function.
The second structural change is the reorientation of residues comprising the nascent base pair-binding pocket, as illustrated by the differences in the average structures of the WT and H273R RdRp-RNA complexes derived from MD simulations (Fig. 10A and supplemental Movie S5). As discussed above, flexibility at this site will diminish the fidelity of nucleotide selection. With this change complete for a correct nucleotide, the architecture of the active site should support catalysis: closure of motif D, primer alignment, and binding of metal A (supplemental Movie S3). Structural perturbations caused by the binding or incorporation of an incorrect nucleotide will therefore have an impact on active-site architecture and catalytic efficiency.
Thus far our discussion has emphasized WT and H273R, the low-fidelity enzyme. What about G64S, the high fidelity enzyme? Our studies suggest that the mechanism for the increased fidelity of this enzyme is related to the creation of dysfunctional dynamics, for example converting the positive correlations observed for WT (Fig. 8) into negative correlations (Fig. 11D). Also, simulations of the high fidelity mutant G64S revealed active-site residues, in contrast to the H273R mutator (Fig. 7), to be more rigid compared with WT (Fig. 11C). These dysfunctional dynamics exhibit the greatest adverse effect on an incorrect nucleotide. The concept of creating dysfunctional dynamics to perturb RdRp fidelity may have merit and may be readily extrapolated to other RdRp. Gly-64 resides in a region of the Ramachandran plot that can only be occupied by glycine residues (18, 27). There are only a handful of these residues in PV RdRp and other RdRp. Changing one or more of these glycine residues to alanine or serine may create the same effect as observed for G64S, a stably folded enzyme with dysfunctional dynamics.
We have used MD simulations complemented by kinetics and NMR to study a mutator RdRp to further elaborate our understanding of the physical/structural basis for correct nucleotide selection by this class of polymerases and, perhaps, polymerases in general. Average structures from MD simulations differed from crystal structures, revealing a set of rapidly interconverting interactions among the residues of the active site participating in nucleotide binding, divalent cation binding, and/or catalysis (supplemental Movie S1). The WT RdRp-RNA complex favors a nucleotide binding-occluded state, and that of H273R favors a nucleotide binding-competent state, with the full suite of conformations providing a new perspective on the structural changes that occur during nucleotide selection. We propose that each moiety of the nucleotide (triphosphate, ribose, and base pair) will sequentially disrupt specific interactions and at the same time enable the interrogation of the next element of the nucleotide. For example, the triphosphate interacts with motif F, of which Arg-174 is a part; therefore, its presence will disrupt the nucleotide binding-occluded state and release Asp-238 (supplemental Movie S2). The presence of the ribose 2′-hydroxyl will trap the motif-B loop in the catalytically competent conformation by interacting with Ser-288, Asn-297, and Asp-238 (supplemental Movies S1 and S2). These changes lead to formation of a rigid nascent base pair-binding pocket, which is a checkpoint for the appropriate base pair geometry (supplemental Movie S5). Finally, all is in place for completion of active-site catalysis; in the case of the PV RdRp, this involves making Asp-233, the ligand for the catalytic metal, accessible and positioning Lys-359, the general acid, for protonation of the pyrophosphate leaving group (supplemental Movie S3). Consistent with recent work on DNA polymerases, our data support a model for fidelity of nucleotide selection that is governed by nucleotide-dependent stabilization of the catalytically competent state (56). The application of MD simulations to the other classes of nucleic acid polymerases is an important next step in determining the universality of the concepts presented here.
Supplementary Material
Acknowledgment
We thank the Statistical Consulting Center at The Pennsylvania State University for expert assistance with statistical analysis of data.
This work was supported by National Institutes of Health Grant R01 AI045818 from NIAID (to C. E. C.).

This article contains supplemental Movies S1–S5.
I. M. Moustafa, C. M. Colina, and C. E. Cameron, unpublished data.
J. J. Arnold, D. D. Boehr, and C. E. Cameron, unpublished observations.
- RdRp
- RNA-dependent RNA polymerase
- PV
- poliovirus
- SDKIE
- solvent deuterium kinetic isotope effect
- MD
- molecular dynamics
- r.m.s.d.
- root-mean square deviation
- PCA
- principal component analysis
- DCCM
- dynamic cross-correlation map
- PDB
- Protein Data Bank.
REFERENCES
- 1. Graci J. D., Cameron C. E. (2008) Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 3, 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weeks S. A., Lee C. A., Zhao Y., Smidansky E. D., August A., Arnold J. J., Cameron C. E. (2012) A Polymerase mechanism-based strategy for viral attenuation and vaccine development. J. Biol. Chem. 287, 31618–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smidansky E. D., Arnold J. J., Cameron C. E. (2008) Nucleic acid polymerase fidelity and viral population fitness, in Origin and Evolution of Viruses (Domingo E., Parrish C. R., Holland J. J., eds) pp. 135–160, Academic Press, London [Google Scholar]
- 4. Smith E. C., Denison M. R. (2013) Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 9, e1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korboukh V. K., Lee C. A., Acevedo A., Vignuzzi M., Xiao Y., Arnold J. J., Hemperly S., Graci J. D., August A., Andino R., Cameron C. E. (2014) RNA virus population diversity: an optimum for maximal fitness and virulence. J. Biol. Chem. 289, 29531–29544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnold J. J., Vignuzzi M., Stone J. K., Andino R., Cameron C. E. (2005) Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280, 25706–25716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graci J. D., Cameron C. E. (2006) Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 16, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pfeiffer J. K., Kirkegaard K. (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U.S.A. 100, 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sierra M., Airaksinen A., González-López C., Agudo R., Arias A., Domingo E. (2007) Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 81, 2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vignuzzi M., Stone J. K., Andino R. (2005) Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107, 173–181 [DOI] [PubMed] [Google Scholar]
- 11. Eckerle L. D., Becker M. M., Halpin R. A., Li K., Venter E., Lu X., Scherbakova S., Graham R. L., Baric R. S., Stockwell T. B., Spiro D. J., Denison M. R. (2010) Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gnädig N. F., Beaucourt S., Campagnola G., Bordería A. V., Sanz-Ramos M., Gong P., Blanc H., Peersen O. B., Vignuzzi M. (2012) Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, E2294–E2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham R. L., Becker M. M., Eckerle L. D., Bolles M., Denison M. R., Baric R. S. (2012) A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 18, 1820–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith E. C., Blanc H., Vignuzzi M., Denison M. R. (2013) Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9, e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vignuzzi M., Wendt E., Andino R. (2008) Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 14, 154–161 [DOI] [PubMed] [Google Scholar]
- 16. Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnold J. J., Gohara D. W., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43, 5138–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thompson A. A., Peersen O. B. (2004) Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23, 3462–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong P., Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang X., Welch J. L., Arnold J. J., Boehr D. D. (2010) Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry 49, 9361–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X., Yang X., Lee C. A., Moustafa I. M., Smidansky E. D., Lum D., Arnold J. J., Cameron C. E., Boehr D. D. (2013) Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J. Biol. Chem. 288, 32753–32765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cameron C. E., Moustafa I. M., Arnold J. J. (2009) Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase? Curr. Opin Struct. Biol. 19, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moustafa I. M., Shen H., Morton B., Colina C. M., Cameron C. E. (2011) Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 410, 159–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen H., Moustafa I. M., Cameron C. E., Colina C. M. (2012) Exploring the dynamics of four RNA-dependent RNA polymerases by a coarse-grained model. J. Phys. Chem. B 116, 14515–14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen H., Sun H., Li G. (2012) What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus? PLoS Comput. Biol. 8, e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tzeng S. R., Kalodimos C. G. (2011) Protein dynamics and allostery: an NMR view. Curr. Opin Struct. Biol. 21, 62–67 [DOI] [PubMed] [Google Scholar]
- 27. Marcotte L. L., Wass A. B., Gohara D. W., Pathak H. B., Arnold J. J., Filman D. J., Cameron C. E., Hogle J. M. (2007) Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 81, 3583–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gohara D. W., Ha C. S., Kumar S., Ghosh B., Arnold J. J., Wisniewski T. J., Cameron C. E. (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D. Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Case D. A., Darden T., Cheatham T. E., 3rd, Simmerling C., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., et al. (2012) Amber 12, University of California, San Francisco [Google Scholar]
- 37. Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T. E., 3rd, Laughton C. A., Orozco M. (2007) Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 92, 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Darden T., York D., Pedersen L. (1993) Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 40. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 41. Ryckaert J. P., Ciccotti G., Berendsen H. J. C. (1977) Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 42. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 43. Berendsen H. J. C., Postma J. P., Vangunsteren W. F., Dinola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 44. Roe D. R., Cheatham T. E. (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 [DOI] [PubMed] [Google Scholar]
- 45. Shao J., Tanner S. W., Thompson N., Cheatham T. E. (2007) Clustering Molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3, 2312–2334 [DOI] [PubMed] [Google Scholar]
- 46. Amadei A., Linssen A. B., Berendsen H. J. (1993) Essential dynamics of proteins. Proteins 17, 412–425 [DOI] [PubMed] [Google Scholar]
- 47. García A. E. (1992) Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 68, 2696–2699 [DOI] [PubMed] [Google Scholar]
- 48. Ichiye T., Karplus M. (1991) Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins 11, 205–217 [DOI] [PubMed] [Google Scholar]
- 49. Yang X., Smidansky E. D., Maksimchuk K. R., Lum D., Welch J. L., Arnold J. J., Cameron C. E., Boehr D. D. (2012) Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 51. Wilcoxon F. (1945) Individual comparisons by ranking methods. Biometrics Bull. 1, 80–83 [Google Scholar]
- 52. R Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 53. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bakhtina M., Lee S., Wang Y., Dunlap C., Lamarche B., Tsai M. D. (2005) Use of viscogens, dNTPαS, and rhodium(III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase β. Biochemistry 44, 5177–5187 [DOI] [PubMed] [Google Scholar]
- 55. Johnson K. A. (2010) The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta 1804, 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kellinger M. W., Johnson K. A. (2010) Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 107, 7734–7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smidansky E. D., Arnold J. J., Reynolds S. L., Cameron C. E. (2011) Human mitochondrial RNA polymerase: evaluation of the single-nucleotide addition cycle on synthetic RNA/DNA scaffolds. Biochemistry 50, 5016–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bull J. J., Sanjuán R., Wilke C. O. (2007) Theory of lethal mutagenesis for viruses. J. Virol. 81, 2930–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y., Hong Z., Andino R., Cameron C. E. (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6, 1375–1379 [DOI] [PubMed] [Google Scholar]
- 61. Sholders A. J., Peersen O. B. (2014) Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 426, 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Henzler-Wildman K. A., Thai V., Lei M., Ott M., Wolf-Watz M., Fenn T., Pozharski E., Wilson M. A., Petsko G. A., Karplus M., Hübner C. G., Kern D. (2007) Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 [DOI] [PubMed] [Google Scholar]
- 63. Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. (2007) A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 450, 913–916 [DOI] [PubMed] [Google Scholar]
- 64. Karplus M. (2014) Development of multiscale models for complex chemical systems: from h+h2 to biomolecules (Nobel lecture). Angew. Chem. Int. Ed. Engl. 53, 9992–10005 [DOI] [PubMed] [Google Scholar]
- 65. Arias A., Arnold J. J., Sierra M., Smidansky E. D., Domingo E., Cameron C. E. (2008) Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kineic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 82, 12346–12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gohara D. W., Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43, 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Garriga D., Ferrer-Orta C., Querol-Audí J., Oliva B., Verdaguer N. (2013) Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 425, 2279–2287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






