Background: Beta cell apoptosis is a key factor in diabetes, but the mechanisms are not well understood.
Results: Beta cell miR-200 is induced by thioredoxin-interacting protein (TXNIP) and diabetes, directly targets Zeb1, up-regulates E-cadherin, and promotes apoptosis.
Conclusion: This novel TXNIP/miR-200/Zeb1/E-cadherin signaling pathway links miR-200 to beta cell apoptosis, control of epithelial-mesenchymal transition, and diabetes.
Significance: The results provide new insight into microRNA biology and the regulation of beta cell apoptosis.
Keywords: Apoptosis, Beta Cell (β-Cell), Diabetes, Epithelial-Mesenchymal Transition (EMT), MicroRNA (miRNA), TXNIP
Abstract
Small noncoding microRNAs have emerged as important regulators of cellular processes, but their role in pancreatic beta cells has only started to be elucidated. Loss of pancreatic beta cells is a key factor in the pathogenesis of diabetes, and we have demonstrated that beta cell expression of thioredoxin-interacting protein (TXNIP) is increased in diabetes and causes beta cell apoptosis, whereas TXNIP deficiency is protective against diabetes. Recently, we found that TXNIP also impairs beta cell function by inducing microRNA (miR)-204. Interestingly, using INS-1 beta cells and primary islets, we have now discovered that expression of another microRNA, miR-200, is induced by TXNIP and by diabetes. Furthermore, we found that miR-200 targeted and decreased Zeb1 (zinc finger E-box-binding homeobox 1) and promoted beta cell apoptosis as measured by cleaved caspase-3 levels, Bax/Bcl2 ratio, and TUNEL. In addition, Zeb1 knockdown mimicked the miR-200 effects on beta cell apoptosis, suggesting that Zeb1 plays an important role in mediating miR-200 effects. Moreover, miR-200 increased beta cell expression of the epithelial marker E-cadherin, consistent with inhibition of epithelial-mesenchymal transition, a process thought to be involved in beta cell expansion. Thus, we have identified a novel TXNIP/miR-200/Zeb1/E-cadherin signaling pathway that, for the first time, links miR-200 to beta cell apoptosis and diabetes and also beta cell TXNIP to epithelial-mesenchymal transition. In addition, our results shed new light on the regulation and function of miR-200 in beta cells and show that TXNIP-induced microRNAs control various processes of beta cell biology.
Introduction
Elevated blood glucose and pancreatic beta cell loss are critical features of both type 1 and type 2 diabetes (1, 2). Thioredoxin-interacting protein (TXNIP),2 also known as TBP2 (thioredoxin-binding protein 2) and VDUP1 (vitamin D3-up-regulated protein 1), is a cellular redox regulator and pro-apoptotic factor that we identified as the most up-regulated gene in response to glucose in human pancreatic islets (3, 4). TXNIP is also increased in obese diabetic mice, and this increase is associated with beta cell apoptosis (5). We further determined that TXNIP induces apoptosis through the mitochondrial cell death pathway (6). Moreover, lack of TXNIP protects mice from type 1 as well as type 2 diabetes by preventing beta cell apoptosis and increasing pancreatic beta cell mass (7).
Using microRNA microarray analysis, we recently discovered that TXNIP also regulates the expression of a number of microRNAs (8). MicroRNAs are small noncoding nucleotide sequences that cause down-regulation of their target genes by translational repression and/or degradation of the target mRNAs. Indeed, we have shown that TXNIP increases microRNA (miR)-204, which subsequently regulates insulin transcription and beta cell function (8). Intriguingly, the microRNAs most significantly induced by TXNIP included four of the five members of the miR-200 family, which consists of miR-200a, miR-200b, miR-200c, miR-141, and miR-429. Although the definition is not always applied uniformly, it is generally accepted that a group of microRNAs is categorized as a family based on the high similarity of their precursor sequences, and this clearly applies to the members of the miR-200 family. In addition, the miR-200 family can be broken down into two subgroups based on two distinct genetic loci or based on two unique seed sequences. miR-200a, miR-200b, and miR-429 are clustered together on chromosome 1 in humans, whereas miR-200c and miR-141 are clustered on chromosome 12 (9, 10). When divided into groups based on seed sequence, miR-200b, miR-200c, and miR-429 fall into a group of identical sequence, with miR-141 and miR-200a also sharing an exact seed sequence. However, between these two groups of miR-200 family members, there is only one nucleotide difference in the seed sequence (11).
Many roles for miR-200 family members have been reported (as reviewed in Ref. 12). Prominently among them are the down-regulation of tumor progression and inhibition of epithelial-mesenchymal transition (EMT) (13, 14). The antitumor effects of miR-200 include repression of cancer stem cell self-renewal (15), inhibition of cell division (16), and an increase in apoptosis (17, 18). The process of EMT is also involved in development and progression of cancer (19) and is characterized by epithelial cells losing their cell-cell adherens junctions and becoming migratory mesenchymal cells. The miR-200-induced inhibition of EMT is mediated by direct targeting of the E-cadherin transcriptional repressor Zeb1 (zinc finger E-box-binding homeobox 1), resulting in increased expression of the E-cadherin transmembrane protein, strengthening of the cell-cell adherens junctions, and maintenance of the epithelial phenotype (11). However, although miR-200 has been extensively studied in cancer (20–22), very little is known about this microRNA family in pancreatic beta cells. Also, despite some initial controversy, studies suggest that EMT-like processes are involved in beta cell expansion (23–26). Together, these results raised the possibility that miR-200 may be involved in the control of beta cell mass and survival. The aim of this study was therefore to determine the factors that regulate beta cell miR-200 expression (e.g. TXNIP and diabetes) and to analyze the subsequent effects that miR-200 family members have on beta cell biology.
EXPERIMENTAL PROCEDURES
Tissue Culture
Rat INS-1 beta cells were cultured in RPMI 1640 medium (Invitrogen) with 10% fetal bovine serum, 1% penicillin/streptomycin, 1 mm sodium pyruvate, 2 mm l-glutamine, 10 mm HEPES solution, and 0.05 mm β-mercaptoethanol. Cells were kept at 37 °C in an incubator at 5% CO2. Stably transfected INS-LacZ and INS-hTXNIP (where h is human) cells have been described previously (4) and were cultured using the same medium plus 50 μg/ml Geneticin (Invitrogen). Mouse islets were isolated by collagenase digestion as described previously (5).
Animal Studies
Mouse studies were approved by the University of Alabama at Birmingham Animal Care and Use Committee and complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. C3H/DiSnA control mice and TXNIP-deficient HcB-19 mice harboring a naturally occurring nonsense mutation in txnip have been described previously (7). Obese diabetic C57BL/6lepob/ob (B6-obese) and control lean C57BL/6lep+/+ (B6-control) mice were obtained from The Jackson Laboratory and were analyzed previously for islet TXNIP expression (8). BTBRlep+/+ (BTBR-lean) and severely diabetic BTBRlepob/ob (BTBR-ob) mice, as well as double-mutant congenic BTBRlepob/obtxniphcb/hcb (BTBR-ob/HcB) mice, have been described previously (7).
Transfection of siRNAs, MicroRNA Mimics, and MicroRNA Inhibitors
INS-1 cells were plated in 6-well plates and grown overnight to ∼60% confluence. Human islets (500/tube) were gently dispersed by incubation for 5 min in 200 μl of 0.05% trypsin/EDTA (Invitrogen) at 37 °C, washed, and resuspended in culture medium. Cells were transfected with 25 nm microRNA mimics (Invitrogen), microRNA inhibitors (Thermo Scientific), or 100 nm siRNAs (Thermo Scientific) or with the respective negative controls or scrambled siRNA using DharmaFECT1 transfection reagent (Thermo Scientific). 48 h after transfection, cells were harvested for RNA or protein analysis.
Quantitative Real-time RT-PCR
RNA was isolated using miRNeasy kits (Qiagen) according to the manufacturer's instructions. A first-strand cDNA synthesis kit (Roche Applied Science) was used for reverse transcription. Detection of mRNA was carried out using SYBR Green (Life Technologies), and results were corrected for 18 S. MicroRNA expression was determined using microRNA TaqMan assays (Invitrogen), and results were corrected for U6. For all quantitative RT-PCR (qRT-PCR) experiments, the LightCycler 480 system was used (Roche Applied Science). The following primer sequences were used: rat Zeb1, 5′-CAGGGCGGCCATTCTG (forward) and 5′-TGCCATCCTGATCAACTAAAGGA-3′ (reverse); rat E-cadherin, 5′-GCTGCCACCAGATGACGATAC-3′ (forward) and 5′-TCTCCACCTCCCTCTTCATCA-3′ (reverse); 18 S, 5′-AGTCCTGCCCTTTGTACACA-3′ (forward) and 5′-GATCCGAGGGCCTCACTAAAC-3′ (reverse); rat Bax, 5′-GCCCACCAGCTCTGAGCA-3′ (forward) and 5′-CTCGATCCTGGATGAAACCCT-3′ (reverse); rat Bcl2, 5′-GGGATGCCTTTGTGGAACTATATG-3′ (forward) and 5′-CAGCCAGGAGAAATCAAACAGA-3′ (reverse); human Bax, 5′-CCAAGGTGCCGGAACTGA-3′ (forward) and 5′-CCCGGAGGAAGTCCAATGT-3′ (reverse); and human Bcl2, 5′-CATGTGTGTGGAGAGCGTCAA-3′ (forward) and 5′-TCATCCACAGGGCGATGTT-3′ (reverse).
Immunoblotting
Protein extracts were prepared and analyzed as described previously (5). The following antibodies were used: cleaved caspase-3 (Asp-175, 9661, Cell Signaling), TCF8/ZEB1 (D80D3, 3396, Cell Signaling), and E-cadherin (Ab53033, Abcam). Zeb1 protein was analyzed on an 8% Tris/glycine gel, whereas other proteins were analyzed on 10–20% Tris/glycine gels. After immunoblotting, images were analyzed using ImageQuant Software (GE Healthcare).
TUNEL Assay
The DeadEnd fluorometric TUNEL system (Promega) was used for detection of apoptosis according to the manufacturer's instructions. VECTASHIELD with DAPI mounting solution (Vector Labs, Burlingame, CA) was used for visualization of nuclei.
Immunohistochemistry
E-cadherin staining was performed on pancreatic sections as described previously (7) using anti-E-cadherin antibody (1:150), biotinylated anti-rabbit IgG secondary antibody (1:200), and VECTASTAIN ABC-AP with Vector Blue alkaline phosphatase substrate kit III (Vector Labs).
Statistical Analysis
p values were calculated using Student's t test or by one-way analysis of variance for data sets of more than two groups.
RESULTS
TXNIP Increases Beta Cell Expression of miR-200 Family Members
TXNIP overexpression in INS-1 beta cells induces the expression of a number of microRNAs as demonstrated by our recent microRNA microarray analysis (8). Interestingly, we discovered that of the 11 microRNAs induced by TXNIP with a difference in log median ratio of ≥0.5 (≥1.45-fold), four belong to the five-member miR-200 family of microRNAs (Table 1). Using qRT-PCR, we further confirmed that TXNIP overexpression significantly increased miR-200 in INS-1 beta cells (Fig. 1, A–C).
TABLE 1.
Top TXNIP-induced beta cell microRNAs
MicroRNAs with a difference in log median ratio (dLMR) of ≥0.5 (≥1.45-fold induction) in INS-hTXNIP versus INS-LacZ microRNA microarray are shown. Members of the miR-200 family are shown in boldface.
| MicroRNA | dLMR | -Fold change |
|---|---|---|
| 1. miR-139–5p | 1.76 | 3.38 |
| 2. miR-193 | 0.84 | 1.80 |
| 3. miR-204 | 0.73 | 1.66 |
| 4. miR-200c | 0.73 | 1.66 |
| 5. miR-141 | 0.70 | 1.62 |
| 6. miR-200b | 0.66 | 1.59 |
| 7. miR-194 | 0.66 | 1.58 |
| 8. miR-33 | 0.65 | 1.57 |
| 9. miR-192 | 0.62 | 1.53 |
| 10. miR-32 | 0.54 | 1.46 |
| 11. miR-200a | 0.53 | 1.45 |
FIGURE 1.
Effects of TXNIP on miR-200 levels. A–C, the levels of miR-200 family members were measured by qRT-PCR in TXNIP-overexpressing (INS-hTXNIP) and control (INS-LacZ) beta cells. D, the expression of miR-200 family members was assessed in various tissues of wild-type mice, and the results are shown relative to miR-200a expression in pancreatic islets. E–I, the expression of miR-200 family members was assessed in primary islets of control C3H mice and TXNIP-deficient HcB-19 mice. Error bars represent mean -fold change ± S.E. (n = 3).
Members of the miR-200 family are highly conserved across species, and although our initial findings were in rat INS-1 beta cells, we found that all miR-200 family members exhibited robust expression in primary mouse islets and human islets (Table 2). Moreover, tissue analysis of wild-type C57BL/6 mice revealed that miR-200 family members were highly expressed in pancreatic islets compared with other tissues such as liver, heart, and skeletal muscle (Fig. 1D), suggesting that this microRNA family may play an important role in pancreatic beta cell biology.
TABLE 2.
miR-200 expression in primary human and mouse islets and rat INS-1 beta cells
The expression levels of the different miR-200 family members were assessed by qRT-PCR, and values represent mean cycle threshold ± S.E. from at least three independent experiments. U6 was run as an internal control.
| Human islets | Mouse islets | INS-1 beta cells | |
|---|---|---|---|
| U6 control | 23.27 ± 0.39 | 23.94 ± 0.22 | 23.47 ± 0.11 |
| miR-200a | 24.43 ± 0.54 | 28.10 ± 0.18 | 34.61 ± 0.12 |
| miR-200b | 25.39 ± 0.50 | 28.47 ± 0.16 | 31.18 ± 0.04 |
| miR-200c | 24.07 ± 0.44 | 26.03 ± 0.04 | 25.71 ± 0.09 |
| miR-141 | 24.95 ± 0.50 | 29.73 ± 0.13 | 27.92 ± 0.25 |
| miR-429 | 28.32 ± 0.44 | 30.44 ± 0.19 | 38.96 ± 0.78 |
Expression of miR-200 Family Members Is Decreased in Primary Islets of TXNIP-deficient Mice
Based on the increase in the expression of miR-200 family members in response to TXNIP, we hypothesized that lack of TXNIP would decrease these microRNAs in islets of TXNIP-deficient mice (HcB-19) compared with control mice (C3H). Indeed, expression of each of the five miR-200 family members was decreased in TXNIP-deficient islets (Fig. 1, E–I), further confirming miR-200 regulation by TXNIP in vivo.
Islet Expression of miR-200 Is Elevated in Diabetes
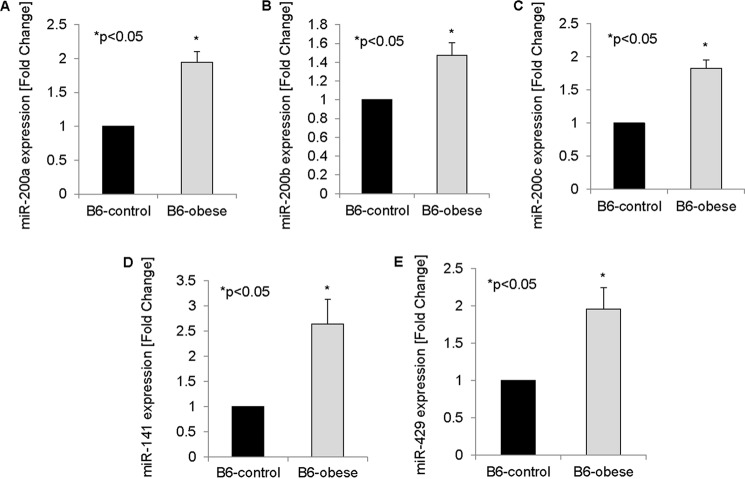
Because TXNIP is up-regulated in diabetic B6-obese mice (5), we further hypothesized that miR-200 family members would also be increased in islets of B6-obese mice. In fact, we found that the expression of all five members of the miR-200 family was elevated in diabetic B6-obese mice relative to B6-control mice as assessed by qRT-PCR (Fig. 2).
FIGURE 2.
miR-200 levels in diabetes. A–E, the expression of miR-200 family members was measured in islets of B6-obese mice by qRT-PCR and compared with islets of B6-control mice. Error bars represent mean -fold change ± S.E. (n = 3).
Of note, the expression of miR-375, the most abundant and best studied microRNA in beta cells (27, 28), did not change in response to TXNIP overexpression in INS-1 beta cells or to TXNIP deficiency in primary HcB-19 islets (Fig. 3, A and B). This provides a strong negative control and underscores the specificity of our findings with regard to the observed regulation of miR-200 expression.
FIGURE 3.

Negative control (miR-375) and effects of TXNIP deletion on diabetes-induced miR-200b. As a negative control, the expression of miR-375 was measured in TXNIP-overexpressing (INS-hTXNIP) and control (INS-LacZ) beta cells (A) and in primary islets of control C3H mice and TXNIP-deficient HcB-19 mice by qRT-PCR (B). C, to assess the effects of TXNIP deletion on diabetes-induced miR-200b, the expression of miR-200b was assessed in islets of control (BTBR-lean), diabetic (BTBR-ob), and double-mutant mice lacking leptin and TXNIP (BTBR-ob/HcB). Error bars represent mean ± S.E. (n = 3). N.S., not significant.
To further test whether TXNIP may impact not only basal miR-200 levels but also diabetes-induced miR-200 expression, we used our double-mutant congenic BTBR-ob/HcB mice (7). On the BTBR background, the ob/ob mutation leads to severe diabetes, and we again saw a significant increase in miR-200b expression in the islets of these mice, consistent with the findings in B6-obese mice. In contrast, lack of TXNIP in the BTBR-ob/HcB mice completely blunted this effect (Fig. 3C), demonstrating that TXNIP deficiency can compensate for diabetes-induced miR-200 expression.
Together, these in vivo results strongly supported the findings in INS-1 beta cells and raised the possibility that the observed increase in miR-200 expression may play an important pathophysiological role in beta cell biology and diabetes. To address this possibility further, we next aimed to determine what role TXNIP-induced miR-200 expression plays in beta cell biology.
miR-200b Induces Beta Cell Apoptosis
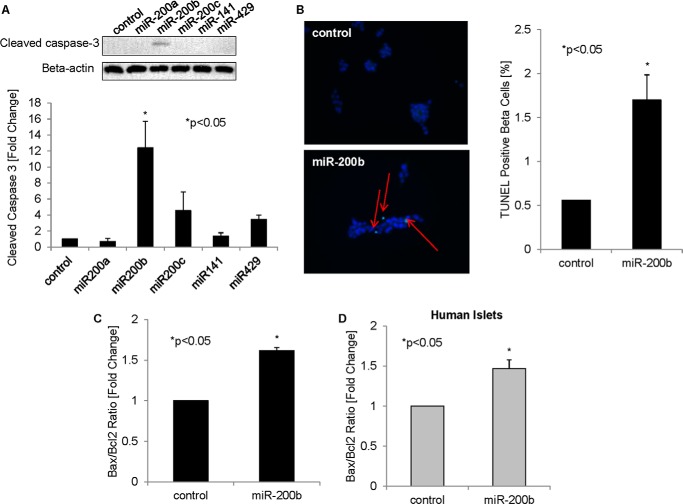
We have previously shown that TXNIP induces beta cell apoptosis (4–6). We therefore hypothesized that TXNIP-induced miR-200 may contribute to these pro-apoptotic effects. Overexpression of the five individual miR-200 family members in INS-1 beta cells and analysis of cleaved caspase-3 indeed revealed that miR-200b (in particular) had a strong effect and induced apoptosis significantly (Fig. 4A). Moreover, miR-200b also led to a significant increase in the percentage of TUNEL-positive beta cells (Fig. 4B). This pro-apoptotic effect of miR-200b was further supported by an increase in the ratio of pro-apoptotic Bax to anti-apoptotic Bcl2 (Fig. 4C). Of note, the Bax/Bcl2 ratio was also significantly increased in human islets in response to miR-200b overexpression (Fig. 4D), suggesting that this pro-apoptotic miR-200b effect also occurs in primary islets. These results are consistent with previous reports of miR-200b causing apoptosis in extrapancreatic tissues (29), but represent the first demonstration of miR-200b promoting beta cell death. Together with the increased miR-200 expression in diabetic islets, they further link the miR-200 family to diabetes and the associated beta cell loss.
FIGURE 4.
Role of miR-200b in beta cell apoptosis. A, members of the miR-200 family were overexpressed in INS-1 beta cells by transfection. 48 h after transfection, cells were harvested, and cleaved caspase-3 was measured by immunoblotting and corrected for β-actin. The results of three independent experiments were used for the quantification shown. B, INS-1 beta cells transfected with negative control or pre-miR-200b for 48 h were analyzed by TUNEL staining. Representative pictures are shown; the red arrows point to TUNEL-positive apoptotic nuclei. For quantification of the percent TUNEL-positive beta cells, >500 nuclei were analyzed per group. C, INS-1 beta cells transfected with miR-200b were analyzed for the Bax/Bcl2 ratio by qRT-PCR. D, human islets were transfected with miR-200b or negative control, and after 72 h, the Bax/Bcl2 ratio was assessed as a marker of apoptosis. Error bars represent mean ± S.E. (n = 3).
miR-200b Targets and Decreases Beta Cell Zeb1
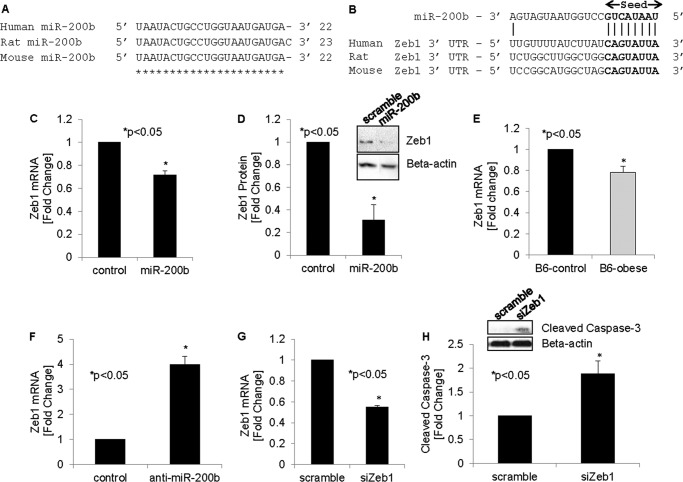
We next wanted to determine the downstream targets of miR-200b and whether they contributed to the apoptosis seen in response to increased miR-200b. Interestingly, the microRNA target prediction software (TargetScan) identified Zeb1 as a top target of miR-200. Indeed, the Zeb1 3′-UTR contains highly conserved CAGUAUUA sequences, providing a perfect match for the miR-200b seed sequence (30). Of note, the sequence of miR-200b is highly conserved across species, as shown by an alignment of human, rat, and mouse miR-200b (Fig. 5A), as is the binding site for the miR-200b seed sequence in the 3′-UTR of human, rat, and mouse Zeb1 (Fig. 5B). Furthermore, miR-200 family members have also been shown experimentally to target Zeb1, as confirmed by Zeb1 3′-UTR luciferase assays (31, 32). Intriguingly, compared with other miR-200 family members, miR-200b showed the greatest effect in these Zeb1 3′-UTR luciferase studies (31), which is consistent with the more pronounced effects we observed with miR-200b in terms of apoptosis.
FIGURE 5.
Effects of miR-200b on Zeb1 in beta cells. A, sequence alignment of miR-200b in human, rat, and mouse. B, alignment of miR-200b with the 3′-UTR of Zeb1 in human, rat, and mouse. The miR-200b seed sequence and its binding site are shown in boldface. Zeb1 mRNA (C) and protein (D) levels were measured in INS-1 cells 48 h after transfection with miR-200b. E, Zeb1 expression was also assessed in primary islets of obese diabetic and control mice. F, miR-200b was knocked down by transfecting INS-1 beta cells with anti-miR-200b, and the effects on Zeb1 expression were assessed by qRT-PCR. INS-1 cells transfected with Zeb1 siRNA (siZeb1) for 48 h were analyzed for Zeb1 knockdown efficiency (G) and cleaved caspase-3 by immunoblotting (H). Error bars represent mean -fold change ± S.E. (n = 3–4).
To specifically determine whether Zeb1 is targeted by miR-200b in beta cells, we overexpressed miR-200b in INS-1 beta cells and measured Zeb1 expression. We discovered that miR-200b significantly decreased Zeb1 mRNA and protein levels (Fig. 5, C–D), indicating that miR-200b indeed targets and inhibits Zeb1 expression in beta cells. Of note, we also observed a significant decrease in Zeb1 expression in primary islets of obese diabetic mice (Fig. 5E). This result is consistent with the demonstrated increase in miR-200b in the same samples (Fig. 2B) and underscores the pathophysiological relevance of this observation. We next performed the inverse experiment, knocking down miR-200b in INS-1 beta cells using anti-miR oligonucleotides. In beta cells, there was marked expression of miR-200b even at base line (Fig. 1D and Table 2), and knockdown with anti-miR-200b resulted in a significant 4.7 ± 0.7-fold decrease in miR-200b expression (p < 0.005). We therefore hypothesized that this would lead to a corresponding increase in Zeb1. Indeed, miR-200b knockdown significantly increased Zeb1 expression (Fig. 5F), further confirming the specificity of the miR-200b effects.
Inhibition of Zeb1 Promotes Beta Cell Apoptosis
Interestingly, in extrapancreatic tissues, Zeb1 has been implicated in the inhibition of apoptosis (18). We therefore hypothesized that a decrease in Zeb1 may also promote apoptosis in beta cells and thereby contribute to the observed induction of apoptosis in response to miR-200b. In fact, knockdown of Zeb1 (Fig. 5G) revealed a significant increase in beta cell apoptosis as assessed by cleaved caspase-3 (Fig. 5H), demonstrating that inhibition of Zeb1 can mimic the pro-apoptotic effects of miR-200b on beta cells.
miR-200b, Zeb1 siRNA, and Diabetes Induce Beta Cell E-cadherin
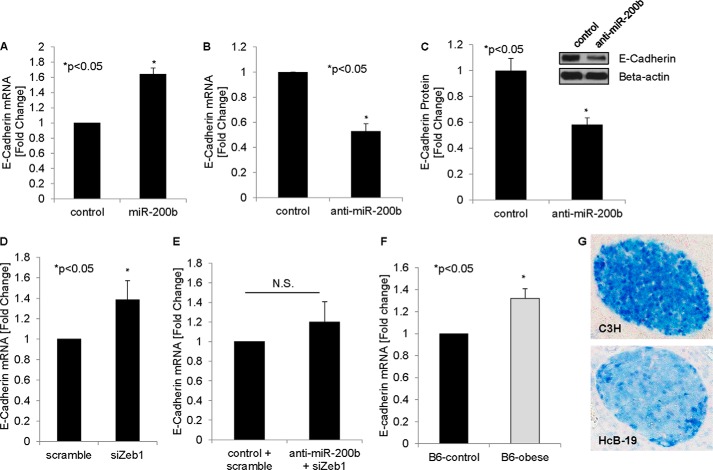
Members of the miR-200 family have also been implicated in EMT (31, 33, 34), and increased Zeb1 expression (as observed in response to miR-200b knockdown (Fig. 5F)) and decreased E-cadherin expression are strong markers for EMT (10, 35, 36).
We therefore next investigated whether miR-200b can also regulate beta cell E-cadherin expression. Indeed, E-cadherin expression was significantly increased in response to miR-200b overexpression (Fig. 6A). Conversely, knockdown of miR-200b resulted in a decrease in E-cadherin mRNA (Fig. 6B) and protein (Fig. 6C) levels, further confirming the regulation of beta cell E-cadherin by miR-200b and suggesting that miR-200b may inhibit EMT in beta cells. Because we demonstrated that miR-200b targets beta cell Zeb1, we hypothesized that Zeb1 knockdown should also induce E-cadherin expression, and this is indeed what we found (Fig. 6D). Moreover, using co-transfection experiments, we were able to demonstrate that Zeb1 siRNA completely blunted the decrease in E-cadherin in response to anti-miR-200b (Fig. 6E), suggesting that the observed miR-200b effects are mediated by Zeb1. Importantly, we also found that primary islets of obese diabetic mice demonstrated an increase in E-cadherin expression (Fig. 6F), consistent with the observed decrease in Zeb1 (Fig. 5E) and increase in miR-200 (Fig. 2B) in the same samples, whereas TXNIP-deficient HcB-19 islets showed a reduction in E-cadherin (Fig. 6G). These findings not only confirm the different steps of this beta cell pathway in primary islets, but also establish a novel link between beta cell E-cadherin expression, diabetes, TXNIP, and miR-200b signaling.
FIGURE 6.
miR-200b, Zeb1 siRNA, and diabetes effects on beta cell E-cadherin. A, INS-1 cells were transfected with miR-200b or negative control. After 48 h, cells were harvested, and the effects on E-cadherin expression were measured by qRT-PCR. INS-1 cells were also transfected with anti-miR-200b, and 48 h after transfection, the effects of miR-200b knockdown on E-cadherin mRNA (B) and protein (C) were determined. E-cadherin expression in INS-1 beta cells was also determined in response to Zeb1 siRNA (siZeb1) (D) and in response to co-transfection with anti-miR-200b and Zeb1 siRNA (E). F, primary islets of obese diabetic mice and lean controls were assessed for E-cadherin expression by qRT-PCR. G, E-cadherin was visualized by immunohistochemistry in islets of control C3H and TXNIP-deficient HcB-19 mice; representative cross-sections are shown. Error bars represent mean -fold change ± S.E. (n = 3). N.S., not significant.
Taken together, the results of our studies reveal that increased beta cell TXNIP expression (as found in diabetes) leads not only to beta cell apoptosis through the mitochondrial death pathway and phosphorylation and activation of ASK1 (apoptosis signal-regulating kinase 1), but also to increased expression of pro-apoptotic miR-200. In turn, miR-200b targets Zeb1, and the resulting decrease in Zeb1 expression promotes beta cell apoptosis and enhances E-cadherin expression, which is known to inhibit EMT. Although the role of EMT in beta cell biology is still not fully resolved, loss of E-cadherin seems to be required for compensatory beta cell expansion (23). Together, these miR-200b induced processes may therefore contribute to progression of beta cell loss and worsening of diabetes (Fig. 7).
FIGURE 7.
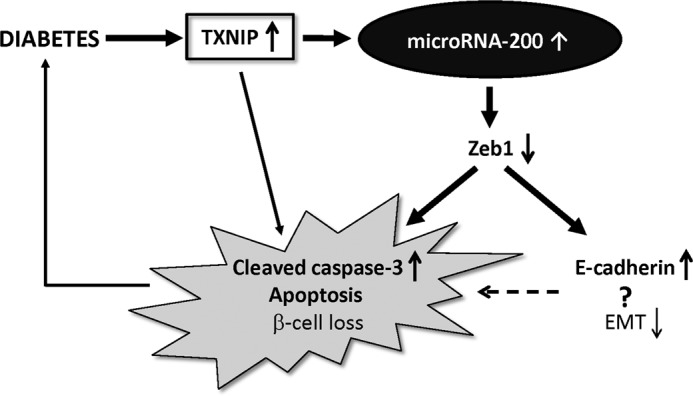
Schematic of proposed TXNIP/miR-200/Zeb1/E-cadherin signaling pathway. Diabetes increases beta cell TXNIP expression, which leads to activation of the mitochondrial death pathway and promotes cleaved caspase-3, apoptosis, and beta cell loss. Increased beta cell TXNIP also induces expression of microRNA-200, which, by targeting and down-regulating anti-apoptotic Zeb1, further contributes to the increase in cleaved caspase-3 and beta cell apoptosis. TXNIP-induced microRNA-200 and Zeb1 reduction also lead to an increase in E-cadherin, a marker of decreased EMT. This inhibition of EMT may further exacerbate beta cell loss by inhibiting beta cell expansion and promoting apoptosis.
DISCUSSION
Our combined in vitro and in vivo data reveal for the first time that TXNIP and diabetes up-regulate miR-200 expression in INS-1 beta cells and primary islets and that miR-200 in turn directly targets Zeb1 and promotes beta cell apoptosis. We discovered that TXNIP increases beta cell expression of all five miR-200 family members, miR-200a, miR-200b, miR-200c, miR-141, and miR-429 (but not miR-375, run as a negative control), and found that miR-200b in particular is highly efficient in inducing beta cell apoptosis. This finding is consistent with the pro-apoptotic effects attributed previously to miR-200 family members in cancer (17, 18), as well as the pro-apoptotic role we have found TXNIP to play in beta cells (4, 5). In this regard, we previously found that TXNIP increases beta cell apoptosis via the mitochondrial death pathway (37) and by binding to mitochondrial Trx2 (thioredoxin-2) and consequently preventing the interaction between Trx2 and ASK1, resulting in ASK1 phosphorylation and activation and the subsequent downstream increase in apoptosis. However, we have now found that miR-200b directly targets Zeb1 and that this inhibition of Zeb1 is sufficient to induce beta cell apoptosis. Moreover, compared with other miR-200 family members and consistent with our data, miR-200b has previously been shown to have the strongest effect on Zeb1 (31), further underscoring the unique role that miR-200b and Zeb1 seem to play and supporting our apoptosis findings. However, considering the multitude of targets and wide-ranging effects of microRNAs, it is obviously conceivable that additional pathways might be involved in the pro-apoptotic effects of miR-200. In any case, our results suggest that TXNIP-induced miR-200b expression represents yet another pathway by which TXNIP promotes beta cell apoptosis. Together with the observed increase in islet miR-200 expression in two models of diabetes and the rescue of this effect by TXNIP deletion, they also, for the first time, link miR-200b to beta cell death and diabetes.
In agreement with the observed increase in miR-200 expression in response to TXNIP observed in this study, we recently also found that TXNIP induces beta cell expression of another microRNA, miR-204 (8). However, miR-204 does not induce beta cell apoptosis and seems to primarily control insulin production and thereby regulate beta cell function. On the other hand, the results of this study suggest that TXNIP-induced miR-200 controls beta cell death and survival and demonstrate that TXNIP can modulate different critical aspects of beta cell biology by regulating distinct microRNAs.
It is interesting to note that miR-200b signaling was also able to affect key EMT-related factors such as Zeb1 and E-cadherin in beta cells. Although the concept of EMT in beta cells has generated some controversy in the past, several lines of evidence now suggest that, under certain circumstances, the process may play an important role in beta cell biology. Human islets were originally reported to give rise to proliferating islet precursor cells via EMT (38), but this work was rebutted with evidence that mouse islets in vitro could not do the same (39). However, more recently, human beta cells were shown to undergo EMT in vitro (24). Moreover, it has been reported that beta cell expansion in response to insulin resistance is associated with an EMT-like process and loss of E-cadherin expression (23). EMT may also play a role in attempts to expand beta cell populations ex vivo for transplantation (24, 25). Thus, although the role of EMT in the beta cell still remains to be fully elucidated, miR-200 family members seem to play a part in its regulation and may therefore provide a potential target for manipulating this process in future beta cell population expansion attempts.
In summary, this study sheds new light on the regulation and function of miR-200b in beta cells and, for the first time, links this microRNA to beta cell apoptosis and diabetes. In addition, the identification of the beta cell miR-200/E-cadherin signaling pathway provides a novel link to EMT in beta cells and demonstrates that TXNIP-induced microRNAs control various processes of beta cell biology.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK-078752. This work was also supported by American Diabetes Association Grant 7-12-BS-167 and JDRF/JNJSI Grant 40-2011-1.
- TXNIP
- thioredoxin-interacting protein
- miR
- microRNA
- EMT
- epithelial-mesenchymal transition
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Poitout V., Robertson R. P. (2002) Minireview: secondary beta cell failure in type 2 diabetes–a convergence of glucotoxicity and lipotoxicity. Endocrinology 143, 339–342 [DOI] [PubMed] [Google Scholar]
- 2. Mathis D., Vence L., Benoist C. (2001) Beta cell death during progression to diabetes. Nature 414, 792–798 [DOI] [PubMed] [Google Scholar]
- 3. Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. (2002) Oligonucleotide microarray analysis of intact human pancreatic islets: identification of glucose-responsive genes and a highly regulated TGFβ signaling pathway. Endocrinology 143, 3695–3698 [DOI] [PubMed] [Google Scholar]
- 4. Minn A. H., Hafele C., Shalev A. (2005) Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta cell apoptosis. Endocrinology 146, 2397–2405 [DOI] [PubMed] [Google Scholar]
- 5. Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. (2008) Thioredoxin-interacting protein: a critical link between glucose toxicity and beta cell apoptosis. Diabetes 57, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J., Fontes G., Saxena G., Poitout V., Shalev A. (2010) Lack of TXNIP protects against mitochondria-mediated apoptosis but not against fatty acid-induced ER stress-mediated beta cell death. Diabetes 59, 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J., Hui S. T., Couto F. M., Mungrue I. N., Davis D. B., Attie A. D., Lusis A. J., Davis R. A., Shalev A. (2008) Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta cell mass and protects against diabetes. FASEB J. 22, 3581–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu G., Chen J., Jing G., Shalev A. (2013) Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 19, 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M. J., Tuschl T., Margalit H. (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 33, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 11. Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Day E., Lal A. (2010) MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 12, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim Y. Y., Wright J. A., Attema J. L., Gregory P. A., Bert A. G., Smith E., Thomas D., Lopez A. F., Drew P. A., Khew-Goodall Y., Goodall G. J. (2013) Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J. Cell Sci. 126, 2256–2266 [DOI] [PubMed] [Google Scholar]
- 14. Schliekelman M. J., Gibbons D. L., Faca V. M., Creighton C. J., Rizvi Z. H., Zhang Q., Wong C. H., Wang H., Ungewiss C., Ahn Y. H., Shin D. H., Kurie J. M., Hanash S. M. (2011) Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 71, 7670–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iliopoulos D., Lindahl-Allen M., Polytarchou C., Hirsch H. A., Tsichlis P. N., Struhl K. (2010) Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uhlmann S., Zhang J. D., Schwäger A., Mannsperger H., Riazalhosseini Y., Burmester S., Ward A., Korf U., Wiemann S., Sahin O. (2010) miR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29, 4297–4306 [DOI] [PubMed] [Google Scholar]
- 17. Schickel R., Park S. M., Murmann A. E., Peter M. E. (2010) miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol. Cell 38, 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magenta A., Cencioni C., Fasanaro P., Zaccagnini G., Greco S., Sarra-Ferraris G., Antonini A., Martelli F., Capogrossi M. C. (2011) miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 18, 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J., Weinberg R. A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 20. Wiklund E. D., Bramsen J. B., Hulf T., Dyrskjøt L., Ramanathan R., Hansen T. B., Villadsen S. B., Gao S., Ostenfeld M. S., Borre M., Peter M. E., Ørntoft T. F., Kjems J., Clark S. J. (2011) Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 128, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 21. Wiklund E. D., Gao S., Hulf T., Sibbritt T., Nair S., Costea D. E., Villadsen S. B., Bakholdt V., Bramsen J. B., Sørensen J. A., Krogdahl A., Clark S. J., Kjems J. (2011) MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE 6, e27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun J., Hoang-Vu C., Dralle H., Hüttelmaier S. (2010) Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 29, 4237–4244 [DOI] [PubMed] [Google Scholar]
- 23. Kulkarni R. N., Jhala U. S., Winnay J. N., Krajewski S., Montminy M., Kahn C. R. (2004) PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J. Clin. Invest. 114, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Russ H. A., Ravassard P., Kerr-Conte J., Pattou F., Efrat S. (2009) Epithelial-mesenchymal transition in cells expanded in vitro from lineage-traced adult human pancreatic beta cells. PLoS ONE 4, e6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Russ H. A., Sintov E., Anker-Kitai L., Friedman O., Lenz A., Toren G., Farhy C., Pasmanik-Chor M., Oron-Karni V., Ravassard P., Efrat S. (2011) Insulin-producing cells generated from dedifferentiated human pancreatic beta cells expanded in vitro. PLoS ONE 6, e25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joglekar M. V., Hardikar A. A. (2010) Epithelial-to-mesenchymal transition in pancreatic islet beta cells. Cell Cycle 9, 4077–4079 [DOI] [PubMed] [Google Scholar]
- 27. Poy M. N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. (2009) miR-375 maintains normal pancreatic alpha and beta cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230 [DOI] [PubMed] [Google Scholar]
- 29. Tang H., Kong Y., Guo J., Tang Y., Xie X., Yang L., Su Q., Xie X. (2013) Diallyl disulfide suppresses proliferation and induces apoptosis in human gastric cancer through Wnt-1 signaling pathway by up-regulation of miR-200b and miR-22. Cancer Lett. 340, 72–81 [DOI] [PubMed] [Google Scholar]
- 30. Adam L., Zhong M., Choi W., Qi W., Nicoloso M., Arora A., Calin G., Wang H., Siefker-Radtke A., McConkey D., Bar-Eli M., Dinney C. (2009) miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 15, 5060–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korpal M., Lee E. S., Hu G., Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Williams L. V., Veliceasa D., Vinokour E., Volpert O. V. (2013) miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS ONE 8, e83991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 35. Brabletz S., Brabletz T. (2010) The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 11, 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hay E. D. (1995) An overview of epithelio-mesenchymal transformation. Acta Anat. 154, 8–20 [DOI] [PubMed] [Google Scholar]
- 37. Saxena G., Chen J., Shalev A. (2010) Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. J. Biol. Chem. 285, 3997–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gershengorn M. C., Hardikar A. A., Wei C., Geras-Raaka E., Marcus-Samuels B., Raaka B. M. (2004) Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science 306, 2261–2264 [DOI] [PubMed] [Google Scholar]
- 39. Atouf F., Park C. H., Pechhold K., Ta M., Choi Y., Lumelsky N. L. (2007) No evidence for mouse pancreatic beta cell epithelial-mesenchymal transition in vitro. Diabetes 56, 699–702 [DOI] [PubMed] [Google Scholar]