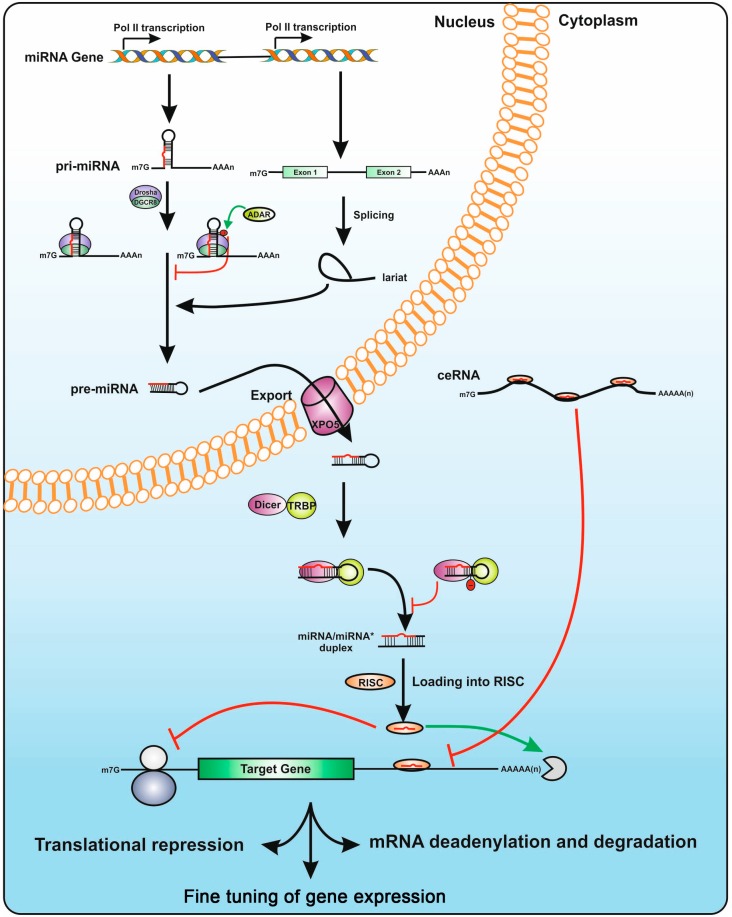
Figure 2.
Overview of the miRNA biogenesis pathway. miRNA genes are transcribed in the nucleus by RNA Pol II as long pri-miRNA transcripts that are 5′ capped and 3′ polyadenylated. The pri-miRNA sequence folds into a hairpin loops structure that is recognized and processed by the Microprocessor complex Drosha-DGCR8, generating a pre-microRNA. Mirtrons, a class of unconventional miRNAs, are encoded in small introns and do not require Drosha processing. The intron lariat excised by the spliceosome is refolded into a pre-miRNA hairpin loop. The pre-miRNA is exported from the nucleus to the cytoplasm by exportin 5 (XPO5), where is further cropped by Dicer in complex with TRBP, yielding a ~22 nt double-stranded RNA called miRNA/miRNA* duplex. The functional mature miRNA is loaded together with Argonaute proteins into the RISC complex, guiding RISC to silence a target mRNA through translational repression or deadenylation. The biogenesis pathway of miRNAs are post-transcriptionally controlled by RNA editing. A-to-I editing of the miRNA’s precursors, a reaction catalyzed by ADAR enzymes, can block Drosha and Dicer processing, and thereby regulates the availability of mature miRNA in the cell. Competing endogenous RNAs (ceRNAs) can regulate mRNA expression levels by competing for shared miRNA binding sites.