Abstract
Nonribosomal peptide synthetases represent the enzymatic assembly lines for the biosynthesis of pharmacologically relevant natural peptides, e.g., cyclosporine, vancomycin, and penicillin. Due to their modular organization, in which every module accounts for the incorporation of a single amino acid, artificial assembly lines for the production of novel peptides can be constructed by biocombinatorial approaches. Once transferred into an appropriate host, these hybrid synthetases could facilitate the bioproduction of basically any peptide-based molecule. In the present study, we describe the fermentative production of the cyclic dipeptide d-Phe-Pro-diketopiperazine, as a prototype for the exploitation of the heterologous host Escherichia coli, and the use of artificial nonribosomal peptide synthetases. E. coli provides a tremendous potential for genetic engineering and was manipulated in our study by stable chromosomal integration of the 4′-phosphopantetheine transferase gene sfp to ensure heterologous production of fully active holoenzmyes. d-Phe-Pro-diketopiperazine is formed by the TycA/TycB1 system, whose components represent the first two modules for tyrocidine biosynthesis in Bacillus brevis. Coexpression of the corresponding genes in E. coli gave rise to the production of the expected diketopiperazine product, demonstrating the functional interaction of both modules in the heterologous environment. Furthermore, the cyclic dipeptide is stable and not toxic to E. coli and is secreted into the culture medium without the need for any additional factors. Parameters affecting the productivity were comprehensively investigated, including various genetic setups, as well as variation of medium composition and temperature. By these means, the overall productivity of the artificial system could be enhanced by over 400% to yield about 9 mg of d-Phe-Pro-diketopiperazine/liter. As a general tool, this approach could allow the sustainable bioproduction of peptides, e.g., those used as pharmaceuticals or fine chemicals.
Nonribosomal peptide (NRP) biosynthesis is a powerful tool of nature for producing a wide variety of pharmacologically relevant peptide products. These include antibiotics, e.g., tyrocidine (30), vancomycin (47), and the penicillin precursor ACV (6); immunosuppressive agents, e.g., cyclosporine (49); cytostatics, e.g., epothilone (27, 46); and iron ion-chelating siderophores, e.g., vibriobactin (20). In all these cases, product formation takes place under the catalytic control of a large multimodular enzyme complex, termed NRP synthetase (NRPS), which serves as a template for the peptide product to be synthesized (32, 39). Each module within this biosynthetic assembly line is responsible for the recognition, activation, and incorporation of a certain amino acid into the nascent peptide product. Specialized catalytic domains accomplish these different tasks and ensure efficient and directed synthesis. The incorporation of unusual amino acids and the often extensive modification of side chains and the peptide backbone result in a structural diversity of the peptide products that is usually incompatible with the large-scale chemical synthesis required for the production of pharmaceuticals or fine chemicals.
Consequently, much of the attention in the field of NRPSs is attracted by the potential to obtain new peptide products with novel biological activities by directed engineering of the biosynthetic genes (32, 48). The modular logic of NRPSs lends itself to biocombinatorial approaches, such as domain and module swapping, as well as fusion and truncation of existing catalytic units (9, 10, 31). However, little is reported on the fermentative production of engineered NRP products (12, 38, 44, 51), which must be the ultimate goal for their affordable synthesis. The natural producer strains are often difficult or impossible to cultivate under laboratory conditions, and many are not amenable to genetic manipulation (5).
We therefore set out to establish Escherichia coli as a general host for the overproduction of engineered NRPSs. The choice of E. coli is obvious, given its favorable genetic accessibility and established culture conditions. E. coli has been used for many years to express heterologous NRPS genes for subsequent protein purification and biochemical characterization (2, 3, 9, 10, 23, 24, 31, 43). Less is known, however, about the in vivo reconstitution of NRPS complexes from several subunits (34, 35) and the cellular stability of the new products, as well as the potential requirement of specific transporters and resistance genes.
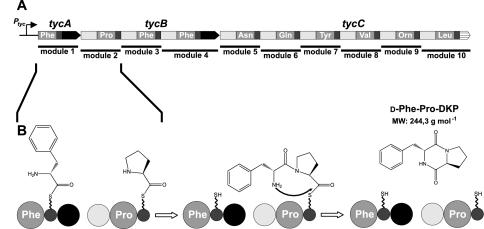
As a model system, we chose the bimodular NRPS complex TycA/TycB1 from Bacillus brevis, which corresponds to the first two modules of the tyrocidine biosynthetic system (see Fig. 1) (30) and which is biochemically well characterized from purified, recombinant proteins (2, 3, 24, 31, 43, 45). The modules involved activate and bind the amino acids l-phenylalanine and l-proline, epimerize l-phenylalanine into the d stereomer, and condense the two amino acids to the enzyme-bound dipeptide d-Phe-l-Pro-4′-phosphopantetheine (Ppant) TycB1. This dipeptide is autocatalytically released to give the cyclic d-Phe-Pro-diketopiperazine (d-Phe-Pro-DKP) (42). DKPs constitute a class of natural products with interesting biological properties (14, 16, 22).
FIG. 1.
Tyrocidine biosynthetic system. The enzymatic assembly line of the cyclic decapeptide antibiotic tyrocidine consists of three peptide synthetases, TycABC, which are encoded by the polycistronic genes tycABC. The proteins are composed of one, three, and six modules, respectively, each one responsible for the integration of one amino acid into the nascent peptide chain (A). In this study, the first two modules (TycA and TycB1) were used as an artificial bimodular NRPS system for the production of d-Phe-Pro-DKP (B). Different catalytic domains are highlighted: condensation domain (light gray), adenylation domains (medium gray), peptidyl carrier protein domain (dark gray; wavy line, cofactor Ppant), epimerization domain (black), and thioesterase domain (striped).
In this work, the genes encoding TycA and TycB1 were coexpressed in an E. coli host carrying the Ppant transferase (PPTase) gene sfp from Bacillus subtilis in the chromosome, whose gene product is required for modification of NRPSs into the active holoform (21). d-Phe-Pro-DKP was produced and secreted into the medium at reasonable levels, demonstrating functional coexpression and interaction of the two modules, as well as product release and secretion without the need for any additional factors, i.e., transporters. The product was stable and not toxic to the E. coli cells. Product yields were compared under different conditions, including various genetic setups, as well as variation of medium composition and temperature.
MATERIALS AND METHODS
Strains, culture media, and general methods.
E. coli strains used in this study are listed in Table 1. Cells were grown in Luria broth (LB) rich medium or M9 minimal medium (26) supplemented, if applicable, with 50 μg of ampicillin, 40 μg of spectinomycin, 15 μg of tetracycline, and/or 20 μg of chloramphenicol/ml (final concentrations). Growth conditions and additional supplements varied, and exact fermentation parameters are indicated for the individual experiments conducted. Batch fermentation was carried out in 1-liter Erlenmeyer flasks containing 200 ml of medium in an incubator-shaker (New Brunswick, Nuertingen, Germany) with orbital motion at 250 rpm. Tryptone and other ingredients of the media were purchased from Becton Dickinson (Heidelberg, Germany).
TABLE 1.
E. coli strains used in this study
| Strain name | Relevant genotype or property | Origin or reference |
|---|---|---|
| XL1-Blue | Standard cloning strain | Stratagene |
| HSK42 | MC4100, polA | 7 |
| GT869 | Standard cloning strain, K-12 derivative | Laboratory collection |
| HM0079 | GT869 nrdD::sfp Spcr | This study |
| NK5525 | proA81::Tn10 | E. coli Genetic Stock Center |
| MG1655 | Wild-type E. coli K-12 | 4 |
| MG1655 (pro-81::Tn10) | MG1655 pro-81::Tn10 | This study |
Standard procedures were applied for all DNA manipulations (37), P1 phage transduction (26), and plasmid preparation from E. coli XL1-Blue (Stratagene, Amsterdam, The Netherlands) (37). Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany). Partial DNA sequencing on an ABI genetic analyzer confirmed the integrity of all constructs.
PCR amplification and cloning of NRPS genes.
PCR amplification was carried out with the Pfu Turbo polymerase in accordance to the manufacturer's protocol (Stratagene). NRPS genes were amplified by PCR from chromosomal DNA of B. brevis ATCC 8185 with the following oligonucleotides (restriction sites are underlined): HM98-12, 5′-ATA CCA TGG TAG CAA ATC AGG CCA ATC-3′; HM98-1, 5′-ATA CCA TGG GTG TAT TTA GCA AAG AAC AAG TTC-3′; HM98-2, 5′-TAT GGA TCC TTC CAC ATA CGC TGC CAG-3′.
Construction of ptycA/tycB1 (one-plasmid system).
tycA and the coding region of the first module of tycB were collectively amplified from the polycistronic tyrocidine biosynthetic gene cluster with primers HM98-12 and HM98-2. The resulting 6.5-kb fragment was terminally modified with the restriction endonucleases NcoI and BamHI and ligated into pQE61 (24), previously cut in the same manner to give ptycA/tycB1(one-plasmid system).
Construction of pSU18-tycA and pTrc99a-tycB1 (two-plasmid system).
Stable maintenance of two plasmids in the same host requires compatible origins of replications and a complementary set of selection markers. Consequently, both genes, tycA and tycB1, had to be recloned. To this end, pTycA (31) was digested with EcoRI and NheI to yield an approximately 3.4-kb fragment containing the tycA gene along with the ribosomal binding site (RBS) and the hexahistidine (His6) tag coding sequence of the pQE60 vector. The fragment was ligated into the chloramphenicol resistance-conferring, medium-copy-number vector pSU18 (1), which was cut beforehand with the restriction enzymes EcoRI and XbaI, to yield pSU18-tycA-His-tag. Subsequently, the coding sequence of the Strep tag (primary sequence: WSHPQFEK) was introduced into the His6 tag expression plasmid. For this purpose, the following oligonucleotides were annealed, phosphorylated with T4 polynucleotide kinase (New England Biolabs, Frankfurt, Germany), and cloned into BamHI-digested plasmid pSU18-tycA-His-tag to give pSU18-tycA (modified bases underlined): SG31, 5′-GA TCT TGG AGC CAC CCG CAG TTC GAA AAA TAA A-3′, and SG32, 5′-GA TCT TTA TTT TTC GAA CTG CGG GTG GCT CCA A-3′. Likewise, pTycB1 (31) was digested with NcoI and XbaI to yield an approximately 4.1-kb fragment containing tycB1 along with the RBS and the His6 tag coding sequence of pQE60. The fragment was ligated into the ampicillin resistance-conferring, high-copy-number vector pTrc99a, which was cut in the same manner to give pTrc99a-tycB1.
Construction of the fusion system ptycA-tycB1(fusion).
Construction of the TycA-TycB1 fusion construct ptycA-tycB1(fusion) was described elsewhere (24). The encoded fusion protein represents an in-frame fusion in which the C terminus of TycA is directly joined to the N terminus of TycB1, replacing the TycB1 initiator Met residue by the artificial linker Gly-Ser.
Construction of an E. coli strain carrying the sfp gene in the chromosome.
The sfp gene, encoding the PPTase Sfp, was first linked to a spectinomycin resistance cassette (spc) to monitor integration into the chromosome in later steps. The spc fragment was excised from pDG1726 (18) with endonucleases EcoRV and HincII and cloned into pUC18-sfp (33) linearized with HincII. A fragment containing spc and sfp under the control of the lac promoter was excised from the resulting plasmid with PvuII and ligated into the EcoRV-digested vector pUC18-nrdD to give pUC18-nrdD::sfp-spc. The 5′ and 3′ fragments of the nonessential nrdD gene served as flanking regions for homologous recombination with the chromosomal allele of nrdD (28). To this end, E. coli HSK42 was transformed with pUC18-nrdD::sfp-spc and transformants were selected on agar plates supplemented with spectinomycin. Due to a polA mutation, HSK42 is unable to replicate ColE1-based plasmids, and thus integration into the chromosome can be enforced (7). Transformants resulting from a double-crossover event were identified by testing for sensitivity against ampicillin and confirmed by PCR analysis of the nrdD locus using chromosomal DNA as a template and primers derived from the nrdD gene. Finally, a P1 phage lysate of strain HSK42-nrdD::sfp-spc was prepared to transduce the nrdD::sfp-spc allele into the standard E. coli strain GT869 to give the strain HM0079 (26).
Production and preparation of d-Phe-Pro-DKP.
E. coli strain HM0079 was transformed with the NRPS expression plasmids constructed to give rise to (i) the one-plasmid system HM0079[ptycA/tycB1], (ii) the fusion system HM0079[ptycA-tycB1(fusion)], and (iii) the two-plasmid system HM0079[pSU18-tycA/pTrc99a-tycB1]. The different strains were grown in either LB or M9 medium and at different temperatures. At certain time points, 1.5-ml samples were taken and centrifuged at 16,000 × g. The supernatant was transferred to fresh tubes and immediately extracted with 1 volume of butanol-chloroform (4:1 [vol/vol]). The organic layer was washed once with 1 volume of 0.1 M sodium chloride and then transferred to a fresh tube. After removal of the solvent under vacuum, the residue was dissolved in 50 μl of 25% methanol for further analysis by high-performance liquid chromatography (HPLC) or HPLC-mass spectrometry (MS).
Detection and analysis of d-Phe-Pro-DKP by HPLC and HPLC-MS.
DKP samples were applied to a C18 reverse-phase column (Nucleosil; 3 by 70 mm; pore size, 120 Å; particle size, 3 μm; Macherey-Nagel, Dueren, Germany) and analyzed by HPLC on a Beckman Coulter System Gold instrument, with simultaneous monitoring at detector wavelengths of 214 and 254 nm. Samples were separated under isocratic conditions (25% methanol) at a constant flow rate of 0.6 ml min−1. HPLC-MS analysis was carried out under the same conditions on an HP series 1100 system (Agilent Technologies), with the only exception that probes were acidulated by adding 0.1% trichloroacetic acid (TCA) to the solvent.
Expression of NRPS genes and purification of the gene products.
For the biochemical characterization of the recombinant NRPSs, TycA and TycB1, coexpression experiments were performed using the two-plasmid system HM0079[pSU18-tycA/pTrc99a-tycB1]. Cells were grown at various temperatures in M9 medium, supplemented with 0.1% Casamino Acids, until the optical density at 600 nm (OD600) reached about 0.6. At this point, expression of the NRPS genes was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration), and cells were allowed to grow for another 3.5 h. The cells were harvested and broken by three passages through a French press at 20 MPa. Cleared cell lysates were applied to a Ni2+ affinity chromatography column, in order to purify the His6-tagged TycB1 protein as described previously (31). Alternatively, cleared cell lysates were applied to a Strep-Tactin Sepharose column, in order to purify the Strep-tagged TycA protein in accordance with the manufacturer's protocol (IBA, Göttingen, Germany). Fractions containing the recombinant proteins were identified by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE), pooled, and subjected to anion-exchange chromatography using a MonoQ HR 5/5 column (Pharmacia, Uppsala, Sweden). Again, samples containing the recombinant proteins were identified by SDS-7.5% PAGE, pooled, and dialyzed against Tris buffer (10 mM Tris-HCl, pH 7.5). Protein concentrations were determined by using the calculated extinction coefficients for the absorbance at 280 nm (A280): 139,270 M−1 cm−1 for TycA and 92,230 M−1 cm−1 for TycB1.
Quantification of in vivo TycA and TycB1 production by SDS-PAGE analysis.
In vivo production of TycA and TycB1 proteins under different growth conditions was investigated by using the one-plasmid and two-plasmid systems. SDS-10% polyacrylamide gels of total cell extracts were Coomassie stained and densitometrically analyzed with National Institutes of Health Image, version 1.6.1, software. The amounts of total cell extracts loaded onto each lane were normalized to the OD600 in order to use equal cell numbers.
ATP pyrophosphate exchange assay.
Reaction mixtures in assay buffer (50 mM HEPES [pH 8.0], 100 mM sodium chloride, 10 mM magnesium chloride, 1 mM EDTA) contained 0 or 1 mM substrate amino acid and 500 nM enzyme (final volume, 100 μl). The reaction was initiated by addition of 2 mM ATP, 0.2 mM tetrasodium pyrophosphate, and 0.15 μCi of tetrasodium [32P]pyrophosphate (Perkin-Elmer Life Science, Rodgau-Jügesheim, Germany). After 10 min at 37°C, the reaction was quenched by the addition of 0.5 ml of a stop mixture containing 1.2% (wt/vol) activated charcoal, 0.1 M tetrasodium pyrophosphate, and 0.35 M perchloric acid. The charcoal was pelleted by centrifugation, washed twice with 1 ml of water, and resuspended in 0.5 ml of water. After addition of 3.5 ml of liquid scintillation fluid (Roth, Karlsruhe, Germany), the charcoal-bound radioactivity was determined by liquid scintillation counting in a Packard Tri-Carb 2100 TR liquid scintillation analyzer.
Radioassay for the detection of thioester formation.
Reaction mixtures in assay buffer (final volume, 100 μl) contained 500 nM enzyme, as well as 1 μCi of l-[14C]phenylalanine (469 mCi mmol−1) or 2 μCi of l-[14C]proline (242 mCi mmol−1). The reactions were initiated by the addition of 2 mM ATP, the reaction mixtures were incubated for 15 min at 37°C, and the reactions were quenched by the addition of 800 μl of ice-cold TCA (10% [wt/vol]). The TCA precipitate was collected by centrifugation (15 min, 4°C, 16,000 × g), washed once with 800 μl of 10% TCA (wt/vol), and dissolved in 400 μl of formic acid. Acid-stable, enzyme-bound radioactivity was quantified by liquid scintillation counting as described above.
In vitro product assay for d-Phe-Pro-DKP formation.
Reaction mixtures in assay buffer (final volume, 500 μl) contained 1 mM l-phenylalanine and 1 mM l-proline, as well as 1 μM TycA and 0.5 to 40 μM TycB1, corresponding to molar ratios (TycA to TycB1) of 1:0.5 to 1:40. The reactions were started by the addition of 2 mM ATP. The reaction mixtures were incubated for 1 h at 37°C, and the reactions were stopped by immediate extraction with butanol-chloroform (4:1 [vol/vol]). Samples were further prepared as described above and analyzed by HPLC.
RESULTS
Strategy.
The overall goal of this study was to explore the gram-negative bacterium E. coli as a heterologous host for the fermentative high-yield production of nonribosomally produced peptides. A special focus for these studies was the exploitation of the genetic potential of E. coli that allows easy manipulation of the chromosomal DNA, as well as the stable maintenance of up to three compatible plasmids with a variety of inducible promoters of different strengths. For the prototype nonribosomal peptide chosen in this study, d-Phe-Pro-DKP, the requirement for additional factors such as transporters or resistance genes was tested, and various culture conditions such as medium composition, temperature, and the genetic setup were optimized. The cyclic dipeptide d-Phe-Pro-DKP represents a by-product of tyrocidine biosynthesis and is formed by the action of the initiation module TycA, featuring the domain organization PheATE, and the first elongation module TycB1, composed of the domains C-APro-PCP (Fig. 1 and 2). This system has already been used in previous in vitro studies (2, 3, 24, 31, 43, 45). After formation of the peptide bond, the enzyme-bound dipeptide d-Phe-Pro-S-Ppant is autocatalytically released from its biosynthetic NRPS template to give cyclic DKP.
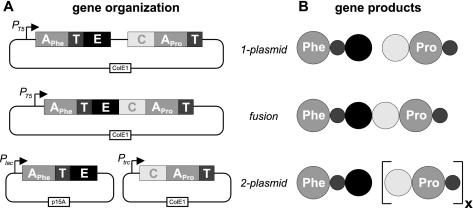
FIG. 2.
Bimodular hybrid NRPS system TycA/TycB1. Shown are the different genetic setups investigated in this study (A) and their presumed consequences on the protein level (B). Catalytic domains are highlighted as in Fig. 1.
Integration of the PPTase gene sfp into E. coli.
A requirement for the establishment of E. coli as a heterologous host for NRP production is to produce the complex NRPS assembly lines in the active holoform. Apo-to-holo conversion of heterologous NRPSs in E. coli requires the coexpression of a specialized PPTase. In order not to sacrifice one plasmid for its coexpression, we decided to stably integrate the gene of the B. subtilis PPTase Sfp (21, 28, 33) into the chromosome of K-12 wild-type strain E. coli MG1655. Sfp is the PPTase of choice, because it is known to be promiscuous and to phosphopantetheinylate any given NRPS carrier protein (21, 28). Expression of NRPS genes in this novel E. coli sfp+ strain, designated HM0079, yielded fully functional holo-NRPSs, as determined by the ability of the enzymes to covalently bind their substrate amino acids as Ppant thioesters. In these assays, activity of recombinant proteins derived from HM0079 was equivalent to that of those proteins obtained by in vitro modification with Sfp and coenzyme A and could not be enhanced by additional subsequent in vitro modification (data not shown). Expression of sfp in E. coli HM0079 was verified by Western blot analysis using specific anti-Sfp antibodies (data not shown).
Stability and toxicity of d-Phe-Pro-DKP in E. coli.
A general problem in using a heterologous host for the production of a given peptide is the potential instability or toxicity of the product. To investigate the effect of d-Phe-Pro-DKP on E. coli, a genetic approach combined with feeding experiments was applied. The proline auxotroph E. coli K-12 derivative MG1655 (pro-81::Tn10) and the wild-type strain E. coli MG1655 were grown on M9 agar plates supplemented with 1 mM (final concentration) l-proline, l-Phe-Pro, d-Phe-Pro, l-Phe-Pro-DKP, or d-Phe-Pro-DKP. After 24 h of incubation at 37°C, growth of the cells was determined. According to this study, the mutant strain E. coli MG1655 (pro-81::Tn10) grew only in the presence of l-proline and linear l-Phe-Pro, indicating that neither linear d-Phe-Pro nor the cyclic dipeptides l-Phe-Pro-DKP and d-Phe-Pro-DKP are metabolized and used as exogenic sources of proline. The wild-type control strain E. coli MG1655, in contrast, was able to grow under all conditions tested, indicating that the linear, as well as the cyclic, dipeptides are not toxic. From these experiments, we cannot rule out the possibility that the linear d-Phe-Pro and both cyclic peptides simply might not be taken up by the bacterium. However, the results clearly indicate that our expected product, d-Phe-Pro-DKP, is nontoxic to E. coli at exogenic concentrations of at least 1 mM and is not metabolized.
In vivo production and secretion of d-Phe-Pro-DKP.
Initial experiments for the in vivo production of the cyclic dipeptide d-Phe-Pro-DKP were carried out with the tycA/tycB1 (one-plasmid) system (Fig. 2), featuring the genes of the first two modules of the tyrocidine biosynthetic gene cluster under the control of the same IPTG-inducible T5 promoter. The resulting polycistronic assembly resembles the situation within the natural biosynthetic system in B. brevis, where all tyrocidine synthetase genes, tycABC, are under the control of the same growth phase-dependent promoter, Ptyc (30).
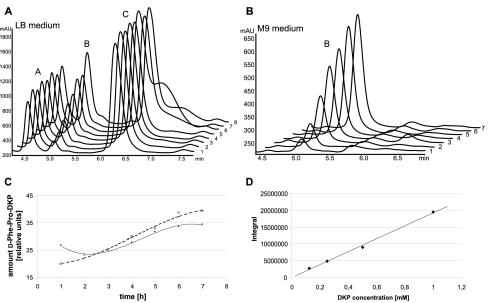
E. coli HM0079 was transformed with ptycA/tycB1 (one-plasmid system) and grown in LB medium at 37°C. At certain time points, samples were taken, prepared as described in Material and Methods, and analyzed by HPLC and HPLC-MS. As shown in Fig. 3A, the HPLC chromatograms revealed three distinct peaks with retention times of 4.5 (compound A), 5.1 (compound B), and 6.4 min (compound C). Concentrations of compounds A and C remained constant over time. Further analysis revealed that both compounds could also be extracted from the cultured broth of a control strain (E. coli HM0079[pQE61]), and even from LB medium alone, indicating that compounds A and C represent inherent ingredients of the medium (data not shown). The intensity of peak B, in contrast, increased over time and depended on the presence and expression of the bimodular NRPS system tycA/tycB1 (one-plasmid system). Strikingly, masses of both compounds B and C corresponded to the calculated mass of d-Phe-Pro-DKP (see below).
FIG. 3.
d-Phe-Pro-DKP formation in the heterologous host E. coli. Samples taken at certain time points (1 to 8 h after induction) and derived from cultures grown in LB rich medium (A) and M9 minimal medium (B) were analyzed by HPLC. Compound B coeluted with the chemical standard and corresponds to the expected d-Phe-Pro-DKP as verified by HPLC-MS (data not shown). The time-dependent production of d-Phe-Pro-DKP in LB (dots and solid line) and M9 (open circles and dashed line) media was normalized to OD600 (C). Defined quantities of d-Phe-Pro-DKP (chemical standard) were analyzed by HPLC in order to establish a calibration curve (D). All samples were taken and analyzed in triplicate.
Final characterization of the compounds was carried out by comparison with chemically synthesized standards (d-Phe-Pro-DKP and l-Phe-Pro-DKP) using HPLC and HPLC-MS analyses (data not shown). These studies showed that compound C coeluted with l-Phe-Pro-DKP, which obviously represents an inherent ingredient of the LB medium used. To rule out the possibility of an artifact, the ingredients of the rich medium, tryptone, yeast extract, and sodium chloride, were individually subjected to an organic extraction followed by HPLC analysis. These tests unequivocally revealed that compound C was derived from tryptone (data not shown). Compound B, on the other hand, the only compound whose concentration increased over time, coeluted with the chemical standard of d-Phe-Pro-DKP, the expected product of the integrated bimodular NRPS system. The mass spectrum revealed signals at m/z values of 245, 267, and 283 (data not shown), which can be assigned to the expected [M + H+], [M + Na+], and [M + K+], respectively, thereby confirming the calculated mass of 244,28 g mol−1 for the cyclic dipeptide d-Phe-Pro-DKP.
This experiment unambiguously demonstrated that the initial steps in the biosynthesis of the decapeptide antibiotic tyrocidine A could be reconstituted in the heterologous host E. coli on the basis of an engineered NRPS system. Interestingly, identical quantities of d-Phe-Pro-DKP could be obtained from the cultured broth of lysed and intact cells (data not shown), indicating that biosynthesized product did not accumulate within the cells but rather was efficiently secreted into the cultured broth. Apparently, the cyclic dipeptide either freely diffuses through the cell's membrane or is transported by the bacterium by a yet-unknown mechanism.
Influence of the medium on product formation.
As shown in Fig. 3A, preparations from the cultured broth of cells grown in LB medium basically contained three compounds: an unknown compound A and l-Phe-Pro-DKP, both derived from the LB medium, as well as the expected product d-Phe-Pro-DKP. With regard to experimental cleanliness and to avoid possible problems in interpretation of the results that may be caused by the presence of the diastereomer l-Phe-Pro-DKP as an ingredient of the LB medium, we used M9 minimal medium instead, which does not contain any tryptone. In an initial test, we ensured that extracts of M9 medium alone did not contain any compound A or C (data not shown).
Subsequently, E. coli HM0079[ptycA/tycB1] (one-plasmid system) was grown in M9 medium at 37°C, and at certain time points, samples were taken, prepared by organic extraction, and analyzed by HPLC-MS. The HPLC chromatograms revealed only one compound whose intensity increased over time (Fig. 3B). This compound coeluted with the d-Phe-Pro-DKP standard. The identity of this compound was verified by HPLC-MS analysis (data not shown).
To determine the absolute product yields, the amounts of d-Phe-Pro-DKP produced by E. coli HM0079[ptycA/tycB1] (one-plasmid system) under the different medium conditions were quantified. To this end, a calibration curve was recorded, with chemically synthesized d-Phe-Pro-DKP as a reference (Fig. 3D). The analysis revealed that within 7 h after induction the amount of DKP produced in LB medium (19 μM or 4.26 mg/liter) was about twice that produced in M9 medium (11 μM or 2.69 mg/liter). Interestingly, however, standardization to the cell's wet weight showed that the amount of d-Phe-Pro-DKP produced per OD unit is independent of the medium used, and thus only the higher cell density obtained in LB medium than in M9 minimal medium accounted for the differences in productivity observed (Fig. 3C). Since utilization of M9 medium (i) circumvented possible problems in interpretation of the results that may be caused by the presence of the l-Phe-Pro-DKP in LB medium and (ii) allowed more-stringent control of the experimental conditions, e.g., by controlling the availability of nutrients and substrates, all following experiments were performed in M9 medium.
Influence of the availability of substrates and nutrients.
To extend our experiments to targeted substrate feeding, M9 medium was supplemented with either 0.1% Casamino Acids or a 1 mM final concentration of substrate amino acids l-phenylalanine and l-proline. None of the supplements had any significant effect on the productivity of the NRPS system per OD unit (data not shown). However, since volumetric productivity was significantly improved by the utilization M9 supplemented with 0.1% Casamino Acids, we decided to routinely enrich the minimal medium with this ingredient.
Strategy for the construction of artificial bimodular NRPS systems.
To further optimize d-Phe-Pro-DKP production, we next varied the bimodular NRPS system itself. The experiments described above were performed with the one-plasmid system tycA/tycB1, which essentially copied the polycistronic wild-type situation, where all NRPS genes are under the control of the same promoter. In addition to this system, we decided to create two novel systems, which are shown in Fig. 2 and described below.
(i) ptycA-tycB(fusion).
NRP biosynthetic gene clusters derived from bacteria usually consist of two or more individual NRPSs, which have to interact in a coordinated fashion. Little is known about the structural elements that facilitate this specific interaction. In particular, this interaction might be less efficient in the heterologous host E. coli because of different cellular conditions, unknown factors, or improper localization to the cell membrane, which has been found for many NRPSs in their natural producer strains (19). We therefore used a construct in which the structural genes of TycA and TycB1 are fused to one open reading frame.
The resulting hybrid gene is under the control of the IPTG-inducible T5 promoter and encodes a single bimodular NRPS TycA-TycB1 with a size of 241 kDa. The interaction of the two modules in this construct should be relatively independent of the different conditions. d-Phe-Pro-DKP production mediated by the fusion protein in vitro was previously investigated by Linne et al. and was shown to drop to about 50% in comparison to that for the monomodular wild-type system, TycA/TycB1 (24). A similar drop in product yield in our in vivo studies would indicate that the productive interaction between TycA and TycB1 is not disturbed in E. coli.
(ii) pSU18-tycA/pTrc99a-tycB1 (two-plasmid system).
In the course of biosynthesis of the decapeptide antibiotic tyrocidine, activated d-phenylalanine is translocated from the initiation module d-Phe-S-Ppant TycA onto the elongation module l-Pro-S-Ppant TycB1, leading to the formation of the linear, enzyme-bound intermediate d-Phe-Pro-S-Ppant TycB1. In the absence of succeeding modules or substrates, the dipeptide is autocatalytically released under formation of the cyclic d-Phe-Pro-DKP (apparent rate constant [kapp] ≤ 1 min−1) (2, 3, 24, 31, 43, 45). The latter reaction represents the rate-limiting step in the formation of d-Phe-Pro-DKP, indicating that the elongation module is blocked, until the dipeptide has been released from the enzyme. This circumstance prevents the d-Phe-S-Ppant TycA donor from translocating the next activated d-phenylalanine onto TycB1. We attempted to overcome this kinetic bottleneck by increasing the ratio between acceptor and donor, such that every TycA donor could interact with more than one TycB1 acceptor within a certain period of time.
This hypothesis was first challenged by using purified proteins in vitro (next section). To test this theory in vivo, the two-plasmid system pSU18-tycA/pTrc99a-tycB1 was constructed; this system featured tycA on the medium-copy-number vector pSU18 and tycB1 on the high-copy-number vector pTrc99a. Both genes are under the control of IPTG-inducible promoters lac and trc, respectively. Given the different copy numbers of the compatible plasmids and the different strengths of the promoters (trc > lac), the overall ratio between the TycB1 acceptor and TycA donor should be elevated in E. coli.
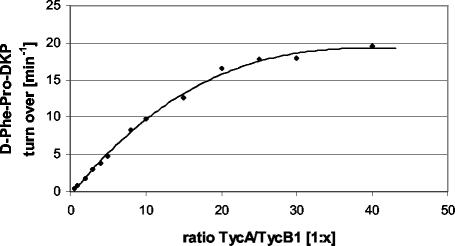
Determination of the optimal ratio of TycA and TycB1 for DKP formation.
The reflections above suggested that increasing the ratio between the TycB1 acceptor and the TycA donor might enhance the productivity of the bimodular NRPS system. To determine the optimal ratio between both proteins, an in vitro assay for d-Phe-Pro-DKP formation was carried out (42, 45). To this end, enzymes at different ratios, ranging from 1:0.5 to 1:40 (TycA to TycB1), were incubated at 37°C. After 1 h, reactions were quenched by the addition of butanol-chloroform (4:1, vol/vol) and the products were prepared as described in Material and Methods and analyzed by HPLC. As shown in Fig. 4, the productivity could be enhanced by a factor of 20 by increasing the ratio between the TycB1 acceptor and TycA donor (relative to a constant amount of TycA). The optimal ratio was found to be approximately 1:20, while a further excess of the TycB1 acceptor did not significantly improve the productivity. With this observation, the two-plasmid system pSU18-tycA/pTrc99a-tycB1 seemed to be a promising route for improved in vivo d-Phe-Pro-DKP production.
FIG. 4.
Determination of the optimal ratio of TycA to TycB1 for d-Phe-Pro-DKP formation. Both enzymes were incubated in the presence of l-phenylalanine, l-proline, ATP, and magnesium chloride, and the reactions were quenched after 1 h by immediate organic extraction. DKP was prepared as described in Materials and Methods and analyzed by HPLC. All samples were taken and analyzed in triplicate.
Quantification of TycA and TycB1 production by SDS-PAGE analysis.
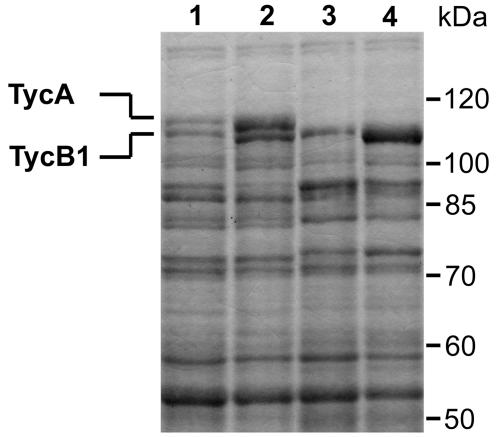
We next determined protein concentrations from coproduced proteins to see whether the two-plasmid system resulted in an elevated ratio between TycB1 and TycA. The amounts of TycA and TycB1 produced by the one-plasmid and two-plasmid systems under different conditions were determined. Figure 5 shows an analysis of a Coomassie-stained SDS-10% polyacrylamide gel that served for the calculation of protein ratios by densitometrical analysis. The bands corresponding to TycA and TycB1 were well separated at about 115 and 110 kDa (calculated, 123 and 119 kDa, respectively). As expected for the polycistronic organization of the NRPS genes, approximately equal amounts of both proteins could be observed in the one-plasmid system. The lanes of uninduced samples already showed relatively strong production of TycA and TycB1, which can be ascribed to the known leakiness of the promoters (one plasmid: T5; two plasmid: lac and trc) used. Importantly, the protein pattern of cells from the two-plasmid system clearly indicated a high excess of TycB1 relative to TycA, which was calculated to be ≥30-fold. On the other hand, the amount of TycA was strongly reduced compared to that associated with the one-plasmid system, in agreement with the lower-copy-number plasmid used for its expression. Interestingly, also the amount of TycB1 was elevated about fourfold in the two-plasmid system compared to that for the one-plasmid system.
FIG. 5.
SDS-PAGE analysis for the quantification of TycA and TycB1 levels, as produced by the one-plasmid and two-plasmid systems. Amounts of total cell extracts applied to each lane were normalized to the OD600. Lane 1, one-plasmid system without induction; lane 2, one-plasmid system with induction; lane 3, two-plasmid system without induction; lane 4, two-plasmid system with induction.
Effect of the different genetic arrangements on in vivo d-Phe-Pro-DKP production.
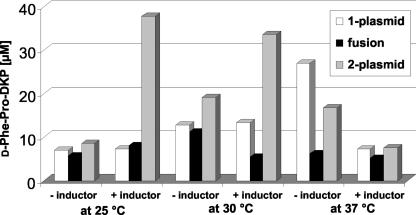
To investigate the effects of the different genetic arrangements of the bimodular NRPS system TycA/TycB1, E. coli HM0079 was transformed with the one-plasmid, fusion, and two-plasmid systems constructed. The resulting DKP production strains were grown in M9 medium supplemented with 0.1% Casamino Acids. At certain time points, samples were taken, prepared as described above, and analyzed by HPLC. We also studied the effects of different growth temperatures, 25, 30, and 37°C, as well as the presence and absence of the inducer IPTG, because of the known leakiness of the promoters used in the expression plasmids.
The results of this comprehensive study are recapitulated in Fig. 6 and can be summarized as follows. (i) Of the three different genetic arrangements tested, the two-plasmid system was the most productive one under nearly all conditions tested. The overall ranking between all systems can be rated as follows: two plasmid > one plasmid > fusion. The lower productivity of the artificial fusion protein is in agreement with the decreased activity of this system under in vitro conditions (24) and suggests that the interaction between the subunits TycA and TycB1 in the other two systems is not hampered under the in vivo conditions in the heterologous host. The higher productivity of the two-plasmid system than of the one-plasmid system is in agreement with the increased total amount of TycB1 and the increased ratio between TycB1 and TycA. This result suggests that our insights into the optimal ratio between the two modules, as determined with purified proteins, are also valid for the in vivo system. This finding further underlines the advantages of using E. coli, which makes it possible to independently control expression levels by simple means such as choice of plasmid copy number and promoter strength.
FIG. 6.
Effect of different genetic setups on in vivo d-Phe-Pro-DKP production. The one-plasmid system, fusion system, and two-plasmid system were individually studied at different growth temperatures, as well as with and without IPTG induction. All samples were taken and analyzed in triplicate.
(ii) Variation of the temperature had distinctive consequences for the productivity of the different tycA-tycB1 systems under investigation. The one-plasmid system performed best at higher temperatures (25°C < 30°C ≈ 37°C), whereas the artificial fusion system produced the most DKP at lower temperatures (37°C < 25°C < 30°C). Although the results were less pronounced, basically the same applied to the two-plasmid system (37°C ≪ 25°C ≈ 30°C).
(iii) To trigger DKP production, addition of inducer was especially required at lower temperatures. Apparently, leakiness of the IPTG-inducible promoters is somehow temperature dependent, with a tighter regulation at lower temperatures. The lowest productivity of all three NRPS systems tested was observed for our starting conditions, the one-plasmid approach at 37°C with induction. Omitting IPTG resulted in an increase in productivity by a factor of 2 to 3, both for the one-plasmid and the two-plasmid systems, at 37°C.
(iv) By using the superior two-plasmid system and by shifting the temperature from 37 to 25°C, the overall productivity of the heterologous host E. coli HM0079 for the synthesis of the NRP product d-Phe-Pro-DKP could be enhanced by approximately 400% (from 7.5 to 37.7 μM, corresponding to a change from 1.8 to 9.2 mg/liter).
Investigation of the biochemical properties of NRPSs.
Higher cellular concentrations of TycA and TycB1 should translate into larger amounts of d-Phe-Pro-DKP product, and thus the highest productivity should be obtained at high temperature (37°C) and after induction. However, the exact opposite was found for the fusion and two-plasmid systems, where maximal productivity was observed at a lower temperature (25°C). A possible explanation for this phenomenon could be that the quality, activity, or even solubility of NRPS proteins was affected at higher temperatures, e.g., due to improper folding and/or incomplete apoform-to-holoform conversion (see below). In fact, heterologous expression of NRPSs is usually carried out at lower temperatures in this and other laboratories to increase the amount of soluble protein (8-10, 24, 31, 41-43, 45).To validate this theory, tycB1 was expressed at different temperatures and the gene product was purified and biochemically characterized.
The ATP-pyrophosphate exchange assay was performed to determine the activity of the adenylation domain. The highest value obtained (TycB1 produced at 25°C without inducer) was used as reference and set to 100%. The activities of TycB1 prepared under the various conditions in general did not differ significantly. Only under expression conditions involving 37°C and induction did the activity strongly decrease, likely due to a higher degree of improper folding under these conditions. The overall ranking between all systems can be rated as follows: 25°C without induction (100%) > 25°C with induction > 30°C without induction > 30°C with induction (80%) > 37°C without induction ≫ 37°C with induction (23%) (data not shown).
This observation was (partially) carried forward to the covalent loading assay, where binding of the substrate amino acid as a Ppant thioester was measured. TycB1 prepared from induced samples showed a steady decrease in covalent binding activity with increasing temperature, whereas TycB1 prepared from uninduced samples grown at 30°C reached a maximum activity in this assay (data not shown). These results are in agreement with the outcome of the d-Phe-Pro-DKP production assay (Fig. 6) determined under the same conditions. Apparently, product formation at higher temperatures is limited by improper folding and/or insufficient apo-to-holo conversion of the NRPSs produced.
DISCUSSION
In the present study, we describe the fermentative production of the cyclic dipeptide d-Phe-Pro-DKP, as a prototype for the nonribosomal biosynthesis of complex peptide products in the heterologous host E. coli. The approach is a first step for the large-scale production of novel peptides obtained by exploitation of the biocombinatorial potential of NRPSs through genetic engineering. There is a significant interest in the sustainable and affordable bioproduction of such peptides, e.g., those used as pharmaceuticals or fine chemicals.
In general, fermentative production of complex peptides has advantages over chemical synthesis in many aspects, since it uses renewable primary products and does not require expensive reactants and poisonous chemicals and solvents. In particular, many NRPs with interesting biological functions are too complex to be synthesized chemically on a reasonable scale (13). Consequently, the artificial engineering of NRPS assembly lines is an attractive route to prepare analogs of complex natural products (32, 40). The practicability of this general concept has been demonstrated in various examples, ranging from targeted modifications of existing products to the constitution of entirely artificial NRPSs from scratch (9, 24, 29, 31, 38, 44, 51). For example, an artificial NRPS that catalyzes formation of the dipeptide precursor of the high-intensity sweetener aspartame was constructed de novo (10). In all these cases, production of the expected peptides could be demonstrated in vitro. The goal of this study was to establish E. coli as a heterologous host for the fermentative production of such products.
To bring about the targeted nonribosomal biosynthesis of d-Phe-Pro-DKP, we took advantage of the bimodular NRPS system TycA/TycB1 (43). Both modules are part of the tyrocidine biosynthetic system of the gram-positive host B. brevis ATCC 8185 (30). A major requirement to engineer and establish a coexpression system in the gram-negative bacterium E. coli was to produce the complex NRPSs in a properly folded, active holoform. Apo-to-holo conversion occurs posttranslationally under the catalytic control of a specialized PPTase (21). Since E. coli PPTases are incapable of phosphopantetheinylating heterologous NRPS carrier domains, we stably integrated the gene of the B. subtilis PPTase Sfp, which is known to be promiscuous and to modify any given carrier protein (15, 21), into the chromosome of E. coli. This way, the commonly used approach for coexpressing a PPTase gene from a compatible plasmid was avoided to provide full flexibility with compatible plasmids for the expression of NRPS gene fragments or other additional genes (8-10, 24, 31, 41, 42). The resulting E. coli sfp+ strain HM0079 proved to be competent for the production of fully active holo-NRPSs.
We next expressed the bimodular NRPS system TycA/TycB1 from one plasmid and showed formation of the desired dipeptide product, d-Phe-Pro-DKP. This experiment demonstrated for the first time that an engineered peptide product could be efficiently produced in vivo by the artificial NRPS system within a heterologous environment. Interestingly, the experiment revealed that the biosynthesized d-Phe-Pro-DKP was also efficiently secreted into the cultured broth, indicating that the cyclic dipeptide can freely diffuse through the cell's membrane or, maybe less likely, is transported by the bacterium by an unknown mechanism (22). The membrane permeability for a passive diffusion mechanism might be explained by the relatively hydrophobic nature of the cyclic d-Phe-Pro-DKP, without free hydrophilic N or C termini.
Importantly, the study showed that the initial steps of the biosynthesis of the decapeptide antibiotic tyrocidine A could be reconstituted in vivo. This implies a proper interaction between the two modules TycA and TycB1 under the conditions of the heterologous host E. coli. Little is known about the structural elements that facilitate the specific interaction between two NRPSs and prevent the unproductive contact between nonpartner NRPSs. Studies on polyketide synthases that follow a similar assembly line logic revealed so-called intermodular linkers that facilitate protein-protein communication (50). The presence of similar specificity-conferring linkers has also been postulated for NRPSs. The robust in vivo interaction observed in this study could smooth the path for a reconstitution of more-complex NRP systems, such as the entire tyrocidine biosynthetic gene cluster (30). Such a reconstitution would most likely also require the presence of associated genes (11, 34, 35), e.g., those involved in the export of the cyclic decapeptide antibiotic.
The formation of productive bacterial NRPS complexes is likely to be supported by the native polycistronic organization of the involved genes, which ensures a coordinated production of all enzymes required at the same period of growth, and in defined relative quantities. However, given the particularities of the artificial NRPS system under investigation (TycA/TycB1), in particular the fact that autocatalytic cyclization represents the rate-limiting step in the formation of d-Phe-Pro-DKP, we postulated that the productivity is limited by the availability of aminoacylated TycB1. The acceptor module loaded with the dipeptidyl intermediate is blocked for the next elongation step until product cleavage and recharging with l-proline occur (2, 43, 45). We attempted to overcome this kinetic bottleneck by increasing the ratio between acceptor and donor modules. Indeed, the optimal ratio between TycA and TycB1 was determined in in vitro studies with purified enzymes to be about 1:20. This suggests that the protein-protein interaction between NRPSs is actually a dynamic process of fast association and disassociation, indicating that the common perception of a rigid assembly line and biosynthetic complex may have to be reconsidered. Comparable results were obtained for the enterobactin and yersiniabactin biosynthetic systems, which, however, possess a distinctively different organization of the subunit involved (17, 25).
We next implemented these insights in our in vivo studies by exploiting an artificial two-plasmid system that expressed the tycB1 and tycA genes in an elevated ratio; this resulted in a fivefold increase in d-Phe-Pro-DKP productivity to 9.2 mg per liter. These manipulations underline the advantages of E. coli as a heterologous host, which provides a tremendous potential for genetic engineering, including the stable maintenance of up to three compatible plasmids and many different promoters of various strengths. Further optimization might be possible along these lines, as well as expression of other factors that might increase the activity of NPRS, for example, coexpression of a type II thioesterase gene, which was shown in in vitro studies to liberate NRPS blocked by misacylation (41). In contrast, we found that nutrient availability played only a negligible role in the productivity per OD unit. It is likely, however, that the importance of feeding certain nutrients or substrates will require a case-to-case study and will be important especially when rare or unusual amino acid monomers are incorporated by the NRP. The effect of different temperatures for the cultivation of E. coli mainly seemed to follow the previously noted better solubility of NRPS heterologously expressed at lower temperatures.
The prototype peptide d-Phe-Pro-DKP belongs to a family of secondary metabolites with a DKP core structure. Many of these compounds exhibit useful biological properties, such as antibacterial (e.g., bicyclomycin), immunosuppressive (e.g., gliotoxin), and antitumoral (e.g., verticillin) activity, and have been noted in all kinds of organisms, including Streptomyces spp. and bacilli, as well as animals and humans (14, 16, 22). DKPs can be formed after proteolytic digestion of precursor molecules or synthesized from dedicated NRPS systems, as was shown for the biosynthetic clusters for ergotamine (36) and albonoursin (22). Thus, our engineered TycA/TycB1 system may serve as a starting point for the production of engineered variants of this class of compounds (16).
In summary, we have presented here the first example for the successful utilization of an artificial hybrid NRPS assembly line for the in vivo production of a complex peptide product in the heterologous host E. coli. Engineering NRPS to artificial assembly lines takes advantage of their modularity and can be exploited for the production of basically any small peptide product. Thus this approach may be generally useful for the sustainable bioproduction of industrially relevant, peptide-based pharmaceuticals or fine chemicals.
Acknowledgments
We thank Mohamed A. Marahiel for supporting the work. We are obliged to Uwe Linne for discussions and assistance with HPLC-MS analysis, we thank Mary Berlyn from the E. coli Genetic Stock Center for providing strains, and we acknowledge the technical assistance of Daniel Stein.
The BioFuture program from the Federal Ministry of Education and Research sponsored this work.
REFERENCES
- 1.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, P. J., C. T. Walsh, and T. Stachelhaus. 1999. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284:486-489. [DOI] [PubMed] [Google Scholar]
- 3.Bergendahl, V., U. Linne, and M. A. Marahiel. 2002. Mutational analysis of the C-domain in nonribosomal peptide synthesis. Eur. J. Biochem. 269:620-629. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 6.Byford, M. F., J. E. Baldwin, C. Y. Shiau, and C. J. Schofield. 1997. The mechanism of ACV synthetase. Chem. Rev. 97:2631-2650. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doekel, S., K. Eppelmann, and M. A. Marahiel. 2002. Heterologous expression of nonribosomal peptide synthetases in B. subtilis: construction of a bi-functional B. subtilis/E. coli shuttle vector system. FEMS Microbiol. Lett. 216:185-191. [DOI] [PubMed] [Google Scholar]
- 9.Doekel, S., and M. A. Marahiel. 2000. Dipeptide formation on engineered hybrid peptide synthetases. Chem. Biol. 7:373-384. [DOI] [PubMed] [Google Scholar]
- 10.Duerfahrt, T., S. Doekel, T. Sonke, P. J. Quaedflieg, and M. A. Marahiel. 2003. Construction of hybrid peptide synthetases for the production of α-L-aspartyl-L-phenylalanine, a precursor for the high-intensity sweetener aspartame. Eur. J. Biochem. 270:4555-4563. [DOI] [PubMed] [Google Scholar]
- 11.Eppelmann, K., S. Doekel, and M. A. Marahiel. 2001. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 276:34824-34831. [DOI] [PubMed] [Google Scholar]
- 12.Eppelmann, K., T. Stachelhaus, and M. A. Marahiel. 2002. Exploitation of the selectivity-conferring code of nonribosomal peptide synthetases for the rational design of novel peptide antibiotics. Biochemistry 41:9718-9726. [DOI] [PubMed] [Google Scholar]
- 13.Evans, D. A., M. R. Wood, B. W. Trotter, T. I. Richardson, J. C. Barrow, and J. L. Katz. 1998. Total synthesis of vancomycin and eremomycin A glycons. Angew. Chem. Int. Ed. Engl. 37:2700-2704. [DOI] [PubMed] [Google Scholar]
- 14.Faden, A. I., G. B. Fox, X. Di, S. M. Knoblach, I. Cernak, P. Mullins, M. Nikolaeva, and A. P. Kozikowski. 2003. Neuroprotective and nootropic actions of a novel cyclized dipeptide after controlled cortical impact injury in mice. J. Cereb. Blood Flow Metab. 23:355-363. [DOI] [PubMed] [Google Scholar]
- 15.Finking, R., J. Solsbacher, D. Konz, M. Schobert, A. Schafer, D. Jahn, and M. A. Marahiel. 2002. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:50293-50302. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, P. M. 2003. Diketopiperazines in peptide and combinatorial chemistry. J. Pept. Sci. 9:9-35. [DOI] [PubMed] [Google Scholar]
- 17.Gehring, A. M., I. Mori, and C. T. Walsh. 1998. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37:2648-2659. [DOI] [PubMed] [Google Scholar]
- 18.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 19.Hoppert, M., C. Gentzsch, and K. Schorgendorfer. 2001. Structure and localization of cyclosporin synthetase, the key enzyme of cyclosporin biosynthesis in Tolypocladium inflatum. Arch. Microbiol. 176:285-293. [DOI] [PubMed] [Google Scholar]
- 20.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 21.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily-the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 22.Lautru, S., M. Gondry, R. Genet, and J. L. Pernodet. 2002. The albonoursin gene cluster of S. noursei. Biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 9:1355-1364. [DOI] [PubMed] [Google Scholar]
- 23.Linne, U., and M. A. Marahiel. 2000. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry 39:10439-10447. [DOI] [PubMed] [Google Scholar]
- 24.Linne, U., D. B. Stein, H. D. Mootz, and M. A. Marahiel. 2003. Systematic and quantitative analysis of protein-protein recognition between nonribosomal peptide synthetases investigated in the tyrocidine biosynthetic template. Biochemistry 42:5114-5124. [DOI] [PubMed] [Google Scholar]
- 25.Miller, D. A., L. Luo, N. Hillson, T. A. Keating, and C. T. Walsh. 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9:333-344. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Molnar, I., T. Schupp, M. Ono, R. Zirkle, M. Milnamow, B. Nowak-Thompson, N. Engel, C. Toupet, A. Stratmann, D. D. Cyr, J. Gorlach, J. M. Mayo, A. Hu, S. Goff, J. Schmid, and J. M. Ligon. 2000. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 7:97-109. [DOI] [PubMed] [Google Scholar]
- 28.Mootz, H. D., R. Finking, and M. A. Marahiel. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289-37298. [DOI] [PubMed] [Google Scholar]
- 29.Mootz, H. D., N. Kessler, U. Linne, K. Eppelmann, D. Schwarzer, and M. A. Marahiel. 2002. Decreasing the ring size of a cyclic nonribosomal peptide antibiotic by in-frame module deletion in the biosynthetic genes. J. Am. Chem. Soc. 124:10980-10981. [DOI] [PubMed] [Google Scholar]
- 30.Mootz, H. D., and M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179:6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2000. Construction of hybrid peptide synthetases by module and domain fusions. Proc. Natl. Acad. Sci. USA 97:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 33.Nakano, M. M., N. Corbell, J. Besson, and P. Zuber. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232:313-321. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer, B. A., C. C. Wang, C. T. Walsh, and C. Khosla. 2003. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 69:6698-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimmann, C., H. M. Patel, L. Serino, M. Barone, C. T. Walsh, and D. Haas. 2001. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 183:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riederer, B., M. Han, and U. Keller. 1996. d-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. J. Biol. Chem. 271:27524-27530. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schneider, A., T. Stachelhaus, and M. A. Marahiel. 1998. Targeted alteration of the substrate specificity of peptide synthetases by rational module swapping. Mol. Gen. Genet. 257:308-318. [DOI] [PubMed] [Google Scholar]
- 39.Schwarzer, D., R. Finking, and M. A. Marahiel. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275-287. [DOI] [PubMed] [Google Scholar]
- 40.Schwarzer, D., and M. A. Marahiel. 2001. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88:93-101. [DOI] [PubMed] [Google Scholar]
- 41.Schwarzer, D., H. D. Mootz, U. Linne, and M. A. Marahiel. 2002. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. USA 99:14083-14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarzer, D., H. D. Mootz, and M. A. Marahiel. 2001. Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetases. Chem. Biol. 8:997-1010. [DOI] [PubMed] [Google Scholar]
- 43.Stachelhaus, T., H. D. Mootz, V. Bergendahl, and M. A. Marahiel. 1998. Peptide-bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J. Biol. Chem. 273:22773-22781. [DOI] [PubMed] [Google Scholar]
- 44.Stachelhaus, T., A. Schneider, and M. A. Marahiel. 1995. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 269:69-72. [DOI] [PubMed] [Google Scholar]
- 45.Stachelhaus, T., and C. T. Walsh. 2000. Mutational analysis of the epimerization domain in the initiation module PheATE of gramicidin S synthetase. Biochemistry 39:5775-5787. [DOI] [PubMed] [Google Scholar]
- 46.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 47.van Wageningen, A. M., P. N. Kirkpatrick, D. H. Williams, B. R. Harris, J. K. Kershaw, N. J. Lennard, M. Jones, S. J. Jones, and P. J. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5:155-162. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, C. T. 2002. Combinatorial biosynthesis of antibiotics: challenges and opportunities. Chembiochem 3:125-134. [DOI] [PubMed] [Google Scholar]
- 49.Weber, G., K. Schorgendorfer, E. Schneider-Scherzer, and E. Leitner. 1994. The peptide synthetase catalyzing cyclosporin production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 26:120-125. [DOI] [PubMed] [Google Scholar]
- 50.Wu, N., S. Y. Tsuji, D. E. Cane, and C. Khosla. 2001. Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J. Am. Chem. Soc. 123:6465-6474. [DOI] [PubMed] [Google Scholar]
- 51.Yakimov, M. M., L. Giuliano, K. N. Timmis, and P. N. Golyshin. 2000. Recombinant acylheptapeptide lichenysin: high level of production by Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol. 2:217-224. [PubMed] [Google Scholar]