Summary
Brain networks contain a large diversity of functionally distinct neuronal elements, each with unique properties, enabling computational capacities and supporting brain functions. Understanding their functional implications for behavior requires the precise identification of the cell types of a network and in vivo monitoring of their activity profiles. Here, we developed a channelrhodopsin-assisted patching method allowing the efficient in vivo targeted recording of neurons identified by their molecular, electrophysiological and morphological features. The method has a high yield, does not require visual guidance, and thus can be applied at any depth in the brain. This approach overcomes limitations of present technologies. We validate this strategy by in vivo recordings of identified subtypes of GABAergic and glutamatergic neurons in deep cortical layers, subcortical cholinergic neurons and neurons in the thalamic reticular nucleus in anesthetized and awake mice. We propose this method as an important complement to existing technologies to relate specific cell type activity to brain circuitry, function and behavior.
Introduction
Most brain structures consist of a large diversity of neuronal subtypes differing in their precise anatomical location, morphology, connectivity, molecular composition, biophysical properties and activity patterns. To understand network function and the neural basis of behavior, it is necessary to measure the activity of precisely identified cell types in the intact brain. However, studying the activity of specific cell types in vivo has been limited, especially when optical access is restricted or when the cells of interest do not represent the majority within a brain structure. Here we report the development and validation of a high-yield method for the in vivo targeted recording and labeling of genetically identified neurons throughout the brain, which provides their activity and precise anatomical location, morphological and molecular properties.
Several approaches have been employed to record the in vivo activity of different cell types in the brain. Extracellular blind unit recordings can take advantage of spike waveform properties to separate putative excitatory and inhibitory cells (Csicsvari et al., 1999). However, since both populations contain cell types exhibiting overlapping spectra of spike shapes, assignment of recorded units to particular cell types remains difficult (Fuentealba et al., 2008, Vigneswaran et al., 2011). The use of glass electrodes and blind patch technology enable dye loading of recorded neurons solving this ambiguity (Pinault and Deschenes, 1998). In principle, the full morphology and anatomical position can be recovered and the expression of molecular markers determined through post hoc immunohistochemistry (Klausberger et al., 2003). Despite the high degree of precision of such an approach, its low yield makes its use impractical when studying infrequent cell types.
The recent development of optical and genetic technologies has dramatically facilitated the in vivo targeting and recording of the activity of identified cell populations. Using fluorescent protein expression in specific cell types in transgenic animals, two-photon targeted patching (TPTP) enables the visualization of genetically defined neurons in order to direct the recording electrode to the cell type of interest for extracellular recording of spiking activity or intracellular membrane potential measurements (Margrie et al., 2003). However, light scattering through brain tissue has largely restricted the use of two-photon guided methodologies to superficial brain structures, such as upper cortical layers.
The expression of channelrhodopsin-2 (ChR2) under specific promoters, in combination with tetrodes or silicon probes coupled to a light source overcomes this limitation (Lima et al., 2009, Anikeeva et al., 2012, Roux et al., 2014). Through this strategy, genetically defined cells that conditionally express ChR2 can be identified through their tight temporal responsiveness to light stimulation, allowing the ‘tagging’ of extracellularly recorded units and monitoring of their activity regardless of recording depth. This method has been successfully applied to record the activity of genetically tagged neuronal populations in freely behaving animals (Kvitsiani et al., 2013, Stark et al., 2013). However, unlike TPTP, this technique cannot offer information on the morphology, precise anatomical position and membrane potential dynamics of the recorded cells, and as such relies largely on the genetic identification provided by the conditional expression of ChR2 and spike waveform isolation.
Strict dependence on genetic identification is not sufficient for the unambiguous identification of cell types. Many widely used transgenic mouse lines do not target single neuronal subtypes, but rather neuronal subpopulations that include substantial and physiologically relevant heterogeneity (Huang, 2014). For instance, cortical GABAergic interneurons expressing parvalbumin (PV) can exhibit basket and chandelier morphologies (Rudy et al., 2011) with potentially opposite postsynaptic impacts (Szabadics et al., 2006). Thus, the ambiguities created by the partial specificity obtained with the genetic identifier alone calls for a combinatorial approach in which electrophysiological and morphological properties are obtained, for more consistent and robust cell type identification.
Here, we report a strategy allowing the targeted recording and morphological recovery of genetically defined neurons. Our method employs the conditional expression of ChR2 in genetically defined cell populations in combination with optical fiber coupled patch electrodes and neurobiotin loading to record and label neurons of interest. This approach does not require visual guidance, and thus enables robust identification and targeting of ChR2-expressing neurons regardless of recording depth. Our method has a high yield as compared to traditional blind recording, even for rare neuronal subtypes. Moreover, it allows the characterization of genetic, morphological, and electrophysiological properties of recorded and labeled neurons, surpassing the rate and precision of current approaches regarding cell type identification. As a proof of principle and to illustrate the merit of our strategy, we targeted different types of GABAergic neurons for juxtacellular recording and labeling in deep cortical layers. The observed physiologically relevant heterogeneity within cells sharing genetic and electrophysiological features demonstrates the importance of the combinatorial approach for cell type identification. Such experiments were easily integrated with functional studies in anesthetized and awake, head-fixed preparations. We also demonstrate the potential of this method to perform optogenetically guided whole-cell recordings. Finally, we performed targeted juxtasomal recording and labeling of thalamic reticular neurons and cholinergic neurons of the striatum and basal forebrain. We propose this efficient and robust technique as a valuable complement to existing tools to monitor the activity of distinct cellular elements composing brain circuits and accelerate our understanding of their functional implications for network operations and behavior.
Results
Channelrhodopsin-assisted patching: Strategy for the recording and labeling of genetically defined neurons in vivo
Our goal was to develop a method for the efficient in vivo recording and labeling of genetically defined cell types independent of recording depth in order to characterize their activity, morphology and other identifying features. Since most brain areas are composed of heterogeneous neuronal populations, the probability of recording from a particular cell type using the blind approach is a function of the density distribution of the neurons of interest and the success rate of post hoc identification. This can render the study of specific neuronal types impractical when they represent a minority of the population. We reasoned that accelerating the cell sampling and a priori identification of cells would sufficiently increase the number of recorded and labeled neurons of interest, making such studies amenable for in vivo interrogation of precisely identified neurons.
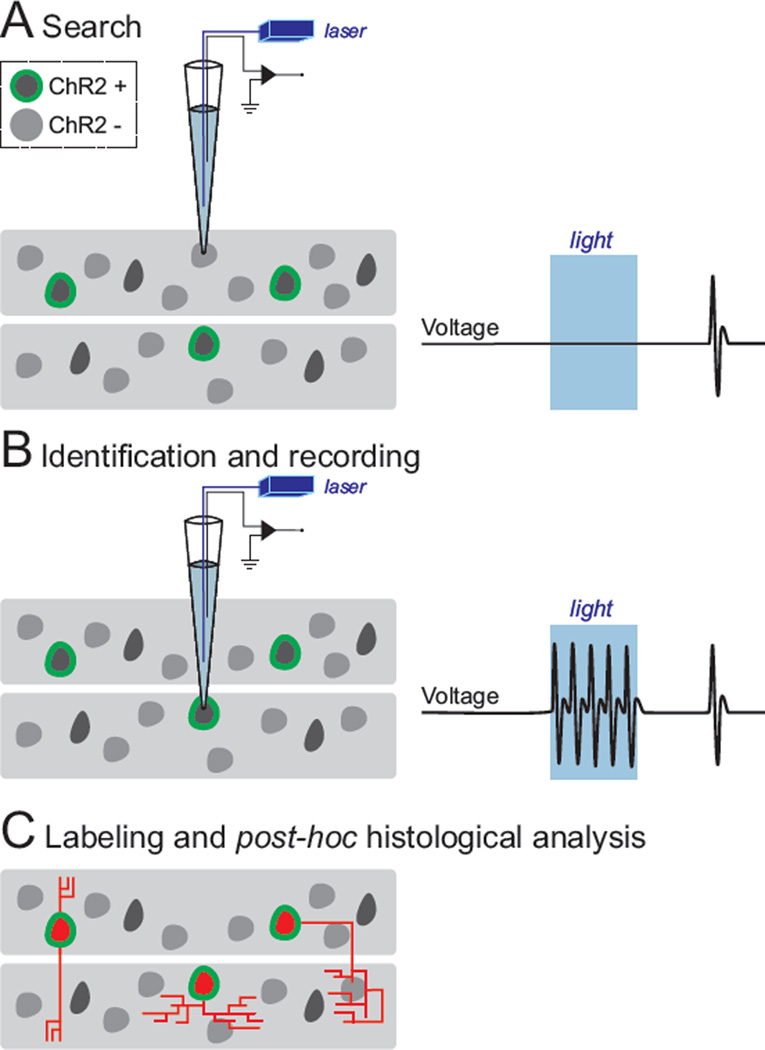
To achieve this goal, we relied on the following strategy (Figure 1). First, we established genetic access to cell types of interest by the conditional expression of ChR2 in Cre driver lines, either by viral transduction with Cre-conditional ChR2 vectors or by breeding Cre driver mice to conditional ChR2 reporter lines (Taniguchi et al., 2011, Madisen et al., 2012). Then, using a glass electrode containing an optical fiber coupled to a blue light laser (λ = 473 nm, Figure 1), we initiated the standard blind patching procedure by lowering pipettes into the brain area of interest with small steps until an increase in pipette resistance was observed, indicating that a cell was encountered. A train of light pulses was then immediately presented to test for ChR2-mediated spiking activity as a proxy for the genetic identifier. If the tested cell did not show light-evoked activity (Figure 1A), the pipette was withdrawn from the cell by transiently increasing the positive pressure through the patch pipette and/or by applying a high voltage zap pulse and the search for another neuron was resumed. This cycle could be repeated quickly (~5 seconds per cell) with the same pipette until encountering a cell with robust light-evoked activation (Figure 1B). The positive pressure was then released to obtain either the loose-patch configuration to record the spiking activity of the cell of interest, or to establish a gigaohm seal for whole-cell recording and to gain intracellular access. Following the recording, the cell was labeled with neurobiotin, either by diffusion from the pipette in whole-cell configuration or by applying current pulses through the pipette in the case of loose-patch recordings. This allowed post hoc histological analysis and reconstruction of recorded cells providing information on their precise anatomical location, morphology, immunochemical identity, and confirmation of ChR2 expression (Figure 1C).
Figure 1. Strategy for in vivo recording and labeling of genetically defined cell types in the brain.
The conditional expression of ChR2 in cell types of interest renders genetically defined neurons within a network responsive to light, which can be exploited for their identification. (A) Employing the standard blind patching approach, glass electrodes (carrying an optical fiber inside for light stimulus delivery) are slowly advanced within the structure of interest until a cell is encountered. (B) ChR2-expressing cells fire reliable, time-locked spikes, upon light stimulation (right). Once a ChR2-expressing cell has been identified, its activity profile can be studied. (C) ChR2-expressing cells are labeled with neurobiotin, and post hoc histological analysis is employed to study its morphology (e.g., axonal projection patterns) and precise anatomical localization (e.g., layer, column, nucleus, etc.).
Optical activation and identification of genetically defined, ChR2-expressing neurons
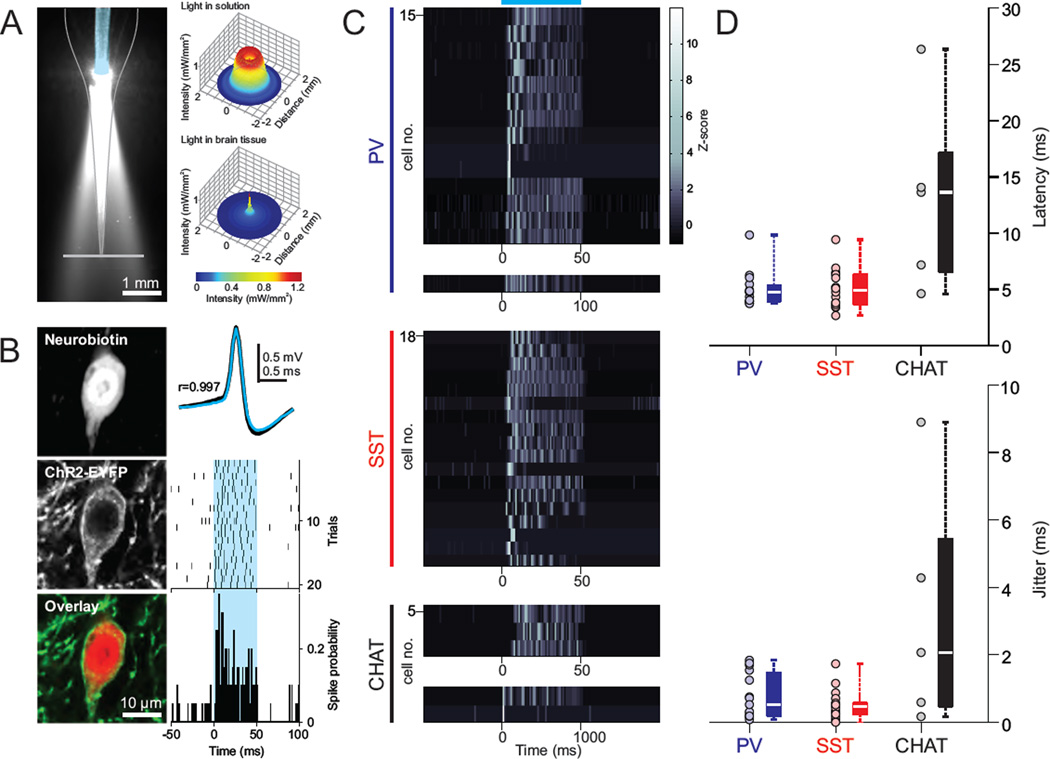
In order to use our approach at any location in the brain, a consistent and sufficient light delivery at the recording location is required for the proper stimulation and identification of ChR2-expressing cells. Since light intensity steeply decays as it travels through brain tissue (Stark et al., 2012) surface illumination is not suited for stimulation of deep brain regions because it does not allow sufficient light delivery to target cells. To circumvent this issue, focal light sources have been coupled to extracellular recording probes (Lima et al., 2009, Royer et al., 2010, Anikeeva et al., 2012) or to glass pipettes (Stuhmer and Almers, 1982, LeChasseur et al., 2011, Katz et al., 2013), and have been shown effective to stimulate ChR2-expressing cells. Following this strategy, we inserted an optical fiber coupled light source into our recording pipette, which ensured the consistent delivery of sufficient light intensities at the tip of the pipette where cells are contacted (Figure 2A, left). Measurements of the light distribution profile from this source inside of the pipette exhibited an expected geometric dispersion in solution (13° from vertical axis; (Stark et al., 2012)). Considering the effect of light divergence, as well as scattering in brain tissue as a function of distance, estimations of light intensity reaching the pipette tip suggested that contacted cells are exposed to 0.9–1.4 mW·mm−2 of blue light (at λ = 473 nm) during our standard stimulations (Figure 2A, right; (Stark et al., 2012)). This pipette tip intensity range was sufficient to drive ChR2-expressing cells to spike (Figure 2B).
Figure 2. Optical activation of genetically defined, ChR2-expressing neurons.
(A) Cone of blue light emitted by the optical fiber (blue false color) located inside of the glass pipette. Right top, cross-sectional light intensity profile (source: blue 473 nm laser light) at the pipette tip, estimated from the light power measured along the gray plane in the left image (75 mW·mm−2 at tip of fiber). Right bottom, estimation of light intensity at the tip of pipette inside the brain. Light intensity is corrected for power attenuation due to scattering, geometric dispersion and distance from light source (Stark et al., 2012). (B) Representative example of light-evoked activity for a post hoc histologically verified ChR2-expressing cortical interneuron in the PV-Cre driver line. Left, fluorescence and merged images of the neurobiotin staining and ChR2 expression of the recorded cell. Right top, spontaneous (black) and light-evoked (light blue) spike waveforms are highly correlated (Pearson correlation coefficient, r = 0.997). Raster (middle) and peristimulus histogram of spike probability (bottom; bin: 1 ms) for 20 trials of light stimulation of the recorded cell. Blue shaded area represents light stimulus. (C–D) Z-scored peristimulus histograms, latency and jitter to first spike of all histologically verified ChR2-expressing cells in the PV, SST and CHAT Cre driver lines. Light stimulation intensity (at pipette tip): 0.9–1.4 mW·mm−2. Box plot central bar represents the median, and edges represent the 25th and 75th percentiles of the data set. Blue horizontal line represents light stimulus.
In other extracellular recording configurations, such as fiber-coupled tetrodes and silicon probes, multiple cells can be recorded simultaneously from the same electrode. In such configurations, brief light stimuli of weak intensities are preferred so as to minimize the chances of activating multiple ChR2-expressing cells, which could distort recorded signals and interfere with unit isolation protocols (Roux et al., 2014). However, as the patch recording configuration isolates the activity from a single recorded cell, we were able to exploit the dependency of light-evoked responses on the duration and intensity of the light stimulus for the robust identification of different ChR2-expressing neuronal populations (Figure 2 and S1). Importantly, as we illustrate below for different cell types, our recordings of ChR2-expressing cells retained high spike waveform correlations between baseline and light stimulation epochs, even when the intensity of light stimulation was increased (Pearson correlation coefficient, r > 0.98, Figure S3–S5), demonstrating that our light stimulus did not distort the recorded signals and that a single cell is identified, recorded and labeled in these experiments.
We first tested the utility of our method in mice conditionally expressing ChR2 in various subtypes of neocortical GABAergic interneurons (INs). We focused on INs since, despite being present throughout the cortex, they represent a minority of cortical neurons (10–15% in rodents; (Meyer et al., 2011)), and consist of highly specialized subpopulations (Rudy et al., 2011). Major efforts are focusing on studying the functional ‘division of labor’ among cortical INs in vivo using recently developed genetic tools. However, most studies have focused on superficial layers or have strictly used genetic tagging as sole identifier, which is insufficient for the unambiguous identification of specific INs (Huang, 2014).
We targeted ChR2-expressing neocortical neurons in the deep layers of PV or SST Cre driver mice. Together these IN groups represent 70–90% of deep neocortical INs (Rudy et al., 2011). To identify ChR2-expressing neurons online, we first focused our analysis on three characteristics of their light-evoked activity: (a) large and reliable light-evoked increase in firing rate (PV: 5.8 ± 2.4 max z-scored peristimulus spike probability, n = 15; SST: 5.6 ± 1.9 max z-scored peristimulus spike probability, n=18), (b) short latency light-evoked responses (PV: 5.0 ± 1.6 ms, n = 15; SST: 5.0 ± 1.7 ms, n=18), and (c) low jitter of first light-evoked spike (PV: 0.8 ± 0.7 ms, n = 15; SST: 0.5 ± 0.4 ms, n=18; see Figure 2C–D, for all histologically confirmed ChR2 expressing cells and Figure S1A, for all recorded cells, including cells that were not histologically recovered). Importantly, we found that observing at least one of these criteria was enough to rapidly and reliably identify ChR2-expressing GABAergic neurons (Figure S1B), as confirmed by the histological examination of ChR2 expression of all labeled and successfully recovered cells (Figure S3–S5). PV INs exhibit fast spiking (FS) electrophysiological phenotypes. Consistent with this, we found that all cells in which we could reliably elicit light-evoked spiking had brief spike waveforms (Figure 2B and 6). On the other hand, SST cells, largely non-overlapping with PV, are more heterogeneous in terms of electrophysiological properties (Ma et al., 2006). In line with this, we observed light activation of ChR2-expressing SST cells with a larger variety of spike waveforms than PV cells (Figure 6). Both genetic groups had comparable light evoked responses (Figure 2C–D).
Figure 6. Comparison of spike waveform properties of genetically and morphologically identified cortical neurons.
(A) Normalized spike waveforms of all genetically and morphologically identified cortical neurons in the PV and SST Cre driver lines compared to putative pyramidal regular spiking cells recorded blindly (range: 350–1100 µm below pia, presumed layers 2/3–6). (B) Cumulative plot of peak-to-trough times of PV and SST subtypes and putative pyramidal cells and (C) comparison of peak-to-trough time and voltage ratios. Note that although PV and regular spiking putative pyramidal cells have narrow and mutually exclusive distributions, SST neurons have widespread and roughly continuous distribution with significant overlap with PV distribution. Also note that while SST spiny bipolar cells (red triangles) are similar to regular spiking cells, PV pyramidal cells (blue triangles) are hardly distinguishable from some PV basket cells (blue squares). Basket, Martinotti, non-Martinotti, spiny bipolar, and pyramidal cells were identified according to morphological criteria (see text). Non-pyramidal cells are histologically confirmed cells lacking apical dendrite and dendritic spines in which not enough axonal information was available for clear morphological assignment. Non-identified cells are histologically confirmed cells where only the soma was recovered.
Next, we tested whether we could generalize these observations to the targeting of other cell types. We recorded identified cholinergic neurons, which compose sparse populations in different subcortical nuclei (0.3% in striatum and 1.83–5% in the basal forebrain complex; (Peterson et al., 1999, Gritti et al., 2006)) using choline acetyltransferase (CHAT) Cre mice. In these experiments, we observed that light responses of some cholinergic cells could exhibit larger latency (13.2 ± 8.4 ms, n = 5) and jitter (3.2 ± 3.6 ms, n = 5) to first light-evoked spike, compared to PV and SST neurons (Figure 2C–D). Such light responses could lead to erroneously classify an encountered cell as an indirectly activated ChR2-negative neuron. We thus sought to understand the source of this different light response pattern, in order to define the light stimulation parameters by which this cell type could be reliably identified online. The larger latency of light-evoked responses could be due to insufficient expression of ChR2 and/or biophysical properties of cholinergic cells countering membrane depolarization and delaying spiking. In brain slices we observed that basal forebrain cholinergic neurons, carrying one or two copies of the ChR2 transgene in the Ai32 reporter mouse line, also had long delay responses to light stimulation (Figure S2A). Moreover, whole-cell recordings showed that cholinergic cells have high input resistance (262 ± 72 MΩ, n = 7), required little current injection to reach spike threshold (64 ± 38 pA, n = 7), and light-evoked photocurrents were much larger than rheobase currents (for 1.4–1.8 mW/mm2 blue light, peak current: ~600 pA, and steady-state current: ~400 pA; measured at −70 mV), ruling out low expression levels of ChR2 as the explanation for the observed latencies and jitters. However, the latency to first spike in response to both, intracellular current injection and light stimulation of ChR2-expressing cholinergic cells was dramatically affected by the resting membrane potential (Figure S2B–C). This suggests that, in our in vivo experimental conditions (mice under isoflurane anesthesia), the resting membrane potential range of cholinergic cells might allow for the subthreshold activation of a dampening conductance upon depolarization (Figure S2C; (Unal et al., 2012)). Based on these observations, we identified cholinergic neurons in vivo by relying on the larger magnitude and tighter temporal characteristics of their activity in response to light pulses of longer duration (1 second instead of 50 ms) and/or greater intensity (1.8 mW/mm2 instead of 0.9; Figure 2 and S1–S2). Cholinergic cells identified with these stimulation parameters were histologically confirmed as ChR2-expressing neurons (Figures S5).
We also investigated whether we could apply our strategy to reliably identify ChR2-expressing cortical excitatory neurons, a case in which, due to recurrent excitation, light stimulation can produce indirect synaptic activation of non-ChR2-expressing (ChR2−) cells, leading to false positives. We recorded and histologically confirmed neocortical ChR2+ and nearby ChR2− pyramidal neurons in the Thy1 Cre driver mouse line (Figure 3), which allows genetic access to a subpopulation of cortical pyramidal neurons mainly located in L5 (Dewachter et al., 2002). We observed distinct light activation patterns of these two cell groups by varying the light stimulus parameters. Notably, while the ChR2+ group showed sustained spiking throughout light pulses, only transient spiking was observed in ChR2− cells, presumably as a result of synaptic depression and/or strong recruitment of inhibition (Figure 3A). Consequently, short light pulses evoked similar responses in both groups and increasing pulse duration reliably disambiguates the two populations. Furthermore, the light intensity required to drive indirect responses of ChR2− neurons was slightly higher than that needed to reliably activate ChR2-expressing neurons (Figure 3B). Additionally, although the average latency of directly and indirectly evoked first spikes differed across the tested intensities, significant overlap was observed between their ranges (ChR2+: for 1.4 mW/mm2, 3.0 ± 1.1 ms, for 1.8, 2.1 ± 0.6 ms, n = 6; ChR2−: for 1.4 mW/mm2, 8.7 ± 9.0 ms, for 1.8, 4.9 ± 1.2 ms, n = 4, Figure 3C). The jitter of the first light-evoked spike could not clearly distinguish these two cell groups (Figure 3D). These observations stress the importance of observing multiple aspects of the magnitude and temporal characteristics of light-evoked activity of specific ChR2-expressing neuron types and the usefulness of our recording configuration that enables testing different stimulation conditions (intensity and duration) in order to derive a reliable set of parameters for their in vivo identification.
Figure 3. Varying light stimulus duration disambiguates ChR2-expressing glutamatergic and synaptically driven neurons.
(A) Left, example of histological confirmation of a recorded ChR2-expressing (top) and a ChR2-negative (bottom) glutamatergic neocortical Thy1 pyramidal neurons. Right, raster of light-evoked activity of corresponding cells for 5 and 100 ms light pulses (tip intensity: 1.4 mW/mm2). (B) While histologically confirmed ChR2-expressing (n = 6) and ChR2-negative (n=4) cells show similar response magnitude to brief (5–10 ms) light stimulation, only ChR2-expressing cells fire continuously during pulses of longer duration (50–100 ms). (C) Population latency of first light-evoked spike of ChR2-expressing neurons shows a trend for shorter values, although these do not clearly segregate from ChR2-negative group values. (D) Population jitter values of first light-evoked spike of ChR2-expressing and ChR2-negative neurons are small and overlap significantly.
Success rate of the proposed method
To assess the efficiency of our strategy in encountering and identifying sparsely distributed populations of neurons, we quantified the proportion of recorded ChR2-expressing neurons relative to the number of sampled cells in deep neocortical layers of both PV and SST Cre mouse lines. We could typically obtain targeted recordings in every experiment. On average we were able to record from one PV IN for every 2 ± 1 pipette insertions (n = 6 animals), and for every 3 ± 1 pipette insertions (n = 5 animals) for SST INs. On average, 10 ± 7 penetrations per craniotomy (n = 13 animals) were possible while maintaining good histological conditions for morphological reconstructions (average pipette penetration separation = 150µm, craniotomy diameter = 0.5–1 mm, generally 1 craniotomy per animal, Figure S6). Overall, we were able to record up to 7 PV- or SST-expressing INs per animal (PV: 5 ± 1 ChR2-expressing cells/animal, n = 5 animals; SST: 3 ± 2 ChR2-expressing cells/animal, n = 8 animals), by sampling on average 61 ± 31 and 160 ± 53 cells/animal from PV and SST Cre mice, respectively.
An important aspect for consideration is that while multiple pipette penetrations are necessary for our strategy, excessive number of penetrations in a small area (such as one specific cortical column) can lead to tissue damage. We point to strategies that can be undertaken to minimize or eliminate these effects. For instance, in our experiments, pipettes with long and narrow shanks were used to minimize damage. A typical pipette used for cortical recording had an outer diameter of ≤40 and 100 µm, when measured 300 and 1000 µm from the tip, respectively. Moreover, in the case of attempting targeted recordings in a single barrel column, we were able to reach the target area diagonally (rather than entering the pipette in an angle normal to the pia), in which case the craniotomy was made next to the target area (Figure S6). In such a preparation, only the narrow portion of the pipette enters the area of interest, leading to virtually no damage of the target site after multiple pipette penetrations (Figure S6).
The probability of encountering specific INs was roughly proportional to their share of the total cortical population (PV: 8.6 ± 3.7%, n = 5, in our study versus ~5.8% in layers 4–6; SST: 2.8 ± 0.5%, n = 4, in our study versus ~2.6%; (Meyer et al., 2011)). Notably, the fast sampling and detection of light-responsiveness of a neuron (generally occurring within 5 seconds for both PV and SST INs) enabled us to rapidly resume the search until a ChR2 expressing cell was found, resulting in a significantly higher yield than that of the traditional blind patching approach. Encountered interneurons could be recorded for several minutes to hours allowing further characterization of light responsiveness, spontaneous and/or sensory evoked activity (Figure 4–7), as well as performing filling protocols (see below).
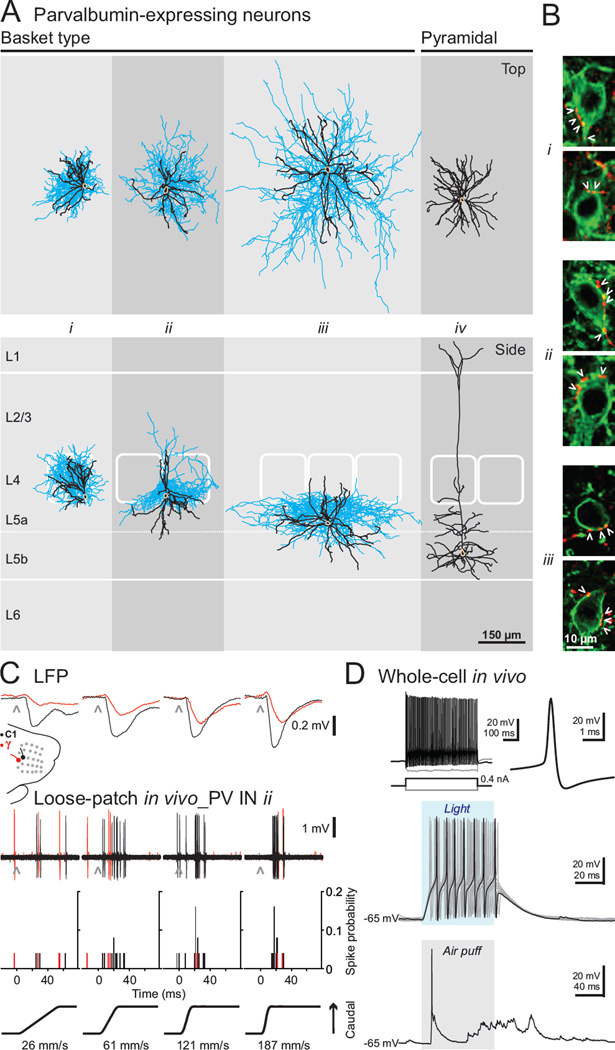
Figure 4. Morphological and functional characterization of cortical PV neurons.
(A) Top and side perspectives of the morphology of PV INs and thick-tufted pyramidal cell (beige: somas, black: dendrites, blue: axons). For illustrative purpose, laminar and columnar positions and sizes of morphologies where proportionally adjusted to fit a background prototypical barrel cortex of 1 mm thick from pia to white matter; barrels have been outlined from multiple histological sections containing and flanking somatic position. Only dendrites are shown for cell iv. (B) Basket terminals of labeled cells i–iii (red) show multiple axonal varicosities (arrowheads) in the vicinity of somas of neighboring ChR2-expressing unrecorded cells (green, two examples per filled neuron). (C) Example of sensory responses of a morphologically identified PV-expressing basket cell (cell ii in panel A) of the primary somatosensory cortex. C1 (black traces) and gamma (red traces) whiskers were deflected caudally (black) at varying velocities (26–187 mm/s) while simultaneously monitoring whisker-evoked LFP (position: ~500 µm deep, presumed layer 4, and ~200–250 µm laterally from the cell ii recording site) and recording the spiking activity of basket cell ii in the loose-patch configuration. (D) Intracellular recording of a fast spiking neuron in the primary somatosensory cortex. Top, firing pattern (left) and spike waveform (right) in response to intracellular current injections. Middle, light-evoked depolarization and spiking of the recorded cell (blue shaded region represents light stimulus). Bottom, membrane potential fluctuations in response to a whisker pad air puff (gray shaded region represents the 100 ms, 20 psi air puff, delivered over the C and D whisker rows).
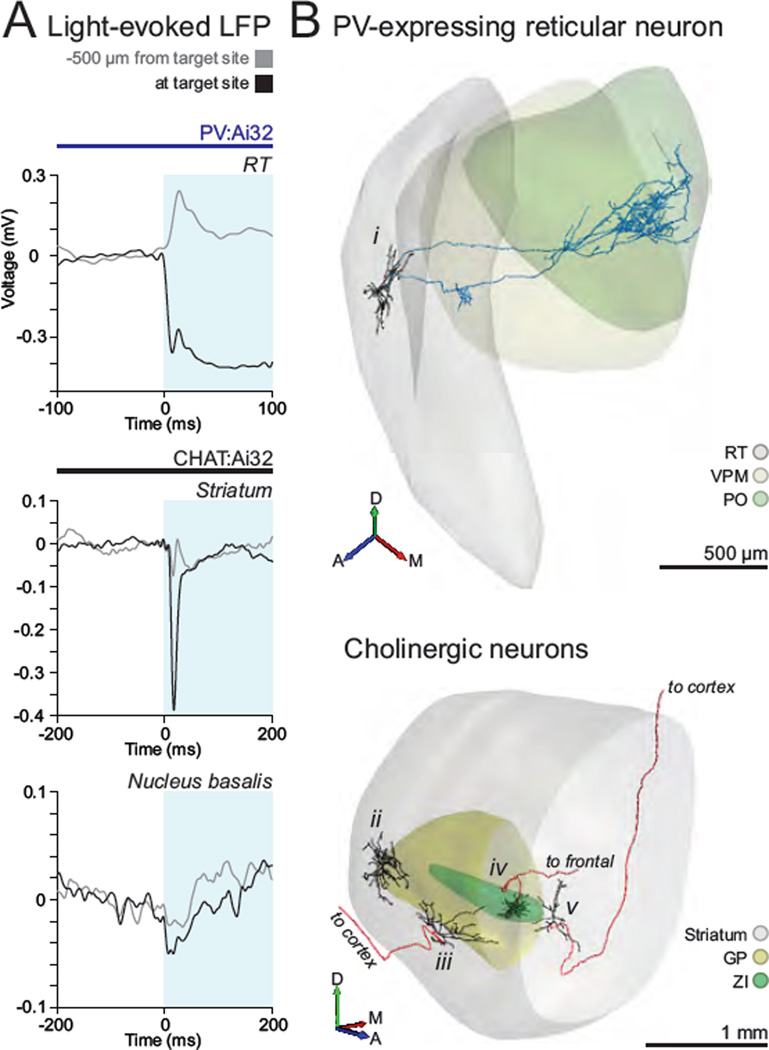
Figure 7. Recording and labeling of genetically defined cell types in several subcortical structures.
(A) A combination of stereotaxic procedures with mapping of light-evoked LFP can be exploited to pinpoint the location of structures of interest, such as reticular nucleus of the thalamus (RT) (top), the striatum (middle), and the cholinergic nucleus basalis (bottom). Note the large sustained or transient light-evoked field potentials at the site where genetically defined cells have subsequently been recorded and labeled (black traces), and how it evolves from what is observed at a distal, more superficial recording location (grey traces). Blue shaded area represents light stimulation. (B) Top, 3D rendition of the somatodendritic (black) and axonal (blue) projections of a GABAergic neuron of the thalamic reticular nucleus in the PV Cre driver line (A, anterior, M, medial, D, dorsal). Bottom, 3D rendition of somatodendritic and axonal projections of cholinergic neurons of the striatum (ii), nucleus basalis (iii and v), and zona incerta (iv) in the CHAT Cre driver line.
Morphological characterization is important to our strategy because of the heterogeneity that exists within current genetically based categories. However, in our early attempts, GABAergic INs were difficult to label by the juxtacellular labeling protocol, as compared to neocortical pyramidal neurons (data not shown). We therefore investigated conditions to increase the probability of histologically recovering INs and improving the quality of the morphological labeling for reconstructions. We found that tightly regulating the DC current to control the total number of elicited spikes, improved the iontophoresis of neurobiotin, the recovery rate and the morphological quality of filled INs (see Experimental Procedures and Figure S6–S7). By employing this strategy, we were able to histologically recover ~75% of our recorded PV and SST neurons of which half could be accurately reconstructed (for axons, length ranging from 20 to 80 millimeters, 34.7 ± 20 mm, n = 12; for dendrites, 3.9 ± 1 mm, n = 12). Altogether, these results demonstrate that our method enables efficient, reliable and unambiguous targeted recording and labeling of sparsely distributed neuronal populations.
In vivo recording of morphologically identified neocortical PV neurons
To evaluate the use of our approach to study specific cell types, we recorded and reconstructed a subset of PV neurons in the somatosensory barrel cortex in vivo (Figure 4). The presence of numerous perisomatic axonal puncta, or baskets, onto neighboring cell bodies (Figure 4B) identifies all our PV INs as basket cells, consistent with the previous observation that the vast majority of PV neurons in layers 4–6 are basket cells (Kisvarday, 1992). We observed that cells with similar somatic position could exhibit very different axonal and dendritic territories in both the laminar and the columnar axes (Figure 4A; compare the location of axons and dendrites of cell i versus ii in relation to their principal versus surrounding barrels). Dendritic and axonal fields define circuit affiliation, emphasizing the merit of obtaining detailed morphological information in addition to somatic position, molecular and electrophysiological properties. Moreover, given the stability of the recordings, it was possible to perform functional studies on these sparse cell types. For example, we used the loose-patch configuration to record the whisker-evoked responses of PV basket INs in the somatosensory barrel cortex in response to single whisker deflections of different orientations and velocities (Figure 4C; showing cell ii from Figure 4A) before proceeding to the filling protocol. The recordings could also be combined with local field potential (LFP) measurement using another electrode placed nearby for more global assessment of local network state. This highlights the potential of our strategy to bridge our understanding of the contribution of precisely identified cell types to local network activity and function.
We were also able to extend our strategy to carry out intracellular recordings in the whole-cell configuration. Figure 4D shows the intrinsic properties, light-evoked membrane depolarization and sensory-evoked synaptic responses to whisker pad stimulation of a deep layer PV FS IN. Establishing the requisite gigaohm seal for whole-cell configuration requires a clean pipette tip. However, testing multiple cells for ChR2 expression can compromise this condition. We found that maintaining positive pressure on the pipette while searching and testing multiple cells for ChR2 expression allowed us to successfully establish gigaohm seals even after 10 encountered and tested cells (see Experimental Procedures). Thus, our approach also allows investigation of the contributions of synaptic mechanisms and intrinsic membrane properties of genetically identified neurons to in vivo network activity, at locations beyond the TPTP recording range.
Importantly, our method eliminates identification ambiguities inherent to relying on spike waveform characteristics of the recorded cell or on molecular-genetic identification alone. For instance, we recorded and labeled light-responsive, PV-expressing thick- and slender-tufted pyramidal cells in neocortical layer 5 (Figure 4A, cell iv; (Tanahira et al., 2009)). These pyramidal cells exhibited spike waveforms that were undistinguishable in peak-to-trough times and ratios to those of some morphologically identified basket INs, making the morphological characterization in this case critical (Figure 6).
In vivo recording of morphologically identified neocortical SST neurons
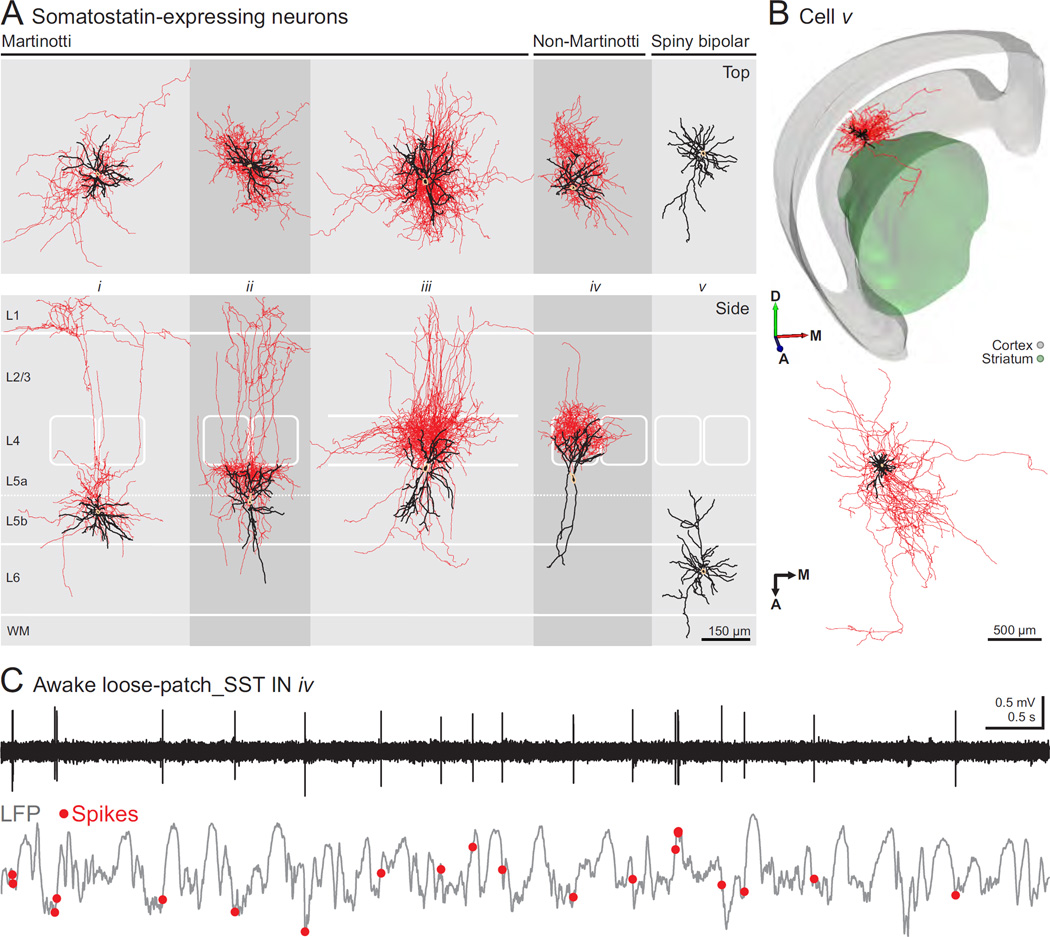
Applying our approach to neocortical SST neurons, a group exhibiting large morphological and electrophysiological diversity, we were able to further validate the relevance of our approach as a robust method for dissecting the heterogeneity of cell types within this molecular group. These experiments are the first recordings of morphologically identified SST INs of deep cortical layers in vivo. Based on morphological reconstructions, we identified Martinotti and non-Martinotti cell types within our recorded and labeled sample of SST INs in deep cortical layers (Figures 5). Here, Martinotti INs were defined as having ascending axonal projections and branching in cortical layer 1, in some cases with an additional plexus in layer 5 (Figure 5A, cells i–ii; (Xu et al., 2013)), or in layer 4 (Figure 5A, cells iii; (Wang et al., 2004)). Non-Martinotti SST INs were defined as cells without axonal projections to layer 1 (Figure 5A, cell iv; (Ma et al., 2006)). In our sample, cells of these two categories were identified in roughly equal proportions in neocortical layers 5–6 (Figure S6E). Consistent with previous reports in vitro (Ma et al., 2006) and in vivo (Kvitsiani et al., 2013), SST neurons had a broad range of electrophysiological features (Figure 6). Based on the established morphological identity of the recorded cells, we observed that Martinotti cells tended to exhibit broader spikes than non-Martinotti neurons, although there was not a complete segregation of these two morphological groups according to the peak-to-trough times and ratios (Figure 6).
Figure 5. Morphological and functional characterization of SST-expressing INs.
(A) Top and side perspectives of the morphology of SST INs and a spiny bipolar neuron (beige: somas, black: dendrites, red: axons). Both Martinotti and non-Martinotti cells are shown (see text for definition). (B) Top, 3D rendition of spiny bipolar cell v showing intracortical and striatal axonal arbor. All of the intracortical axon was located in infragranular layers. (A, anterior, M, medial, D, dorsal). Bottom, top view (normal to pia) of the same cell showing anterodorsal and mediolateral span of the axon (beige: soma, black: dendrites, red: axon). (C) Spontaneous activity of morphologically identified SST-expressing, non-Martinotti cortical interneuron iv (top) and simultaneous LFP recording in awake, head-restrained mouse.
Additionally, we recorded and labeled layer 6 SST spiny bipolar excitatory neurons (Figure 5A, cell v), consistent with a previous report of the expression of somatostatin mRNA in this neuronal type (Andjelic et al., 2009). These cells showed characteristic vertically and diagonally oriented apical dendrites and extensive intracortical axonal arborization, as well as branches reaching the striatum and corpus callosum (Figure 5A–B).
We also tested whether our strategy could be used to dissect the activity of different circuit elements of a genetic group in an awake preparation. We established the loose-patch configuration in vivo and recorded the spontaneous activity of SST INs and LFP in awake head-fixed animals (Figure 5C shows a representative trace from cell iv in Figure 5A). We found our success rate of establishing such recordings to be similar to that of anesthetized preparations (anesthetized: 4.3 ± 3 recorded cells/experiment, n = 3 animals; awake: 2.8 ± 1.8 recorded cells/experiment, n = 5 animals). As previously described for superficial SST INs, we observed that spiking activity of deep layer SST INs was lower in anesthetized versus awake conditions, (anesthetized: 0.28 ± 0.18 spikes·s−1, n = 11; awake: 4.20 ± 1.78 spikes·s−1, n = 11, mean ± SEM; (Adesnik et al., 2012)).
In vivo recording and labeling of genetically defined subcortical neurons
We sought to extend our strategy for the study of defined cell types located in deep subcortical structures. In order to optimize the search for ChR2-expressing neurons at such distant and structurally complex sites, we took advantage of a combination of stereotaxical and electrophysiological mapping approaches. Under stereotaxic guidance, we recorded the light-evoked LFP responses with a patch pipette throughout subcortical space to precisely circumscribe the target structures where ChR2-expressing cells are located (Figure 7A and S6F). Light-evoked LFPs were recorded as fast-onset negativities associated with a group of ChR2-expressing activated neurons. Such LFP signals could be sustained or transient depending on the genetic group being activated and the structure where LFPs were recorded (Figure 7A). Once a target structure was located, we proceeded to search and reliably encountered ChR2-expressing cells within the circumscribed region using the same approach as above.
Following this mapping strategy, we recorded and labeled PV cells of the reticular nucleus of the thalamus (Figure 7B, cell i). The example cell shown in Figure 7 primarily innervated the posteromedial nucleus of the thalamus, and to a lesser extent, the ventroposterior medial complex, as described previously for this morphological type of the somatosensory group in this complex (Pinault and Deschenes, 1998). Additionally, we recorded and labeled cholinergic cells of the striatum (cell ii), at the boundary between globus pallidus (GP) and the dorsal striatum (cell iii), the zona incerta (cell iv), and the nucleus basalis (cell v, Figure 7B). As described above, cholinergic cells constitute a minute proportion of the neuronal populations in each of these structures. Our targeted approach significantly enhanced the yield of recording and labeling these cell types (1–2 ChR2-expressing CHAT cells/animal versus 0.1 CHAT cells/animal in (Lee et al., 2005)). Knowledge of the precise anatomical position of CHAT cells is critical in this region (Munoz and Rudy, 2014), as adjacent cholinergic cells in the striatum and at the GP border are morphologically distinct and integrate into different brain circuitry; while cholinergic cells of the striatum project locally (Figure 7b, cell ii); cholinergic cells at the border with GP project to the cortex (Figure 7B, iii).
Discussion
In the present study, we described the development and implementation of a robust and efficient technique for the targeted recording of the activity of cortical and subcortical neurons in vivo allowing the association of their genetic, morphological, anatomical, and electrophysiological features. Such an integrated analysis enables enhanced precision in the identification of the neuronal type being recorded and the detailed dissection of cell-type specific activity patterns during different network processing and behavioral contexts.
We demonstrated its utility by in vivo recording and labeling PV and SST neurons of deep layers of the neocortex, PV-expressing thalamic reticular neurons, and cholinergic cells of several subcortical structures, and we showed that our method can be used to study subpopulations of cortical excitatory neurons. Through a combination of slice and in vivo recordings, we established protocols and parameters allowing reliable identification of different types of ChR2-expressing neurons with standard light responses (PV and SST neurons), delayed light responses (cholinergic neurons), as well as distinguishing between directly and indirectly activated neurons in the context of light-evoked recurrent excitation (Thy1 excitatory neocortical neurons). We demonstrated the feasibility of applying this approach for in vivo awake and anesthetized functional studies that characterize the activity of distinct circuit elements. Furthermore, we were able to obtain intracellular recordings of genetically defined types, extending the spatial range for such recordings beyond that afforded heretofore by visual guidance. Importantly, sparse genetic groups could be targeted efficiently. After having established optimal conditions, we had a very high success rate generally leading to the recording of many cells per animal (e.g., obtaining up to 7 genetically defined GABAergic INs in deep cortical layers), of which more than half were successfully labeled. Using our approach, we observed a considerable morphological diversity of functional relevance for deep neocortical PV and SST INs, which has been suggested to serve important computational functions (Kisvarday, 1992, Xu et al., 2013).
Comparison to current strategies for the recording of identified cell types in the brain
Previous efforts combining blind recordings with histological identification have led to significant insights regarding the association of the activity of specific neuronal types to network oscillations, state-dependent engagement and their contribution to sensory processing and behavioral responses (Lee et al., 2005, Klausberger et al., 2003, Burgalossi et al., 2011). However, the blind recording strategy poses serious disadvantages for the study of sparsely distributed neuronal subtypes, requiring laborious and time consuming efforts to obtain a numerically meaningful sample of the neuronal types of interest. In addition to attaining similar results to this method, our approach represents a dramatic yield improvement, facilitating accessibility to biologically important questions attempting to link cellular physiology, network activity and behavior.
Targeted approaches like calcium imaging and TPTP have been the most direct ways of recording the activity of genetically defined neurons in vivo, yet are limited in their spatial working range. As these methods rely on two-photon microscopy to visualize target neurons, only superficial structures in the brain have been investigated. In order to perform imaging in deeper brain structures, highly invasive procedures have to be undertaken such as large device implantation into the recording site or removal of the overlying brain tissue (Marshel et al., 2012, Andermann et al., 2013). In the case of TPTP, an additional factor limiting its recording depth range is the challenge of positioning the recording electrode in the path leading to deep target neurons. Our strategy is not subject to these limitations. On the other hand, visually guided recordings allow the selection of particular neurons or even subcellular compartment to be probed (Kitamura et al., 2008), which would be difficult using our technique. Furthermore, unlike calcium imaging, our method is not applicable to population activity recordings and longitudinal studies (Margolis et al., 2012). Also, while our approach opens the possibility for whole-cell recordings at deeper recording sites than TPTP, we should emphasize that this implementation remains a low yield effort. For instance, in the case of PV cells, considering the PV IN distribution, the success rate of gigaohm seals per encountered PV IN, the rate of conversion from gigaohm seal to whole-cell configuration, and the number of tests possible with a single pipette, we required on average 7 pipettes to obtain a whole cell recording from a PV IN.
Another generation of methods for targeted recordings is based on the principle of optical tagging, where optic fiber-coupled tetrodes and silicon probes are used to identify and record light-responsive neurons conditionally expressing ChR2 (Lima et al., 2009, Anikeeva et al., 2012). This approach made possible to simultaneously survey the activity profiles of genetically defined neuronal populations throughout the brain in freely behaving animals. However, this approach does not offer the morphology nor precisely informs on the anatomical location of the recorded cells. The use of metal electrodes provides coarse spatial resolution, with recorded cells being located up to 60 µm from the recording site (Buzsaki, 2004). Considering the morphological heterogeneity described here and elsewhere and the structural complexity of most synaptic pathways and microcircuits, this represents a major limitation. Although these methods have made great use of the combination of genetic identity with spike waveform analysis (Kvitsiani et al., 2013), our results show that further refinement is often needed (Figure 6). Additional limitations, such as spike sorting accuracy and bursting cell types exhibiting drastic spike waveform changes, pose a general challenge for the assignment of light-evoked spike properties to a given recorded unit (Buzsaki, 2004). Moreover, identification under conditions in which light stimulation drives multiple ChR2-expressing cells (detected as a compound spike), indirect activation in the context of excitatory cells or rebound spikes can impede the robust identification of light responsive cells and require special attention (Roux et al., 2014). As we have shown, our approach can circumvent these issues by providing higher precision in the identification of ChR2-expressing cells based on their activation profiles in response to a range of light stimulation parameters, as well as providing their anatomical position, morphological and electrophysiological characteristics. Importantly, a systematic analysis of morphological identity vis-à-vis electrophysiological characteristics of cell types with our method can lead to a better scheme of correlations that could facilitate the identification of blindly recorded or optogenetically tagged units.
Outlook
As we continue to document the cellular and synaptic organization of neural networks, the challenges remain in ascribing computational contributions to the distinct cellular elements that compose them, and understanding how these participate in network processing, behavior and cognition. Given the high yield and robustness of our approach for more comprehensive identification of cell types, it is now possible to efficiently survey the activity patterns of precisely identified neurons in vivo. Additionally, it might be possible in the future to expand our method to freely behaving preparations by combining our strategy with recent developments in miniaturized head-anchored microdrives and mini-preamplifiers (Long and Lee, 2012). Our strategy could also be combined with single-cell electroporation for genetic manipulations that permit single-cell analyses of input-output connectivity (Marshel et al., 2010).
Experimental Procedures
Mice
For in vivo experiments, 7 PVcre; Ai32flox/+ (JAX, stock numbers 008069 and 012569), 8 SSTcre; Ai32flox/+ (JAX, stock numbers 013044), 4 CHATcre; Ai32flox/+ and 1 CHATcre; Ai32flox/flox (JAX, stock numbers 006410), 2 Thy1cre (JAX, stock number 006143) male and female mice were used (2–6 months old). For in vitro experiments, 4 CHATcre; Ai32flox/+ and 1 CHATcre; Ai32flox/flox mice were used (2–6 months old). Heterozygous mice for the Cre allele were used for experiments. Mice were housed under reversed light-dark (12 hours-12 hours) cycles. Surgical procedures for anesthetized, awake and brain slice preparations are detailed in Supplemental experimental procedures. All procedures were carried out in accordance the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and with the approval of the Institutional Animal Care and Use Committee of the New York University School of Medicine.
In vivo electrophysiology and data acquisition
For loose patch recordings, borosilicate glass pipettes (Sutter Instruments, tip outer diameter: 1.5–3 µm, resistance: 2.7–9.5 MΩ) were used and filled with a solution containing 1.5–3% neurobiotin (Vector Laboratories) in normal rat ringer (NRR, in mM: 135 NaCl, 5.4 KCl, 5 HEPES, 1.8 CaCl2 dihydrate, 1 MgCl2 hexahydrate, pH 7.3 with NaOH, osmolarity: ~300 mmol/kg). For whole cell recordings, glass pipettes (resistance: 5–7 MΩ) were used and filled with a solution containing (in mM): 135 potassium gluconate, 4 KCl, 10 HEPES, 10 phosphocreatine, 4 MgATP, 0.3 NaGTP, pH 7.3 with KOH, osmolarity: 290–295 mmol/kg. Pipettes were pulled with long, narrow shanks (typically 4 mm taper for cortical recordings, 7 mm taper for subcortical recordings) to minimize damage. To couple light stimulation with recordings, we used a two port pipette holder (Warner Instruments) in order to insert an optical fiber inside the patch pipette in addition to the AgCl wire while being able to maintain constant pressure. The optical fiber (BFL37, 200 µm diameter, Thorlabs) was positioned in nearest proximity to the tip of the pipette (internal sleeve), while avoiding obstruction. The optical fiber was coupled to a 473 nm laser (source power: 1.6–2.4 mW, adjusted depending on the shank length to match the estimated tip intensity range of 0.9–1.4 mW·mm−2; Shangai Dream Laser), and illumination duration was controlled by a TTL-gated shutter (LS3, Vincent Associates).
Electrophysiological measurements were carried out with an Axopatch 200B amplifier (Molecular Devices) for single cell recordings. Carbon fiber microelectrodes (Carbostar-1, Kation Scientific) and a microelectrode AC amplifier (1800, A-M Systems) were used for LFP recordings. Extracellular spikes and intracellular membrane potentials were low-pass filtered at 10 kHz and digitized at 20 kHz (Digidata 1440A, Molecular Devices). Broadband LFP signals were band-pass filtered at 0.1 Hz–10 kHz and digitized at 20 kHz. Electrodes, angled 60–90° relative to horizontal, were introduced into the brain with positive pressure (50 mbar) and advanced slowly (~7 µm·s−1) into the target recording area. Positive pressure was then reduced to the searching value (20–30 mbar) until a cell was encountered, detected as an increase in pipette resistance on the voltage-clamp configuration. Then, 50 ms to 1 s pulses of blue light were presented at rate of 0.5–10Hz for the detection of light-evoked activity of encountered neurons, depending on the cell type being stimulated. If light-evoked spiking was observed, immediate release of positive pressure and slight suction were applied followed by electrode position adjustments (~5 µm retraction) to establish either a stable loose-seal (10–40 MΩ) for juxtacellular recordings or a gigaohm seal (1–3 GΩ) followed by the whole-cell configuration for intracellular recordings.
Pipettes were changed if an increase in pipette resistance measured in the bath (>3 MΩ and >0.2 MΩ from the baseline resistance for loose-patch and whole-cell configurations, respectively) was observed upon entering brain tissue or during the search. These changes in pipette resistance are indicative of tip clogging, which can be minimized by maintaining a small, constant positive pressure (20–30 mbar). This was particularly important for whole-cell recording, for which a clean pipette tip is necessary for the formation of a gigaohm seal. When attempting to get a gigaohm seal, if no light-evoked activity was observed for a given encountered cell, the pipette was retracted (~10 µm) and positive pressure transiently increased (40 mbar) to minimize chances of further contact with the encountered cell that might affect pipette tip conditions. Then, we resumed the search by advancing the electrode past the encountered cell by applying a voltage zap pulse after which the positive pressure was returned to searching value. Once a light responsive cell was encountered, positive pressure was released leading to the formation of the gigaohm seal. Under such conditions, the probability of gigaohm formation after sampling 1–3 encountered cells (46%; n=13) was comparable to the probability after sampling 5–10 encountered cells (50%; n=6), confirming the suitability of this approach to probe multiple cells while maintaining proper pipette conditions for whole-cell recordings. The success rate in the conversion from gigaohm seal to whole-cell configuration was ~50% (n = 5). The probability of encountering PV INs for whole-cell was similar to that observed with loose-patch recordings, suggesting that both pipette solutions did not differently affect our ability to detect ChR2-expressing neurons (7.9% for whole-cell versus 8.6% for loose-patch recordings).
Loose-patch recordings were considered for analysis if a stable seal was maintained, without substantial voltage drift on current clamp and if extracellular spike amplitude was at least 0.5 mV. Whole-cell recordings were pursued if the recording was stable, the cell had a resting membrane potential during down-states below −50 mV and overshooting action potentials.
Juxtacellular labeling
Following loose-patch recordings, the pipette was retracted (~3–6µm), with a consequent ~10% decrease in seal resistance. Then, 200 ms current pulses steps of 0.5 nA increments were injected every 2.5 seconds until the juxtasomal current injection threshold driving the cell to spike was found (1–40 nA). Current pulses (200 ms every 2.5 sec) representing 80% of this current threshold value were then used to fill the cell. Evoked firing rate was adjusted by carefully and constantly adjusting a DC current of positive or negative values to maintain reliable evoked spiking without damaging the cell, as seen by dramatic changes in spontaneous activity and/or action potential amplitude (Figure S7). Once filling protocol was completed (approximately 500 total trial sweeps, with substantial proportion of modulated sweeps amounting to ≤1000 current-evoked spikes), pipettes were slowly retracted while monitoring pipette resistance to ensure cell viability. Each pipette position was noted on the blood vessel map.
Data analysis
Analyses were conducted in MATLAB (MathWorks). For the analysis of light intensity in brain tissue, similar factors and equations as in (Stark et al., 2012) were considered. Briefly, the estimated transformation of light intensity from source to the tip of pipette was a function of: (a) the geometric dispersion of light observed in solution, and (b) light scattering and attenuation as a function of distance traveled in brain tissue (scattering factor of blue light as in (Stark et al., 2012)). For the analysis of light- and sensory-evoked spiking, recorded traces were band-pass filtered and spike times were determined as the peaks of well-isolated threshold crossings (≥ 3 standard deviations above background noise). These were then segmented into spike histograms or rasters (bin: 1 ms), according to the onset of light stimulus or whisker deflection. Peristimulus time histograms were computed as the probability of spike per bin over the total number of trials. In these analyses, we only considered light-evoked activity occurring during the window of light pulses, in order to avoid the interference that light-evoked rebound spikes pose for the reliable identification of ChR2-expressing cells. Data are presented as mean ± standard deviation, unless otherwise specified.
Supplementary Material
Acknowledgements
We are grateful to Lisa Roux, Illya Kruglikov, Soohyun Lee, Michael Long, György Buzsáki, Wen-Biao Gan, Gordon Fishell, and other members of the Rudy and Fishell labs for helpful discussions and comments on the manuscript, and Joseph Cichon for providing the Thy1Cre mice. This work was supported by NIH Grants R01NS30989 and P01NS074972 to BR and NRSA fellowship F31NS087919 to WM and a Graduate Research Fellowship to R.T. from NSERC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
W.M., R.T. and B.R. conceived the project. W.M. and R.T. performed all the experiments and analyzed data. W.M., R.T. and B.R. wrote the manuscript.
References
- Adesnik H, Bruns W, Taniguchi H, Huang ZJ, Scanziani M. A neural circuit for spatial summation in visual cortex. Nature. 2012;490:226–231. doi: 10.1038/nature11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann ML, Gilfoy NB, Goldey GJ, Sachdev RN, Wolfel M, Mccormick DA, Reid RC, Levene MJ. Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron. 2013;80:900–913. doi: 10.1016/j.neuron.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjelic S, Gallopin T, Cauli B, Hill EL, Roux L, Badr S, Hu E, Tamas G, Lambolez B. Glutamatergic nonpyramidal neurons from neocortical layer VI and their comparison with pyramidal and spiny stellate neurons. J Neurophysiol. 2009;101:641–654. doi: 10.1152/jn.91094.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anikeeva P, Andalman AS, Witten I, Warden M, Goshen I, Grosenick L, Gunaydin LA, Frank LM, Deisseroth K. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgalossi A, Herfst L, Von Heimendahl M, Forste H, Haskic K, Schmidt M, Brecht M. Microcircuits of functionally identified neurons in the rat medial entorhinal cortex. Neuron. 2011;70:773–786. doi: 10.1016/j.neuron.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Large-scale recording of neuronal ensembles. Nat Neurosci. 2004;7:446–451. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- Csicsvari J, Hirase H, Czurko A, Mamiya A, Buzsaki G. Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving Rat. J Neurosci. 1999;19:274–287. doi: 10.1523/JNEUROSCI.19-01-00274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewachter I, Reverse D, Caluwaerts N, Ris L, Kuiperi C, Van Den Haute C, Spittaels K, Umans L, Serneels L, Thiry E, Moechars D, Mercken M, Godaux E, Van Leuven F. Neuronal deficiency of presenilin 1 inhibits amyloid plaque formation and corrects hippocampal long-term potentiation but not a cognitive defect of amyloid precursor protein [V717I] transgenic mice. J Neurosci. 2002;22:3445–3453. doi: 10.1523/JNEUROSCI.22-09-03445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba P, Begum R, Capogna M, Jinno S, Marton LF, Csicsvari J, Thomson A, Somogyi P, Klausberger T. Ivy cells: a population of nitric-oxide-producing, slow-spiking GABAergic neurons and their involvement in hippocampal network activity. Neuron. 2008;57:917–929. doi: 10.1016/j.neuron.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritti I, Henny P, Galloni F, Mainville L, Mariotti M, Jones BE. Stereological estimates of the basal forebrain cell population in the rat, including neurons containing choline acetyltransferase, glutamic acid decarboxylase or phosphate-activated glutaminase and colocalizing vesicular glutamate transporters. Neuroscience. 2006;143:1051–1064. doi: 10.1016/j.neuroscience.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ. Toward a Genetic Dissection of Cortical Circuits in the Mouse. Neuron. 2014;83:1284–1302. doi: 10.1016/j.neuron.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Yizhar O, Staiger J, Lampl I. Optopatcher--an electrode holder for simultaneous intracellular patch-clamp recording and optical manipulation. J Neurosci Methods. 2013;214:113–117. doi: 10.1016/j.jneumeth.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Kisvarday ZF. GABAergic networks of basket cells in the visual cortex. Prog Brain Res. 1992;90:385–405. doi: 10.1016/s0079-6123(08)63623-7. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Magill PJ, Marton LF, Roberts JD, Cobden PM, Buzsaki G, Somogyi P. Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature. 2003;421:844–848. doi: 10.1038/nature01374. [DOI] [PubMed] [Google Scholar]
- Kvitsiani D, Ranade S, Hangya B, Taniguchi H, Huang JZ, Kepecs A. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature. 2013;498:363–366. doi: 10.1038/nature12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechasseur Y, Dufour S, Lavertu G, Bories C, Deschenes M, Vallee R, De Koninck Y. A microprobe for parallel optical and electrical recordings from single neurons in vivo. Nat Methods. 2011;8:319–325. doi: 10.1038/nmeth.1572. [DOI] [PubMed] [Google Scholar]
- Lee MG, Hassani OK, Alonso A, Jones BE. Cholinergic basal forebrain neurons burst with theta during waking and paradoxical sleep. J Neurosci. 2005;25:4365–4369. doi: 10.1523/JNEUROSCI.0178-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Hromadka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MA, Lee AK. Intracellular recording in behaving animals. Curr Opin Neurobiol. 2012;22:34–44. doi: 10.1016/j.conb.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26:5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ, 3rd, Gu X, Zanella S, Kidney J, Gu H, Mao Y, Hooks BM, Boyden ES, Buzsaki G, Ramirez JM, Jones AR, Svoboda K, Han X, Turner EE, Zeng H. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Lutcke H, Schulz K, Haiss F, Weber B, Kugler S, Hasan MT, Helmchen F. Reorganization of cortical population activity imaged throughout long-term sensory deprivation. Nat Neurosci. 2012;15:1539–1546. doi: 10.1038/nn.3240. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Meyer AH, Caputi A, Monyer H, Hasan MT, Schaefer AT, Denk W, Brecht M. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Marshel JH, Kaye AP, Nauhaus I, Callaway EM. Anterior-posterior direction opponency in the superficial mouse lateral geniculate nucleus. Neuron. 2012;76:713–720. doi: 10.1016/j.neuron.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HS, Schwarz D, Wimmer VC, Schmitt AC, Kerr JN, Sakmann B, Helmstaedter M. Inhibitory interneurons in a cortical column form hot zones of inhibition in layers 2 and 5A. Proc Natl Acad Sci U S A. 2011;108:16807–16812. doi: 10.1073/pnas.1113648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz W, Rudy B. Spatiotemporal specificity in cholinergic control of neocortical function. Curr Opin Neurobiol. 2014;26:149–160. doi: 10.1016/j.conb.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, Dickinson-Anson HA, Leppert JT, Lee KF, Gage FH. Central neuronal loss and behavioral impairment in mice lacking neurotrophin receptor p75. J Comp Neurol. 1999;404:1–20. doi: 10.1002/(sici)1096-9861(19990201)404:1<1::aid-cne1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Pinault D, Deschenes M. Projection and innervation patterns of individual thalamic reticular axons in the thalamus of the adult rat: a three-dimensional, graphic, and morphometric analysis. J Comp Neurol. 1998;391:180–203. doi: 10.1002/(sici)1096-9861(19980209)391:2<180::aid-cne3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Roux L, Stark E, Sjulson L, Buzsaki G. In vivo optogenetic identification and manipulation of GABAergic interneuron subtypes. Curr Opin Neurobiol. 2014;26C:88–95. doi: 10.1016/j.conb.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Barbic M, Losonczy A, Buzsaki G, Magee JC. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur J Neurosci. 2010;31:2279–2291. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Eichler R, Roux L, Fujisawa S, Rotstein HG, BUZSAKI G. Inhibition-induced theta resonance in cortical circuits. Neuron. 2013;80:1263–1276. doi: 10.1016/j.neuron.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Koos T, Buzsaki G. Diode probes for spatiotemporal optical control of multiple neurons in freely moving animals. J Neurophysiol. 2012;108:349–363. doi: 10.1152/jn.00153.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhmer W, Almers W. Photobleaching through glass micropipettes: sodium channels without lateral mobility in the sarcolemma of frog skeletal muscle. Proc Natl Acad Sci U S A. 1982;79:946–950. doi: 10.1073/pnas.79.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadics J, Varga C, Molnar G, Olah S, Barzo P, TAMAS G. Excitatory effect of GABAergic axo-axonic cells in cortical microcircuits. Science. 2006;311:233–235. doi: 10.1126/science.1121325. [DOI] [PubMed] [Google Scholar]
- Tanahira C, Higo S, Watanabe K, Tomioka R, Ebihara S, Kaneko T, Tamamaki N. Parvalbumin neurons in the forebrain as revealed by parvalbumin-Cre transgenic mice. Neurosci Res. 2009;63:213–223. doi: 10.1016/j.neures.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal CT, Golowasch JP, Zaborszky L. Adult mouse basal forebrain harbors two distinct cholinergic populations defined by their electrophysiology. Front Behav Neurosci. 2012;6:21. doi: 10.3389/fnbeh.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneswaran G, Kraskov A, Lemon RN. Large identified pyramidal cells in macaque motor and premotor cortex exhibit "thin spikes": implications for cell type classification. J Neurosci. 2011;31:14235–14242. doi: 10.1523/JNEUROSCI.3142-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jeong HY, Tremblay R, Rudy B. Neocortical somatostatin-expressing GABAergic interneurons disinhibit the thalamorecipient layer 4. Neuron. 2013;77:155–167. doi: 10.1016/j.neuron.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.