Evidence is provided that chromosome doubling induces A genome rRNA gene silencing in the synthetic tetraploid wheat. Elimination of the silenced A genome rDNA loci occurred in the S7 generation, and the rDNA sequences from the A and D genomes for the most part have been lost during the evolution of common wheat.
Abstract
rRNA genes consist of long tandem repeats clustered on chromosomes, and their products are important functional components of the ribosome. In common wheat (Triticum aestivum), rDNA loci from the A and D genomes were largely lost during the evolutionary process. This biased DNA elimination may be related to asymmetric transcription and epigenetic modifications caused by the polyploid formation. Here, we observed both sets of parental nucleolus organizing regions (NORs) were expressed after hybridization, but asymmetric silencing of one parental NOR was immediately induced by chromosome doubling, and reversing the ploidy status could not reactivate silenced NORs. Furthermore, increased CHG and CHH DNA methylation on promoters was accompanied by asymmetric silencing of NORs. Enrichment of H3K27me3 and H3K9me2 modifications was also observed to be a direct response to increased DNA methylation and transcriptional inactivation of NOR loci. Both A and D genome NOR loci with these modifications started to disappear in the S4 generation and were completely eliminated by the S7 generation in synthetic tetraploid wheat. Our results indicated that asymmetric epigenetic modification and elimination of rDNA sequences between different donor genomes may lead to stable allopolyploid wheat with increased differentiation and diversity.
INTRODUCTION
Allopolyploidization is mainly derived from interspecific or intergeneric hybridization and functions as a major force in plant evolution (Wendel, 2000; Feldman and Levy, 2005; Otto, 2007; Leitch and Leitch, 2008). Chromosome imbalances and genome instability induced by allopolyploidization lead to “genome shock” (McClintock, 1984) of the newly formed amphidiploids, which causes extensive genetic and epigenetic changes in the nascent hybrids (Comai, 2000; Chen, 2007). Complicated changes that occur during the evolution of natural and synthetic allopolyploids include chromosomal rearrangements, chromatin remodeling, alteration in gene expression, DNA methylation and histone modification, and reactivation of transposable elements (Kashkush et al., 2003; Pontes et al., 2003, 2004; Adams and Wendel, 2005; Gaeta et al., 2007; Ha et al., 2009).
Originating from one or more allopolyploidization events, tetraploid and hexaploid wheat (Triticum aestivum) experienced rapid alterations and sporadic genomic changes in the process of evolution (Feldman and Levy, 2005, 2012). Both low-copy sequences (Liu et al., 1998) and repetitive DNA sequences (Salina et al., 2004; Han et al., 2005) were largely eliminated in synthetic wheat. Such nonrandom elimination of genes and noncoding sequences induced differentiation and diversity of different genomes or chromosomes in subsequent generations, which is required for successful homology recognition and meiotic pairing (Feldman et al., 1997). Extensive changes in gene expression (transcriptome shock) and epigenetic modifications (epigenome changes) were accompanied by sequence deletion during the process of allopolyploid formation (Shaked et al., 2001; Kashkush et al., 2002, 2003; Levy and Feldman, 2004; Feldman and Levy, 2005, 2009). However, these genomic and epigenomic alterations show genome-specific bias in wheat (Ozkan et al., 2001; Ma et al., 2004). In the hybrid of Aegilops sharonensis (SlSl) × Aegilops umbellulata (SuSu), 14% of the loci from Ae. sharonensis, but only 0.5% of the loci from Ae. umbellulata, were lost and twice as many sequences from Triticum monococcum were affected compared with Ae. sharonensis in the amphiploids of Ae. sharonensis × T. monococcum (Shaked et al., 2001). Analysis of genome-wide transcription revealed that the patterns of partial gene expression in synthetic hexaploid wheat are homoeolog specific, nonadditive, and parentally dominant (Akhunova et al., 2010; Chagué et al., 2010; Qi et al., 2012). In addition, morphological traits and ecological adaptations are regulated only by one of the parental genomes in allopolyploid wheat (Peng et al., 2003; Nalam et al., 2006; Feldman and Levy, 2012). However, the mechanisms underlying the establishment and maintenance of genomic asymmetry in the allopolyploid species remain unclear.
rRNA genes are long tandem repeats that experienced drastic changes such as sequence loss, structural rearrangement, and modified expression during the course of plant evolution (Schubert and Kunzel, 1990; Wendel et al., 1995; Mishima et al., 2002; Pontes et al., 2004; Książczyk et al., 2011). In six tetraploid Oryza species, the internal transcribed spacer (ITS) sequence of rRNA genes exhibits coexistence and maintenance of paralogs, generation of novel sequence variants, loss of arrays, or interarray sequence homogenization (Bao et al., 2010). rRNA genes with the shorter intergenic spacer (IGS) sequences of Nicotiana sylvestris show elimination and rearrangement in the allotetraploid Nicotiana tabacum (Volkov et al., 1999; Kovarik et al., 2004, 2008). Expression of rRNA genes in allopolyploids are mainly regulated via “nucleolar dominance,” an epigenetic phenomenon in which rRNA genes inherited from one parent are transcribed while those from the other parent are silenced (Navashin, 1928; Pikaard, 2000; McStay, 2006). In both synthetic and natural polyploid species of Brassica napus, strong uniparental silencing of rRNA genes from Brassica oleracea is accompanied by hypermethylation of polymerase I promoters (Książczyk et al., 2011). The concerted actions of DNA methylation, histone H3K9 dimethylation, and H3K4 trimethylation induce silencing of NORs only from Arabidopsis thaliana in the newly formed allotetraploid Arabidopsis suecica (Lawrence et al., 2004). Due to the rapid response of the rRNA genes to the genetic stress triggered by allopolyploidization, they are used as models for studying genomic changes in allopolyploids (Baum and Feldman, 2010).
The major 45S rDNA loci in diploid wheat are distributed on the 1A, 5A, 1B, 6B, and 5D chromosomes, and some of these loci, also called the nucleolus organizing region (NOR), became minor loci in tetraploid and hexaploid wheat during the course of evolution (Flavell and O'Dell, 1979; Appels et al., 1980; Miller et al., 1983; Frankel et al., 1987). The activities of NORs are related to the size of the intergenic regulatory region and the status of cytosine methylation in wheat (Sardana et al., 1993; Dubcovsky and Dvorák, 1995; Neves et al., 1995; Houchins et al., 1997; Akhunov et al., 2001, 2010; Silva et al., 2008). However, details regarding the mechanism of rDNA sequence loss and epigenetic changes remain unknown.
We successfully synthesized tetraploid hybrids from three parental A, B (also referred to as S) (Sarkar and Stebbins, 1956; Blake et al., 1999), and D genomes. We simulated consistent changes in sequence elimination, gene expression, and epigenetic modifications of the rRNA genes corresponding to their natural counterparts. In addition, several newly formed hexaploids (AABBDD) were used to study rRNA genes changes in comparison to the natural evolution between common and semi-wild wheat varieties such as Xinjiang and Tibet wheat. In this work, we provide insights that asymmetric genetic, DNA methylation, and histone modification variations are possible mechanisms of rDNA evolution that may function in three homoeologous A, B, and D genomes.
RESULTS
rDNA Loci from A and D Genomes Are Lost in Natural and Synthesized Allotetraploid and Hexaploid Wheat
rDNA loci were detected by fluorescence in situ hybridization (FISH) in synthetic and natural wheat (Tables 1 and 2). Two pairs of NOR loci are found in each parental species: Aegilops longissima TL05 (chromosomes 1Sl and 6Sl) and Triticum urartu TMU06 (chromosomes 1A and 5A) (Figures 1A and 1B). In the first self-pollinated generation (S1) of the newly formed amphidiploid TL05 × TMU06, there were four pairs of NOR loci derived from both parents (Figure 1C). After self-pollinating for four generations, one NOR locus disappeared in ∼5.3% (16/299) of the amphidiploid individuals. Furthermore, 25% of the plants with NOR variation had only three pairs of NOR loci in the S5 generation (Figure 1D). Using the pHvG38 probe cloned from barley (Hordeum vulgare; Pedersen and Langridge, 1997), the parental Sl and A genomes could be distinguished by distinct signal patterns on the chromosomes (Supplemental Figures 1A and 1B). Interestingly, one pair of NOR loci on chromosome 5A was deleted first (Supplemental Figure 1D). We confirmed that NOR loci on the 5A chromosome were eliminated and total copy number of A genome NOR loci was significantly reduced by dot-blot and quantitative PCR (qPCR) results (Supplemental Figure 2). Furthermore, A genome NORs were continuously eliminated and the locus on chromosome 1A was lost in the S6 generation of the amphidiploids (∼0.5%, 3/521) (Figure 1E). Consequently, after successive self-pollinations (S7 generation) in the synthetic amphidiploid TL05 × TMU06, nearly all individuals had lost the rDNA loci from the A genome, and only the two pairs of NORs from the Sl genome remained. The pattern of NOR loci caused by elimination in the synthetic tetraploids was consistent with that of natural tetraploids, in which the two dominant NOR loci from the B genome, and few or no copies from the A genome were present in the genome over long evolutionary histories (Figure 1F; Supplemental Figures 3 and 4).
Table 1. Synthesized Allopolyploids Derived From Triticum and Aegilops Used in This Study.
| Cross Combination | Genome Constitution |
|---|---|
| Tetraploid (2n=4×=28) | |
| Ae. longissima (TL05) × T. urartu (TMU06) | SlSlAA |
| Ae. sharonensis (TH02) × T. monococcum ssp aegilopoides (TMB02) | SlSlAmAm |
| A. bicornis (TB01) × Ae. tauschii (TQ27) | SbSbDD |
| T. urartu (TMU38) × Ae. tauschii (TQ27) | AADD |
| T. urartu (TMU06) × Ae. tauschii (TQ27) | AADD |
| Ae. tauschii (TQ27) × T. urartu (TMU38) | DDAA |
| Hexaploid (2n=6×=42) | |
| 960 [T. turgidum ssp Durum (13-1) × Ae. tauschii (30A)] | BBAADD |
| AT5 [T. turgidum ssp Durum (TTR04) × Ae. tauschii (TQ27)] | BBAADD |
Table 2. Species and Lines of Diploid, Tetraploid, and Hexaploid Wheat Used in This Study.
| Species and Cultivars | Line Designation | Genome |
|---|---|---|
| Diploids (2n=2×=14) | ||
| T. urartu | TMU06 | AA |
| TMU38 | AA | |
| T. monococcum ssp aegilopoides | TMB02 | AmAm |
| Ae. speltoides | AE739 | SS |
| Ae. bicornis | TB01 | SbSb |
| Ae. longissima | TL01 | SlSl |
| TL05 | SlSl | |
| Ae. sharonensis | TH02 | SlSl |
| Ae. tauschii | 30A | DD |
| TQ27 | DD | |
| Tetraploid (2n=4×=28) | ||
| T. turgidum ssp dicoccoides | TTD14 | BBAA |
| T. turgidum | AS2255 | BBAA |
| T. turgidum ssp durum | Langdon | BBAA |
| 13-1 | BBAA | |
| T. polonicum | PI286547 | BBAA |
| Hexaploid (2n=6×=42) | ||
| T. aestivum | Chinese Spring | BBAADD |
| Jing411 | BBAADD | |
| T. aestivum ssp tibetanum Shao | AS329 | BBAADD |
| AS330 | BBAADD | |
| T. petropavloski Udats et Migusch | XJ358 | BBAADD |
| XJ356 | BBAADD | |
| XM1341 | BBAADD | |
| 1B/1R translocation lines | ||
| T. aestivum (1RS/1BL) | K-salmon | BBRRAADD |
| T. aestivum (1RS/1BL) | 41004 | BBRRAADD |
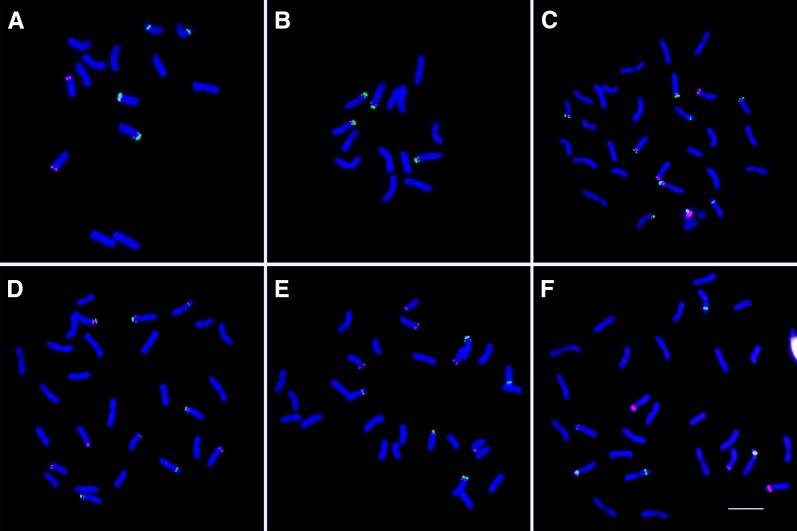
Figure 1.
FISH Analysis of rDNA Distribution on Somatic Metaphase Chromosomes of the New Amphidiploid TL05 × TMU06.
45S rDNA sequences are labeled in green, 5S rDNA sequences are labeled in red, and 4′,6-diamidino-2-phenylindole (DAPI) is blue. Bar = 10 μm.
(A) The diploid maternal line TL05 (four 45S signals and four 5S signals).
(B) The diploid paternal line TMU06 (four 45S signals and four 5S signals).
(C) The S1 generation of the amphidiploid TL05 × TMU06. There are eight 45S signals and eight 5S signals.
(D) The S5 generation of amphidiploid TL05 × TMU06. There are six 45S signals (two from the A genome are lost) but no change in the number of 5S signals.
(E) The S6 generation of amphidiploid TL05 × TMU06. There are five 45S signals (three from the A genome are lost) but no change in the number of 5S signals.
(F) The natural tetraploid wheat Langdon. There are four 45S signals (most 45S rDNA loci from the A genome are lost) and only six 5S signals.
Another newly formed amphidiploid TMU38 × TQ27 (T. urartu × Ae. tauschii) also showed elimination of A genome rDNA sequences. There are six NOR loci in this new amphidiploid: four from the A genome and two from the D genome (Figures 2A to 2C). However, a small fraction (∼0.4%, 3/700) of amphidiploids had only five NOR loci in the S4 generation (Figure 2D). A single locus from the A genome was lost, which affected the total number of A genome NOR loci, as detected by the pAsI probe (Pedersen and Langridge, 1997) (Supplemental Figures 2 and 5).
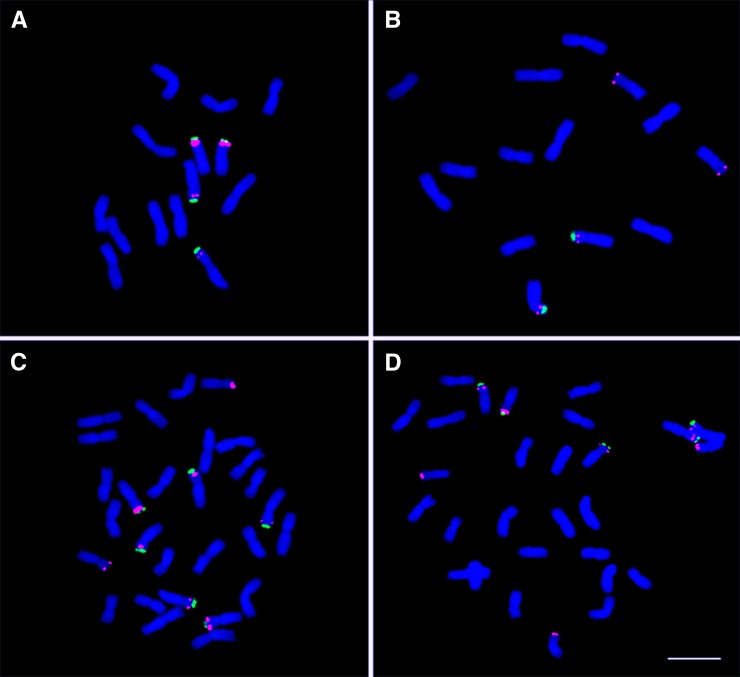
Figure 2.
FISH Analysis of the rDNA Distribution on Somatic Metaphase Chromosomes of the New Amphidiploid TMU38 × TQ27.
45S rDNA sequences are labeled in green, 5S rDNA sequences are labeled in red, and DAPI is blue. Bar = 10 μm.
(A) The diploid maternal line TMU38 (four 45S signals and four 5S signals).
(B) The diploid paternal line TQ27 (two 45S signals and four 5S signals).
(C) The S1 generation of amphidiploid TMU38 × TQ27. There are six 45S signals and eight 5S signals.
(D) The S4 generation of amphidiploid TMU38 × TQ27. There are five 45S signals (one from the A genome is lost) but no change in the number of 5S signals.
As in tetraploids, the copy number of rDNA sequences from the D genome was also reduced during the process of hexaploid wheat formation. There were two pairs of NOR loci from the B genome, one pair from the D genome, and few copies from the A genome in the synthetic and natural hexaploid wheat (Figure 3; Supplemental Figure 6). However, in contrast to synthetic hexaploids AT5 and 960 (Tables 1 and 2, Figures 3A and 3B; Supplemental Figures 6A and 6B), the semi-wild hexaploids T. aestivum ssp tibetanum Shao (AS329) and Triticum petropavloski Udats et Migusch (XJ356) had weak NOR signals from the D genome and no significant changes in the B genome NORs (Table 2, Figures 3C and 3D; Supplemental Figures 6C and 6D). Furthermore, compared with the synthetic hexaploid wheat AT5 and 960, very weak signals from D genome NORs were detected in the common wheat varieties Chinese Spring and Jing 411 (Table 2, Figures 3E and 3F; Supplemental Figures 6E and 6F). Quantitative analysis by dot-blot and qPCR showed increased copy number of B genome NORs in the common wheat Chinese Spring, but remarkably decreased copy number of D genome NORs in both XJ356 and Chinese Spring (Supplemental Figure 2).
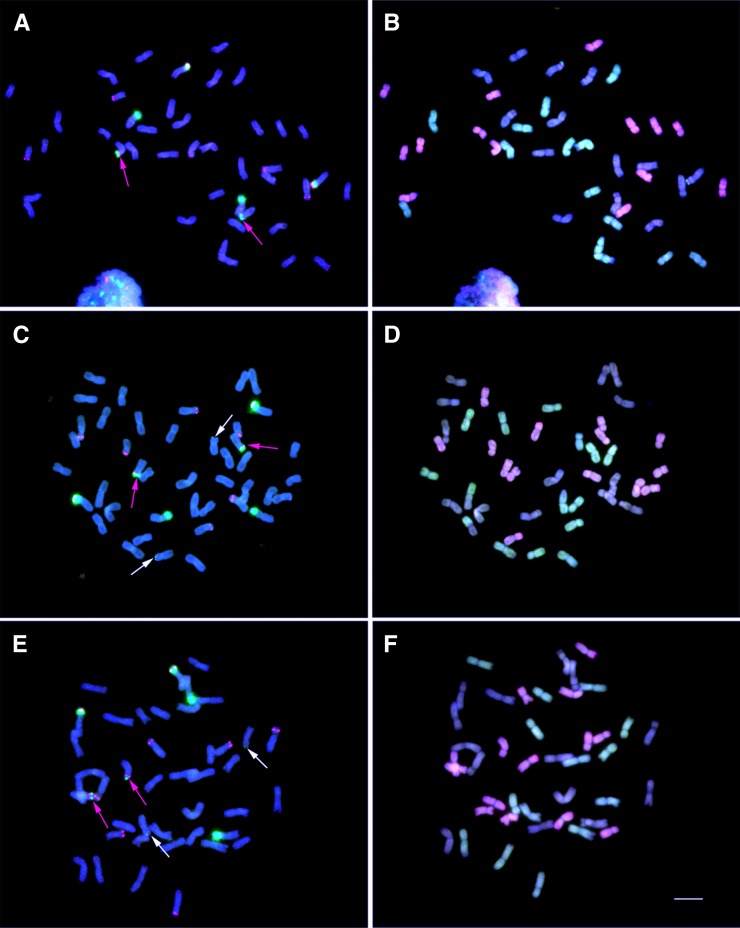
Figure 3.
FISH and Multicolor FISH Analysis of rDNA Distribution on Somatic Metaphase Chromosomes of Synthetic, Semi-Wild, and Natural Hexapolyploid Wheat.
In (A), (C), and (E), 45S rDNA sequences are labeled in green, and 5S rDNA sequences are labeled in red. DAPI is blue. The red and white arrows indicate the 45S signals from the D genome and the A genome, respectively. The other 45S signals are from the B genome. In (B), (D), and (F), the A genome DNA is labeled in green, the D genome DNA is labeled in red, and the B genome DNA is used as a block. Bar = 10 μm.
(A) and (B) FISH and multicolor FISH of synthetic hexaploid wheat AT5. The AT5 line shows strong 45S signals from the B and D genomes.
(C) and (D) FISH and multicolor FISH of semi-wild hexaploid wheat XJ356. This line has major 45S loci in the B genome and minor 45S loci in the D and A genomes.
(E) and (F) FISH and multicolor FISH of Natural hexaploid wheat CS. This line shows major 45S loci in the B genome and minor 45S loci in the D and the A genomes.
Deletion of rDNA loci from the D genome, rather than the Sb genome, was detected in the new tetraploid Ae. bicornis (TB01) × Ae. tauchii (TQ27) (Supplemental Figures 7 and 8). All of these results indicated that the NORs of the B genome are dominant to the NORs from the A and D genomes in tetraploid and hexaploid varieties.
Silencing of rRNA Genes from the A Genome Can Be Induced by Chromosome Doubling
Sequence polymorphisms in the ITS region allow the identification of distinct NORs by their recognition sites for specific restriction enzymes in different parental and amphidiploid lines (Figure 4). Transcripts of rRNA genes were analyzed in polyploid wheat via reverse transcription-cleaved amplified polymorphic sequence (RT-CAPS) (Sardana et al., 1993; Pikaard et al., 2005).
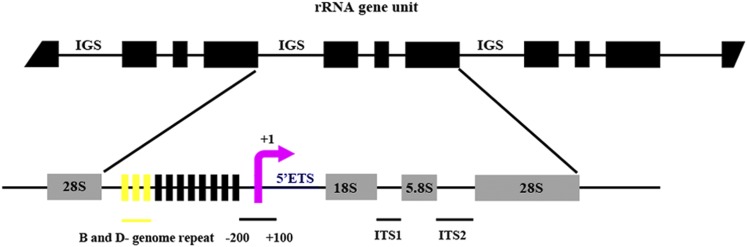
Figure 4.
Representative Structure of rRNA Genes.
The upper figure shows that each 45S rRNA gene unit contains 18S, 5.8S, and 28S rRNAs, which are separated by IGS and ITS sequences. A detailed schematic structure of the rRNA gene unit is below. The yellow region represents a specific repeated sequence, nearly 425 bp in length, existing only in the B and D genomes. The red arrow indicates the TIS. The flanking region from 200 bp upstream and 100 bp downstream of TIS was used for analysis of DNA methylation with bisulfite sequencing. Because of polymorphisms in the ITS sequences, the region containing the ITS1, 5.8S, and ITS2 (659 bp) was amplified to analyze rRNA gene expression.
To study the rRNA gene transcription in amphidiploid TL05-TMU06, the DrdI restriction site was used, as it is found in the ITS region of the maternal line TL05, but not in the paternal line TMU06. A 659-bp fragment was amplified from partial 18S to 28S rRNA genes that contained the complete ITS region. Two smaller bands (585 and 74 bp) in TL05 were obtained after incubation with DrdI, while an intact 659-bp band was seen in TMU06 (the 74-bp band is not shown in all the figures). In F1 hybrids of the amphidiploid TL05-TMU06, digestion with DrdI of the ITS sequences (two lanes per set), which were amplified from both DNA and cDNA templates, resulted in two bands: one was 659 bp from TMU06 and the other was 585 bp from TL05 (Figure 5A). This indicated that the NORs from both parental lines were expressed. However, after chromosome doubling (the S0 generation), only one band of ∼585 bp from cDNA template was observed after digestion, similar to the pattern of TL05. The disappearance of the other band (659 bp) from TMU06 indicated that NORs from TMU06 were silenced (Figure 5A). As a control, the patterns after digestion of the ITS products amplified from DNA template showed no difference between the F1 and S0 generations, which confirmed that the paternal rDNAs remained in the amphidiploid genomes.
Figure 5.
Analysis of the Expression Pattern of rRNA Genes in Synthetic, Semi-Wild, and Natural Wheat by RT-CAPS.
There are two lanes per line: DNA template (the first lane) and RNA template (cDNA, the second lane). The restriction enzymes used for analysis are indicated on the left in different lines.
(A) The digested product by DrdI from parental lines TL05, TMU06, F1 hybrid, and amphidiploid in S0 generation.
(B) The digested product by DrdI from amphidiploid TL05 × TMU06 in S3 and S4 generations. T. durum Langdon is a natural tetraploid wheat used as a control.
(C) The digested product by DrdI from parental lines TMU38, TQ27, F1 hybrid, and amphidiploid in S0 generation. AADD and DDAA indicated the hybrids of TMU38 × TQ27 and TQ27 × TMU38.
(D) The digested product by DrdI from amphidiploid TMU38 × TQ27 in S3 and S4 generations.
(E) The digested product by DrdI from synthetic hexaploid AT5 and 960.
(F) The digested product by PvuII from synthetic hexaploid AT5 and 960.
(G) The digested product by DrdI from semi-wild hexaploid XJ356, XJ358, XM1341, AS329, AS330, parental line TMU38, and natural hexaploid CS and Jing411.
(H) The digested product by PvuII from semi-wild hexaploid XJ356, XJ358, XM1341, AS329, AS330, parental line TMU38, and natural hexaploid CS and Jing411.
Similar results were found in two other crosses: TMU38 × TQ27 and TQ27 × TMU38. The NORs derived from both parents were actively transcribed in the F1 hybrids, but in the S0 generation the A genome NORs were silenced (Figure 5C). The NORs from A genomes were consistently silenced in the reciprocal hybrids AADD and DDAA (Figure 5C). The A genome rRNA genes in different amphidiploids were silenced prior to D genome rRNA genes during the chromosome doubling process.
Silenced rRNA Genes Maintain Functional Inactivity after Self-Pollination
In TL05 × TMU06 (SSAA) and TMU38 × TQ27 (AADD), both NORs from the A genome, were silenced in the S0 generation. After successive self-pollinations for three or four generations, the progenies, especially those that lost partial rDNA loci from the A genomes (Figure 1D), showed that the A genome NORs remain silenced (Figures 5B and 5D). The silenced A genome NORs also maintained functional inactivity after self-pollination in another amphidiploid, Ae. sharonensis (TH02) × T. monococcum (TMB02), and in several natural tetraploids (Supplemental Figure 9). Furthermore, rRNA gene transcription was detected by a nucleolin protein that was shown to bind only to transcriptionally active rRNA genes (Pontvianne et al., 2010). A unique 425-bp repetitive sequence from the B and the D genome IGS regions was identified and used as a specific probe to distinguish the NOR loci from different genomes (Figure 4; Supplemental Figure 10). Most of the rDNA loci from the parental lines TL05 and TMU06 were observed to colocalize with the nucleolin protein that binds only transcriptionally active rRNA genes (Figure 6). However, in the S4 generation of the hybrids, nucleolin was bound to Sl genome but not to A genome NORs (Figure 6). These results further confirmed the pattern of transcript accumulation that was obtained by RT-CAPS as described above (Figure 5B).
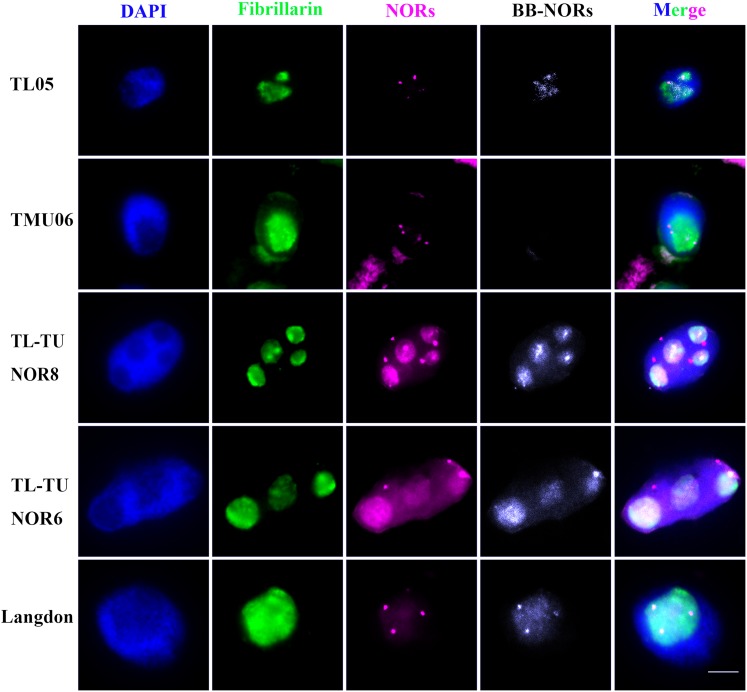
Figure 6.
Immunolocalization Analysis of Antifibrillarin in Interphase Nuclei of the Amphidiploid TL05 × TMU06.
Antifibrillarin antibody is labeled in green, all NORs are labeled in red, and B genome NORs are labeled in white. Bar = 10 μm. First line, diploid maternal line TL05. Second line, diploid paternal line TMU06. Third line, amphidiploid TL05 × TMU06 containing eight NOR loci. Fourth line, amphidiploid TL05 × TMU06 containing six NOR loci. Fifth line, natural tetraploid wheat Langdon.
The nucleolar dominance of rRNA genes among the A, B, and D genomes was also observed in the synthetic, semi-wild, and natural hexaploids. In three parental genomes, the DrdI restriction site was absent only from the A genome ITS region, and the PvuII restriction site was found only in the B genome ITS region (Supplemental Figure 9). Therefore, it revealed that the NORs from the A and D genomes were silenced, and only the B genome NORs were active in the S6 generation of the synthetic, semi-wild, and natural hexaploids (Figures 5E to 5H).
Haploidization Cannot Reactivate Silenced rRNA Genes from the A and D Genomes
To determine whether NOR silencing can reversed by inducing haploidy, we checked the NOR activity in 1RS/1BL translocation lines K-salmon (2n=42) and its haploid derivatives (2n=21) (Tsunewaki, 1964) (Table 2).
In K-salmon, FISH results showed two pairs of B genome NORs, one pair of R and D genome NORs, and two weak signals of A genome NORs (Figure 7A). In the haploid, only half of the signals were observed (Figure 7B). According to sequence polymorphisms in the ITS region, the NOR activity from the A, D, R, and B genomes in the hybrids can be distinguished by DrdI, BssSI, BsrBI, and PvuII restriction enzymes (Figures 7C and 7D). After digestion with DrdI, one band was visible, and no expression of NORs from the A genome was detected in both K-salmon and its haploids (Figure 7E). NORs from the D and R genomes were silenced as indicated by digestion of ITS regions with BssSI and BsrBI (Figures 7F and 7G). Only B genome NORs were shown to be active by digestion with PvuII in both diploids and haploids (Figure 7H). Crossing of K-salmon with other lines (Chinese Spring and 41004; Table 2) can also induce haploid plants, but no differences in transcription were found in these diploids and haploids (Supplemental Figure 11). While chromosome doubling can cause NOR silencing, the silencing cannot be reversed by haploidization.
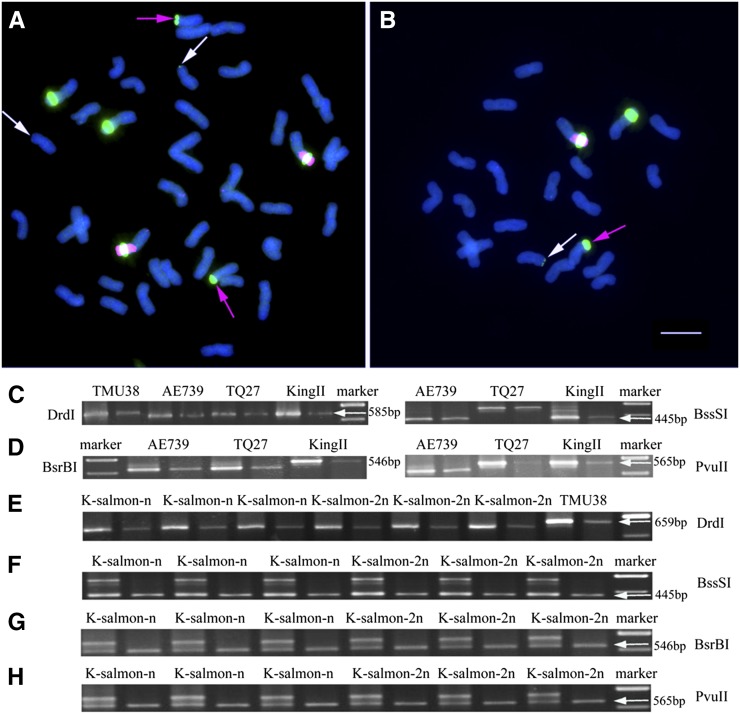
Figure 7.
Analysis of the Distribution and Expression of rRNA Genes in K-Salmon and Its Haploid Lines.
In (A) and (B), the 45S rDNA sequences are labeled in green, and rye genomic DNA is labeled in red. DAPI is blue. The red and white arrows indicate the 45S signals from the D and A genomes, respectively. Bar = 10 μm.
(A) The K-salmon 1BL/1RS translocation line (2n=42). There are four strong 45S signals from the B genome, two strong signals from the R genome, two weaker signals from the D genome, and two very weak signals from the A genome.
(B) The haploid K-salmon 1BL/1RS derivative (n = 21). The signal number is half of the diploid line K-salmon.
In each set (two lanes per set) of the (C) to (H), DNA template (the first lane) and RNA template (cDNA, the second lane). The restriction enzymes used for analysis are indicated on the left or right in different lines.
(C) The digested product by DrdI and BssSI from the paternal lines TMU38, AE739, TQ27, and KingII.
(D) The digested product by BsrBI and PvuII from the paternal lines TMU38, AE739, TQ27, and KingII.
(E) The digested product by DrdI from K-salmon and its haploid lines. The patterns of digestion by DrdI show that no 45S rRNA genes from the A genome are expressed in K-salmon and its haploid lines.
(F) The digested product by BssSI from K-salmon and its haploid lines. The patterns of digestion by BssSI show that no 45S rRNA genes from the D genome are expressed in K-salmon and its haploid lines.
(G) The digested product by BsrBI from K-salmon and its haploid lines. The patterns of digestion by BsrBI show that no 45S rRNA genes from the R genome are expressed in K-salmon and its haploid lines.
(H) The digested product by PvuII from K-salmon and its haploid lines. The patterns of digestion by PvuII show that only 45S rRNA genes from the B genome expressed in K-salmon and its haploid lines.
rRNA Genes of the Donor Genomes Show Differential DNA Methylation in Newly Formed Amphiploids
The proportion of methylated cytosine residues at CCGG sites at promoters is negatively correlated with transcription of rRNA genes (Sardana et al., 1993; Earley et al., 2010). We chose a 298-bp segment in the promoter region that contains the transcription initiation site (TIS) and its flanking regions for bisulfite sequencing (Figure 4). In F1 hybrids of TL05 × TMU06, the level of DNA methylation (CG, CHG, and CHH) at the promoter region of rDNA loci, which are derived from the A genome, was kept at the same level (or slightly decreased) as in the paternal line TMU06. After chromosome doubling (S0 generation), A genome CHG and CHH DNA methylation was strongly increased in the newly formed amphiploids, while CG methylation was unchanged (Figure 8A; Supplemental Figure 12A). The percentage of CHH methylation in total methylation (relative CHH methylation content) was highest in S0 generation (Supplemental Figure 12B). However, the increase of A genome CHH methylation disappeared in the S4 generation (Figure 8A; Supplemental Figure 12), but CHG methylation still showed a significant increase when compared with the parental A genome (Figure 8A; Supplemental Figure 12). For the S genome in the amphidiploid TL05 × TMU06, CHG methylation rose from 29.81 to 38.48% in the F1 hybrids and returned to 34.21% during self-pollination. The CG and CHH methylation (except in the S0 generation) decreased or remained the same as the parental S genome from the F1 to S4 generation (Figure 8A). Overall, only the relative proportion of CG methylation continually decreased during subsequent self-pollinations (Supplemental Figure 12B).
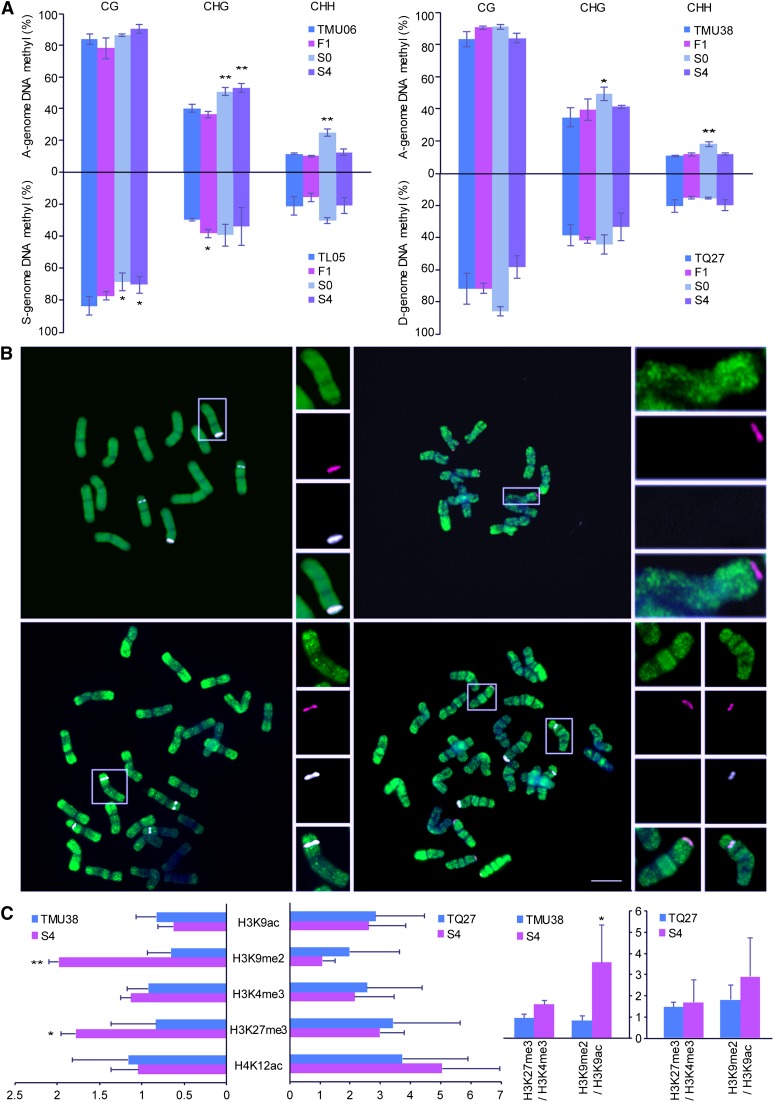
Figure 8.
Analysis of DNA Methylation and Histone Modification in the Parental A Genome and Amphidiploids TL05 × TMU06 and TMU38 × TQ27.
(A) Bisulfite sequencing analysis was used to detect DNA methylation on the NOR promoters in amphidiploids TL05 × TMU06 and TMU38 × TQ27 (left and right histograms respectively). The DNA methylation changes of the A genome lines TMU06 and TMU38 compared with different generations in the amphidiploids are shown in the positive y axis. Comparisons to the S genome (TL05) and D genome (TQ27) are present in the negative y axis. In (A) and (C), the columns and error bars represent the mean relative level and sd, respectively. Each line had three biological replications. Differences between different lines were compared by Student’s t test. P values:*P < 0.05 and **P < 0.01, respectively.
(B) Immunolocalization analysis of anti-5-methyl cytosine in the parental lines TL05 (left in upper lane), TMU06 (right in upper lane), natural tetraploid wheat Langdon (left in bottom lane), and the amphidiploid TL05 × TMU06 (right in bottom lane). In the amphidiploid TL05 × TMU06, insets show higher magnification view of the chromosomes from A genome (left) and S genome (right). The anti-5-methyl cytosine antibody is labeled in green, 45S rDNA sequence is labeled in red, the specific repeated sequence from the B is labeled in white. DAPI is blue in all panels. Bar = 10 μm.
(C) ChIP-qPCR analysis of five histone modification in the NOR promoters of the parental lines and the amphidiploid TMU38 × TQ27 (left).Changes of the ratio of H3K27me3/H3K4me3 and H3K9me2/H3K9ac in the parental lines and the amphidiploids TMU38 × TQ27 (right).
Immunolocalization with an antibody that recognizes methylated DNA (anti-5-methyl cytosine) revealed that most A genome rDNA loci had stronger DNA methylation in the amphidiploid TL05 × TMU06 than in the paternal line TMU06, whereas signals from S genome NORs were not obviously changed (Figure 8B).
In the newly formed amphidiploid TMU38 × TQ27, an increase in the three types of DNA methylation in A genome NORs was observed in the S0 generation. Only the increased CHG methylation consistently remained in the S4 generation (Figure 8A; Supplemental Figure 12). With the exception of CG methylation in the S4 generation, no drastic alterations of DNA methylation were found in D genome NORs (Figure 8A; Supplemental Figure 12).
Histone Modification at Silenced rRNA Genes Changes during Polyploidization
Histone deacetylation as well as H3K27 and H3K9 methylation are important epigenetic modifications involved in silencing rRNA genes (Lawrence et al., 2004; Probst et al., 2004; Pontvianne et al., 2012). We detected five histone modifications (H4K12 acetylation, H3K27 trimethylation, H4K3 trimethylation, H3K9 dimethylation, and H3K9 acetylation) at the rDNA promoter of amphidiploid TMU38-TQ27 and its progenitors through chromatin immunoprecipitation (ChIP)-qPCR.
In the amphidiploid, we found that the heterochromatin marks H3K27me3 and H3K9me2 were greatly increased, while H3K9 acetylation and H4K12ac were slightly decreased (Figure 8C). H3K27me3 and H3K4me3, as well as H3K9me2 and H3K9ac, represent markers with inverse functional implications regarding gene expression and chromatin status. A remarkable increase of the ratio of H3K9me2/H3K9ac (the increase of H3K27me3/H3K4me3 was not significant) was correlated with heterochromatinization in the A genome promoter of amphidiploid TMU38-TQ27 compared with its progenitor (Figure 8C). In the D genome, most histone modifications maintained the same enrichment, except for an increase of H4K12ac (Figure 8C). However, the higher level of active marker H3K9ac versus the repressive marker H3K9me2 (decrease of H3K9me2/H3K9ac) indicated that D genome rDNA loci are associated with more euchromatin marks in the amphidiploids than in the paternal D genome (Figure 8C).
We also detected H4K12ac on metaphase chromosomes through immunostaining. In the parental lines TL05 and TMU06, NORs were more enriched for H4K12ac than other regions on the chromosome (Supplemental Figures 13A and 13B). However, this euchromatic modification disappeared on the silenced NORs from the A genome in the hybrid line TL05 × TMU06, which contains two pairs of A genome NORs (Supplemental Figure 13C). Only four NORs from the B genome still maintained high levels of H4K12ac in amphidiploid TL05 × TMU06 and natural tetraploids (Supplemental Figures 13C and 13D). In the amphidiploid TMU38 × TQ27, which has four A genome NORs, the silenced A genome NORs lost the euchromatin marker H4K12ac (Supplemental Figure 14).
DISCUSSION
Asymmetric Sequence Elimination Follows Repression of rRNA Genes
The elimination of rDNA loci from specific genomes in allopolyploid plants has been reported in many allotetraploid species (Brettell et al., 1986; Volkov et al., 1999; Pontes et al., 2004; Shcherban et al., 2008; Xiong et al., 2011). For natural allopolyploid wheat, previous studies reported that NOR loci on chromosome 5A were lost (Miller et al., 1983; Jiang and Gill, 1994). In this study, we found an interaction between A, B, and D genome NORs. As only A genome NORs were lost, we suggest that the rDNA elimination was not random but rather was directed. Loci from the S or D genomes did not undergo obvious changes within the synthetic amphidiploids TL05 × TMU06 (SlSlAA) and TMU38 × TQ27 (AADD) (Figures 1 and 2). However, NORs of the D, rather than of Sb or B genomes, experienced sequence elimination and copy number reduction in the amphidiploid TB01 × TQ27 (SbSbDD) and the common wheat Chinese Spring and Jing411 (AABBDD) (Figure 3; Supplemental Figures 6 to 8). These results indicate that NORs of the B (also called S) genomes were the most dominant during rRNA directed evolution of wheat. Such genomic asymmetry was also manifested in various morphological traits, agronomical traits, and DNA sequence alterations during the origination of allopolyploid wheat (Feldman et al., 1997; Ozkan et al., 2001; Baum and Feldman, 2010; Feldman and Levy, 2012).
Genes of A, B, and D genomes experience unequal regulation and show parental expression level dominance (ELD) in allopolyploid wheat (Akhunova et al., 2010; Chagué et al., 2010; Qi et al., 2012; Li et al., 2014). A recent report suggests that ELD of AB genome genes participates in plant development and that ELD of D genome genes is associated with plant adaptation to stress in young spikes of allopolyploid wheat (Li et al., 2014). We show that ELD of B and S genes mainly involves rRNA gene transcription and ribosome function in synthetic and natural tetraploid and hexaploid wheat (Figure 5). A genome NORs were the most recessive, independent of the crossing direction. They were rapidly silenced in early generations and were lost in later generations of the amphidiploids TL05 × TMU06 (SlSlAA) and TMU38 × TQ27 (AADD). Similarly, the D genome NORs initially lost their activity before the copy number was reduced in the hexaploid AABBDD (Figures 3 and 5E to 5H). Therefore, we propose that asymmetric rDNA sequence loss is possibly related to the repressed gene activity in early generations.
The loss of NOR loci after their transcriptional silencing was also found in other newly formed allotetraploids such as A. suecica and Nicotiana tomentosiformis (Volkov et al., 1999; Pontes et al., 2003, 2004; Kovarik et al., 2008). However, putative correlation between rRNA gene silencing and sequence elimination has yet to be shown in allopolyploid plants. Previous research indicated that transcription-induced cohesin dissociation might provide a general mechanism for determining rDNA copy number via recombination regulation in the yeast Saccharomyces cerevisiae (Kobayashi and Ganley, 2005). In fact, gene silencing and differential expression of parental genomes can be rapidly established in early generations of plant allopolyploids. The winning parental genome depends on the expression state of genes in allotetraploid Arabidopsis and cotton (Gossypium hirsutum; Wang et al., 2004, 2006; Chaudhary et al., 2009; Rapp et al., 2009). Furthermore, the silenced genes are gradually lost after gene silencing for long periods of time, which leads to homoeologous blocks with different degrees of gene retention (Thomas et al., 2006; Soltis et al., 2010). This sequence downsizing, which is important for species evolution, is common in polyploid plants and is believed to counteract genome expansion after polyploidization (Leitch and Bennett, 2004; Baack et al., 2005). These biased eliminations of rDNA and other DNA sequences in the allopolyploid wheat may facilitate genome size reduction after allopolyploidization (Feldman et al., 1997).
NOR Silencing Depending on DNA Methylation May Be Triggered by Alloploidization and Cannot Be Reversed by Haploidization
Silencing of rRNA genes in the S0 generation might be related to increased DNA methylation after chromosome doubling. Chromosome doubling events were shown to promote evolutionary success by altering gene expression during the formation of allopolyploids (Hegarty and Hiscock, 2007). Genes involved in metabolism, disease resistance, and cell cycle regulation of the newly formed tetraploid wheat altered their activities and DNA methylation level in the S1 generation (after chromosome doubling) (Kashkush et al., 2002). In F1 hybrids of TL05 × TMU06 and TMU38 × TQ27, most types of DNA methylation in the promoter of A genome NORs remained at the same level as the parental lines TMU06 and TMU38, and no changes in NOR activity occurred (Figures 5A, 5C, and 8A). However, in the S0 generation, both CHG and CHH methylation were significantly increased and may induce the silencing of A genome NORs (Figures 5A, 5C, and 8A). Previous works indicated that CHH methylation, which is mediated by de novo cytosine methyltransferase DRM2 together with the methylcytosine binding domain proteins MBD6 and MBD10, may play an important role in the large-scale silencing of rRNA genes in A. thaliana (Preuss et al., 2008). CHH DNA is first methylated by DRM2, and the DNA methylation is subsequently recognized by MBD6, which helps to maintain heterochromatin by CG or CHG DNA methylation (Preuss et al., 2008). An increase of CG methylation in the promoter region results in silencing of rRNA genes from Brassica rapa in new synthetic and natural polyploid species B. napus (Książczyk et al., 2011). In the synthetic tetraploid wheat TL05 × TMU06 and TMU38 × TQ27, both CHG and CHH methylation may be required for repressing A genome NORs and maintaining hypermethylation in the promoter regions similar to A. thaliana (Figure 8A). However, CHG methylation was dominant over CHH methylation, which may be the reason for consistent A genome NOR silencing. In wheat, we assumed that CHH methylation functioned as a rapid response to the “genome shock” and CHG methylation acted as a “holder” to maintain the response (Figure 8A). However, either the S or D genome NORs, mostly with low CG methylation, remained active as suggested for B. napus (Książczyk et al., 2011) (Figure 8A). In hexaploid wheat, earlier work revealed that drastic regulation of siRNA corresponding to transposable elements occurred only upon allopolyploidization (S1 generation) but not immediately after hybridization in F1 hybrids (Kenan-Eichler et al., 2011).
Allopolyploidization is a critical step for NOR silencing, and haploidization cannot reverse the process in wheat. The hexaploid wheat K-salmon and its haploids showed no difference in transcription of the A, B, D, and R genome rRNA genes (Figures 7C to 7H). These data imply that certain gene transcription and epigenetic modifications cannot be altered simply by changing the ploidy. Allopolyploidization is always accompanied by genome-wide rearrangements, changes in genes expression, and epigenetic modification that are involved in the formation of stable epialleles (Lee and Chen, 2001; Kashkush et al., 2002). A previous report showed that segregation of inactivated hygromycin phosphotransferase epialleles from a tetraploid A. thaliana, which were activated in the diploid, could lead to persistent gene silencing even when the ploidy was changed, suggesting that the stable epigenetic modification on alleles cannot be reversed by reducing the ploidy level (Mittelsten Scheid et al., 2003), similar to our current results for the rRNA genes in wheat.
Epigenetic Modifications May Contribute to Sequence Elimination of Silenced rRNA Genes
Nucleolar dominance and rRNA gene dosage are regulated by concerted changes in DNA methylation and histone modification in Arabidopsis (Lawrence et al., 2004). Increased DNA and H3K4 methylation and decreased histone acetylation are important for establishing inactive states of rRNA genes (Probst et al., 2004). Histone deacetylase HDT1 was reported to be a key component for repressing rRNA activity in A. thaliana (Lawrence et al., 2004). HDA6, another histone deacetylase, suppresses intergenic Pol II transcription and mediates different effects regarding symmetric (CG and CHG) and asymmetric (CHH) cytosine methylation for silencing rRNA genes (Earley et al., 2010). DNA methylation and repressive histone modifications regulate the silenced rRNA genes by promoting each other in a self-reinforcing cycle (Lawrence et al., 2004; Earley et al., 2006). However, for allopolyploid wheat, histone modification and differential DNA methylation have not been reported for rDNAs to date.
A significant increase of H3K27me3and H3K9me2 mainly maintained the heterochromatin state and might be associated with A genome NOR silencing in the S4 generation of amphidiploid TMU38-TQ27 (Figure 8C). Disruption of the H3K27 methyltransferases ATXR5 and ATXR6 and knockdown of H3K9 methyltransferase genes affected both the transcript accumulation of rRNA genes and the relative abundance of rDNA in A. thaliana (Pontvianne et al., 2012). We hypothesize that the increased H3K27me3/H3K4me3 and H3K9me2/H3K9ac content might drive the elimination of inactive A genome NOR loci after gene silencing had been established. Furthermore, the changes of the ratio of H3K9me2/H3K9ac might maintain both the inactive A genome NORs and the active D genome NORs (Figure 8C). Two chromodomain proteins Pdd1p and Pdd3p that bind H3(Lys9)Me were sufficient to promote DNA excision in Tetrahymena, suggesting a link between heterochromatin formation and DNA elimination (Taverna et al., 2002). In addition, H3K27 methylation is also required for heterochromatin formation and DNA elimination by regulating H3K9 methylation (Liu et al., 2007). Similar mechanisms might regulate H3K27me3 and H3K9me2 to affect heterochromatin and rDNA sequence deletion in wheat.
Different from H3K27me3 and H3K9me2, the H4K12ac, H3K9ac, and H3K4me3 modifications were not obviously changed in the A genome promoters (Figure 8C). Previously, it was reported that HDA6 mutations could induce enrichment of H3K4 methylation and modify patterns of DNA methylation (Probst et al., 2004). However, in the amphidiploid TMU38-TQ27, no significant interaction was observed among H3, H4 histone acetylation, H3K4me3, and DNA methylation (Figures 8A and 8C). Although the enrichment of H4K12ac was not obviously changed in NOR promoters of the A genome, it was drastically reduced in whole region (Supplemental Figures 13 and 14), suggesting that this modification still affected NOR activity. Thus, DNA methylation and histone modifications both had differential function in the inactive and active NORs, and they were important for regulating the dosage of rRNA genes at both DNA and RNA transcript levels in allopolyploid wheat.
Mechanisms Underlying the Evolution of Repetitive Sequences in Wheat
Repetitive sequences comprise a large portion (80%) of the wheat genome and often experience evolutionary alterations in allopolyploid wheat, which includes rapid variation of copy number, structure, function, and epigenetic profile (Smith and Flavell, 1975; Wicker et al., 2001; Jurka et al., 2007).
Rapid and repeated eliminations of the repetitive DNA sequence pGc1R-1a were found in synthetic wheat allopolyploids (Han et al., 2005). Twenty microsatellite sequences in 15 populations of Triticum dicoccoides from Israel and Turkey showed that different distribution patterns of simple sequence repeat alleles (Fahima et al., 2002). The sequences of Stowaway-like transposable element families displayed high conservation, genomic diversification, and significant differences in the methylation status of the insertion sites in diploid, tetraploid, and hexaploid wheat (Yaakov et al., 2013). These reports suggest that evolution of repetitive sequences can be affected by both the genomic and environmental context. However, our results indicate that the expression and evolution of rDNA repeats is associated with epigenetic alterations (especially increased DNA methylation) and polyploidization rather than hybridization per se and exclude an effect of the origin of sequences in the sense of a “maternal effect” (Figure 9). These discoveries provide insight into the function of repetitive sequence in wheat evolution.
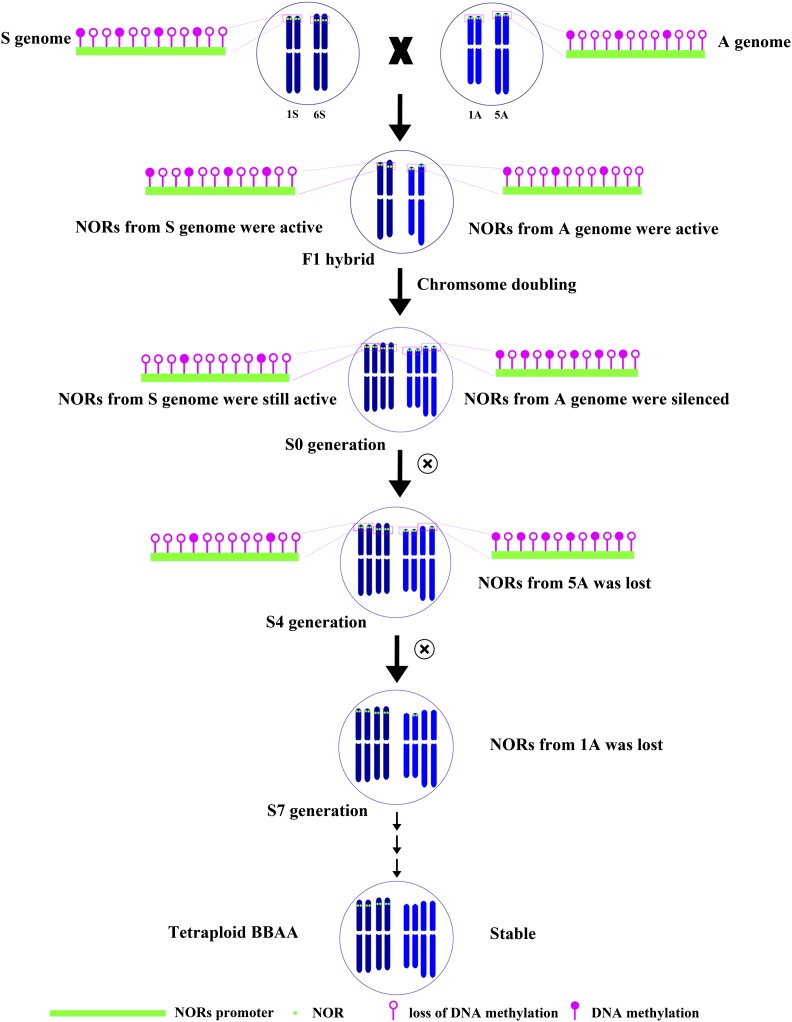
Figure 9.
Model for the Evolution of rRNA Genes in Newly Formed Tetraploid Wheat.
A schematic course of the evolution of rRNA genes is shown. Solid red circles indicate methylation, and the open circles indicate lack of methylation. In the parental A genome, transcriptionally active NORs are accompanied by certain types of DNA methylation. NORs from the A genome remain active in F1 hybrids due to unchanged DNA methylation. After chromosome doubling, A genome NORs show increased DNA methylation and become silenced. In the S4 generation, the increase of DNA methylation on A genome NORs may induce the elimination of NOR loci on chromosome 5A. The A genome NORs lose another locus on chromosome 1A in the S6 generation. The S genome NORs with unchanged or decreased DNA methylation retain their activities and copy numbers in the self-pollinated generations. The newly formed tetraploid wheat becomes stable, although they have lost most NOR loci from the A genome.
METHODS
Plant Materials
The F1 hybrids of TL05 × TMU06 (SSAA), TMU06 × TQ27 (AADD), and TQ27 × TMU38 (DDAA) were generated in our laboratory. Embryo rescue was performed as described (Chu et al., 2008). Two days after pollination, the stigma was sprayed with a mixture of 213.05 mg/L 2,4-dichlorophenoxyacetic acid (Sigma-Aldrich), 50 mg/L gibberellic acid (Sigma-Aldrich), and 80 μL/L Tween-80 (Sigma-Aldrich). Twelve days later, the immature embryos were dissected out and cultured on Murashige and Skoog medium (Sigma-Aldrich). When the embryos grew to three-leaf plants, the hybrids were treated with a mixture of 0.04% colchicine (Sigma-Aldrich) and 1% dimethyl sulfoxide (Sigma-Aldrich) for 12 h. Residual colchicine was washed away under running water for 24 h. The plants were then moved into an incubator for growth. The rest of the synthetic allopolyploids (except TL05 × TMU06, TMU06 × TQ27, and TQ27 × TMU38) were kindly provided by Moshe Feldman (see Tables 1 and 2).
Cytological Preparation and Probe Labeling
Root tips for FISH experiments were fixed with 90% acetic acid, and metaphase chromosome preparations were performed as published (Kato et al., 2004). For immunostaining, nuclei and metaphase chromosomes were prepared as described (Houben et al., 1999) with the modification that the root tips were fixed with 4% formaldehyde in 1× PBS for 1 h. The probes were prepared by direct labeling (Kato et al., 2004). The 45S, 5S, IGS-B repeat, and PhvG38 DNAs were labeled with different fluorescent modified nucleotides: Alexa Fluor-488-dUTP, Alexa Fluor-546-14-dCTP, Alexa Fluor-594-5-dUTP, and Alexa- Fluor-647-aha-dCTP (Invitrogen). FISH images were acquired using an epifluorescence Olympus BX61 microscope equipped with a cooled charge-coupled device camera operated with MetaMorph software and processed with Adobe Photoshop CS 3.0.
Dot-Blot Analysis
One hundred nanograms of DNA for each sample was used as the DNA target, and specific PCR products from A, B, and D genome NOR promoters were used as probes. The specific sequences of A, B, and D genome promoters were amplified using the primers TIS-A, TIS-B, and TIS-D (Supplemental Table 1). The dot-blot protocol was performed as previously described with the slight modification that the hybridization was continued for at least 15 h at 65°C (Malinen et al., 2003).
Transcriptional Analysis of NORs from Different Genomes of Hybrids
RNA was isolated from plants of three-leaf synthetic and natural hybrids using an RNA isolation kit (ZYMO Research) and then reverse-transcribed into cDNA using M-MLV reverse transcriptase (Promega) with random primers (New England Biolabs). The ITS sequences from different genomes were amplified using ITS primers (Supplemental Table 1) and cDNA as template. Unique cut sites for restriction enzymes (DrdI, PvuII, NspI, BssSI, and BsrBI; New England Biolabs) were found in these ITS sequences from different genomes using sequence alignment (GeneTool). The differential expression of NORs in the hybrids was analyzed using RT-CAPS as reported (Sardana et al., 1993; Pikaard et al., 2005).
Isolation of IGS Sequences from A, B, and D Genomes
PCR amplification of IGS regions from the A, B, and D genomes using specific primers (Supplemental Table 1) was as described (Sardana et al., 1993; Pikaard et al., 2005). The full-length sequences of IGSs were submitted to GenBank. The specific repetitive sequences from the B and D genomes were identified by comparing to the A genome IGS sequences (Supplemental Figure 10). The specific part of each IGS sequence was amplified using the primers IGS-B repeat-F and IGS-B repeat-R (Supplemental Table 1).
Immunostaining on the Nuclei and Chromosomes
Prepared slides were preincubated with 3% BSA and 1% Triton X-100 in 1× PBS for 1 h at 37°C and then incubated with primary antibodies overnight at 4°C. The primary antibodies used were rabbit anti-H4K12ac (07-595; Millipore) and mouse antifibrillarin (ab4566; Abcam). For DNA methylation detection with anti-5-mc antibody (ab10805; Abcam), the slides were prepared as described above. The fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch) was used as a secondary antibody.
Sodium Bisulfite Treatment and Sequencing of Promoter Regions
For DNA methylation analysis, 500 ng genomic DNA from each sample was treated with a sodium bisulfite treatment kit (ZYMO Research). The treated DNA was recovered and amplified with a converted promoter primer set (Supplemental Table 1). All the converted sequences were aligned (GeneTool), and the DNA methylation state was assessed with bioinformatic software CyMATE (Hetzl et al., 2007). Significant differences were calculated with the SPSS 19.0 software.
ChIP-qPCR and Relative qPCR Quantification of Copy Number
ChIP with the antibody H4K12ac, H3K27me3, H4K3me3, H3K9me2, and H3K9ac were performed as described (Nagaki et al., 2003) with a small modifications. Nearly 20g fresh leaf tissue was treated in 1% formaldehyde and the isolated cross-linked chromatin was cut into mono- or dinucleosomes by the micrococcal nuclease (Sigma-Aldrich). Prior to further qPCR analysis, DNAs isolated by ChIP were purified using a PCR purification kit (Qiagen). Chromatin precipitated without antibody, and isolated chromatin before precipitation, were used as negative control and input control, respectively. Three biological replicates were used per experiment. For copy number quantification, each 20-μL PCR assay contained 100 ng DNA. The actin gene in wheat (Triticum aestivum) was used as a control (Supplemental Table 1). The qPCR protocol was as previously described (Livak and Schmittgen, 2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: T. urartu TMU38 ITS sequence, KF482074; T. urartu TMU06 ITS sequence, KF482075; Ae. longissima TL05 ITS sequence, KF482076; Ae. speltoides AE739 ITS sequence, KF482077; Ae. sharonensis TH02 ITS sequence, KF482078; T. monococcum TMB02 ITS sequence, KF482079; T. durum 13-1 ITS sequence, KF482080; Ae. tauschii 30A ITS sequence, KF482081; T. aestivum 960 ITS sequence clone1, KF482082; T. aestivum 960 ITS sequence clone 2, KF482083; T. durum TTR04 ITS sequence, KF482084; Ae. tauschii TQ27 ITS sequence, KF482085; T. aestivum AT5 ITS sequence clone 1, KF482086; T. aestivum AT5 ITS sequence clone 2, KF482087; T. polonicum PI286547 ITS sequence, KF482088; T. dicoccoides TTD04 ITS sequence, KF482089; T. durum Langdon ITS sequence, KF482090; T. turgidum AS2255 ITS sequence, KF482091; T. petropavloski Udats et Migusch XJ356 ITS sequence clone 1, KF482092; T. petropavloski Udats et Migusch XJ356 ITS sequence clone 2, KF482093; T. petropavloski Udats et Migusch XJ358 ITS sequence clone 1, KF482094; T. petropavloski Udats et Migusch XJ358 ITS sequence clone 2, KF482095; T. aestivum Chinese Spring ITS sequence clone 1, KF482096; T. aestivum Chinese Spring ITS sequence clone 2, KF482097; T. aestivum ssp tibetanum Shao AS329 ITS sequence clone 1, KF482098; T. aestivum ssp tibetanum Shao AS329 ITS sequence clone 2, KF482099; T. aestivum ssp tibetanum Shao AS330 ITS sequence clone 1, KF482100; T. aestivum ssp tibetanum Shao AS330 ITS sequence clone 2, KF482101; T. petropavloski Udats et Migusch XM1341 ITS sequence clone 1, KF482102; T. petropavloski Udats et Migusch XM1341 ITS sequence clone 2, KF482103; T. aestivum Jing411 ITS sequence clone 1, KF482104; T. aestivum Jing411 ITS sequence clone 2, KF482105; Secale cereal KingII 106 ITS sequence, KF482106; T. aestivum K-salmon ITS sequence clone 1, KF482107; T. aestivum K-salmon ITS sequence clone 2, KF482108; T. urartu TMU38 IGS sequence clone 1, KF482109; T. urartu TMU38 IGS sequence clone 2, KF482110; Ae. longissima TL05 IGS sequence, KF482111; and Ae. tauschii TQ27 IGS sequence, KF482112.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FISH Analysis of pHvG38 in the Amphidiploid TL05 × TMU06.
Supplemental Figure 2. Dot Blot and qPCR Analysis of rDNA Copy Number in Different Natural and Synthetic Amphidiploid Wheat.
Supplemental Figure 3. FISH Analysis of rDNA Distribution on Somatic Metaphase Chromosomes in Natural Tetraploid Wheat.
Supplemental Figure 4. FISH Analysis of pHvG38 in Natural Tetraploid Wheat.
Supplemental Figure 5. FISH Analysis of pAs1 in the Amphidiploid TMU38 × TQ27.
Supplemental Figure 6. FISH and Multicolor FISH Analysis of Somatic Metaphase Chromosomes in Hexaploid Wheat.
Supplemental Figure 7. FISH Analysis of rDNA Distribution on Somatic Metaphase Chromosomes in Amphidiploid TB01 × TQ27.
Supplemental Figure 8. FISH Analysis of pHvG38 in the Amphidiploid TB01 × TQ27.
Supplemental Figure 9. Analysis of the Expression Pattern of rRNA Genes in Natural Tetraploid Wheat.
Supplemental Figure 10. Sequence Alignments of IGS Regions from Different Parental Genomes.
Supplemental Figure 11. Analysis of the Distribution and Expression of rRNA Genes in K-Salmon × CS, K-Salmon × 41,004, and Their Haploid Lines.
Supplemental Figure 12. Analysis of DNA Methylation and Relative DNA Methylation Ratio in Amphidiploids TL05 × TMU06 and TMU38 × TQ27.
Supplemental Figure 13. Immunolocalization Analysis of H4K12 Acetylation in the Amphidiploid TL05 × TMU06.
Supplemental Figure 14. Immunolocalization Analysis of H4K12 Acetylation on Metaphase Chromosomes of the Amphidiploid TMU38 × TQ27.
Supplemental Table 1. Primers Used in This Study
Supplementary Material
Acknowledgments
We thank Moshe Feldman (The Weizmann Institute of Sciences) for supplying seed samples, and Ingo Schubert and James Birchler for critical reading of the article. We also thank Martin Lysk, Bao Liu, Jiming Jiang, Khalili Kushkush, and Xu Jia for comments and suggestions. We thank Ryan Douglas and Nathan D. Han for editing the article. This work was supported by 863 Grant (2010AA100101) and State Key Laboratory of Plant Cell and Chromosome Engineering.
AUTHOR CONTRIBUTIONS
X.G. performed the research. X.G. and F.H. designed the research, analyzed the data, and wrote the article.
Glossary
- NOR
nucleolus organizing region
- IGS
intergenic spacer
- ITS
internal transcribed spacer
- FISH
fluorescence in situ hybridization
- qPCR
quantitative PCR
- RT-CAPS
reverse transcription-cleaved amplified polymorphic sequence
- TIS
transcription initiation site
- ChIP
chromatin immunoprecipitation
- ELD
expression level dominance
- DAPI
4′,6-diamidino-2-phenylindole
- RT-CAPS
reverse transcription-cleaved amplified polymorphic sequence
Footnotes
Online version contains Web-only data.
References
- Adams K.L., Wendel J.F. (2005). Novel patterns of gene expression in polyploid plants. Trends Genet. 21: 539–543. [DOI] [PubMed] [Google Scholar]
- Akhunov E.D., et al. (2010). Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11: 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov E.D., Chemeris A.V., Kulikov A.M., Vakhitov V.A. (2001). Functional analysis of diploid wheat rRNA promoter by transient expression. Biochim. Biophys. Acta 1522: 226–229. [DOI] [PubMed] [Google Scholar]
- Akhunova A.R., Matniyazov R.T., Liang H., Akhunov E.D. (2010). Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R., Gerlach W., Dennis E., Swift H., Peacock W. (1980). Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78: 293–311. [Google Scholar]
- Baack E.J., Whitney K.D., Rieseberg L.H. (2005). Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 167: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Wendel J.F., Ge S. (2010). Multiple patterns of rDNA evolution following polyploidy in Oryza. Mol. Phylogenet. Evol. 55: 136–142. [DOI] [PubMed] [Google Scholar]
- Baum B.R., Feldman M. (2010). Elimination of 5S DNA unit classes in newly formed allopolyploids of the genera Aegilops and Triticum. Genome 53: 430–438. [DOI] [PubMed] [Google Scholar]
- Blake N.K., Lehfeldt B.R., Lavin M., Talbert L.E. (1999). Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: the B genome of wheat. Genome 42: 351–360. [PubMed] [Google Scholar]
- Brettell R.I., Pallotta M.A., Gustafson J.P., Appels R. (1986). Variation at the Nor loci in triticale derived from tissue culture. Theor. Appl. Genet. 71: 637–643. [DOI] [PubMed] [Google Scholar]
- Chagué V., Just J., Mestiri I., Balzergue S., Tanguy A.-M., Huneau C., Huteau V., Belcram H., Coriton O., Jahier J., Chalhoub B. (2010). Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 187: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R.M., Udall J.A., Verma N., Springer N.M., Wendel J.F. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium). Genetics 182: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.G., Xu S., Friesen T., Faris J. (2008). Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol. Breed. 22: 251–266. [Google Scholar]
- Comai L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43: 387–399. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorák J. (1995). Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Pontvianne F., Wierzbicki A.T., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C.S. (2010). Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahima T., Röder M.S., Wendehake K., Kirzhner V.M., Nevo E. (2002). Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel. Theor. Appl. Genet. 104: 17–29. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2005). Allopolyploidy—a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109: 250–258. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2009). Genome evolution in allopolyploid wheat—a revolutionary reprogramming followed by gradual changes. J. Genet. Genomics 36: 511–518. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2012). Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A.A., Vega J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R.B., O'Dell M. (1979). The genetic control of nucleolus formation in wheat. Chromosoma 71: 135–152. [Google Scholar]
- Frankel O., Gerlach W., Peacock W. (1987). The ribosomal RNA genes in synthetic tetraploids of wheat. Theor. Appl. Genet. 75: 138–143. [Google Scholar]
- Gaeta R.T., Pires J.C., Iniguez-Luy F., Leon E., Osborn T.C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.-J., Chen Z.J. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Fedak G., Guo W., Liu B. (2005). Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170: 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M., Hiscock S. (2007). Polyploidy: doubling up for evolutionary success. Curr. Biol. 17: R927–R929. [DOI] [PubMed] [Google Scholar]
- Hetzl J., Foerster A.M., Raidl G., Mittelsten Scheid O. (2007). CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51: 526–536. [DOI] [PubMed] [Google Scholar]
- Houben A., Wako T., Furushima-Shimogawara R., Presting G., Künzel G., Schubert I., Fukui K. (1999). Short communication: the cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J. 18: 675–679. [DOI] [PubMed] [Google Scholar]
- Houchins K., O’Dell M., Flavell R.B., Gustafson J.P. (1997). Cytosine methylation and nucleolar dominance in cereal hybrids. Mol. Gen. Genet. 255: 294–301. [DOI] [PubMed] [Google Scholar]
- Jiang J., Gill B.S. (1994). Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support diphyletic origin of polyploid wheat. Chromosome Res. 2: 59–64. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Kohany O., Jurka M.V. (2007). Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 8: 241–259. [DOI] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A.A. (2002). Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A.A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J.C., Birchler J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan-Eichler M., Leshkowitz D., Tal L., Noor E., Melamed-Bessudo C., Feldman M., Levy A.A. (2011). Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A.R. (2005). Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kovarik A., Dadejova M., Lim Y.K., Chase M.W., Clarkson J.J., Knapp S., Leitch A.R. (2008). Evolution of rDNA in Nicotiana allopolyploids: A potential link between rDNA homogenization and epigenetics. Ann. Bot. 101: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A., Matyasek R., Lim K.Y., Skalicka K., Koukalova B., Knapp S., Chase M., Leitch A.R. (2004). Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol. J. Linn. Soc. Lond. 82: 615–625. [Google Scholar]
- Książczyk T., Kovarik A., Eber F., Huteau V., Khaitova L., Tesarikova Z., Coriton O., Chèvre A.-M. (2011). Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural forms of a polyploid species Brassica napus. Chromosoma 120: 557–571. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Chen Z.J. (2001). Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98: 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A.R., Leitch I.J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Leitch I., Bennett M. (2004). Genome downsizing in polyploid plants. Biol. J. Linn. Soc. Lond. 82: 651–663. [Google Scholar]
- Levy A.A., Feldman M. (2004). Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. Lond. 82: 607–613. [Google Scholar]
- Li A., et al. (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Vega J.M., Feldman M. (1998). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41: 535–542. [DOI] [PubMed] [Google Scholar]
- Liu Y., Taverna S.D., Muratore T.L., Shabanowitz J., Hunt D.F., Allis C.D. (2007). RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 21: 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ma X.F., Fang P., Gustafson J.P. (2004). Polyploidization-induced genome variation in triticale. Genome 47: 839–848. [DOI] [PubMed] [Google Scholar]
- Malinen E., Kassinen A., Rinttilä T., Palva A. (2003). Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149: 269–277. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- McStay B. (2006). Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 20: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Miller T.E., Hutchinson J., Reader S.M. (1983). The identification of the nucleolus organiser chromosomes of diploid wheat. Theor. Appl. Genet. 65: 145–147. [DOI] [PubMed] [Google Scholar]
- Mishima M., Ohmido N., Fukui K., Yahara T. (2002). Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110: 550–558. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Afsar K., Paszkowski J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34: 450–454. [DOI] [PubMed] [Google Scholar]
- Nagaki K., Talbert P.B., Zhong C.X., Dawe R.K., Henikoff S., Jiang J. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam V.J., Vales M.I., Watson C.J.W., Kianian S.F., Riera-Lizarazu O. (2006). Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.). Theor. Appl. Genet. 112: 373–381. [DOI] [PubMed] [Google Scholar]
- Navashin, M.S. (1928). Amphiplastic, eine neue karyologische Erscheinung. Proc. Int. Conf. Genet. 5: 1148–1152. [Google Scholar]
- Neves N., Heslop-Harrison J.S., Viegas W. (1995). rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat x rye hybrids. Theor. Appl. Genet. 91: 529–533. [DOI] [PubMed] [Google Scholar]
- Otto S.P. (2007). The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A.A., Feldman M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen C., Langridge P. (1997). Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40: 589–593. [DOI] [PubMed] [Google Scholar]
- Peng J., Ronin Y., Fahima T., Röder M.S., Li Y., Nevo E., Korol A. (2003). Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 100: 2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C.S. (2000). The epigenetics of nucleolar dominance. Trends Genet. 16: 495–500. [DOI] [PubMed] [Google Scholar]
- Pikaard C.S., Preuss S., Earley K., Lawrence R.J., Lewis M.S., Chen Z.J. (2005). Detecting differential expression of parental or progenitor alleles in genetic hybrids and allopolyploids. Methods Enzymol. 395: 554–569. [DOI] [PubMed] [Google Scholar]
- Pontes O., Lawrence R.J., Neves N., Silva M., Lee J.H., Chen Z.J., Viegas W., Pikaard C.S. (2003). Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S. (2004). Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 101: 18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Feng W., Stroud H., Jacobsen S.E., Michaels S.D., Pikaard C.S. (2012). Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26: 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S.B., Costa-Nunes P., Tucker S., Pontes O., Lawrence R.J., Mosher R., Kasschau K.D., Carrington J.C., Baulcombe D.C., Viegas W., Pikaard C.S. (2008). Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 32: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B., Huang W., Zhu B., Zhong X., Guo J., Zhao N., Xu C., Zhang H., Pang J., Han F., Liu B. (2012). Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R.A., Udall J.A., Wendel J.F. (2009). Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina E.A., Numerova O.M., Ozkan H., Feldman M. (2004). Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat. Genome 47: 860–867. [DOI] [PubMed] [Google Scholar]
- Sardana R., O’Dell M., Flavell R. (1993). Correlation between the size of the intergenic regulatory region, the status of cytosine methylation of rRNA genes and nucleolar expression in wheat. Mol. Gen. Genet. 236: 155–162. [DOI] [PubMed] [Google Scholar]
- Sarkar P., Stebbins G.L. (1956). Morphological evidence concerning the origin of the B genome in wheat. Am. J. Bot. 43: 297–304. [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban A.B., Badaeva E.D., Amosova A.V., Adonina I.G., Salina E.A. (2008). Genetic and epigenetic changes of rDNA in a synthetic allotetraploid, Aegilops sharonensis x Ae. umbellulata. Genome 51: 261–271. [DOI] [PubMed] [Google Scholar]
- Silva M., Pereira H.S., Bento M., Santos A.P., Shaw P., Delgado M., Neves N., Viegas W. (2008). Interplay of ribosomal DNA loci in nucleolar dominance: dominant NORs are up-regulated by chromatin dynamics in the wheat-rye system. PLoS ONE 3: e3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Flavell R. (1975). Characterisation of the wheat genome by renaturation kinetics. Chromosoma 50: 223–242. [Google Scholar]
- Soltis D.E., Buggs R.J., Doyle J.J., Soltis P.S. (2010). What we still don't know about polyploidy. Taxon 59: 1387–1403. [Google Scholar]
- Taverna S.D., Coyne R.S., Allis C.D. (2002). Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110: 701–711. [DOI] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunewaki K. (1964). Genetic studies of a 6x-derivative from an 8x Triticale. Can. J. Genet. Cytol. 6: 1–11. [Google Scholar]
- Volkov R.A., Borisjuk N.V., Panchuk I.I., Schweizer D., Hemleben V. (1999). Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol. 16: 311–320. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Madlung A., Lee H.-S., Chen M., Lee J.J., Watson B., Kagochi T., Comai L., Chen Z.J. (2004). Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.-S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wendel J.F., Schnabel A., Seelanan T. (1995). Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 92: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Stein N., Albar L., Feuillet C., Schlagenhauf E., Keller B. (2001). Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26: 307–316. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R.T., Pires J.C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B., Ben-David S., Kashkush K. (2013). Genome-wide analysis of Stowaway-like MITEs in wheat reveals high sequence conservation, gene association, and genomic diversification. Plant Physiol. 161: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.