Acetate is a primary Chlamydomonas fermentative product and is linked to dark, anoxic ATP biosynthesis. Chlamydomonas ack/pat mutants were isolated to further characterize fermentation networks, revealing that chloroplast pathways are dominant in this alga, and that despite blocking the primary ATP-generating routes to acetate, Chlamydomonas retains the metabolic flexibility to produce acetate.
Abstract
Chlamydomonas reinhardtii insertion mutants disrupted for genes encoding acetate kinases (EC 2.7.2.1) (ACK1 and ACK2) and a phosphate acetyltransferase (EC 2.3.1.8) (PAT2, but not PAT1) were isolated to characterize fermentative acetate production. ACK1 and PAT2 were localized to chloroplasts, while ACK2 and PAT1 were shown to be in mitochondria. Characterization of the mutants showed that PAT2 and ACK1 activity in chloroplasts plays a dominant role (relative to ACK2 and PAT1 in mitochondria) in producing acetate under dark, anoxic conditions and, surprisingly, also suggested that Chlamydomonas has other pathways that generate acetate in the absence of ACK activity. We identified a number of proteins associated with alternative pathways for acetate production that are encoded on the Chlamydomonas genome. Furthermore, we observed that only modest alterations in the accumulation of fermentative products occurred in the ack1, ack2, and ack1 ack2 mutants, which contrasts with the substantial metabolite alterations described in strains devoid of other key fermentation enzymes.
INTRODUCTION
Oxygen continually fluctuates in numerous terrestrial and aquatic environments, especially over the diel cycle. Soil-dwelling algae like Chlamydomonas reinhardtii can experience hypoxia and/or anoxia during the late afternoon and evening when respiration dominates over photosynthesis in the soil biosphere. Under hypoxic/anoxic conditions, Chlamydomonas generates energy by substrate level phosphorylation, a process which requires the glycolytic catabolism of fixed carbon (polysaccharides and sugars) to pyruvate. The reductant that accumulates during anoxic glycolysis is eliminated by metabolizing pyruvate to a number of different fermentative end products (predominately formate, acetate, ethanol, H2, and CO2) that are secreted from the cell (Gfeller and Gibbs, 1984, 1985; Mus et al., 2007; Dubini et al., 2009; Philipps et al., 2011; Magneschi et al., 2012; Catalanotti et al., 2012, 2013; Yang et al., 2014).
Aspects of Chlamydomonas metabolism appear to have significant flexibility, as established by physiological/metabolic studies with wild type and mutants aberrant for aspects of fermentation metabolism (Gfeller and Gibbs, 1984, 1985; Kreuzberg, 1984; Gibbs et al., 1986; Ohta et al., 1987; Hemschemeier and Happe, 2005; Atteia et al., 2006; Mus et al., 2007; Dubini et al., 2009; Timmins et al., 2009; Grossman et al., 2011; Philipps et al., 2011; Burgess et al., 2012; Catalanotti et al., 2012, 2013; Magneschi et al., 2012; Meuser et al., 2012; Yang et al., 2014; Figure 1), global examination of gene expression as cells acclimate to anoxic conditions (Mus et al., 2007), and analysis of the Chlamydomonas genome (Grossman et al., 2007, 2011; Merchant et al., 2007). The two dominant fermentative pathways that putatively consume pyruvate under anoxic, laboratory conditions involve the enzymes pyruvate formate lyase (PFL1) and pyruvate ferredoxin oxidoreductase (PFR1). Chlamydomonas PFL1, located in both mitochondria and chloroplasts (Kreuzberg et al., 1987; Atteia et al., 2006), converts pyruvate and CoASH to acetyl-CoA and formate (Philipps et al., 2011; Burgess et al., 2012; Catalanotti et al., 2012), while PFR1, specifically in chloroplasts (Atteia et al., 2009; Terashima et al., 2010; van Lis et al., 2013), converts pyruvate and CoASH to acetyl-CoA, CO2, and reduced ferredoxin. The reduced ferredoxin can be reoxidized by hydrogenases (Mus et al., 2007; Dubini et al., 2009; Grossman et al., 2011; Meuser et al., 2012), while the acetyl-CoA is either reduced to ethanol by alcohol/aldehyde dehydrogenase (ADH1) (Atteia et al., 2006; Grossman et al., 2011; Magneschi et al., 2012) or converted to acetate through the sequential action of phosphate acetyltransferase (PAT) and acetate kinase (ACK) (Wolfe, 2005; Ingram-Smith et al., 2006; Mus et al., 2007; Grossman et al., 2011; Catalanotti et al., 2013; Yang et al., 2014). Recently, Chlamydomonas fermentative mutants have been isolated, including hydEF, which is unable to assemble a functional hydrogenase (Posewitz et al., 2004) and under anoxic conditions biosynthesizes lower levels of CO2, extracellular formate, acetate, and ethanol relative to wild-type cells, but shows increased carboxylation of pyruvate to generate extracellular succinate and recycle NADH (Dubini et al., 2009); pfl1 mutants (Philipps et al., 2011; Burgess et al., 2012; Catalanotti et al., 2012) that exhibit increased pyruvate decarboxylation, extracellular ethanol, and lactate accumulation, as well as elevated intracellular levels of alanine, succinate, malate, and fumarate relative to wild-type cells (Catalanotti et al., 2012); and an adh1 mutant that is unable to biosynthesize either ethanol or CO2 and accumulates lower levels of formate and higher levels of acetate, lactate, and especially glycerol relative to the control strain (Magneschi et al., 2012). These metabolic differences among the mutants suggest an ability of Chlamydomonas to exploit a variety of fermentative pathways for recycling NADH to sustain glycolytic ATP production as the cells become hypoxic/anoxic.
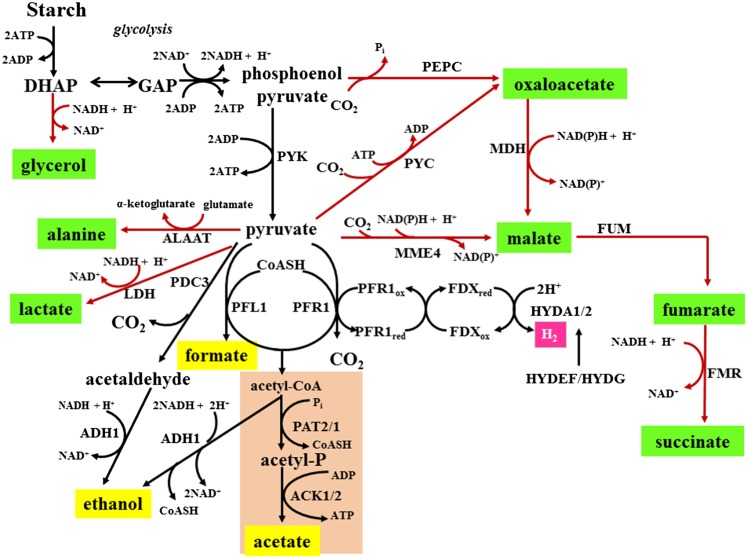
Figure 1.
Chlamydomonas Dark Fermentative Metabolism.
Photosynthetically derived starch that accumulates in chloroplasts during the day is degraded in the dark via glycolysis to pyruvate. Under anoxic conditions, pyruvate can be metabolized to acetyl-CoA, which is the substrate for the acetate-generating or ethanol-generating pathways. Pyruvate can also be used as a substrate to produce ethanol via the PDC3 (pyruvate decarboxylase)-ADH1 pathway. The compounds highlighted in yellow represent the major external metabolites produced by anoxic wild-type cells (Mus et al., 2007), while the compounds in green represent metabolites that accumulate in various mutant strains (externally or internally) under anoxic conditions. ALAAT, alanine aminotransferase; DHAP, dihydroxyacetone phosphate; FMR, fumarate reductase; FUM, fumarase; GAP, glyceraldehyde-3-phosphate; HYDEF, hydrogenase assembly factor EF; HYDG, hydrogenase assembly factor G; LDH, lactate dehydrogenase; MDH, malate dehydrogenase; MME4, malic enzyme 4; PEPC, phosphoenolpyruvate carboxylase; PYC, pyruvate carboxylase; PYK, pyruvate kinase.
To further characterize metabolic adjustments when major fermentation pathways are disrupted, we isolated mutants in the acetate biosynthesis pathways. Acetate biosynthesis and utilization have several functions in green algal cells under both aerobic and anaerobic conditions. In Chlamydomonas, acetate can serve as a sole energy source through respiratory metabolism. Acetate has also been found to contribute to the maintenance of anoxic conditions in the light since its oxidation promotes the utilization of O2 (Kosourov et al., 2007; Morsy, 2011) and, under certain nutrient deprivation conditions, serves as the building block for storing fixed carbon in the form of triacylglycerides (Johnson and Alric, 2013). During anaerobic metabolism, the production of acetate (and its subsequent secretion) through the PAT (Pta in bacteria)-ACK (Ack in bacteria) pathway can recycle the cofactor CoASH from acetyl-CoA and also generate ATP (Mus et al., 2007; Tielens et al., 2010; Atteia et al., 2013).
Bacterial and archaeal systems (Ingram-Smith et al., 2006) have retained a number of different pathways for acetate generation. In the methanoarchaeon Methanosarcina thermophila, Ack and Pta were found to be critical for acetate production (Ingram-Smith et al., 2000, 2005, 2006; Lawrence and Ferry, 2006) and for activation of acetate to acetyl-CoA when using acetate as the sole carbon/growth substrate (Rother and Metcalf, 2004). In Escherichia coli, acetate can be generated either from acetyl-CoA by the AckA-Pta system or from pyruvate catalyzed by pyruvate oxidase (Yang et al., 1999, 2001; Abdel-Hamid et al., 2001; Dittrich et al., 2005a, 2005b). In Corynebacterium glutamicum, all acetate biosynthesis in anoxic cultures is a consequence of the production of acetyl-CoA by pyruvate dehydrogenase (Blombach et al., 2007; Yasuda et al., 2007). In other organisms, acetate can be generated by the oxidative catabolism of pyruvate to acetate and CO2 by pyruvate quinone oxidoreductase and pyruvate oxidoreductase (Lin et al., 2003; Lorquet et al., 2004; Schreiner et al., 2006; Phue et al., 2010).
Putative ACK genes have been identified in some eukaryotes, including algae, fungi, and Entamoeba histolytica, although no PAT sequence has been found in amoeba or fungi (Ingram-Smith et al., 2006). In non-yeast fungi, ACK can partner with xylulose-5-phosphate/fructose-6-phosphate phosphoketolase, which catalyzes the production of acetyl phosphate (acetyl-P) from the breakdown of xylulose-5-phosphate or fructose-6-phosphate (Wolfe, 2005; Ingram-Smith et al., 2006). In some anaerobic parasites, acetyl-CoA can be converted into acetate, with concomitant production of succinyl-CoA by acetate:succinate CoA-transferase. Succinyl-CoA synthase catalyzes the conversion of succinyl-CoA back to succinate, which generates ATP (van Grinsven et al., 2008; Atteia et al., 2013; Catalanotti et al., 2013). Other enzymes that may influence acetate/acetyl-CoA levels are the evolutionarily and mechanistically distinct ADP-forming and AMP-forming acetyl-CoA synthetases. ADP-forming acetyl-CoA synthetase (ACS; EC 6.2.1.13) has been implicated in acetate production in amitochondriate protists (e.g., E. histolytica) as well as some archaea (Ingram-Smith et al., 2006; Fowler et al., 2012). The evolutionarily distinct AMP-forming ACS (EC 6.2.1.1) has generally been considered to operate solely in the direction of acetyl-CoA formation. However, two instances have been reported indicating that some AMP-forming acetyl-CoA synthetases may operate in the acetate-forming direction (Takasaki et al., 2004; Yoshii et al., 2009).
Interconversion between acetate and acetyl-CoA is also governed by differential subcellular localization of enzymes. In vascular plants such as spinach (Spinacia oleracea), acetyl-CoA hydrolase converts acetyl-CoA to acetate and CoASH in mitochondria, and acetate is transported to the chloroplast where acetyl-CoA can be regenerated by acetyl-CoA synthetase. The acetyl-CoA hydrolase Ach1p was shown to regulate intracellular acetyl-CoA or CoASH pools in yeast (Buu et al., 2003).
In Chlamydomonas, PAT-ACK activities appear to comprise the predominant pathways for acetate biosynthesis under dark anoxic conditions. This alga has two parallel ACK-PAT pathways, with previous work indicating that ACK2-PAT1 are localized to mitochondria, while ACK1-PAT2 are in chloroplasts (Atteia et al., 2006, 2009; Terashima et al., 2010). Chlamydomonas acetate accumulation can be influenced by altering fermentation pathways through the generation of mutants (Dubini et al., 2009; Philipps et al., 2011; Burgess et al., 2012; Catalanotti et al., 2012; Magneschi et al., 2012). In addition to the activities of PAT-ACK, the Chlamydomonas genome encodes other enzymes that may play a role in the synthesis of acetate, including four enzymes with homology to AMP-forming ACS (ACS1-4) and eight with homology to aldehyde dehydrogenase (ALDH) (Kirch et al., 2004, 2005; Brocker et al., 2013), which may catalyze the conversion of acetaldehyde to acetate with concomitant NAD(P)H production (Kirch et al., 2004, 2005; Brocker et al., 2013).
Currently, there is no experimental evidence demonstrating the contributions of the various biochemical paths outlined above to acetate metabolism in Chlamydomonas, under either oxic or anoxic conditions. If the combined ACK-PAT activities of Chlamydomonas represent the sole activities responsible for acetate production under anoxic conditions, suppression of these activities could potentially result in a total loss of acetate accumulation and rerouting of fermentation pathways to accommodate both the redox and energetic conditions of the cultures. To explore these issues, we used random insertional mutagenesis coupled with a PCR-based reverse genetic screen (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011) to identify Chlamydomonas strains disrupted for ACK and PAT genes. Characterization of these mutants has allowed us to evaluate the effect of these enzymes on algal cells experiencing dark anoxic conditions, expanding our understanding of the intricate relationships among the metabolic circuits associated with acetate metabolism.
RESULTS
ACK and PAT Genes in Chlamydomonas
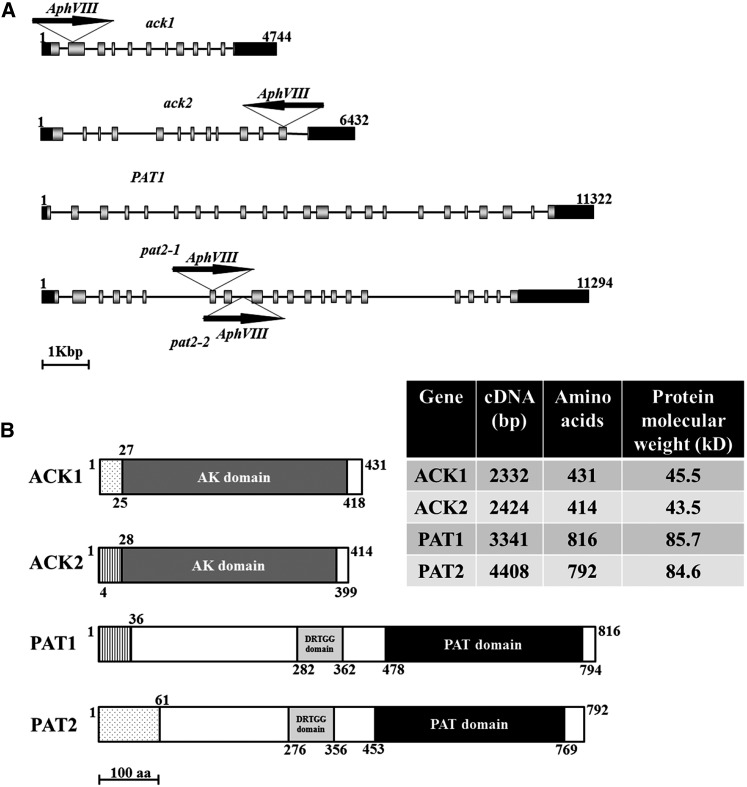
The Chlamydomonas genome has two genes encoding acetate kinase (ACK1 and ACK2) and two genes encoding phosphate acetyltransferase (PAT1 and PAT2). Based on Phytozome 9.1, ACK1 and PAT2 are contiguous on chromosome 9, while ACK2 and PAT1 are separate on chromosome 17 (Figure 2A). The predicted ACK1 and ACK2 proteins contain similar acetate kinase domains that span nearly the entire polypeptide sequence of each (although some residues at the beginning of this domain are in the presequence). PAT1 and PAT2 both contain a DRTGG domain and a phosphate acetyltransferase/butyryl transferase domain, the latter is found in a number of PAT (also designated Pta) proteins, although its function is not known (Figure 2B) (Campos-Bermudez et al., 2010). In the regions that align, the predicted amino acid sequences of ACK1 and ACK2 are 74% identical (304 of 412 amino acids), while those of PAT1 and PAT2 are 75% identical (548 of 734 amino acids).
Figure 2.
ACK and PAT Genomic Sequences and Protein Domains.
(A) ACK1, ACK2, PAT1, and PAT2 genes and the positions of insertions in the ack1, ack2, pat2-1, and pat2-2 mutants. Arrows indicate the orientation of the insertion. Black boxes represent 5′ and 3′UTRs, and gray boxes and black lines represent exons and introns, respectively. The numbers represent the size, in base pairs, of the genomic DNA.
(B) Predicted ACK1, ACK2, PAT1, and PAT2 protein domains. The white boxes indicate the segments of ACK and PAT that have not been specifically associated with a function/domain. The acetate kinase (AK), phosphate acetyl/butyryl transferase (PAT), and DRTGG domains are indicated. The lightly stippled boxes in ACK1 and PAT2 represent chloroplast transit peptides, whereas in ACK2 and PAT1, vertical lines represent mitochondrial presequences. The numbers along the length of the proteins represent amino acid positions. aa, amino acids.
ACK1 and PAT2 Localize to Chloroplasts, While ACK2 and PAT1 Localize to Mitochondria
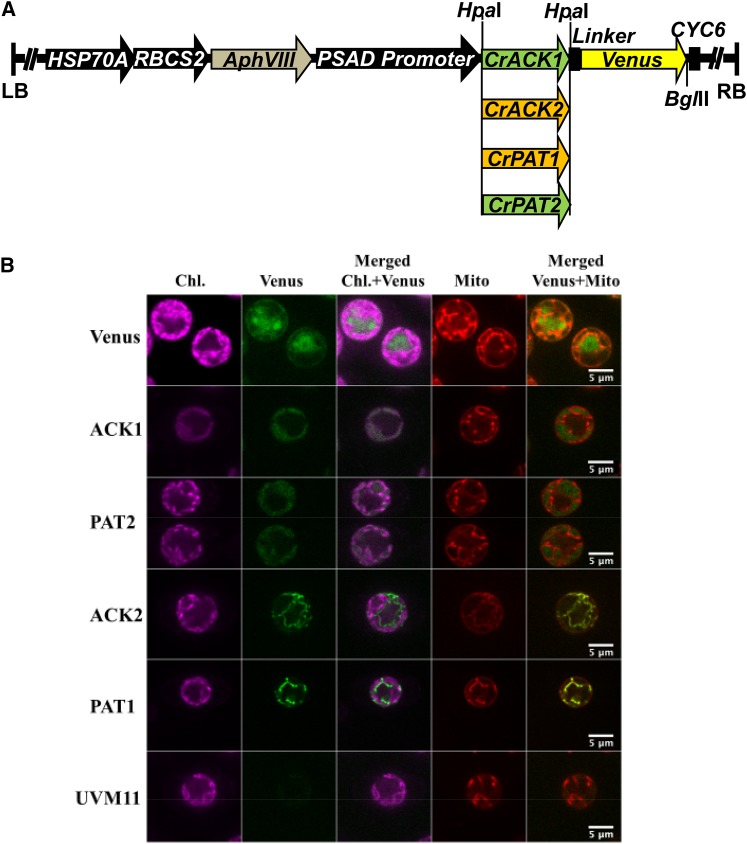
Previous biochemical and proteomic work with Chlamydomonas has suggested that the PAT2 and ACK1 proteins are localized to chloroplasts and the PAT1 and ACK2 proteins to mitochondria (Atteia et al., 2006, 2009; Terashima et al., 2010). The program PredAlgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main) (Tardif et al., 2012) predicted chloroplast transit peptides at the N termini of PAT2 and ACK1 (Figure 2B) and mitochondrial targeting presequences for PAT1 and ACK2 (Figure 2B). To experimentally determine the subcellular localization of these enzymes, the ACK1, ACK2, PAT1, and PAT2 genes were cloned for expression in Chlamydomonas as Venus fusion proteins (Figure 3A), transformed into Chlamydomonas cells, and transformants were screened for Venus fluorescence (Supplemental Figures 1A to 1D). Selected transformants were then imaged by spinning disk confocal microscopy. No Venus fluorescence was detected in control cells (Figure 3B, UVM11), while those expressing free Venus protein exhibited green Venus fluorescence exclusively in the cytosol (Figure 3B, Venus). By contrast, the ACK1 and PAT2 fusion proteins were localized to chloroplasts, while the ACK2 and PAT1 fusion proteins were localized in mitochondria (Figure 3B). We also prepared ACK2 and PAT1 fusion constructs with mCherry (Supplemental Figures 1E, 1F and 2A). Confocal microscopy again confirmed mitochondrial localization for ACK2 and PAT1, whereas the unfused mCherry was apparent only in the cytosol (Supplemental Figure 2B, mCherry). For some of the transformants (with both Venus and mCherry fluorophores), multiple cells for each strain were observed by spinning disk confocal microscopy; they all showed identical localization patterns (Supplemental Figures 3A and 3B).
Figure 3.
Localization of ACK and PAT Isozymes.
(A) Constructs for expression of ACK or PAT fused to Venus. Black arrows represent promoters; AphVIII confers paromomycin resistance, and CYC6 is a terminator. LB and RB, left and right border, respectively. Green represents genes encoding predicted chloroplast-localized proteins, and brown denotes those predicted to be mitochondrial.
(B) Localizations of ACK and PAT Venus fusion proteins in Chlamydomonas. For the indicated proteins, chlorophyll autofluorescence (Chl.), Venus fluorescence, merged chlorophyll and Venus signals, fluorescence from staining with Mitotracker to reveal mitochondria (Mito), and merged mitochondrial and Venus signals are shown. UVM11 is the Chlamydomonas strain used as the transformation host. Bars = 5 μm.
Identification of ack and pat Mutants in Chlamydomonas
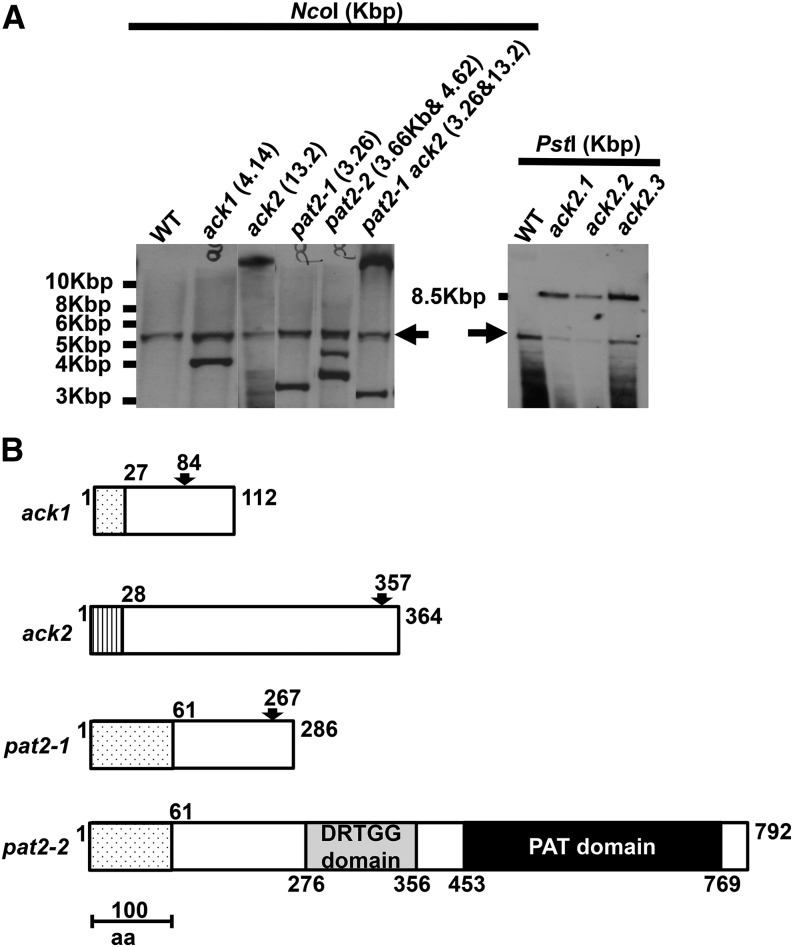
A PCR-based method (Gonzalez-Ballester et al., 2011) was used to identify strains specifically disrupted in the ACK1, ACK2, and PAT2 genes after random insertion of a AphVIII cassette conferring paromomycin resistance (Figure 2A; Supplemental Figure 4). No mutant disrupted in PAT1 was identified. DNA gel blot hybridizations demonstrated a single transgene insertion in the ack1, ack2, and pat2-1 mutants, whereas pat2-2 harbored two insertions and was therefore excluded from further analysis (Figure 4A). Molecular analyses demonstrated that the ack1, ack2, pat2-1, and pat2-2 mutants contained either small fragments of introduced Chlamydomonas DNA or small genomic deletions/rearrangements at the site of cassette insertion. These genomic alterations resulted in shifts of the open reading frames and changes in the sizes of the final gene products in ack1, ack2, and pat2-1 (Supplemental Figures 5 and 6; Figure 4B). By contrast, the insertion in the pat2-2 mutant would be spliced out as part of intron 8 (although the splicing is not nearly as efficient as for the wild-type intron), resulting in a predicted wild-type gene product of 792 amino acids (Supplemental Figure 5; Figure 4B).
Figure 4.
Genetic Analyses of Insertions in the Mutant Strains.
(A) DNA gel blot analyses showing insertions of the paromomycin-resistance cassette into the PAT/ACK genes in mutants ack1, ack2, pat2-1, pat2-2, and pat2-1 ack2. The strains and predicted sizes of the insertions are indicated above the blot. ack2.1, ack2.2, and ack2.3 are progeny from a backcross of the ack2 single mutant with the wild-type strain. NcoI and PstI were used to digest the genomic DNA. Marker sizes are given to the left of the blot. The arrows represent the hybridization to native PSAD promoter.
(B) Schematic representation of the proteins predicted to be synthesized in the ack1, ack2, pat2-1, and pat2-2 mutants. The arrows mark the positions of the insertions, and the numbers both above and below the representations are the amino acid positions in the protein.
A nearly homogeneous genetic background for all of the mutants was established with four consecutive backcrosses of ack1, ack2, and pat2-1 with wild-type strains (CC-124 and CC-125; same genetic background but minus and plus mating types, respectively). The backcrossed strains were then crossed with each other to generate the double mutants ack1 ack2 and pat2-1 ack2 (Supplemental Figure 4B; Figure 4A). Because the ACK1 and PAT2 genes are contiguous (within 300 bp) on chromosome 9, we were unable to generate the ack1 pat2 double mutant.
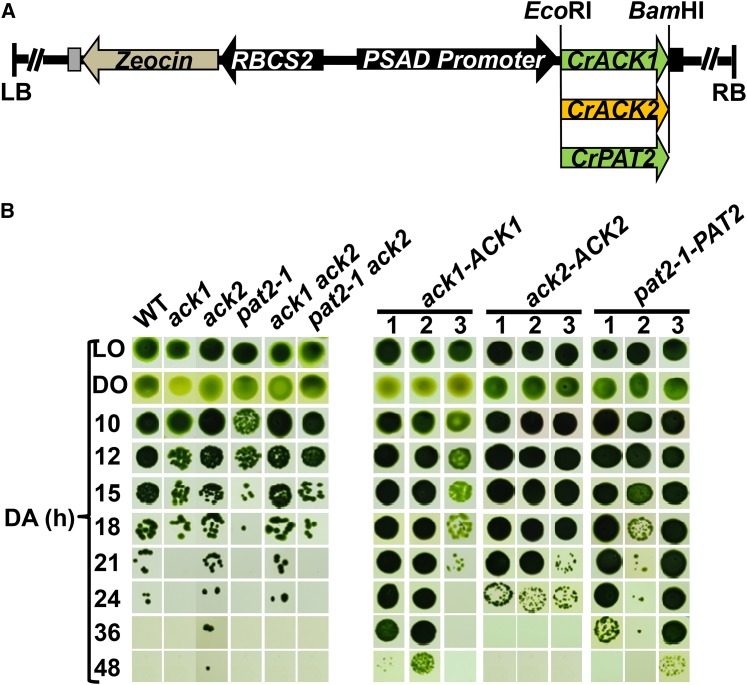
Fitness of ack and pat Mutants Following Exposure to Anoxic Conditions
ACK and PAT are thought to be critical for acetate and ATP production under anoxic conditions. To address whether the inability to synthesize acetate coupled to ATP production might make the mutant strains more sensitive to anoxia relative to the wild-type cells, we examined the viability of both single and double mutants during exposure to anoxic conditions. The ack1 and pat2-1 mutants were always the most vulnerable to anoxia; in no case were we able to recover these strains after 24 h of anoxia (Figure 5B). Interestingly, both the ack2 and ack1 ack2 mutants survived for at least as long as wild-type cells. These results suggest that acetate production in the chloroplast mediated by ACK1 and PAT2 is important for survival and for the production of ATP under anoxic conditions. The increased survival (e.g., over 24 h) of the ack1 ack2 double mutant relative to the ack1 single mutant (Figure 5B) suggests that other metabolic factors/pathways are induced when both the mitochondrial and chloroplast PAT-ACK pathways are blocked.
Figure 5.
Cell Viability Following Exposure to Anoxic Conditions.
(A) Constructs used to complement the mutant strains. The black arrows represent promoters and the black box following the introduced ACK or PAT gene indicates the PSAD terminator. Green represents genes encoding the chloroplast-localized ACK1 and PAT2 proteins, and brown denotes the mitochondrion-localized ACK2 protein.
(B) Viability of the wild-type, mutant, and complemented strains. Cells were concentrated in HS medium and exposed to dark anoxic conditions. After the indicated times under dark anoxic conditions, the cells were spotted onto solid TAP medium under relatively low illumination conditions (50 µmol photons m−2 s−1) and allowed to grow for 7 d. The initial number of cells spotted was ∼10,000 for oxic growth and ∼100,000 for anoxic growth. DA, LO, and DO represent dark anoxic, light oxic, and dark oxic conditions, respectively. Three isolates for each of the complementation strains were used for viability tests. A single representative experiment is shown; similar results were obtained in six repetitions.
To confirm that the ack1 and pat2 lesions were responsible for the loss of viability during anoxia, the same viability assay was performed in complemented strains containing a wild-type copy of ACK1 and PAT2 (Figure 5A). Two ack1-ACK1 transformants recovered the anoxia tolerance phenotype and survived longer than the wild-type strain under anoxic conditions, while the third transformant survived for approximately the same amount of time as wild-type cells (Figure 5B). The ack2-ACK2 strain maintained anoxia tolerance with robust survival to 24 h (the original mutant was not impaired in survival). The pat2-1-PAT2 strains survived longer than wild-type cells during anoxia (Figure 5B). Collectively, these results indicate that loss of activity of the chloroplast-localized ACK1 and PAT2 proteins causes increased sensitivity to anoxia, which is not observed for strains disrupted in the mitochondrion-localized ACK2.
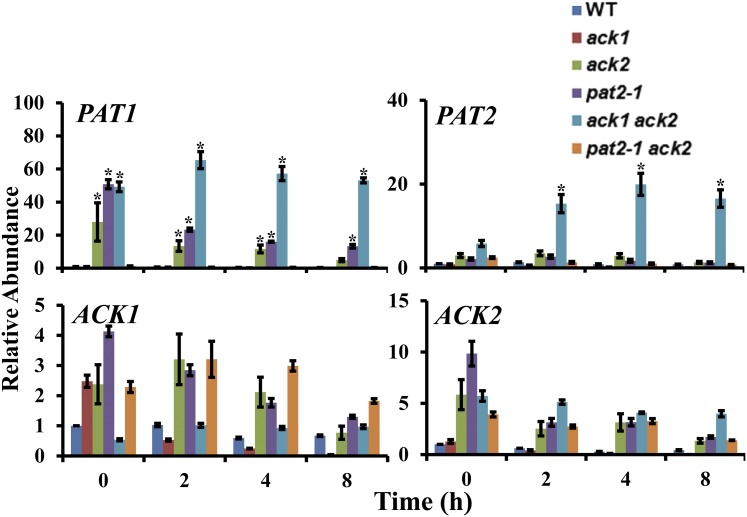
Transcription of Fermentative Enzymes
We examined how disruption of specific PAT and ACK genes affected the levels of transcripts encoding PAT1, PAT2, ACK1, and ACK2. PAT1 mRNA was elevated in the ack2, pat2-1, and ack1 ack2 mutant strains relative to the wild-type cells under dark anoxic conditions (Figure 6); the most pronounced increase was in the ack1 ack2 double mutant. PAT2 mRNA was significantly increased only in the ack1 ack2 double mutant (Figure 6). The expression of ACK1 and ACK2 (analyzed with primers that bind upstream of the site of the insertion in the ack2 mutant) was not significantly different in the ack2, pat2-1, and pat2-1 ack2 mutant strains compared with wild-type cells under dark anoxia (Figure 6).
Figure 6.
Changes in ACK and PAT Transcript Abundances in ack and pat Mutants.
RT-qPCR was used to assess ACK and PAT transcript levels in the wild type, the ack1, ack2, and pat2-1 single mutants, and the ack1 ack2, pat2-1 ack2 double mutants under dark anoxic conditions. The T0 time point represents the time prior to anaerobic induction (oxic samples). Absolute quantifications were performed; expression levels are presented relative to gene expression at 0 h. Errors bars represent sd. Asterisks represent significance (Student's t test, P < 0.01).
To investigate how disruption of PAT and ACK genes affects expression of genes involved in fermentative metabolism under anoxia, we analyzed levels of transcripts from HYDA1, HYDA2, PFR1, PFL1, ADH1, and PDC3 under anoxia (Supplemental Figure 7). In all of the strains, except the ack1 mutant, but including wild-type cells, the HYDA1 and HYDA2 transcripts were elevated, with the changes ranging between 2- and 7-fold under the conditions used. Any differences between the mutant and wild-type cells were not significant. The differences in the levels of PFR1, PFL1, ADH1, and PDC3 transcripts in the mutant and wild-type cells were generally less than 2-fold, although the highest levels of the PFR1 and PFL1 transcripts were observed (throughout the time course of anoxia) in the ack2 mutant. Overall, these results suggest that mutations in ACK and PAT can influence levels of transcripts from the various ACK and PAT genes but do not markedly affect expression of other genes associated with fermentation metabolism.
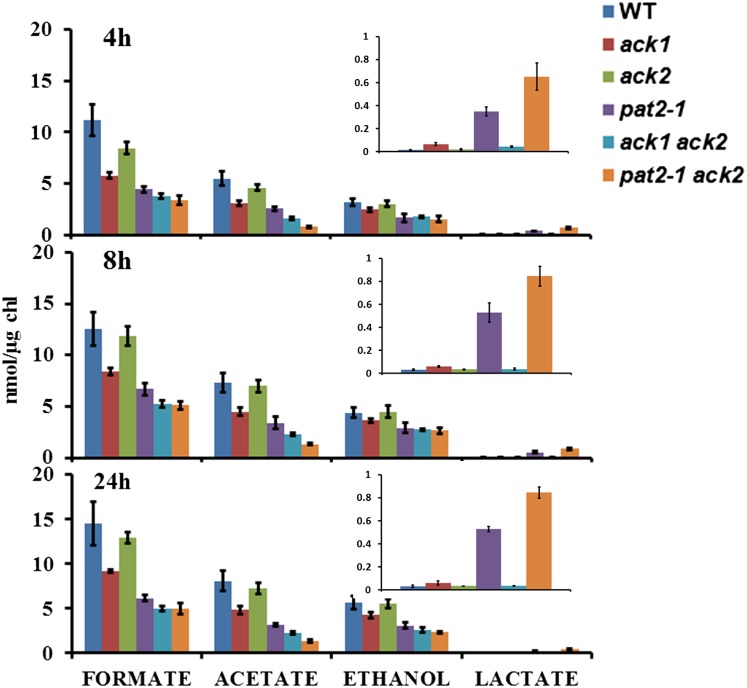
Extracellular Metabolite Analyses
To further explore the functions of ACK and PAT with respect to fermentative metabolism in Chlamydomonas, we analyzed the accumulation of metabolites excreted into the medium when the wild-type and mutant strains were transitioned from oxic to anoxic conditions (Figure 7). Overall, there was less formate, acetate, and ethanol produced in all of the strains relative to wild-type cells at 4, 8, and 24 h after the imposition of anoxia, except for ack2, for which there was essentially no difference from the wild type (Figure 7). This once again suggests that disruption of the PAT-ACK pathway in the chloroplast (the ack1, pat2-1, ack1 ack2, and pat2-1 ack2 mutants would all be affected in acetate production in chloroplasts) has a larger influence relative to mitochondrial mutations on the overall fermentative responses of the cells to anoxia. Lactate production was elevated in pat2-1 and pat2-1 ack2 mutants (Figure 7, insets), although to a relatively small extent, indicating that some rerouting of metabolites to lactate did occur, which would facilitate NADH reoxidation. Complementation of the pat2-1 mutant with a wild-type copy of PAT2 yielded strains with slightly higher levels of metabolites, but the transformed cell lines tested did not fully recover wild levels of fermentative metabolites (Supplemental Figure 8). Interestingly, there was still acetate generated in the ack1 ack2 double mutant under dark anoxia. NMR was used to confirm that acetate was being made in this mutant (Supplemental Figure 9). These results suggest that some other acetate producing pathway(s) becomes active (or more apparent) in the ack1 ack2 double mutant (which would be unable to synthesize acetate by the PAT-ACK pathway in either organelle) during dark anoxia. Overall, our findings demonstrate that when no acetate is synthesized through the PAT-ACK pathway, particularly through the chloroplast pathway, the cells exhibit a downregulation of fermentation metabolism and that activities/pathways other than those of PAT-ACK must be active in producing acetate under dark anoxic conditions in these mutants.
Figure 7.
Accumulation of External Metabolites in Mutant Cultures.
HPLC quantitation of extracellular metabolites secreted by the indicated mutants after 4, 8, or 24 h of dark anoxic acclimation. Lactate production is shown as an inset with an adjusted scale. Errors bars represent se of the mean.
H2 Evolution
Fermentative H2 levels were measured by gas chromatography to determine the effect of disrupting acetate biosynthesis in the various mutant strains on H2 production. In general, there was little difference in H2 production between wild-type cells and the mutant strains (Supplemental Figure 10). Furthermore, no substantial differences were found in the production of CO2 between the wild type and the mutant strains (Supplemental Figure 11), indicating that ethanol production through the PDC3 pathway does not appear to be activated in these mutants, which is consistent with the finding that these strains exhibit little change in the levels of PDC3 and ADH1 transcripts (Supplemental Figure 7).
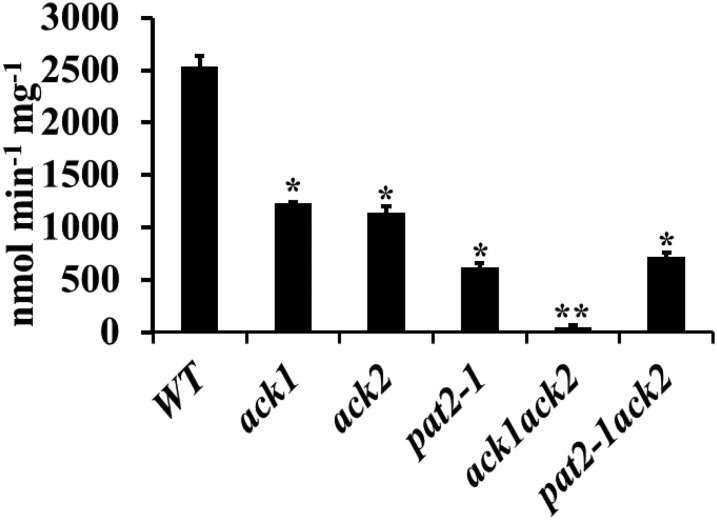
Decreased Acetate Kinase Activities Were Detected in Various Single and Double Mutants
To evaluate the contribution of ACK to acetate production, in vitro acetate kinase activity was measured in wild-type, ack1, ack2, pat2-1, ack1 ack2, and pat2-1 ack2 mutant strains (Figure 8). The ack1 and ack2 single mutants each had ∼50% of the activity of wild-type, whereas the pat2-1 single mutant and the pat2-1 ack2 double mutant showed ∼25% of the activity of the wild type. The ACK activity in the ack1 ack2 mutant was near background levels but could be restored by complementation with either the ACK1 or ACK2 gene (Figure 8; Supplemental Figure 12). In spite of the near complete loss of ACK enzyme activity, the ack1 ack2 double mutant still secretes acetate (Figure 7).
Figure 8.
In Vitro Acetate Kinase Activity in the Mutants.
Acetate kinase activity was assayed as the biosynthesis of acetate from acetyl-CoA in the wild type and the pat and ack mutants. Errors bars represent se of the mean. Asterisks represent significant difference from the wild type (*P < 0.05; **P < 0.01).
Alternative Enzymatic Routes to Acetate Production under Anoxic Conditions
The finding that the ack1 ack2 double mutant still produces acetate suggests that the cells have mechanisms outside the PAT-ACK pathway for acetate biosynthesis. We therefore used informatic tools to identify genes in the Chlamydomonas genome potentially associated with acetate production (Figure 9, Table 1). The ADP-forming acetyl-CoA synthetases, present in certain parasites and a limited number of archaea, are absent in Chlamydomonas. However, there are four Chlamydomonas gene models encoding proteins with similarity to characterized AMP-forming acetyl-CoA synthetases. Among these, ACS1 and ACS2 possess typical AMP-forming ACS sequence motifs and likely specifically use acetate as a substrate, whereas ACS3 has Ala instead of Val at position 1521 and may be able to use propionate as a substrate (Ingram-Smith et al., 2006). ACS4 is more similar to an acyl-CoA synthetase (Table 1). Alignments and phylogenetic analyses suggest significant divergence of these genes (Supplemental Figures 13A and 13C and Supplemental Data Set 1). Although AMP-forming acetyl-CoA synthetases are not typically associated with acetate production, there are two reports of acetate production from such enzymes (Takasaki et al., 2004; Yoshii et al., 2009).
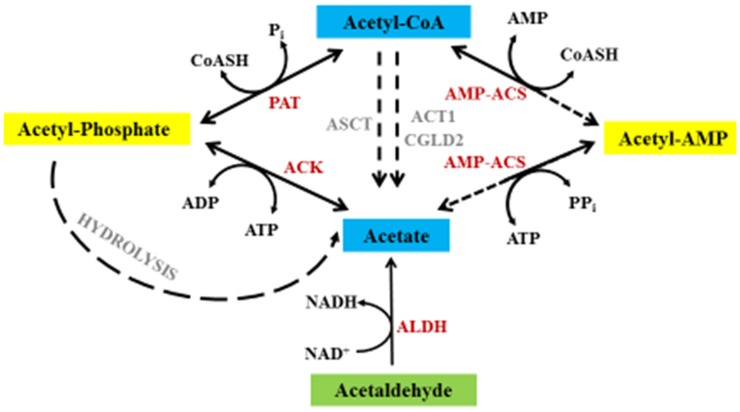
Figure 9.
Potential Metabolic Routes to Acetate Biosynthesis.
The various potential routes for acetate production from acetyl-CoA in Chlamydomonas. Red denotes enzymes encoded by high-confidence gene models in the Chlamydomonas genome, and gray represents gene models for which the function is not entirely clear. Solid lines represent reactions confirmed in Chlamydomonas, while dashed lines indicate proposed reactions based on gene model analyses and homology searches. ACT1, Acyl-CoA thioesterase; CGLD2, Acyl-CoA thioesterase.
Table 1. Gene Models Encoding Uncharacterized Enzymes Potentially Involved in Acetate Production.
| Gene | Phytozome v10.0.4 | NCBI | Annotation | Localizationa |
|---|---|---|---|---|
| ACS1 | g1290.t1 | XP_001700210.1 | AMP-forming acetyl-CoA synthase | O |
| ACS2 | g1224.t1 | XP_001700230.1 | AMP-forming acetyl-CoA synthase | C |
| ACS3b | Cre07.g353450 | XP_001702039.1 | AMP-forming acetyl-CoA synthase | O |
| ACS4 | Cre01.g055500 | XP_001700230.1 | AMP-forming acetyl-CoA synthase | M or SP |
| ALDH1 | g13400.t1 | XP_001694180.1 | Aldehyde dehydrogenase | O |
| ALDH2 | Cre16.g675650 | XP_001695943.1 | Aldehyde dehydrogenase | M |
| ALDH3 | Cre12.g500150 | XP_001690955.1 | Aldehyde dehydrogenase | M |
| ALDH4c | Cre12.g520350 | XP_001696928.1 | Aldehyde dehydrogenase | M |
| ALDH5 | Cre01.g033350 | XP_001690075.1 | Aldehyde dehydrogenase | SP |
| ALDH6 | g8982.t1 | XP_001694332.1 | Aldehyde dehydrogenase | M |
| ALDH7 | g16809 | XP_001698924.1 | Aldehyde dehydrogenase | C |
| ALDH8c | Cre13.g605650 | XP_001699134.1 | Aldehyde dehydrogenase | O or SP |
| SCLA1c | Cre03.g193850 | XP_001693108.1 | Succinate-CoA ligase | M |
| SCLB1ac | g17060.t1 | XP_001691581.1 | Succinate-CoA ligase | M |
| ACT1 | g16435.t1 | XP_001692073.1 | Acyl-CoA thioesterase | O |
| CGLD2 | g837.t1 | XP_001690113.1 | Acyl-CoA thioesterase | M |
Predicted localizations of ACS, ALDH, and other possible acetate metabolism related proteins using PredAlgo software (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main). Names, protein IDs, NCBI accession numbers, and predicted localizations are indicated. C, chloroplast; M, mitochondrion; SP, secretory pathway; O, other. ACS3 was shown to be localized to mitochondria (Atteia et al., 2009; Terashima et al., 2010). ALD1 and ALD2 were shown to be localized to mitochondria (Atteia et al., 2009), which correspond to ALDH4 and ALDH8 in our study. SCLA1 and SCLB1a were shown to be localized to mitochondria (Atteia et al., 2009).
The locations of the proteins are either predicted or were experimentally determined.
Another potential pathway for acetate generation involves acetate:succinate CoA-transferase (ASCT) and succinyl-CoA ligase (SCL; also called succinyl-CoA synthetase). ASCT transfers the CoA moiety of acetyl-CoA to succinate, and SCL converts succinyl-CoA back to succinate (this latter reaction is not shown in Figure 9). In the SCL reaction, the energy in the thioester bond of succinyl-CoA is used for the biosynthesis of ATP. There are two gene models on the Chlamydomonas genome that encode putative SCLs. However, no gene model for ASCT has been identified, which could mean that there is no Chlamydomonas ASCT, that lack of similarity to characterized ASCT enzymes does not allow for its easy identification, or that the gene is present in a region of the genome that has not been sequenced or where the sequence quality is poor.
Eight gene models encode putative ALDHs, which may convert acetaldehyde to acetate, with the concomitant production of NADH (Figure 9, Table 1). Alignments and phylogenetic analyses showed that ALDH3 and ALDH4 have some similarity to atypical ALDH enzymes, which brings their biological function into question (Supplemental Figures 13B and 13D and Supplemental Data Set 2). The Chlamydomonas ALDH proteins are predicted to be in various cellular locations (Table 1); ALDH1-4, ALDH6, and ALDH8 are predicted to be in mitochondria, while ALDH5 and ALDH7 are predicted to be targeted to the secretory pathway and chloroplasts, respectively (Table 1). Furthermore, the Chlamydomonas genome also encodes acetyl-CoA hydrolases (indicated as ACT1 and CGLD2 in Figure 9, Table 1), but there is no experimental evidence to show that they function in the conversion of acetyl-CoA to acetate.
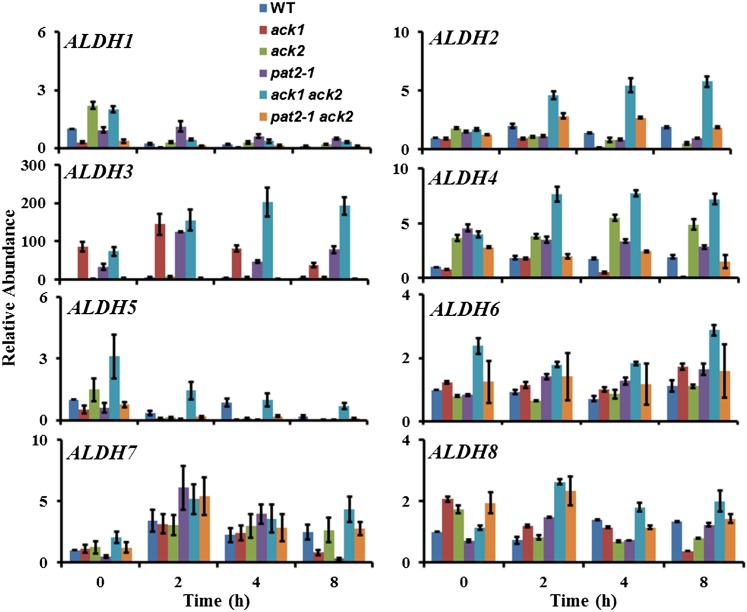
To elucidate the potential origin of acetate production in the ack1 ack2 double mutant, reverse transcription-quantitative PCR (RT-qPCR) was performed to monitor expression of the eight ALDH genes and the four ACS genes under dark anoxic conditions (Figure 10; Supplemental Figure 14). The ACS genes were not highly upregulated during dark anoxia in wild-type or mutant strains, suggesting that AMP-forming ACS does not function in dark anoxia in Chlamydomonas (Supplemental Figure 14), although posttranscriptional regulation allowing such a function remains possible. By contrast, ALDH2, ALDH3, and ALDH4 transcripts showed significantly higher accumulation in the ack1 ack2 double mutant, with the ALDH3 transcript upregulated by almost 200-fold. ALDH3 was also expressed highly in the ack1 and pat2-1 single mutants under anoxic conditions, raising the possibility that alternative acetate-producing pathways may be activated in mutants defective in the PAT/ACK pathway and especially in those strains defective for the chloroplast isoforms of the enzymes.
Figure 10.
ALDH Transcript Abundance during Acclimation to Anoxia.
RT-qPCR was used to quantify transcripts for eight distinct ALDH gene models in the wild type and the indicated mutant strains. Data are from two biological replicates, each with three technical replicates. Errors bars represent sd.
Intracellular Metabolites
Intracellular levels of pyruvate and acetyl-phosphate were measured to determine whether the accumulation of these metabolites, which are potentially influenced by acetate production (Figure 1), were affected by ACK/PAT disruption. Intracellular levels of pyruvate increased slightly (∼2-fold) in the ack1 ack2 and pat2-1 backgrounds (Supplemental Table 1), whereas the levels of acetyl-phosphate decreased slightly in all mutants (Supplemental Figures 15).
DISCUSSION
Photosynthetic microorganisms rely on dark-phase metabolism for approximately half of the diel cycle. Elucidating the metabolic networks activated during the night is key to understanding net carbon cycling and cellular physiology when light is not available to drive photosynthetic processes. Such studies provide valuable information regarding the specific roles of enzymes predicted to be associated with dark metabolism and to elucidate metabolic networks that are available to sustain ATP production in the dark. From a biofuel perspective, it is critical to understand catabolic processes in the absence of light, how much fixed carbon is directed toward respiratory processes over the course of the day, and how these processes influence overall net carbon assimilation. Furthermore, “dark” metabolism has ecological consequences as it is now clear that many algae and cyanobacteria secrete reduced energy carriers (e.g., organic acids/alcohols and H2) under fermentation conditions (Mus et al., 2007; Ananyev et al., 2008; Dubini et al., 2009; Carrieri et al., 2010), providing reducing equivalents and carbon substrates to the heterotrophic consortia of organisms that coexist in the same ecological niche. Such secreted products likely play an important role in influencing the biota present in a variety of aquatic and soil ecosystems (Hoehler et al., 2002; Spear et al., 2005).
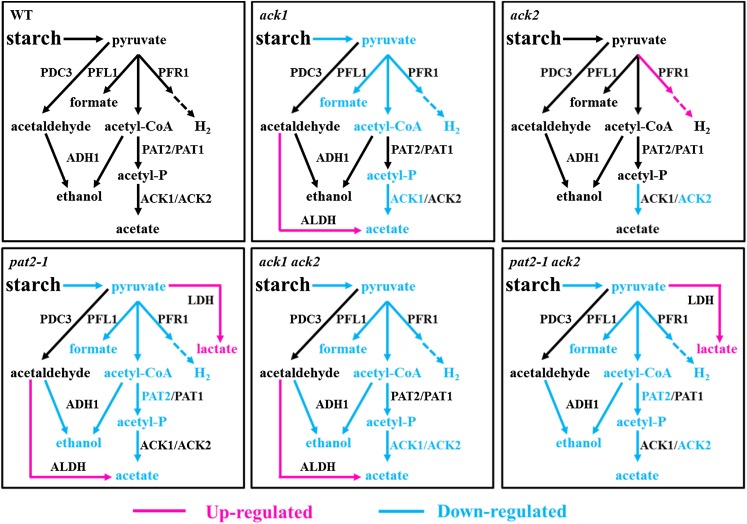
Chlamydomonas wild-type strains typically secrete formate, acetate, and ethanol, as the predominant organic products, in addition to evolving CO2 and H2 during anoxia in the dark. In recent years, mutant analysis has shown that an extraordinary suite of fermentation pathways are available to Chlamydomonas and that this alga has a remarkable ability to reorganize fermentation pathways when specific metabolic nodes are blocked by genetic disruption (Dubini et al., 2009; Philipps et al., 2011; Burgess et al., 2012; Catalanotti et al., 2012; Magneschi et al., 2012). This flexibility might also serve to allow Chlamydomonas to overcome enzyme inhibition by chemicals/compounds present in the natural environments. Here, we elucidated the roles of the PAT and ACK enzymes, which are putatively responsible for anoxic acetate production (Figure 1).
Our localization analyses (Figure 2; Supplemental Figures 2 and 3) support the previous assignment of PAT1 and ACK2 as mitochondrial and PAT2 and ACK1 as chloroplastic (Atteia et al., 2006, 2009; Terashima et al., 2011). The presence of two distinct PAT/ACK pathways in Chlamydomonas suggests that fermentation occurs in both the mitochondrion and chloroplast. Analysis of mutants disrupted in PAT2, ACK1, or ACK2, and the pat2-1 ack2 and ack1 ack2 double mutants, revealed that the chloroplast is the dominant contributor to overall cellular fermentation metabolism and that Chlamydomonas is able to retain the ability to produce acetate even when both the chloroplast and mitochondrial PAT/ACK pathways are compromised. In contrast to mutations affecting H2, formate, or ethanol metabolism, the ack mutations did not result in a significant rerouting of metabolites, whereas disruptions of pat2-1 led to increased lactate secretion relative to the control strain (Figure 7). The most severe attenuation of extracellular acetate accumulation was in the pat2-1 ack2 mutant, which is blocked at different steps in the respective chloroplast and mitochondrial acetate production pathways; however, even this mutant still showed significant levels of acetate secretion. To eliminate acetate carryover from the medium as a potential source of acetate, fermentative metabolite experiments were performed using cells cultured in acetate-free minimal medium. In vitro ACK enzyme assays showed that the ack1 ack2 double mutant lacked acetate kinase activity, eliminating ACK activity as the mechanism for acetate production. The in vitro acetate kinase data also showed that ACK1 and ACK2 were approximately equal in their contributions to whole-cell activity (Figure 8; Supplemental Figure 11), indicating that both mitochondrial and chloroplast routes could contribute to acetate production, provided availability of sufficient substrate.
The finding that the ack1 ack2 and pat2-1 ack2 mutants (in which both chloroplast and mitochondrial pathways are disrupted) secrete low levels of acetate after being cultured in minimal medium led us to consider routes other than the PAT/ACK pathway that potentially function in acetate production (Figure 9, Table 1). These include: (a) the spontaneous hydrolysis of acetate phosphate to acetate and Pi (Koshland, 1952; Di Sabato and Jencks, 1961); (b) aldehyde dehydrogenase activity that oxidizes acetaldehyde (from pyruvate decarboxylation) to acetate (Kirch et al., 2004, 2005; Brocker et al., 2013); (c) acetyl-CoA hydrolase, which hydrolyzes acetyl-CoA to acetate and CoA without the production of ATP (Tielens et al., 2010); (d) acetate formation by AMP-forming acetyl-CoA synthetase (Takasaki et al., 2004; Yoshii et al., 2009); and (e) ASCT and SCL activities (van Grinsven et al., 2008; Millerioux et al., 2012). ASCT transfers the CoA moiety of acetyl-CoA to succinate and SCL converts succinyl-CoA back to succinate. In the SCL reaction, the energy in the thioester bond of succinyl-CoA is used for the synthesis of ATP (van Grinsven et al., 2008). Although SCL homologs are encoded on the Chlamydomonas genome, no ASCT homologs were identified, and it is unlikely that this pathway represents a viable alternative for acetate production in Chlamydomonas. Acetate production by AMP-forming acetyl-CoA synthetase cannot be ruled out, as described above (Takasaki et al., 2004; Yoshii et al., 2009); however, it should be noted that despite these reports, the majority of these enzymes typically function in the direction of acetyl-CoA synthesis (Tielens et al., 2010).
For the other three potential pathways, an important consideration in the interpretation of the metabolite data is that different metabolites would have accumulated in the respective pat and ack mutants as a consequence of the enzymatic block resulting from the gene disruption. In the ack mutants, metabolism can proceed to the level of acetyl-P prior to enzymatic disruption, whereas in the pat2-1 mutant, the acetate pathway is blocked at the level of acetyl-CoA in the chloroplast. In the ack1 ack2 double mutant, acetyl-P biosynthesis should occur. Acetyl-P can undergo spontaneous hydrolysis in aqueous solvents to generate acetate and Pi (Koshland, 1952; Di Sabato and Jencks, 1961), and it is possible that this leads to the observed low levels of acetate secreted from the ack1 ack2 and pat2-1 ack2 mutants in the medium. Presumably, a functional ACK enzyme in wild-type cells would capture the acetyl-P prior to spontaneous hydrolysis. The production of acetate by nonenzymatic acetyl-P hydrolysis would deprive the cell of the ATP produced by the action of ACK. In the absence of ATP production by ACK activity, cellular ATP levels may be diminished below a threshold level that would be needed to efficiently prime glycolysis. Analysis of intracellular metabolites indicated that pyruvate does accumulate to slightly higher levels (Supplemental Table 1) relative to the control at several points of anoxic acclimation, consistent with a metabolic block downstream of pyruvate. Increased levels of acetyl-P were not observed as a consequence of disruptions in the PAT-ACK pathway, which may indicate a kinetically facile conversion of this substrate (Supplemental Figure 15). The ack1, pat2-1, ack1 ack2, and pat2-1 ack1 mutants secrete lower levels of all fermentation products, which is consistent with a reduced rate of glycolysis in these mutant strains (Figure 11; Supplemental Figure 16). Relative to the ack1 ack2 double mutant, the pat2-1 ack2 mutant produces slightly less acetate. As the PAT2 disruption blocks acetyl-P production in the chloroplast, this lesion would be expected to have diminished acetyl-P flux, which may explain why acetate production is more attenuated in the pat2-1 ack2 mutant. It is possible that nonenzymatic acetyl-P hydrolysis explains the observed residual production of acetate in the double mutants, the lower overall yields of all fermentation products (presumably due to a slower rate of glycolysis) in the double mutants and ack1, and the more severely diminished levels of acetate secretion in pat2-1 ack2 relative to ack1 ack2.
Figure 11.
Modulation of Anoxic Pathway Activities in Single and Double ack/pat Mutants.
Summary of altered metabolic pathway activities in the indicated mutants relative to the wild-type control strain. Blue represents attenuated pathway activities in the respective mutants; red represents increased pathway activities in the indicated mutants; dashed arrows represent catalytic steps that were omitted in the transfer of electrons from PFR1 to the hydrogenases.
Another possibility that would explain acetate secretion from the ack1 ack2 and pat2-1 ack2 double mutants is that the metabolites acetyl-CoA and pyruvate, which are directly upstream of acetyl-P in fermentation metabolism, are also accumulated due to metabolic backup when the PAT/ACK pathway is blocked and that these substrates are converted to acetate. Pyruvate could be decarboxylated by PDC3 to generate acetaldehyde, which may then be oxidized to acetate by ALDH activity, a mechanism that is frequently used in some yeast (Remize et al., 2000). This reaction would form NADH, which if accumulated would slow glycolysis (as is observed) unless the cell reoxidized the NADH to maintain redox balance. We do not observe the secretion of any additional metabolites that would serve as this reductant sink; however, some of the ALDH transcripts, particularly ALDH3, show significant increases in abundance and this pathway cannot be dismissed. Despite this, no significant aldehyde dehydrogenase activities were detected among mutant and wild-type strains at different times after dark anoxia, reflecting the complexity of the physiology of the ALDH enzymes and substrates in vivo (Supplemental Figure 17).
Alternatively, acetyl-CoA could be converted to acetate by acetyl-CoA hydrolase. Genes encoding homologs of acetyl-CoA hydrolase are found on the Chlamydomonas genome. If acetyl-CoA accumulated on the cell and was not metabolized by alternative pathways, cellular CoA would be committed and unavailable for other cellular processes. Hydrolysis of acetyl-CoA would free CoA and produce acetate without generating ATP. As in the case of spontaneous acetyl-P hydrolysis, this would deny the cell ATP production by the action of ACK, potentially diminishing the rate of glycolysis and leading to the reduction of fermentation metabolite secretion. If the AMP-forming acetyl-CoA synthetase was used for acetate production, ATP production would be maintained; we did not observe significant changes in ACS transcript levels in any of the mutant cell lines, but this activity is reported to be posttranslationally regulated (Takasaki et al., 2004). This alternative pathway would be functionally equivalent to the PAT/ACK pathway in that ATP production would be retained; consequently, it is questionable whether a diminished glycolysis and acetate-secretion phenotype would emerge if AMP-forming ACS was able to functionally substitute for PAT/ACK.
While multiple potential mechanisms for generating acetate in ack1 ack2 and pat2-1 ack2 have been identified, we are unable to determine which of the pathways (or potentially which combination of pathways) is used to produce the acetate secreted in the double mutants or if there are additional options to biosynthesize acetate. However, it is clear that several potential mechanisms exist to explain residual acetate production in the absence of ACK activity. At this point, we do not know whether these alternative routes are active in wild-type cells or if they only become active when the mutants accumulate activated acetyl units. Interestingly, robust acetate production is also retained by unknown mechanisms in mutants of Clostridium species in which Ack activity is eliminated by a gene knockout (Sillers et al., 2008; Kuit et al., 2012).
Regarding the single gene disruptions, ack1 had a more pronounced reduction of acetate secretion relative to ack2 (Figure 7). This implies that the chloroplast is the dominant location of fermentative acetate production and perhaps fermentative pathways in general. The pat2-1 mutant consistently produced less acetate than the ack1 mutant (both chloroplast enzymes), which would be expected if nonenzymatic acetyl-P hydrolysis contributed to the observed levels of secreted acetate in the mutants. Interestingly, both the pat2-1 and the pat2-1 ack2 mutants showed an increase in lactate secretion (Figure 11; Supplemental Figure 16), suggesting that metabolic rerouting occurs exclusively in the pat2-1 genetic background. This would be expected if the metabolic block occurred at the level of acetyl-CoA contributing to the increased accumulation of pyruvate, which could be readily redirected toward lactate production for redox balancing, as observed in the adh1 and pfl1-1 mutants (Catalanotti et al., 2012; Magneschi et al., 2012). Metabolic engineering approaches seeking to eliminate the flux of pyruvate to the PAT/ACK pathway so that pyruvate can be directed to other pathways typically target PAT rather than ACK for gene disruption/deletion (Atsumi et al., 2008a, 2008b). The absence of metabolic rerouting to lactate in the ack mutants indicates that after the production of acetyl-P by PAT, the cell uses an alternative route to produce acetate that is secreted, thereby eliminating metabolite accumulation and the opportunity to redirect metabolism to alternative pathways. To reintegrate acetate into central metabolism via the glyoxylate cycle would establish a futile cycle that would consume ATP and produce reducing equivalents, two unsustainable outcomes during anoxia. Therefore, the data suggest that once acetyl-P is produced, the cell’s options for metabolic redirection are more limited relative to mutations that result in the accumulation of metabolites at earlier steps in the fermentation pathway.
In summary, our insertional mutagenesis/reverse genetics analyses of primary fermentation networks underscore the remarkable ability of Chlamydomonas to overcome deletions of preferred enzymatic activity and to sustain fermentation metabolism, often using pathways that are completely uncharacterized. In contrast to mutants in the other steps of fermentation, we observed no major metabolic rerouting in the ack mutants and limited metabolic rerouting to lactate in the pat2-1 mutant. The generally diminished accumulation of all fermentative metabolites in the ack and pat mutants is indicative of a diminished rate of glycolysis and pyruvate supply to fermentation. Finally, our results suggest that mitochondrial fermentation pathways are less active than chloroplast fermentation under the conditions analyzed.
METHODS
Strains, Mutant Isolation, and Growth Conditions
The parental Chlamydomonas reinhardtii strains used in these studies were CC-124 (nit2−, mt−) and CC-125 (nit2−, mt+), which are designated as “wild-type” cells throughout the article. The various mutant strains used are the ack1, ack2, pat2-1, and pat2-2 single mutants and the ack1 ack2, pat2-1 ack2, and pat2-2 ack2 double mutants. The single mutants were generated in the D66 (CC-4425; nit2−, cw15, mt+) genetic background (Pollock et al., 2003) by a PCR-based screen that was previously described (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011), while the double mutants were generated by mating pairs of single mutants. The primers used for the mutant screen are listed in Supplemental Table 2A. All of the mutants were backcrossed four times with CC-124 and CC-125, alternatively, including the ack1 ack2, pat2-1 ack2, and pat2-2 ack2 double mutants. Both parental and mutant strains were maintained on solid (agar plates) Tris-acetate-phosphate (TAP) medium (pH 7.2) and inoculated into liquid TAP medium, minimal medium, or high-salt medium (HS) and grown at different light intensities as indicated. The UVM11 strain was kindly provided by Ralph Bock (Neupert et al., 2009) and was specifically used for the subcellular localization of the ACK and PAT polypeptides.
DNA Gel Blot Analyses
DNA gel blot hybridizations were performed as previously described (Catalanotti et al., 2012; Magneschi et al., 2012). Genomic DNA was isolated from 50 mL liquid cultures of wild-type Chlamydomonas, various mutants (ack1, pat2-1, pat2-2, and pat2-1 ack2) and from four tetrads derived from a cross between ack2 single mutants. Genomic DNA was isolated using a standard phenol-chloroform extraction protocol (Sambrook et al., 1989) and 10 μg extracted DNA was digested for 2 h with 10 units PstI or NcoI (New England Biolabs). The digested DNA was resolved by agarose (0.8%) gel electrophoresis, blotted overnight in 20× SSC onto nylon membranes (Bio-Rad), and then cross-linked to the membranes by UV illumination. An alkaline phosphatase-labeled DNA probe was synthesized by chemically cross-linking a thermostable alkaline phosphatase to the 1.7-kb PSAD-AphVIII PCR fragment (Sizova et al., 2001), which also contains the PSAD promoter and a 3′ sequence from the Cyc6 gene (Fischer and Rochaix, 2001). The probe was amplified from plasmid DNA using a forward primer designated Paro-up and reverse primer designated Paro-dw. The synthesis of the probe, hybridizations to the membrane-associated DNA fragments, and detection of the hybridization signal were performed using an Amersham AlkPhos Direct Labeling and Detection System according to the manufacturer’s protocol (Amersham Biosciences).
Construction of the pLM004_Venus and pLM004_mCherry Vectors
The pLM004_Venus and pLM004_mCherry vectors were constructed identically (see Supplemental Figure 18 for maps). pLM004 was made from six fragments assembled via Gibson assembly (Gibson et al., 2009). The HSP70A/RbcS2 promoter region driving the AphVIII paromomycin resistance gene followed by the RbcS2 3′UTR (untranslated region) was amplified from pJR38 (Neupert et al., 2009). The PSAD promoter from pMJ013b was inserted in the same direction as the resistance elements. Downstream of the PSAD promoter and flanked by HpaI restriction sites is the LacZ alpha fragment cloned in the opposite direction (amplified from pUC19) to allow blue/white selection during plasmid construction. Directly after the second HpaI site is a flexible linker region encoding the sequence GDLGGSGGR, with a BglII site at the start to allow removal of the fluorophore-encoding sequence. Downstream of the flexible linker is the sequence encoding either Venus (amplified from pJR39; Neupert et al., 2009) or mCherry; each is followed by a GGG flexible linker, a 6x His tag, a BglII site, and a stop codon. Downstream of the stop codon is a Cyc6 intron prior to the PSAD 3′UTR, followed by the inverted Rpl12 3′UTR to terminate transcription in the opposite direction (amplified from pMJ013b). Escherichia coli propagation elements, including the ampicillin resistance gene, were cloned from pUC19. EcoRV sites flanking the Chlamydomonas elements in the vector were added to allow for removal of the pUC19 backbone prior to transformation.
Subcellular Localization of ACK1, ACK2, PAT1, and PAT2 Proteins
The coding sequences of ACK1, ACK2, PAT1, and PAT2 were amplified from random primer retrotranscribed cDNAs using the forward and reverse primers ACK1-Loc, ACK2-Loc, PAT1-Loc, and PAT2-Loc with HpaI adapters at both of the 5′ and 3′ ends (Supplemental Table 2B). The ACK1, ACK2, PAT1, and PAT2 genes were cloned into the pLM004_Venus vector, and the ACK2 and PAT1 genes were also cloned into the pLM004_mCherry vector. After sequencing, the plasmids were transformed by electroporation using a Bio-Rad GenePulser II Electroporator into the Chlamydomonas UVM11 strain grown to a cell density of 0.5 to 1 × 108 per mL (Neupert et al., 2009). The parameters used for electroporation were a capacitance of 25 μF and a voltage of 800 V (using a standard 0.4-cm cuvette; Bio-Rad). Transformants were plated onto solid TAP medium containing 100 µg/mL ampicillin and 10 μg/mL paromomycin. Following transformation, 11 positive Chlamydomonas colonies (paromomycin resistant) were selected for each construct. Colonies were replicated and transferred to liquid TAP medium in a 96-well plate format and grown to late exponential/early stationary phase. After sequential growth for 3 d each, triplicate aliquots of cells (150 μL each) were transferred to individual wells of 96-well flat bottom microtiter plates (Greiner Bio-One 655101) also containing 6 to 12 wells of untransformed UVM11 and two wells of TAP only. The plates were analyzed using a Tecan infinite M1000 PRO plate reader (Tecan Group) for Venus (excitation 515/12 and emission 550/12), mCherry (excitation 585/12 and emission 615/12), and chlorophyll (excitation 440/9 and emission 680/20) fluorescence. The fluorophore signal was determined by normalizing the fluorophore emission of the cells from individual wells to their chlorophyll fluorescence and then subtracting the mean fluorescence of the untransformed UVM11 cells over the range of Venus and mCherry emissions (Rasala et al., 2013). Strains with the highest fluorescence were used for imaging by point scanning (Leica SP5) or spinning disk confocal microscopy.
Anaerobic Induction of Liquid Cell Suspensions and Sampling
Chlamydomonas precultures were grown in liquid TAP medium and then transferred to 1-liter Roux culture bottles (HS medium), stirred with a magnetic stir bar at 25°C, 80 μmol photon m−2 s−1 of continuous PAR irradiance (cool, white fluorescence lights, Sylvania, F34W), and vigorously bubbled with air enriched with 3% CO2. Cultures were grown to a density of 15 to 20 μg chlorophyll mL−1 (measured three times), and cells were pelleted by centrifugation at 2500g for 5 min. For external metabolite analysis, cell precultures and cultures were grown in HS medium in a Multitron II incubated shaker (Infors HT) at 25°C, 90 μmol photon m−2 s−1 of continuous PAR irradiance, 3% CO2, and collected at mid-log phase (∼15 to 20 μg chlorophyll−1 mL−1) by centrifugation (3700g for 10 min). Pelleted cells were concentrated by resuspension in one-tenth volume of HS medium (normally 60 mL), and then transferred to a sealed anaerobic vial (final chlorophyll was 150 to 200 μg chlorophyll mL−1), flushed with argon for 30 min and incubated at room temperature under anaerobic conditions (Coy Laboratory Products anaerobic chamber) in the dark. Aliquots of the culture were sampled at different times (0, 2, 4, and 8 h for proteins and nucleic acid; 4, 8, and 24 h for external metabolites) over a 24-h period after making the cultures anaerobic. The samples were centrifuged (10,000g for 1 min) to pellet the cells, and supernatants and pellets were separated, frozen in liquid N2, and stored at −80°C for later analyses (e.g., external metabolites, protein, and nucleic acid). To monitor survival of cells during anaerobic incubations, cultures were grown to a density of 2 × 106 cells mL−1 (counted three times using a hemocytometer) and cells were pelleted as described above (final cell numbers were 2 × 107 cells mL−1). Five-microliter aliquots of the cultures were collected at specific intervals, spotted onto the surface of solid TAP medium, and exposed to ambient levels of O2 (in air under standard growth conditions; 50 μmol photons m−2 s−1 at 25°C) to allow for the growth of viable cells. Control samples were maintained under aerobic conditions in the light and dark.
Chlorophyll Extraction and Growth Rate
Chlamydomonas cells were pelleted by centrifuging 1 mL of culture at 21,113g for 5 min, and the pelleted cells resuspended in 1 mL 100% methanol. The tubes were immediately wrapped in aluminum foil to exclude light, vortexed 10 times for 10 s each, and then allowed to rest for 5 to 10 min. The cell debris was pelleted by centrifugation (21,113g for 5 min). The absorbance of the supernatant was then quantified at OD652 and OD665 and total chlorophyll was calculated as previously described (Heinnickel et al., 2013).
Extraction of RNA
Total RNA was isolated from frozen cell pellets (no thawing) using a standard phenol-chloroform extraction protocol (Sambrook et al., 1989). The RNA was precipitated overnight at 4°C from the aqueous extract by adding an equal volume of 8 M lithium chloride (4 M LiCl, final concentration), which eliminates most of the DNA. To completely remove DNA from the samples, ∼40 μg of the RNA was treated with 5 units of RNase-free DNase I (Qiagen) for 1 h at room temperature; this treatment was repeated when necessary. A Qiagen RNeasy MinElute kit was used to further purify the DNase-treated total RNA and to remove degraded DNA, tRNA, 5S rRNA, DNase, contaminating proteins, and potential inhibitors of the reverse transcriptase reaction. The A260 of the eluted RNA was measured and the integrity of the RNA checked by electrophoresis in agarose-formaldehyde gels (Catalanotti et al., 2012).
RT-qPCR
The abundance of specific transcripts in total mRNA of each sample was quantified by RT-qPCR using the Light Cycler 480 (Roche Applied Science). First-strand cDNA synthesis was primed from purified RNA templates using gene-specific primers (Supplemental Table 2C. Reverse transcription reaction conditions were previously described (Mus et al., 2007; Dubini et al., 2009). Amplifications were performed using the following cycling parameters: an initial step at 95°C for 5 min (denaturation) and then 40 cycles of (1) 95°C for 10 s (denaturation), (2) 60°C for 15 s (annealing), and (3) 72°C for 15 s (elongation). Melting curves (65 to 100°C, heating rate of 0.2°C s−1 with continuous fluorescence measurements) were evaluated for PCR products to ensure that a single DNA species was amplified. Both the absolute (Steunou et al., 2006) and relative levels of each specific RNA (normalized to the wild-type T0 sample corresponding to oxic conditions) were determined. All reactions were performed in triplicate with at least two biological replicates.
Extracellular Metabolite Analysis
Organic acid analysis was performed by liquid chromatography using a Surveyor Plus (Thermo Scientific) HPLC. Dark-adapted cells were collected at indicated time points following the imposition of anoxic conditions and centrifuged (17,000g for 3 min). The resulting supernatant was filtered with a 0.45-μm filter and either directly injected (25 μL) onto a fermentation monitoring 150 × 7.8-mm column (Bio-Rad) or transferred to a new vial and frozen in liquid N2 for subsequent analysis. Metabolites in the supernatant were separated by the column at 45°C at a flow rate of 0.5 mL min−1, with 8 mM sulfuric acid serving as the mobile phase. Retention peaks for the various organic acids, detected by a refractive index detector at the operating temperature 50°C, in parallel with a photodiode array detector, at 210 nm, were recorded and analyzed using Agilent ChemStation software; quantifications were performed by comparisons with the absorption of known amounts of a standard for each of the organic acids.
Acetate Confirmation by NMR
Acetate content was confirmed by NMR using the same samples that were used for HPLC measurements. A total volume of 0.5 mL, which contained 2/3 sample supernatant and 1/3 water (99.9% D2O), was analyzed by NMR. 1H NMR spectra were obtained using an ECA-500 NMR spectrometer (Jeol) and analyzed by Jeol/Delta software.
CO2 and H2 Measurement
Cells acclimated to dark, anoxic conditions (4, 8, and 24 h) were assayed for CO2 production by adding 1 mL of 1 M HCl to sealed vials containing 1 mL of the anoxic cells. The acidified cell suspension was shaken vigorously to liberate CO2 into the vial headspace, and CO2 (0.2 mL injection) was quantified by gas chromatography (Hewlett Packard 5890 series II) using a Supelco column (80/100 PORAPAK N 6FT × 1/8 IN × 2.1 mm) coupled to a thermal conductivity detector; helium was used as the carrier gas. The resulting signal was integrated using ClassVP software, and the gas was quantified according to standard curves generated from known quantities of CO2. Fermentative H2 production was measured after 4, 8, and 24 h of dark anoxic conditions by withdrawing and analyzing 0.2 mL of headspace gas by gas chromatography using a Hewlett Packard Series II 5890 (60/80 mole sieve 5A 6FT × 1/8 IN) instrument fitted with a Restek 5Å Molecular Sieve 80/100 6’ 1/8” column and a thermal conductivity detector with argon as the carrier gas. The resulting signal was integrated using ChemStation software, and H2 was quantified using a standard curve generated from known quantities of H2 (Posewitz et al., 2004; Mus et al., 2007).
Complementation
The 1935-bp Chlamydomonas ACK1 cDNA was amplified using the specific primers ACK1-COM-F1 and ACK1-COM-R1. The 1658-bp Chlamydomonas ACK2 cDNA was amplified using the specific primers ACK2-COM-F1 and ACK2-COM-R1. The 2728-bp Chlamydomonas PAT2 cDNA was amplified using the specific primers PAT2-COM-F1 and PAT2-COM-R1. The ACK1, ACK2, and PAT2 products were cloned into pGEM T-easy (Promega) and sequenced. The pGEM T-easy constructs were digested with EcoRI and BamHI, which allowed for directional cloning of the ACK1, ACK2, and PAT2 products into plasmid pJM43Ble (Invitrogen) for constitutive expression of the resistance gene under the control of the RBCS2 promoter. Nuclear transformations were performed by introducing 1 μg plasmids pJM43Ble-ACK1, pJM43Ble-ACK2, and pJM43Ble-PAT2 (all linearized with NotI) into ack1, ack2, and pat2-1 mutants (in the CC-124 background), respectively, by electroporation as described above. Additionally, pJM43Ble-ACK1 and pJM43Ble-ACK2 were introduced into the ack1 ack2 double mutant to generate ack2 (ack1 ack2-ACK1) and ack1 (ack1 ack2-ACK2) single mutant rescued strains. The plasmids pJM43Ble-ACK2 and pJM43Ble-PAT2 were introduced into the pat2-1 ack2 double mutant to obtain pat2-1 (pat2-1 ack2-ACK2) and ack2 (pat2-1 ack2-PAT2) single mutant rescued strains. Transformants were spread onto solid TAP medium containing 100 μg/mL ampicillin, 5 μg/mL paromomycin, and 6 μg/mL bleomycin.
Acetate Kinase Activity
The cells were grown to mid-log phase (1 × 106 cells mL−1), and 100 mL culture was pelleted by centrifugation at 3200g. The pellet was resuspended in 20 mL extraction buffer (25 mM HEPES, pH 7.5 KOH, 5 mM MgCl2, and 0.3 M sucrose) containing protease inhibitors (100 mM PMSF, 200 mM benzamidine, and 400 mM ε-aminocapoic acid). The cell suspension was then passed through the French Pressure Cell twice at 2000 p.s.i. (this results in complete breakage of the cells, which was checked microscopically). The lysed suspension was centrifuged at 18,000g for 15 min, and the supernatant containing the soluble proteins was transferred to a Falcon tube and then assayed for protein concentration using the Bradford assay (Bio-Rad) with BSA as the standard. Acetate kinase enzymatic activity in the acetate/ATP-forming direction was determined using a coupled enzyme assay in which ATP production was coupled to the reduction of NAD(P)+ to NAD(P)H by hexokinase and glucose-6-phosphate dehydrogenase activities. Reactions (200 μL total) contained 100 mM Tris (pH 7.5), 20 mM MgCl2, 5.5 mM glucose, 0.5 mM DTT, 1 mM NADP+, 5 mM ADP, and 10 mM acetyl phosphate. Hexokinase and glucose-6-phosphate dehydrogenase (5 units; Sigma-Aldrich H8629) were added and the reaction mixture was allowed to equilibrate at ambient temperature. Reactions were initiated by the addition of the cell extract, and the amount of extract added was determined empirically to give a linear response under the conditions of the assay. The change in absorbance at 340 nm was monitored over the 5-min reaction time. The results shown are the mean and se of three independent assays for each sample.
ALDH Activity Assay
Samples were prepared under anoxic conditions. The cells were prepared as described above (Anaerobic Induction of Liquid Cell Suspensions and Sampling). Cells were disrupted by bead-beating (4 × 30 s, full speed) with 200 μg of 0.4-mm glass beads inside the anoxia chamber. The ALDH activity in the cell extract was measured under oxic conditions using the Aldehyde Dehydrogenase Activity Colorimetric Assay Kit (BioVision), with some modifications (http://www.biovision.com/article_info.php?articles_id=79).
Pyruvate Detection and Quantification by Gas Chromatography-Mass Spectrometry
Anoxic cell samples were prepared and collected as described above for external metabolite analyses at 4, 8, and 24 h after dark anoxia. The cells were stored at −80°C until assayed. The analysis was performed with at least two biological and four technical replicates. To perform the assay, frozen cells were resuspended in 1 mL ice-cold 80% methanol, sonicated for 15 min, vigorously shaken for 2 h at room temperature, and then centrifuged at 3000g for 10 min. Supernatant (500 μL) was transferred to a separate tube and dried under a vacuum using a SpeedVac. Metabolites were resuspended in 50 μL pyridine containing 15 mg/mL methoxyamine hydrochloride, incubated at 60°C for 45 min, sonicated for 10 min, and incubated for an additional 45 min at 60°C. Fifty microliters of N-methyl-N-trimethylsilyltrifluoroacetamide with 1% trimethylchlorosilane (Thermo Scientific) was then added to the samples, which were further incubated at 60°C for 30 min, centrifuged at 3000g for 5 min, and cooled to room temperature. Then, 80 μL supernatant was transferred to a 150-μL glass insert in a gas chromatography-mass spectrometry autosampler vial. Pyruvate was detected using a Trace GC Ultra coupled to a Thermo ISQ mass spectrometer (Thermo Scientific). Each sample was injected twice at a 1:10 split ratio in discrete randomized blocks. Separation occurred using a 30-m TG-5MS column (Thermo Scientific; 0.25 mm i.d., 0.25-μm film thickness) with a 1.2 mL/min helium gas flow rate, and a separation program of 80°C for 30 s, a ramp of 15°C per min to 330°C, and an 8-min hold. Masses between 50 and 650 m/z were scanned at five scans/second after electron impact ionization. Pyruvate was identified by matching retention time and m/z values to a commercial standard and quantified by calibrating peak areas to a five-point standard curve. Finally, pyruvate concentrations in the extracts were normalized to the total number of cells harvested.
Acetyl-Phosphate Quantification
The assay was performed as described by Prüss and Wolfe (1994) with modifications. Anoxic samples were collected as described above for external metabolite analyses at 4, 8, and 24 h after dark anoxia. Lysis buffer (1% SDS, 200 mM NaCl, 20 mM EDTA, and 50 mM Tris-HC, pH 8.0) was added to the cell pellets on ice. After vortexing for 20 s, the samples were transferred into the tubes with one-fifth volume of glass beads (0.45 μm; Sigma-Aldrich G8722) and disrupted by bead beating for 30 s at 4°C. Activated charcoal (0.5 g) was then added to the samples, which were vortexed for 2 min and centrifuged at 18,407g for 25 min at 4°C. The supernatant was transferred to storage vials (2 mL; National Scientific). The charcoal was removed by centrifugation and filtration; AcP was used to catalyze the formation of ATP by adding 1 μL 1 M MgCl2, 250 μL 30 mM ADP (A2754-1G; Sigma-Aldrich), and 25 μg Ack (25 μL) (from E. coli; A7437; Sigma-Aldrich) to 1 mL charcoal-treated extract followed by incubation at 30°C for 90 min. The concentration of ATP was then determined using the luciferase assay (E1500; Promega). ATP levels in each charcoal-treated extract (assayed but without the addition of Ack) were determined as background. The acetyl-P content of each sample was calculated using standards (0, 10 μM, 100 μM, 500 μM, 1 mM, 2 mM, and 5 mM acetyl-P; A0262; Sigma-Aldrich) that were subjected to the entire extraction and conversion procedure. The intracellular concentration of acetyl-P was expressed as μM, normalized by cell number or chlorophyll. The concentration was calculated by the equation (A2 sample-A1 sample)-(A2 blank-A1 blank)/(A2 standard-A1 standard)-(A2 Blank-A1 blank).
Accession Numbers
Sequence data from this article can be found in the Phytozome 10.0 and NCBI databases under the following accession numbers: the ACK1 (Cre09.g396700, XP_001694505.1), PAT2 (Cre09.g396650, XP_001694504.1), ACK2 (Cre17.g709850, XP_001691682.1), and PAT1 (Cre17.g699000, XP_001691787.1). The accession numbers of the other proteins mentioned in this study are listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Relative Fluorescence Intensity of Chlamydomonas Cells Expressing Fluorescent Fusion Proteins.
Supplemental Figure 2. Localization of ACK2 and PAT1 Using mCherry Fusion Proteins.
Supplemental Figure 3. Localization of ACK 1/2 and PAT 1/2 in Multiple Cells.
Supplemental Figure 4. Genetic Analyses of ack and pat Single and Double Mutants.
Supplemental Figure 5. Genomic and CDS Sequences of ack1, ack2, pat2-1, and pat2-2 Mutants.
Supplemental Figure 6. Alignments of Wild-Type and Truncated ACK and PAT Proteins.
Supplemental Figure 7. Changes in Abundance of Transcripts Encoding Fermentative Enzymes in pat and ack Mutants.
Supplemental Figure 8. Accumulation of External Metabolites in the Complemented Strains of pat2-1.
Supplemental Figure 9. Nuclear Magnetic Resonance to Confirm Acetate Production in ack1 ack2 Double Mutants.
Supplemental Figure 10. Fermentative H2 Production by Indicated Mutants.
Supplemental Figure 11. Fermentative CO2 Production by Indicated Mutants.
Supplemental Figure 12. In Vitro Acetate Kinase Activity in Mutant and Complemented Strains.
Supplemental Figure 13. Alignments and Phylogenetic Analyses of Chlamydomonas ACS and ALDH Proteins.
Supplemental Figure 14. Changes in Levels of Putative Acetyl-CoA Synthetase Transcripts.
Supplemental Figure 15. Accumulation of Intracellular Acetyl-Phosphate.
Supplemental Figure 16. Proposed Anoxic Metabolisms in Single and Double Mutants.
Supplemental Figure 17. In Vitro Aldehyde Dehydrogenase Activity.
Supplemental Figure 18. Maps of Plasmids used for Protein Subcellular Localization.
Supplemental Table 1. Intracellular Pyruvate Levels.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment of ACS Proteins Used for the Phylogenetic Analysis Shown in Supplemental Figure 13C.
Supplemental Data Set 2. Text File of the Alignment of ALDH Proteins Used for the Phylogenetic Analysis Shown in Supplemental Figure 13D.
Supplementary Material
Acknowledgments
This work was supported by the Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division, U.S. Department of Energy Grants to A.R.G. (DE-FG02-12ER16338) and M.C.P. (DE-FG02-12ER16339) and by the National Science Foundation Award 0920274 and South Carolina Experiment Station Project SC-1700340 to K.S.S. and is designated as technical contribution number 6207 of the Clemson University Experiment Station. Aspects of the work were also funded by National Science Foundation grants awarded to A.R.G. (MCB0824469 and MCB0235878). The costs of publication for this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. We thank Heather Cartwright for her help on imaging and Ralph Bock for providing the UVM11 strain. We also thank Michelle Davison for phylogenetic analyses.
AUTHOR CONTRIBUTIONS
W.Y., M.C.P., and A.R.G. designed the research. W.Y., C.C., S.D., T.M.W., C.J.I.-S., L.M., G.P., A.L.H., and T.M. performed research. W.Y., C.C., S.D., T.M.W., M.C.P., C.J.I.-S., G.P., K.S.S., and M.C.J. analyzed the data. W.Y., M.C.P., and A.R.G wrote the article.
Glossary
- ACS
acetyl-CoA synthetase
- ALDH
aldehyde dehydrogenase
- ASCT
acetate:succinate CoA-transferase
- SCL
succinyl-CoA ligase
- RT-qPCR
reverse transcription-quantitative PCR
- TAP
Tris-acetate-phosphate
- HS
high-salt
- UTR
untranslated region
Footnotes
Online version contains Web-only data.
References
- Abdel-Hamid A.M., Attwood M.M., Guest J.R. (2001). Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Ananyev G., Carrieri D., Dismukes G.C. (2008). Optimization of metabolic capacity and flux through environmental cues to maximize hydrogen production by the cyanobacterium “Arthrospira (Spirulina) maxima.” Appl. Environ. Microbiol. 74: 6102–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Cann A.F., Connor M.R., Shen C.R., Smith K.M., Brynildsen M.P., Chou K.J., Hanai T., Liao J.C. (2008a). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10: 305–311. [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J.C. (2008b). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89. [DOI] [PubMed] [Google Scholar]
- Atteia A., et al. (2009). A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26: 1533–1548. [DOI] [PubMed] [Google Scholar]
- Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W. (2006). Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281: 9909–9918. [DOI] [PubMed] [Google Scholar]
- Atteia A., van Lis R., Tielens A.G., Martin W.F. (2013). Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim. Biophys. Acta 1827: 210–223. [DOI] [PubMed] [Google Scholar]
- Blombach B., Schreiner M.E., Holátko J., Bartek T., Oldiges M., Eikmanns B.J. (2007). L-valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl. Environ. Microbiol. 73: 2079–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C., Vasiliou M., Carpenter S., Carpenter C., Zhang Y., Wang X., Kotchoni S.O., Wood A.J., Kirch H.H., Kopečný D., Nebert D.W., Vasiliou V. (2013). Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237: 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.J., Tredwell G., Molnàr A., Bundy J.G., Nixon P.J. (2012). Artificial microRNA-mediated knockdown of pyruvate formate lyase (PFL1) provides evidence for an active 3-hydroxybutyrate production pathway in the green alga Chlamydomonas reinhardtii. J. Biotechnol. 162: 57–66. [DOI] [PubMed] [Google Scholar]
- Buu L.M., Chen Y.C., Lee F.J. (2003). Functional characterization and localization of acetyl-CoA hydrolase, Ach1p, in Saccharomyces cerevisiae. J. Biol. Chem. 278: 17203–17209. [DOI] [PubMed] [Google Scholar]
- Campos-Bermudez V.A., Bologna F.P., Andreo C.S., Drincovich M.F. (2010). Functional dissection of Escherichia coli phosphotransacetylase structural domains and analysis of key compounds involved in activity regulation. FEBS J. 277: 1957–1966. [DOI] [PubMed] [Google Scholar]
- Carrieri D., Momot D., Brasg I.A., Ananyev G., Lenz O., Bryant D.A., Dismukes G.C. (2010). Boosting autofermentation rates and product yields with sodium stress cycling: application to production of renewable fuels by cyanobacteria. Appl. Environ. Microbiol. 76: 6455–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C., Dubini A., Subramanian V., Yang W., Magneschi L., Mus F., Seibert M., Posewitz M.C., Grossman A.R. (2012). Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell 24: 692–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C., Yang W., Posewitz M.C., Grossman A.R. (2013). Fermentation metabolism and its evolution in algae. Front. Plant Sci. 4: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sabato G., Jencks W.P. (1961). Mechanism and catalysis of reactions of acyl phosphates. II. Hydrolysis 1. J. Am. Chem. Soc. 83: 4400–4405. [Google Scholar]
- Dittrich C.R., Bennett G.N., San K.Y. (2005a). Characterization of the acetate-producing pathways in Escherichia coli. Biotechnol. Prog. 21: 1062–1067. [DOI] [PubMed] [Google Scholar]
- Dittrich C.R., Vadali R.V., Bennett G.N., San K.Y. (2005b). Redistribution of metabolic fluxes in the central aerobic metabolic pathway of E. coli mutant strains with deletion of the ackA-pta and poxB pathways for the synthesis of isoamyl acetate. Biotechnol. Prog. 21: 627–631. [DOI] [PubMed] [Google Scholar]
- Dubini A., Mus F., Seibert M., Grossman A.R., Posewitz M.C. (2009). Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284: 7201–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Rochaix J.D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265: 888–894. [DOI] [PubMed] [Google Scholar]
- Fowler M.L., Ingram-Smith C., Smith K.S. (2012). Novel pyrophosphate-forming acetate kinase from the protist Entamoeba histolytica. Eukaryot. Cell 11: 1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller R.P., Gibbs M. (1984). Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75: 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller R.P., Gibbs M. (1985). Fermentative metabolism of Chlamydomonas reinhardtii: II. Role of plastoquinone. Plant Physiol. 77: 509–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M., Gfeller R.P., Chen C. (1986). Fermentative metabolism of Chlamydomonas reinhardii: III. Photoassimilation of acetate. Plant Physiol. 82: 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J.J., Prior M., Galván A., Fernandez E., Grossman A.R. (2011). Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Croft M., Gladyshev V.N., Merchant S.S., Posewitz M.C., Prochnik S., Spalding M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10: 190–198. [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Catalanotti C., Yang W., Dubini A., Magneschi L., Subramanian V., Posewitz M.C., Seibert M. (2011). Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol. 190: 279–288. [DOI] [PubMed] [Google Scholar]
- Heinnickel M.L., Alric J., Wittkopp T., Yang W., Catalanotti C., Dent R., Niyogi K.K., Wollman F.A., Grossman A.R. (2013). Novel thylakoid membrane GreenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J. Biol. Chem. 288: 7024–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Happe T. (2005). The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33: 39–41. [DOI] [PubMed] [Google Scholar]
- Hoehler T.M., Albert D.B., Alperin M.J., Bebout B.M., Martens C.S., Des Marais D.J. (2002). Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems. Antonie van Leeuwenhoek 81: 575–585. [DOI] [PubMed] [Google Scholar]
- Ingram-Smith C., Barber R.D., Ferry J.G. (2000). The role of histidines in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 275: 33765–33770. [DOI] [PubMed] [Google Scholar]
- Ingram-Smith C., Gorrell A., Lawrence S.H., Iyer P., Smith K., Ferry J.G. (2005). Characterization of the acetate binding pocket in the Methanosarcina thermophila acetate kinase. J. Bacteriol. 187: 2386–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Smith C., Martin S.R., Smith K.S. (2006). Acetate kinase: not just a bacterial enzyme. Trends Microbiol. 14: 249–253. [DOI] [PubMed] [Google Scholar]
- Johnson X., Alric J. (2013). Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints between carbon partitioning between oil and starch. Eukaryot. Cell 12: 776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch H.H., Bartels D., Wei Y., Schnable P.S., Wood A.J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9: 371–377. [DOI] [PubMed] [Google Scholar]
- Kirch H.H., Schlingensiepen S., Kotchoni S., Sunkar R., Bartels D. (2005). Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 57: 315–332. [DOI] [PubMed] [Google Scholar]
- Koshland D.E. (1952). Effect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J. Am. Chem. Soc. 74: 2286–2292. [Google Scholar]
- Kosourov S., Patrusheva E., Ghirardi M.L., Seibert M., Tsygankov A. (2007). A comparison of hydrogen photoproduction by sulfur-deprived Chlamydomonas reinhardtii under different growth conditions. J. Biotechnol. 128: 776–787. [DOI] [PubMed] [Google Scholar]
- Kreuzberg K. (1984). Starch fermentation via formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant. 61: 87–94. [Google Scholar]
- Kreuzberg K., Klöck G., Grobheiser D. (1987). Subcellular distribution of pyruvate-degrading enzymes in Chlamydomonas reinhardtii studied by an improved protoplast fractionation procedure. Physiol. Plant. 69: 481–488. [Google Scholar]
- Kuit W., Minton N.P., López-Contreras A.M., Eggink G. (2012). Disruption of the acetate kinase (ack) gene of Clostridium acetobutylicum results in delayed acetate production. Appl. Microbiol. Biotechnol. 94: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S.H., Ferry J.G. (2006). Steady-state kinetic analysis of phosphotransacetylase from Methanosarcina thermophila. J. Bacteriol. 188: 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.C., Yang Y.L., Witman W.B. (2003). The anabolic pyruvate oxidoreductase from Methanococcus maripaludis. Arch. Microbiol. 179: 444–456. [DOI] [PubMed] [Google Scholar]
- Lorquet F., Goffin P., Muscariello L., Baudry J.B., Ladero V., Sacco M., Kleerebezem M., Hols P. (2004). Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 186: 3749–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Mus F., Posewitz M.C., Seibert M., Perata P., Grossman A.R. (2012). A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 158: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuser J.E., D’Adamo S., Jinkerson R.E., Mus F., Yang W., Ghirardi M.L., Seibert M., Grossman A.R., Posewitz M.C. (2012). Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: Insight into the role of HYDA2 in H₂ production. Biochem. Biophys. Res. Commun. 417: 704–709. [DOI] [PubMed] [Google Scholar]
- Millerioux Y., et al. (2012). ATP synthesis-coupled and -uncoupled acetate production from acetyl-CoA by mitochondrial acetate:succinate CoA-transferase and acetyl-CoA thioesterase in Trypanosoma. J. Biol. Chem. 287: 17186–17197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsy F.M. (2011). Acetate versus sulfur deprivation role in creating anaerobiosis in light for hydrogen production by Chlamydomonas reinhardtii and Spirulina platensis: two different organisms and two different mechanisms. Photochem. Photobiol. 87: 137–142. [DOI] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486. [DOI] [PubMed] [Google Scholar]
- Neupert J., Karcher D., Bock R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57: 1140–1150. [DOI] [PubMed] [Google Scholar]
- Ohta S., Miyamoto K., Miura Y. (1987). Hydrogen evolution as a consumption mode of reducing equivalents in green algal fermentation. Plant Physiol. 83: 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G., Krawietz D., Hemschemeier A., Happe T. (2011). A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66: 330–340. [DOI] [PubMed] [Google Scholar]
- Phue J.N., Lee S.J., Kaufman J.B., Negrete A., Shiloach J. (2010). Acetate accumulation through alternative metabolic pathways in ackA (-) pta (-) poxB (-) triple mutant in E. coli B (BL21). Biotechnol. Lett. 32: 1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock S.V., Colombo S.L., Prout D.L. Jr., Godfrey A.C., Moroney J.V. (2003). Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO2 atmosphere. Plant Physiol. 133: 1854–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A.R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz M.C., King P.W., Smolinski S.L., Zhang L., Seibert M., Ghirardi M.L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279: 25711–25720. [DOI] [PubMed] [Google Scholar]
- Prüss B.M., Wolfe A.J. (1994). Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12: 973–984. [DOI] [PubMed] [Google Scholar]
- Rasala B.A., Barrera D.J., Ng J., Plucinak T.M., Rosenberg J.N., Weeks D.P., Oyler G.A., Peterson T.C., Haerizadeh F., Mayfield S.P. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74: 545–556. [DOI] [PubMed] [Google Scholar]
- Remize F., Andrieu E., Dequin S. (2000). Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 66: 3151–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M., Metcalf W.W. (2004). Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. USA 101: 16929–16934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, E.F., Fritsch, T., and Eikmanns, B.J. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Schreiner M.E., Riedel C., Holátko J., Pátek M., Eikmanns B.J. (2006). Pyruvate:quinone oxidoreductase in Corynebacterium glutamicum: molecular analysis of the pqo gene, significance of the enzyme, and phylogenetic aspects. J. Bacteriol. 188: 1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillers R., Chow A., Tracy B., Papoutsakis E.T. (2008). Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab. Eng. 10: 321–332. [DOI] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229. [DOI] [PubMed] [Google Scholar]
- Spear J.R., Walker J.J., McCollom T.M., Pace N.R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102: 2555–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A.S., Bhaya D., Bateson M.M., Melendrez M.C., Ward D.M., Brecht E., Peters J.W., Kühl M., Grossman A.R. (2006). In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 103: 2398–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki K., Shoun H., Yamaguchi M., Takeo K., Nakamura A., Hoshino T., Takaya N. (2004). Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. J. Biol. Chem. 279: 12414–12420. [DOI] [PubMed] [Google Scholar]
- Tardif M., Atteia A., Specht M., Cogne G., Rolland N., Brugière S., Hippler M., Ferro M., Bruley C., Peltier G., Vallon O., Cournac L. (2012). PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29: 3625–3639. [DOI] [PubMed] [Google Scholar]
- Terashima M., Specht M., Naumann B., Hippler M. (2010). Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9: 1514–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens A.G., van Grinsven K.W., Henze K., van Hellemond J.J., Martin W. (2010). Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 40: 387–397. [DOI] [PubMed] [Google Scholar]
- Timmins M., Thomas-Hall S.R., Darling A., Zhang E., Hankamer B., Marx U.C., Schenk P.M. (2009). Phylogenetic and molecular analysis of hydrogen-producing green algae. J. Exp. Bot. 60: 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grinsven K.W., Rosnowsky S., van Weelden S.W., Pütz S., van der Giezen M., Martin W., van Hellemond J.J., Tielens A.G., Henze K. (2008). Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J. Biol. Chem. 283: 1411–1418. [DOI] [PubMed] [Google Scholar]
- van Lis R., Baffert C., Couté Y., Nitschke W., Atteia A. (2013). Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate:ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 161: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A.J. (2005). The acetate switch. Microbiol. Mol. Biol. Rev. 69: 12–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.T., Bennett G.N., San K.Y. (1999). Effect of inactivation of nuo and ackA-pta on redistribution of metabolic fluxes in Escherichia coli. Biotechnol. Bioeng. 65: 291–297. [PubMed] [Google Scholar]
- Yang Y.T., Bennett G.N., San K.Y. (2001). The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab. Eng. 3: 115–123. [DOI] [PubMed] [Google Scholar]
- Yang W., Catalanotti C., Posewitz M., Alric J., Grossman A.R. (2014). Insights into algal fermentation. In Low-Oxygen Stress in Plants: Oxygen Sensing and Adaptive Responses to Hypoxia, J.T. van Dongen and F. Licausi, eds (Wien, Heidelberg, New York, Dortrecht, London: Springer), pp. 135–163. [Google Scholar]
- Yasuda K., Jojima T., Suda M., Okino S., Inui M., Yukawa H. (2007). Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl. Microbiol. Biotechnol. 77: 853–860. [DOI] [PubMed] [Google Scholar]
- Yoshii Y., Furukawa T., Yoshii H., Mori T., Kiyono Y., Waki A., Kobayashi M., Tsujikawa T., Kudo T., Okazawa H., Yonekura Y., Fujibayashi Y. (2009). Cytosolic acetyl-CoA synthetase affected tumor cell survival under hypoxia: the possible function in tumor acetyl-CoA/acetate metabolism. Cancer Sci. 100: 821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.