The cell walls of flax bast fibers contain high cellulose and low lignin levels, imparting tensile strength and flexibility. To learn more about the mechanisms responsible for this type of cell wall structure, a collection of ectopic lignin mutants was identified. Characterization of the lbf1 mutant provided key information on lignification in flax that is also relevant to other plants.
Abstract
Histochemical screening of a flax ethyl methanesulfonate population led to the identification of 93 independent M2 mutant families showing ectopic lignification in the secondary cell wall of stem bast fibers. We named this core collection the Linum usitatissimum (flax) lbf mutants for lignified bast fibers and believe that this population represents a novel biological resource for investigating how bast fiber plants regulate lignin biosynthesis. As a proof of concept, we characterized the lbf1 mutant and showed that the lignin content increased by 350% in outer stem tissues containing bast fibers but was unchanged in inner stem tissues containing xylem. Chemical and NMR analyses indicated that bast fiber ectopic lignin was highly condensed and rich in G-units. Liquid chromatography-mass spectrometry profiling showed large modifications in the oligolignol pool of lbf1 inner- and outer-stem tissues that could be related to ectopic lignification. Immunological and chemical analyses revealed that lbf1 mutants also showed changes to other cell wall polymers. Whole-genome transcriptomics suggested that ectopic lignification of flax bast fibers could be caused by increased transcript accumulation of (1) the cinnamoyl-CoA reductase, cinnamyl alcohol dehydrogenase, and caffeic acid O-methyltransferase monolignol biosynthesis genes, (2) several lignin-associated peroxidase genes, and (3) genes coding for respiratory burst oxidase homolog NADPH-oxidases necessary to increase H2O2 supply.
INTRODUCTION
Lignin is a major component of many plant cell walls and is essential for water transport in vascular tissue, mechanical support, and resistance to pathogens in higher land plants (Baucher et al., 1998; Boerjan et al., 2003; Weng and Chapple, 2010). This phenolic polymer also contributes to the recalcitrance of lignocellulosic biomass for biofuel production and the regulation of lignin biosynthesis has therefore been intensely studied (Whetten and Sederoff, 1995; Fu et al., 2011; Vanholme et al., 2012a). Much information about this process has been obtained by biochemical and genetics studies on mutants showing modified lignification profiles (Anterola and Lewis, 2002; Bonawitz and Chapple, 2010; Vanholme et al., 2012b).
Generally, lignin mutants can be divided into two main groups: (1) those showing reduced cell wall lignin levels and (2) ectopic lignification mutants where the secondary cell wall developmental program is activated. In the first group, lignin is often reduced and/or modified via the downregulation of genes involved in lignin monomer biosynthesis and/or the oxidation of monomers for subsequent polymerization (laccases and peroxidases) (Vanholme et al., 2010; Weng and Chapple, 2010; Zhao et al., 2013). Reduced lignin content is often accompanied by modifications to other cell wall polymers, suggesting the existence of a dynamic relationship between the cell wall matrix and the lignification process (Hu et al., 1999; Van Acker et al., 2013). In the second group, upregulation/downregulation of different transcription factors leads to the activation of the secondary cell wall developmental program and the biosynthesis and deposition of cellulose, hemicellulose, and lignin in parenchyma-type cells that normally only produce nonlignified primary cell walls (Mitsuda et al., 2007; Zhong et al., 2007; Zhao and Dixon, 2011). Alternatively, ectopic lignification can also result from perturbations in the biosynthesis of other cell wall polymers (Zhong et al., 2002; Caño-Delgado et al., 2003).
Interestingly, the stems of certain fiber plants (e.g., flax, hemp, ramie, etc.) naturally contain two populations of cells showing highly contrasted secondary cell wall compositions. In outer-stem tissues, specialized cells (bast fibers) possess hypolignified and cellulose-rich thick secondary cell walls, whereas the xylem cells from inner-stem tissues have a more typical lignified secondary cell wall structure. Analyses in flax (Linum usitatissimum), for example, show that bast fiber secondary cell walls are almost completely filled with cellulose (∼70%) and contain 5 to 15% of noncellulosic polysaccharide (NCP) mainly composed of β-1-4 galactan and arabinogalactan, but only 2 to 4% lignin (Davis et al., 1990; Girault et al., 1997; Day et al., 2005; Gorshkova and Morvan, 2006). By contrast, xylem cell walls contain lower amounts of cellulose (∼40%) and much higher amounts (30%) of lignin (Day et al., 2005). Bast fibers are elongated cells that provide mechanical support and allow relatively tall plants with small stem diameters to maintain an erect state (Neutelings, 2011; Guerriero et al., 2013). In flax, the outer stem tissues enriched in hypolignified primary bast fibers can be easily peeled away from the central xylem core-containing lignified cells, and this plant therefore appears to be an excellent model to study secondary cell wall formation and lignification.
We have previously shown that lignification in flax appears to be partially modulated through transcriptional regulation of genes encoding lignin monomer biosynthesis and polymerization (Fenart et al., 2010; Huis et al., 2012). More recently, we generated and characterized a flax ethyl methanesulfonate (EMS) mutant population that has allowed us to obtain mutants for the CAD and C3H lignin genes through a TILLinG reverse genetics approach (Chantreau et al., 2013). In this article, we report the screening of this population and the identification of 319 independent mutants showing altered lignification profiles in bast fibers. We believe that this collection of flax lignin mutants represents a valuable biological resource for plant cell wall biologists. The detailed characterization of individual mutants should provide information on the different regulatory mechanisms and signaling pathways used by plants to regulate lignin biosynthesis. In addition, the identification of novel key genes involved in this process could provide targets for engineering improved lignocellulosic quality in other plant species. Cell wall analyses of mutants containing bast fibers with variable lignin content will also lead to a better understanding of the dynamic relationship between lignin and other cell wall polymers. As a proof of concept, we report the detailed characterization of a highly lignified flax bast fiber mutant.
RESULTS
Identification and Visual Phenotyping of the Flax Lignified Bast Fiber Mutant Core Collection
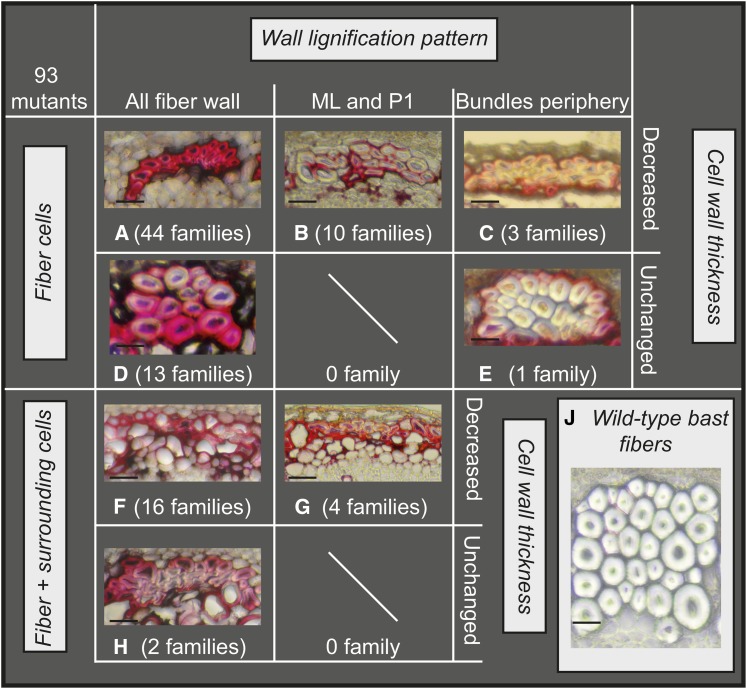
To identify mutants showing increased lignification in bast fibers, we first screened 8999 plants from 3391 M2 families (Chantreau et al., 2013). Examination of transversal hand-sections of stem from individual plants by UV microscopy allowed us to identify 540 families showing increased autofluorescence in bast fibers. Families were assigned to three different classes based on a visual estimation of modified autofluorescence: strong, class 1; moderate, class 2; and weak, class 3 (Table 1). In a second round of screening, thin freehand stem sections were prepared from the previously identified 540 families and stained with phloroglucinol-HCl. Families showing a red coloration of bast fibers (indicating potential lignin deposition) were once again assigned to three different classes (strong, moderate, and weak) (Table 1). Two hundred and twenty-one families previously identified in the first round of screening showed only little/no differences when compared with wild-type plants and were therefore not retained for further analyses. Altogether, 319 families showed increased coloration of bast fibers and 93 families showed strong coloration (class 1). We named this core collection of class 1 mutant families lignified bast fiber (lbf) mutants. The lbf core collection was then subclassified into eight different groups according to the type of modified lignification pattern (Figure 1). For example, the groups A to E show increased lignification uniquely in fiber cells, whereas the groups F to H also show increased lignification in surrounding cells.
Table 1. Number of M2 Families Assigned to Different Bast Fiber Lignification Classes.
| Bast Fiber Lignification Class | ||||
|---|---|---|---|---|
| Screen | Class 3 | Class 2 | Class 1 | Total |
| UV | 252 | 156 | 132 | 540 |
| P-HCl | 150 | 176 | 93 | 319 |
Figure 1.
Classification of Flax lbf Mutants into Eight Different Groups According to Modified Lignification Pattern.
Groups A to C: Only fiber cells lignified, cell wall thickness decreased. Groups D and E: Only fiber cells lignified, cell wall thickness unchanged. Groups F and G: fibers and surrounding cells lignified, cell wall thickness decreased. Group H: fibers and surrounding cells lignified, cell wall thickness unchanged. Group J: wild-type bast fibers. Number of families in each group is given in brackets. Bar = 10 μm, phloroglucinol-HCl staining of stem cross sections (lignified walls are colored red).
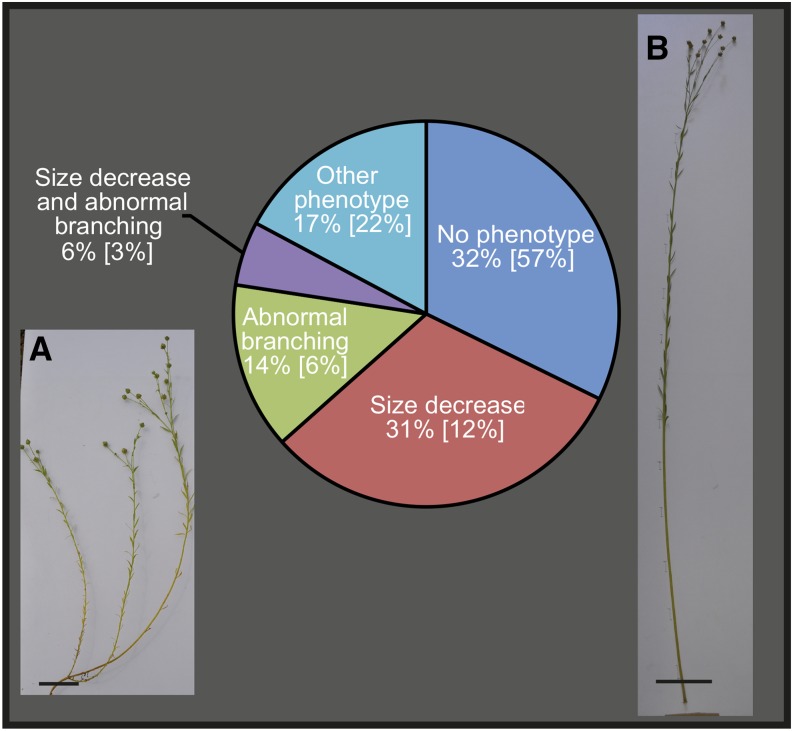
We then classified the flax lbf mutants into different categories based on previously established visual phenotypes of the flax EMS mutant population (Supplemental Table 1; Chantreau et al., 2013). Thirty-two percent of the core collection families showed no obvious morphological phenotype, 31% were smaller to wild-type plants, and 14% showed increased stem branching (Figure 2). Other observed phenotypes included early death and nonerect stems. Comparison of these values with the corresponding values for the overall mutant population (Figure 2) would suggest that lbf mutants are generally smaller, show increased branching, and have thinner stems when compared with either wild-type plants or other mutants.
Figure 2.
Proportion of Flax lbf Mutants in Different Categories Based on Visual Phenotype.
Percentage values refer to the proportion of lbf mutants showing a given phenotype. Values in brackets correspond to the proportion of all flax mutants (PT-flax collection) showing the phenotype.
(A) Photo of typical lbf mutant showing reduced size and increased branching. Bar = 5 cm.
(B) Photo of wild-type flax plant. Bar = 10 cm.
[See online article for color version of this figure.]
The Flax lbf1 Mutant Has a Modified Lignin Content
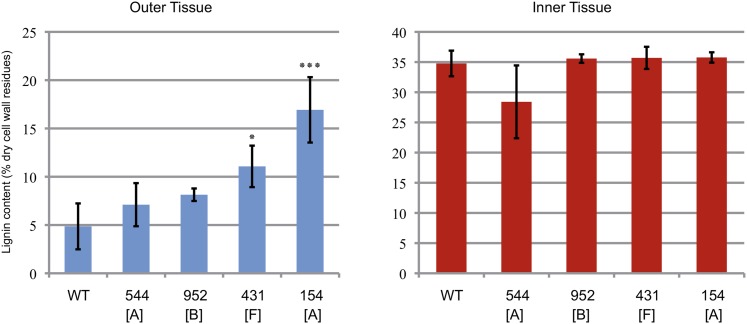
To confirm that the red coloration of bast fibers observed in phloroglucinol-stained sections of lbf mutants corresponded to lignin, we determined the acetyl bromide lignin content (Iiyama and Wallis, 1990) in stem tissues from four independent lbf mutants belonging to three different groups (A, B, and F). Our results (Figure 3) show that the lignin content of outer stem tissues from two of these mutants was significantly greater than in wild-type plants. In contrast, the lignin content of inner stem tissues (xylem) from all mutants was not significantly increased. We then focused on a single family (154) that showed the largest increase out of the four mutant lines evaluated for further detailed characterization. We named this mutant lbf1 for lignified bast fiber1. This mutant line shows a typical core collection phenotype (reduced plant size and stem diameter and increased basal stem ramification) (Figure 2).
Figure 3.
Acetyl Bromide Lignin Content of Inner- and Outer-Stem Tissues of Four lbf Mutants and Wild-Type Plants.
Letters in brackets refer to the subclassification presented in Figure 1. Significant differences (Student’s t test) between wild-type and mutant tissues were observed at P < 0.001 (***) and P < 0.05 (*); error bars = sd.
[See online article for color version of this figure.]
Thioacidolysis of M4 lbf1 outer tissues revealed significant increases in all three lignin units (H, G, and S) (Table 2). As in wild-type flax lignin, the G unit is the major monomer present in the lbf1 ectopic lignin and constitutes ∼70% of total released lignin units (Table 2, Figure 4A). No significant changes in the S/G ratio were observed, indicating that the ectopic lignin is similar to that previously analyzed in flax (Day et al., 2005; del Río et al., 2011). Thioacidolysis only releases lignin units from noncondensed intermonomeric bonds (β-O-4 and α-O-4); therefore, calculation of the amount of S and G units released, divided by the amount of acetyl bromide lignin values (Table 2; S+G/lignin values), provides an estimate of the relative condensation of the lignin polymer (i.e., the proportion of condensed bonds, such as β-5:phenylcoumaran and β-β:resinol). No significant changes in these values were seen (Table 2, Figure 4B), suggesting that increased lignification in the lbf1 mutant is not associated with changes in the degree of lignin condensation.
Table 2. Lignin Monomeric Composition Determined by Thioacidolysis in Inner- and Outer-Stem Tissues of Flax lbf1 Mutants and Wild-Type Plants.
| Outer Tissues | Inner Tissues | |||
|---|---|---|---|---|
| Wild Type | lbf1 | Wild Type | lbf1 | |
| H | 1.14 ± 0.45 | 3.33 ± 2.07 | 3.11 ± 0.82 | 2.96 ± 0.70 |
| G | 10.10 ± 5.80 | 26.60 ± 10.31 | 243.37 ± 41.09 | 243.05 ± 28.09 |
| S | 3.25 ± 1.34 | 6.94 ± 2.64 | 49.02 ± 3.73 | 32.85 ± 7.98 |
| S/G | 0.34 ± 0.05 | 0.26 ± 0.11 | 0.20 ± 0.03 | 0.13 ± 0.02 |
| S+G | 13.35 ± 7.14 | 35.03 ± 11.53 | 292.39 ± 42.14 | 275.90 ± 33.73 |
| S+G/lignin | 329.90 ± 203.75 | 301.49 ± 81.87 | 872.00 ± 79.94 | 968.70 ± 147.75 |
H, G, S = yields of the thioethylated products of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units expressed as μmoles per gram of CWR. S/G = ratio of G to S lignin units. S+G = total S plus G lignin units expressed as μmoles per gram of CWR. S+G/lignin = yields of total lignin (S+G) expressed as μmoles per gram of lignin. For lbf1, values represent the mean values obtained with 16 and 33 individual plants for inner and outer tissues, respectively. For the wild type, inner and outer values are the mean ± sd from three individual plants. Values significantly different from the wild type at P < 0.05 are indicated in bold.
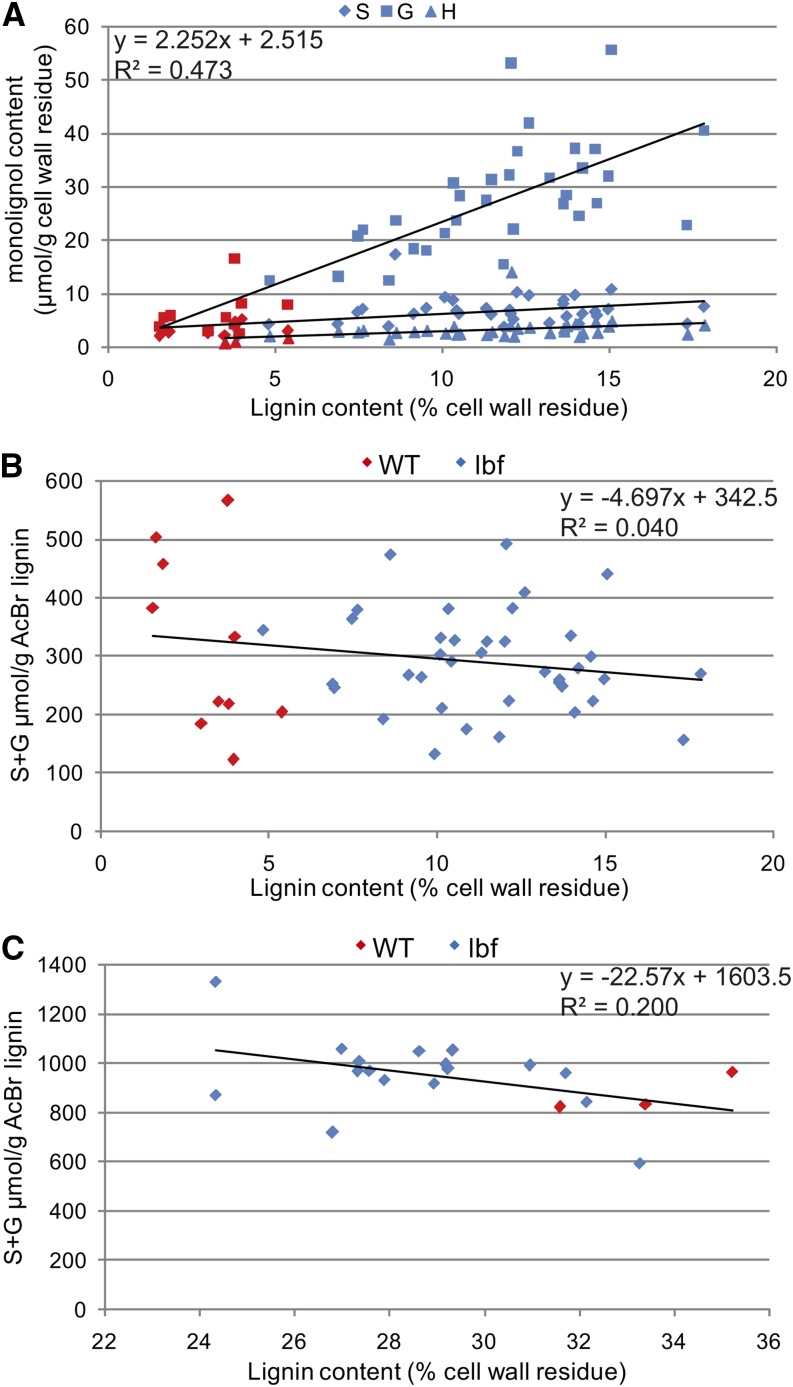
Figure 4.
Lignin Analyses in lbf1 Inner- and Outer-Stem Tissues.
(A) Relationship between lignin content and the composition in H (triangle), G (square), and S (diamond) lignin units in outer tissues of wild-type and lbf1 mutants. Correlation coefficient given for G units only.
(B) Relationship between lignin condensation and lignin content in outer tissues.
(C) Relationship between lignin condensation and lignin content in inner tissues. Lignin content was determined by acetyl bromide analyses; S/G ratios and lignin composition were determined by thioacidolysis. Blue dots correspond to individual lbf1 mutants and red dots correspond to wild-type plants.
[See online article for color version of this figure.]
Our previous results (Figure 3) had shown no increase in the lignin content of lbf1 stem inner tissues. Thioacidolysis showed that lignin composition was much less affected than in outer tissues (Table 2). A slight but significant decrease in S units was observed resulting in a lower S/G ratio. As for lbf1 outer tissues, no modification in lignin condensation was observed (Figure 4C).
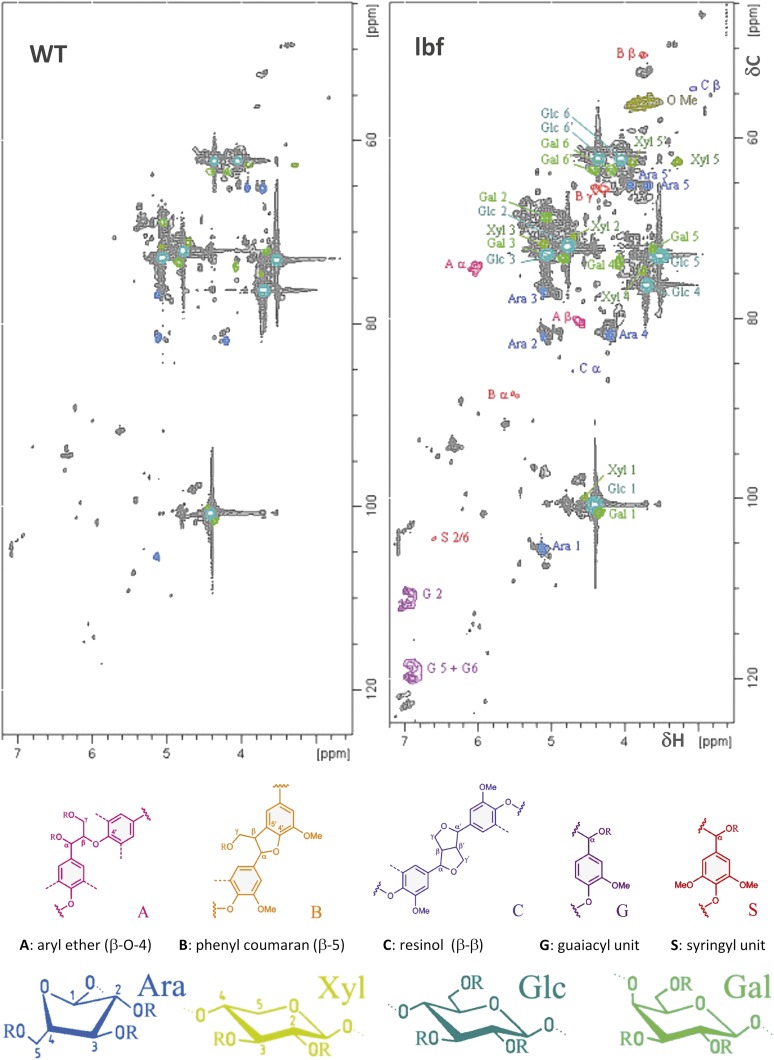
Flax lignin is generally highly condensed, and we therefore used 2D NMR analyses to provide complementary information on the condensed fraction of the lignin polymer. The heteronuclear single quantum coherence (HSQC) spectra (δC/δH 45 – 125/2.5 – 7.2) of the acetylated cell wall from flax fibers are shown in Figure 5. Our results show the presence of G and S signals in the outer stem tissues of the lbf1 mutant, whereas barely any lignin signals were detected in the spectra of the wild-type plants. Various signals from cellulose and polysaccharides were also detected in both spectra. For quantification of the different interlinkages, we used the guaiacyl (G2) C2-H signal as internal standard. Then all signals assigned to the various interunit linkage types were integrated: β-O-4’ alkyl-aryl ether linkages (Aα and Aβ), phenyl coumaran β-5′/α-O-4’ linkages (Bα and Bβ), and resinol β-β’/α-O-γ’/γ-O-α’ linkages (Cα and Cβ). The main lignin substructures observed in lbf1 outer tissues correspond to the β-O-4’ alkyl-aryl ether, with 52.3% (Aα) and 47.9% (Aβ) relative abundance signals, respectively, followed by phenylcoumaran and resinol with 14.5% (Bα) and 18% (Bβ), and 13.6% (Cα) and 14.7% (Cβ), respectively. These values are in good agreement with previous HSQC analyses of flax fiber milled wood lignin (del Río et al., 2011). Signals were also assigned to other minor substructures (Aγ, Βγ, and Cγ, dibenzodioxocin). The S/G ratio (0.12) estimated by NMR was lower than the molar ratio (0.28) determined by thioacidolysis. H units (data not shown) were difficult to quantify in lbf1 lignin because of masking by polysaccharide signals in the HSQC spectra. For lbf1 inner-stem tissues, the HSQC spectra confirmed chemical analyses, suggesting that the inner tissue lignin structure of lbf1 mutants was very similar to that of wild-type plants. No changes in the proportion of lateral chains were observed with aryl ether, phenyl coumaran, and resinol bonds, representing 58.6, 16.0, and 14.2%, respectively. In contrast to outer-stem tissues, the S/G ratio (0.12) estimated from lbf12D NMR spectra (Supplemental Figure 1) was very similar to that estimated by thioacidolysis (0.13).
Figure 5.
2D NMR Spectra Revealing Lignin Monomers, Interunit Distribution, and Sugar Signals.
Partial short-range (HSQC) spectra (dC/dH 45 – 125/2.5 – 7.2) of the acetylated cell wall from wild-type (left) and lbf1 (right) outer tissues.
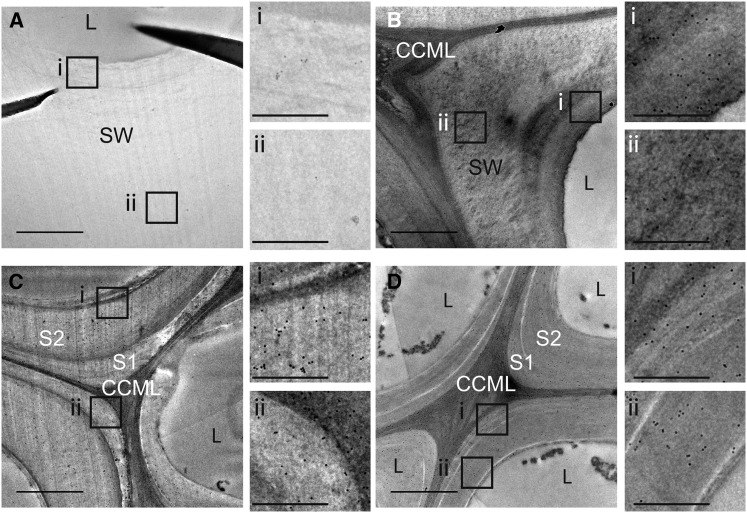
Further information on lbf1 lignin was obtained using the KM1 antibody targeted against lignin phenylcoumaran linkages (β-5) (Kiyoto et al., 2013). TEM observation showed that labeling was present in the whole cell wall (primary cell wall, secondary cell wall) of lbf1 fibers, but was conspicuously absent in wild-type fibers (Figure 6). By contrast, a similar comparison of lbf1 and wild-type xylem tissues did not show any differences with labeling being detected in the middle lamella and primary and secondary cell walls of both mutant and wild-type plants (Figure 6).
Figure 6.
Immunogold Silver Staining of a Transverse Section of the Flax Stem Median Region with KM1 Antibody.
Bast fibers ([A] and [B]), xylem fibers ([C] and [D]), wild-type ([A] and [C]), and lbf1 ([B] and [D]). CCML, cell corner middle lamella; SW, secondary wall; L, lumen. Smaller photos (i and ii) on the right side of each main photo ([A] to [D]) show a zoom of the corresponding regions indicated on main photo. Bars = 1 μm (main photos) and 0.15 μm (small photos).
Liquid Chromatography-Mass Spectrometry Profiling Reveals Modifications of the Oligolignol Pool in the lbf1 Mutant
Oligolignol liquid chromatography-mass spectrometry (LC-MS) profiling provides information about the availability and nature (e.g., glycosylated versus nonglycosylated) of lignin monolignols and di/trilignols present in lignifying tissues. The composition of the oligolignol pool is closely related to polymeric lignin structure and gives complementary insight into the lignification process and metabolic flow (Morreel et al., 2010a). Changes in lignin content are often accompanied by modifications in the soluble oligolignol pool, and we therefore performed an LC-MS-based metabolite profiling of ethanol extracts of inner- and outer-stem tissues of lbf1 mutants and wild-type plants (Table 3; Supplemental Figure 2). Because of the strong variation in intensities of many of the phenolic compounds observed, only the compounds that were consistently above the detection limit in all samples, or consistently below the detection limit in all samples, within each of the four categories (wild-type inner, mutant inner, wild-type outer, and mutant outer), were taken into account. In the outer tissues, four nonhexosylated dilignols and eight nonhexosylated trilignols that were consistently detected in the wild-type plants were not detected in the mutants. In addition, one nonhexosylated dilignol was significantly less abundant in lbf1 outer tissues compared with corresponding wild-type samples. In stem inner tissues, six compounds (coniferin, syringin, and four hexosylated dilignols) were only detected in the mutants, and two compounds (two hexosylated dilignols) were significantly more abundant in the mutants than in the wild type. In these eight compounds, coniferyl alcohol moieties were overrepresented with respect to sinapyl alcohol moieties: The intensity of coniferin was 27 times higher than that of syringin, and sinapyl alcohol moieties were present in only two out of the eight upregulated compounds. The MS data and structure of five oligolignols identified for the first time in flax are shown in Supplemental Figure 2.
Table 3. Identified Differentially Accumulating Phenolics in Inner- and Outer-Stem Tissues of the lbf1 Mutant as Revealed by LC-MS.
| Outer Tissues | Inner Tissues | ||||
|---|---|---|---|---|---|
| tR (min) | Compound | Wild Type | lbf1 | Wild Type | lbf1 |
| 7.75 | Coniferin | n.d. | n.d. | n.d. | 131,603 ± 28,678 |
| 10.48 | Syringin | n.d. | n.d. | n.d. | 4,788 ± 695 |
| 12.88 | *G(8-O-4)G' hex | n.d. | n.d. | n.d. | 4,294 ± 2,419 |
| 13.30 | *G(8-O-4)G' hex | n.d. | n.d. | n.d. | 6,187 ± 3,826 |
| 14.90 | *G(8-O-4)G' hex | n.d. | n.d. | 401 ± 201 | 11,431 ± 3,315 |
| 15.12 | G(e8-O-4)S hex | n.d. | n.d. | n.d. | 25,94 ± 866 |
| 15.79 | G(8-5)G hex | n.d. | n.d. | 9,062 ± 4,375 | 266,534 ± 46,161 |
| 15.86 | G(t8-O-4)G | 1,466 ± 1,078 | n.d. | 13,168 ± 2,265 | 19,750 ± 10,882 |
| 15.87 | Lariciresinol hex | n.d. | n.d. | n.d. | 5,803 ± 1,963 |
| 16.36 | G(e8-O-4)G | 1,738 ± 1,060 | n.d. | 14,266 ± 2,556 | 23,642 ± 9,543 |
| 16.70 | *G(e8-O-4)FA | 50,087 ± 22,282 | 7,725 ± 3,722 | 4,327 ± 1,503 | 11,461 ± 5,925 |
| 21.48 | Lariciresinol | 3,533 ± 980 | n.d. | 2,808 ± 1,866 | 6,582 ± 2,210 |
| 21.57 | G(t8-O-4)secoisolariciresinol | 2,153 ± 441 | n.d. | 342 ± 73 | 985 ± 369 |
| 22.17 | G(e8-O-4)lariciresinol | 3,208 ± 32 | n.d. | 8,056 ± 2,475 | 5,673 ± 1,576 |
| 22.79 | *G(t8-O-4)lariciresinol | 764 ± 83 | n.d. | 1,429 ± 465 | 925 ± 263 |
| 23.65 | *G(8-5)FA | 2,474 ± 327 | n.d. | 129 ± 53 | 1,089 ± 362 |
| 24.54 | G(t8-O-4)S(8-5)G | 873 ± 237 | n.d. | 29,468 ± 5,586 | 28,725 ± 8,515 |
| 24.93 | G(t8-O-4)Sred/S(8-8/5)Gred/G | 1,377 ± 614 | n.d. | 2,184 ± 522 | 1,322 ± 373 |
| 25.80 | G(e?8-O-4)G(8-5)G' | 338 ± 3 | n.d. | 9,551 ± 1,938 | 4,253 ± 1,477 |
| 27.55 | *G(t8-O-4)S(8-8)S | 1,554 ± 457 | n.d. | 2,457 ± 802 | 4,462 ± 1,308 |
| 28.86 | *G(e8-O-4)S(8-8)S | 232 ± 52 | n.d. | 593 ± 144 | 963 ± 272 |
Values represent the relative abundance based on the extracted ion chromatogram, expressed as per mg dry weight tissue; values significantly different (Student’s t test) from the wild type at P < 0.05 are indicated in bold or italics, respectively, when they are higher or lower in abundance. n.d., not detected. Nomenclature is based on Morreel et al. (2004). Guaiacyl units, syringyl units, and units derived from ferulic acid and coniferaldehyde are referred to as G, S, FA, and G’, respectively. The linkage type is indicated in parentheses. “red,” reduced unit or adjacent linkage (Morreel et al., 2010a). A forward slash indicates that two units or two linkage positions are equally possible at this position in the shorthand name. hex, hexose or hexoside; tR, retention time. Asterisks indicate compounds that have not been described previously in flax stem tissue. Spectral data and structures of these compounds are given in Supplemental Figure 2. The spectral data of the other compounds are described by Huis et al. (2012). Three wild-type and six lbf1 plants were analyzed. Values are means ± se. Only the differentially accumulating metabolites with known identities are shown.
lbf1 Ectopic Lignification Is Associated with Modifications to Other Cell Wall Polymers
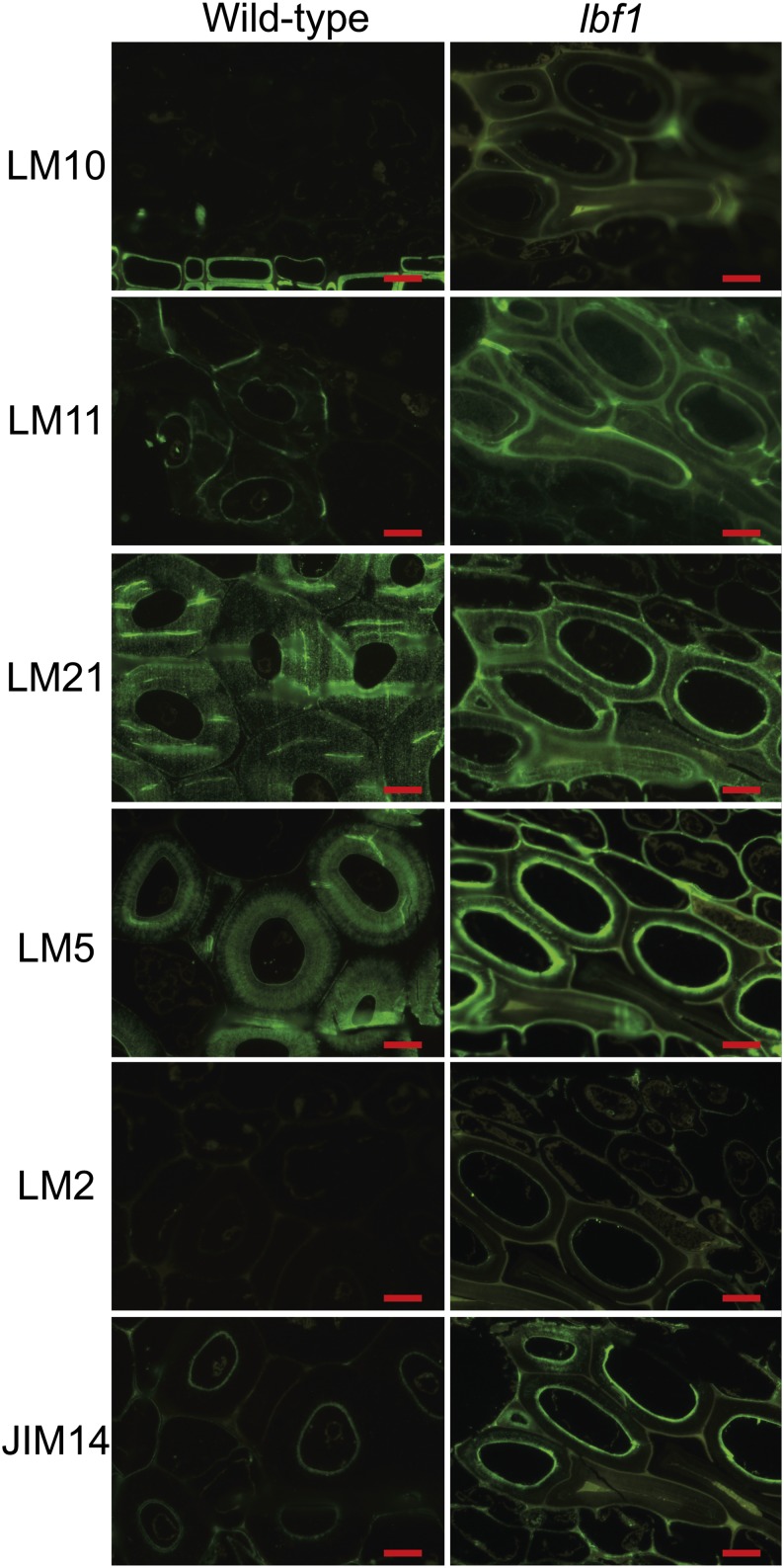
To investigate whether the modified lignin content in flax outer tissues was also associated with changes to other cell wall polymers, we first used antibodies targeted against the main classes of cell wall polymers: (1) pectin (galactan)-LM5 (Jones et al., 1997); (2) hemicellulose-LM10, LM11 (McCartney et al., 2005), and LM21 (Marcus et al., 2010); and (3) arabinogalactan proteins (AGPs)-LM2 (Smallwood et al., 1996; Yates et al., 1996) and JIM14 (Knox et al., 1991; Yates and Knox, 1994; Yates et al., 1996). LM5, JIM14, LM10, and LM11 showed stronger fluorescence in lbf1 outer tissues than in wild-type outer tissues (Figure 7), suggesting that cell walls in this mutant are enriched in pectin, hemicellulose, and glycoprotein compared with the wild type. LM5 labeling appeared strongly on the inner part of the lbf1 secondary wall, whereas labeling appeared weakly on the whole secondary cell wall of the wild type. JIM14 labeling is restricted to the inner part of the secondary wall in the wild type, whereas epitope distribution in the mutant seems more diffuse in the secondary wall. Increased LM10 and LM11 labeling was apparent on the whole secondary cell wall. LM21 gave fluorescence labeling in the thick secondary cell walls and in the inner secondary wall layer in the wild type and lbf1, respectively, suggesting modest changes in mannan hemicelluloses content. By contrast, almost no labeling was observed with LM2 antibodies in the bast fibers of both lbf1 and the wild type. Measurements of bast fiber cell wall thickness also indicated that the cell walls of lbf1 mutants are generally thinner (3.8 ± 1.1 μm) than corresponding cell walls in wild-type plants (9.9 ± 2.2 μm). When lbf1 and wild-type inner stem tissues were compared, no differences in antibody labeling were seen (Supplemental Figure 3).
Figure 7.
Fluorescent Microscopy Immunolocalization of Cell Wall NCPs with LM10, LM11, LM21, LM5, LM2, and JIM14 Antibodies.
Bars = 10 μm.
[See online article for color version of this figure.]
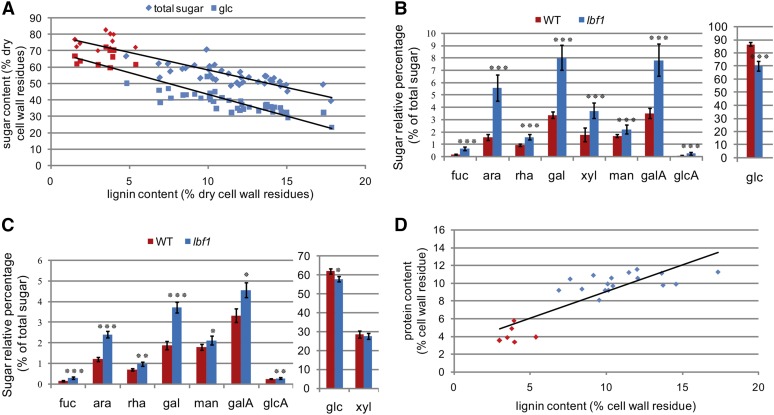
Further information on cell wall polymer modifications in the lbf1 mutant was obtained by analyzing total sugar content in stem outer tissues. Our results (Figure 8A) showed that increased lignification was correlated with a reduction in total sugar (mainly glucose) content when the latter was expressed as a percentage of the dry cell wall residue content. When the quantities of individual sugars were expressed as a percentage of the total sugar content (Figure 8B), glucose decreased from 87% (wild-type) to 70% (lbf1) total sugars, suggesting that cellulose content was reduced in lbf1 mutants. Further analyses with trifluoroacetic acid that does not degrade crystalline cellulose (Crônier et al., 2005) showed that the amounts of trifluoroacetic acid-released glucose accounted for 10.0% ± 1.5% and 9.8% ± 0.3% of glucose released by total hydrolysis of lbf1 and wild-type cell walls, respectively, thereby suggesting that the proportion of crystalline to noncrystalline cellulose is unchanged in mutant outer tissues. By contrast, the relative proportion of sugar monomers (Fuc, Ara, Rha, Gal, Xyl, Man, GalA, and GlcA) from other NCPs increased in the outer tissues of lbf1 mutants (Figure 8B) in agreement with outer tissue NMR data (Figure 5). For lbf1 inner tissues, there was no significant decrease in total sugar content and only a slight decrease in the relative proportion of glucose (Figure 8C). The relative proportions of other sugars increased, but less significantly than in the outer tissues. Our JIM14 results (Figure 7) indicated increased AGP content in lbf1 outer stem tissues, and we therefore quantified nitrogen levels in order to estimate relative protein content. Our results (Figure 8D) revealed that increased lignification was correlated with increased protein content.
Figure 8.
Sugar and Protein Analyses in lbf1 Inner- and Outer-Stem Tissues.
(A) Relationship between sugar content and lignin content in outer tissues of wild-type and mutant plants. Diamond-shaped dots correspond to total sugar content and square dots correspond to glucose content (red, wild-type plants; blue, lbf1 mutants).
(B) Relative content of different sugars in outer tissues of wild-type and lbf1 mutants. Glucose content is separated from other sugars due to the scale difference.
(C) Relative amounts of different sugars in inner tissues of wild-type and lignified mutants. Glucose and xylose are separated from others sugars due to the scale difference.
(D) Relationship between lignin and protein content. Lignin content was determined by acetyl bromide and protein content by nitrogen dosage (red, wild-type plants; blue, lbf1 mutant plants).
For (B) and (C), significant differences (Student’s t test) between the wild type and mutant were observed at P < 0.001 (***), P < 0.01 (**), and P < 0.05 (*). Error bars = sd.
Transcriptomics Suggests a Role for Lignin-Related Peroxidases in the lbf1 Phenotype
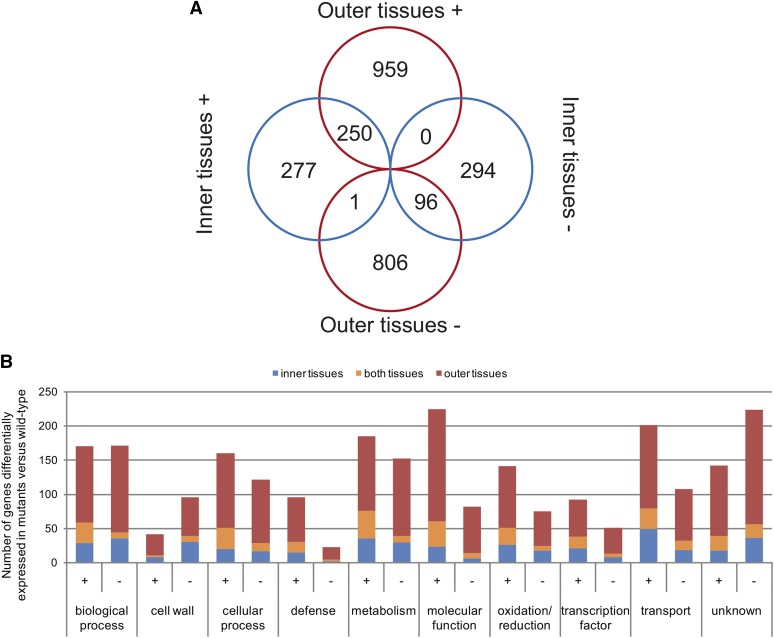
To obtain information about modifications in gene expression associated with the lbf1 phenotype, we performed whole-genome transcriptomics using flax-specific Agilent microarrays. Gene expression patterns in inner- and outer-stem tissues from six individual lignified lbf1 mutants were compared with corresponding tissues from wild-type plants. Our results (Figure 9A; Supplemental Data Set 1) show that transcripts of 1487 genes were significantly more abundant (P value < 0.05) in the lbf1 mutants as compared with wild-type plants. Of these 1487 transcripts, 959 were specifically more abundant in stem outer tissues and 277 were specifically more abundant in stem inner tissues; transcripts for 250 genes were more abundant in both tissues (Figure 9A). A total of 1197 transcripts were less abundant in the lbf1 mutants (Figure 9A; Supplemental Data Set 6), of which 806 were specifically less abundant in stem outer tissues, 294 were specifically less abundant in stem inner tissues, and 96 were less abundant in both inner- and outer-stem tissues of lbf1 mutants when compared with wild-type plants.
Figure 9.
Differentially Accumulated Transcripts in lbf1 Mutants versus the Wild Type in Separated Tissues.
(A) Venn diagram; ±, over/underaccumulated (P value < 0.05, Bonferroni method) transcripts in lbf1 stem tissues versus corresponding tissues of the wild type.
(B) GO classification of differentially accumulated transcripts in outer (red), inner (blue), and both (orange) tissues. GO classification was determined using blast2go on protein sequences (NCBI) and verified by expert curation.
Functional classification using Gene Ontology (GO) (Figure 9B) showed that the differentially accumulated transcripts are implicated in diverse biological processes, molecular functions, and transport. For example, 5.6% (149 genes) of the differential transcript abundance is related to genes involved in biosynthesis and maintenance of the plant cell wall. Examination of the 20 most abundant transcripts in outer tissues showed that the most represented class corresponded to defense genes (5) and oxidation/reduction genes (5) (Table 4). For inner tissues, the two most represented classes were transport and cellular process with four genes in each class. Among the 20 least abundant transcripts in outer-stem tissues (Table 5), the classes biological process and unknown were the two most represented. In mutant inner tissues, the least abundant transcript (Lus10038721) corresponds to a homolog of CCD8 belonging to the carotenoid cleavage dioxygenase family (Leyser, 2008). In Arabidopsis thaliana, a mutation in this gene is associated with a decrease in strigolactone content and increased axillary bud production (Sorefan et al., 2003). It is possible that the reduced transcript accumulation of the flax putative CCD8 ortholog is related to the branched phenotype of the lbf1 mutants.
Table 4. List of the 20 Most Highly Abundant Transcripts in lbf1 Mutants versus the Wild Type.
| Reference | Name | Delta | P Value | GO Annotation | Arabidopsis Correspondence |
|---|---|---|---|---|---|
| 20 Most Abundant Transcripts in Outer Tissues | |||||
| Lus10020493 | Pathogenesis-related gene 1 | 7.41 | 0.00E+00 | Defense | AT2G14610.1 |
| Lus10003264 | Pathogenesis-related 4 | 6.98 | 0.00E+00 | Defense | AT3G04720.1 |
| Lus10006925 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 6.70 | 0.00E+00 | Metabolism | AT4G02780.1 |
| Lus10028898 | Cytochrome P450, family 76, subfamily C, polypeptide 4 | 6.64 | 2.22E-16 | Oxidation/reduction | AT2G45550.1 |
| Lus10022642 | LYS/HIS transporter 7 | 6.36 | 4.44E-16 | Transport | AT4G35180.1 |
| Lus10003339 | Transmembrane amino acid transporter family protein | 6.35 | 2.22E-16 | Transport | AT1G47670.1 |
| Lus10004958 | Somatic embryogenesis receptor-like kinase 2 | 6.35 | 4.44E-16 | Molecular function | AT1G34210.1 |
| Lus10012684 | Peroxidase superfamily protein | 6.29 | 0.00E+00 | Oxidation/reduction | AT2G41480.1 |
| Lus10020826 | Peroxidase superfamily protein | 6.15 | 1.78E-15 | Oxidation/reduction | AT2G41480.1 |
| Lus10032178 | Unknown | 6.14 | 2.22E-16 | Unknown | |
| Lus10014508 | Unknown | 6.08 | 3.77E-15 | Unknown | |
| Lus10030945 | Nitrate transporter 1.5 | 6.03 | 4.46E-14 | Transport | AT1G32450.1 |
| Lus10015339 | Unknown | 5.96 | 1.11E-15 | Defense | |
| Lus10035241 | Glutathione S-transferase tau 7 | 5.95 | 5.33E-15 | Molecular function | AT2G29420.1 |
| Lus10039454 | MLP-like protein 423 | 5.91 | 3.49E-14 | Defense | AT1G24020.1 |
| Lus10022415 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 5.90 | 4.44E-16 | Oxidation/reduction | AT1G06620.1 |
| Lus10035221 | Matrixin family protein | 5.54 | 2.75E-14 | Cellular process | AT1G24140.1 |
| Lus10004410 | Pathogenesis-related thaumatin superfamily protein | 5.51 | 3.88E-11 | Defense | AT1G20030.2 |
| Lus10008173 | Peroxidase superfamily protein | 5.47 | 4.88E-15 | Oxidation/reduction | AT5G06730.1 |
| Lus10025253 | Protein of unknown function (DUF567) | 5.45 | 1.39E-11 | Transport | AT5G01750.2 |
| 20 Most Abundant Transcripts in Inner Tissues | |||||
| Lus10000453 | Homolog of carrot EP3-3 chitinase | 6.38 | 4.44E-16 | Cell wall | AT3G54420.1 |
| Lus10016323 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | 5.87 | 2.22E-16 | Transport | AT5G48490.1 |
| Lus10002741 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | 5.79 | 0.00E+00 | Transport | AT5G48490.1 |
| Lus10031759 | Plant natriuretic peptide A | 5.72 | 0.00E+00 | Defense | AT2G18660.1 |
| Lus10007270 | P-loop-containing nucleoside triphosphate hydrolases superfamily protein | 5.67 | 8.44E-15 | Molecular function | AT3G28540.1 |
| Lus10005395 | Unknown | 5.14 | 1.11E-11 | Unknown | |
| Lus10021102 | Glutathione S-transferase TAU 8 | 5.12 | 1.78E-15 | Cellular process | AT3G09270.1 |
| Lus10032930 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 5.11 | 6.66E-16 | Oxidation/reduction | AT5G24530.1 |
| Lus10010702 | MLP-like protein 423 | 4.87 | 1.22E-14 | Defense | AT1G24020.1 |
| Lus10034484 | Chaperone DnaJ-domain superfamily protein | 4.82 | 2.27E-13 | Cellular process | AT4G36040.1 |
| Lus10015350 | Disease resistance protein (TIR-NBS-LRR class) family | 4.82 | 5.46E-14 | Defense | AT4G12010.1 |
| Lus10005523 | Unknown | 4.75 | 6.20E-12 | Unknown | |
| Lus10025060 | Chaperone DnaJ-domain superfamily protein | 4.71 | 2.11E-12 | Cellular process | AT4G36040.1 |
| Lus10023142 | Cytochrome P450, family 79, subfamily B, polypeptide 2 | 4.51 | 4.67E-09 | Oxidation/reduction | AT4G39950.1 |
| Lus10033041 | Leucine-rich repeat protein kinase family protein | 4.40 | 8.88E-16 | Molecular function | AT2G31880.1 |
| Lus10022547 | Phosphate transporter 1;5 | 4.38 | 1.09E-13 | Transport | AT2G32830.1 |
| Lus10016635 | Phosphate transporter 1;7 | 4.37 | 3.82E-13 | Transport | AT3G54700.1 |
| Lus10015933 | Unknown | 4.36 | 6.88E-11 | Metabolism | AT5G61820.1 |
| Lus10040328 | α/β-Hydrolases superfamily protein | 4.34 | 1.01E-11 | Cellular process | AT2G39420.1 |
| Lus10034312 | NIM1-interacting 2 | 4.34 | 2.22E-12 | Molecular function | AT3G25882.1 |
Table 5. List of the 20 Least Abundant Transcripts in lbf1 Mutants versus the Wild Type.
| Reference | Name | Delta | P Value | GO Annotation | Arabidopsis Correspondence |
|---|---|---|---|---|---|
| 20 Least Abundant Transcripts in Outer Tissues | |||||
| Lus10011872 | Tetratricopeptide repeat (TPR)-like superfamily protein | −5.08 | 1.58E-10 | Biological process | AT5G48850.1 |
| Lus10022806 | Ethylene-dependent gravitropism-deficient and yellow-green-like 2 | −5.08 | 1.47E-09 | Cellular process | AT5G05740.2 |
| Lus10012353 | Unknown | −5.02 | 1.69E-09 | Unknown | |
| Lus10041133 | Purine permease 3 | −4.90 | 2.75E-10 | Transport | AT1G28220.1 |
| Lus10009917 | Expansin A8 | −4.48 | 1.29E-07 | Cell wall | AT2G40610.1 |
| Lus10006996 | Unknown | −4.33 | 4.43E-11 | Unknown | AT2G27830.1 |
| Lus10006759 | Gibberellin 2-oxidase 8 | −4.30 | 1.97E-12 | Oxidation/reduction | AT4G21200.1 |
| Lus10038566 | Dynein light chain type 1 family protein | −4.29 | 2.04E-11 | Biological process | AT4G27360.1 |
| Lus10000385 | Unknown | −4.16 | 4.72E-10 | Unknown | AT2G27830.1 |
| Lus10003913 | Urophorphyrin methylase 1 | −4.12 | 3.55E-09 | Molecular function | AT5G40850.1 |
| Lus10023289 | Unknown | −3.90 | 6.35E-11 | Unknown | AT1G30260.1 |
| Lus10025278 | Aluminum sensitive 3 | −3.86 | 2.41E-11 | Transport | AT2G37330.1 |
| Lus10009069 | Aluminum sensitive 3 | −3.84 | 2.41E-13 | Transport | AT2G37330.1 |
| Lus10038821 | Nodulin MtN3 family protein | −3.78 | 7.64E-09 | Biological process | AT5G53190.1 |
| Lus10010529 | Unknown | −3.72 | 5.15E-11 | Unknown | AT5G19340.1 |
| Lus10008485 | Protein of unknown function (DUF567) | −3.72 | 6.79E-12 | Unknown | AT3G14260.1 |
| Lus10028947 | Xyloglucan endotransglucosylase/hydrolase 15 | −3.65 | 1.03E-11 | Cell wall | AT4G14130.1 |
| Lus10038517 | Unknown | −3.64 | 5.59E-10 | Unknown | AT1G30260.1 |
| Lus10037476 | Urophorphyrin methylase 1 | −3.64 | 8.61E-08 | Molecular function | AT5G40850.1 |
| Lus10023377 | Sec14p-like phosphatidylinositol transfer family protein | −3.56 | 4.58E-11 | Transport | AT1G30690.1 |
| 20 Least Abundant Transcripts in Inner Tissues | |||||
| Lus10038721 | Carotenoid cleavage dioxygenase 8 | −5.34 | 3.59E-12 | Metabolism | AT4G32810.1 |
| Lus10002073 | Protein of unknown function, DUF584 | −4.08 | 3.00E-08 | Unknown | AT1G61930.1 |
| Lus10023311 | Gibberellin 2-oxidase | −3.98 | 7.66E-09 | Oxidation/reduction | AT1G30040.1 |
| Lus10017253 | RING/U-box superfamily protein | −3.77 | 3.03E-13 | Molecular function | AT5G42200.1 |
| Lus10034238 | NAD-dependent glycerol-3-phosphate dehydrogenase family protein | −3.77 | 6.99E-07 | Metabolism | AT2G40690.1 |
| Lus10025771 | Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase A protein | −3.74 | 1.31E-10 | Cell wall | AT3G14920.1 |
| Lus10034206 | Major facilitator superfamily protein | −3.72 | 9.36E-11 | Transport | AT2G40460.1 |
| Lus10005617 | RING/U-box superfamily protein | −3.65 | 2.94E-12 | Molecular function | AT5G42200.1 |
| Lus10008304 | Pathogenesis-related thaumatin superfamily protein | −3.60 | 3.75E-09 | Defense | AT5G40020.1 |
| Lus10035519 | HXXXD-type acyl-transferase family protein | −3.56 | 6.39E-08 | Molecular function | AT5G01210.1 |
| Lus10043404 | Unknown | −3.55 | 3.63E-10 | Unknown | AT3G11600.1 |
| Lus10017817 | Major facilitator superfamily protein | −3.50 | 6.18E-11 | Transport | AT1G68570.1 |
| Lus10013489 | Late embryogenesis abundant protein (LEA) family protein | −3.50 | 2.54E-10 | Biological process | AT1G52690.1 |
| Lus10025278 | Aluminum sensitive 3 | −3.43 | 1.45E-09 | Transport | AT2G37330.1 |
| Lus10030457 | Glucose-methanol-choline (GMC) oxidoreductase family protein | −3.43 | 1.47E-08 | Metabolism | AT1G14185.1 |
| Lus10013401 | Branched-chain α-keto acid decarboxylase E1 β-subunit | −3.42 | 7.33E-09 | Oxidation/reduction | AT1G55510.1 |
| Lus10029063 | Major facilitator superfamily protein | −3.37 | 5.23E-10 | Transport | AT2G40460.1 |
| Lus10023189 | Laccase 11 | −3.33 | 3.46E-08 | Oxidation/reduction | AT5G03260.1 |
| Lus10037164 | Expansin A1 | −3.32 | 6.67E-08 | Cell wall | AT1G69530.1 |
| Lus10041338 | Serine carboxypeptidase-like 48 | −3.31 | 9.73E-08 | Cellular process | AT3G45010.1 |
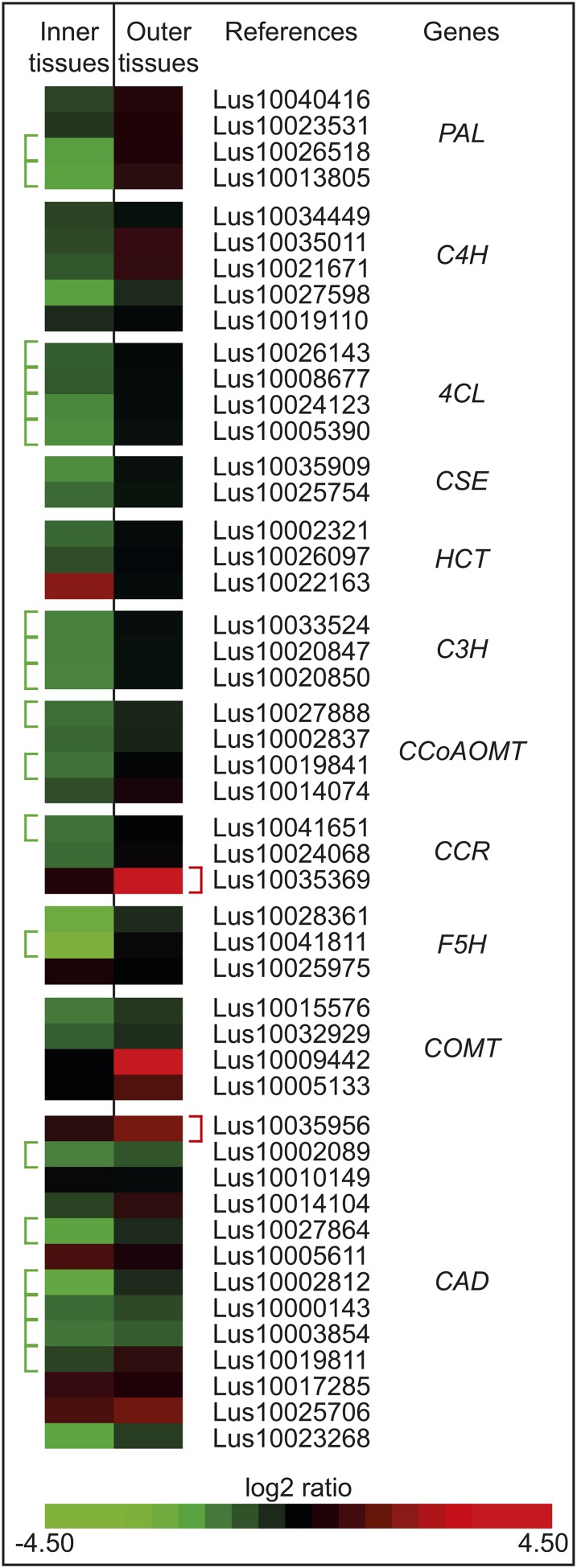
Increased lignification is the major observed cell wall phenotype in lbf1 mutants, and we therefore focused our attention on transcripts corresponding to two major control points in the lignification process: (1) monolignol biosynthesis and (2) monolignol polymerization. Following interrogation of the Arabidopsis database, sequence alignment, and phylogenetic analyses, we identified a total of 48 putative genes involved in monolignol biosynthesis in the flax genome. Transcripts corresponding to 22 of these genes were differentially accumulated between lbf1 mutants and the wild type. In outer tissues, transcripts corresponding to a CCR, a COMT, and a CAD gene were significantly more abundant. For inner tissues, transcripts corresponding to 19 lignin genes (2 × PAL, 4 × 4CL, 3 × C3H, 1 × F5H, 2 × CCoAOMT, 1 × CCR, and 6 × CAD) genes were significantly less abundant (Figure 10).
Figure 10.
Heat Map Representing Comparative Accumulation of Monolignol Biosynthetic Transcripts in Outer- and Inner-Stem Tissues of lbf1.
Transcripts annotated by a bracket are significantly overaccumulated (red) or underaccumulated (green) (P value < 0.05, Bonferroni method) in the mutant compared with the wild type.
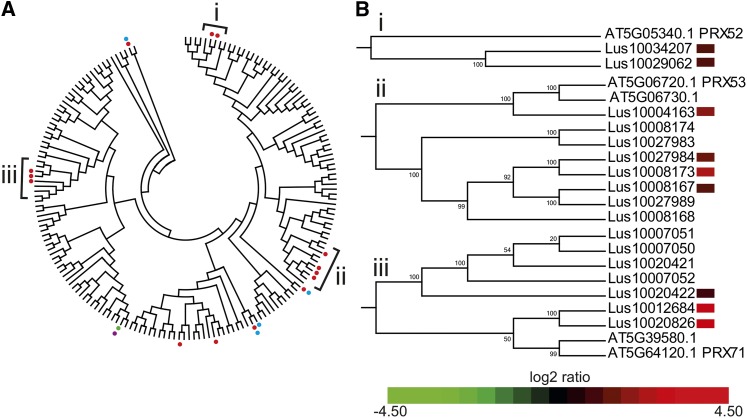
During lignification, the synthesized monolignols are exported to the cell wall where they are oxidized by laccases and/or peroxidases prior to polymerization into the lignin polymer. Analyses of transcriptomics data showed that no laccase transcripts were differentially accumulated between lbf1 and wild-type outer tissues. By contrast, transcripts corresponding to the laccase11 (LAC11) gene were significantly less abundant in lbf1 inner tissues. Transcripts corresponding to 16 peroxidase genes showed significant differential accumulation between lbf1 mutants and wild-type plants (Figure 11A; Supplemental Data Set 2). Transcripts for 11 of these genes were more abundant uniquely in outer stem tissues, transcripts for one gene were more abundant uniquely in inner stem tissues, and transcripts for three genes were more abundant in both tissues. Transcripts for one peroxidase gene were significantly less abundant in the lbf1 mutant. A phylogenetic tree (Figure 11B; Supplemental Data Set 2) based on an alignment of protein sequences of both flax and Arabidopsis peroxidases shows that 9 of the 11 flax peroxidase transcripts specifically more abundant in lbf1 outer tissues are phylogenetically close to three distinct At-PRXs (At-PRX52, At-PRX53, and At-PRX71) known to oxidize monolignols and therefore are potentially involved in lignin polymerization (Østergaard et al., 2000; Nielsen et al., 2001; Herrero et al., 2013; Shigeto et al., 2013).
Figure 11.
Phylogenetic and Expression Analyses of Peroxidases in lbf1 Mutants.
(A) Phylogenetic unrooted tree of Arabidopsis and flax peroxidase proteins. Branches marked by a dot correspond to individual peroxidase transcripts significantly more abundant (P value < 0.05, Bonferroni method) in inner (blue dot) or outer tissues (red dot) or less abundant in inner (pink dot) or outer tissues (green) of lbf1 mutant compared with the wild type.
(B) Heat map of transcript accumulation corresponding to flax peroxidases in clades A, B, and C containing known Arabidopsis lignin-related peroxidases.
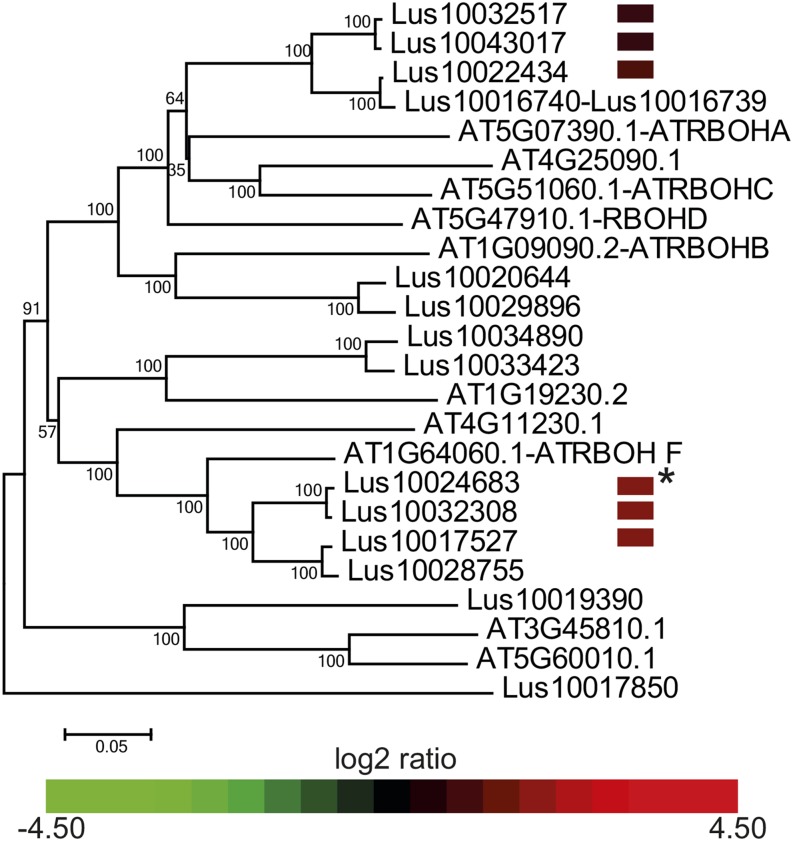
Peroxidases require H2O2 to oxidize monolignols in order to make lignin. H2O2 is produced through the action of two types of enzyme: (1) NADPH-oxidase enzymes, and more specifically RBOH enzymes, that produce superoxide ions; and (2) superoxide dismutase, which converts superoxide ions into H2O2 (Karpinska et al., 2001; Karlsson et al., 2005). We identified 14 flax orthologs of the 10 RBOH genes identified in Arabidopsis (Torres, 2010). Transcripts corresponding to five of these genes were specifically more abundant in lbf1 outer-stem tissues. Phylogenetic analyses indicated that two of these genes are closely related to At-RBOH-F (Figure 12), recently shown to be involved in Casparian strip lignification (Lee et al., 2013). The other three flax RBOH genes are orthologs of AtRBOH-A and C genes involved in defense related apoplastic H2O2 production (Schweizer, 2008). Our analyses also showed that transcripts corresponding to another At-RBOH-F ortholog were significantly more abundant in both inner- and outer-stem tissues of the mutant compared with wild-type plants (Figure 12; Supplemental Data Set 3). Finally, our data (Supplemental Data Set 1) indicated that a transcript corresponding to a superoxide dismutase gene was specifically more abundant in lbf1 outer tissues.
Figure 12.
Phylogenetic and Expression Analyses of RBOH Proteins in lbf1 Mutants.
Phylogenetic unrooted tree of Arabidopsis and flax RBOH proteins. Heat map expression data of the six flax genes overexpressed in lbf1 mutants compared with the wild type are given in front of their corresponding Phytozome references. Transcripts of all genes are specifically more abundant in lbf1 outer tissues except where transcripts are more abundant in outer and inner lbf1 tissues (asterisk).
DISCUSSION
Lignification plays an important role in plant biology and has a major impact on the quality of a wide range of different products derived from plants. In timber, the presence of lignin is positive as it provides rigidity and mechanical support to fiber cell walls. In contrast, the presence of lignin inhibits saccharification during biofuel production and therefore has a negative effect on the quality of lignocellulosic biomass. The lignin polymer is initially deposited in the preexisting middle lamella and primary wall of cells during the formation of the secondary cell wall. Lignin deposition then continues in the secondary wall with the result that most secondary plant cell walls contain relatively high amounts of lignin. This type of lignification is typical of the cell walls of xylem fibers, vessels, and tracheids. By contrast, bast fiber plants, such as flax, ramie, and jute, have been exploited by man for many thousands of years precisely because their stems also contain elongated fiber cells with thick cellulose-rich secondary cell walls but only low amounts of lignin (Day et al., 2005; del Río et al., 2011). It therefore appears that certain plant species possess particular regulatory mechanisms that allow them to construct thick nonlignified secondary cell walls. A better understanding of these mechanisms could provide novel targets for engineering of plant biomass. In flax stems, the outer tissues containing the cellulose-rich bast fibers can be easily separated from the inner tissues containing the lignified secondary xylem cells, thereby allowing comparative studies of cell wall formation in these two tissues (Fenart et al., 2010; Huis et al., 2012). To learn more about the mechanisms regulating cell wall biosynthesis in flax, we used a combination of UV autofluorescence and phloroglucinol-HCl staining to screen a flax EMS mutant population for mutants showing altered bast fiber lignification patterns (Chantreau et al., 2013). This approach allowed us to identify 93 families showing increased lignification in bast fibers, and we then went on to characterize one of these mutants (lbf1) in detail.
Characterization of the Flax lbf1 Mutant
(1) Lignin and Oligolignols
Chemical analyses of bast fiber ectopic lignin monomeric composition in lbf1 mutants showed significant increases in the amounts of all three lignin monomers with no significant modification in the S/G ratio, indicating that lignin structure was unchanged. Flax lignin is particularly condensed and therefore only ∼10% of outer tissue lignin and 20% of inner tissue lignin are probably accessible via thioacidolysis disruption of noncondensed alkyl-aryl ether linkages (Day et al., 2005). We therefore used NMR analysis of solubilized cell wall samples to complete the chemical data. These results confirmed that the chemical composition of bast fiber ectopic lignin was rich in G units. NMR data provided a lower S/G ratio than that obtained with thioacidolysis, suggesting a preferential involvement of S units in alkyl-aryl ether in agreement with previous NMR analysis of milled wood lignin (del Río et al., 2011). Immunolabeling of phenylcoumaran in lbf1 mutant bast fiber walls was in good agreement with lignin analysis showing a noticeable amount of side chains involved in this structure.
Oligolignol profiling indicated that ectopic lignification in the outer stem tissues of the lbf1 mutant was accompanied by a strong decrease in the accumulation of nonhexosylated oligolignols in that tissue. We have previously shown that a wide range of (mono)oligolignols normally accumulates in this tissue in wild-type flax, and it is possible that their levels decrease in the mutant because they are incorporated into the lignin polymer (Huis et al., 2012). This hypothesis was supported by the observation that the depleted lignin oligolignols in lbf1 bast fibers were mainly composed of G units and several contained phenylcoumaran linkages in agreement with the chemical, NMR, and immunological analyses. These results would suggest that hypolignification in wild-type flax bast fibers is not so much caused by a lack of lignin precursors but is rather due to insufficient polymerization.
The polymerization of lignin occurs via radical coupling of monolignol and oligolignol radicals, which are formed by peroxidase and/or laccase activity (Zhao et al., 2013). In support of peroxidase involvement in ectopic bast fiber lignification, we observed increased transcript levels of nine lignin-related peroxidase genes specifically in the outer tissues of lbf1 mutants compared with wild-type plants. Peroxidase activity was previously reported to be associated with the onset of lignification in flax fibers (McDougall, 1991, 1992) and peroxidase ESTs/genes are highly represented/expressed in flax outer stem cDNA libraries (Day et al., 2005; Roach and Deyholos, 2007) and tissues (Fenart et al., 2010; Huis et al., 2012). Based on microarray data, laccase genes are probably more closely associated with lignification of flax xylem tissues, but not bast fibers (Huis et al., 2012). The transcriptomics data from the lbf1 mutant suggest that flax outer-stem peroxidases and not laccases are responsible for the increased lignification. It would obviously be interesting to characterize other flax lbf mutants and/or create laccase overexpressors to investigate whether lignified bast fibers could be induced by upregulating laccase gene expression.
Peroxidases, but not laccases, require H2O2 for radical production, and we also observed increased transcript levels in lbf1 outer-stem tissues of five NADPH oxidase genes. Interestingly, two of these flax NADPH-oxidase genes are homologs of Arabidopsis type RBOH-F NADPH-oxidases recently shown to be involved in the polymerization of lignin within the Casparian strip of the endodermis (Lee et al., 2013). The highly localized Casparian strip lignification in Arabidopsis occurs through docking proteins, called Casparian strip domain proteins, which are targeted to the area of the Casparian strip and recruit both an NADPH oxidase and a peroxidase. Such enzyme assemblies then direct localized oligolignol polymerization to form the Casparian strip. The coordinated overexpression of NADPH oxidases and peroxidases specifically in lbf1 outer tissues would suggest a similar concerted action of these two enzymes. However, no evidence exists as yet, based on the comparative microarray data set of lbf1 and wild-type flax, of the involvement of a scaffolding protein homologous to the Arabidopsis endodermis Casparian strip domain proteins in lignification in flax stem tissues. Further evidence for a potential role of peroxidases and NADPH oxidases in lbf1 lignification was provided by the observation that the other three flax NADPH oxidase genes are all orthologs of the Arabidopsis RBOH-A and RBOH-C genes involved in the generation of apoplastic H2O2 during the defensive oxidative burst (Schweizer, 2008). Increased accumulation of RBOH-A and -C transcripts could therefore also contribute to apoplastic H2O2 content and stimulate lignification. Somewhat intriguingly, we also observed a significant accumulation of transcripts corresponding to another flax RBOH-F ortholog in both outer- and inner-stem tissues of the lbf1 mutant despite the fact that increased lignification was only observed in outer tissues. Further work is necessary to understand the significance of increased NADPH oxidase accumulation in lbf1 inner stem tissues.
Although our results suggest that ectopic lignification in lbf1 bast fibers is related to modified polymerization, the increased transcript abundance of the monolignol biosynthesis genes COMT, CCR, and CAD suggests that the supply of monolignols to these fibers is also increased. The Lu-CCR gene is phylogenetically close to At-CCR involved in developmental lignification in Arabidopsis and CCR downregulation drastically reduces lignin biosynthesis (Lacombe et al., 1997; Dauwe et al., 2007; Leplé et al., 2007). By contrast, the Lu-COMT and Lu-CAD genes are not part of the bona fide lignin group responsible for developmental lignification (Supplemental Figure 3 and Supplemental Data Sets 4 to 6) but rather belong to gene groups involved in the response to stress or pathogen attack (Barakat et al., 2010, 2011). This observation is interesting since among the 20 most abundant transcripts in the inner- and outer-stem tissues, several are potentially involved in defense, raising the possibility that increased lignification in the lbf1 mutant could be caused by a mutation affecting the defense signaling and/or response pathway.
The idea that bast fiber ectopic lignification in lbf1 outer tissues could be associated with increased monolignol production and/or availability is also supported by the decreased accumulation of transcripts corresponding to a UGT (UDP-glucosyltransferase) gene. This gene encodes a putative ortholog of the Arabidopsis UGT72E1 protein that glucosylates monolignols (Lanot et al., 2006, 2008). Monolignol glucosylation is believed to play a role in detoxifying monolignols and is also involved in addressing monolignols to the vacuole for storage (Miao and Liu, 2010; Tsuyama et al., 2013). This process therefore represents a potential control point in lignification and the observed reduction in UGT transcript abundance in outer tissues of the flax lbf1 mutant could be expected to increase monolignol availability for subsequent lignification. Alternatively, increased incorporation of monolignols into the lignin polymer could reduce the necessity for detoxification and/or vacuolar storage leading to UGT downregulation. Interestingly, decreased lignification in Arabidopsis triple laccase mutants is associated with increased expression of genes encoding the 72E2 and 72E3 UGT proteins (Zhao et al., 2013), suggesting the existence of a relationship between modified lignification and regulation of monolignol supply via glycosylation.
In contrast to outer-stem tissues, levels of lignin and nonglycosylated oligolignols remained unchanged in lbf1 inner-stem tissues compared with wild-type plants. Nevertheless, quantities of the hexosylated monolignols coniferin and syringin as well as of several hexosylated dilignols were significantly higher in mutant xylem tissues. Transcriptomics data indicated that transcripts of four peroxidase genes were more abundant in lbf1 inner tissues. Whereas none of these peroxidase genes belong to the same clades as the lignin-related peroxidase genes, they exhibit increased transcript abundance in lbf1 outer tissues, suggesting that they are not involved in lignification. In the absence of a significant increase in the capacity to oxidize monolignols for polymerization, it is possible that monolignols cannot be incorporated into the lignin polymer and must be detoxified by other mechanisms such as glycosylation. The observed decrease in the abundance of transcripts corresponding to a laccase gene (LAC11) implicated in lignification also suggests a reduced monolignol oxidizing capacity (Zhao et al., 2013). Although no significant change in UGT expression was observed, transcript abundance was reduced for 19 genes in the lignin biosynthetic pathway. This massive decrease in transcripts corresponding to 7 of the 11 lignin gene families could be interpreted as an attempt to regulate monolignol production and cellular toxicity. Further work is necessary to clarify this point.
Altogether our observations indicate that the lbf1 mutation results in contrasted tissue-specific effects on transcript abundance of a range of lignin-related genes (i.e., genes encoding enzymes involved in monolignol biosynthesis, a UGT, peroxidases, NADPH-oxidases, and a superoxide dismutase), oligolignol content, and lignin quantity in flax stems. These observations not only suggest the existence of complex tissue-specific regulation mechanisms, but also underline the importance of taking into account organ and tissue specificity when interpreting expression data.
(2) Other Cell Wall Polymers
The thick secondary walls of mature flax bast fibers largely consist of cellulose and pectic galactan as the main incrusting NCP together with AGPs (His et al., 2001; Morvan et al., 2003). Both chemical analyses and immunolabeling suggested that lbf1 bast fibers contain less cellulose and significantly higher amounts of NCPs and AGPs compared with the wild type and provide strong evidence that increased lignification is accompanied by changes in polysaccharide architecture as previously observed in different Arabidopsis lignin mutants (Van Acker et al., 2013).
Although at first view such changes could be due to the higher lignin content in the mutant, another intriguing possibility is that crosstalk between cell wall polymers during biosynthesis may favor the formation of a cell wall matrix more favorable to lignification. Higher hemicellulose deposition concomitant with lower cellulose content, for example, would lead to a looser cell wall structure and/or less crystalline cellulose, both of which could facilitate monolignol transport and subsequent lignin polymerization within a xylan hemicellulose matrix. In agreement with this idea is the observation that disruption of cellulose biosynthesis, either by chemical inhibition with isoxaben, or by mutations in the CESA3 gene leads to ectopic lignification in Arabidopsis wild type and eli1 mutants (Caño-Delgado et al., 2003). Similarly, ectopic lignification in the elp1 Arabidopsis mutant is due to a mutation in a chitinase-like (CTL) gene (Zhong et al., 2002). While only a small number of transcripts corresponding to genes directly involved in cell wall biosynthesis were differentially accumulated between flax lbf1 mutant and wild-type plants, these included several glucosyltransferases, possibly accounting for the observed changes in cell wall matrix polysaccharides. In addition, transcripts for a COBRA4-like extracellular glycosyl-phosphatidyl inositol-anchored protein were less abundant in the outer tissues of the flax mutant. This protein has been proposed to modulate cellulose assembly through interaction with cellulose microfibrils (Liu et al., 2013), and reduced expression of this gene is associated with lower cellulose content and higher lignification in mature stem tissues of the maize (Zea mays) bk2 mutant (Sindhu et al., 2007).
In addition to modifications in cell wall chemical composition, lbf1 fiber cell walls were also significantly thinner than wild-type ones and could be related to the differential transcript abundance of putative flax orthologs corresponding to Arabidopsis expansin (AtEXP8) and xyloglucan endotransglycosylase/hydrolase (AtXTR7) genes (Cosgrove, 2005; Sasidharan et al., 2008).
Further analyses, not only of the spatial distribution of different cell wall components, but also fiber morphology and cell wall thickness at different stages of fiber development, are needed to obtain better insight into the relationship between polysaccharides and lignin deposition in the growing cell wall.
Conclusions and Perspectives
In conclusion, we generated a core collection of flax lbf mutants that represent an interesting biological resource for investigating the regulatory mechanisms used by fiber plants to produce poorly lignified, thick, secondary cell walls. As a proof of concept, we undertook a detailed characterization of the lbf1 mutant. Our results suggest that the main regulatory point occurs at the oxidative polymerization step and that the typical low lignification observed in wild-type bast fibers is related to the absence of different actors necessary for monolignol oxidation. Recent analyses of peroxidase gene promoters have suggested that these genes are regulated by a number of different transcription factors (NAC, MYB, AP2, and class I and III HD-ZIP) previously associated with vascular tissue formation and/or secondary cell wall formation (Herrero et al., 2014), and it is possible that increased bast fiber lignification is associated with a mutation in such a gene(s). Alternatively, peturbations in the biosynthesis of other cell wall polymers affecting cell wall integrity and/or activation of defense signaling could also be responsible for the ectopic lignification in the lbf1 mutant. The flax genome at ∼390 Mb is relatively small, and recent advances in NGS technology should allow the development of a “mapping by sequencing” approach (Wang et al., 2012; Allen et al., 2013; Wijnen and Keurentjes, 2014) in this species and the subsequent identification of the gene(s) associated with increased lignification and other interesting phenotypes. We observed that for lbf1 the lignified phenotype is heritable over several generations, and we are currently generating F2 backcrossed material for such an approach. Heritability of the lignified phenotype has also been confirmed for 6 out of 10 other lbf families that we are multiplying.
The systematic exploitation of the flax lbf collection will allow us to improve our understanding of the functional relationship between lignin and other cell wall polymers, thereby leading to a better understanding of cell wall dynamics. Finally, our collection can be used to gain a better knowledge of how increased lignification modifies different fiber mechanical and physical properties. For example, preliminary saccharification analyses using a commercial cellulase cocktail (Novozymes) indicate that 30% less glucose is released from flax lbf mutant outer-stem tissues when compared with wild-type tissues.
METHODS
Plant Material
Flax (Linum usitatissimum) EMS mutants used in this study come from the PT-Flax Collection (Chantreau et al., 2013). M2 to M5 plants were grown in greenhouses or outside at the University of Lille, France. For chemical, metabolomic, and transcriptomic analyses, stem outer tissues were separated from inner tissues by peeling as previously described (Day et al., 2005). For transcriptomics, tissues were harvested before flowering and were immediately frozen in liquid nitrogen. For chemistry and metabolomics, tissues were harvested at grain maturity and lyophilized before analyses.
Microscopy
UV-based lignin screening was made on thick freehand cross sections from the median part of M2 mutant stems. Outer-tissue fluorescence was determined using an inverted microscope (Nikon Eclipse TS100) coupled with an UV irradiation system (λ excitation, 365 nm; λ emission, 420 nm). Phloroglucinol-HCl staining was made on semi-thin freehand cross sections and examined with a Nikon Eclipse TS100 and/or a LEICA DM2000 microscope. Photographs were taken with a Nikon D5000 camera.
Immunohistochemical Analyses
Ethanol-fixed specimens of the median region of flax stems were dehydrated using an ethanol series and acetone prior to epoxy resin impregnation and embedding (epoxy embedding medium, EEM hardener DDSA, and EEM hardener NMA; Fluka). Immunolabeling was done on semithin (0.5 μm) and ultrathin (200 nm) transverse sections of resin-embedded block prior to observations by fluorescence microscopy (Nikon Eclipse TE300) and transmission electron microscopy at 200 kV (JEM2100F; JEOL), respectively.
Immunogold labeling of 8-5′ Linked Lignin Structure for Transmission Electron Microscopy
Transverse ultrathin sections were cut from the Epoxy resin-embedded block and mounted on nickel grids (200 mesh). Sections were floated on a drop of blocking buffer (1% BSA, and 0.1% NaN3 in TBS) for 30 min at room temperature and then floated on a drop of KM1 ascites fluid diluted 1:100 in blocking buffer for 2 d at 4°C. Following washing thrice for 15 min on drops of blocking buffer, sections were incubated with immunogold conjugate EM goat anti-mouse IgG, 10 nm (EM.GAM10; BB International), diluted 1:100 in blocking buffer for 4 h at room temperature. Finally, the sections were washed six times for 15 min on drops of blocking buffer and then washed with ultra pure water. Sections were observed under a JEM2100F transmission electron microscope (JEOL) without poststaining.
Immunolabeling for Fluorescence Microscopy
Sections were mounted on silanized slides and incubated with 3% protein milk in PBS (0.1 M phosphate containing 0.9% NaCl, pH 7.6) for 30 min at room temperature to avoid nonspecific binding of antibody. Sections were then washed with PBS and incubated with LM10, LM11, or LM21 diluted 1:20 in blocking buffer (PBS containing 1% BSA and 0.01% sodium azide) or LM5, LM2, or JIM14 diluted 1:10 in blocking buffer for 3 h at room temperature and 1 d at 4°C. After washing twice for 5 min with PBS, the sections were incubated at room temperature for 4 h with Alexa Fluor 488 goat anti-rat IgG (H+L) (Life Technologies) diluted 1:100 in TBS. They were again washed three times for 5 min with PBS and washed with ultrapure water. Sections were mounted in Eukit (Sigma-Aldrich).
Chemical Analyses and NMR
Cell Wall Residue Preparation
All chemical analyses were performed on extractive-free cell wall residue (CWR) obtained from manually separated outer- and inner-stem tissues. CWR was obtained by extracting tissues (7-fold) with 80% ethanol (6 mL/100 mg CWR) prior to grinding.
Lignin, Sugar, and Protein Determination
Acetyl bromide lignin was determined by measuring absorbance at 280 nm as previously described (Iiyama and Wallis, 1990). Thioacidolysis and subsequent gas chromatography-mass spectrometry analyses of β-O-4 ether-linked lignin monomers (analyzed as their trimethylsilylated derivatives) were performed as previously described using a Hewlett-Packard HP6890 Series gas chromatograph-flame ionization detector and a Thermo Focus gas chromatograph coupled with a Polaris Q gas chromatograph-mass spectrometer (Day et al., 2005).
Sugar analysis was performed by high-performance anion-exchange chromatography (Dionex DX 500; Thermo Scientific) after a two-step sulfuric acid hydrolysis of CWR using 2 deoxyribose as internal standard (Belmokhtar et al., 2013).
Protein content was determined in triplicate by measuring the total N contents (N*6.25) of 3 mg of ball-milled samples using an elemental analyzer (NA 1500; Carlo Erba) coupled to a mass spectrometer (Euro EA elemental analyzer).
NMR Analysis
Approximately 200 mg of CWR was ball-milled in a 25-mL jar with 20 × 20-mm ZrO2 ball bearings using a Retsch MM2000 mixer mill, for 1 h and 50 min using 20-min milling intervals with 10-min breaks. DMSO (1.8 mL) and N-methylimidazole (0.9 mL) were added to 100 mg of each ball-milled cell wall sample for cell wall dissolution (Hedenström et al., 2009). After acetylation and precipitation into water, samples were centrifuged in a Beckman JLA-10.500 rotor at 18,600g for 10 min. The pellets were washed twice with water and then centrifuged as previously. Around 80 mg of acetylated cell wall was dissolved in 0.6 mL of CDCl3 in a 5-mm NMR tube prior to NMR acquisition. NMR spectra were acquired on a Bruker Biospin Avance III 600 MHz spectrometer, using a 5-mm TCI cryoprobe equipped with cold preamplifiers for 1H, 13C, and 15N. Adiabatic HSQC (hsqcedetgpsisp2.2) spectra widths were 5102 and 24,147 Hz for the 1H- and 13C-dimensions, respectively. The number of collected complex points was 1024 for the 1H-dimension using a relaxation delay of 1 s. The number of scans was 64, and 386 time increments were always recorded in the 13C-dimension. The spectra were processed using Topspin 3.1 Bruker Biospin. All spectra were manually phase corrected and calibrated with CDCl3peak (δC, 77.2; δH, 7.26 ppm) used as internal reference. Signals were assigned by comparison with 2D NMR spectra reported in the literature (del Río et al., 2011; Mansfield et al., 2012; Ralph et al., 2012) and recorded on acetylated standards dehydrogenation polymers (Cathala et al., 1998), galactan (lupin) P-GALLU; 1,5-α-l-arabinan (sugar beet) P-LARB (Megazyme).
Oligolignol Profiling
Sample Preparation
Phenolic profiling was independently performed on inner- and outer-stem tissues of three wild-type plants and six lbf1 mutants. For wild-type plants, three inner- and three outer-stem tissues were analyzed, and for lbf1 mutants, five inner- and six outer-stem tissues were analyzed. Ethanolic extracts from CWR preparations were mixed and filtered through a paper filter then evaporated at 40°C to dryness under reduced pressure. Dry extracts were resuspended with ∼1.5 mL of a mix of diethyl ether and Milli-Q Water and then transferred into 2.5-mL vials. Vials were kept open and diethyl ether evaporated at room temperature under a stream of ambient air. Vials were stored at 4°C and then evaporated with Centrivap LABONCO at 50°C prior to analysis.
Oligolignol Profiling by HPLC-High-Resolution Mass Spectrometry
Phenolic profiling was performed using 10 μL of the water phase. Extracts were analyzed with a DionexUltiMate 3000 LC module equipped with a LPG-3400 pump, UV-Vis detector (model VWD-3400), and an autosampler (model WPS-3000 SL) and further hyphenated to an LTQ Orbitrap XL hybrid FTMS mass spectrometer (MS) (Thermo Electron) consisting of a linear ion trap MS connected with a Fourier transform Orbitrap MS. The separation was performed on a reversed phase Sunfire C18 column (150 mm × 3 mm, 3.5 μm; Waters) with aqueous 0.1% acetic acid and acetonitrile/water (99/1, v/v, acidified with 0.1% acetic acid) as solvents A and B. A gradient of 0 min 5% B, 40 min 45% B, and 45 min 100% B was applied using a flow rate of 300 μL/min and a column temperature of 40°C. The autosampler temperature was 10°C. Analytes were negatively ionized with an electrospray source using the following parameter values: source voltage 5.00 kV, source current 100.00 μA, capillary temperature 300°C, sheath gas 20 (arb), aux gas 10 (arb), and sweep gas 2 (arb). Full Fourier transform-mass spectrometry spectra between 120 and 1400 m/z were recorded at a resolution of 100,000. In parallel, three data-dependent MSn spectra were recorded on the ion trap MS using the preliminary low-resolution data obtained during the first 0.1 s of the previous full Fourier transform-mass spectrometry scan: a MS2 scan of the most abundant m/z ion of the full Fourier transform-mass spectrometry scan, followed by two MS3 scans of the most abundant first product ions. MSn scans were obtained with 35% collision energy.
Elucidation of MS2 Spectra
Elucidation of the MS2 spectra and the sequencing terminology of the first product ions was based on the lignin oligomer/(neo)lignan sequencing approach mentioned by Morreel et al. (2010a) and on the fragmentation rules of the different linkage types described by Morreel et al. (2010b). Briefly, the three types of linkages, i.e., 8-O-4 (β-aryl ether), 8-5 phenylcoumaran), and 8-8 (resinol), either loose, small, neutral molecules that are indicative of the type of linkage (referred to as pathway I, I) or are cleaved, hence yielding information on the units that are connected by the linkage (referred to as pathway II, II). In the case of a β-aryl ether, pathway II cleavage leads to first product ions corresponding with the phenolic 8-end (A- ion) and aliphatic 4-end (B- ion) moieties. The structures of the described compounds are given in Supplemental Figure 2.
Agilent Microarray Transcriptomics
Total RNA was extracted from separated inner- and outer-stem tissues of six individual lbf1 mutants and three wild-type plants using the TriReagent method (Molecular Research Center). RNA integrity and concentration were evaluated with RNA StdSens Chips using the Experion automated capillary electrophoresis system (Bio-Rad). RNA processing and hybridization were performed following the manufacturer’s instruction for One-Color Microarray-Based Gene Expression Analysis (Agilent Technologies). Samples were hybridized to the Agilent-045382 UGSF flax 45K v1.0 array based upon flax genome coding sequence (Wang et al., 2012) available at Phytozome (http://phytozome.org). The array contains 45,220 60-mer in situ oligonucleotides per block. All nine samples were analyzed independently. Following hybridization, washing was performed following the manufacturer’s instruction, and slides were immediately scanned at 5-mm pixel−1 resolution using an Axon GenePix 4000B scanner (Molecular Devices) piloted by GenePix Pro 6.0 software (Axon). Grid alignment and expression data analyses were made with the same software. After background noise elimination, median values of overall hybridization were normalized by robust local regression (Yang et al., 2002). Artifact spots were manually eliminated. Differential analysis was performed with the method varmixt (Delmar et al., 2005), available in the package anapuce of the software R. A double-sided, unpaired t test was computed for each gene between the two conditions. Variance of the difference in gene expression (transcript abundance) was split between subgroups of genes with homogeneous variance (Delmar et al., 2005). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate (Ge et al., 2003). A gene is declared differentially expressed if the Bonferroni-corrected P value is <0.05.
Bioinformatics
Phylogenetic trees were made using a neighbor-joining method implemented in MEGA5. Bootstrap consensus tree were inferred from 1000 replicates. Branches corresponding to partitions reproducing <50% bootstrap replicates are collapsed. The evolutionary distances were computed using the p-distance method.
Accession Numbers
All data are available through the Gene Expression Omnibus repository at NCBI (Barrett et al., 2007) under accession numbers GSE61311 and GPL19181.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. 2D NMR Spectra of Lignin from Flax Wild-Type and lbf1 Inner Tissues.
Supplemental Figure 2. Structures of Oligolignols Previously Unidentified in Flax.
Supplemental Figure 3. Immunolocalization of Xylem Cell Wall NCPs.
Supplemental Figure 4. Phylogenetic Trees of Lignin Genes Overexpressed in lbf1 Outer Tissues.
Supplemental Table 1. Visual Phenotyping Classes for Flax lbf Mutants, as Previously Described (Chantreau et al., 2013).
Supplemental Data Set 1. Over- and Underaccumulated Transcripts in Outer Tissues.
Supplemental Data Set 2. Alignments Used to Generate the Peroxidase Phylogenies Presented in Figure 11.
Supplemental Data Set 3. Alignments Used to Generate the RBOH Phylogenies Presented in Figure 12.
Supplemental Data Set 4. Alignments Used to Generate the CCR Phylogenies Presented in Supplemental Figure 4A.
Supplemental Data Set 5. Alignments Used to Generate the COMT Phylogenies Presented in Supplemental Figure 4B.
Supplemental Data Set 6. Alignments Used to Generate the CAD Phylogenies Presented in Supplemental Figure 4C.
Supplementary Material
Acknowledgments
M. Chantreau gratefully acknowledges the University Lille1 and the Nord-Pas de Calais Region for a PhD fellowship. S.K. gratefully acknowledges the financial support of the Kyoto University Foundation. This work was carried in the context of and financed by the French national project PT-Flax (ANR-09-GENM-020). We thank Marie-Laure Martin-Magniette (URGV France) for her advice on transcriptomics data analyses. Authors acknowledge the technical support of the PICT IBiSA biological imaging center (transmission electron microscopy) and the PLANET analytical platform (NMR) at the University of Reims Champagne-Ardenne.
AUTHOR CONTRIBUTIONS
S.H. and B.C. conceived the project and decided on the scientific strategy. A.P. performed cell wall chemical analyses, and D.C. realized the NMR analyses. R.D. performed the oligolignol analyses and interpreted all MS data together with K.M. S.K. performed lignin and cell wall light microscopy and transmission electron microscopy immunolocalization. M. Chantreau, S.G., B.C., S.H., and G.N. collected plant material. M. Chantreau produced plant material, screened the mutant population, and undertook all bioinformatic analyses and transcriptomics. S.A. and M. Chabi assisted with transcriptomics and data analyses. G.N. validated microarray data. W.B., A.Y., and F.M. provided important scientific criticism and input during the writing of this article. This article was written by M. Chantreau and S.H. with important contributions from B.C. and R.D. All authors read, reviewed, and approved the final article.
Glossary
- NCP
noncellulosic polysaccharide
- EMS
ethyl methanesulfonate
- HSQC
heteronuclear single quantum coherence
- LC-MS
liquid chromatography-mass spectrometry
- AGP
arabinogalactan protein
- GO
Gene Ontology
- CWR
cell wall residue
- MS
mass spectrometer
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allen R.S., Nakasugi K., Doran R.L., Millar A.A., Waterhouse P.M. (2013). Facile mutant identification via a single parental backcross method and application of whole genome sequencing based mapping pipelines. Front. Plant Sci. 4: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola A.M., Lewis N.G. (2002). Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294. [DOI] [PubMed] [Google Scholar]
- Barakat A., Bagniewska-Zadworna A., Frost C.J., Carlson J.E. (2010). Phylogeny and expression profiling of CAD and CAD-like genes in hybrid Populus (P. deltoides x P. nigra): evidence from herbivore damage for subfunctionalization and functional divergence. BMC Plant Biol. 10: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A., Choi A., Yassin N.B.M., Park J.S., Sun Z., Carlson J.E. (2011). Comparative genomics and evolutionary analyses of the O-methyltransferase gene family in Populus. Gene 479: 37–46. [DOI] [PubMed] [Google Scholar]
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. (2007). NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 35: D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Monties B., Montagu M.V., Boerjan W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17: 125–197. [Google Scholar]
- Belmokhtar N., Habrant A., Ferreira N.L., Chabbert B. (2013). Changes in phenolics distribution after chemical pretreatment and enzymatic conversion of Miscanthus x giganteus internode. BioEnergy Res. 6: 506–518. [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44: 337–363. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A., Penfield S., Smith C., Catley M., Bevan M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Cathala B., Saake B., Faix O., Monties B. (1998). Evaluation of the reproducibility of the synthesis of dehydrogenation polymer models of lignin. Polym. Degrad. Stabil. 59: 65–69. [Google Scholar]
- Chantreau M., et al. (2013). PT-Flax (phenotyping and TILLinG of flax): development of a flax (Linum usitatissimum L.) mutant population and TILLinG platform for forward and reverse genetics. BMC Plant Biol. 13: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Crônier D., Monties B., Chabbert B. (2005). Structure and chemical composition of bast fibers isolated from developing hemp stem. J. Agric. Food Chem. 53: 8279–8289. [DOI] [PubMed] [Google Scholar]
- Dauwe R., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52: 263–285. [DOI] [PubMed] [Google Scholar]
- Davis E.A., Derouet C., Herve Du Penhoat C., Morvan C. (1990). Isolation and an NMR study of pectins from flax (Linum usitatissimum L.). Carbohydr. Res. 197: 205–215. [Google Scholar]
- Day A., Ruel K., Neutelings G., Crônier D., David H., Hawkins S., Chabbert B. (2005). Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222: 234–245. [DOI] [PubMed] [Google Scholar]
- Delmar P., Robin S., Daudin J.J. (2005). VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics 21: 502–508. [DOI] [PubMed] [Google Scholar]
- del Río J.C., Rencoret J., Gutiérrez A., Nieto L., Jiménez-Barbero J., Martínez Á.T. (2011). Structural characterization of guaiacyl-rich lignins in flax (Linum usitatissimum) fibers and shives. J. Agric. Food Chem. 59: 11088–11099. [DOI] [PubMed] [Google Scholar]
- Fenart S., et al. (2010). Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics 11: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Mielenz J.R., Xiao X., Ge Y., Hamilton C.Y., Rodriguez M. Jr., Chen F., Foston M., Ragauskas A., Bouton J., Dixon R.A., Wang Z.Y. (2011). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. USA 108: 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Dudoit S., Speed T.P. (2003). Resampling-based multiple testing for microarray data analysis. Test 12: 1–77. [Google Scholar]
- Girault R., Bert F., Rihouey C., Jauneau A., Morvan C., Jarvis M. (1997). Galactans and cellulose in flax fibres: putative contributions to the tensile strength. Int. J. Biol. Macromol. 21: 179–188. [DOI] [PubMed] [Google Scholar]
- Gorshkova T., Morvan C. (2006). Secondary cell-wall assembly in flax phloem fibres: role of galactans. Planta 223: 149–158. [DOI] [PubMed] [Google Scholar]
- Guerriero G., Sergeant K., Hausman J.-F. (2013). Integrated -omics: a powerful approach to understanding the heterogeneous lignification of fibre crops. Int. J. Mol. Sci. 14: 10958–10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenström M., Wiklund-Lindström S., Öman T., Lu F., Gerber L., Schatz P., Sundberg B., Ralph J. (2009). Identification of lignin and polysaccharide modifications in Populus wood by chemometric analysis of 2D NMR spectra from dissolved cell walls. Mol. Plant 2: 933–942. [DOI] [PubMed] [Google Scholar]
- Herrero J., Esteban-Carrasco A., Zapata J.M. (2013). Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol. Biochem. 67: 77–86. [DOI] [PubMed] [Google Scholar]
- Herrero J., Esteban Carrasco A., Zapata J.M. (2014). Arabidopsis thaliana peroxidases involved in lignin biosynthesis: in silico promoter analysis and hormonal regulation. Plant Physiol. Biochem. 80: 192–202. [DOI] [PubMed] [Google Scholar]
- His I., Andème-Onzighi C., Morvan C., Driouich A. (2001). Microscopic studies on mature flax fibers embedded in LR white: immunogold localization of cell wall matrix polysaccharides. J. Histochem. Cytochem. 49: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Huis R., Morreel K., Fliniaux O., Lucau-Danila A., Fénart S., Grec S., Neutelings G., Chabbert B., Mesnard F., Boerjan W., Hawkins S. (2012). Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol. 158: 1893–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.J., Harding S.A., Lung J., Popko J.L., Ralph J., Stokke D.D., Tsai C.J., Chiang V.L. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17: 808–812. [DOI] [PubMed] [Google Scholar]
- Iiyama K., Wallis A.F. (1990). Determination of lignin in herbaceous plants by an improved acetyl bromide procedure. J. Sci. Food Agric. 51: 145–161. [Google Scholar]
- Jones L., Seymour G.B., Knox J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1 [->] 4)-[beta]-D-galactan. Plant Physiol. 113: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M., Melzer M., Prokhorenko I., Johansson T., Wingsle G. (2005). Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J. Exp. Bot. 56: 2085–2093. [DOI] [PubMed] [Google Scholar]
- Karpinska B., Karlsson M., Schinkel H., Streller S., Süss K.-H., Melzer M., Wingsle G. (2001). A novel superoxide dismutase with a high isoelectric point in higher plants. expression, regulation, and protein localization. Plant Physiol. 126: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyoto S., Yoshinaga A., Tanaka N., Wada M., Kamitakahara H., Takabe K. (2013). Immunolocalization of 8-5′ and 8-8′ linked structures of lignin in cell walls of Chamaecyparis obtusa using monoclonal antibodies. Planta 237: 705–715. [DOI] [PubMed] [Google Scholar]
- Knox J.P., Linstead P.J., Peart J., Cooper C., Roberts K. (1991). Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1: 317–326. [DOI] [PubMed] [Google Scholar]
- Lacombe E., Hawkins S., Van Doorsselaere J., Piquemal J., Goffner D., Poeydomenge O., Boudet A.M., Grima-Pettenati J. (1997). Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 11: 429–441. [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Jackson R.G., George G.L., Elias L., Lim E.-K., Vaistij F.E., Bowles D.J. (2006). The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48: 286–295. [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Lim E.-K., Vaistij F.E., Bowles D.J. (2008). Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 228: 609–616. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Leplé J.-C., et al. (2007). Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2008). Strigolactones and shoot branching: a new trick for a young dog. Dev. Cell 15: 337–338. [DOI] [PubMed] [Google Scholar]
- Liu L., Shang-Guan K., Zhang B., Liu X., Yan M., Zhang L., Shi Y., Zhang M., Qian Q., Li J., Zhou Y. (2013). Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 9: e1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.D., Kim H., Lu F., Ralph J. (2012). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7: 1579–1589. [DOI] [PubMed] [Google Scholar]
- Marcus S.E., et al. (2010). Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 64: 191–203. [DOI] [PubMed] [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546. [DOI] [PubMed] [Google Scholar]
- McDougall G.J. (1991). Cell-wall-associated peroxidases and lignification during growth of flax fibres. J. Plant Physiol. 139: 182–186. [Google Scholar]
- McDougall G.J. (1992). Changes in cell wall-associated peroxidases during the lignification of flax fibres. Phytochemistry 31: 3385–3389. [Google Scholar]
- Miao Y.-C., Liu C.-J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 107: 22728–22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Dima O., Kim H., Lu F., Niculaes C., Vanholme R., Dauwe R., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W. (2010a). Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 153: 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Kim H., Lu F., Dima O., Akiyama T., Vanholme R., Niculaes C., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W. (2010b). Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal. Chem. 82: 8095–8105. [DOI] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. (2004). Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 136: 3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan C., Andème-Onzighi C., Girault R., Himmelsbach D.S., Driouich A., Akin D.E. (2003). Building flax fibres: more than one brick in the walls. Plant Physiol. Biochem. 41: 935–944. [Google Scholar]
- Neutelings G. (2011). Lignin variability in plant cell walls: contribution of new models. Plant Sci. 181: 379–386. [DOI] [PubMed] [Google Scholar]
- Nielsen K.L., Indiani C., Henriksen A., Feis A., Becucci M., Gajhede M., Smulevich G., Welinder K.G. (2001). Differential activity and structure of highly similar peroxidases. Spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 40: 11013–11021. [DOI] [PubMed] [Google Scholar]
- Østergaard L., Teilum K., Mirza O., Mattsson O., Petersen M., Welinder K.G., Mundy J., Gajhede M., Henriksen A. (2000). Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 44: 231–243. [DOI] [PubMed] [Google Scholar]
- Ralph J., Akiyama T., Coleman H.D., Mansfield S.D. (2012). Effects on lignin structure of coumarate 3-hydroxylase downregulation in poplar. BioEnergy Res. 5: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach M.J., Deyholos M.K. (2007). Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics 278: 149–165. [DOI] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Voesenek L.A., Pierik R. (2008). The regulation of cell wall extensibility during shade avoidance: a study using two contrasting ecotypes of Stellaria longipes. Plant Physiol. 148: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P. (2008). Tissue-specific expression of a defence-related peroxidase in transgenic wheat potentiates cell death in pathogen-attacked leaf epidermis. Mol. Plant Pathol. 9: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J., Kiyonaga Y., Fujita K., Kondo R., Tsutsumi Y. (2013). Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J. Agric. Food Chem. 61: 3781–3788. [DOI] [PubMed] [Google Scholar]
- Sindhu A., Langewisch T., Olek A., Multani D.S., McCann M.C., Vermerris W., Carpita N.C., Johal G. (2007). Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 145: 1444–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood M., Yates E.A., Willats W.G., Martin H., Knox J.P. (1996). Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 198: 452–459. [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17: 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A. (2010). ROS in biotic interactions. Physiol. Plant. 138: 414–429. [DOI] [PubMed] [Google Scholar]
- Tsuyama T., Kawai R., Shitan N., Matoh T., Sugiyama J., Yoshinaga A., Takabe K., Fujita M., Yazaki K. (2013). Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 162: 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., Vanholme R., Storme V., Mortimer J.C., Dupree P., Boerjan W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Darrah C., Oyarce P., Grabber J.H., Ralph J., Boerjan W. (2012a). Metabolic engineering of novel lignin in biomass crops. New Phytol. 196: 978–1000. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J.H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W. (2012b). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., et al. (2012). The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 72: 461–473. [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187: 273–285. [DOI] [PubMed] [Google Scholar]
- Whetten R., Sederoff R. (1995). Lignin biosynthesis. Plant Cell 7: 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen C.L., Keurentjes J.J. (2014). Genetic resources for quantitative trait analysis: novelty and efficiency in design from an Arabidopsis perspective. Curr. Opin. Plant Biol. 18: 103–109. [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates E.A., Knox J.P. (1994). Investigations into the occurrence of plant cell surface epitopes in exudate gums. Carbohydr. Polym. 24: 281–286. [Google Scholar]
- Yates E.A., Valdor J.-F., Haslam S.M., Morris H.R., Dell A., Mackie W., Knox J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6: 131–139. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Dixon R.A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16: 227–233. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.-Y., Dixon R.A. (2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Kays S.J., Schroeder B.P., Ye Z.-H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.