This work dissects the intriguing biotrophic interaction between Theobroma cacao and the fungus Moniliophthora perniciosa in the devastating witches’ broom disease. Infection by M. perniciosa leads to massive genetic reprogramming in cacao tissues, which culminates in the onset of premature senescence. A detailed molecular model of this peculiar plant-pathogen interaction is presented.
Abstract
Witches’ broom disease (WBD), caused by the hemibiotrophic fungus Moniliophthora perniciosa, is one of the most devastating diseases of Theobroma cacao, the chocolate tree. In contrast to other hemibiotrophic interactions, the WBD biotrophic stage lasts for months and is responsible for the most distinctive symptoms of the disease, which comprise drastic morphological changes in the infected shoots. Here, we used the dual RNA-seq approach to simultaneously assess the transcriptomes of cacao and M. perniciosa during their peculiar biotrophic interaction. Infection with M. perniciosa triggers massive metabolic reprogramming in the diseased tissues. Although apparently vigorous, the infected shoots are energetically expensive structures characterized by the induction of ineffective defense responses and by a clear carbon deprivation signature. Remarkably, the infection culminates in the establishment of a senescence process in the host, which signals the end of the WBD biotrophic stage. We analyzed the pathogen’s transcriptome in unprecedented detail and thereby characterized the fungal nutritional and infection strategies during WBD and identified putative virulence effectors. Interestingly, M. perniciosa biotrophic mycelia develop as long-term parasites that orchestrate changes in plant metabolism to increase the availability of soluble nutrients before plant death. Collectively, our results provide unique insight into an intriguing tropical disease and advance our understanding of the development of (hemi)biotrophic plant-pathogen interactions.
INTRODUCTION
Crop diseases caused by fungi severely limit agricultural productivity, leading to drastic economic losses each year (Fisher et al., 2012). In general, the outcome of a disease process depends on the molecular interplay between the pathogen and its host and involves substantial transcriptional changes in both organisms. In response to microbial invasion, plants activate an array of defense mechanisms (e.g., reinforcement of cell walls, production of reactive oxygen species, and accumulation of antimicrobial compounds). By contrast, pathogens have devised different strategies to evade plant immunity as well as to feed on and reproduce in the host tissues (Dodds and Rathjen, 2010). Dissecting the molecular mechanisms governing plant-pathogen interactions is an important step toward developing novel and effective strategies to control diseases and improve food security.
Tropical areas have a suitable climate for the cultivation of a large number of crops. However, they are also highly prone to the development of a variety of pathogenic microorganisms. Despite the importance of tropical plants in maintaining the growing human population, plant diseases of tropical regions have traditionally received little attention. The basidiomycete Moniliophthora perniciosa causes witches’ broom disease (WBD) in Theobroma cacao, the chocolate tree (Aime and Phillips-Mora, 2005). This highly destructive disease has negatively affected cacao productivity in many American countries and is a threat to global cocoa production (Purdy and Schmidt, 1996; Meinhardt et al., 2008). WBD is a challenging disease to study, as it results from the interaction between two nonmodel organisms that exhibit complex life cycles. Furthermore, genetic tools that would be suitable for analyzing these organisms are limited. On the other hand, the economic importance of WBD and the limited knowledge of tropical plant-pathogen interactions render the study of this disease of great relevance.
The M. perniciosa lifestyle is classified as hemibiotrophic (Evans, 1980). However, it has some distinctive characteristics that distinguish WBD from other well studied pathosystems. In general, hemibiotrophic pathogens (e.g., Magnaporthe oryzae and Colletotrichum spp) display an initial transient and asymptomatic biotrophic stage that is rapidly followed by a destructive necrotrophic phase, wherein the most prominent disease symptoms develop (Perfect and Green, 2001; Münch et al., 2008). Conversely, the M. perniciosa biotrophic stage is extremely prolonged, lasting even longer than typical biotrophic interactions (e.g., those exhibited by powdery mildews, rusts, and smut diseases). During this stage, M. perniciosa slowly grows between living cacao cells and is found at very low density within the infected tissues. Even so, biotrophic colonization induces drastic physiological and morphological alterations in the plant (Purdy and Schmidt, 1996; Meinhardt et al., 2008). After 2 to 3 months of biotrophic development, the infected tissues become necrotic as WBD enters the necrotrophic stage of development (Evans, 1980).
Although M. perniciosa can also infect flowers and young fruits, the infection of shoots is responsible for the typical symptoms of the disease, namely, the formation of abnormal structures called “green brooms” (Griffith et al., 2003). Infected shoots become swollen, with irregular growth and loss of apical dominance, suggesting the occurrence of hormonal imbalances during disease development. Moreover, important biochemical alterations were verified to occur in cacao plants during WBD (Scarpari et al., 2005). Changes in soluble sugar, amino acid, secondary metabolite, ethylene, and glycerol content were observed in cacao shoots upon infection, indicating that a remarkable degree of genetic reprogramming took place in the infected tissue (Scarpari et al., 2005). Notably, the green broom stage is considered a commitment point in WBD progression, and cacao plants are not able to stall fungal development once these structures have formed.
Despite considerable advances in our understanding of WBD over the past few years (Scarpari et al., 2005; Mondego et al., 2008; Rincones et al., 2008), our knowledge of the intriguing biotrophic interaction established between M. perniciosa and cacao is still very limited. Previous studies investigated cacao responses to M. perniciosa infection at the transcriptional level (Gesteira et al., 2007; Leal et al., 2007; da Hora Junior et al., 2012). However, they mainly involved comparisons between resistant and susceptible genotypes, with the aim of identifying resistance mechanisms. Consequently, little emphasis has been placed on the molecular events underlying the complexity of compatible interactions in WBD. Moreover, the number of genes analyzed in these studies was restricted by the use of Sanger-based cDNA sequencing and/or nucleic acid hybridization techniques. Such methodologies were also applied in large-scale gene expression analyses in M. perniciosa, providing initial insight into the pathogen’s biology (Rincones et al., 2008; Pires et al., 2009; Leal et al., 2010). Nevertheless, these studies addressed the transcriptional regulation of M. perniciosa exclusively under artificial conditions, and the genetic program governing the fungal interaction with cacao remains unexplored.
Advances in next-generation sequencing technologies have facilitated high-throughput analyses with the potential of uncovering key aspects of host-microbe interactions (Studholme et al., 2011). The RNA-seq approach allows the simultaneous inspection of transcriptomes from both the pathogen and its host with a high level of accuracy and depth, providing the big picture of an infective process (Westermann et al., 2012). With the purpose of dissecting the M. perniciosa-cacao interaction and establishing a foundation for the study of this disease, a comprehensive transcriptomic analysis of WBD was recently initiated with the construction of the WBD Transcriptome Atlas (www.lge.ibi.unicamp.br/wbdatlas). Based on RNA-seq data, this database allows the analysis of a wide range of developmental stages, growth conditions, and stress responses of the fungus, both under in vitro and in planta conditions.
This work provides a detailed transcriptomic analysis of the biotrophic interaction between M. perniciosa and cacao, which corresponds to the green broom stage of WBD. We identified important alterations in the transcriptome of infected cacao plants that suggest the induction of a premature senescence process triggered by depletion of nutrients in the infected tissues. The RNA-seq data were corroborated by complementary histological and biochemical assays. Moreover, the pathogen’s transcriptome was analyzed in unprecedented depth, allowing the characterization of the fungal nutritional and infection strategies during WBD and the identification of putative virulence effectors. Overall, these data provide novel insight into the complex hemibiotrophic interaction between M. perniciosa and cacao and represent a major advance in our understanding of a tropical plant disease.
RESULTS
Symptoms of Infected Plants
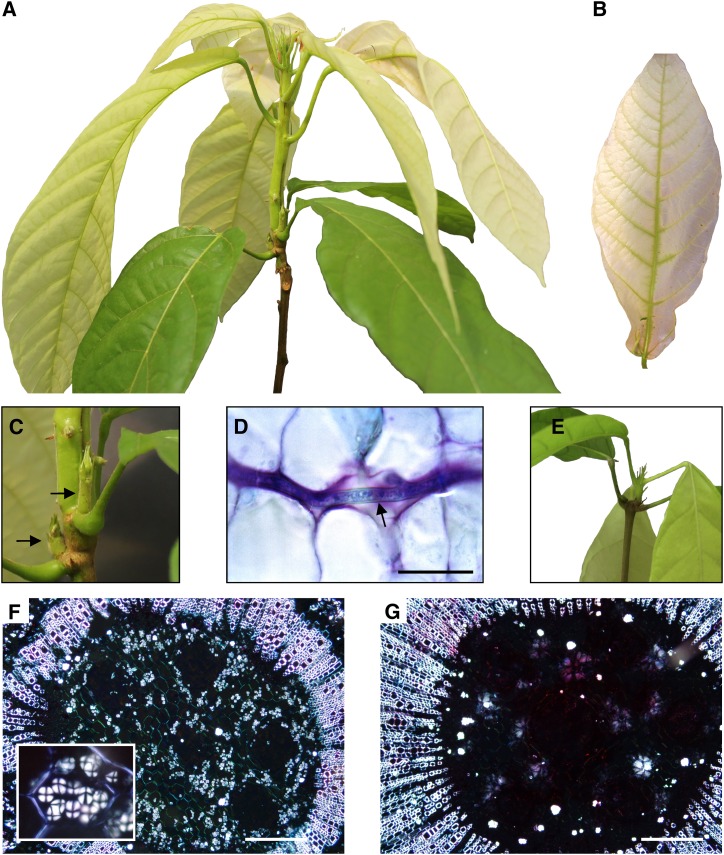
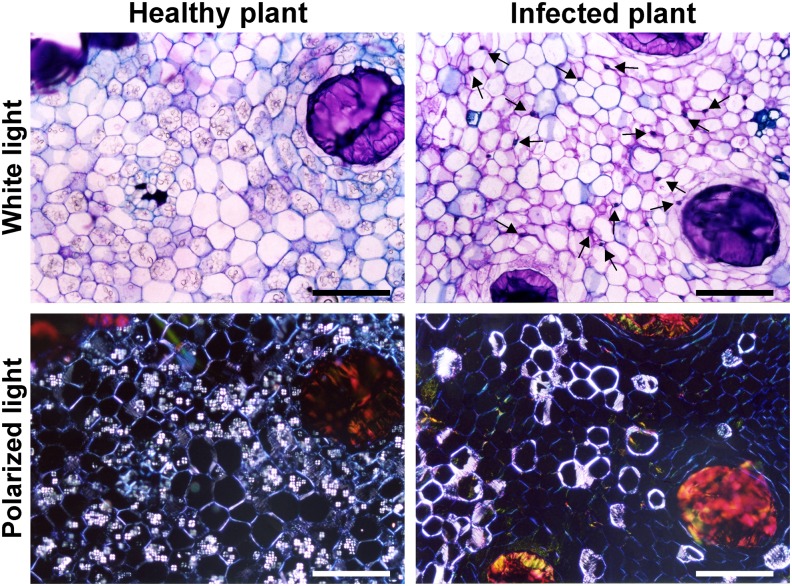
We inspected cacao seedlings inoculated with basidiospores of the pathogen M. perniciosa for the development of the typical symptoms of WBD. The first visible symptom, a slight swelling of the plant apical meristem, was detected ∼20 d postinfection. Within 30 d of inoculation, typical symptoms of the green broom stage of WBD were observed (Figure 1A): Diseased leaves were chlorotic (Figure 1B) and stems exhibited hypertrophic and exacerbated growth and loss of apical dominance, as evidenced by the proliferation of lateral buds (Figure 1C). Moreover, in this stage of WBD, the fungus was restricted to the narrow space between cacao cells (Figure 1D). A healthy plant is shown in Figure 1E for comparison. Interestingly, histological analysis revealed a clear reduction in the starch content of green brooms in comparison to the corresponding shoot tissues of healthy noninfected plants (Figures 1F and 1G). A detailed view of WBD progression and the major symptoms observed in cacao plants during disease development are shown in Supplemental Figure 1.
Figure 1.
The Green Broom Stage of Witches’ Broom Disease.
(A) Apical portion of representative infected plant illustrating the major morphological changes caused by M. perniciosa infection.
(B) Leaf chlorosis observed in green brooms.
(C) Detail of the loss of apical dominance observed in infected plants (arrows).
(D) Histological analysis showing fungal growth (arrow), which is restricted to the cacao intercellular space. A longitudinal section of the infected cacao plant was stained with toluidine blue and observed under white light. Bar = 25 μm.
(E) Apical region of a healthy cacao plant.
(F) Cortical region of shoots from healthy plants visualized under polarized light showing numerous starch grains (inset). Bar = 200 μm.
(G) Infected plants are depleted in starch grains. Starch grains are visualized as bright structures inside the cells (Maltese cross) in these transverse sections of cacao plants. All images of infected plants correspond to the green broom stage of WBD (30 d after infection). Bar = 200 μm.
Sequencing the Green Broom Transcriptome
Five biological replicates of each condition (healthy seedlings and seedlings at 30 d after infection) were harvested for transcriptome sequencing using RNA-seq. For this experiment, the entire green broom structure was collected, including cacao leaves and shoots (Supplemental Figure 2). Quantitative real-time PCR (qPCR) assays using sectioned brooms showed that the pathogen is uniformly distributed along the infected tissue (Supplemental Figure 3). Considering that five biological replicates were performed for each condition, we produced a total of 562 million and 436 million paired reads for infected and healthy (control) plants, respectively (Table 1). All reads were aligned against 34,997 gene models of cacao, which are available at www.cacaogenomedb.org. For each RNA-seq library, ∼80% of the reads mapped to the cacao reference. Also, reads were mapped against 17,008 gene models of M. perniciosa for the analysis of the fungal transcriptome. Whereas an insignificant fraction of reads (0.0002%) from control libraries mapped to M. perniciosa genes, ∼0.3% of the reads derived from infected seedlings mapped to the fungal reference (Table 1). The small proportion of M. perniciosa reads in green brooms is consistent with the low density of fungal cells in the biotrophic stage of WBD (Penman et al., 2000).
Table 1. Sequencing Metrics of the 10 RNA-seq Libraries.
| Source | Library | Total Paired Reads | Mapping to T. cacao | Mapping to M. perniciosa |
|---|---|---|---|---|
| Infected plants | I1 | 77,308,238 | 56,884,442 (73.58%) | 265,238 (0.34%) |
| I2 | 207,441,756 | 167,016,562 (80.51%) | 904,727 (0.44%) | |
| I3 | 83,953,292 | 67,870,238 (80.84%) | 196,086 (0.23%) | |
| I4 | 90,591,723 | 71,473,561 (78.90%) | 186,831 (0.21%) | |
| I5 | 103,196,353 | 82,127,996 (79.58%) | 158,021 (0.15%) | |
| Total | 562,491,362 | 445,372,799 (79.18%) | 1,710,903 (0.30%) | |
| Healthy plants | H1 | 89,659,731 | 73,930,327 (82.46%) | 27 (0.00%) |
| H2 | 82,560,659 | 68,560,121 (83.04%) | 633 (0.00%) | |
| H3 | 86,454,964 | 68,962,874 (79.77%) | 208 (0.00%) | |
| H4 | 80,950,589 | 65,521,727 (80.94%) | 91 (0.00%) | |
| H5 | 96,856,054 | 73,928,395 (76.33%) | 12 (0.00%) | |
| Total | 436,481,997 | 350,903,444 (80.39%) | 971 (0.00%) |
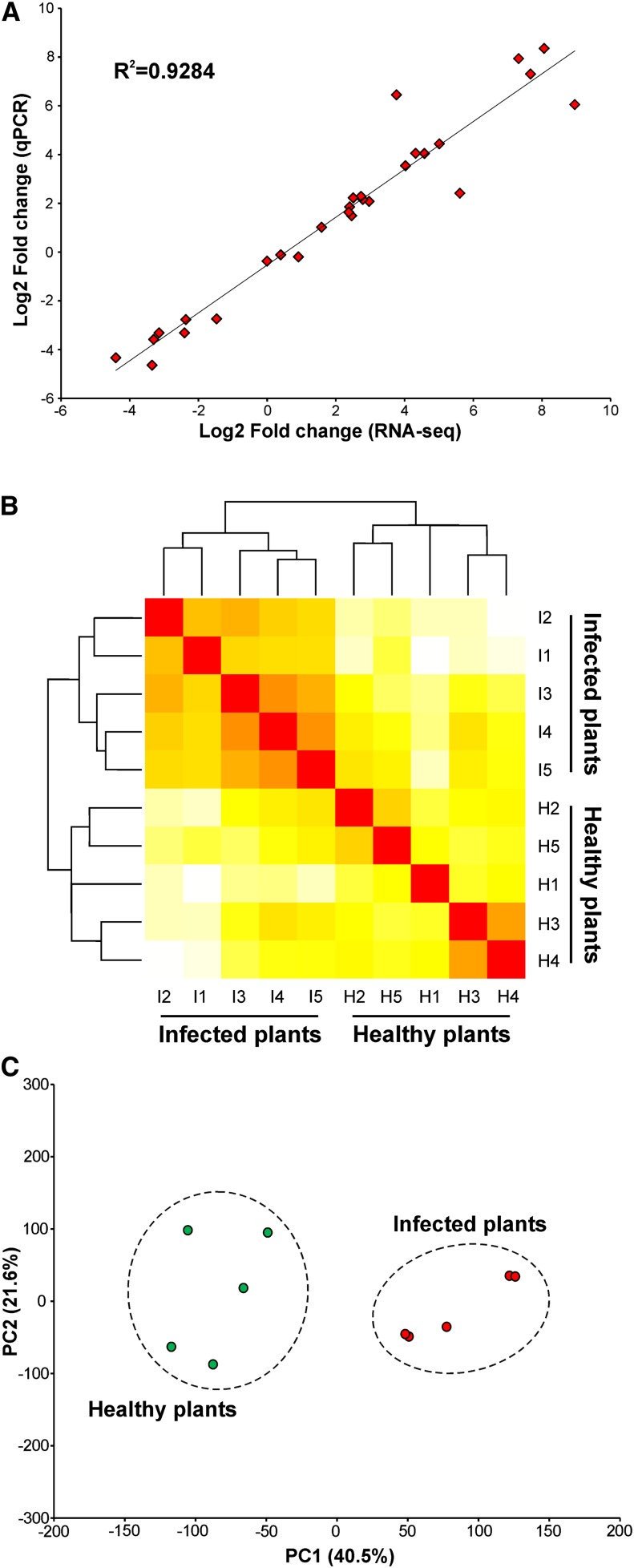
Genes differentially expressed between infected and healthy plants (false discovery rate [FDR] ≤ 0.01) were defined using the edgeR software package (Robinson et al., 2010). Of the 34,997 cacao genes, 17,713 were expressed in the conditions analyzed. Among them, 1269 were upregulated in green brooms and 698 were downregulated, totaling 1967 differentially expressed genes (Supplemental Data Set 1). In addition, we performed qPCR assays to validate the gene expression values obtained by RNA-seq. To this purpose, we analyzed the expression of 28 cacao genes (Supplemental Table 1) in two plants that were also sequenced by RNA-seq. We observed a strong correlation (R2 > 0.92) between the results obtained using the two techniques (Figure 2A), demonstrating the reliability of the data produced. Finally, we performed hierarchical clustering and principal component analysis (PCA) to assess the biological variability among all samples. The results indicated that most of the variation in gene expression is a consequence of the infection process. Two distinct groups were formed in the hierarchical clustering; one group was comprised of infected plants and the other of healthy plants (Figure 2B). Similarly, PCA showed that the transcriptomes of infected and healthy plants were clearly different from each other (Figure 2C).
Figure 2.
Global Evaluation of the RNA-seq Experiment.
(A) Comparison of gene expression values obtained by qPCR and RNA-seq. Fold changes were calculated for 28 cacao genes and a high correlation (R2 > 0.92) was observed between the results obtained using the two techniques.
(B) Hierarchical clustering of the 10 samples used in this study showing two distinct clades: one comprised of infected plants and the other of healthy plants. Individual plants were identified according to the nomenclature presented in Table 1.
(C) PCA displaying the intrinsic biological variation among samples. The result confirms the clear distinction between the transcriptomes of infected and healthy plants.
[See online article for color version of this figure.]
Metabolic Pathways and Protein Domain Enrichment Analyses: An Overview of the Transcriptional Responses of Infected Plants
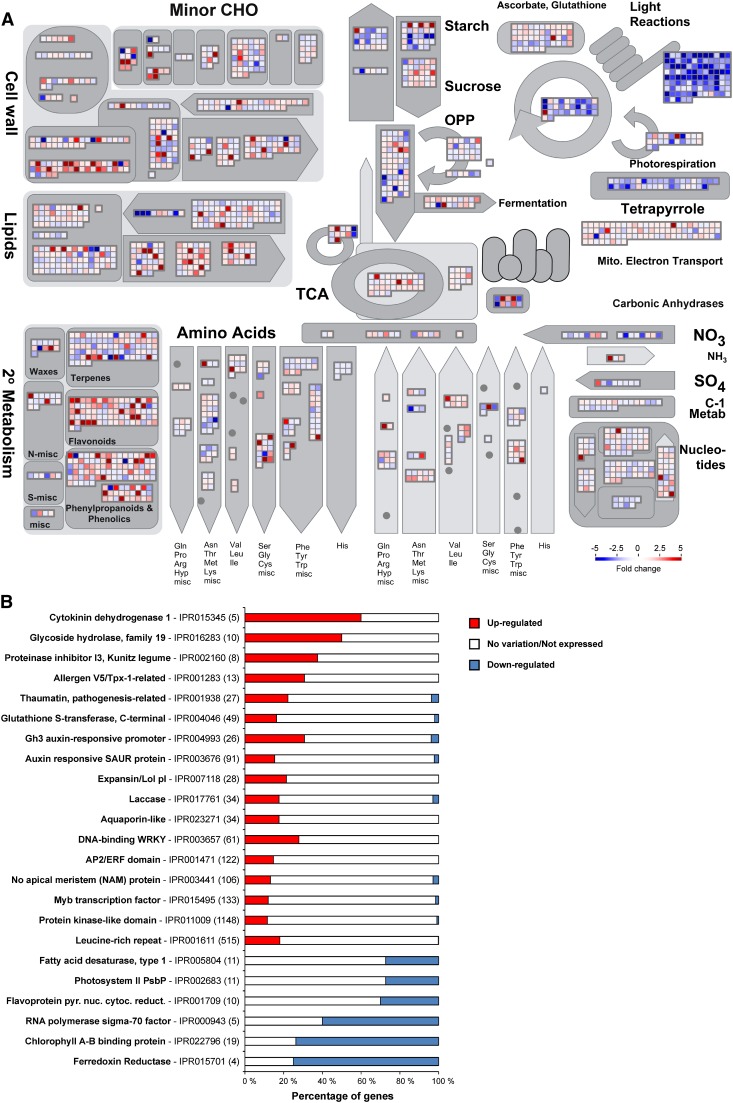
Global analyses of diseased cacao plants were performed to provide an overview of the physiological alterations caused by M. perniciosa infection. Individual gene responses in metabolic pathways were visualized using the MapMan tool (Figure 3A). A remarkable repression of genes related to photosynthesis, tetrapyrrole (chlorophyll chromophore) biosynthesis, starch biosynthesis, desaturation of fatty acids, and nitrogen assimilation was verified. By contrast, genes involved in secondary metabolism, cell wall modification, and lipid degradation were induced. A complete list of MapMan pathways differentially represented in green brooms is provided in Supplemental Data Set 2. Complementing these findings, enrichment analysis of InterPro terms showed specific domains and families differentially represented in green brooms (Figure 3B; Supplemental Data Set 3). Protein domains/families related to defense, hormonal metabolism, signaling pathways, stress responses, and cell wall modification were overrepresented. Conversely, underrepresented terms included proteins associated with photosynthesis, chlorophyll biosynthesis, chloroplastic RNA polymerase, and desaturation of fatty acids. In general, these results indicate important changes that occur in the green broom stage of WBD, which will be explored in the following sections.
Figure 3.
Overview of the Transcriptional Changes Occurring in Green Brooms.
(A) MapMan representation of metabolic pathways. Each box depicts an individual gene. Upregulated and downregulated genes are shown in red and blue, respectively. The scale bar represents fold change values.
(B) Selected InterPro terms differentially represented in infected plants. The total number of cacao genes in each category is shown in parentheses. The proportion of upregulated and downregulated genes within a category is represented by red and blue bars, respectively. See Supplemental Data Set 3 for the complete result of the InterPro enrichment analysis.
Transcriptional Alterations in Infected Plants
Infected Cacao Tissues Undergo Carbon Starvation during WBD
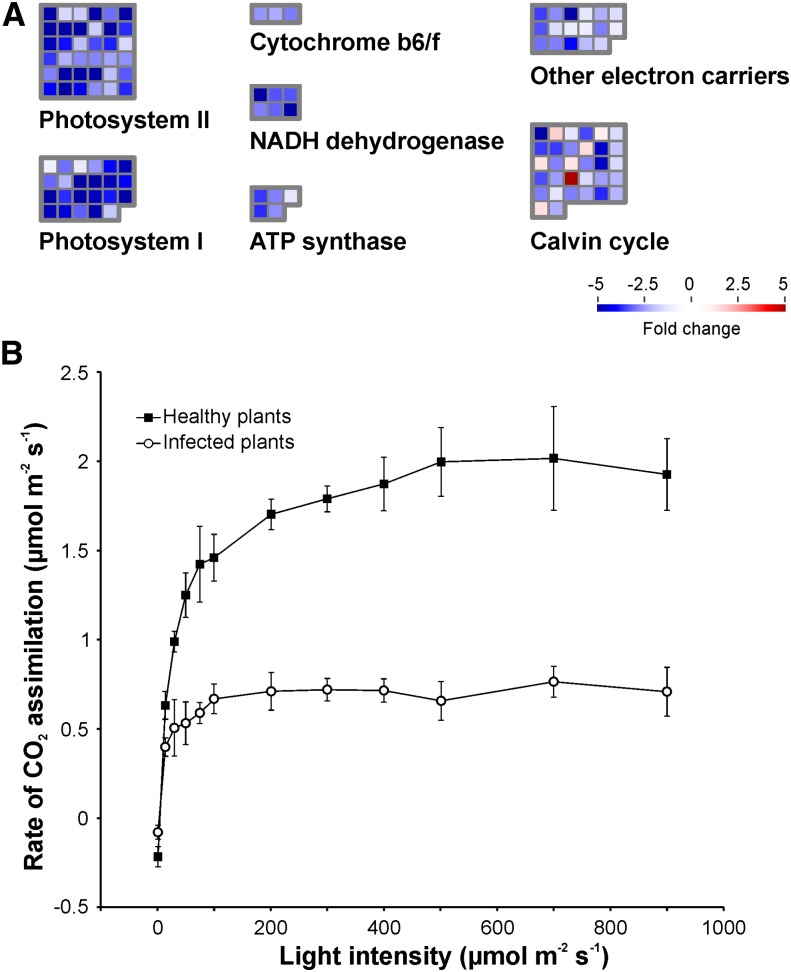
Remarkable changes in the primary metabolism of cacao plants occur in the green broom stage of WBD. Thirty-day-old leaves developed from apical meristems were expected to be photosynthetically active. However, as a consequence of the disease, genes related to the photosynthetic apparatus were downregulated (Figure 4A; Supplemental Data Set 4). Leaves developed from infected tissues exhibited severe chlorosis (Figure 1B; Supplemental Figure 1) and had lower rates of CO2 assimilation (Figure 4B) than did those from healthy plants. By contrast, genes required for sucrose breakdown (cell wall invertase) and hexose transport (hexose transporters) were upregulated, indicating that the disease interferes with the normal physiology of cacao meristematic tissues, resulting in the maintenance of their primary sink capacity.
Figure 4.
Photosynthetic Metabolism Is Impaired in Infected Cacao Plants.
(A) Genes involved in photosynthesis are downregulated in green brooms when compared with healthy plants. Gene classes were defined according to the MapMan ontology.
(B) Consistent with the RNA-seq results, infected plants presented lower rates of CO2 assimilation. Measurements were performed in triplicate under different light intensities. Error bars represent standard errors.
In parallel, genes associated with the degradation of carbohydrate and lipid storage molecules (starch and triacylglycerol, respectively) were also upregulated, including genes encoding amylases and lipases (Supplemental Data Set 4). Indeed, reduced amounts of starch granules were clearly observed in infected tissues (Figures 1F and 1G), suggesting that, upon infection, the plant mobilizes these storage molecules as complementary sources of energy. In addition, genes related to the fatty acid β-oxidation pathway (Supplemental Data Set 4), a process by which fatty acids are broken down to produce energy, were overexpressed. The final product of fatty acid β-oxidation is acetyl-CoA, which can directly enter the glyoxylate cycle. In the glyoxylate cycle, lipids are converted into carbohydrates through the enzymes isocitrate lyase and malate synthase. Notably, these enzymes are highly transcribed in the green broom stage of WBD. Even the remaining glycerol, derived from lipid degradation, seems to be used as an energy source. The transcription of a gene encoding a FAD-dependent glycerol-3-phosphate dehydrogenase is increased, suggesting that the phosphorylated form of glycerol (glycerol-3-phosphate) is directly oxidized in the mitochondrial electron transport chain. Overall, the general disarrangement in plant metabolism caused by M. perniciosa infection appears to culminate in the deprivation of carbon skeletons, compelling the cacao plant to obtain energy from alternative sources.
Host nitrogen Metabolism Is Directed toward the Production of Carbon Skeletons
In addition to carbohydrates and lipids, plant amino acids also seem to be energetically consumed in green brooms. Genes involved in the catabolism of several amino acids were upregulated (Supplemental Data Set 4), which may result in the release of ammonium and intermediate compounds to the tricarboxylic acid cycle or to other metabolic processes. By contrast, nitrogen assimilation (from nitrate/ammonium to glutamine), which is a high-energy-consuming process, is impaired in green brooms. Diseased plants showed decreased expression of nitrate/nitrite reductases and glutamine synthetase genes, which encode critical enzymes for primary nitrogen assimilation. Remarkably, despite the overall degradation of amino acids, asparagine biosynthesis was transcriptionally induced (Supplemental Data Set 4). Along with genes that are directly involved in nitrogen biosynthesis/catabolism, several transporters that mediate the uptake of extracellular amino acids and peptides were upregulated (Supplemental Data Set 3). Additionally, a cacao gene that is orthologous to the tonoplast intrinsic gene subfamily of Arabidopsis thaliana (TIP2;1) was overexpressed in green brooms. TIP2;1 was shown to mediate the extracytosolic transport of ammonium across the tonoplast and may participate in vacuolar compartmentalization of this toxic compound (Loqué et al., 2005).
Insight into the Hormonal Metabolism of Green Brooms
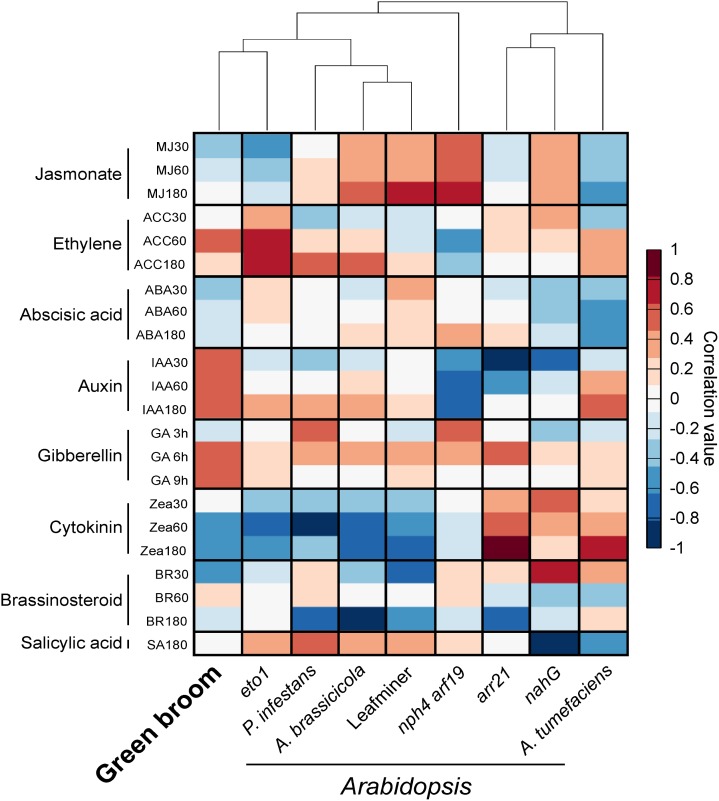
The symptoms observed in infected cacao tissues suggest the occurrence of significant and essentially unexplored hormonal imbalances during WBD. To identify signatures of hormonal responses in infected cacao plants, we used the HORMONOMETER software, which compares the variation in gene expression of a query experiment with indexed data sets of hormone treatments (Volodarsky et al., 2009). Since this software requires Arabidopsis genes as input, we used 8870 putative orthologs of cacao genes that were expressed in our experiment. To evaluate the HORMONOMETER performance using this gene set, we analyzed public gene expression data of Arabidopsis mutants with known alterations in hormone levels (i.e., eto1, arr21, nahG, and nph4 arf19). We also included transcriptomes of Arabidopsis interacting with other organisms (i.e., Agrobacterium tumefaciens, Phytophthora infestans, Alternaria brassicicola, and the leaf miner insect Liriomyza huidobrensis). The HORMONOMETER profile of each Arabidopsis mutant was highly representative of the global hormonal alterations that occur in these plants (Figure 5), thus validating the use of this set of genes.
Figure 5.
Identification of Hormonal Signatures Based on Transcriptomic Data.
The analysis was conducted using the HORMONOMETER software, which compares gene expression data of a query experiment with data sets of hormone responses. Transcriptomes of Arabidopsis mutants with known alterations in hormonal responses were used as controls (arr21, increased cytokinin response; eto1, increased ethylene response; nahG, reduced salicylic acid levels; nph arf19/IAA, reduced auxin signaling). We also included transcriptomes of Arabidopsis interacting with other organisms (Agrobacterium, 6 d after infection; P. infestans, 24 h after infection; A. brassicicola, 24 h after infection; and the leaf miner insect L. huidobrensis, locally damaged leaves). A positive correlation between the query transcriptome and a hormone treatment is denoted in red, whereas a negative correlation is represented by blue.
Auxins.
The HORMONOMETER profile of green brooms showed a strong correlation with auxin responses (Figure 5). Primary auxin-responsive genes fall into three major classes: GH3, Aux/IAA (auxin/indole-3-acetic acid protein), and SAUR (small auxin-up RNA). In accordance with the HORMONOMETER results, InterPro terms representing proteins encoded by these three classes of genes were significantly enriched in infected plants (Figure 3B; Supplemental Data Set 3). Furthermore, members of the PIN and PILS (PIN-LIKES) class of auxin transporters were differentially expressed (Supplemental Data Set 5), suggesting the existence of altered levels of auxin in green brooms. Genes involved in auxin biosynthesis were not differentially expressed.
Gibberellins.
The HORMONOMETER analysis also indicated a high correlation between green brooms and gibberellin treatments (Figure 5). Several genes involved in the gibberellin response were upregulated in green brooms, including those encoding α-amylases, invertases, GAST-like (gibberellic acid stimulated transcript-like), pectinesterases, XTHs (xyloglucan endotransglycosylase/hydroxylases), and expansins. Many of these gibberellin-responsive genes mediate cell elongation/expansion and their increased expression is in agreement with the morphological alterations observed in infected cacao shoots (Figure 1; Supplemental Figure 1). Genes encoding enzymes involved in gibberellin perception (GID1-like), biosynthesis (GA3ox), and inactivation (GA2ox) were also identified among the upregulated genes (Supplemental Data Set 5).
Cytokinins.
Although some components of the cytokinin signaling circuitry were upregulated in infected plants (Supplemental Data Set 5), a clear signature of cytokinin responses was not evident in the green broom stage of WBD (Figure 5). In accordance, a gene involved in cytokinin biosynthesis (IPT, isopentenyltransferase) was repressed in infected plants. Also, other genes related to the cytokinin response and biosynthesis were not differentially expressed. Instead, many genes related to cytokinin degradation (cytokinin oxidases) or inactivation (cytokinin glucosyltransferases) showed increased transcript levels (Supplemental Data Set 5), indicating that cytokinins are being degraded/inactivated in this stage of WBD.
Salicylic Acid, Jasmonic Acid, and Ethylene.
The HORMONOMETER analysis showed no correlation between green brooms and salicylic acid or jasmonic acid responses. By contrast, higher correlation values were verified for ethylene (Figure 5). In particular, transcripts related to ethylene biosynthesis were more abundant in green brooms (Supplemental Data Set 5). Moreover, a large number of genes encoding ethylene response factor (ERF) transcription factors were clearly overexpressed in infected plants, supporting the notion that ethylene participates in WBD.
M. perniciosa Triggers Plant Defense Responses during the Biotrophic Interaction with Cacao
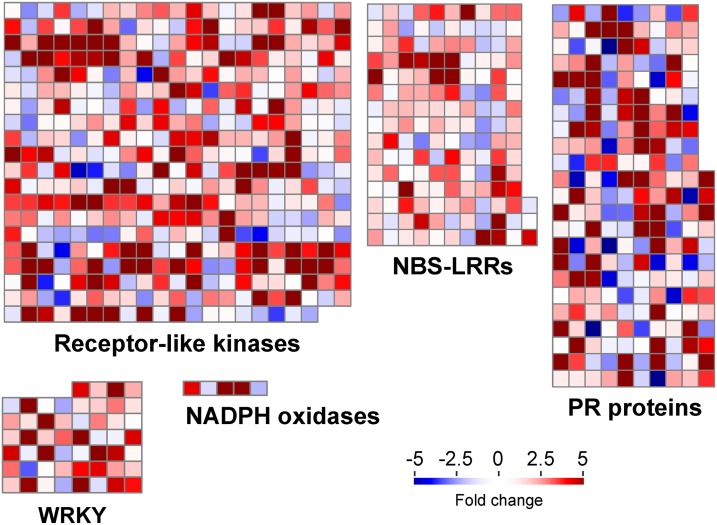
A remarkable characteristic of the green broom transcriptome is the prevalence of transcripts related to defense responses (Figure 6). Several genes encoding putative immune receptors were differentially expressed in infected plants (Supplemental Data Set 6). Remarkably, of the 1269 upregulated genes in green brooms, at least 151 genes (11.9%) belong to the receptor-like kinase or receptor-like protein classes. These proteins are transmembrane receptors that perceive extracellular molecules, including microbe-associated molecular patterns (Monaghan and Zipfel, 2012). InterPro terms related to kinases as well as to different extracellular domains commonly found in these proteins (e.g., LRR, lectin, malectin, and WAK) were significantly enriched in infected plants (Figure 3B; Supplemental Data Set 3). In addition to receptor-like kinases and receptor-like proteins, the expression of at least 30 cacao receptors belonging to the NB-LRR family was also induced by M. perniciosa infection (Supplemental Data Set 6). These genes encode intracellular proteins that directly or indirectly recognize pathogen effectors, leading to strong resistance responses (Jones and Dangl, 2006).
Figure 6.
Cacao Defense Mechanisms Are Activated during the Green Broom Stage of Witches’ Broom Disease.
Selected classes of genes with known functions in plant defense responses are shown. Each square represents an individual gene within a category. Upregulated and downregulated genes in infected plants are shown in red and blue, respectively. The scale bar represents fold change values.
Along with immune receptors, many genes encoding antimicrobial proteins were upregulated in green brooms (Figure 6). At least 67 of these genes belong to the pathogenesis-related (PR) superfamily, which comprises a heterogeneous group of 17 families that are part of the plant-inducible defense mechanisms against pathogens. Transcripts of genes encoding members of the PR-1 (unknown activity), PR-2 (β-1-3-glucanase), PR-3, PR-4, PR-8, PR-11 (chitinase), PR-5 (thaumatin), PR-6 (protease inhibitor), PR-9 (peroxidase), and PR-10 (ribonuclease) families accumulated during WBD. Other upregulated defense genes include 16 WRKY transcription factors, three Kunitz protease inhibitors, and three NADPH oxidases, which are responsible for the production of superoxide ions (Supplemental Data Set 6). These results support the hypothesis that cacao defense responses are active in the green broom stage of WBD.
Genes Associated with Secondary Metabolism and Cell Wall Modification Are Upregulated in Green Brooms
Despite the evident impairment in the primary metabolism of green brooms, infected cacao tissues seem to allocate carbon skeletons and energy to the production of secondary metabolites (Supplemental Data Set 7). Such metabolites are well known stress indicators and are implicated in defense responses against microbes and animals. Several genes related to the biosynthesis of flavonols, anthocyaninins, terpenoids, and alkaloids were strongly upregulated in green brooms (Supplemental Data Set 7). Moreover, genes involved in lignin biosynthesis were also induced, indicating that lignin is deposited in infected tissues, possibly as part of an attempt to limit pathogen colonization. Indeed, histological analyses revealed the existence of cell wall reinforcement in infected tissues (Figure 7), supporting the idea that the plant perceives the pathogen and activates defense mechanisms during infection. Remarkably, numerous genes encoding cell-wall-modifying enzymes, including expansins, endoglucanases, glycosyl transferases, and xyloglucan endotransglycosylases/hydrolases, were upregulated in green brooms. Transcripts of genes associated with the biosynthesis of cellulose and hemicellulose also accumulated in infected tissues.
Figure 7.
Histological Analysis Showing Plant Cell Wall Reinforcement in Green Brooms.
In addition to the reduction in starch content (granules inside the cells), infected plants show reinforcement of cell walls (brighter walls under polarized light), which is in line with the idea that the cacao host perceives the pathogen and mounts defense responses during the green broom stage of WBD. Arrows point to fungal cells in the infected plant. Samples were stained with toluidine blue and visualized under white and polarized lights. Bars = 200 μm.
[See online article for color version of this figure.]
Identification of M. perniciosa Transcripts
As mentioned above, only a small proportion (0.3%) of the transcripts detected in infected plants were of fungal origin. Sequencing of the five infected plants resulted in a total of 562 million paired reads, 1.7 million of which mapped to M. perniciosa genes (Table 1). Using a threshold of 1 RPKM (reads per kilobase per million mapped reads) in all five replicates to designate a gene as being expressed, we detected 8617 M. perniciosa genes in our experiment, corresponding to 51% of the predicted fungal gene models. A less stringent analysis revealed that 13,529 genes (79%) had a minimum expression level of one RPKM in at least one of the five replicates. To evaluate the effect of sequencing depth on our ability to detect M. perniciosa transcripts, we pooled all replicates and generated different subsets of our RNA-seq data using randomly selected reads. We verified that only genes with the highest expression values were detected at low sequencing depths. However, the number of expressed genes started to stabilize with ∼0.5 million mapped paired reads, and few additional genes were detected by increasing the sequencing depth (Supplemental Figure 4). This result indicates that most of the M. perniciosa genes that are expressed in green brooms were sampled in our experiment. Supplemental Data Set 8 presents the expression levels of each of the 17,008 M. perniciosa genes.
Searching for Fungal Genes That Are Distinctively Expressed in Green Brooms
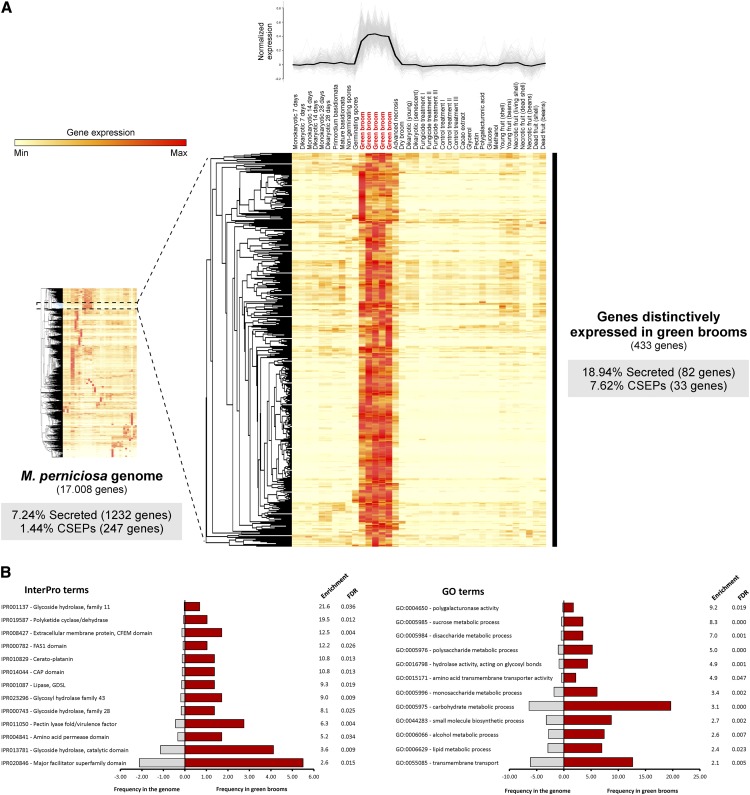
We utilized the WBD Transcriptome Atlas as a starting point to identify the most prominent fungal transcripts expressed in green brooms. This atlas comprises a set of transcriptomes representing M. perniciosa growth in vitro and in planta under several biological conditions (e.g., fungal response to diverse carbon sources or drugs, WBD progression in fruits, WBD progression in shoots, nongerminating and germinating spores, and basidiomata). Hierarchical clustering analysis of the M. perniciosa genes was performed and groups of genes with correlated expression profiles (i.e., coregulated genes) were obtained (Figure 8A). Remarkably, we identified a cluster of 433 genes that were distinctively expressed in the green broom stage (Supplemental Data Set 10). In general, these genes show very high expression values in green brooms, which support a major role for the encoded proteins during the biotrophic interaction with cacao. Indeed, the median expression value of the genes enriched in green brooms was 10 times higher than that of the remaining genes detected in the same conditions (Supplemental Figure 5).
Figure 8.
Hierarchical Clustering Analysis of the Set of M. perniciosa Gene Models.
(A) Genes with correlated expression profiles were grouped, and a cluster of 433 genes that are distinctively expressed in the green broom stage of WBD was obtained. Closer inspection of the green-broom-specific cluster showed that 82 (18.94%) of the 433 genes encode potential secreted proteins and 33 (7.62%) encode CSEPs. Whereas 7.24% of the 17,008 genes present in the M. perniciosa genome encode secreted proteins and 1.44% encode CSEPs, the green broom gene cluster is clearly enriched in these classes of genes (P value < 0.001).
(B) Selected InterPro and GO terms enriched in the set of 433 M. perniciosa genes that are distinctively expressed in green brooms. A list with all enriched terms is shown in Supplemental Data Set 11.
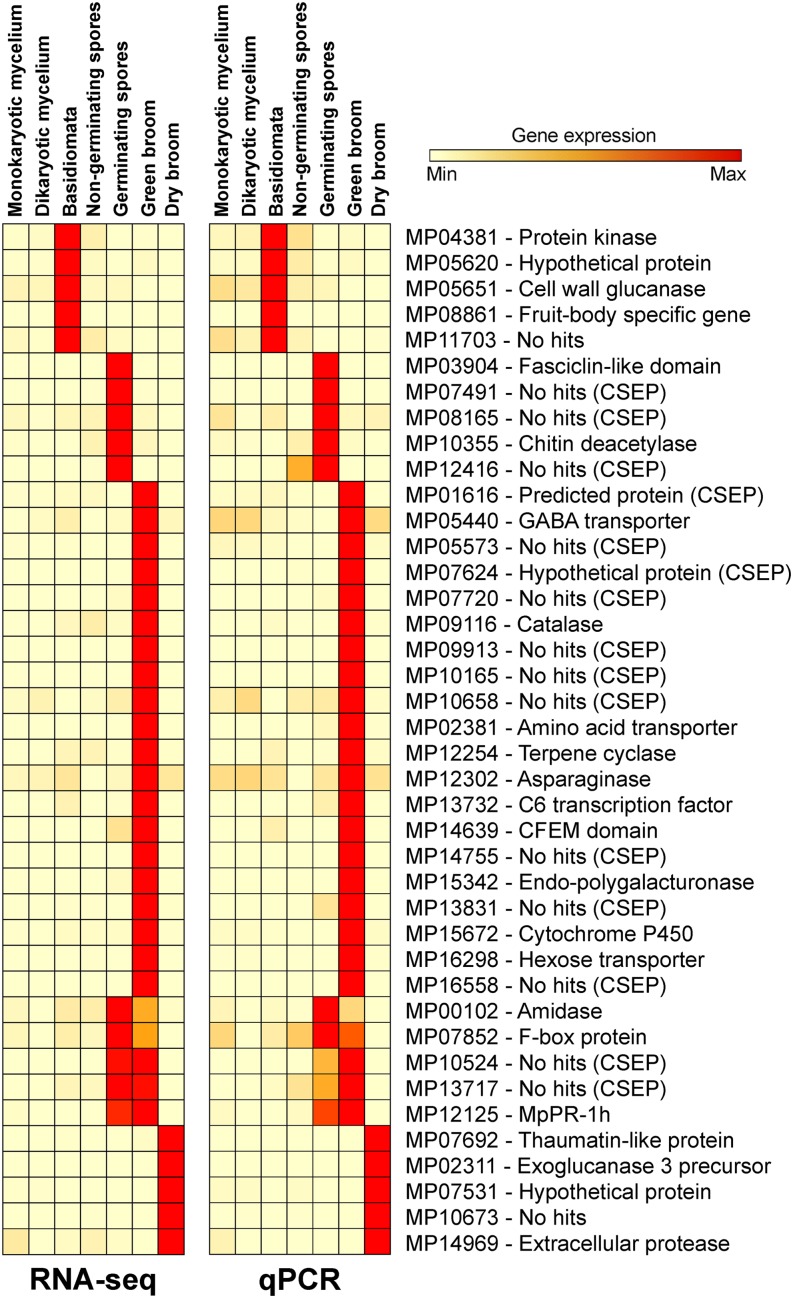
Closer inspection of the green-broom-specific cluster showed that 84 (19.4%) of the 433 genes encoded potential secreted proteins and 33 genes (7.4%) encoded candidate secreted effector proteins (CSEPs; secreted proteins that lack clearly characterized homologs in other sequenced organisms). Whereas 7.24% of the 17,008 genes present in the M. perniciosa genome encode secreted proteins and 1.44% encode CSEPs, the green broom gene cluster is clearly enriched in these classes of genes (19.4% secreted and 7.4% CSEP; P value < 0.001). We selected 20 genes from this cluster for validation by qPCR assays and confirmed that the gene expression profile obtained from the WBD Transcriptome Atlas was highly consistent (Figure 9). Furthermore, we also validated the expression profile of 20 additional genes that were predicted to be expressed in different M. perniciosa life stages. On the basis of these results, we verified that the use of the WBD Transcriptome Atlas is a very efficient approach for identifying genes that are involved in the biotrophic colonization and pathogenesis of M. perniciosa. The WBD Transcriptome Atlas is publicly available at www.lge.ibi.unicamp.br/wbdatlas.
Figure 9.
Expression Profile of M. perniciosa Genes Based on the WBD Transcriptome Atlas.
Forty fungal genes that exhibited different expression patterns were selected for validation using qPCR. Data for each gene are shown in relation to the mean expression values across all samples. RNA-seq expression values are presented as RPKMs. qPCR results are presented as 2−ΔCt using the M. perniciosa β-actin and the IF3b (transcription initiation factor) genes as normalizers. Raw results are shown in Supplemental Data Set 9.
[See online article for color version of this figure.]
Functional Gene Set Enrichment Analysis: Categorizing Significant Metabolic Processes of M. perniciosa in WBD
We performed enrichment analyses of InterPro and Gene Ontology (GO) terms to identify statistically significant overrepresentation of particular protein families/domains and pathways in the green-broom-specific cluster of fungal genes (Figure 8B; Supplemental Data Set 11). Overrepresented InterPro terms were mostly related to nutrient acquisition, microbial pathogenesis, and degradation/modification of components of the plant cell wall (particularly pectin). Other enriched terms included a tyrosinase domain, which is present in proteins required for the formation of polyphenolic compounds (e.g., melanin), and a polyketide cyclase/dehydrase domain, which mediates the biosynthesis of polyketides (e.g., toxins). In addition, GO term enrichment analysis revealed that processes associated with carbohydrate metabolism (e.g., metabolism of starch, hexoses, and sucrose), biosynthesis of cellular nitrogen, transport of amino acids, oxidoreduction, and the metabolism of components of the plant cell wall were overrepresented in the green-broom-specific cluster. Overall, the InterPro and GO enrichment results are highly complementary and present an overview of the pathways/processes that are potentially important for M. perniciosa during the green broom stage of the disease.
Characterizing the Green Broom Cluster: Insights from Homologs of Other Fungal Species
BLASTx analysis of the 433 genes of the green broom cluster was performed against the NCBI nonredundant database (e-value cutoff of 10−3). We verified that 68 of the 433 M. perniciosa genes that were distinctively expressed in green brooms had no significant similarities to sequences in organisms outside the Moniliophthora genus. Furthermore, at least 60 of the remaining genes had no clearly assigned function (i.e., they were annotated as hypothetical or predicted proteins and had no InterPro or GO terms). Based on sequence similarity, we defined putative functions for 305 of the 433 genes that were distinctively expressed in green brooms. Importantly, some of these genes encoded proteins with recognized roles in fungal pathogenesis, such as members of the PR-1 and cerato-platanin gene families. Moreover, fasciclin-encoding genes were induced (RPKM > 400) in M. perniciosa and possibly mediate the adhesion between cacao and fungal cells. Genes encoding CFEM domain-containing proteins (RPKM > 90) and chitin deacetylases (RPKM > 75) were also highly expressed by M. perniciosa during its biotrophic interaction with cacao. Interestingly, the green broom cluster included seven genes that encode transcription factors. These transcription factors are members of the basic-leucine zipper, Zn(2)-C6 fungal-type DNA binding protein, and C2H2-type zinc finger families, and they constitute potential regulators of virulence genes in M. perniciosa. A selection of M. perniciosa genes potentially involved in pathogenesis is provided in Supplemental Data Set 12.
Genes Required for Pectin Breakdown Are Distinctively Expressed during the Biotrophic Development of M. perniciosa
Our analysis of the green broom transcriptome revealed a strikingly high level of expression of genes encoding pectinolytic enzymes in M. perniciosa (mean RPKM > 1000; Supplemental Data Set 12). During its biotrophic stage, the pathogen seems to produce a complete set of enzymes necessary for the breakdown of pectin, including pectin methylesterases, polygalacturonases, and pectate lyases. Remarkably, several of these genes are part of the green broom-specific cluster and might function in fungal growth within the host living tissues. Moreover, transcripts of genes involved in the metabolism of subproducts of pectin degradation (i.e., genes encoding methanol oxidase, formaldehyde dehydrogenase, and formate dehydrogenase) were identified. Some of these genes were also highly expressed in other RNA-seq libraries that comprise the WBD Transcriptome Atlas, such as infected cacao pods, which are tissues with high pectic content. In addition, M. perniciosa seems to be capable of degrading cellulose and hemicellulose during the green broom stage of WBD. Many genes encoding putative cellulases, xylanases, arabinofuranosidases, manosidases, and acetylesterases were identified among the distinctively expressed fungal genes that comprise the green broom-specific cluster (Supplemental Data Set 12).
M. perniciosa Has a Great Potential for Cellular Detoxification and Stress Tolerance
By inspecting the green broom transcriptome, we found that M. perniciosa expresses an arsenal of genes encoding enzymes involved in detoxification and stress tolerance. Reactive oxygen species (ROS), such as superoxide anions (O2·−), hydroxyl radicals (·OH), and hydrogen peroxide (H2O2), are produced by plants to halt pathogen invasion (Apel and Hirt, 2004). M. perniciosa genes encoding the antioxidant enzymes superoxide dismutase and catalase were distinctively expressed (RPKM > 650) in green brooms (Figure 8; Supplemental Data Set 10), indicating that the fungus might deal with ROS toxicity in this stage of infection. In agreement with this finding, a glutathione synthetase, which catalyzes the final step in the glutathione biosynthesis pathway, was also induced (RPKM = 58). Additionally, 12 fungal genes encoding cytochrome P450 enzymes were specifically identified in the green broom cluster (Figure 8; Supplemental Data Set 10). These proteins oxidize a vast array of metabolic intermediates and environmental compounds and are important for adaptation to hostile ecological niches (Moktali et al., 2012). Genes encoding transporters of the major facilitator superfamily were also distinctively expressed in the green broom stage (RPKM > 480). Moreover, although not exclusive to this stage, transporters of the ATP binding cassette superfamily (ABC) showed high expression values in green brooms (RPKM > 45). Interestingly, some members of these classes of transporters have been associated with fungal pathogenesis, acting through the extrusion of host defense molecules and through the secretion of fungal virulence factors (Coleman and Mylonakis, 2009). Taken together, our results indicate that M. perniciosa is exposed to highly stressful conditions during cacao infection and that a distinctive ability to counteract such conditions is a central aspect of its virulence.
Evidence of the Nutritional Strategy of the M. perniciosa Biotrophic Mycelium
Colonizing the plant apoplast and being devoid of specialized feeding structures (i.e., haustoria), the M. perniciosa biotrophic mycelium might have the ability to take up soluble compounds that are transported through the apoplast. To understand M. perniciosa nutrition during the green broom stage, we sought to identify genes related to the uptake of nutrients, particularly of nitrogen and carbon derivatives. Many fungal proteinases along with an oligopeptide transporter were highly expressed (RPKM > 290) during infection (Supplemental Data Set 12), indicating that this fungus has the apparatus to degrade plant proteins secreted in the apoplast and capture the resulting peptides. Remarkably, an asparaginase, which catalyzes the release of ammonia from asparagine, showed distinctive expression levels (RPKM = 91) during M. perniciosa biotrophic interaction with cacao. A transporter of the nonprotein amino acid gamma amino butyric acid was also highly induced in green brooms (RPKM = 223). Furthermore, numerous transcripts related to sugar transporters were specifically identified in green brooms (Figure 8; Supplemental Data Set 10). It is likely that saccharides derived from the degradation of complex sugars of the plant cell wall, such as pectin, cellulose, and hemicellulose, and sugars translocated to green brooms (i.e., sucrose) can be absorbed by M. perniciosa through these transporters and used as energy sources.
The Green Broom Transcriptome Reveals Candidate Effectors in M. perniciosa
The current version of the M. perniciosa genome encodes 247 CSEPs, which were defined as secreted proteins that lack clearly characterized homologs in other sequenced organisms (Supplemental Data Set 13). Remarkably, several of the most highly expressed fungal genes in green brooms encoded CSEPs (Table 2), supporting an important role for these proteins during infection. In total, 48 CSEP-encoding genes with RPKM > 35 were identified in infected plants. In general, these genes encode small cysteine-rich proteins, with a mean size of 157 amino acids and a Cys content of 3.95%, both of which are typical of virulence effectors (Stergiopoulos and de Wit, 2009). Furthermore, inspection of the WBD Transcriptome Atlas revealed that 33 of these 48 CSEP genes were part of the green-broom-specific cluster (Figure 8; Supplemental Data Set 10), thus providing additional evidence that they encode potential effectors.
Table 2. Most Highly Expressed M. perniciosa Genes during the Green Broom Stage of WBD.
| Gene Expression Values (RPKM) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Annotation | Length (aa)a | I1 | I2 | I3 | I4 | I5 | Mean |
| MP13831 | No hits (CSEP) | 185 | 47,106 | 73,411 | 92,642 | 81,894 | 99,895 | 78,990 |
| MP14755 | No hits (CSEP) | 246 | 81,666 | 50,657 | 67,109 | 81,134 | 63,814 | 68,876 |
| MP14757 | No hits (CSEP) | 270 | 22,364 | 19,546 | 25,939 | 27,095 | 36,987 | 26,386 |
| MP15869 | No hits | 243 | 36,079 | 10,330 | 17,487 | 24,296 | 19,698 | 21,578 |
| MP13352 | MpPR-1g | 252 | 9,121 | 16,141 | 17,621 | 20,775 | 20,990 | 16,930 |
| MP15868 | Hypothetical protein | 143 | 17,204 | 5,903 | 10,031 | 12,341 | 12,508 | 11,597 |
| MP16558 | No hits (CSEP) | 269 | 12,680 | 9,664 | 9,827 | 10,544 | 12,103 | 10,964 |
| MP09913 | No hits (CSEP) | 84 | 2,166 | 13,141 | 13,579 | 9,870 | 11,194 | 9,990 |
| MP14724 | No hits | 203 | 8,858 | 7,721 | 11,358 | 8,550 | 11,281 | 9,554 |
| MP13834 | No hits (CSEP) | 183 | 5,449 | 6,958 | 11,154 | 10,006 | 12,669 | 9,247 |
| MP13136 | No hits | 97 | 2,741 | 10,858 | 10,471 | 8,982 | 9,493 | 8,509 |
| MP09729 | Alcohol dehydrogenase | 347 | 5,849 | 13,822 | 8,228 | 6,722 | 6,577 | 8,240 |
| MP09537 | Glycoside hydrolase 18 | 438 | 8,276 | 6,000 | 6,013 | 7,077 | 6,662 | 6,805 |
| MP15315 | MpCP12 | 155 | 3,881 | 5,014 | 6,892 | 4,963 | 5,948 | 5,339 |
| MP15025 | Endoglucanase v-like | 157 | 2,618 | 5,578 | 5,479 | 5,764 | 4,372 | 4,762 |
| MP15342 | Endo-polygalacturonase | 385 | 3,198 | 1,998 | 5,065 | 5,894 | 5,545 | 4,340 |
| MP16511 | MpCP11 | 186 | 4,228 | 4,687 | 3,871 | 3,288 | 4,170 | 4,049 |
| MP04874 | Elongation factor ef1-α | 460 | 4,877 | 4,243 | 4,033 | 3,799 | 3,172 | 4,025 |
| MP12125 | MpPR-1 h | 245 | 6,069 | 2,605 | 3,137 | 4,132 | 4,178 | 4,024 |
| MP15312 | MpCP4 | 192 | 3,966 | 3,115 | 4,043 | 4,003 | 4,172 | 3,860 |
aa, amino acids.
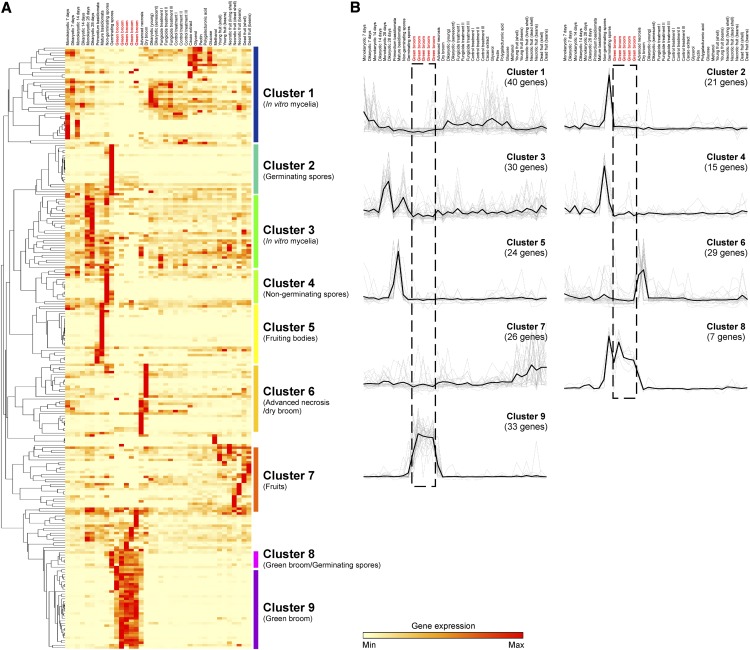
To explore the expression profile of the M. perniciosa CSEPs, we performed a specific clustering analysis of the 247 CSEPs using the diverse biological conditions of the WBD Transcriptome Atlas. Nine clusters of coregulated genes were identified (Figure 10). As expected, we identified a cluster of CSEP genes that was distinctively expressed in green brooms (Cluster 9). In addition, two other clusters emerged. Cluster 2 included genes expressed in germinating spores, whereas Cluster 8 was composed of genes expressed in both germinating spores and green brooms (Figure 10). Together, these three clusters (61 CSEPs) represent conditions associated with cacao shoot infection and are likely to play a fundamental role in M. perniciosa pathogenicity. Inspection of the protein sequences of the M. perniciosa CSEPs did not reveal the presence of motifs that were previously found in effectors of other filamentous pathogens (e.g., RXLR, RSIDELD, CHXC, and Y/F/WXC motifs) (Godfrey et al., 2010; Kale et al., 2010; Kemen et al., 2011; Zuccaro et al., 2011).
Figure 10.
RNA-seq Expression Profiling of M. perniciosa Candidate Effectors.
(A) The 247 CSEP genes were clustered using expression data from the WBD Transcriptome Atlas. Nine clusters representing different fungal life stages are highlighted.
(B) Visualization of the average expression of each cluster (black line). Cluster 9 contains a set of 33 CSEPs distinctively expressed in green brooms. Genes specifically expressed in stages not directly related to pathogenesis (e.g., fruiting bodies) might not encode true effectors. Libraries representing green brooms are marked with red font.
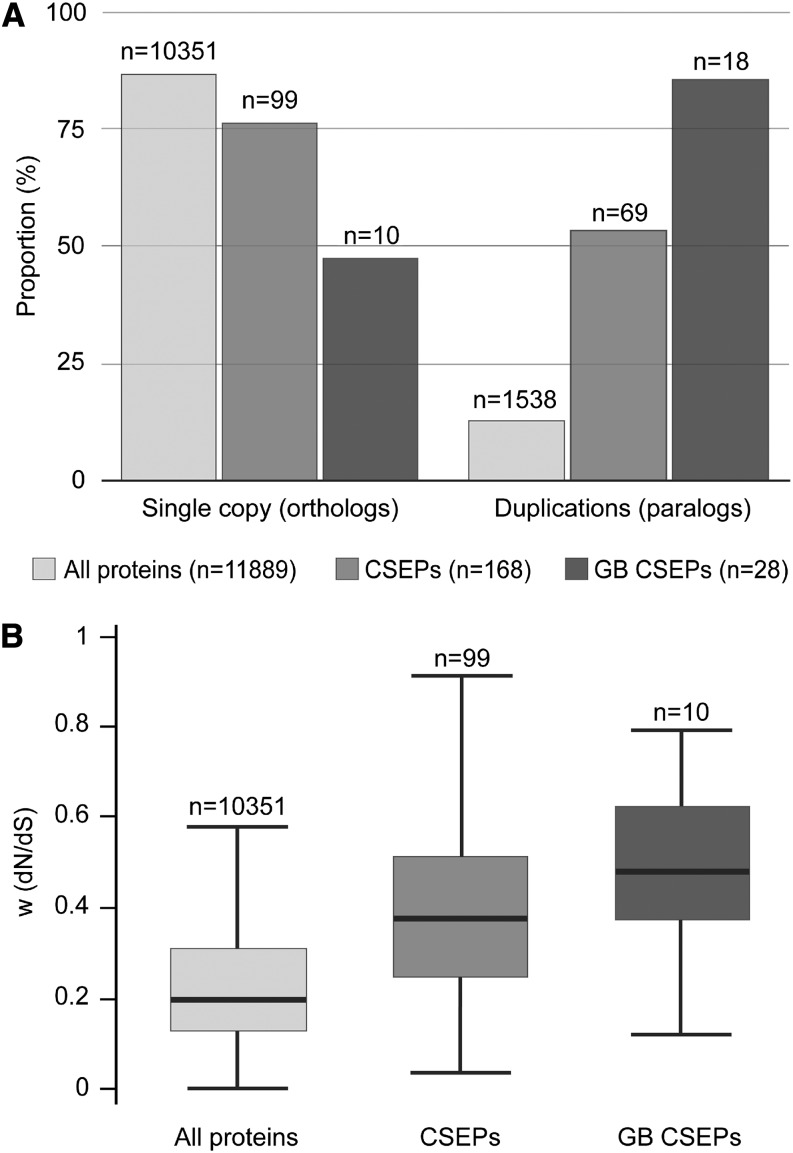
We also evaluated the evolutionary pattern of the M. perniciosa CSEPs by performing a comparative analysis with Moniliophthora roreri, a recently sequenced cacao pathogen that is closely related to M. perniciosa (Meinhardt et al., 2014). Using the OrthoMCL program, we defined 11,889 homolog families in these fungi, of which 10,351 had a single gene in each species (orthologs) and 1538 contained more than one gene in at least one of the species (paralogs). Curiously, we identified a lower proportion of ortholog families in CSEPs in comparison to all coding genes (P = 3.9E-20) and in green broom CSEPs in comparison to CSEPs (P = 9.4E-6) (Figure 11A), indicating higher levels of gene duplications among CSEPs. We next estimated the ratio of nonsynonymous and synonymous substitution rates (dN/dS) in all ortholog pairs to quantify the evolutionary pressure on these genes. Interestingly, CSEP genes exhibited high dN/dS rates, which is a hallmark of rapid evolution, whereas the remaining protein-encoding genes did not (P = 1.4E-10; Figure 11B). This enrichment was even higher when only green-broom-specific CSEPs (the 10 pairs of orthologs) were considered (P = 0.037). Altogether, these results support the hypothesis that our list of CSEP genes does indeed encode effectors of pathogenicity.
Figure 11.
Evolutionary Analysis of M. perniciosa Candidate Effectors.
(A) Percentage of orthologs and paralogs in all protein-encoding genes, CSEPs, and green broom (GB) CSEPs. CSEP genes are significantly enriched for paralogs in relation to all protein-encoding genes, and green broom CSEP genes are significantly enriched for paralogs in relation to CSEP genes.
(B) dN/dS ratio box plots showing medians and quartiles of values for the set of orthologous protein-encoding genes, CSEPs, and green broom CSEPs. CSEP genes are significantly enriched for higher dN/dS rates in relation to all protein-encoding genes, and green broom CSEP genes are significantly enriched for higher dN/dS rates in relation to CSEP genes.
DISCUSSION
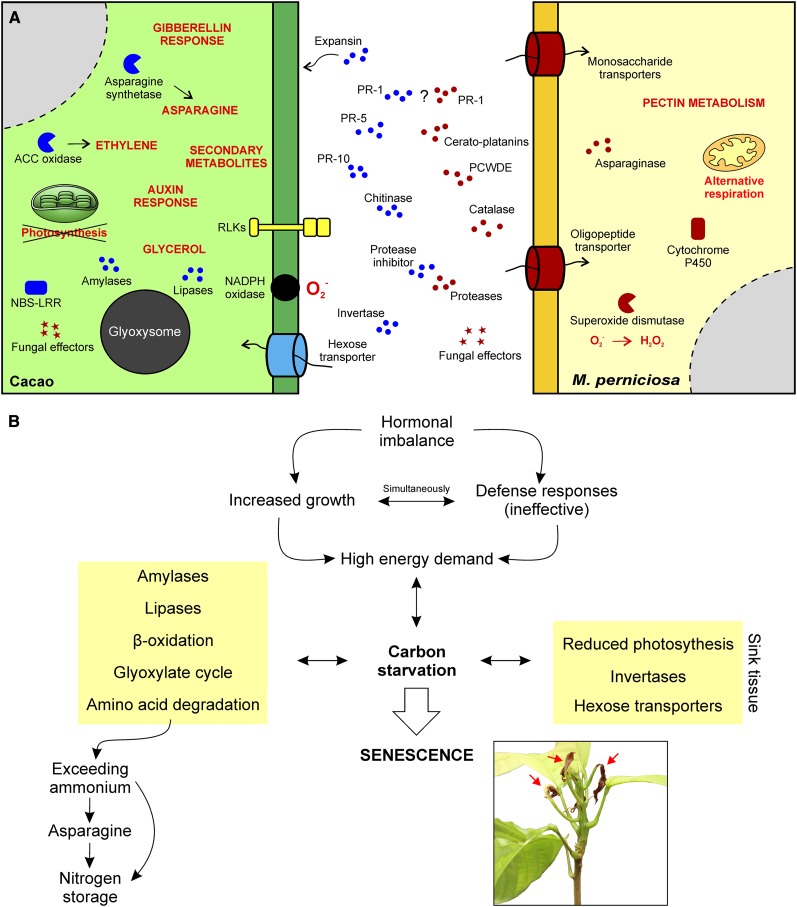
Plants and microorganisms constantly confront each other in a battle for growth and survival, and the outcome of such encounters directly interferes with human agricultural production. Dynamic and tightly regulated alterations in gene expression occur in both interacting organisms. In this regard, large-scale gene expression analyses of plant-pathogen interactions are of great relevance in unveiling the molecular basis of a specific disease (Wise et al., 2007). Recently, the term “dual RNA-seq” was coined to refer to the use of RNA-seq in transcriptomic analyses in which gene expression changes in both the pathogen and the host are analyzed simultaneously (Westermann et al., 2012). Whereas numerous studies have focused on either the pathogen or the host (Xu et al., 2011; Kunjeti et al., 2012; Petre et al., 2012; Weßling et al., 2012; Garnica et al., 2013; Link et al., 2014), only a few have involved the comprehensive analysis of both organisms (Kawahara et al., 2012; Tierney et al., 2012; Fernandez et al., 2012; Lowe et al., 2014; Yamagishi et al., 2014). Here, we present an in-depth transcriptomic analysis of the M. perniciosa-cacao interaction, which vastly expands our current knowledge of WBD and also contributes to our general understanding of plant-pathogen interactions. Based on these results, we present a model that describes the major molecular and physiological aspects of the devastating WBD (Figure 12).
Figure 12.
A Representative Model of the Biotrophic Interaction between Cacao and M. perniciosa.
(A) Representation of proteins and metabolic processes with important roles in WBD. Plant and fungal molecules are depicted in blue and red, respectively.
(B) A summary of the identified physiological alterations in green brooms that culminate in the establishment of a senescence process triggered by nutrient starvation. The plant in the inset shows the first signs of necrosis (arrows), which are similar to those of regular plant senescence.
High-Throughput RNA-seq Analysis as a Means of Identifying Fungal Transcripts That Are Specifically Expressed during Infection
Until now, the study of genes associated with M. perniciosa virulence was mainly restricted to extrapolations from in vitro studies. Using the power of RNA-seq, we applied a robust strategy to identify fungal transcripts that are important during the biotrophic interaction between M. perniciosa and cacao. This strategy is based on the analysis of a comprehensive database denominated WBD Transcriptome Atlas (www.lge.ibi.unicamp.br/wbdatlas), which is comprised of several transcriptomes representing different fungal growth conditions. Using this approach, we identified 433 fungal genes that are specifically upregulated in the green broom stage of WBD.
Of the ∼1 billion paired-end reads generated in this work, 1.7 million reads mapped to fungal gene models, which allowed the analysis of M. perniciosa genes in an unprecedented depth. Despite recent advances in the understanding of WBD, little is known about the transcriptional alterations of this pathogen during its interaction with cacao. Therefore, this study significantly deepens our knowledge on this organism, and the approach presented here might be applied to the study of other important plant-pathogen interactions, in particular, those involving nonmodel organisms that are poorly understood.
Since this work highlights a set of M. perniciosa genes that are likely involved in virulence, it constitutes a starting point for more specific studies. For instance, our results may serve as an initial filter in a structural genomics approach aiming to expand our knowledge of unknown proteins, including candidate effectors. More importantly, the advent of genome-editing methodologies (e.g., the CRISPR/Cas9-based system) paves the way for the genetic manipulation of many intractable organisms (Cong et al., 2013), such as M. perniciosa. In this regard, fungal genes that are highly and/or specifically expressed in green brooms are attractive candidates for further functional analyses.
A Catalog of the Potential Pathogenicity Factors of M. perniciosa
Plant pathogens possess a plethora of proteins that participate in many aspects of the disease process, including fungal penetration/colonization, host manipulation and microbial resistance. Based on our GO and InterPro enrichment analyses, we identified specific pathways and protein families/domains of M. perniciosa that are involved in these different facets of fungal parasitism (Figure 8B; Supplemental Data Set 10). Of particular interest, protein classes that have been associated with pathogenesis in other organisms are overrepresented in green brooms. Among them, we found proteins harboring the CAP domain (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1), which is a signature of plant PR-1 proteins. Although PR-1s are considered markers of plant defense against pathogen attack, the CAP domain is also found in many other organisms, including fungal pathogens (Gibbs et al., 2008; Cantacessi et al., 2009). In the M. perniciosa genome, the PR-1 gene family is expanded and contains 11 members, four of which are distinctively expressed during the biotrophic interaction with cacao (Teixeira et al., 2012). In addition, two other PR-1 genes are highly expressed in both green brooms and germinating basidiospores, supporting a role for these proteins in pathogenesis (Teixeira et al., 2012). Remarkably, CAP proteins recently emerged as potential virulence factors in fungal pathogens. In particular, depletion of CAP-encoding genes from the ascomycetes Fusarium oxysporum and Candida albicans leads to reduced virulence on animals (Braun et al., 2000; Prados-Rosales et al., 2012). Whereas the precise molecular function of these proteins during infection is still unknown, the Saccharomyces cerevisiae homologs were shown to bind sterols and protect the yeast from eugenol, a plant hydrophobic compound able to affect fungal membranes (Choudhary and Schneiter, 2012). Moreover, a CAP protein from humans (CRISP2) complemented the yeast mutant phenotype, indicating that the sterol binding activity is conserved among different CAP family members. Strikingly, we found that at least five of the 11 M. perniciosa PR-1s are also able to complement the yeast sterol binding phenotype (R. Schneiter, unpublished data). Therefore, a presumed role for the M. perniciosa PR-1 proteins is the detoxification of lipid toxins produced by cacao during infection and, hence, protection of the fungus.
Another class of proteins linked to microbial pathogenesis that is overrepresented in green brooms is the cerato-platanin (CP) protein family. In phytopathogenic fungi, CP proteins act as virulence factors or elicitors of plant defense responses (Pazzagli et al., 2014). The M. perniciosa genome holds 12 copies of CP-coding sequences, four of which are highly expressed in green brooms. Interestingly, the tridimensional structure of M. perniciosa CPs revealed binding sites for free chitin fragments, and these proteins are thought to act as scavengers of N-acetylglucosamine molecules released from the M. perniciosa cell wall (de O Barsottini et al., 2013). In this context, CP activity would interfere with pathogen recognition by plant receptors, disturbing cacao defense responses and favoring biotrophic colonization. Furthermore, at least four putative chitin deacetylases were overexpressed in green brooms (Supplemental Data Set 12). These enzymes catalyze the modification of chitin to chitosan and can mask/protect the fungal cell wall from hydrolytic enzymes produced by the host plant (e.g., chitinases) (El Gueddari et al., 2002). In this regard, like cerato-platanins, chitin deacetylases may impair chitin-induced immunity, thus promoting M. perniciosa virulence.
Green broom genes potentially involved in microbial pathogenesis also include five genes encoding CFEM-domain-containing proteins. CFEM is a cysteine-rich domain found specifically in fungi and described in proteins with proposed roles in pathogenesis (Kulkarni et al., 2003). For instance, this domain has already been described in pathogenicity factors of M. oryzae (DeZwaan et al., 1999) and in haustoria-expressed proteins of Melampsora spp (Joly et al., 2010). More recently, Zhang et al. (2012) showed that a CFEM gene is required for full virulence of Fusarium graminearum in wheat (Triticum aestivum). Nevertheless, CFEM proteins are also present in nonpathogenic species, which suggests that these proteins can play other roles in fungi in addition to pathogenesis.
M. perniciosa Biotrophic Infection Involves the Transcriptional Regulation of a Specific Set of Effector-Encoding Genes
The gene expression profile of cacao indicates that plant defenses are activated in green brooms (Figure 6; Supplemental Data Set 6). However, these defenses are not effective in preventing M. perniciosa biotrophic growth. Based on the definition proposed by Jones and Dangl (2006), the defense response observed in green brooms can be classified as a basal defense response, i.e., one that is activated by virulent pathogens on susceptible hosts. This weaker form of defense is usually a consequence of the activity of effector proteins deployed by the pathogen with the aim of manipulating host immunity and physiology, thus favoring the biotrophic colonization and the proper establishment of a compatible interaction.
To date, no effector has been functionally characterized in M. perniciosa, but inspection of the fungal genome led to the identification of 247 CSEPs. Our RNA-seq data showed that at least 33 of these CSEPs are distinctively expressed in the green broom stage (Figure 10). Moreover, we demonstrate that the majority of these CSEPs are among the most highly expressed fungal genes in green brooms (Table 2), whereas essentially no expression of these effectors is observed under in vitro conditions or in the necrotrophic stage of the disease (Figure 10). In agreement with these findings, high transcript levels in planta have been used as a filter to identify effectors in other filamentous pathogens (Pedersen et al., 2012; Guyon et al., 2014). This high abundance of effectors may allow these proteins to efficiently interact and interfere with their host targets. Collectively, these data provide substantial evidence that the 33 genes identified in our analysis encode real virulence effectors during the green broom stage of WBD. Other M. perniciosa CSEPs may play a role in other stages of the disease, as evidenced by their expression profile (Figure 10). For instance, a set of candidate effectors is strongly expressed in germinating spores. Remarkably, there is little overlap between the set of CSEPs expressed during the colonization of shoots (brooms) and fruits, indicating that particular plant organs require different transcriptional programs of the fungus. Indeed, tissue-specific transcription of virulence factors has been reported in other interactions, such as maize (Zea mays)-Ustilago maydis and rice (Oryza sativa)-M. oryzae (Marcel et al., 2010; Skibbe et al., 2010).
The continuous pressure imposed by the host immune system on effectors might lead to the rapid evolution of effector gene families. To gain insight into the evolutionary features of the set of M. perniciosa CSEPs, we performed a comparative analysis with M. roreri, a sister species of M. perniciosa that causes Frosty Pod Rot in cacao. Although most genes in these fungi exhibit a high degree of sequence conservation, the rate of sequence divergence (dN/dS) is significantly higher in CSEPs (Figure 11). Our comparative analysis revealed that 28 of the 33 green broom CSEPs have homologs in M. roreri. Interestingly, most of these genes (18) have multiple copies in at least one of the species (Figure 11), suggesting the occurrence of recent gene duplication events. Duplicated genes can rapidly evolve and acquire novel functions, which can help the pathogen evade the plant immune system and increase its fitness. Importantly, variations in the set of effectors of M. perniciosa and M. roreri may account for the differences in their infection strategies, in spite of their overall genomic similarity.
M. perniciosa Infection Induces Hormonal Imbalances in Cacao Tissues
The distinctive symptoms of WBD include intense growth of infected stems, hypertrophy and hyperplasia of tissues, and loss of apical dominance (Figure 1; Supplemental Figure 1). Although these characteristics have long been ascribed to the occurrence of hormonal imbalances, the precise contribution of each plant hormone to the development of green brooms was hitherto unclear. In this study, we verified that the transcription of cacao genes related to plant hormone signaling is noticeably altered in response to M. perniciosa infection (Figure 5; Supplemental Data Set 5). Gene expression data point to the existence of increased levels of gibberellins in green brooms, which is supported by the considerable stem elongation observed in infected plants (Figure 1). Additionally, auxin-responsive genes were upregulated, but no genes related to auxin biosynthesis were differentially expressed. Although the importance of auxin during WBD remains to be confirmed, the production of this hormone by M. perniciosa had already been described (Kilaru et al., 2007). Therefore, it is possible that the upregulation of auxin-responsive genes in infected cacao plants is a consequence of the biosynthesis of this hormone by M. perniciosa. Various phytopathogens are indeed known to produce auxin (e.g., Agrobacterium, Pseudomonas syringae, and U. maydis), and this hormone has been associated with the attenuation of plant defense responses mediated by salicylic acid, thus favoring biotrophic development (Navarro et al., 2006; Wang et al., 2007). Another remarkable finding is the upregulation of genes involved in the biosynthesis of and response to ethylene in green brooms (Figure 5; Supplemental Data Set 5). Indeed, increased concentrations of this hormone have been previously detected in infected cacao plants (Scarpari et al., 2005). Given that ethylene is usually involved in defense responses against necrotrophs (Glazebrook, 2005), it is possible that the successful establishment of WBD is favored by the development of inadequate plant defenses against M. perniciosa biotrophic hyphae.
Notably, we found that genes related to the degradation and inactivation of cytokinins are activated in green brooms. Increased expression of cytokinin oxidases (CKXs) is known to occur as a feedback response to the presence of cytokinins (Rashotte et al., 2003). Since we did not encounter any transcriptional evidence of cytokinin biosynthesis by the plant or the pathogen in green brooms, the upregulation of cacao CKXs is a possible response to the cytokinin produced at very specific sites or at earlier stages of infection. In agreement with this, Orchard et al. (1994) reported that zeatin riboside contents are high in the early stages of WBD infection (i.e., when the first symptoms become apparent) and decrease at later stages. Moreover, high expression of cacao CKXs was also identified in flower cushions infected by M. perniciosa (Melnick et al., 2012), which are characterized by the development of parthenocarpic fruits and the ectopic formation of vegetative shoots. Similar to other plant-pathogen interactions (Walters and McRoberts, 2006), manipulation of the cacao cytokinin pathway by M. perniciosa could favor nutrient mobilization toward the infection site and could explain the loss of apical dominance observed in WBD.
WBD Involves Significant Transcriptional Alterations in Carbon Metabolism and Triggers Premature Senescence in the Plant
During the prolonged biotrophic phase of WBD, cacao plants appear to expend large amounts of energy to sustain the exaggerated growth of infected tissues; indeed, we found that remarkable transcriptional alterations related to plant carbohydrate metabolism take place in green brooms. Genes encoding a cell wall invertase, which breaks sucrose into fructose and glucose, and hexose transporters are upregulated in cacao infected with WBD. In addition, the transcription of genes related to photosynthesis is reduced (Figure 4; Supplemental Data Set 4). These characteristics are typical of developing sink tissues, which reallocate nutrients from other plant organs to sustain their own growth and development (Berger et al., 2007). Since M. perniciosa can only infect meristematic tissues, which are generally sinks, it is likely that the fungus reprograms the regular development of cacao meristems to prevent the transition from sink to source tissues. Similarly, other pathogens also induce the formation of local sinks in the diseased parts, impairing plant photosynthesis and increasing the activity of invertases and hexose transporters (Chou et al., 2000; Fotopoulos et al., 2003; Bonfig et al., 2006; Deeken et al., 2006; Horst et al., 2008; Chandran et al., 2010). Although WBD severely impacts the development of the infected parts and significantly reduces the number of fruits per tree, the disease is not lethal to the whole cacao tree. It is possible that the decrease in tree productivity is, at least in part, a consequence of nutrient allocation to the infected parts.
In comparison to healthy tissues, green brooms have decreased starch contents (Figure 1); accordingly, amylase-encoding genes are overexpressed, thus indicating that this storage carbohydrate may be used as an energy source. Moreover, the higher number of transcripts related to lipases, β-oxidation, and glyoxylate cycle enzymes indicates that lipids are also metabolized as a source of energy by the infected tissue. A subproduct of lipid catabolism is glycerol, which was previously found to occur at higher concentrations in infected cacao plants (Scarpari et al., 2005). Remarkably, the glyoxylate cycle is associated with two major developmental processes in plants, i.e., seed germination and senescence. During germination, lipid reserves are used to fuel the growth and development of the growing embryo (Graham, 2008). In addition, glyoxysomal activity is involved in the nutrient recycling of senescent tissues (Buchanan-Wollaston, 1997). Therefore, the expression of genes encoding enzymes of the glyoxylate cycle in green brooms is in line with the significant metabolic reprogramming that takes place during WBD progression.
Along with carbohydrates and lipids, the carbon skeleton of amino acids seems to be used to sustain green broom development and metabolism. However, the use of amino acids increases ammonium levels in cells, which can be toxic to the plant. To handle ammonium toxicity, green brooms seem to employ at least two different strategies: (1) the expression of a gene encoding a tonoplast transporter (tonoplast intrinsic protein [TIP]), which mediates ammonia storage in vacuoles (Loqué et al., 2005), and (2) the expression of an asparagine synthetase (ASN), which is homologous to the Asn from Arabidopsis and mediates the incorporation of nitrogen into aspartate to produce asparagine (Lam et al., 1994). This amino acid is an optimal nitrogen transport and reserve molecule due to its high nitrogen/carbon ratio and stability. Therefore, its production is typically associated with situations of low carbon availability (Lam et al., 1994). Supporting our findings, increased amounts of asparagine were previously reported in green brooms (Scarpari et al., 2005). Interestingly, Asn is also called Din6 (Dark Inducible gene 6), and its expression is induced in senescing tissues and after treatment with exogenous photosynthesis inhibitors (Fujiki et al., 2001).
Altogether, these results indicate that infected tissues appear to have a paucity of carbon skeletons and thus use alternative sources (i.e., starch, lipids, and amino acids) to obtain energy. However, considering that green brooms appear to develop as nutrient sinks, the carbon deprivation signature in infected tissues is unexpected. In fact, with the progression of WBD, a callus-like structure is formed on the basal region of the infected growing shoot (Supplemental Figure 6). This structure was shown to disrupt the communication between healthy and infected parts of the plant, intercepting the translocation of nutrients to the diseased shoot (J.G. Barau, unpublished data). Consequently, infected tissues are isolated from the plant, being forced to use their own stores and structures for energy production. As a result, carbon deprivation might occur, culminating in the onset of a senescence process triggered by nutrient starvation. Therefore, senescence, which is a developmentally regulated and highly controlled process that leads to the death of the plant tissue, appears to be responsible for the initial death of infected parts during WBD development.
Indeed, a typical characteristic of WBD is the death of infected tissues beginning on the edges of leaves (Figure 12; Supplemental Figures 1 and 6), which is similar to the pattern of foliar senescence (Gan and Amasino, 1997). Also, Ceita et al. (2007) showed that programmed cell death precedes the necrotrophic stage in WBD. Therefore, it is quite possible that the onset of necrosis in WBD is a physiological process of cacao caused by the metabolic disarrangement of WBD rather than a direct action of the pathogen. Consistent with this idea, M. perniciosa NEP (NECROSIS AND ETHYLENE-INDUCING PROTEIN) genes, which are involved in the necrosis of cacao tissues, are not expressed when the first signs of death are observed in infected plants (Zaparoli et al., 2011). However, as necrosis progresses, the fungus expresses its NEP2 gene, thus actively contributing to the death of the cacao tissue. A pioneer report by Evans (1980) suggested two possible causes for the death of infected tissues in WBD: (1) accelerated host senescence or (2) the action of fungal toxins. Interestingly, our results provide evidence that these two processes might occur consecutively in the disease.
The Peculiar Hemibiotrophic Life Cycle of M. perniciosa in Cacao
WBD is a complex tropical disease, resulting from the interaction between a perennial tree and a fungal pathogen with a peculiar hemibiotrophic life cycle. Usually, plant diseases caused by hemibiotrophic fungi involve a transient and asymptomatic biotrophic stage followed by destructive plant necrosis. However, the hemibiotrophic life cycle of M. perniciosa is characterized by a long-lasting and symptomatic biotrophic phase, which can endure for more than 60 d in the living cacao tissues. Remarkably, it can be even longer than the typical life cycles of obligate and nonobligate biotrophs.
To feed on the host tissues, some hemibiotrophs (e.g., M. oryzae and Colletotrichum graminicola) invade a few plant cells and employ nutrient-absorbing structures (e.g., haustorium and invasive hyphae), whereas others, such as M. perniciosa, grow intercellularly and depend on nutrients derived from the plant apoplast. Once living in the apoplast for a prolonged period (30 to 60 d), M. perniciosa seems to manipulate the host metabolism to increase nutrient availability. Therefore, the maintenance of the sink capacity of infected branches and the formation of the vigorous green broom structure appear to favor the availability and acquisition of nutrients by the fungus. In agreement with this, fungal genes encoding oligopeptide and monosaccharide transporters, proteases, and an asparaginase are highly induced in green brooms (Supplemental Data Set 12). Furthermore, M. perniciosa colonizes the middle lamella, which is highly rich in the polysaccharide pectin (Albersheim et al., 1960). Given that many genes associated with pectinolytic metabolism are upregulated in green brooms (Supplemental Data Set 12), M. perniciosa might have a large capacity to degrade plant pectin during biotrophic colonization. Although high pectinolytic activity is usually associated with necrotrophic pathogens (e.g., Botrytis cinerea), genes associated with pectin degradation have undergone expansion in the genome of the biotrophic fungus Cladosporium fulvum compared with other biotrophs (de Wit et al., 2012). Similarly to M. perniciosa, C. fulvum inhabits the intercellular space of its host (i.e., tomato); thus, a high pectinolytic activity may facilitate its colonization and plant infection. Interestingly, a by-product of pectin catabolism is methanol, and a recent report by de Oliveira et al. (2012) showed that M. perniciosa is able to grow on methanol as the sole carbon source, even in elevated concentrations of this alcohol. Therefore, we reason that methanol might be an optional carbon source for M. perniciosa during the green broom stage of WBD.
As mentioned above, the gene expression profile of cacao indicates that plant defenses, albeit ineffective, are activated in green brooms (Figure 6; Supplemental Data Set 6). Therefore, even during a very prolonged biotrophic stage, M. perniciosa does not seem to fully suppress plant defenses as is observed for many biotrophic pathogens (Wäspi et al., 2001; Panstruga, 2003; Caldo et al., 2006). For instance, maize infection by the smut pathogen U. maydis causes a transient induction of plant defense mechanisms (e.g., upregulation of genes encoding chitinases, glucanases, and PR proteins), which is strongly suppressed a few hours after the infection began (Doehlemann et al., 2008). A similar virulence strategy is also adopted by the hemibiotroph Mycosphaerella graminicola (Adhikari et al., 2007). Conversely, analogous to M. perniciosa, the hemibiotrophic fungi C. graminicola and M. oryzae do not fully suppress plant defenses as a virulence strategy during plant infection (Marcel et al., 2010; Vargas et al., 2012). These pathogens display a very rapid biotrophic phase (that lasts from hours to a few days), in which a progressive increase in the expression of plant defense genes occurs. Remarkably, these genes will later contribute to the onset of necrotrophic development and death of plant tissues.
In response to the upregulation of plant defense genes in green brooms, M. perniciosa holds an arsenal of genes encoding protective enzymes. Several genes encoding enzymes involved in the detoxification of host toxins (e.g., efflux pump transporters and cytochrome P450s) and protection against oxidative stress (e.g., superoxide dismutase, glutathione synthetase, catalase, and peroxiredoxin) are highly expressed, supporting the hypothesis that M. perniciosa deals with a hostile environment during infection. In agreement with this notion, Thomazella et al. (2012) reported that an alternative respiratory pathway mediated by the enzyme alternative oxidase is activated as a protective mechanism of M. perniciosa biotrophic hyphae against the plant oxidative environment. Moreover, other studies have already suggested that ROS are produced during the compatible interaction between cacao and M. perniciosa (Scarpari et al., 2005; Ceita et al., 2007; Dias et al., 2011). Supporting these findings, our transcriptomic data showed that a plant NADPH oxidase gene is overexpressed in green brooms (Figure 6; Supplemental Data Set 6). The enzyme NADPH oxidase produces the free radical anion superoxide and contributes to the generation of oxidative stress in response to pathogen invasion.
As discussed above, after this long-lasting interplay between M. perniciosa biotrophic hyphae and cacao cells, the infected tissues collapse and a senescence-like process is installed. However, plant death seems to favor M. perniciosa, which switches to its necrotrophic stage. Following the first signs of cacao senescence, the fungus expresses NEP2 and actively kills the remaining cacao tissues. As a consequence, soluble nutrients derived from dead host cells become available to the necrotrophic hyphae, which later produce basidiomata, thus completing the disease cycle. It is noteworthy that the M. perniciosa NEP genes were acquired by horizontal gene transfer from oomycete species, possibly from other cacao pathogens of the Phytophthora genus that coexist with M. perniciosa in its natural environment (Tiburcio et al., 2010). Moreover, M. perniciosa and its sister species M. roreri are the only pathogenic basidiomycetes in which NEP genes have been described (Supplemental Table 3). Therefore, it is plausible that the acquisition of NEP genes might have conferred or at least improved the ability of M. perniciosa to cause WBD in cacao. Overall, the findings and peculiarities reported for the intriguing hemibiotrophic interaction between M. perniciosa and cacao make WBD a very attractive and innovative model for further in-depth studies in the field of plant pathology.
Conclusions
Recently, the genome sequences of both M. perniciosa (Mondego et al., 2008) and T. cacao (Argout et al., 2011; Motamayor et al., 2013) were published, significantly opening the possibility for more detailed molecular studies of these organisms. Here, we explore the molecular mechanisms of WBD and present novel resources for the study of this important disease that threatens the world’s cacao production. This work also provides novel perspectives and establishes bases for questions that should be explored in future research. For instance, it will be important to determine how biotrophy is maintained for so long in M. perniciosa and how cacao senescence signals the end of this fungal stage. Given that nutrient availability and stress conditions seem to be tightly associated with M. perniciosa virulence and development (Alvim et al., 2009; Pungartnik et al., 2009; Thomazella et al., 2012; Argôlo Santos Carvalho et al., 2013), cacao nutritional status (e.g., carbon starvation) and/or specific plant molecules (e.g., ROS) possibly orchestrate WBD infection. It will be relevant to establish to what extent and how the alterations identified in cacao metabolism/physiology correlate with direct fungal action. In this regard, the study of specific pathogen proteins, particularly candidate effectors, will shed light on the molecular mechanisms that M. perniciosa uses to manipulate the host and favor virulence. Considering that hormonal imbalance is a major feature in WBD development, understanding how M. perniciosa interferes with the plant hormonal pathways throughout disease progression is also a pertinent topic for future research. Overall, the results presented in this work establish an unprecedented model for this peculiar plant disease and provide a robust basis for the study of an important tropical crop.
METHODS
Biological Material
Theobroma cacao cv ‘Comum’ (Forastero genotype) and the Moniliophthora perniciosa isolate BP10 were used in this study. Cacao seedlings were grown for∼3 months in a greenhouse under controlled temperature (26°C) and humidity (>80%) conditions and a photoperiod of 12 h. Active apical meristems were inoculated with 30 μL of a basidiospore suspension (105 spores mL−1), following the procedures described by Frias et al. (1995). After 30 d, infected tissues representing the mature green broom stage were excised and used for RNA isolation. As controls, the corresponding tissues of mock-inoculated seedlings were collected at the same time point. Five biological replicates were used for each condition. Tissues harvested for gene expression analysis are shown in Supplemental Figure 2.
RNA Isolation and RNA-seq Library Preparation
RNA isolation was performed as described by Azevedo et al. (2003) with minor modifications. For each sample, 5 μg total RNA was used to prepare the mRNA-seq library according to the TrueSeq RNA Sample Prep Kit protocol (Illumina). Library quality control and quantification were performed with an Experion DNA 1K Chip (Bio-Rad) and a Qubit fluorometer (Invitrogen), respectively. For each library, ∼75 million 48-bp paired-end sequences were generated using an Illumina HiSeq 2000 sequencer.
Read Mapping and Gene Expression Analysis
After removing low-quality sequences containing uncalled bases (Ns), we used the software Bowtie (Langmead et al., 2009) to align the RNA-seq reads against 34,997 and 17,008 gene models of T. cacao and M. perniciosa, respectively. Cacao gene models were downloaded from the Cacao Genome Database (version 0.9, www.cacaogenomedb.org; Motamayor et al., 2013), and M. perniciosa gene models were obtained from the Witches’ Broom Genome Project (www.lge.ibi.unicamp.br/vassoura). Bowtie alignment parameters were set to allow a maximum of two mismatches and to exclude reads mapping to more than one position on the reference. Moreover, only reads for which both pairs successfully aligned were considered. Differential expression analyses were performed for cacao genes with the edgeR package (Robinson et al., 2010) using the Benjamini-Hochberg method for correction of multiple comparisons. To filter out weakly expressed genes, only those genes with a minimum expression level of 1 count per million in at least five of the 10 libraries were included in the analysis. Genes with a FDR of below 0.01 were considered differentially expressed between conditions. To assess the variability among samples, we performed hierarchical clustering and PCA using cacao genes. Hierarchical clustering was performed based on Euclidean distances. PCA was conducted using the prcomp command with default parameters in the R software package. Reads that mapped to the fungal reference were used to define expression values for M. perniciosa genes according to the RPKM method (Mortazavi et al., 2008). Hierarchical clustering of fungal genes was performed with the GenePattern software (Reich et al., 2006) using RPKM values calculated in green brooms and in 33 additional RNA-seq libraries from the WBD Transcriptome Atlas. Genes were clustered using the Pearson correlation distance measure and the pairwise average-linkage method. Expression values were normalized for each gene, and data were centered by subtracting the mean expression value of all conditions. Data were visualized using the Gene-E tool (http://www.broadinstitute.org/cancer/software/GENE-E/).
Functional Classification Based on MapMan, Gene Ontology, and InterPro Terms
MapMan software (Thimm et al., 2004) was used to visualize cacao gene expression data in the context of metabolic pathways. MapMan uses a plant-specific ontology that classifies genes into well defined hierarchical categories, denominated BINs. Cacao genes were assigned to BINs using the Mercator automated annotation pipeline (http://mapman.gabipd.org/web/guest/mercator). Differentially represented MapMan pathways were defined by a two-tailed Wilcoxon rank sum test corrected by the Benjamin-Hochberg method (FDR ≤ 0.05). Additionally, enriched InterPro and GO terms were identified using the BiNGO plugin for Cytoscape (Maere et al., 2005). To this purpose, a customized annotation file was created according to the software instructions to allow the use of InterPro terms in enrichment analyses. A hypergeometric test combined with a Benjamin and Hochberg FDR correction with a cutoff of 0.05 was applied.
HORMONOMETER Analysis
HORMONOMETER software (Volodarsky et al., 2009) was used to evaluate transcriptional similarities between green brooms and hormone responses. Since this software only accepts Arabidopsis thaliana gene IDs as input, we defined the putative Arabidopsis orthologs of cacao genes by bidirectional BLAST analyses. Bidirectional best hits having an e-value ≤ 1e-5 in both directions were considered orthologous gene pairs. The 8870 resulting genes expressed in our experiment were then used to investigate the hormonal profile of green brooms. To validate the analysis with this set of genes, we used public gene expression data of Arabidopsis mutants with known hormonal alterations: arr21 (increased cytokinin response; GEO, GSE5699), eto1 (increased ethylene response; GEO, GSE20897), nahG (reduced salicylic acid levels; GEO, GSE5727), and nph arf19/IAA (reduced auxin signaling; GEO, GSE627). We also evaluated transcriptomes of Arabidopsis interacting with other organisms: Agrobacterium tumefaciens (6 d after infection by strain C58; GSE14106), Phytophthora infestans (24 h after infection; GSE5616), Alternaria brassicicola (24 h after infection; GSE50526), and the leaf miner insect Liriomyza huidobrensis (locally damaged leaves, GSE38281).
Comparing RNA-seq and qPCR Results
qPCR assays were performed to confirm RNA-seq results with an independent technique. To this end, the expression of 28 cacao genes was analyzed in plants that were also sequenced by RNA-seq (infected plant I2 and healthy plant H1, Table 1). Additionally, 40 M. perniciosa genes were evaluated in seven different conditions (monokaryotic mycelium, dikaryotic mycelium, basidiomata, spores, germinating spores, green broom, and dry broom) to validate the results from the WBD Transcriptome Atlas. qPCRs were conducted on a StepOne Plus Real-Time PCR system (Applied Biosystems) using SYBR Green I for the detection of PCR products. Each reaction was performed in a final volume of 16 μL, containing 8 μL SYBR Green PCR Master Mix (Applied Biosystems), 250 nM each primer, and 50 ng cDNA template. No-template reactions were included as negative controls for each set of primers used. The thermal cycling conditions were 94°C for 10 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 60°C for 1 min, with fluorescence detection at the end of each cycle. The amplification of a single product per reaction was confirmed by melting curve analysis. All reactions were performed in technical triplicates. Relative fold differences in mRNA abundance were defined by the mathematical model described by Livak and Schmittgen (2001). Cacao data were normalized using four reference genes that showed little variation in the RNA-seq analysis (α-tubulin, actin, 60S ribosomal L35, and 60S ribosomal L11). Expression levels of M. perniciosa genes are given in relation to the fungal β-actin and IF3b (transcription initiation factor). Primers used in these experiments are listed in Supplemental Tables 1 and 2.
Determination of Photosynthetic Rates
Photosynthetic rates were determined using the LI-6400 portable gas exchange system (LI-COR). Measurements in infected and healthy cacao plants were performed in biological triplicates on the most recent fully expanded stem leaf. The CO2 assimilation rate was measured at 25°C under light intensity ranging from 0 to 900 μmol photons m−2 s−1. The CO2 concentration of the air entering the chamber was maintained at 350 μmol mol−1. Plants analyzed in this experiment were similar to those used for transcriptome sequencing.
Histological Analyses
Green brooms and the corresponding tissues of healthy plants were fixed in Karnovsky fixative (50 mL 1% glutaraldehyde, 20 mL 4% paraformaldehyde, and 30 mL 0.2 M phosphate buffer, pH 7.2; Karnovsky, 1965). Samples were left in the fixative solution for 48 h, washed, and stored in 70% ethanol. After dehydration using a graded ethanol series (30, 50, 70, 90, and 100%), tissues were embedded in plastic resin (Leica Historesin; Gerrits and Smid, 1983), and transverse sections (12- to 16-μm thick) were cut with a steel knife. Sections were then incubated in a toluidine blue solution (0.01%, dissolved in citrate buffer; O' Brien et al., 1964). Samples were observed under white and polarized lights using an Olympus BX51 microscope coupled to an Olympus DP72 camera. All analyses were repeated at least three times with consistent results.
Evolutionary Analysis of CSEPs
To evaluate the dN/dS ratio of orthologous genes, all predicted proteins from M. perniciosa, and from its sister species Moniliophthora roreri (Meinhardt et al., 2014), were clustered into homologous families using OrthoMCL (Li et al., 2003) with a minimum BLAST e-value of 1E-5 and inflation index of 1.5. Genes were considered orthologs if clustered in families containing only one copy from each species. Genes clustered in families with more than one member from each species were considered paralogs.
Each pair of orthologous genes was aligned using MACSE (Ranwez et al., 2011), and the dN/dS ratio was estimated by maximum likelihood using the codon-based model of Goldman and Yang (1994) implemented in the codeml program of the PAML 4 package (Yang, 2007). The divergence among M. perniciosa, M. roreri, and their most recent common ancestor was considered equal in the codeml analysis. The dN/dS ratios were assessed separately for all orthologs, including non-CSEPs, CSEPs, and green-broom-specific CSEPs. Differential dN/dS rates among these groups were tested statistically using the hypergeometric distribution.
Accession Numbers
Raw sequencing data are available at the NCBI Sequence Read Archive under accession number SRA066232.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Detailed View of WBD Progression and the Major Symptoms Observed in Cacao Plants during Disease Development.
Supplemental Figure 2. Plant Tissue Harvested for Gene Expression Analysis.
Supplemental Figure 3. M. perniciosa Is Uniformly Distributed along the Infected Shoot.
Supplemental Figure 4. Number of M. perniciosa Genes Detected at Different Sequencing Depths.
Supplemental Figure 5. Distribution of Expression Levels of M. perniciosa Genes in Green Brooms.
Supplemental Figure 6. Callus-Like Structure Formed on the Basal Region of Infected Shoots.
Supplemental Table 1. Primer Sequences Used for Cacao qPCR Analyses.
Supplemental Table 2. Primer Sequences Used for M. perniciosa qPCR Analyses.
Supplemental Table 3. Identification of NEP Genes in Basidiomycetes.
Supplemental Data Set 1. Expression Analysis of Cacao Genes Based on edgeR Software.
Supplemental Data Set 2. MapMan BINs Enriched in Green Brooms.
Supplemental Data Set 3. InterPro Terms Enriched in Green Brooms.
Supplemental Data Set 4. Differentially Expressed Cacao Genes Related to Photosynthetic, Carbon, and Nitrogen Metabolism.
Supplemental Data Set 5. Differentially Expressed Cacao Genes Related to Hormonal Metabolism.
Supplemental Data Set 6. Differentially Expressed Cacao Genes Related to Defense Responses.
Supplemental Data Set 7. Differentially Expressed Cacao Genes Related to Secondary Metabolism and Cell Wall Modification.
Supplemental Data Set 8. Expression Values Defined for M. perniciosa Genes during Cacao Infection.
Supplemental Data Set 9. qPCR Validation of the WBD Transcriptome Atlas.
Supplemental Data Set 10. M. perniciosa Genes Distinctively Expressed in Green Brooms.
Supplemental Data Set 11. InterPro and GO Terms Enriched in Green Broom-Specific Genes.
Supplemental Data Set 12. Selected M. perniciosa Genes Thought to Be Involved in Cacao Infection.
Supplemental Data Set 13. List of M. perniciosa CSEP-Encoding Genes.
Supplementary Material
Acknowledgments
We thank Halley Caixeta de Oliveira and Li Yang for valuable suggestions and critical reading of the article, Marc Lohse for his assistance with the MapMan annotation, Ramon Vidal for assistance with the bioinformatics, and Ricardo Silverio Machado for helping with the photosynthesis measurements. We thank the University of North Carolina High Throughput Sequencing Facility team for intense support with RNA-seq. This work was supported by the São Paulo Research Foundation (FAPESP, Grants 2009/51018-1, 2009/50119-9, 2006/59843-3, 2008/54527, and 2012/09136-0).
AUTHOR CONTRIBUTIONS
P.J.P.L.T. designed the research. P.J.P.L.T., P.F.V.P., M.C.S.R., and G.L.F. performed experimental procedures. O.R., G.G.L.C., and J.J. performed bioinformatics analyses. P.M. contributed reagents/materials/analysis tools. P.J.P.L.T. and D.P.T.T. interpreted the data. J.M.C.M. participated in the analysis of fungal genes, PR genes, and plant receptors. P.J.P.L.T. and D.P.T.T. wrote the article. G.A.G.P. supervised the project.
Glossary
- WBD
witches’ broom disease
- qPCR
quantitative real-time PCR
- PCA
principal component analysis
- FDR
false discovery rate
- RPKM
reads per kilobase per million mapped reads
- CSEP
candidate secreted effector protein
- GO
Gene Ontology
- ROS
reactive oxygen species
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adhikari T., Balaji B., Breeden J., Goodwin S. (2007). Resistance of wheat to Mycosphaerella graminicola involves early and late peaks of gene expression. Physiol. Mol. Plant Pathol. 71: 55–68. [Google Scholar]
- Aime M.C., Phillips-Mora W. (2005). The causal agents of witches’ broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 97: 1012–1022. [DOI] [PubMed] [Google Scholar]
- Albersheim P., Muhlethaler K., Frey-Wyssling A. (1960). Stained pectin as seen in the electron microscope. J. Biophys. Biochem. Cytol. 8: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim F.C., Mattos E.M., Pirovani C.P., Gramacho K., Pungartnik C., Brendel M., Cascardo J.C., Vincentz M. (2009). Carbon source-induced changes in the physiology of the cacao pathogen Moniliophthora perniciosa (Basidiomycetes) affect mycelial morphology and secretion of necrosis-inducing proteins. Genet. Mol. Res. 8: 1035–1050. [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Argôlo Santos Carvalho H., de Andrade Silva E.M., Carvalho Santos S., Micheli F. (2013). Polygalacturonases from Moniliophthora perniciosa are regulated by fermentable carbon sources and possible post-translational modifications. Fungal Genet. Biol. 60: 110–121. [DOI] [PubMed] [Google Scholar]
- Argout X., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43: 101–108. [DOI] [PubMed] [Google Scholar]
- Azevedo H., Lino-Neto T., Tavares R.M. (2003). An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol. Biol. Rep. 21: 333–338. [Google Scholar]
- Berger S., Sinha A.K., Roitsch T. (2007). Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 58: 4019–4026. [DOI] [PubMed] [Google Scholar]
- Bonfig K.B., Schreiber U., Gabler A., Roitsch T., Berger S. (2006). Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta 225: 1–12. [DOI] [PubMed] [Google Scholar]
- Braun B.R., Head W.S., Wang M.X., Johnson A.D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. (1997). The molecular biology of leaf senescence. J. Exp. Bot. 48: 181–199. [Google Scholar]
- Caldo R.A., Nettleton D., Peng J., Wise R.P. (2006). Stage-specific suppression of basal defense discriminates barley plants containing fast- and delayed-acting Mla powdery mildew resistance alleles. Mol. Plant Microbe Interact. 19: 939–947. [DOI] [PubMed] [Google Scholar]
- Cantacessi C., Campbell B.E., Visser A., Geldhof P., Nolan M.J., Nisbet A.J., Matthews J.B., Loukas A., Hofmann A., Otranto D., Sternberg P.W., Gasser R.B. (2009). A portrait of the “SCP/TAPS” proteins of eukaryotes—developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27: 376–388. [DOI] [PubMed] [Google Scholar]
- Ceita G.D., et al. (2007). Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora pemiciosa. Plant Sci. 173: 106–117. [Google Scholar]
- Chandran D., Inada N., Hather G., Kleindt C.K., Wildermuth M.C. (2010). Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc. Natl. Acad. Sci. USA 107: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.M., Bundock N., Rolfe S.A., Scholes J.D. (2000). Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Mol. Plant Pathol. 1: 99–113. [DOI] [PubMed] [Google Scholar]
- Choudhary V., Schneiter R. (2012). Pathogen-Related Yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. USA 109: 16882–16887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.J., Mylonakis E. (2009). Efflux in fungi: la pièce de résistance. PLoS Pathog. 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Hora Junior B.T., Poloni Jde.F., Lopes M.A., Dias C.V., Gramacho K.P., Schuster I., Sabau X., Cascardo J.C., Mauro S.M., Gesteira Ada.S., Bonatto D., Micheli F. (2012). Transcriptomics and systems biology analysis in identification of specific pathways involved in cacao resistance and susceptibility to witches’ broom disease. Mol. Biosyst. 8: 1507–1519. [DOI] [PubMed] [Google Scholar]
- de O Barsottini M.R., et al. (2013). Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol. Plant Microbe Interact. 26: 1281–1293. [DOI] [PubMed] [Google Scholar]
- de Oliveira B.V., Teixeira G.S., Reis O., Barau J.G., Teixeira P.J., do Rio M.C., Domingues R.R., Meinhardt L.W., Paes Leme A.F., Rincones J., Pereira G.A. (2012). A potential role for an extracellular methanol oxidase secreted by Moniliophthora perniciosa in Witches’ broom disease in cacao. Fungal Genet. Biol. 49: 922–932. [DOI] [PubMed] [Google Scholar]
- de Wit P.J., et al. (2012). The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8: e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R., Engelmann J.C., Efetova M., Czirjak T., Müller T., Kaiser W.M., Tietz O., Krischke M., Mueller M.J., Palme K., Dandekar T., Hedrich R. (2006). An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18: 3617–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. (1999). Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C.V., Mendes J.S., dos Santos A.C., Pirovani C.P., da Silva Gesteira A., Micheli F., Gramacho K.P., Hammerstone J., Mazzafera P., de Mattos Cascardo J.C. (2011). Hydrogen peroxide formation in cacao tissues infected by the hemibiotrophic fungus Moniliophthora perniciosa. Plant Physiol. Biochem. 49: 917–922. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kühnemann J., Sonnewald U., Kahmann R., Kämper J. (2008). Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56: 181–195. [DOI] [PubMed] [Google Scholar]
- El Gueddari N.E., Rauchhaus U., Moerschbacher B.M., Deising H.B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156: 103–112. [Google Scholar]
- Evans H.C. (1980). Pleomorphism in Crinipellis perniciosa, causal agent of Witches' broom disease of cocoa. Trans. Br. Mycol. Soc. 74: 515–526. [Google Scholar]
- Fernandez D., Tisserant E., Talhinhas P., Azinheira H., Vieira A., Petitot A.S., Loureiro A., Poulain J., Da Silva C., Silva Mdo.C., Duplessis S. (2012). 454-pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta-expressed pathogen-secreted proteins and plant functions in a late compatible plant-rust interaction. Mol. Plant Pathol. 13: 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.C., Henk D.A., Briggs C.J., Brownstein J.S., Madoff L.C., McCraw S.L., Gurr S.J. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotopoulos V., Gilbert M.J., Pittman J.K., Marvier A.C., Buchanan A.J., Sauer N., Hall J.L., Williams L.E. (2003). The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atbetafruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol. 132: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias G., Purdy L.H., Schmidt R.A. (1995). An inoculation method for evaluating resistence of cacao to Crinipellis perniciosa. Plant Dis. 79: 787–791. [Google Scholar]
- Fujiki Y., Yoshikawa Y., Sato T., Inada N., Ito M., Nishida I., Watanabe A. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111: 345–352. [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica D.P., Upadhyaya N.M., Dodds P.N., Rathjen J.P. (2013). Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS ONE 8: e67150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits P.O., Smid L. (1983). A new, less toxic polymerization system for the embedding of soft tissues in glycol methacrylate and subsequent preparing of serial sections. J. Microsc. 132: 81–85. [DOI] [PubMed] [Google Scholar]
- Gesteira A.S., Micheli F., Carels N., Da Silva A.C., Gramacho K.P., Schuster I., Macêdo J.N., Pereira G.A., Cascardo J.C. (2007). Comparative analysis of expressed genes from cacao meristems infected by Moniliophthora perniciosa. Ann. Bot. (Lond.) 100: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs G.M., Roelants K., O’Bryan M.K. (2008). The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr. Rev. 29: 865–897. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Godfrey D., Böhlenius H., Pedersen C., Zhang Z., Emmersen J., Thordal-Christensen H. (2010). Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N., Yang Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- Griffith G.W., Nicholson J., Nenninger A., Birch R.N., Hedger J.N. (2003). Witches' brooms and frosty pods: two major pathogens of cacao. N.Z. J. Bot. 41: 423–435. [Google Scholar]
- Guyon K., Balagué C., Roby D., Raffaele S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R.J., Engelsdorf T., Sonnewald U., Voll L.M. (2008). Infection of maize leaves with Ustilago maydis prevents establishment of C4 photosynthesis. J. Plant Physiol. 165: 19–28. [DOI] [PubMed] [Google Scholar]
- Joly D.L., Feau N., Tanguay P., Hamelin R.C. (2010). Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.). BMC Genomics 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295. [DOI] [PubMed] [Google Scholar]
- Karnovsky M.J. (1965). A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27: 137–139. [Google Scholar]
- Kawahara Y., Oono Y., Kanamori H., Matsumoto T., Itoh T., Minami E. (2012). Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 7: e49423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E., Gardiner A., Schultz-Larsen T., Kemen A.C., Balmuth A.L., Robert-Seilaniantz A., Bailey K., Holub E., Studholme D.J., Maclean D., Jones J.D. (2011). Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru A., Bailey B.A., Hasenstein K.H. (2007). Moniliophthora perniciosa produces hormones and alters endogenous auxin and salicylic acid in infected cocoa leaves. FEMS Microbiol. Lett. 274: 238–244. [DOI] [PubMed] [Google Scholar]
- Kulkarni R.D., Kelkar H.S., Dean R.A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28: 118–121. [DOI] [PubMed] [Google Scholar]
- Kunjeti S.G., Evans T.A., Marsh A.G., Gregory N.F., Kunjeti S., Meyers B.C., Kalavacharla V.S., Donofrio N.M. (2012). RNA-Seq reveals infection-related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 13: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.M., Peng S.S., Coruzzi G.M. (1994). Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 106: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal G.A. Jr., Albuquerque P.S., Figueira A. (2007). Genes differentially expressed in Theobroma cacao associated with resistance to witches’ broom disease caused by Crinipellis perniciosa. Mol. Plant Pathol. 8: 279–292. [DOI] [PubMed] [Google Scholar]
- Leal G.A., Gomes L.H., Albuquerque P.S., Tavares F.C., Figueira A. (2010). Searching for Moniliophthora perniciosa pathogenicity genes. Fungal Biol. 114: 842–854. [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link T.I., Lang P., Scheffler B.E., Duke M.V., Graham M.A., Cooper B., Tucker M.L., van de Mortel M., Voegele R.T., Mendgen K., Baum T.J., Whitham S.A. (2014). The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Mol. Plant Pathol. 15: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loqué D., Ludewig U., Yuan L., von Wirén N. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R.G., Cassin A., Grandaubert J., Clark B.L., Van de Wouw A.P., Rouxel T., Howlett B.J. (2014). Genomes and transcriptomes of partners in plant-fungal-interactions between canola (Brassica napus) and two Leptosphaeria species. PLoS ONE 9: e103098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Marcel S., Sawers R., Oakeley E., Angliker H., Paszkowski U. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22: 3177–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt L.W., et al. (2014). Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. BMC Genomics 15: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt L.W., Rincones J., Bailey B.A., Aime M.C., Griffith G.W., Zhang D., Pereira G.A. (2008). Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao: what’s new from this old foe? Mol. Plant Pathol. 9: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick R.L., Marelli J.P., Sicher R.C., Strem M.D., Bailey B.A. (2012). The interaction of Theobroma cacao and Moniliophthora perniciosa, the causal agent of witches' broom disease, during parthenocarpy. Tree Genet. Genomes 8: 1261–1279. [Google Scholar]
- Moktali V., Park J., Fedorova-Abrams N.D., Park B., Choi J., Lee Y.H., Kang S. (2012). Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics 13: 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15: 349–357. [DOI] [PubMed] [Google Scholar]
- Mondego J.M., et al. (2008). A genome survey of Moniliophthora perniciosa gives new insights into Witches’ Broom Disease of cacao. BMC Genomics 9: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Motamayor J.C., et al. (2013). The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 14: r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch S., Lingner U., Floss D.S., Ludwig N., Sauer N., Deising H.B. (2008). The hemibiotrophic lifestyle of Colletotrichum species. J. Plant Physiol. 165: 41–51. [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. [DOI] [PubMed] [Google Scholar]
- O' Brien T.P., Feder N., Mccully M.E. (1964). Polychromatic staining of plant cell walls by Toluidine blue O. Protoplasma 59: 368. [Google Scholar]
- Orchard J., Collin H.A., Hardwick K., Isaac S. (1994). Changes in morphology and measurement of cytokinin levels during the development of witches' brooms on cocoa. Plant Pathol. 43: 65–72. [Google Scholar]
- Panstruga R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6: 320–326. [DOI] [PubMed] [Google Scholar]
- Pazzagli L., Seidl-Seiboth V., Barsottini M., Vargas W., Scala A., Mukherjee P. (2014). Cerato-platanins: Elicitors and effectors. Plant Sci. http://dx.doi.org/10.1016/j.plantsci.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Pedersen C., et al. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman D., Britton G., Hardwick K., Collin H.A., Isaac S. (2000). Chitin as a measure of biomass of Crinipellis perniciosa, causal agent of witches' broom disease of Theobroma cacao. Mycol. Res. 104: 671–675. [Google Scholar]
- Perfect S.E., Green J.R. (2001). Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2: 101–108. [DOI] [PubMed] [Google Scholar]
- Petre B., Morin E., Tisserant E., Hacquard S., Da Silva C., Poulain J., Delaruelle C., Martin F., Rouhier N., Kohler A., Duplessis S. (2012). RNA-Seq of early-infected poplar leaves by the rust pathogen Melampsora larici-populina uncovers PtSultr3;5, a fungal-induced host sulfate transporter. PLoS ONE 7: e44408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires A.B., Gramacho K.P., Silva D.C., Góes-Neto A., Silva M.M., Muniz-Sobrinho J.S., Porto R.F., Villela-Dias C., Brendel M., Cascardo J.C., Pereira G.A. (2009). Early development of Moniliophthora perniciosa basidiomata and developmentally regulated genes. BMC Microbiol. 9: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R.C., Roldán-Rodríguez R., Serena C., López-Berges M.S., Guarro J., Martínez-del-Pozo Á., Di Pietro A. (2012). A PR-1-like protein of Fusarium oxysporum functions in virulence on mammalian hosts. J. Biol. Chem. 287: 21970–21979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungartnik C., Melo S.C., Basso T.S., Macena W.G., Cascardo J.C., Brendel M. (2009). Reactive oxygen species and autophagy play a role in survival and differentiation of the phytopathogen Moniliophthora perniciosa. Fungal Genet. Biol. 46: 461–472. [DOI] [PubMed] [Google Scholar]
- Purdy L.H., Schmidt R.A. (1996). Status of cacao witches’ broom: biology, epidemiology, and management. Annu. Rev. Phytopathol. 34: 573–594. [DOI] [PubMed] [Google Scholar]
- Ranwez V., Harispe S., Delsuc F., Douzery E.J. (2011). MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE 6: e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Carson S.D., To J.P., Kieber J.J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132: 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. (2006). GenePattern 2.0. Nat. Genet. 38: 500–501. [DOI] [PubMed] [Google Scholar]
- Rincones J., et al. (2008). Differential gene expression between the biotrophic-like and saprotrophic mycelia of the witches’ broom pathogen Moniliophthora perniciosa. Mol. Plant Microbe Interact. 21: 891–908. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpari L.M., Meinhardt L.W., Mazzafera P., Pomella A.W., Schiavinato M.A., Cascardo J.C., Pereira G.A. (2005). Biochemical changes during the development of witches’ broom: the most important disease of cocoa in Brazil caused by Crinipellis perniciosa. J. Exp. Bot. 56: 865–877. [DOI] [PubMed] [Google Scholar]
- Skibbe D.S., Doehlemann G., Fernandes J., Walbot V. (2010). Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., de Wit P.J. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47: 233–263. [DOI] [PubMed] [Google Scholar]
- Studholme D.J., Glover R.H., Boonham N. (2011). Application of high-throughput DNA sequencing in phytopathology. Annu. Rev. Phytopathol. 49: 87–105. [DOI] [PubMed] [Google Scholar]
- Teixeira P.J., Thomazella D.P., Vidal R.O., do Prado P.F., Reis O., Baroni R.M., Franco S.F., Mieczkowski P., Pereira G.A., Mondego J.M. (2012). The fungal pathogen Moniliophthora perniciosa has genes similar to plant PR-1 that are highly expressed during its interaction with cacao. PLoS ONE 7: e45929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Thomazella D.P., Teixeira P.J., Oliveira H.C., Saviani E.E., Rincones J., Toni I.M., Reis O., Garcia O., Meinhardt L.W., Salgado I., Pereira G.A. (2012). The hemibiotrophic cacao pathogen Moniliophthora perniciosa depends on a mitochondrial alternative oxidase for biotrophic development. New Phytol. 194: 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio R.A., Costa G.G., Carazzolle M.F., Mondego J.M., Schuster S.C., Carlson J.E., Guiltinan M.J., Bailey B.A., Mieczkowski P., Meinhardt L.W., Pereira G.A. (2010). Genes acquired by horizontal transfer are potentially involved in the evolution of phytopathogenicity in Moniliophthora perniciosa and Moniliophthora roreri, two of the major pathogens of cacao. J. Mol. Evol. 70: 85–97. [DOI] [PubMed] [Google Scholar]
- Tierney L., Linde J., Müller S., Brunke S., Molina J.C., Hube B., Schöck U., Guthke R., Kuchler K. (2012). An interspecies regulatory network inferred from simultaneous RNA-seq of Candida albicans invading innate immune cells. Front. Microbiol. 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas W.A., Martín J.M., Rech G.E., Rivera L.P., Benito E.P., Díaz-Mínguez J.M., Thon M.R., Sukno S.A. (2012). Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 158: 1342–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volodarsky D., Leviatan N., Otcheretianski A., Fluhr R. (2009). HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol. 150: 1796–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.R., McRoberts N. (2006). Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci. 11: 581–586. [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17: 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wäspi U., Schweizer P., Dudler R. (2001). Syringolin reprograms wheat to undergo hypersensitive cell death in a compatible interaction with powdery mildew. Plant Cell 13: 153–161. [PMC free article] [PubMed] [Google Scholar]
- Weßling R., Schmidt S.M., Micali C.O., Knaust F., Reinhardt R., Neumann U., Ver Loren van Themaat E., Panstruga R. (2012). Transcriptome analysis of enriched Golovinomyces orontii haustoria by deep 454 pyrosequencing. Fungal Genet. Biol. 49: 470–482. [DOI] [PubMed] [Google Scholar]
- Westermann A.J., Gorski S.A., Vogel J. (2012). Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10: 618–630. [DOI] [PubMed] [Google Scholar]
- Wise R.P., Moscou M.J., Bogdanove A.J., Whitham S.A. (2007). Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 45: 329–369. [DOI] [PubMed] [Google Scholar]
- Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., Long L., Zhang X. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62: 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J., Natori A., Tolba M.E., Mongan A.E., Sugimoto C., Katayama T., Kawashima S., Makalowski W., Maeda R., Eshita Y., Tuda J., Suzuki Y. (2014). Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res. 24: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zaparoli G., Barsottini M.R., de Oliveira J.F., Dyszy F., Teixeira P.J., Barau J.G., Garcia O., Costa-Filho A.J., Ambrosio A.L., Pereira G.A., Dias S.M. (2011). The crystal structure of necrosis- and ethylene-inducing protein 2 from the causal agent of cacao’s Witches’ Broom disease reveals key elements for its activity. Biochemistry 50: 9901–9910. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Jia L.J., Zhang Y., Jiang G., Li X., Zhang D., Tang W.H. (2012). In planta stage-specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell 24: 5159–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccaro A., et al. (2011). Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 7: e1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.