Abstract
Whiteflies (Hemiptera: Sternorrhyncha: Aleyrodidae) are plant sap-sucking insects that harbor prokaryotic primary endosymbionts (P-endosymbionts) within specialized cells located in their body cavity. Four-kilobase DNA fragments containing 16S-23S ribosomal DNA (rDNA) were amplified from the P-endosymbiont of 24 whiteflies from 22 different species of 2 whitefly subfamilies. In addition, 3-kb DNA fragments containing mitochondrial cytB, nd1, and large-subunit rDNA (LrDNA) were amplified from 17 whitefly species. Comparisons of the P-endosymbiont (16S-23S rDNA) and host (cytB-nd1-LrDNA) phylogenetic trees indicated overall congruence consistent with a single infection of a whitefly ancestor with a bacterium and subsequent cospeciation (cocladogenesis) of the host and the P-endosymbiont. On the basis of both the P-endosymbiont and host trees, the whiteflies could be subdivided into at least five clusters. The major subdivision was between the subfamilies Aleyrodinae and Aleurodicinae. Unlike the P-endosymbionts of may other insects, the P-endosymbionts of whiteflies were related to Pseudomonas and possibly to the P-endosymbionts of psyllids. The lineage consisting of the P-endosymbionts of whiteflies is given the designation “Candidatus Portiera” gen. nov., with a single species, “Candidatus Portiera aleyrodidarum” sp. nov.
Whiteflies are members of the suborder Sternorrhyncha that also contains aphids, psyllids, and mealybugs, insects that use plant sap as their diet (13). All of these insects have an association with bacterial endosymbionts that are located in vesicles within specialized insect cells (bacteriocytes) that form an aggregate within the body cavity called a bacteriome (3, 4). Early studies, by use of light microscopy, indicated that each of these insect groups contains a morphologically similar endosymbiont (called the primary endosymbiont [P-endosymbiont]) that is present in all members of the group (4). Some members may have additional endosymbionts that are morphologically different and which are designated the secondary endosymbionts (S-endosymbionts) (3, 4). Both endosymbiont types are transmitted maternally. More recent studies, by use of molecular methods, have confirmed and extended these conclusions (reviewed in references 3, 20, 21, and 40). Trees representing the molecular phylogeny of the P-endosymbionts and their hosts are generally congruent. This observation has been interpreted as indicating an ancient infection of an insect ancestor by a bacterium followed by its vertical transmission, resulting in cospeciation (or cocladogenesis) of the host and the P-endosymbiont. In contrast to these results, the lack of congruence between the trees of the S-endosymbionts and the insect hosts (or the P-endosymbionts) suggests multiple infections of the host by the S-endosymbiont and/or its horizontal transmission. In the case of the P-endosymbionts, it has been shown that the rate of sequence change of their ribosomal DNA (rDNA) is considerably more rapid than that of free-living bacteria (14, 19, 27).
Plant sap, the diet of whiteflies, aphids, psyllids, and mealybugs is rich in carbohydrates and deficient in essential amino acids. In aphids, there is evidence that one of the functions of the endosymbionts is the synthesis of essential amino acids for the aphid host (reviewed in references 3, 11, 21, and 23). Since plant sap is the diet common to all of the members of the Sternorrhyncha, it is probable that all of the P-endosymbionts of these insects have a similar function.
Whiteflies derive their name from a powdery, white waxy secretion found spread over the body and wings of most adults (13). There are approximately 1,450 species of whiteflies; some are major agricultural pests causing plant debilitation and the transmission of plant viruses (13). Reproduction of whiteflies is usually sexual; following emergence from the egg, the first instar (crawler) is capable of movement while the second, third, and fourth instars are sessile, being attached to the plant surface. By convention, the fourth instar is called the pupa, and its morphology is generally used as the basis of whitefly classification (17, 18). Winged adults emerge from the pupa and reproduce.
Studies, by means of the electron microscope, have indicated that whitefly bacteriocytes contain a pleomorphic bacterium within host derived vesicles (7, 8, 33). This organism differs from that of the P-endosymbionts of aphids (3), psyllids (36), and mealybugs (38) by the absence of the outer membrane of the gram-negative cell wall (7, 8, 33). This pleomorphic organism appears to be present in all whiteflies that have been studied and is designated the P-endosymbiont. Whiteflies have a unique method of transmission of endosymbionts to progeny. In the other members of Sternorrhyncha, the endosymbionts leave the bacteriocytes and enter the germ cells; in whiteflies, intact bacteriocytes migrate to the ovaries (4, 10, 33). Treatment of whiteflies with antibiotics that affect bacterial protein synthesis and transcription had a negative effect on whitefly growth and development, suggesting that endosymbionts play a nutritional role in this association (9).
Limited studies on the phylogeny of endosymbionts of whiteflies by use of 16S rDNA have indicated that the P-endosymbionts constitute a lineage different from the P-endosymbionts of aphids, psyllids, and mealybugs (3, 5, 31, 35, 36). Some whitefly species have additional S-endosymbionts related to Arsenophonus or to other members of the Enterobacteriaceae (5, 31, 34, 41). Recently, it has been found that the whitefly Bemisia tabaci may contain chlamydia (37), a relative of the Encarsia bacterium (a member of the Bacteroidetes phylum) (2, 39),as well as Wolbachia (26). In this study, we extend the previous observations on the phylogeny of whitefly P-endosymbionts by increasing the number of whitefly species studied and by the use of both 16S and 23S rDNA sequences. In addition, to compare the phylogeny of the endosymbiont and the host, we have derived trees based on the analysis of host mitochondrial cytB, nd1, and large-subunit rDNA (LrDNA) sequences. A phylogenetic analysis of 11 whitefly species, based on insect 18S rDNA, has been performed previously (4a).
MATERIALS AND METHODS
Collection and preservation of biological material.
Table 1 presents the sources of the whiteflies, together with the available information on host plants and locations and dates of collections. Upon collection, the whiteflies were stored in 100% ethanol and sent to the Davis laboratory, where they were stored at 4°C. Samples contained between 5 and 60 whiteflies.
TABLE 1.
Information concerning the whiteflies used in this study and the accession numbers of the 16S-23S rDNAs of the P-endosymbionts and cytB-nd1-LrDNA of the host mitochondria
| Whitefly species | GenBank accession no. (P-endosymbiont/host mitochondria) | Host plant/location/date (mo, yr)/collector |
|---|---|---|
| Acanthaleyrodes styraci | AY266092 | Rubus reflexus/Hong Kong/12, 2001/J. H. Martin |
| Aleurochiton aceris | AY266093/AY521266, AY521267 | Acer platanoides/England/9, 2001/J. H. Martin |
| Aleurodicus dugesii 1 | AY266094 | Unavailable/Belize/6, 2002/J. H. Martin |
| Aleurodicus dugesii 2 | AY266095/AY521251 | Laboratory strain/Riverside, Calif./9, 2002/H. S. Costa |
| Aleurodicus dispersus 1 | AY266096/AY521252 | Psidium guava/New Caledonia/1, 2003/J. P. Gullan |
| Aleurodicus dispersus 2 | AY266097 | Palm/Canary Islands/7, 2002/K. Bourtzis |
| Aleuroparadoxus arctostaphylli | AY266098 | Arbutus menziesii/California/4, 2001/P. S. Ward |
| Aleuroplatus gelatinosus | AY266112/AY521254 | Quercus sp./California/4, 2001/R. Dowell |
| Aleuroplatus sp. | AY266100/AY521256 | Maesa perlarius/Hong Kong/12, 2001/J. H. Martin |
| Aleyrodes elevatus | AY266101/AY521253 | Ficus carica/Corsica/10, 2001/J. H. Martin |
| Aleyrodes proletella | AY266102/AY521255 | Lathyrus sp./England/9, 2001/J. H. Martin |
| Bemisia argentifolii | AF211870a/AY521257 | Reference 5 |
| Bemisia sp. | AY266103/AY521258 | Unavailable/Belize/6, 2002/J. H. Martin |
| Bemisia tabaci | AY268081b/AY521259 | Laboratory strain/Davis, Calif./6, 2002/B. W. Falk |
| Dialeurodes group A | AY266105 | Unavailable/Belize/6, 2002/J. H. Martin |
| Dialeurodes group B | AY266106/AY521260 | Unavailable/Belize/6, 2002/J. H. Martin |
| Dialeurodes hongkongensis | AY266107/AY521261 | Dendrotrophe frutescens/Hong Kong/12, 2001/J. H. Martin |
| Neomaskellia andropogonis | AY266108 | Saccharum spontaneum/Hong Kong/12, 2001/J. H. Martin |
| Siphoninus phillyreae | AY266109/AY521268, AY521269 | Olea europea/Corsica/10, 2001/J. H. Martin |
| Tetraleurodes acaciae | AY266110/AY521262 | Erythrina speciosa/Hong Kong/12, 2001/J. H. Martin |
| Tetraleurodes mori | AY266111/AY521263 | Arbutus menziesii/California/4, 2001/P. S. Ward |
| Trialeurodes hutchingsi | AY266099/AY521264 | Manzanita/California/4, 2001/P. J. Gullan |
| Trialeurodes vaporariorum | AY266113/AY521265 | Strawberries/California/8, 2002/F. Zalom |
| Vasdavidius concursus | AY266114 | Saccharum spontaneum/Hong Kong/12, 2001/J. H. Martin |
Molecular biology methods.
The methods used have been described in detail in studies of psyllid and mealybug endosymbionts (35, 36). These include methods for the purification of DNA, amplification of the 16S-23S rDNA genes by PCR, cloning into pBluescript (Stratagene, La Jolla, Calif.), and determination of the DNA sequence by using a variety of primers to conserved regions of 16S-23S rDNA.
Based on ongoing studies of mitochondria of members of the Sternorrhyncha, oligonucleotide primers were designed for the PCR amplification of approximately 3.1-kb mitochondrial DNA (mtDNA) fragments containing a part of cytB, the full nd1, LrDNA, and a small portion of small-subunit rDNA (5′-GCA GGA TCC GCG GCC WTG RGG HCA AAT ATC WTT TTG RGG DGC-3′ [BamHI and SacI] and 5′-GCA GGT ACC TCG AGT ATG TAC AMA TYG CCC GTC AYT CTT-3′ [KpnI and XhoI]). The subsequent procedures are similar to those used for amplification and cloning of 16S-23S rDNA. The optimal annealing conditions in the PCR were determined in a temperature gradient and were between 55 and 60°C. Since the mtDNA sequences were rather poorly conserved, different oligonucleotide primers were designed to obtain the sequence of the inserts.
Analysis of the sequence data.
The methods used for analysis of the sequence data were previously described (35, 36). The intergenic space between 16S and 23S rDNA was removed, and the resulting sequences were aligned by using Pileup of SeqWeb, version 2 (Genetics Computer Group, Madison, Wis.). In the case of the mtDNA, sequences corresponding to Trialeurodes vaporariorum (AY521265) nucleotides (nt) 2 to 700 (cytB) and 1660 to 794 (nd1) were translationally aligned and joined to aligned nt 2844 to 1781 (LrDNA). Phylogenies were reconstructed by using maximum-likelihood and -parsimony methods of PAUP, version 4 (32). Either 500 or 1,000 bootstrap replicates were used to assess support for individual nodes.
Nucleotide sequence accession number.
All of the 16S-23S rDNA sequences and the cytB-nd1-LrDNA sequences were deposited in the GenBank database, and the accession numbers are given in Table 1. In addition, the sequences of similar mtDNA fragments were determined for Pachypsylla venusta (AY521271) and Schizaphis graminum (AY521270). Additional 16S-23S rDNA sequences used in the analyses are for the following free-living organisms and insect P-endosymbionts (accession numbers given in parentheses): Aeromonas hydrophila (AF099022 and X67943), Acetobacter intermedius (Y14694 and Y14680), Burkholderia mallei (NC002970), Citrobacter freundii (AJ233408 and U77928), Escherichia coli (AE000474), Klebsiella pneumoniae (AJ233420 and X87284), Neisseria meningitidis (AE002098), Pseudomonas aeruginosa (AE004091), Salmonella enterica (X80681 and U77919), Vibrio cholerae (NC002525), Yersinia enterocolitica (Z47828 and U77925), Zymobacter palmae (AF211871), Baumannia cicadenillinicola (AF489427), Blochmannia floridanus (NC005061), Buchnera aphidicola (NC002548, NC004061, and NC004545), and Wigglesworthia brevipalpis (NC004344).
RESULTS
General properties of the P-endosymbiont and host mtDNA fragments.
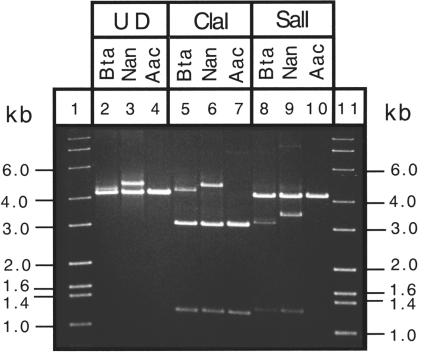
PCR amplification of the 16S-23S rDNA from total whitefly DNA, followed by agarose gel electrophoresis of undigested and restriction enzyme-digested DNA fragments, indicated that 16 of the 22 whitefly species contained two types of DNA corresponding to the P- and S-endosymbionts. This is illustrated in Fig. 1 where the undigested PCR-amplified DNA of B. tabaci and Neomaskellia andropogonis is shown to consist of bands of two sizes (lanes 2 and 3). Only one band of 4.2 kb is present in the DNA of Aleurochiton aceris (lane 4). The 4.2-kb band corresponds to the P-endosymbiont; in most cases, ClaI digests this DNA, giving fragments of 3.0 and 1.2 kb (lanes 5, 6, and 7) while leaving the S-endosymbiont DNA intact (lanes 5 and 6). Similarly, SalI does not digest most P-endosymbionts (lanes 8, 9, and 10) while digesting the S-endosymbiont (lanes 8 and 9).
FIG. 1.
Agarose gel electrophoresis of 16S-23S rDNA PCR products amplified from total DNA of three species of whiteflies. Lanes 1 and 11, molecular size standards; UD, undigested DNA; ClaI and SalI, restriction enzymes; Bta, B. tabaci; Nan, N. andropogonis; Aac, A. aceris. Bands of 0.1 kb (lanes 5, 6, and 7) and 0.2 kb (lanes 5 and 6) are not visible in the photograph.
In this paper, only the P-endosymbionts are considered, the S-endosymbionts are considered in a separate publication (34). The length of the 16S-23S rDNA fragments corresponding to the P-endosymbionts was 4,166 to 4,229 nt, and the guanine plus cytosine (G+C) content was 45.4 to 46.7 mol%. The intergenic space between the 16S and 23S rDNA had a length of 130 to 189 nt and a G+C content of 36.7 to 43.9 mol%. In all cases, the intergenic space contained a sequence encoding tRNAIle (codon ATC). The 16S rDNA portion was 1,503 to 1,520 nt in length and had a G+C content of 47.8 to 49.7 mol% while the 23S rDNA portion was 2,516 to 2,558 nt in length and had a G+C content of 45.4 to 46.7 mol%.
Clones containing portions of mitochondrial cytB and small-subunit rDNA and the complete mitochondrial nd1 and LrDNA were obtained from 17 whitefly species (Table 1). The DNA amounts in the seven remaining whitefly samples were insufficient and could not be included in this part of the study. The DNA fragments had a length of 3,044 to 3,248 nt and G+C contents of 13.7 to 30.2 mol%.
Phylogenetic analyses.
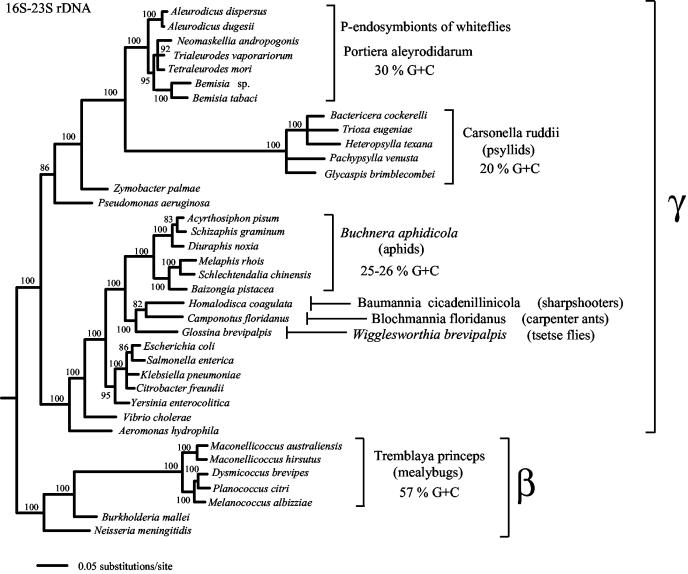
Phylogenetic analyses of the combined 16S-23S rDNA sequences of the P-endosymbionts from 24 whiteflies along with Z. palmae and P. aeruginosa, with A. intermedius as an out-group, were consistent with past results (5, 31). The P-endosymbionts were a monophyletic group (100% bootstrap values), with Z. palmae as the closest free-living organism that was related to P. aeruginosa. The largest difference between the nucleotide sequences of the 16S-23S rDNAs of two endosymbionts was 8.4%. The differences between the P-endosymbionts and Z. palamae was 14 to 15%, and the difference between them and P. aeruginosa was 17 to 18%. A phylogenetic tree showing the relationship of representative whitefly P-endosymbionts as well as other insect P-endosymbionts and related free-living bacteria is shown in Fig. 2. In both this tree and in the phylogenetic tree that included all of the whitefly P-endosymbionts, the branch leading to the P-endosymbionts was of considerable length. Consequently, in the case of the latter tree, we only present the portion of the tree that shows the whitefly P-endosymbionts (Fig. 3A). Nearly identical results were obtained with the maximum-likelihood and -parsimony analyses.
FIG. 2.
Phylogenetic relationships of representative P-endosymbionts of plant sap-sucking insects, other insect P-endosymbionts, and related free-living bacteria. Trees are from maximum-likelihood analyses of the combined 16S-23S rDNAs. Numbers at nodes are for bootstrap percentages from 500 replicates; only nodes supported by 70% or greater are shown. P-endosymbionts that are designated by genus and species names in regular letters have “Candidatus” status. Italicized P-endosymbiont genus and species names were named prior to the “Candidatus” proposal; hence, they bear the usual species designations. Names within brackets refer to insect species from which the endosymbiont 16S-23S rDNA sequence was determined; other names refer to bacterial species. Greek letters by brackets indicate the subdivisions of the Proteobacteria. %G+C indicates the moles percent guanine-plus-cytosine content of either fragments of P-endosymbiont DNA or the whole genome (aphids). GenBank accession numbers are given in this paper or in references 35 and 36).
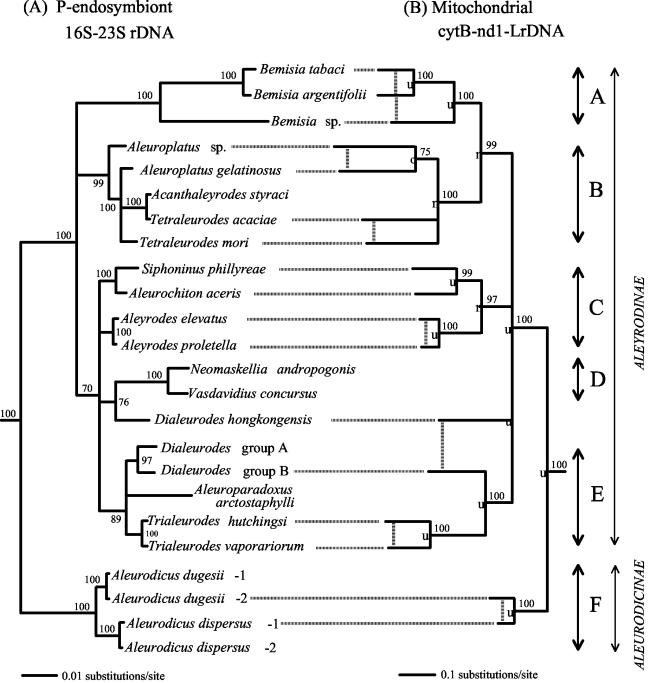
FIG. 3.
Phylogenetic tree from maximum-likelihood analyses of whitefly P-endosymbiont-combined 16S-23S rDNA nucleotide sequences (A) and host mitochondrial combined cytB-nd1-LrDNA (B). Designations refer to whitefly species. In panel B, vertical striped lines are used to join organisms within the same genus. Filled diamonds in panel B indicate nodes identical to those in panel A, filled squares indicate consistent nodes, an open square indicates an inconsistent node, striped vertical lines indicate species within the same genus, thick double-headed arrows indicate major clusters, and thin double-headed arrows indicate species in different subfamilies. Numbers at nodes are for bootstrap percentages from 500 replicates; only nodes supported by 70% or greater are shown.
Phylogenetic analyses of the combined cytB-nd1-LrDNA sequences of 17 whitefly species along with the sequences from S. graminum (aphid) and P. venusta (psyllid), with the latter as the out-group, indicated that the whiteflies were a monophyletic group (100% bootstrap value). Figure 3B presents the portion of the tree that includes only the whiteflies. Only very minor differences were observed when analyses were performed with the maximum-likelihood or -parsimony method. The largest difference between the combined cytB-nd1-LrDNAs of a pair of whitefly species was 36.9%.
On the basis of both the P-endosymbiont tree and the host mtDNA tree, the endosymbionts and their hosts could be divided into six clusters (Fig. 3). Dialeurodes hongkongensis probably constitutes a separate cluster, as is also the case with N. andropogonis and Vasdavidius concursus (mtDNA sequences for the latter two species were not obtained). The major subdivision differentiated clusters A, B, C, D, and E from cluster F. This corresponds to the assignment of whiteflies into the subfamilies Aleyrodinae (clusters A, B, C, D, and E) and Aleurodicinae (cluster F) (17, 18). The P-endosymbionts of the Aleurodicinae differed from the P-endosymbionts of Aleyrodinae in having the sequence TAAATAACTTATTTTGC inserted in its 16S rDNA (positions 53 to 69 in accession no. AY266094) as well as sequences CTAACTTATTAAAGT and ATAACAAG inserted in the 23S rDNA (positions 3195 to 3209 and 3541 to 3548, respectively). The division of the whiteflies into two subfamilies was also confirmed by mtDNA analysis (Fig. 3B). There was general agreement between the phylogenetic trees based on P-endosymbiont DNA and mtDNA (Fig. 3). Thirteen nodes were identical or consistent while one node was contradictory.
DISCUSSION
The results of our studies are consistent with a monophyletic origin of whitefly P-endosymbionts. The similarity of the phylogenetic trees derived from P-endosymbiont 16S-23S rDNA and host mitochondrial cytB-nd1-LrDNA (Fig. 3), suggests a single infection of a whitefly ancestor with a bacterium followed by vertical transmission to progeny. Similar results, consistent with cospeciation (cocladogenesis), have also been observed in the case of a number of other P-endosymbiont-insect associations. This is the case of the P-endosymbionts of plant phloem-sucking insects such as psyllids (Candidatus Carsonella ruddii, Gammaproteobacteria) (36), mealybugs (Candidatus Tremblaya princeps, Betaproteobacteria) (35), and aphids (B. aphidicola, Gammaproteobacteria) (2); the xylem-sucking sharpshooters (Candidatus Baumannia cicadellinicola, Gammaproteobacteria) (24), carpenter ants (Candidatus Blochmannia floridanus, Gammaproteobacteria) (30), termites, and cockroaches (Blattabacterium cuenoti, Bacteroidetes phylum) (15); and blood-sucking tsetse flies (Wigglesworthia brevipalpis, Gammaproteobacteria) (1). The match of the P-endosymbiont and the host trees is not always perfect, as would be expected from a variety of biological and analytical factors which may cause some incongruence (6, 16, 28). The results with the Arsenophonus-like whitefly S-endosymbionts are in marked contrast with those obtained with the P-endosymbionts. There is no congruence between the S-endosymbiont trees and the host or P-endosymbiont trees, suggesting multiple infections of the host and/or horizontal transmission of the S-endosymbiont (34, 41).
The P-endosymbionts of whiteflies appear to be related to the P-endosymbionts of psyllids (12, 31), and both of these differ from the remaining insect endosymbionts of the Gammaproteobacteria in being more closely related to members of the genus Pseudomonas than to members of the Enterobacteriaceae (1, 3, 24, 30). Both the 16S-23S rDNA and mtDNA support the subdivision of the whiteflies into the two subfamilies Aleyrodinae and Aleurodicinae (Fig. 3). The subdivision of the whiteflies into two subfamilies was also indicated by the host 18S rDNA study (4a), which grouped whitefly specis into clusters corresponding to our clusters A, B, C, and F (Fig. 3B). Based on the largest sequence divergence between the P-endosymbionts of these two subgroups and the previously determined rate of sequence change of insect endosymbionts of 1 to 2% per 50 million years (22, 27), the time of divergence between these two groups is estimated to have occurred about 100 to 200 million years ago. This is similar to the estimated time of origin of the endosymbiotic association of other plant sap-sucking insects (27, 35, 36).
At the present, partially characterized bacteria that have not been cultivated on laboratory media are given the designation Candidatus (25). We propose to name the lineage corresponding to the P-endosymbionts of whiteflies “Candidatus Portiera” (por.ti.e′ra. N.L. fem. n.) in honor of Paul Portier, a French biologist who made major contributions to the studies and concepts of endosymbiosis (29). “Candidatus Portiera” consists of pleomorphic bacteria having only the cell membrane and lacking the outer membrane of the gram-negative cell wall, housed within host-derived vesicles in the bacteriocytes of whiteflies (7, 8, 33). The order of the rRNA genes is 16S-23S rDNA, with tRNAIle between these two genes. The G+C contents of the 16S rDNA and 23S rDNA are 47.8 to 49.7 mol% and 45.4 to 46.7 mol%, respectively. Only one copy of these genes is present in the genome (2). Based on the sequence of 16S-23S rDNA, these organisms are assigned to the Gammaproteobacteria. The nearest free-living relative is Z. palmae. “Candidatus Portiera” appears to be related to Candidatus Carsonella, the endosymbiont of psyllids.
“Candidatus Portiera” contains a single species, “Candidatus Portiera aleyrodidarum” (a.ley.ro.di.da′rum. N.L. gen. plur. fem. n. aleyrodidarum, of the Aleyrodidae [whiteflies]). The P-endosymbiont of B. tabaci is proposed as the type strain (GenBank accession no. AY268082). The G+C content of a 31-kb “Candidatus P. aleyrodidarum” DNA fragment is 30.2 mol% (2). The following sequences are unique to “Candidatus P. aleyrodidarum”: 16S rDNA, 5-TCT TAC GAG ATA AAG-3′; 23S rDNA, 5′-CAG TAT CTG TA-3′ and 5′-CAT ATT GAA AGT G-3′. The phylogenetic relationship of “Candidatus P. aleyrodidarum” to other insect P-endosymbionts as well as to their related free-living bacteria is shown in Fig. 2.
Acknowledgments
This material is based on work supported by National Science Foundation Awards MCB-9807145 (P.B.) and DEB-9978518 (N. A. Moran, P.B.) and the University of California Experiment Station (P.B.).
We are grateful to L. Baumann for technical assistance; to K. Bourtzis, H. S. Costa, J. P. Gullan, J. H. Martin, and F. Zalom for the collection of samples; to Hans G. Trüper for advice on nomenclature; and to an anonymous reviewer whose suggestions improved the manuscript.
REFERENCES
- 1.Aksoy, S., X. Chen, and V. Hypsa. 1997. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera: Glossinidae). Insect Mol. Biol. 6:183-190. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, L., M. L. Thao, C. J. Funk, B. W. Falk, J. C. K. Ng, and P. Baumann. 2004. Sequence analysis of DNA fragments from the genome of the primary endosymbiont of Bemisia tabaci. Curr. Microbiol. 48:77-81. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., N. A. Moran, and L. Baumann. 2000. Bacteriocyte-associated endosymbionts of insects. In M. Dworkin (ed.), The prokaryotes. [Online.] Springer, New York, N.Y. http://link.springer.de/link/service/books/10125.
- 4.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms, p. 332-338. John Wiley and Sons Interscience, New York, N.Y.
- 4a.Campbell, B. C., J. D. Steffen-Campbell, and R. J. Gill. 1996. Origin and radiation of whiteflies: an initial molecular phylogenetic assessment, p. 29-51. In D. Gerling and R. T. Mayer (Ed.), Bemisia 1995: taxonomy, biology, damage, control and management. Intercept, Andover, United Kingdom.
- 5.Clark, M. A., L. Baumann, M. A. Munson, P. Baumann, B. C. Campbell, J. E. Duffus, L. S. Osborne, and N. A. Moran. 1992. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodoidea) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr. Microbiol. 25:119-123. [Google Scholar]
- 6.Clark, M. A., N. A. Moran, P. Baumann, and J. Wernegreen. 2000. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution 54:517-525. [DOI] [PubMed] [Google Scholar]
- 7.Costa, H. S., D. M. Westcot, D. E. Ullman, and M. W. Johnson. 1993. Ultrastructure of the endosymbionts of the whitefly, Bemisia tabaci and Trialeurodes vaporarorum. Protoplasma 176:106-115. [Google Scholar]
- 8.Costa, H. S., D. M. Westcot, D. E. Ullman, R. Rosell, J. K. Brown, and M. W. Johnson. 1995. Morphological variation in Bemisia endosymbionts. Protoplasma 189:194-202. [Google Scholar]
- 9.Costa, H. S., T. J. Henneberry, and N. C. Toscano. 1997. Effects of antibacterial materials on Bemisia argentifolii (Homoptera: Aleyrodidae) oviposition, growth, survival, and sex ratio. Ann. Entomol. Soc. Am. 90:333-339. [Google Scholar]
- 10.Costa, H. S., N. C. Toscano, and T. J. Henneberry. 1996. Mycetocyte inclusion in the oocytes of Bemisia argentifolii (Homoptra: Aleyrodidae). J. Econ. Entomol. 89:694-699. [Google Scholar]
- 11.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 12.Fukatsu, T., and N. Nikoh. 2000. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). Appl. Environ. Microbiol. 66:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullan, P. J., and J. H. Martin. 2002. Sternorrhyncha (jumping plant-lice, whiteflies, aphids and scale insects), p. 1079-1089. In V. H. Resh and R. T. Cardé (ed.), Encyclopedia of insects. Academic Press/Elsevier Science, New York, N.Y.
- 14.Itoh, T., W. Martin, and M. Nei. 2002. Acceleration of genomic evolution caused by enhanced mutation rate in endocellular symbionts. Proc. Natl. Acad. Sci. USA 99:12944-12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo, N., C. Bandi, H. Watanabe, C. Nalepa, and T. Beninati. 2003. Evidence for cocladogenesis between diverse Dictyopteran lineages and their intracellular endosymbionts. Mol. Biol. Evol. 20:907-913. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 17.Martin, J. H. 2003. Whiteflies (Hemiptera: Aleyrodidae)—their systematic history and resulting problems of conventional taxonomy, with special reference to descriptions of Aleyrodes proletella (Linnaeus 1758) and Bemisia tabaci (Gennadius, 1889). Entomol. Gazette 54:125-136. [Google Scholar]
- 18.Martin, J. J. 1999. The whitefly fauna of Australia (Sternorrhyncha: Aleyrodidae). Commonwealth Scientific and Industrial Research Organization, Canberra, Australia.
- 19.Moran, N. A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran, N. A., and A. Telang. 1998. Bacteriocyte-associated symbionts of insects: a variety of insect groups harbor ancient prokaryotic endosymbionts. BioScience 48:295-304. [Google Scholar]
- 21.Moran, N. A., and P. Baumann. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270-275. [DOI] [PubMed] [Google Scholar]
- 22.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B 253:167-171. [Google Scholar]
- 23.Moran, N. A., G. R. Plague, J. P. Sandström, and J. L. Wilcox. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 100:14543-14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, N. A., C. Dale, H. Dunbar, W. A. Smith, and H. Ochman. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5:116-126. [DOI] [PubMed] [Google Scholar]
- 25.Murray, R. G. E., and K. H. Schleifer. 1994. Taxonomic notes: a proposal for recording the properties of putative taxa of prokaryotes. Int. J. Syst. Bacteriol. 44:174-176. [DOI] [PubMed] [Google Scholar]
- 26.Nirgianaki, A., G. K. Banks, D. R. Frohlich, Z. Veneti, H. R. Braig, T. A. Miller, I. D. Bedford, P. G. Markham, C. Savakis, and K. Bourtzis. 2003. Wolbachia infections of the whitefly Bemisia tabaci. Curr. Microbiol. 47:93-101. [DOI] [PubMed] [Google Scholar]
- 27.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 96:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rokas, A., B. L. Williams, N. King, and S. B. Carroll. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798-804. [DOI] [PubMed] [Google Scholar]
- 29.Sapp, J. 1994. Evolution by association. Oxford University Press, New York, N.Y.
- 30.Sauer, C., E. Stackebrandt, J. Gadau, B. Hölldobler, and R. Gross. 2000. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int. J. Syst. Evol. Microbiol. 50:1877-1886. [DOI] [PubMed] [Google Scholar]
- 31.Spaulding, A. W., and C. D. von Dohlen. 2001. Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol. Biol. 10:57-67. [DOI] [PubMed] [Google Scholar]
- 32.Swofford, D. L. 1999. PAUP: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, Mass.
- 33.Szklarzewicz, T., and A. Moskal. 2001. Ultrastructure, distribution, and transmission of endosymbionts in the whitefly Aleurochiton aceris Modeer (Insecta, Hemiptera, Aleyrodinea). Protoplasma 218:45-53. [DOI] [PubMed] [Google Scholar]
- 34.Thao, M. L., and P. Baumann. 2004. Evidence of multiple acquisition of Arsenophonus by whitefly species (Sternorryncha: Aleyrodidae). Curr. Microbiol. 48:140-144. [DOI] [PubMed] [Google Scholar]
- 35.Thao, M. L., P. J. Gullan, and P. Baumann. 2002. Secondary (γ) endosymbionts infect the primary (β) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl. Environ. Microbiol. 68:3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thao, M. L., N. A. Moran, P. Abbot, E. B. Brennan, D. H. Burckhardt, and P. Baumann. 2000. Cospeciation of psyllids and their prokaryotic endosymbionts. Appl. Environ. Microbiol. 66:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thao, M. L., L. Baumann, J. M. Hess, B. W. Falk, J. C. K. Ng, P. J. Gullan, and P. Baumann. 2003. Phylogenetic evidence for two new insect-associated chlamydia of the family Simkaniaceae. Curr. Microbiol. 47:46-50. [DOI] [PubMed] [Google Scholar]
- 38.von Dohlen, C. D., S. Kohler, S. T. Alsop, and W. R. McManus. 2001. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature (London) 412:433-436. [DOI] [PubMed] [Google Scholar]
- 39.Weeks, A. R., and J. A. J. Breeuwer. 2003. A new bacterium from the Cytophaga-Flavobacterium-Bacteroides phylum that causes sex-ratio distortion, p. 165-176. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis. CRC Press, Boca Raton, Fla.
- 40.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Gen. 3:850-861. [DOI] [PubMed] [Google Scholar]
- 41.Zchori-Fein, E., and J. K. Brown. 2002. Diversity of prokaryotes associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 95:711-718. [Google Scholar]