RAC/ROP GTPases are import regulators of polar cell growth and morphogenesis. RISAP is shown to be a RAC/ROP effector that functions as myosin receptor and is associated with a subapical trans-Golgi network compartment in tip-growing pollen tubes. RISAP modulates the membrane traffic underlying directional cell expansion by mediating dynamic interactions between endomembrane components and F-actin.
Abstract
RAC/ROP GTPases coordinate actin dynamics and membrane traffic during polar plant cell expansion. In tobacco (Nicotiana tabacum), pollen tube tip growth is controlled by the RAC/ROP GTPase RAC5, which specifically accumulates at the apical plasma membrane. Here, we describe the functional characterization of RISAP, a RAC5 effector identified by yeast (Saccharomyces cerevisiae) two-hybrid screening. RISAP belongs to a family of putative myosin receptors containing a domain of unknown function 593 (DUF593) and binds via its DUF593 to the globular tail domain of a tobacco pollen tube myosin XI. It also interacts with F-actin and is associated with a subapical trans-Golgi network (TGN) compartment, whose cytoplasmic position at the pollen tube tip is maintained by the actin cytoskeleton. In this TGN compartment, apical secretion and endocytic membrane recycling pathways required for tip growth appear to converge. RISAP overexpression interferes with apical membrane traffic and blocks tip growth. RAC5 constitutively binds to the N terminus of RISAP and interacts in an activation-dependent manner with the C-terminal half of this protein. In pollen tubes, interaction between RAC5 and RISAP is detectable at the subapical TGN compartment. We present a model of RISAP regulation and function that integrates all these findings.
INTRODUCTION
Polar cell growth plays a key role in organ morphogenesis, particularly in plants (Kost et al., 1999a; Kost and Chua, 2002). Pollen tube elongation is widely employed as a model system to investigate cellular und regulatory mechanisms underlying polar cell growth. Pollen tubes are large cells formed by germinating pollen grains and expand extremely rapidly (at rates of several μm/min) exclusively in one direction by a process known as tip growth (Hepler et al., 2001). In situ, elongating pollen tubes transport male generative cells enclosed within their cytoplasm through female flower tissue to enable fertilization. Tip growth of pollen tubes requires massive fusion of secretory vesicles containing Golgi-derived cell wall material with the plasma membrane specifically at the apex, a process that needs to be balanced by endocytic recycling of membrane material (Derksen et al., 1995). The spatial organization and molecular control of apical membrane traffic underlying tip growth is poorly understood.
A 10- to 15-μm-long cytoplasmic region at the extreme pollen tube tip (clear zone [CZ]) is filled with apical vesicles and does not contain any other organelles. The CZ presumably contains mostly secretory vesicles (Derksen et al., 1995; Hepler et al., 2001; Campanoni and Blatt, 2007), although recent reports suggest that endocytic vesicles may also accumulate in this zone (Moscatelli et al., 2007; Zonia and Munnik, 2008). The granular cytoplasm adjacent to the CZ contains all other pollen tube organelles. Most of these organelles are rapidly moving within the granular cytoplasm from one end of the protoplast to the other, a process known as “cytoplasmic streaming” (Cheung and Wu, 2007). In the granular cytoplasm close to the CZ, cytoplasmic streaming follows a typical “reverse fountain” pattern, with organelles moving forward along the cortex, changing direction right behind the CZ and moving backward in the center of the cell (Hepler et al., 2001; Cheung and Wu, 2007). Cytoplasmic streaming is presumably required to ensure effective supply of material required for cell expansion, including vesicles filled with cell wall material, to the growing apex. The cellular mechanisms responsible for the retention and accumulation of such vesicles in the CZ are unclear.
The actin cytoskeleton in pollen tubes is essential for cytoplasmic streaming and cell expansion, presumably because it mediates myosin motor protein-dependent organelle and vesicle motility. Fine actin filaments proposed to be present within the CZ (Fu et al., 2001), as well as a subapical cortical F-actin fringe located at the interface between the CZ and the granular cytoplasm (Kost et al., 1998; Lovy-Wheeler et al., 2005), are thought to have important functions in apical membrane traffic, which include the transport of secretory vesicles toward sites of secretion at the plasma membrane. Longitudinally oriented massive F-actin cables in the granular cytoplasm behind the CZ appear to provide tracks for myosin-mediated organelle motility (cytoplasmic streaming) (Hepler et al., 2001; Cheung and Wu, 2007). Treatment with low doses of F-actin disrupting drugs inhibits pollen tube growth without blocking cytoplasmic streaming, indicating that apical actin filaments and/or the subapical cortical F-actin fringe are selectively affected. At higher concentrations, these drugs also interfere with the functions of F-actin cables and stop cytoplasmic streaming (Gibbon et al., 1999; Vidali et al., 2001).
RHO family small GTPases coordinately regulate F-actin reorganization and membrane traffic in animal and yeast cells to bring about cell migration or directional cell growth (Harris and Tepass, 2010; Hall, 2012). In plants, RHO GTPases are represented by the RAC/ROP (RHO of plants) GTPases, which are most closely related to the RAC subfamily of non-plant RHO GTPases (Winge et al., 1997; Li et al., 1998). RAC/ROP GTPases, such as the tobacco (Nicotiana tabacum) isoform RAC5 (Klahre et al., 2006), have key functions in the regulation of pollen tube tip growth. Excess RAC/ROP activity results in depolarized pollen tube growth and induces the formation of large, anisotropically expanding balloons instead of tips that elongate in only one direction. By contrast, RAC/ROP inactivation strongly inhibits pollen tube growth (Kost et al., 1999b; Li et al., 1999).
In the active GTP-bound conformation, most RHO GTPases are membrane-associated based on posttranslational lipid modification (generally C-terminal prenylation) (Wright and Philips, 2006). In this conformation, RHO GTPases typically interact with multiple effectors to initiate different downstream signaling pathways. After GTP hydrolysis stimulated by GTPase-activating proteins, RHO GTPases are inactive as signaling molecules and can be transferred to the cytoplasm by guanine nucleotide dissociation inhibitors (GDIs), with which they form soluble heterodimers. GDI displacement by GDI dissociation factors (GDFs) promotes membrane reassociation of RHO GTPases, which allows guanine nucleotide exchange factor-mediated GDP for GTP exchange resulting in RHO reactivation (Hall, 2012).
Pollen tube RAC/ROP GTPases, such as RAC5, are active at the plasma membrane, specifically at the apex (Hwang et al., 2005), and are inactivated by membrane-associated GTPase-activating proteins at the flanks of the tip (Klahre and Kost, 2006). GDI-mediated recycling of inactive pollen tube RAC/ROP from the flanks of the tip to the apical plasma membrane, where guanine nucleotide exchange factors are present (Gu et al., 2006), ensures maintenance of polarized RAC/ROP activity at this location despite massive vesicle traffic causing constant plasma membrane remodeling (Klahre et al., 2006; Kost, 2008).
The signaling network that coordinates F-actin organization and membrane traffic in pollen tubes downstream of apical RAC/ROP activity is not well understood (Yalovsky et al., 2008). A RAC/ROP-associated phosphatidylinositol-4-phosphate 5-kinase is responsible for the accumulation of the signaling lipid phosphatidylinositol 4,5-bisphosphate (PI4,5P2) in the plasma membrane, specifically at the apex (Kost et al., 1999b; Ischebeck et al., 2008). Membrane-bound phospholipase C activity at the flanks of the tip hydrolyzes PI4,5P2, prevents lateral spreading of this lipid, and thereby maintains the polarity of RAC/ROP signaling and of cell expansion (Dowd et al., 2006; Helling et al., 2006). PI4,5P2 may act as RAC/ROP downstream effector and regulate membrane traffic and/or actin organization at the pollen tube tip, as it does in other cell types (Janmey and Lindberg, 2004; Liu et al., 2007; Martin, 2012). At the same time, PI4,5P2 can function as a GDF (Fauré et al., 1999) and promote RAC/ROP activity (Ischebeck et al., 2011), potentially creating a positive feedback circuit that enhances apical polarization of this activity (Kost, 2008). Arabidopsis thaliana RIC3 and 4, members of a plant-specific family of CRIB (CDC42/RAC interactive binding) domain-containing RAC/ROP effectors (Wu et al., 2001), have been shown to mediate RAC/ROP dependent regulation of the pollen tube actin cytoskeleton (Lee et al., 2008). Another plant-specific RAC/ROP effector called ICR1 (interactor of constitutive active ROPs) has been identified, which interacts with the exocyst component SEC3 and appears to link RAC/ROP activation to the promotion of vesicle fusion with the apical plasma membrane (Lavy et al., 2007).
To enhance our understanding of the RAC/ROP-dependent signaling network that controls F-actin organization, membrane traffic, and polar cell growth in pollen tubes, we functionally characterized tobacco RISAP, a member of a plant-specific family of poorly characterized DUF593 (domain of unknown function 593)-containing proteins. RISAP is specifically expressed in tobacco pollen tubes and interacts with the pollen tube RAC/ROP GTPase RAC5 in an activation-dependent manner. Furthermore, RISAP binds via its DUF593 domain to the globular tail domain (GTD) of the tobacco pollen tube myosin XI MYOXIpt and strongly interacts with F-actin. Interestingly, RISAP accumulates at a subapical trans-Golgi network (TGN) compartment, whose localization within a region of rapidly streaming cytoplasm is maintained by the actin cytoskeleton. This subapical TGN compartment is ideally positioned to integrate endocytic and secretory vesicle traffic at the pollen tube tip. Overexpression of RISAP appears to disrupt the function of this TGN compartment and blocks pollen tube growth by interfering with apical membrane traffic. All together, these observations are consistent with a model suggesting that RISAP acts as a RAC5 effector that regulates apical membrane traffic by modulating dynamic interactions between endomembrane compartments and the actin cytoskeleton.
RESULTS
Identification of RISAP as a RAC5 Interaction Partner in Pollen Tubes
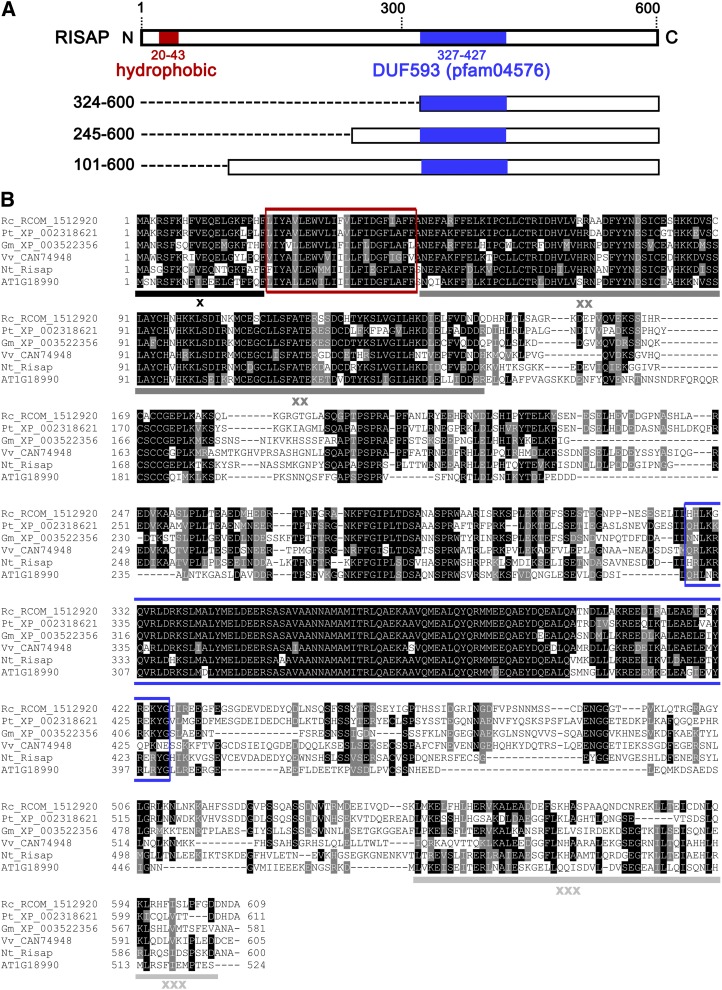
To discover RAC5 interaction partners, mutant RAC5G15V, which is unable to hydrolyze GTP and, hence, constitutively active, was used as bait to search a tobacco pollen tube cDNA library by yeast (Saccharomyces cerevisiae) two-hybrid screening. In addition to important regulators of RAC5 activity (Klahre et al., 2006; Klahre and Kost, 2006), putative RAC5 downstream effectors were identified, including a protein of which three N-terminally truncated fragments with different sizes (277, 356, and 500 amino acids) were isolated in independent screens (Figure 1A). Colony hybridization of the tobacco pollen tube cDNA library resulted in the cloning of a corresponding full-length cDNA with stop codons in all reading frames upstream of an open reading frame encoding a 600-amino acid (68-kD) protein. This protein is called RISAP (RAC5 interacting subapical pollen tube protein) hereafter. Different sequence analysis tools (Sosui, TMPred, DAS, TMHMM, HMMTOP, Psipred, and Dompred) consistently identified a hydrophobic region (amino acids 20 to 43) with similarity to transmembrane domains close to the N terminus of RISAP, in addition to a DUF593 (pfam04576, amino acids 327 to 427) located in the C-terminal half of this protein (Figure 1A). No sequence homology was detected between cleavable signal peptides and the short RISAP N terminus upstream of the hydrophobic region (amino acids 1 to 19).
Figure 1.
RISAP Domain Structure and Amino Acid Sequence Alignment with Closest Homologs.
(A) RISAP domain structure: full-length protein (top) and three truncated forms identified as RAC5 interaction partners in yeast two-hybrid screens. Numbers on the left: first and last RISAP amino acid at the N and C terminus, respectively, of each truncated form. Red box, hydrophobic domain; blue box, DUF593; dashed lines, N-terminal deletions.
(B) ClustalW alignment (http://www.ebi.ac.uk./Tools/msa/clustalw2) (Higgins et al., 1992; Larkin et al., 2007) of RISAP with homologs displaying highest amino acid sequence identities. Rc_RCOM_1512920 (Ricinus communis), 53% identity; Vv_CAN74948 (Vitis vinifera), 52% identity; Pt_XP_002318621 (Populus trichocarpa), 51% identity; Gm_XP_003522356 (Glycine max), 49% identity; AT1G18990 (Arabidopsis), 43% identity. Black shading, sequence identity; gray shading, sequence similarity; red box, hydrophobic domain; blue box, DUF593; underlined regions (x, xx, and xxx), additional highly conserved regions.
RNA gel blot analysis established that RISAP (Figure 2), like RAC5 (Klahre et al., 2006), is highly and specifically expressed in tobacco pollen and pollen tubes. Transcripts coding for both proteins were also detected at low levels in pollen-containing mature flowers and anthers. Therefore, RISAP and RAC5 not only interact with each other, but they also share the same expression pattern.
Figure 2.
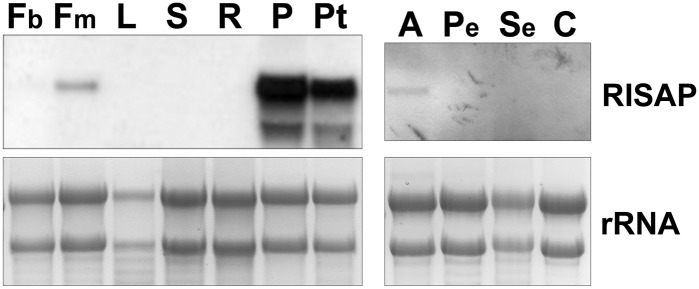
Specific Expression of RISAP in Tobacco Pollen and Pollen Tubes.
RNA gel blot analysis of RISAP RNA levels in different wild-type tobacco tissues. Blots were probed with a DIG-labeled PCR fragment corresponding to the full-length RISAP cDNA (upper panels). As a loading control, ethidium bromide-labeled ribosomal rRNA is shown (lower panels). Each lane was loaded with 10 μg total RNA. Fb, flower buds; Fm, mature flowers; L, leaves; S, stem; R, roots; P, pollen; Pt, pollen tubes; A, anthers; Pe, petals; Se, sepals; C, carpels.
Full-Length RISAP Interacts Specifically with RAC5 in Yeast Two-Hybrid and Pull-Down Assays
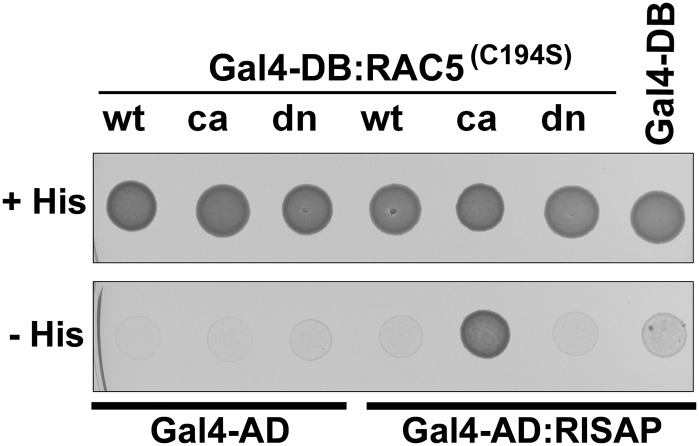
In yeast two-hybrid assays, full-length RISAP fused to the activation domain of the GAL4 transcription factor (GAL4-AD:RISAP) specifically interacted with constitutively active RAC5G15V fused to the GAL4 DNA binding domain (GAL4-DB:RAC5G15V) (Figure 3). By contrast, RISAP did not display detectable two-hybrid interaction with wild-type RAC5 or with dominant-negative mutant RAC5T20N (Figure 3), which binds nucleotides with low affinity and preferentially assumes a conformation corresponding to an inactive, nucleotide-free transition state (Feig, 1999). The three different N-terminally truncated RISAP fragments isolated in the original screens (Figure 1A) also displayed specific interaction with constitutively active RAC5G15V in yeast two-hybrid assays (Supplemental Figure 1), demonstrating the presence of a domain within the C-terminal half of RISAP (RISAP324-600), which mediates specific interaction with activated RAC5.
Figure 3.
Specific Interaction of RISAP with Constitutively Active RAC5 in Yeast Two-Hybrid Assays.
Yeast transformants coexpressing wild type (wt), constitutively active (ca; G15V), or dominant-negative (dn; T20N) RAC5 fused to the DNA binding domain of the GAL4 transcription factor (GAL4-DB) together with RISAP fused to the GAL4 activation domain (GAL4-AD) plated on histidine-containing (+) and on histidine-free (−) culture medium. All RAC5 bait proteins carried a point mutation that enhances nuclear import by preventing posttranslational prenylation (C194S; Klahre et al., 2006). Serving as negative controls were transformants coexpressing RAC5 bait proteins with just the GAL4-AD or the RISAP prey protein with just the GAL4-BD. Growth on histidine-free medium indicated two-hybrid interaction between RISAP and constitutively active RAC5.
To further investigate the specificity of the interaction between RAC5 and RISAP, the tobacco pollen tube cDNA library described above was used to amplify PCR fragments coding for tobacco RAB2 and RAB5.2, which are closely related to RAC5 (35 and 29% identical amino acids, respectively), but belong to the RAB family of small GTPases (Saito and Ueda, 2009). Wild-type, constitutively active, and dominant-negative forms of RAB2 and of RAB5.2 were tested for yeast two-hybrid interaction with full-length RISAP. Apart from a weak, barely detectable interaction between constitutively active RAB2 and RISAP, no interactions were observed in these experiments (Supplemental Figure 1).
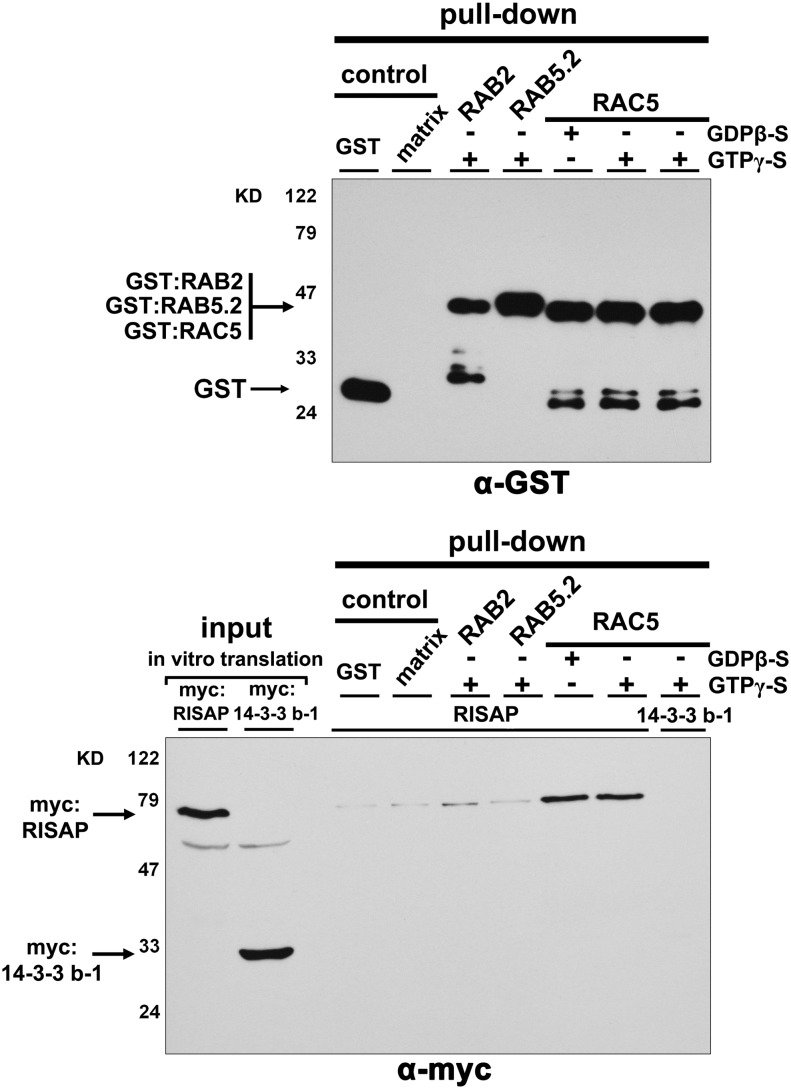
In pull-down assays, myc-tagged RISAP (myc:RISAP) produced by in vitro transcription/translation also specifically interacted with RAC5 fused to GST (glutathione S-transferase; GST:RAC5), which was purified from Escherichia coli. Interestingly, in these assays, RISAP was pulled down equally well with wild-type, constitutively active, or dominant-negative RAC5 (Supplemental Figure 2) and with wild-type RAC5 in the GTP (active) or GDP (inactive) loaded conformation (Figure 4). By contrast, interactions between myc:RISAP and GTP loaded wild-type GST:RAB2 or GST:RAB5.2 were barely detectable (Figure 4). No interaction at all was observed between GTP loaded wild-type GST:RAC5 and myc-tagged 14-3-3 b-1 (Figure 4), a tobacco protein interacting with the RAC5 upstream regulator RHOGAP1 in pollen tubes (Klahre and Kost, 2006), which was used as an additional negative control. In summary, data presented in Figures 3 and 4, and in Supplemental Figures 1 and 2, establish that full-length RISAP specifically interacts with RAC5 both in yeast two-hybrid and in pull-down assays. Interestingly, the interaction between these two proteins depends on RAC5 activation in yeast two-hybrid assays, but not in pull-down experiments.
Figure 4.
Specific Interaction of RISAP with GDP- or GTP-Loaded RAC5 in Pull-Down Assays.
In vitro-transcribed/translated myc-tagged RISAP (lower panel, first lane), or myc-tagged 14-3-3 b-1 serving as a control (lower panel, second lane), was incubated with GDPβ-S or GTPγ-S (nonhydrolyzable) loaded GST-tagged RAC5, which had been purified from E. coli and was immobilized on magnetic beads. Beads carrying immobilized GST, GST:RAB2 (GTPγ-S loaded), or GST:RAB5.2 (GTPγ-S loaded), as well as empty beads (matrix), were used as controls. Proteins associated with purified and washed beads were separated by SDS-PAGE and analyzed by immunoblotting using α-GST (upper panel) or α-myc (lower panel) antibodies.
A Short N-Terminal RISAP Fragment Interacts with RAC5 in Pull-Down Assays
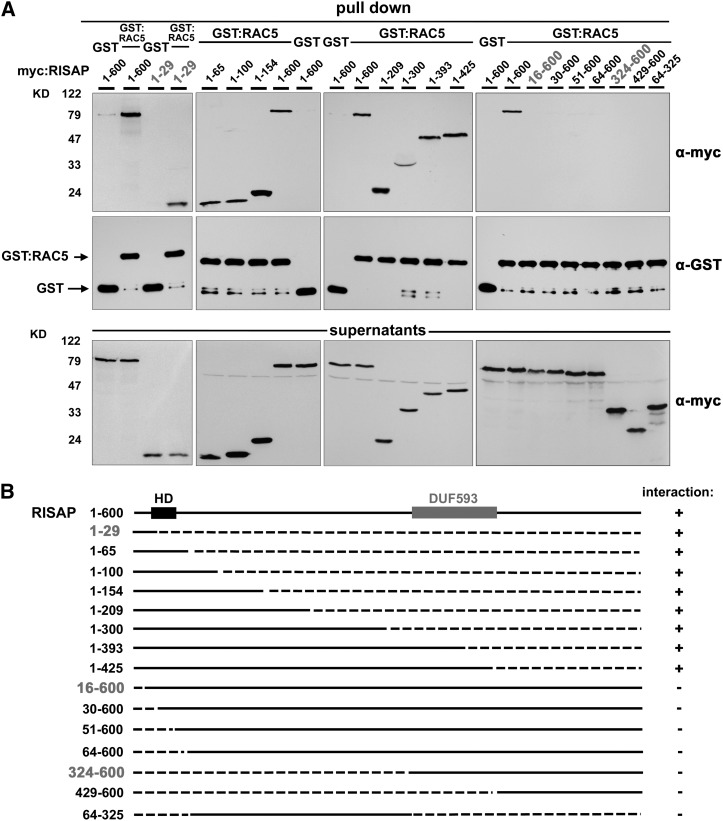
Interactions between wild-type RAC5 fused to GST (GST:RAC5) and myc-tagged N- or C-terminally truncated RISAP fragments of various lengths were analyzed in pull-down assays to identify the RISAP domain responsible for RAC5 binding. Deletion of large C-terminal fragments of RISAP did not affect interaction with RAC5 (Figure 5). An N-terminal RISAP fragment with a length of only 29 amino acids (RISAP1-29) was as effectively pulled down by RAC5 as full-length RISAP (Figure 5A, left-most column). By contrast, deletion of the first 15 amino acids at the RISAP N terminus completely abolished RAC5 interaction (RISAP16-600; Figure 5A, right-most column). This demonstrates that the first N-terminal 29 amino acids of RISAP are necessary and sufficient for RAC5 interaction in pull-down assays. RISAP therefore contains two different RAC5 interacting domains: one positioned within the C-terminal half (RISAP324-600) responsible for specific interaction with active RAC5 in yeast two-hybrid assays (see above; Figures 1A and 3), and another located at the extreme N terminus (RISAP1-29), which mediates constitutive interaction with active and inactive RAC5 in pull-down assays (Figures 4 and 5).
Figure 5.
A Short N-Terminal RISAP Fragment (RISAP1-29) Mediates Interaction with RAC5 in Pull-Down Assays.
(A) In vitro-transcribed/translated C-terminally (columns 1 to 3) and/or N-terminally (column 4) truncated myc-tagged RISAP fragments were incubated with GST-tagged RAC5, which had been purified from E. coli and was immobilized on magnetic beads. Beads carrying immobilized GST were used as controls. Columns 1 to 4 represent independent experiments, each with its own set of controls (incubation of full length RISAP1-600 with GST:RAC5 or with GST immobilized on beads). Proteins associated with purified and washed beads (first and second rows), as well as nonbound supernatant proteins (third row), were separated by SDS-PAGE and analyzed by immunoblotting using α-myc (first and third rows) or α-GST (second row) antibodies.
(B) Schematic representation of full-length RISAP and of RISAP fragments tested for interaction with RAC5 as shown in (A). Top row: full-length RISAP1-600 with a hydrophobic domain (HD; black box) near the N terminus and a DUF593 (gray box) within the C-terminal half. Lower rows: truncated RISAP fragments. Numbers on the left: first and last RISAP amino acid at the N and C terminus of each polypeptide; solid lines: RISAP full-length protein and fragments; dashed lines: N- and C-terminal deletions; column on the right: interaction with RAC5 detectable (+) or not detectable (−).
Yeast two-hybrid assays exclusively revealed activation dependent RAC5 interaction with the RISAP324-600 fragment (Figures 1A and 3; Supplemental Figure 1), suggesting that constitutive RAC5 interaction with the RISAP1-29 domain may be weaker and beyond the detection limit of this technique. By contrast, RAC5 interaction with the RISAP324-600 fragment was not observed in pull-down assays (Figure 5A, right-most panel), possibly because it depends on cofactors that are functional in living cells, but not in cell-free pull-down reactions.
RISAP Belongs to a Plant-Specific Family of DUF593-Containing Proteins
BLAST searches revealed that RISAP is a member of a plant-specific family of poorly characterized, DUF593-containing proteins. This protein family is represented in all land plants, including mosses, and comprises 16 members in Arabidopsis. Detailed phylogenetic analysis has shown that all known DUF593 proteins cluster into six structurally distinct subfamilies, which display characteristic differences in overall protein size and position of the DUF593, as well as in the presence of additional predicted functional domains (Peremyslov et al., 2013). RISAP belongs to the DUF593 protein subfamily IA, which comprises four members in Arabidopsis, including the closest RISAP homolog identified in this plant (AT1G18990; Figure 1B). Genevestigator data indicate that in Arabidopsis all four subfamily 1A members and a number of other DUF593 proteins are expressed in pollen, several of them highly preferentially (Zimmermann et al., 2004).
RISAP and its closest subfamily IA homologs from other plant species (43 to 53% amino acid sequence identity) contain highly conserved regions outside the hydrophobic domain and the DUF593 (Figure 1B), suggesting the presence of additional functional domains that have not been characterized to date. These conserved regions include the short N terminus upstream of the hydrophobic region, which overlaps with the constitutive RAC5 binding domain we have identified (RISAP1-29; Figure 5), a stretch of ∼100 amino acids directly downstream of the hydrophobic region and ∼60 amino acids at the C terminus.
Limited information is available in the literature about the functions of DUF593 proteins. The maize (Zea mays) FLOURY1 protein (NP001106064), which shares 10% identical amino acids with RISAP and belongs to the DUF593 subfamily IIIA, is associated with the endoplasmic reticulum (ER), participates in protein body formation during endosperm development, and interacts with the luminal storage protein Zein (Holding et al., 2007). Recently, three Arabidopsis DUF593 proteins were found to interact in yeast two-hybrid and in pull-down assays with the C-terminal globular cargo binding tail domain (GTD) of myosins and were therefore named myosin binding proteins 1-3 (MYOB1-3). Two of these proteins (MYOB1 and 2) appear to be associated with unidentified endomembrane compartments (Peremyslov et al., 2013). MYOB1 belongs to subfamily IA like RISAP and shares 33% identical amino acids with this protein. However, MYOB1 lacks the short N-terminal constitutive RAC5 interacting domain that we identified in RISAP, a domain that is highly conserved in the closest homologs of this protein (Figure 1B). Furthermore, MYOB1 contains unique insertions between the hydrophobic region and the DUF593, which increase its molecular mass to ∼125 kD. MYOB2 and 3 are members of subfamily IB and share 33 or 32% identical amino acids with RISAP, respectively.
Interestingly, the isolated DUF593 of MYOB2 could be pulled down from extracts of Nicotiana benthamiana leaves, in which this domain was transiently expressed, together with a recombinant myosin GTD purified from E. coli. This indicates that the binding of MYOB2 to the C-terminal GTD of target myosins is mediated by the DUF593 (Peremyslov et al., 2013).
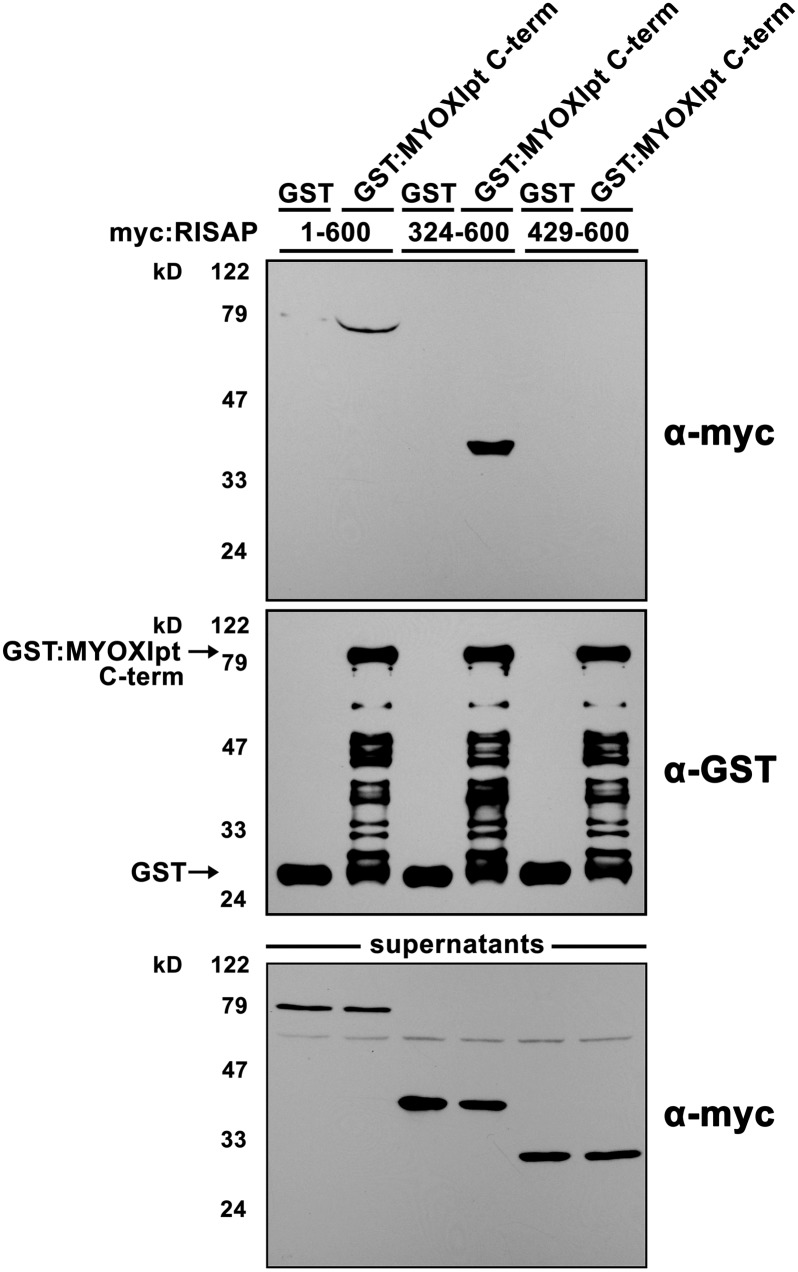
RISAP Binds via Its DUF593 to the C Terminus of the Tobacco Pollen Tube Myosin MYOXIpt
Yeast two-hybrid screening of a tobacco pollen tube cDNA library using as bait truncated RISAP46-600 (lacking the N-terminal hydrophobic region) resulted in the identification of a 608-amino acid C-terminal protein fragment (KM226781) that shares 59.6% identical amino acids with the corresponding C terminus of the well characterized Arabidopsis myosin XI MYA1 (Li and Nebenführ, 2008). The GTDs at the C termini of the identified tobacco RISAP-interacting pollen tube myosin XI (MYOXIpt; last 425 amino acids) and of Arabidopsis MYA1 (last 430 amino acids) display an even higher sequence identity (67.1%).
In pull-down assays, the 608-amino acid C-terminal tobacco MYOXIpt fragment fused to GST strongly interacted with myc-tagged full length RISAP. The interaction between these two proteins remained strong when RISAP was N-terminally truncated immediately upstream of the DUF593 (RISAP324-600) but was completely abolished by further N-terminal truncation removing the DUF593 (RISAP429-600; Figure 6). Yeast two-hybrid assays using these two truncated forms of RISAP as bait confirmed that the DUF593 is required for interaction with the C-terminal MYOXIpt fragment (Supplemental Figure 3).
Figure 6.
The DUF593 of RISAP Mediates Interaction of This Protein with the Tobacco Pollen Tube Myosin XI MYOXIpt in Pull-Down Assays.
In vitro-transcribed/translated myc-tagged full length (lanes 1 and 2) or N-terminally truncated RISAP, either with (lanes 3 and 4) or without (lanes 5 and 6) the DUF593, was incubated with a GST-tagged 608-amino acid C-terminal fragment of MYOXIpt, which had been purified from E. coli and was immobilized on magnetic beads. Beads carrying immobilized GST were used as controls. Proteins associated with purified and washed beads (first and second rows), as well as unbound supernatant proteins (third row), were separated by SDS-PAGE and analyzed by immunoblotting using α-myc (first and third row) or α-GST (second row) antibodies.
These observations establish that RISAP binds via its DUF593 to the GTD-containing C terminus of MYOXIpt. Together with the results of the recent functional characterization of Arabidopsis MYOB1-3, they strongly support the notion that proteins with a DUF593 domain may in general function as myosin receptors, whose interaction with the GTD of target myosins is mediated by this domain.
RISAP Overexpression in Pollen Tubes Disrupts Apical Membrane Traffic and Tip Growth
Three independent transgenic tobacco lines containing an RISAP RNA interference (RNAi) construct under the control of the strong pollen specific Lat52 promoter (Twell et al., 1991) were established. This RNAi construct targets a section of the RISAP gene, which codes for amino acids 204 to 333 and is not highly conserved (Figure 1B). In all three RNAi lines, RISAP expression in pollen tubes was reduced below the limit of detection by quantitative PCR analysis and immunoblotting (data not shown). This did not affect pollen tube growth in culture or in situ, as determined by transgene segregation analysis in offspring obtained after backcrossing heterozygous RNAi plants as male parents (data not shown). Segregation analysis also failed to detect defective pollen tube growth in Arabidopsis double mutants carrying T-DNA insertions in the genes coding for the two pollen tube subfamily 1A DUF593 proteins most closely related to RISAP (AT1G18990 and AT1G74830) (data not shown; T-DNA insertion lines SALK_043134, SALK_040616, and SALK_074471 obtained from the ABRC). As discussed above, in Arabidopsis pollen, two other subfamily 1A members and several DUF593 proteins belonging to different subfamilies appear to be expressed as well. These observations indicate that functional redundancy protects plant reproduction from loss of genes coding for DUF593 proteins.
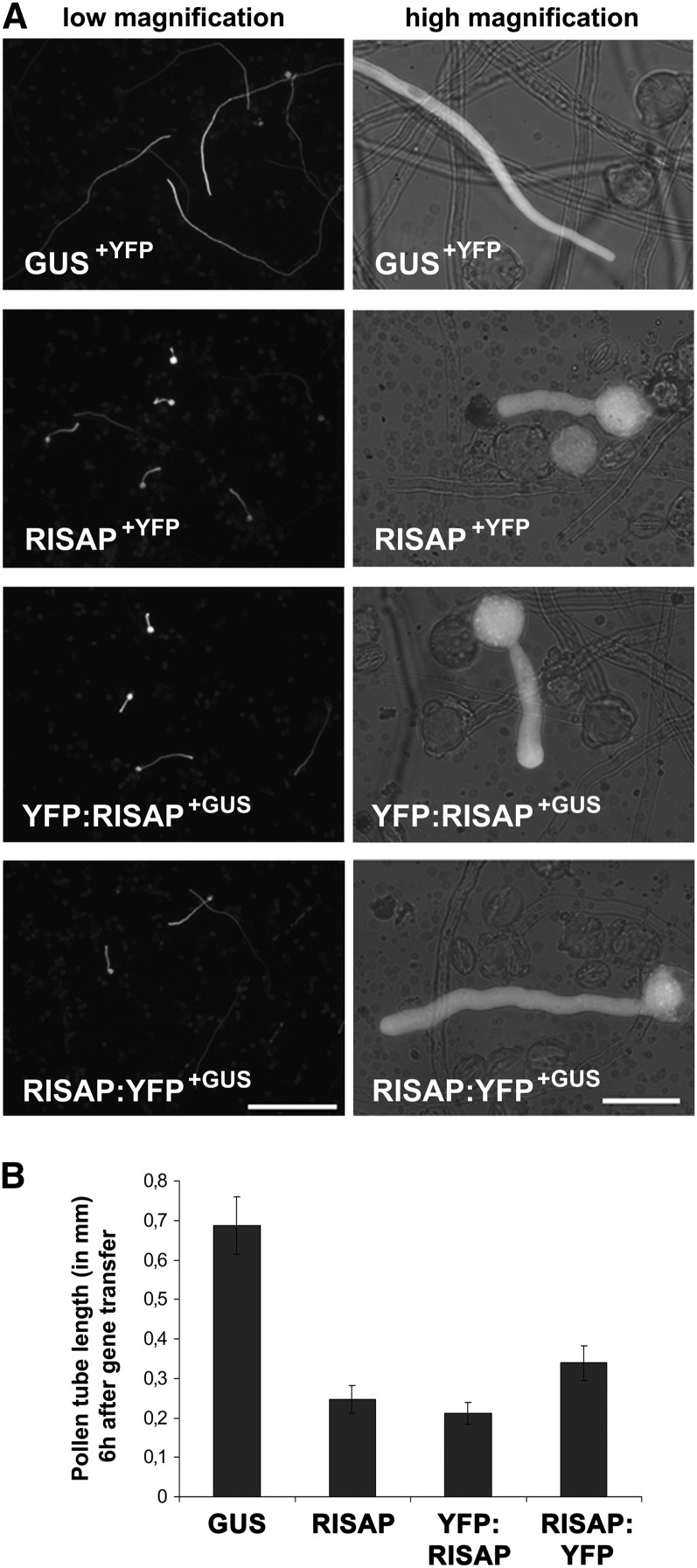
To gain insight into the cellular functions of RISAP, effects of transient overexpression of free RISAP, or of RISAP fused to YFP (yellow fluorescent protein) either at the N terminus (YFP:RISAP) or at the C terminus (RISAP:YFP), in tobacco pollen tubes were analyzed. Six hours after gene transfer the length of pollen tubes was compared, which coexpressed under the control of the Lat52 promoter either free RISAP together with YFP, the noninvasive marker protein GUS (β-glucuronidase) and RISAP YFP fusion proteins or just the two marker proteins YFP and GUS (Figure 7B). Pollen tubes expressing transgenes at high levels were measured. These pollen tubes displayed bright YFP fluorescence and were readily visible on low magnification images (Figure 7A, left column). High-level overexpression of free RISAP and of YFP:RISAP strongly reduced pollen tube growth to the same extent (Figure 7B) and caused an enlargement of the pollen tube diameter (Figure 7A, right column). RISAP:YFP overexpression had slightly, but statistically significantly, weaker effects on pollen tube length (Figure 7B).
Figure 7.
Transient High-Level RISAP Overexpression Interferes with Tobacco Pollen Tube Growth.
(A) Microscopic epifluorescence images of brightly fluorescent pollen tubes expressing the indicated proteins at high levels under the control of the Lat52 promoter 6 h after gene transfer. GUS is a marker protein whose expression does not interfere with pollen tube growth. Left column: low magnification overview images (5× lens; bar = 500 μm); right column: single pollen tubes at higher magnification (40× lens; bar = 50 μm).
(B) Statistical analysis of pollen tube length 6 h after gene transfer. Error bars: 95% confidence interval (n > 35).
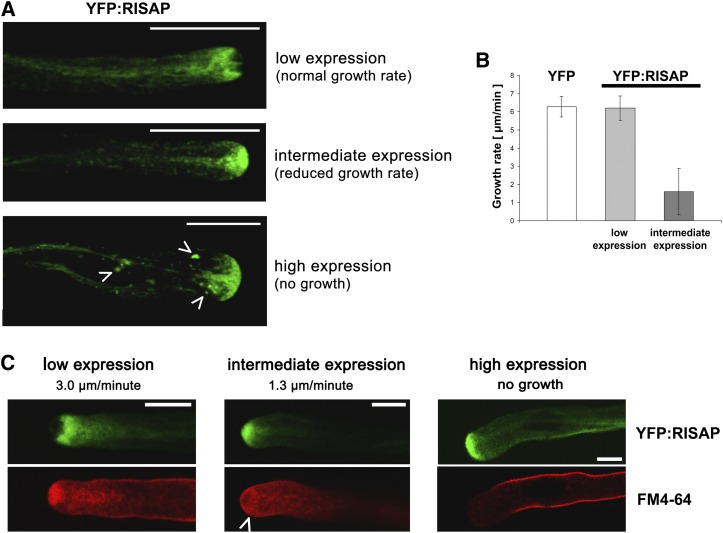
Figure 7 demonstrates that RISAP with YFP attached to its N terminus (YFP:RISAP) displayed the same activity as free RISAP in overexpression experiments. Pollen tubes transiently expressing YFP:RISAP at low levels, at which growth rate was not affected (Figure 8B), were analyzed by confocal microscopy to determine the intracellular distribution of RISAP during normal tip growth (Figure 8A, top image). Interestingly, RISAP accumulated subapically in the pollen tube cytoplasm directly behind the apical CZ. An identical intracellular distribution was displayed by RISAP:YFP (data not shown).
Figure 8.
RISAP Accumulates Subapically Directly Behind the CZ Filled with Apical Vesicles in Normally Elongating Tobacco Pollen Tubes.
(A) Single confocal optical sections through pollen tubes transiently expressing YFP:RISAP at different levels 6 h after gene transfer revealed three clearly discernable distribution patterns. Subapical YFP:RISAP accumulation was seen in pollen tubes elongating at normal growth rates ([A], top image; [B]). Apical YFP:RISAP accumulation was seen in pollen tubes, which were elongating at reduced rates ([A], central image; [B]) or which had stopped growing ([A], bottom image). Pollen tubes that had stopped growing in addition showed tip swelling (cf. Figure 7A, right column) and often contained YFP:RISAP-associated large organelles in the granular cytoplasm ([A], bottom image; arrowheads). Bars = 20 μm.
(B) Statistical analysis of the growth rates of pollen tubes expressing YFP or YFP:RISAP at low (distribution pattern as shown in [A], top image) or intermediate levels (distribution pattern as shown in [A], central image]. Error bars: 95% confidence interval (n > 10).
(C) FM4-64 labeling of apical membrane traffic in pollen tubes transiently expressing YFP:RISAP at different levels 6 h after gene transfer. Simultaneously acquired single confocal optical sections displaying YFP:RISAP (green channel) and FM4-64 (red channel) fluorescence are shown. In normally elongating pollen tubes (left column), YFP:RISAP accumulated subapically directly behind the CZ, which was labeled by FM4-64 as reported in the literature (Parton et al., 2001). In pollen tubes displaying apical YFP:RISAP fluorescence and reduced growth rates, FM4-64-labeled cytoplasmic vesicles accumulated subapically (central column, arrowhead). High-level YFP:RISAP expression, which stopped pollen tube growth and induced tip swelling, also inhibited endocytic FM4-64 internalization (right column). Bars = 10 μm.
The styryl dye FM4-64 accumulates in the plasma membrane, is endocytically internalized, is recycled to the secretory endomembrane system, and labels apical vesicles in pollen tubes (Parton et al., 2001). FM4-64 staining of normally growing tobacco pollen tubes expressing YFP:RISAP confirmed that this fusion protein accumulates directly behind the apical CZ (Figure 8C, left column). At intermediate expression levels, YFP:RISAP invaded the extreme apex and caused the CZ to disappear (Figures 8A, central image, and 8C, central column), apparently by blocking access of FM4-64 labeled vesicles to this zone and by forcing them to accumulate behind the YFP:RISAP filled apex (Figure 8C, central column, arrowhead). Not surprisingly, this correlated with strongly reduced pollen tube growth rates (Figure 8B). Consistent with data shown in Figure 7, pollen tubes expressing YFP:RISAP at high levels stopped elongating completely and displayed an enlarged diameter (Figures 8A, bottom image, and 8C, right column). In addition to accumulating in the apex, YFP:RISAP often associated with large organelles in the granular cytoplasm of such pollen tubes (Figure 8A, bottom image, arrowheads). FM4-64 failed to be internalized under these conditions (Figure 8C, right column), demonstrating that not only cell expansion but also endocytic membrane uptake was inhibited.
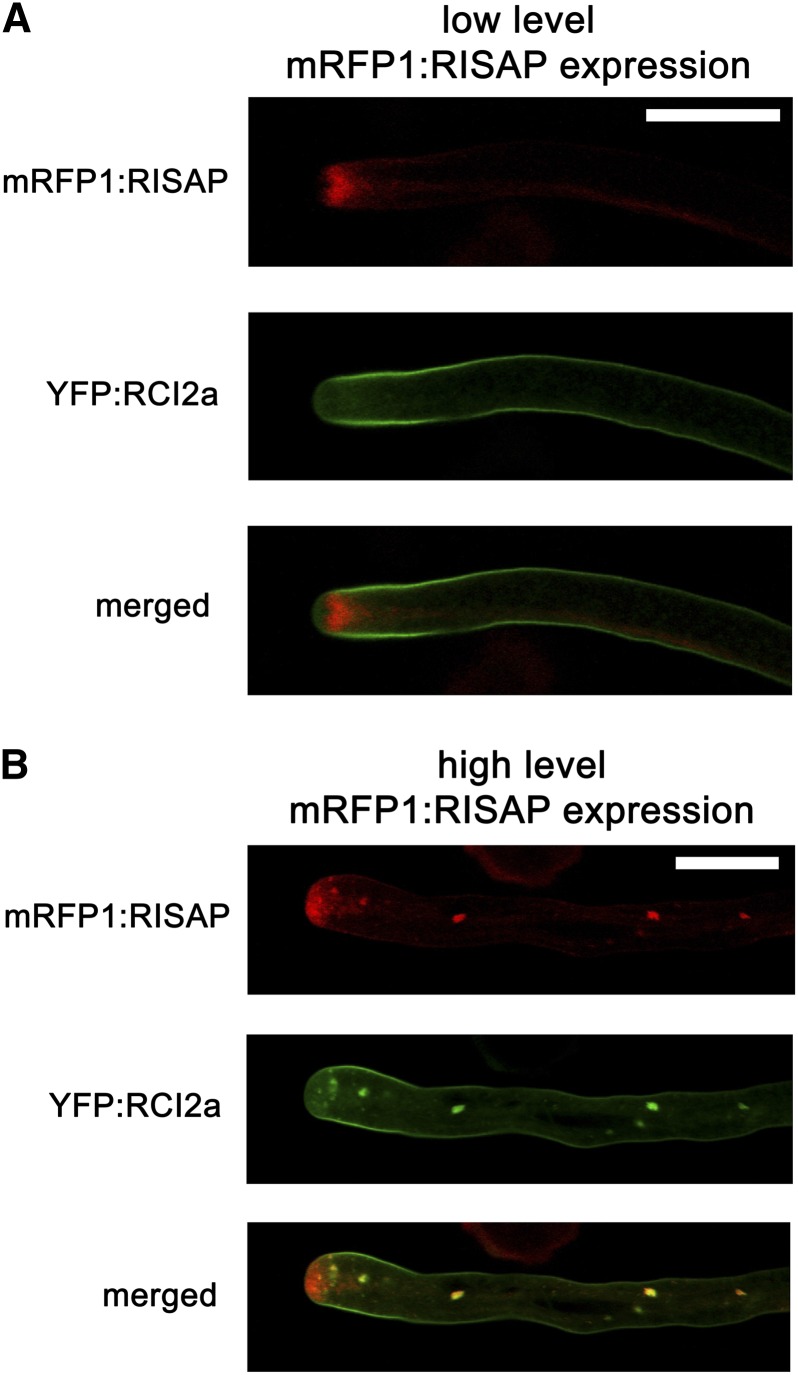
Data presented in Figures 7 and 8 strongly suggest that RISAP overexpression at high levels blocks pollen tube tip growth by interfering with apical membrane traffic. YFP fused to RCI2a, an Arabidopsis protein containing two transmembrane domains (Medina et al., 2007), is transported through the secretory endomembrane system and accumulates in the plasma membrane of normally elongating tobacco pollen tubes coexpressing an mRFP1:RISAP fusion protein at low levels (Figure 9A). When expressed at high levels, mRFP1:RISAP, as discussed above, interferes with pollen tube growth, invades the apex, and often accumulates at organelles in the granular cytoplasm (Figure 9B). YFP:RCI2a colocalized with mRFP1:RISAP at these organelles (Figure 9B), indicating that they are part of the endomembrane system through which membrane proteins are transported. This further supports the notion that RISAP has important functions in membrane traffic in pollen tubes.
Figure 9.
Tobacco RISAP Transiently Overexpressed at High Levels Partially Colocalizes with Arabidopsis RCI2a, an Integral Plasma Membrane Protein Transported through the Endomembrane System.
Simultaneously acquired single confocal optical sections through tobacco pollen tubes coexpressing mRFP1:RISAP (red channel) and YFP:RCI2a (green channel) are displayed separately as well as overlaid (merged).
(A) In normally elongating pollen tubes displaying subapical mRFP1:RISAP labeling (cf. Figure 8A, top image), YFP:RCI2a accumulated in the plasma membrane. Bar = 20 μm.
(B) mRFP1:RISAP expressed at high levels blocked pollen tube growth and often associated with cytoplasmic organelles (cf. Figure 8A, bottom image), where it colocalized with YFP:RCI2a. Bar = 20 μm.
RISAP Is Associated with a Subapical TGN Compartment
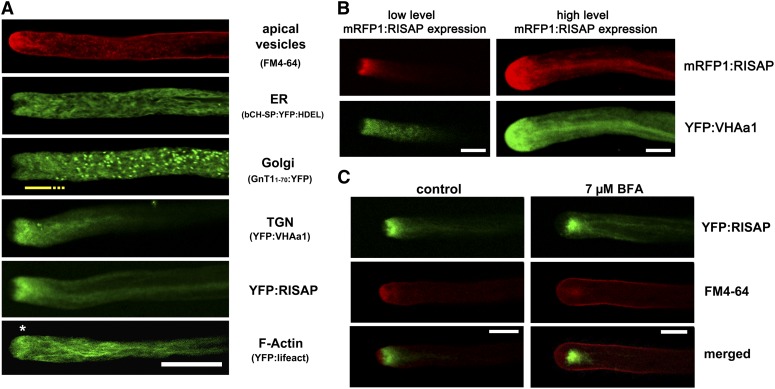
To identify the endomembrane compartment with which RISAP is associated, the intracellular distribution of YFP:RISAP was compared with that of markers for different organelles in transiently transformed or dye-labeled tobacco pollen tubes (Figure 10A). YFP fused to an N-terminal signal peptide and a C-terminal HDEL targeting sequence (At-bCH-SP:YFP:HDEL) accumulates in the pollen tube ER, which forms an interconnected network of membrane tubules that extends throughout most of the cytoplasm, but is excluded from the FM4-64-labeled CZ (Figure 10A). The ER as a whole is rapidly moving with the cytoplasmic streaming (Lovy-Wheeler et al., 2007). Golgi stacks visualized by YFP attached to the membrane anchor of a potato (Solanum tuberosum) N-acetylglucosaminyltransferase (St-GNT11-70:YFP; Wenderoth and von Schaewen, 2000) are also highly motile. Interestingly, they are evenly distributed throughout most of the cytoplasm but are excluded not only from the CZ but also from a small subapical region of granular cytoplasm into which the ER extends (Figure 10A, yellow underlining). YFP fused to the Arabidopsis V-ATPase subunit VHAa1, which serves as a TGN marker (Dettmer et al., 2006), accumulates in this subapical Golgi-free cytoplasmic region and displays a distribution pattern similar to that of YFP:RISAP (Figure 10A). Coexpression of YFP:VHAa1 and mRFP1:RISAP confirmed that the distribution of these two proteins is largely overlapping (Figure 10B), not only in normally elongating pollen tubes expressing mRFP1:RISAP at low levels (left column), but also in pollen tubes that have stopped elongating as a consequence of high-level mRFP1:RISAP expression (right column). Other TGN markers described in the literature (Arabidopsis SYP41 and SYP61; Uemura et al., 2004) were also tested for colocalization with RISAP in tobacco pollen tubes. These markers showed labeling patterns similar to St-GNT1-70:YFP (data not shown) and therefore appear to be associated with TGN subcompartments attached to Golgi stacks, which are distinct from the subapical TGN compartment labeled by fluorescent RISAP and VHAa1 fusion proteins.
Figure 10.
RISAP Is Associated with a Subapical TGN Compartment in Tobacco Pollen Tubes.
(A) Single confocal optical sections through normally growing pollen tubes labeled with FM4-64 or expressing markers for different endomembrane compartments or for F-actin. The ER (At-bCH-SP:YFP:HDEL) was evenly distributed throughout the cytoplasm but was excluded from the FM4-64-labeled CZ. Golgi stacks (St-GnT11-70:YFP) were also evenly distributed in all ER-containing regions of the cytoplasm, with the exception of an ∼10-μm-long supapical region (yellow line). YFP:RISAP and the TGN marker YFP:VHAa1 accumulated exactly in this Golgi-free subapical region, where a cortical F-actin fringe (*; YFP:lifeact; Riedl et al., 2008) was also observed. Bar = 20 μm.
(B) Simultaneously acquired single confocal optical sections through pollen tubes coexpressing mRFP1:RISAP (red channel) and YFP:VHAa1 (green channel). mRFP1:RISAP and YFP:VHAa1 displayed largely overlapping distribution patterns in normally growing pollen tubes expressing mRFP1:RISAP at low levels (left column), as well as in pollen tubes that had stopped elongating due to high level mRFP1:RISAP expression (right column) (cf. Figure 8). Bars = 10 μm.
(C) Simultaneously acquired single confocal optical sections through tobacco pollen tubes expressing YFP:RISAP (green channel) and labeled with FM4-64 (red channel) are displayed separately as well as overlaid (merged). In normally growing untreated pollen tubes, YFP:RISAP accumulated subapically directly behind the FM4-64-labeled CZ (left column, cf. Figure 8C). Both markers translocated to the single subapical BFA body formed in tobacco pollen tubes upon BFA treatment (right column). Bars = 10 μm.
Brefeldin A (BFA) blocks the activity of regulators of membrane trafficking and causes the formation of BFA bodies, which are tubular-vesicular aggregates containing TGN elements (Lam et al., 2009; Langhans et al., 2011). In plant cells, BFA bodies are labeled by FM4-64, which is trapped in these structures together with other plasma membrane components that normally undergo constant endocytic recycling. In tobacco pollen tubes, BFA treatment inhibits cell expansion and causes the formation of a single subapical BFA body in which FM4-64 accumulates (Parton et al., 2003; Helling et al., 2006). When YFP:RISAP-expressing tobacco pollen tubes were incubated with BFA, the YFP:RISAP-labeled TGN compartment translocated to the BFA body, where it colocalized with FM4-64 (Figure 10C).
Together, these results establish that, in normally growing pollen tubes, RISAP is associated with a specialized TGN compartment that is detached from Golgi stacks and is subapically positioned between the Golgi-containing granular cytoplasm and the CZ.
F-Actin Maintains Subapical Localization of the RISAP-Associated TGN Compartment
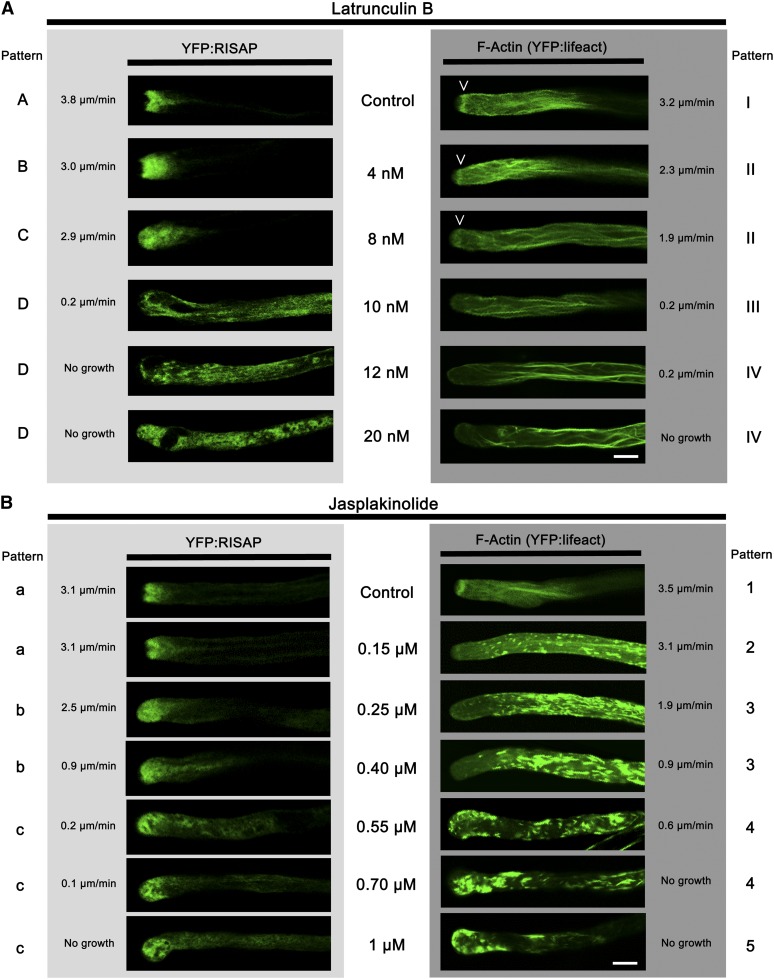
The ER and other organelles (e.g., mitochondria) are rapidly moving with the cytoplasmic streaming through the supapical region in which the RISAP-associated TGN compartment is positioned (Lovy-Wheeler et al., 2007). How can this compartment remain stationary within a region of streaming cytoplasm? F-actin labeling in normally elongating tobacco pollen tubes expressing YFP:lifeact (Riedl et al., 2008; Vidali et al., 2009) established that the subapical cortical F-actin fringe, one of the components of the pollen tube actin cytoskeleton, localizes to the same cytoplasmic region as the YFP:RISAP-associated TGN compartment (Figures 10A and 11). Treatment with latrunculin B (LATB), which interferes with actin polymerization (Morton et al., 2000; Yarmola et al., 2000), inhibited pollen tube growth, disrupted F-actin organization, and caused YFP:RISAP redistribution throughout the cytoplasm (Figure 11A, Table 1). Dose–response experiments showed that increasing the LATB concentration enhanced these effects and that the subapical actin fringe was more sensitive to LATB than longitudinally oriented actin cables in the granular cytoplasm (Gibbon et al., 1999; Vidali et al., 2001; Figure 11A, Table 1). Interestingly, LATB concentrations of 10 nM or higher, which caused the total disappearance of the F-actin fringe, also resulted in a complete loss of apical YFP:RISAP accumulation and in a massive redistribution of this fusion protein throughout the cytoplasm (Figure 11A, Table 1).
Figure 11.
The Actin Cytoskeleton Maintains Subapical Positioning of the RISAP-Associated TGN Compartment.
Single confocal optical sections through stably transformed tobacco pollen tubes expressing YFP:RISAP (SR1YFP:RISAP; left column) or the F-actin marker YFP:lifeact (Riedl et al., 2008; right column) are shown. Drug concentrations used and growth rates of individual analyzed pollen tubes are indicated next to each image. Three to five distinct marker distribution patterns were observed in each experiment (A to D, a to c: left border; I to IV, 1 to 5: right border). For all experimental conditions, the percentage of analyzed pollen tubes displaying specific patterns is listed in Tables 1 and 2. Bars = 10 μm.
(A) In a dose-dependent manner, the F-actin-destabilizing drug LATB disrupted F-actin organization, inhibited pollen tube growth, and caused YFP:RISAP redistribution throughout the cytoplasm. The disappearance of the subapical F-actin fringe (arrowheads), which was more LATB sensitive than longitudinal F-actin bundles, correlated with a complete loss of polarized YFP:RISAP distribution.
(B) The F-actin stabilizing drug JASP also disrupted F-actin organization and inhibited pollen tube growth in a dose-dependent manner. However, polarized YFP:RISAP accumulation at the pollen tube tip was maintained even at highest JASP concentrations.
Table 1. Percentage of Analyzed Pollen Tubes Displaying Specific LATB-Induced Marker Distribution Patterns Shown in Figure 11A.
| YFP:RISAP |
YFP:lifeact |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pattern | A | B | C | D | I | II | III | IV | |
| LATB (nM) | 0 | 100% | 100% | ||||||
| 4 | 50% | 50% | 30% | 70% | |||||
| 8 | 30% | 70% | 30% | 70% | |||||
| 10 | 20% | 80% | 100% | ||||||
| 12 | 100% | 100% | |||||||
| 20 | 100% | 100% | |||||||
A short description of the distinct YFP:RISAP (A to D) and YFP:lifeact (I to IV) distribution patterns displayed in Figure 11A is provided below. These patterns are observed in pollen tubes treated with different concentrations of LATB. The distribution of both markers was investigated in at least 10 pollen tubes at each drug concentration. For each marker and drug concentration, the percentage of analyzed pollen tubes displaying specific distribution patterns is indicated. A, normal subapical accumulation; B, expanded subapical accumulation; C, diffuse apical accumulation; D, redistribution throughout cytoplasm. I, normal subapical actin fringe; II, reduced subapical actin fringe; III, no subapical actin fringe and normal actin filaments in the shank; IV, aggregated actin filaments in the shank.
To exclude the possibility that YFP:RISAP redistribution was an indirect effect of the inhibition of tube growth, rather than a direct consequence of LATB-induced F-actin disruption, pollen tubes were also treated with jasplakinolide (JASP), a drug that stabilizes F-actin (Bubb et al., 1994; Holzinger, 2009). Like LATB, JASP inhibited pollen tube growth and disrupted F-actin organization in a dose-dependent manner (Figure 11B, Table 2). However, even at the highest JASP concentrations, YFP:RISAP remained at the pollen tube tip (Figure 11B, Table 2). F-actin stabilization thus prevented YFP:RISAP redistribution throughout the cytoplasm even in the absence of cell expansion. YFP:RISAP labeling extended into the extreme apex of JASP treated pollen tubes showing reduced or no growth (Figure 11B, Table 2), presumably because the CZ in these cells had shrunk or disappeared.
Table 2. Percentage of Analyzed Pollen Tubes Displaying Specific JASP-Induced Marker Distribution Patterns Shown in Figure 11B.
| YFP:RISAP |
YFP:lifeact |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pattern | a | b | c | 1 | 2 | 3 | 4 | 5 | |
| JASP (μM) | 0 | 100% | 100% | ||||||
| 0.15 | 100% | 100% | |||||||
| 0.25 | 30% | 70% | 100% | ||||||
| 0.4 | 100% | 100% | |||||||
| 0.55 | 20% | 80% | 100% | ||||||
| 0.7 | 100% | 40% | 60% | ||||||
| 1 | 100% | 10% | 90% | ||||||
A short description of the distinct YFP:RISAP (a to c) and YFP:lifeact (1 to 5) distribution patterns displayed in Figure 11B is provided below. These patterns are observed in pollen tubes treated with different concentrations of JASP. The distribution of both markers was investigated in at least 10 pollen tubes at each drug concentration. For each marker and drug concentration, the percentage of analyzed pollen tubes displaying specific distribution patterns is indicated. a, normal subapical accumulation; b, diffuse apical accumulation extending into the shank; c, diffuse apical accumulation. 1, normal subapical actin fringe; 2, reduced subapical actin fringe sometimes shifted towards the apex; 3, no subapical actin fringe with actin filaments and aggregates in the shank; 4, actin aggregates in the shank and at the apex; 5, actin aggregates at the apex.
Treatment with the myosin inhibitor 2,3-butanedione 2-monoxime (BDM), which immobilizes myosins on F-actin by slowing down phosphate release after ATP hydrolysis (Bond et al., 2013), also blocked pollen tube growth and trapped YFP:RISAP at the pollen tube tip (Supplemental Figure 4), confirming that the inhibition of cell expansion is not responsible for LATB-induced redistribution of this fusion protein.
Data described above strongly suggest that myosin-dependent interactions with the F-actin cytoskeleton, in particular with the F-actin fringe, are essential for the maintenance of the normal localization of the RISAP-associated TGN compartment at the pollen tube tip directly behind the CZ.
RISAP Is a Peripheral Membrane Protein That Interacts with F-Actin
The hydrophobic region near the N terminus of RISAP (Figure 1) potentially mediates the association of this protein with the subapical TGN compartment. This notion is strongly supported by the observation that YFP-tagged truncated RISAP composed of only the N terminus with the hydrophobic region and of the DUF593 (YFP:RISAPN-ter/HD:DUF593) displayed essentially the same subapical localization as full-length YFP:RISAP (Supplemental Figure 5).
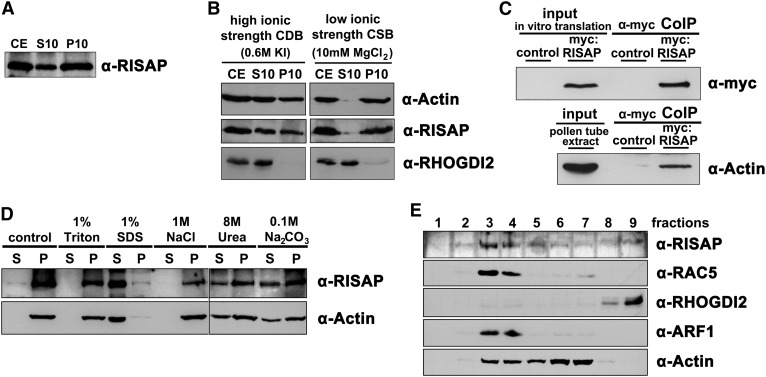
To further investigate the possibility that RISAP via myosin interaction dynamically anchors the subapical pollen tube TGN compartment within the actin cytoskeleton, we employed stably transformed, homozygous tobacco lines (SR1YFP:RISAP), which expressed YFP:RISAP at low levels specifically in pollen tubes. SR1YFP:RISAP pollen tubes displayed subapical YFP:RISAP accumulation and expanded at normal rates (Figures 11A and B, top row, control). Extracts prepared from such pollen tubes were fractionated by high-speed centrifugation and analyzed by immunoblotting using a polyclonal peptide antibody directed against the RISAP C terminus. In addition to endogenous RISAP, this antibody recognizes other pollen tube proteins of the same molecular weight, but allows specific detection of the larger YFP:RISAP fusion protein.
After centrifugation of SR1YFP:RISAP pollen tube extracts at 10,000g (producing the 10K pellet and 10K supernatant), a large proportion of the YFP:RISAP fusion protein partitioned to the 10K pellet (P10: 58% ± 6% [sd], n = 5), which contains cellular fragments including organelles and large cytoskeletal structures. A slightly smaller amount was found in the 10K supernatant (S10: 42% ± 6% [sd], n = 5), in which soluble proteins, short cytoskeletal filaments, and microsomal membrane proteins are enriched (Figure 12A). When this experiment was repeated with extracts prepared in a low ionic strength CSB buffer, which promotes actin polymerization (Abe and Davies, 1991), both actin (S10: 6% ± 4% [sd], n = 3) and YFP:RISAP (S10: 11% ± 4% [sd], n = 3) were almost entirely transferred to the 10K pellet, whereas the soluble protein RHOGDI2 (Klahre et al., 2006) remained in the 10K supernatant (Figure 12B). By contrast, in high ionic strength CDB extracts supplemented with potassium iodide, in which F-actin stability is reduced (Payrastre et al., 1991; Tan and Boss, 1992; Cox and Muday, 1994; S10: 49% ± 4% [sd], n = 3), YFP:RISAP was solubilized to some extent (S10: 60% ± 5% [sd], n = 3; compare Figures 12A and 12B). This demonstrates that F-actin interaction accounts for YFP:RISAP accumulation in the 10K pellet. To confirm that RISAP interacts with actin, a coimmunoprecipitation experiment was performed. Figure 12C shows that actin could be specifically pulled down from pollen tube extracts along with in vitro-transcribed/translated myc-tagged RISAP. Attempts to demonstrate direct in vitro interaction between purified recombinant actin and RISAP failed, indicating that RISAP is likely to indirectly interact with F-actin in vivo, presumably via myosin binding.
Figure 12.
RISAP Is a Peripheral Membrane Protein That Interacts with F-Actin.
To investigate RISAP membrane association and F-actin binding, extracts of normally growing stably transformed tobacco (SR1YFP:RISAP) pollen tubes displaying subapical YFP:RISAP accumulation were fractionated using different high-speed centrifugation techniques. Fractions obtained were analyzed by immunoblotting using the indicated antibodies including a polyclonal peptide antibody directed against the RISAP C terminus (α-RISAP) ([A], [B], [D], and [E]). In addition, actin coimmunoprecipitation from wild-type tobacco pollen tube extracts together with myc-tagged RISAP was investigated (C).
(A) YFP:RISAP partitioning between supernatant (S10) and pellet (P10) after centrifugation of regular crude SR1YFP:RISAP extract (CE) at 10,000g.
(B) Partitioning of YFP:RISAP, actin, and the soluble protein RHOGDI2 between supernatant (S10) and pellet (P10) after 10,000g centrifugation of a crude SR1YFP:RISAP extract (CE) prepared in high ionic strength CDB buffer, which destabilizes F-actin (left column), or in low ionic strength CSB buffer, which promotes actin polymerization (right column).
(C) In vitro-transcribed/translated myc-tagged RISAP was incubated in a wild-type tobacco pollen tube extract and immunoprecipitated using an α-myc antibody coupled to beads. Proteins associated with washed beads (α-myc CoIP), as well as myc:RISAP levels in the in vitro transcription/translation reaction mix and actin levels in pollen tube extracts (input), were analyzed by immunoblotting using α-myc (top panel) or α-actin antibodies (bottom panel). myc:RISAP free in vitro transcription/translation reaction mix added to pollen tube extract was used as a control.
(D) S10 supernatant obtained by centrifugation of crude SR1YFP:RISAP extract at 10,000g was centrifuged at 150,000g to separate membrane-associated proteins in microsomal pellets from soluble proteins in the supernatant. Microsomal pellets were resuspended in buffer containing Triton X-100, SDS, NaCl, urea, or Na2CO3 at the indicated concentrations to solubilize different classes of membrane proteins. After incubation, resuspended microsomal pellets were centrifuged again at 150,000g. RISAP (top panel) and actin (bottom panel) partitioning between supernatants (S) and pellets (P) was analyzed by immunoblotting.
(E) S10 supernatant obtained by centrifugation of crude SR1YFP:RISAP extract at 10,000g was adjusted to 80% sucrose and sequentially overlaid with two layers of buffer containing 65 and 10% sucrose, respectively. After centrifugation at 100,000g, nine fractions were collected starting at top of the gradient. The presence of RISAP as well as of actin in each of these fractions was analyzed by immunoblotting and compared with the distribution patterns of a soluble protein (RHOGDI2) and of two membrane-associated proteins (RAC5 and ARF1).
After centrifugation of the 10K supernatant of regular SR1YFP:RISAP pollen tube extracts at 150,000g, YFP:RISAP was exclusively detected in the 150K pellet, demonstrating that it is not a soluble protein (Figure 12D). After treatment with Na2CO3 or urea, which can solubilize peripheral but not integral membrane or monotopic proteins (Salinas et al., 2011), YFP:RISAP was partially transferred from the 150K pellet to the 150K supernatant (Figure 12D). This strongly suggests that the hydrophobic region of RISAP (Figure 1, amino acids 20 to 43) does not function as a transmembrane domain. YFP:RISAP partitioning to the 150K pellet was completely resistant to NaCl, indicating that membrane association of this protein is not based on electrostatic interactions alone. Most membrane-associated proteins are solubilized by 1% Triton X-100, which dissolves regular microsomal vesicles, whereas 1% SDS denatures and solubilizes all proteins. Interestingly, YFP:RISAP remained in the 150K pellet even in the presence of 1% Triton X-100 (Figure 12D). As F-actin was shown to be highly Triton resistant, but can be destabilized by Na2CO3 or urea (Nagy and Jencks, 1965; Tan and Boss, 1992), we investigated the possibility that F-actin interaction may retain RISAP not only in the 10K pellet (Figure 12B) but also in the 150K pellet. We found that YFP:RISAP and actin partitioning between the supernatant and pellet after 150K centrifugation closely correlated under all conditions tested (Figure 12D).
To directly demonstrate that F-actin interaction promotes YFP:RISAP accumulation in the 150K pellet and to analyze the association of YFP:RISAP with microsomal membranes independently of this interaction, attempts were made to depolymerize pollen tube F-actin using a range of different drugs, buffers, and protocols (including continuous exposure of cultured pollen tubes, as well as of extracts and microsomal fractions prepared from them, to F-actin destabilizing conditions). Like similar efforts by other researchers (e.g., Gibbon et al., 1999), these attempts were unsuccessful. Treatment of microsomal fractions with nonhydrolyzable ATPγS, which potentially detaches myosins from F-actin (Holmes, 1997), partially solubilized not only RISAP but also actin, although ATP binding promotes the polymerization of actin monomers (Carlier, 1990). By contrast, hydrolysable ATP did not affect the accumulation of RISAP or actin in microsomal fractions (data not shown). These observations indicate complex effects of ATP and ATPγS on interactions between microsomal proteins and are therefore not conclusive.
Because it was impossible to determine with certainty whether YFP:RISAP partitioning to the 150K pellet was reflecting membrane association, F-actin interaction, or both, pollen tube extracts were also analyzed by membrane flotation centrifugation. To this end, a SR1YFP:RISAP pollen tube extract adjusted to a sucrose concentration of 80% was sequentially overlaid in a centrifugation tube with two layers of buffer containing 65 and 10% sucrose, respectively. Immunoblotting after high-speed centrifugation revealed that, as expected, the soluble protein RHOGDI2 (Klahre et al., 2006) was present in the 80% sucrose layer at the bottom of the tube (Figure 12E, fractions 8 and 9). By contrast, together with RAC5 and ARF1, two proteins known to be membrane associated (Kost et al., 1999b; Takeuchi et al., 2002), YFP:RISAP had migrated to the interface between the 65 and 10% sucrose layers (Figure 12E, fractions 3 and 4), in which microsomal membranes accumulate. Although F-actin also migrated toward this interface, most of it remained in fractions 5 to 7 closer to the tube bottom. Only very little soluble G-actin was detected in fractions 8 and 9. These data establish that YFP:RISAP is associated with microsomal membranes and cofractionates with only a minor proportion of the pollen tube F-actin.
In summary, the experiments described in this section demonstrate that RISAP interacts with F-actin, presumably indirectly via DUF593-mediated myosin binding. Furthermore, they establish that RISAP is associated with microsomal membranes, although it does not appear to be an integral membrane protein despite the presence of a hydrophobic region near the N terminus. These findings are consistent with a function of RISAP in maintaining the subapical localization of the TGN compartment with which it is associated via myosin mediated F-actin interaction.
In Vivo Interaction between RAC5 and RISAP Is Detectable at the Subapical TGN
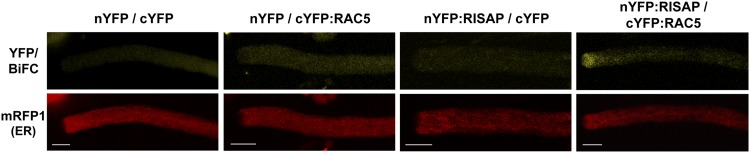
YFP-tagged RAC5 can be detected at the plasma membrane specifically at the pollen tube apex (Klahre et al., 2006), is inactivated at the flanks of the tip by RHOGAP1 (Klahre and Kost, 2006), and is recycled from there through the cytoplasm back to apical plasma membrane by RHOGDI2 (Klahre et al., 2006). Like related animal proteins (Michaelson et al., 2001; Wright and Philips, 2006), newly synthesized RAC5 possibly is targeted to the ER after prenylation and may subsequently be transported to the apical plasma membrane along the secretory pathway on the cytoplasmic surface of the endomembrane system. RISAP accumulates to levels detectable by YFP tagging at the subapical TGN (e.g., Figure 8A, top image) but may be associated with other endomembrane compartments as well. To identify sites of in vivo interaction between RAC5 and RISAP, bimolecular fluorescence complementation (BiFC) experiments were performed. Normally growing tobacco pollen tubes transiently coexpressing nonfluorescent N-terminal (nYFP) and C-terminal (cYFP) YFP fragments fused to the N terminus of RISAP (nYFP:RISAP) or RAC5 (cYFP:RAC5), respectively, displayed YFP fluorescence resulting from BiFC specifically in the subapical region (Figure 13, right-most column), in which the RISAP-associated TGN is positioned. Coexpressed ER-targeted mRFP1 facilitated the identification of transformed cells in these experiments. These data demonstrate that RAC5 and RISAP interact not only in yeast two-hybrid and in pull-down assays but also in situ in elongating pollen tubes. Furthermore, they establish that detectable in vivo interactions between these two proteins occur at the subapical TGN.
Figure 13.
BiFC Showing in Vivo Interaction between RAC5 and RISAP at the Subapical TGN.
Simultaneously acquired single confocal optical sections through transiently transformed tobacco pollen tubes are shown, which coexpressed ER-targeted mRFP1 (At-bCH-SP:mRFP1:HDEL) (red channel), along with nonfluorescent N-terminal (nYFP) and C-terminal (cYFP) YFP fragments either as free proteins, or fused to the N terminus of RISAP (nYFP:RISAP) or of RAC5 (cYFP:RAC5), respectively (green channel). Coexpression of ER-targeted mRFP1 facilitated the identification of transformed pollen tubes. Pollen tubes coexpressing nYFP:RISAP and cYFP:RAC5 displayed subapical YFP fluorescence resulting from BiFC, which indicated in vivo interaction between RAC5 and RISAP at the subapical TGN (right column; of 27 analyzed normally growing pollen tubes, 18 [67%] displayed subapical BiFC, whereas 9 [33%] only showed background fluorescence). Control pollen tubes coexpressing either free nYFP and cYFP (left column; 35 pollen tube analyzed), free nYFP and cYFP:RAC5, or nYFP:RISAP and free cYFP (central columns; 49 and 26 pollen tube analyzed, respectively), never displayed detectable BiFC. Bars = 10 μm.
DISCUSSION
Functions and Regulation of RISAP, a RAC5 Effector
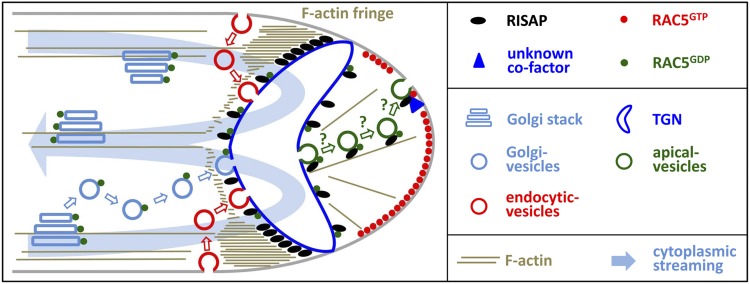
A model of RISAP functions and regulation, which integrates all observations described above, is displayed in Figure 14. RISAP accumulates to the highest levels subapically in pollen tubes, where it appears to interact in a myosin-dependent manner with the F-actin fringe and associates with a specialized TGN compartment to contribute to the stable positioning of this compartment within a region of rapidly streaming cytoplasm. We postulate that RISAP may remain associated with the membrane of secretory vesicles budding from the subapical TGN compartment and may support the transport of these vesicles along fine actin filaments toward the apical plasma membrane. Recent results (Peremyslov et al., 2013) strongly supported by functional characterization of RISAP presented here suggest that DUF593 proteins may constitute a family of myosin receptors, which are associated with the surface of specific plant organelles, bind to the GTD of myosin motor proteins, and thereby promote actin-dependent organelle motility. Myosin-dependent motility of RISAP along F-actin may mediate not only dynamic positioning of the RISAP-associated TGN compartment by the supapical F-actin fringe, but also transport of apical vesicles along fine actin filaments.
Figure 14.
Model of RISAP Functions and Regulation.
The results of this study strongly suggest that RISAP functions as a RAC/ROP effector that contributes to the F-actin-dependent maintenance of the spatial organization of apical membrane traffic, which is essential for pollen tube tip growth. A detailed description of the schematic model shown here is given in the main text in the first part of the discussion. Organelles are not drawn to scale and are represented by sketches that only partially reflect structural organization. In particular the subapical TGN, whose structure remains to be determined, is displayed in a simplified manner.
After prenylation and targeting to the ER, newly synthesized RAC5 may travel on the surface of Golgi stacks and Golgi vesicles to the subapical TGN (Michaelson et al., 2001; Wright and Philips, 2006), where it can constitutively interact with the RISAP N terminus (RISAP1-29; Figures 4 and 5, pull-down data). RAC5 may then continue its journey to the apical plasma membrane on the surface of secretory vesicles, possibly in a complex with RISAP as postulated in the model. Our model predicts that RAC5 activation at the apical plasma membrane (Hwang et al., 2005) induces the recruitment of an unknown cofactor required for activation-dependent RAC5 interaction with the C-terminal half of RISAP (RISAP324-600; Figures 1A and 3, yeast two-hybrid data). This may interfere with myosin binding to RISAP, causing apical vesicles to be released from F-actin and allowing them to fuse with the plasma membrane.
Based on fluorescent protein tagging, RAC5 is detectable at the apical pollen tube plasma membrane (Klahre et al., 2006), whereas RISAP accumulates to its highest levels subapically (e.g., Figure 8A, top image), where BiFC fluorescence indicating interaction between these two proteins was also observed (Figure 13). Our model is consistent with these data and suggests that RAC5, RISAP, as well as complexes of these two proteins are present at additional intracellular localizations, where they may accumulate to levels below the detection limit of fluorescence imaging.
RISAP Contains Multiple Domains Enabling Its Complex Molecular Functions
Apart from the DUF593 and a hydrophobic region recognized by sequence analysis tools, RISAP shares additional highly conserved regions with its closest homologs (Figure 1B). The complex molecular functions of RISAP appear to be based on the ability of these different domains to interact with distinct factors and structures.
The DUF593 of RISAP binds to the GTD-containing C terminus of the tobacco myosin XI MYOXIpt (Figure 6; Supplemental Figure 3) and perhaps to other pollen tube myosins as well. Different RISAP-interacting myosins may mediate subapical TGN positioning and apical vesicle transport. As discussed above, this observation strongly supports the notion that the DUF593 in general may be a GTD binding domain found in proteins that constitute a family of myosin receptors (Peremyslov et al., 2013).
To be able to act as a myosin receptor, RISAP also needs to bind to the membrane of its target organelles. As discussed in the results section, strong interaction of RISAP with stable F-actin structures has prevented the unequivocal demonstration of actin-independent membrane binding of this protein based on classical biochemical assays. Circumventing this problem will require these assays to be repeated with extracts from pollen tubes expressing mutant RISAP capable of membrane, but not of F-actin, association. Efforts are currently underway to develop such RISAP mutants by deleting or modifying the DUF593.
The hydrophobic region of RISAP appears to be responsible for specific association with the subapical TGN compartment (Supplemental Figure 5) and perhaps with secretory vesicles (Figure 14). However, the molecular interactions underlying RISAP membrane association remain to be clarified. In any case, our data strongly suggest that the hydrophobic region of RISAP is not serving as a transmembrane domain. RISAP does not contain an N-terminal signal peptide and is partially solubilized by treatments releasing peripheral, but not integral, proteins from microsomal membranes (Figure 12D).
Consistent with RISAP being a peripheral membrane protein, domains on either side of the hydrophobic region of this protein interact with cytoplasmic RAC5. The short N-terminal fragment of RISAP (RISAP1-29) that we have determined to be responsible for constitutive binding to active and inactive RAC5 represents a small GTPase binding motif with no homology to previously characterized domains interacting with members of this protein family. Such homology is also absent from the C-terminal half of RISAP (RISAP324-600), which we have found to specifically interact with activated RAC5. The last 60 amino acids at the C terminus of RISAP, the most highly conserved region within the RISAP324-600 fragment apart from the DUF593 (Figure 1B), may be responsible for this interaction, which presumably requires a cofactor that remains to be identified. It is conceivable that active RAC5 and this cofactor form a complex that interacts with both RISAP termini and thereby blocks access of the DUF593 to the GTD of myosin motors (Figure 14).
RISAP-Associated TGN: A Central Sorting Station Organizing Apical Membrane Traffic?
Massive secretion required for cell wall biogenesis at the pollen tube tip is thought to result in the incorporation of excess material into the apical plasma membrane, which needs to be retrieved by endocytic membrane recycling (Derksen et al., 1995; Campanoni and Blatt, 2007). Since the classic view that apical secretion is balanced by endocytic membrane uptake at the flanks of the tip has recently been challenged (Moscatelli et al., 2007; Zonia and Munnik, 2008), it is currently unclear where exactly secretion and endocytosis occur at the pollen tube tip. In any case, the subapical RISAP-associated TGN compartment is ideally positioned to play a key role in the coordination of these two processes. In plants, the TGN not only generates secretory vesicles, but also functions as an early endosome, the first organelle with which endocytic vesicles fuse (Dettmer et al., 2006; Richter et al., 2009). Strategically positioned in the gap between Golgi stacks in the granular cytoplasm and the apical CZ (Figure 10A), the subapical RISAP-associated TGN compartment is therefore likely to act as the central sorting station organizing apical membrane traffic during tip growth. In this compartment, newly synthesized and endocytically retrieved membrane material delivered by vesicles derived from Golgi stacks or from endocytic sites at the plasma membrane, respectively, is presumably repacked into secretory vesicles, which are formed at the apical surface and released into the CZ (Figure 14).
Consistent with the above hypothesis, high-level RISAP overexpression causes the CZ to disappear and inhibits pollen tube growth. Excess amounts of RISAP may interfere with the formation of secretory vesicles and/or block other functions of the subapical TGN compartment. Secretion and endocytic membrane recycling converging at this compartment presumably are strictly dependent on each other. Blocking one of these processes is likely to completely inhibit all apical membrane traffic. Interestingly, we observed that endocytically internalized FM4-64 accumulates behind the RISAP-associated TGN in pollen tubes moderately overexpressing this protein (Figure 8C, central column). This is consistent with the idea that endocytic membrane internalization may occur at the flanks of the tip, as suggested in Figure 14. At high levels, overexpressed RISAP completely prevents FM4-64 uptake (Figure 8C, right column) and causes the formation of cytoplasmic organelles with which YFP:RISAP and a plasma membrane marker (RCI2a) are associated (Figure 9). These organelles appear to be aberrant structures derived from the endomembrane system, perhaps from the supapical TGN itself. Conceivably, parts of this organelle may be transported away from its normal subapical location by excess RISAP interacting with the actomyosin system.
Outlook
In summary, data presented here strongly support a model suggesting that RISAP is a RAC5 effector that interacts with F-actin in a myosin-dependent manner, with specific endomembrane compartments and with an unknown cofactor to regulate apical membrane traffic required for pollen tube tip growth. RISAP is likely to act as a myosin receptor, which may promote transport of secretory vesicles to the apical plasma membrane and maintains subapical positioning of a specialized TGN compartment that organizes apical membrane traffic by coordinating secretion and endocytic membrane recycling. To further substantiate this intriguing model of RISAP function and regulation, it will be important to clarify the molecular mechanisms underlying RISAP association with specific endomembrane compartments, to isolate the cofactor postulated to mediate its interaction with activated RAC5, and to determine the structure of the RISAP-associated TGN compartment based on electron microscopy.
METHODS
cDNA Cloning and Recombinant DNA Construction
Isolation of the tobacco (Nicotiana tabacum) RISAP cDNA by colony hybridization was performed as described earlier (Klahre et al., 2006). cDNAs coding for tobacco RAB2 (Cheung et al., 2002) and for the tobacco RAB5 (Dallmann et al., 1992) homolog RAB5.2 were PCR amplified from a pollen tube cDNA library (Klahre et al., 2006) using the following primers: RAB2f, 5′-TCTAGCCGGCGGAACAATGTCGTACGCCTATCTTTTC-3′; RAB2r, 5′-TCTAGAGCTCGAGTCGTTAGCTGCAACAACCTCCTC-3′; RAB5.2f, 5′-TCTAGCCGGCGGAACAATGGCGTCAAGCGGTCACAATAATCTCAATGC-3′; and RAB5.2r, 5′-GTTGGGCCCGAGTCTGTTAAGTACAGCATGATGCAGC-3′.
To construct a cDNA encoding an ER-targeted YFP fusion protein (At-bCH-SP:YFP:HDEL), nucleotide sequences coding for the signal peptide of an Arabidopsis thaliana basic chitinase (Haseloff et al., 1997) and for the ER retention signal HDEL were fused to the 5′ and 3′ end of a YFP cDNA (Clontech Laboratories), respectively. A cDNA encoding a Golgi-targeted YFP fusion protein (St-GnT11-70:YFP) was obtained by fusing a nucleotide sequence coding for the membrane anchor of a potato (Solanum tuberosum) N-acetylglucosaminyltransferase (Wenderoth and von Schaewen, 2000; amino acids 1 to 70) to the 5′ end of the YFP cDNA.
Recombinant DNA construction was performed according to standard techniques (Sambrook and Russell, 2001). cDNAs coding for various polypeptides were cloned into the multiple cloning site (MCS) of the following expression vectors: (1) pGBKT7 (Clontech Laboratories) with modified MCS > bait vectors for yeast two-hybrid screens and assays, templates for recombinant protein production by in vitro transcription/translation; (2) pGADGH (Clontech Laboratories) with modified MCS > prey vectors for yeast two-hybrid assays; (3) pGEX-4T-2 (GE Healthcare) > purification of recombinant GST fusion proteins from Escherichia coli; (4) previously described (Helling et al., 2006; Klahre et al., 2006) pUCAP-based vectors with an MCS between the Lat52 promoter (Twell et al., 1991) and a NOS poly(A)+ signal > expression of free proteins or of fluorescent fusion proteins in pollen tubes; and (5) pSAT1-cEYFP-C1-B and pSAT1-nEYFP-C1 (Citovsky et al., 2006) > transient expression of cYFP:RAC5 and nYFP:RISAP in pollen tubes for BiFC analysis. To generate binary vectors for Agrobacterium tumefaciens-mediated stable tobacco transformation, expression cassettes were transferred into pPZP212 (Hajdukiewicz et al., 1994). PCR-amplified fragments and junctions between ligated fragments were confirmed by Sanger sequencing in all cases.
Software Tools for Sequence Analysis
The following Web-based software tools were employed to analyze the RISAP amino acid sequence: Sosui (http://harrier.nagahama-i-bio.ac.jp/sosui/), TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), DAS (http://www.sbc.su.se/~miklos/DAS/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), HMMTOP (http://www.enzim.hu/hmmtop/), Psipred (http://bioinf.cs.ucl.ac.uk/psipred/), and Dompred (http://bioinf.cs.ucl.ac.uk/psipred/?dompred=1).
Yeast Two-Hybrid Screens and Assays
Yeast two-hybrid screens and assays were performed using the BD Matchmaker system (BD Biosciences, Clontech Laboratories) essentially as described earlier (Klahre et al., 2006; Klahre and Kost, 2006) using the yeast (Saccharomyces cerevisiae) strain AH109.
RNA Gel Blot Analysis
Fresh tobacco tissue samples were frozen in liquid nitrogen and ground. RNA was isolated using the Trizol technique according to the manufacturer’s instructions (Invitrogen). RNA separation, blotting, and detection were performed as described earlier (Klahre et al., 2006; Klahre and Kost, 2006). Blots were probed with PCR-amplified, digoxigenin-labeled (PCR DIG Probe Synthesis Kit; Roche Diagnostics) full-length RISAP cDNA.
Purification of GST-Fusion Proteins and Nucleotide Loading of GST:RAC5
GST-fusion proteins were expressed in E. coli BL21 (DE3) cells transformed with pGEX-4T-2 (GE Healthcare) containing different cDNA inserts. Crude extracts were prepared from cultures (35 mL, 3 × 108 cells/mL) grown in Luria-Bertani media at 37°C for 3 h after adding 1 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested by centrifugation, washed once with sterile water, and resuspended in 600 μL lysis buffer (10 mM Tris-HCl, pH 7.3, 100 mM NaCl, 0.1% Nonidet P-40, 1 mg/mL lysozyme, 5 μg/mL RNase, and 5 μg/mL DNase). To lyse cells, suspensions were sonicated three times for 20 s with a Sonoplus HD2070/SH70G (Bandelin). Cell debris was removed by centrifugation (16,000g, 5 min, 4°C) and 50 μL of MagneGST glutathione particles (V8611; Promega) were added to the supernatant. After incubation for 1.5 h at 7°C on a rotator, the MagneGST glutathione particles were washed four times with 1 mL of lysis buffer and resuspended in 100 μL of lysis buffer.
To load GST:RAC5 with nucleotides, either GTPγ-S (Calbiochem, Merck4Biosciences) or GDPβ-S (Sigma-Aldrich) was added at a concentration of 2 mM to purified recombinant GST:RAC5 in 400 μL loading buffer (50 mM Tris-HCl, pH 7.6, 50 mM NaCl, and 5 mM MgCl2) (Self and Hall, 1995a, 1995b). The reaction mix was incubated for 1 h at 4°C on a rotator, before nucleotide exchange was stopped by increasing the MgCl2 concentration to 10 mM.
Pull-Down Assays
Proteins carrying a myc tag were generated by in vitro transcription/translation using a eukaryotic cell-free protein expression system (TNT T7-coupled wheat germ extract; L4140; Promega). Four micrograms of plasmid DNA (pGBKT7 containing different cDNA inserts) were added to 100 μL wheat germ extract, which was then incubated for 3 h at 30°C. Subsequently, 90 μL of this reaction mix was added to 100 μL of a suspension containing MagneGST glutathione particles loaded with a purified GST-fusion protein. For GDP or GTP loaded GST:RAC5, the MgCl2 concentration of the TNT-wheat germ extract was adjusted to 10 mM prior to adding it to the MagneGST glutathione particles. After a 1.5-h incubation on a rotator at 4°C, MagneGST glutathione particles were washed four times with washing buffer (10 mM Tris-HCl, pH 7.3, 100 mM NaCl, and 0.1% Nonidet P-40) before associated proteins were eluted by boiling in SDS-loading dye. Equal volumes of eluates were analyzed by SDS-PAGE and immunoblotting (chemiluminescence detection). Antibodies used for protein detection were monoclonal α-GST (G1160; Sigma-Aldrich) and polyclonal α-myc sc-789 (A-14; Santa Cruz Biotechnology).
Plant Cultivation
Tobacco (N. tabacum cv Petite Havana SR1) seeds were germinated on soil in a growth chamber at 24°C with 16 h illumination. After 6 weeks, plants were transferred to a greenhouse and maintained under standard conditions until flowering.
Pollen Tube and Plant Transformation
Transient pollen tube transformation by particle bombardment after pollen plating on solid culture medium was performed as previously described (Kost et al., 1998; Klahre and Kost, 2006).
Agrobacterium-mediated stable tobacco transformation was performed essentially as described earlier (Horsch and Klee, 1986). Binary vectors carrying YFP:RISAP or YFP:lifeact expression cassettes were transformed into the Agrobacterium strain AGL-1. Transformants were selected on YEB plates (5 g/L Bacto beef extract, 1 g/L yeast extract, 5 g/L trypton, 5 g/L saccharose, 2 mM MgSO4, and 15 g/L agar) containing 100 mg/L spectinomycin. Single colonies were inoculated in liquid YEB medium containing 100 mg/L spectinomycin and grown overnight at 28°C. Overnight cultures were centrifuged at 1700g for 15 min and resuspended in 10 mL YEB. Fresh tobacco leaf discs were incubated for 5 min in this bacterial suspension and subsequently transferred to solid Murashige and Skoog medium (4.4 g/L Murashige and Skoog salts and vitamins [Duchefa], 20 g/L sucrose, and 8 g/L agar, pH 5.8). After cocultivation for 2 d, leaf discs were transferred to MG-I medium (4.4 g/L Murashige and Skoog salts and vitamins, 16 g/L sucrose, 1 mg/L benzyladenine, 0.1 mg/L naphthalene acetic acid, and 8 g/L agar, pH 5.8) containing 100 mg/L spectinomycin and 100 mg/L kanamycin. Developing shoots were cut and transferred to MG-I medium containing 20 g/L sucrose and 250 mg/L ticarcillin. Fully developed plants were transferred into soil and cultivated in the greenhouse under standard conditions.
Pollen Tube Culture, Drug Treatments, and FM4-64 Labeling
Pollen tubes were cultured at 25°C in the dark either in liquid PTNT medium (1 mM CaCl2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM H3BO3, 30 mM CuSO4, 0.03% casein acid-hydrolysate, 5% sucrose, 12.5% PEG-6000, and 10 mg/mL rifampicin 0.3% MES, pH 5.9, filter sterilized) (Read et al., 1993a, 1993b) or on PTNT medium solidified with 0.25% phytagel (Sigma-Aldrich). To prepare pollen tube cultures, pollen was released from anthers by vortexing for 1 min in liquid PTNT medium. Subsequently, pollen suspensions were filtered through a 1-mm grid to remove anthers and then either cultured directly or transferred onto the surface of solid PTNT medium after vacuum filtration onto membrane discs as previously described (Kost et al., 1998).
Stock solutions containing 10 mM BFA (Molecular Probes, Life Technologies), 5 M BDM (Sigma Aldrich), 100 μM LATB (Calbiochem, Merck4Biosciences), 100 μM JASP (Santa Cruz Biotechnology), or 10 mM FM4-64 (Invitrogen, Life Technologies) were prepared in DMSO and stored at −20°C.
Pollen tubes grown for 3 h in drug-free liquid PTNT medium were treated with LATB, JASP, or BDM by diluting stock solutions directly in the culture medium as required to obtain final concentrations of 4 to 20 nM LATB, 0.15 to 1 μM JASP, or 5 to 35 mM BDM. 1% (v/v) DMSO was added to control cultures. Subsequently, pollen tubes were cultured for another 25 min before imaging to determine the effects on F-actin organization and/or RISAP distribution.
Pollen tubes grown for 3 h on 1.8 mL of solid PTNT medium were treated with BFA or FM4-64 by adding 200 μL liquid PTNT containing 70 μM BFA (final concentration: 7 μM) or 50 μM FM4-64 (final concentration: 5 μM) to the cultures. Two hundred microliters liquid PTNT containing 1% (v/v) DMSO (final concentration: 0.1%) was added to control cultures. Subsequently, pollen tubes were cultured for another 30 to 60 min before imaging effects of BFA treatment and/or FM4-64 distribution.
Preparation of Pollen Tube Extracts and Fractionation by 10,000g Centrifugation
Mature anthers from wild-type or SR1YFP:RISAP tobacco flowers were collected and stored at −80°C. Pooled anthers from 30 flowers were thawed at room temperature for 10 min and used to prepare pollen tube cultures in 10 mL liquid PTNT medium as described above. After 6 h, pollen tubes were collected by centrifugation at 700g for 3 min at 4°C. To remove residual PTNT medium, pollen tubes were washed once in washing buffer (50 mM Tris-HCl, pH 7.3, and 400 mM mannitol) and once in extraction buffer (250 mM sucrose, 25 mM Tris-HCl, pH 7.3, and 50 mM NaCl). Washed pollen tube pellets were frozen in liquid nitrogen and stored at −80°C. Pollen tube pellets were resuspended in 600 μL extraction buffer supplemented with a protease inhibitor cocktail (Hoffmann La-Roche Ltd, Basel, Swiss) containing 1 mM PMSF, 10 μg/mL aprotinin, 1 μg/mL pepstatin, 0.5 μg/mL leupeptin, and 1 mM benzamidin. To lyse cells, suspensions were sheared in the presence of glass beads first on a Vibrax VXR basic (IKA; 10 min, 2500 rpm, 4°C) and then on a Tissue Lyser (Qiagen; 5 min, 30 Hz, 4°C). Subsequently, suspensions were sonicated three times for 20 s using a Sonoplus HD2070/SH70G (Bandelin).
To analyze RISAP partitioning between the S10 supernatant and P10 pellet, cell lysates prepared as described above were centrifuged at 10,000g for 10 min at 4°C. Supernatants were transferred to a fresh tube and supplemented with SDS-loading dye. Pellets were resuspended in SDS-loading dye to obtain the same total volume. Equal volumes of all samples were analyzed by SDS-PAGE and immunoblotting. RISAP was detected using an affinity-purified polyclonal peptide antibody generated against the last 16 amino acids at the C terminus of RISAP (generated by Eurogentec).
Analysis of Effects of Altered F-Actin Stability on RISAP Partitioning between S10 and P10
Frozen (−80°C) pollen tube pellets prepared as described above were resuspended in 600 μL of either a low ionic strength CSB extraction buffer that stabilizes F-actin (5 mM HEPES, pH 7, 10 mM MgCl2, 2 mM EGTA, and 250 mM sucrose) or a high ionic strength CDB extraction buffer that promotes F-actin depolymerization (5 mM HEPES, pH 7, 100 mM KCl, 2 mM EGTA, 250 mM sucrose, and 0.6 M KI). Both buffers were essentially composed as described in the literature (Abe and Davies, 1991; Tan and Boss, 1992; Cox and Muday, 1994) but contained in addition a protease inhibitor cocktail (Hoffmann La-Roche) and 1 mM PMSF. After cell breakage performed as described above, protein extracts were filtered through four layers of Miracloth (Calbiochem, Merck4Biosciences). RISAP, RHOGDI2, and actin partitioning between the S10 supernatant and P10 pellet was analyzed as outlined above. In addition to the α-RISAP antibody described above, a polyclonal α-RHOGDI2 antibody (generated by Eurogentec) and a monoclonal α-Actin11 antibody (Agrisera) were used for protein detection.
Pollen Tube Extract Fractionation by Ultracentrifugation and Protein Solubilization
Pollen tube protein extracts were prepared and fractionated by 10,000g centrifugation as described above. To prepare microsomal fractions, the S10 supernatant was subjected to ultracentrifugation at 150,000g for 45 min at 4°C. Subsequently, the S150 supernatant was transferred to a fresh tube and the P150 pellet was resuspended in pollen tube extraction buffer (same volume as the S150 supernatant) containing either 1% Triton X-100, 1% SDS, 1 M NaCl, 8 M urea, 0.1 M Na2CO3, 10 mM ATP, and 7 mM ATPγS or no additive. Resuspended pellets were incubated for 25 min at 4°C to allow protein solubilization, before they were again centrifuged at 150,000g for 45 min at 4°C. S150b supernatants were transferred to a fresh tube and supplemented with SDS-loading dye. P150b pellets were resuspended in SDS-loading dye to obtain the same total volume. Equal volumes of all samples were analyzed by SDS-PAGE and immunoblotting. α-RISAP and α-actin antibodies described above were used for protein detection.
Actin Coimmunoprecipitation from Pollen Tube Extracts with RISAP
Recombinant myc:RISAP was generated by in vitro transcription/translation. Six hundred microliters of TNT T7-coupled wheat germ extract was supplemented with 36 μg pGBKT7 containing a RISAP cDNA insert and incubated for 3 h at 30°C. Subsequently, 65 μL of a suspension of Dynabeads Pan Mouse IgG (Invitrogen, Life Technologies) preloaded with 3.25 μg of an α-myc antibody (9E11; Dianova) was added to the wheat germ extract. This reaction mix was incubated for 1.5 h at 4°C under constant rotation, before Dynabeads were washed four times with 1 mL lysis buffer (10 mM Tris-HCl, pH 7.3, 100 mM NaCl, and 0.1% Nonidet P-40).
A pollen tube extract prepared as described above was centrifuged at 13,000 rpm in a microcentrifuge for 15 min at 4°C. Six hundred microliters of supernatant was added to the washed Dynabeads. After 1.5 h incubation at 4°C, Dynabeads were washed again four times with 1 mL lysis buffer, and then associated proteins were eluted by boiling in 50 μL SDS-loading dye. Eluates were analyzed by SDS-PAGE and immunoblotting. Proteins were detected using α-myc (A-14; Santa Cruz Biotechnology) and α-actin antibodies described above.
Fractionation by Membrane Flotation Centrifugation
Membrane flotation centrifugation was performed essentially as previously described (Ono and Freed, 1999; Bloch et al., 2005). Pollen tube extracts prepared as described above were adjusted to 25% sucrose. After centrifugation at 10,000g for 10 min, 800 μL supernatant was transferred to a fresh tube, adjusted to 80% sucrose, and sequentially overlaid first with 1500 μL 65% sucrose and then with 700 μL 10% sucrose to generate a density step gradient. The gradient was centrifuged at 100,000g for 5 h at 4°C. Subsequently, 300 μL fractions were collected starting at the top of the gradient, and proteins in each fraction were precipitated. Fractions were incubated for 20 min on ice after adding 75 μL 100% trichloroacetic acid. Precipitated proteins were pelleted by centrifugation at 16,000g for 20 min at 4°C. Acetone washed and dried pellets were dissolved in SDS-loading dye and analyzed by immunoblotting. ARF1 was detected by polyclonal α-Arabidopsis ARF1 antibody (Agrisera). Antibodies described above were used to detect RISAP, RAC5, and RHOGDI2.
Microscopy Analysis of Cultured Tobacco Pollen Tubes
For microscopy analysis, pollen tubes growing on solid PTNT medium were transferred onto a cover slip by cutting a section of the medium and flipping it upside down directly onto the glass surface as described (Kost et al., 1998). Transiently transformed pollen tubes were imaged 3 to 8 h after gene transfer. Pollen tubes growing in liquid PTNT medium were pipetted onto a glass slide within a small droplet of medium and covered with a cover slip.
Epifluorescence imaging was performed using an inverted microscope (DMI4000B; Leica) equipped with X-Cite 200DC fluorescence illumination, a cooled digital black-and-white camera (DFC 365 FX) and N PLAN 5×/0.12 as well as HCX PL FLUOTAR L 40×/0.60 lenses. YFP fluorescence was analyzed through a YFP BP filter block (excitation, band-pass 500/20 nm; dichroic, long-pass 515 nm; emission, band-pass 535/30 nm). The ImageJ software package (http://rsb.info.nih.gov/ij) was employed to measure pollen tube length.
Confocal images were generating using either a LSM510 Meta (Zeiss) or a TCS SP2 (Leica) laser scanning microscope equipped with a PLAN FLUAR 100×/1.45 oil immersion or a HC PL APO 20×/10.7 water immersion lens, respectively. YFP fluorescence was excited at 514 nm and imaged between 530 and 600 nm. mRFP1 and FM4-64 fluorescence was excited at 543 nm and imaged above 560 nm. Simultaneous imaging in both channels was performed in a line-by-line sequential imaging mode. Growth rates of individual pollen tubes were determined by measuring the distance between the positions of the apex on two images taken at a 2-min interval.
Bimolecular Fluorescence Complementation
Tobacco pollen tubes transiently coexpressing nonfluorescent N-terminal (nYFP) and C-terminal (cYFP) YFP fragments under the control of the cauliflower mosaic virus 35S promoter either as free proteins or fused to RAC5 or to RISAP, along with ER-targeted mRFP1 (At-bCH-SP:mRFP1:HDEL) under the control of the Lat52 promoter, were analyzed by confocal microscopy. YFP and mRFP1 fluorescence emitted by these pollen tubes was imaged as described above 5 to 6 h after gene transfer by particle bombardment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: Nt-RAC5, AJ250174; Nt-14-3-3 b-1, AB119467; Nt-RHOGAP1, DQ813657; Nt-RHOGDI2, DQ416769; Nt-RISAP, KJ396934; Nt-MYOXIpt C terminus, KM226781; Nt-RAB2, AF397451; Nt-RAB5.2, KJ439617; At-RCI2a, AF122005; At-ARF1, NM_130285; At-RIC3, NM_100325; At-RIC4, NM_121654; At-VHAa1, AY091008; At-SYP41, AF067789; At-SYP61, AF355754; and Zm-FLOURY1, EF536720. The following Arabidopsis T-DNA insertion lines were used in this study: SALK_043134, SALK_040616 (both with insertions in AT1G18990), and SALK_074471 (insertion in AT1G74830).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Yeast Two-Hybrid Interactions between N-Terminally Truncated RISAP Fragments and Constitutively Active RAC5, as well as between Full-Length RISAP and RAC5-Related Tobacco RAB Proteins.
Supplemental Figure 2. Constitutive Interaction of RISAP with Wild-Type, Constitutively Active, and Dominant-Negative RAC5 in Pull-Down Assays.
Supplemental Figure 3. The DUF593 of RISAP Mediates Interaction of This Protein with the Tobacco Pollen Tube Myosin XI MYOXIpt in Yeast Two-Hybrid Assays.
Supplemental Figure 4. Treatment with the Myosin Inhibitor BDM Blocked Pollen Tube Growth and Trapped the RISAP-Associated TGN Compartment at the Tip.
Supplemental Figure 5. The RISAP N Terminus with the Hydrophobic Domain, together with the DUF593, Are Necessary and Sufficient for Targeting to the Subapical TGN.
Supplementary Material
Acknowledgments
We thank Georg Kreimer, Jan Dettmer, Christopher Staiger, David Robinson, and Christian Koch for helpful discussions. Jennifer Tebart and Stefanie Scholz are acknowledged for excellent technical support. Work presented here was supported by the German Research Council (Deutsche Forschungsgemeinschaft; KO2278/2-2) and the Swedish Research Council (FORMAS; 229-2008-1000).
AUTHOR CONTRIBUTIONS
O.S., S.C., U.K., and B.K. designed the research. O.S., S.C., S.F., D.M.E., and A.M.-R. performed research. S.F. and J.S. contributed new analytic tools. All authors analyzed data. O.S. and B.K. wrote the article.
Glossary
- CZ
clear zone
- GDI
guanine nucleotide dissociation inhibitor
- GDF
GDI dissociation factor
- PI4,5P2
phosphatidylinositol 4,5-bisphosphate
- GTD
globular tail domain
- TGN
trans-Golgi network
- ER
endoplasmic reticulum
- RNAi
RNA interference
- BFA
Brefeldin A
- LATB
latrunculin B
- JASP
jasplakinolide
- BDM
2,3-butanedione 2-monoxime
- BiFC
bimolecular fluorescence complementation
- MCS
multiple cloning site
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abe S., Davies E. (1991). Isolation of F-actin from pea stems. Protoplasma 163: 51–61. [Google Scholar]
- Bloch D., Lavy M., Efrat Y., Efroni I., Bracha-Drori K., Abu-Abied M., Sadot E., Yalovsky S. (2005). Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol. Biol. Cell 16: 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond L.M., Tumbarello D.A., Kendrick-Jones J., Buss F. (2013). Small-molecule inhibitors of myosin proteins. Future Med. Chem. 5: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb M.R., Senderowicz A.M., Sausville E.A., Duncan K.L., Korn E.D. (1994). Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 269: 14869–14871. [PubMed] [Google Scholar]
- Campanoni P., Blatt M.R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58: 65–74. [DOI] [PubMed] [Google Scholar]
- Carlier M.F. (1990). Actin polymerization and ATP hydrolysis. Adv. Biophys. 26: 51–73. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2007). Structural and functional compartmentalization in pollen tubes. J. Exp. Bot. 58: 75–82. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Chen C.Y., Glaven R.H., de Graaf B.H., Vidali L., Hepler P.K., Wu H.M. (2002). Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Cox D.N., Muday G.K. (1994). NPA binding activity is peripheral to the plasma membrane and is associated with the cytoskeleton. Plant Cell 6: 1941–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann G., Sticher L., Marshallsay C., Nagy F. (1992). Molecular characterization of tobacco cDNAs encoding two small GTP-binding proteins. Plant Mol. Biol. 19: 847–857. [DOI] [PubMed] [Google Scholar]
- Derksen J., Rutten T., Lichtscheidl I.K., de Win A.H.N., Pierson E.S., Rongen G. (1995). Quantitative analysis of the distribution of organelles in tobacco pollen tubes: implications for exocytosis and endocytosis. Protoplasma 188: 267–276. [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd P.E., Coursol S., Skirpan A.L., Kao T.H., Gilroy S. (2006). Petunia phospholipase c1 is involved in pollen tube growth. Plant Cell 18: 1438–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré J., Vignais P.V., Dagher M.C. (1999). Phosphoinositide-dependent activation of Rho A involves partial opening of the RhoA/Rho-GDI complex. Eur. J. Biochem. 262: 879–889. [DOI] [PubMed] [Google Scholar]
- Feig L.A. (1999). Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1: E25–E27. [DOI] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hall A. (2012). Rho family GTPases. Biochem. Soc. Trans. 40: 1378–1382. [DOI] [PubMed] [Google Scholar]
- Harris K.P., Tepass U. (2010). Cdc42 and vesicle trafficking in polarized cells. Traffic 11: 1272–1279. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling D., Possart A., Cottier S., Klahre U., Kost B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18: 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Bleasby A.J., Fuchs R. (1992). CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8: 189–191. [DOI] [PubMed] [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.C. (1997). The swinging lever-arm hypothesis of muscle contraction. Curr. Biol. 7: R112–R118. [DOI] [PubMed] [Google Scholar]
- Holzinger A. (2009). Jasplakinolide: an actin-specific reagent that promotes actin polymerization. Methods Mol. Biol. 586: 71–87. [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Klee H.J. (1986). Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of T-DNA borders in the transfer process. Proc. Natl. Acad. Sci. USA 83: 4428–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Gu Y., Lee Y.J., Yang Z. (2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16: 5385–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Heilmann I. (2008). Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 20: 3312–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Hempel F., Jin X., Mosblech A., Heilmann I. (2011). Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J. 65: 453–468. [DOI] [PubMed] [Google Scholar]
- Janmey P.A., Lindberg U. (2004). Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 5: 658–666. [DOI] [PubMed] [Google Scholar]
- Klahre U., Kost B. (2006). Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell 18: 3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Becker C., Schmitt A.C., Kost B. (2006). Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J. 46: 1018–1031. [DOI] [PubMed] [Google Scholar]
- Kost B. (2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 18: 119–127. [DOI] [PubMed] [Google Scholar]
- Kost B., Chua N.H. (2002). The plant cytoskeleton: vacuoles and cell walls make the difference. Cell 108: 9–12. [DOI] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401. [DOI] [PubMed] [Google Scholar]
- Kost B., Mathur J., Chua N.-H. (1999a). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2: 462–470. [DOI] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N.-H. (1999b). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Cai Y., Tse Y.C., Wang J., Law A.H., Pimpl P., Chan H.Y., Xia J., Jiang L. (2009). BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J. 60: 865–881. [DOI] [PubMed] [Google Scholar]
- Langhans M., Förster S., Helmchen G., Robinson D.G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62: 2949–2957. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lavy M., Bloch D., Hazak O., Gutman I., Poraty L., Sorek N., Sternberg H., Yalovsky S. (2007). A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17: 947–952. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Szumlanski A., Nielsen E., Yang Z. (2008). Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J. Cell Biol. 181: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wu G., Ware D., Davis K.R., Yang Z. (1998). Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Nebenführ A. (2008). The tail that wags the dog: the globular tail domain defines the function of myosin V/XI. Traffic 9: 290–298. [DOI] [PubMed] [Google Scholar]
- Liu J., Zuo X., Yue P., Guo W. (2007). Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol. Biol. Cell 18: 4483–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Wilsen K.L., Baskin T.I., Hepler P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221: 95–104. [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Cárdenas L., Kunkel J.G., Hepler P.K. (2007). Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motil. Cytoskeleton 64: 217–232. [DOI] [PubMed] [Google Scholar]
- Martin T.F. (2012). Role of PI(4,5)P(2) in vesicle exocytosis and membrane fusion. Subcell. Biochem. 59: 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Ballesteros M.L., Salinas J. (2007). Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 58: 4333–4346. [DOI] [PubMed] [Google Scholar]
- Michaelson D., Silletti J., Murphy G., D’Eustachio P., Rush M., Philips M.R. (2001). Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton W.M., Ayscough K.R., McLaughlin P.J. (2000). Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2: 376–378. [DOI] [PubMed] [Google Scholar]
- Moscatelli A., Ciampolini F., Rodighiero S., Onelli E., Cresti M., Santo N., Idilli A. (2007). Distinct endocytic pathways identified in tobacco pollen tubes using charged nanogold. J. Cell Sci. 120: 3804–3819. [DOI] [PubMed] [Google Scholar]
- Nagy B., Jencks W.P. (1965). Depolymerization of F-Actin by concentrated solutions of salts and denaturing agents. J. Am. Chem. Soc. 87: 2480–2488. [DOI] [PubMed] [Google Scholar]
- Ono A., Freed E.O. (1999). Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73: 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.M., Fischer-Parton S., Watahiki M.K., Trewavas A.J. (2001). Dynamics of the apical vesicle accumulation and the rate of growth are related in individual pollen tubes. J. Cell Sci. 114: 2685–2695. [DOI] [PubMed] [Google Scholar]
- Parton R.M., Fischer-Parton S., Trewavas A.J., Watahiki M.K. (2003). Pollen tubes exhibit regular periodic membrane trafficking events in the absence of apical extension. J. Cell Sci. 116: 2707–2719. [DOI] [PubMed] [Google Scholar]
- Payrastre B., van Bergen en Henegouwen P.M., Breton M., den Hartigh J.C., Plantavid M., Verkleij A.J., Boonstra J. (1991). Phosphoinositide kinase, diacylglycerol kinase, and phospholipase C activities associated to the cytoskeleton: effect of epidermal growth factor. J. Cell Biol. 115: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Morgun E.A., Kurth E.G., Makarova K.S., Koonin E.V., Dolja V.V. (2013). Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25: 3022–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S.M., Clarke A.E., Bacic A. (1993a). Requirements for division of the generative nucleus in cultured pollen tubes of Nicotiana. Protoplasma 174: 101–115. [Google Scholar]
- Read S.M., Clarke A.E., Bacic A. (1993b). Stimulation of growth of cultured Nicotiana tabacum W38 pollen tubes by poly(ethylene glycol) and Cu(II) salts. Protoplasma 177: 1–14. [Google Scholar]
- Richter S., Voss U., Jürgens G. (2009). Post-Golgi traffic in plants. Traffic 10: 819–828. [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., Sixt M., Wedlich-Soldner R. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5: 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C., Ueda T. (2009). Chapter 4: functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 274: 183–233. [DOI] [PubMed] [Google Scholar]
- Salinas S.R., Bianco M.I., Barreras M., Ielpi L. (2011). Expression, purification and biochemical characterization of GumI, a monotopic membrane GDP-mannose:glycolipid 4-beta-D-mannosyltransferase from Xanthomonas campestris pv. campestris. Glycobiology 21: 903–913. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Self A.J., Hall A. (1995a). Purification of recombinant Rho/Rac/G25K from Escherichia coli. Methods Enzymol. 256: 3–10. [DOI] [PubMed] [Google Scholar]
- Self A.J., Hall A. (1995b). Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256: 67–76. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Ueda T., Yahara N., Nakano A. (2002). Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31: 499–515. [DOI] [PubMed] [Google Scholar]
- Tan Z., Boss W.F. (1992). Association of phosphatidylinositol kinase, phosphatidylinositol monophosphate kinase, and diacylglycerol kinase with the cytoskeleton and F-actin fractions of carrot (Daucus carota L.) cells grown in suspension culture: response to cell wall-degrading enzymes. Plant Physiol. 100: 2116–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., Wing R.A., Ushiba J., McCormick S. (1991). Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 5: 496–507. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Vidali L., McKenna S.T., Hepler P.K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12: 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenderoth I., von Schaewen A. (2000). Isolation and characterization of plant N-acetyl glucosaminyltransferase I (GntI) cDNA sequences. Functional analyses in the Arabidopsis cgl mutant and in antisense plants. Plant Physiol. 123: 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P., Brembu T., Bones A.M. (1997). Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol. Biol. 35: 483–495. [DOI] [PubMed] [Google Scholar]
- Wright L.P., Philips M.R. (2006). Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47: 883–891. [DOI] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S., Yang Z. (2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S., Bloch D., Sorek N., Kost B. (2008). Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147: 1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmola E.G., Somasundaram T., Boring T.A., Spector I., Bubb M.R. (2000). Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 275: 28120–28127. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L., Munnik T. (2008). Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. J. Exp. Bot. 59: 861–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.