This work shows that Arabidopsis EB1b and SPR1, two microtubule plus-end tracking proteins, interact both genetically and biochemically. These proteins have complex interactions as they load onto microtubules and direct polar cell expansion and chiral organ growth in response to directional cues.
Abstract
The microtubule plus-end tracking proteins (+TIPs) END BINDING1b (EB1b) and SPIRAL1 (SPR1) are required for normal cell expansion and organ growth. EB proteins are viewed as central regulators of +TIPs and cell polarity in animals; SPR1 homologs are specific to plants. To explore if EB1b and SPR1 fundamentally function together, we combined genetic, biochemical, and cell imaging approaches in Arabidopsis thaliana. We found that eb1b-2 spr1-6 double mutant roots exhibit substantially more severe polar expansion defects than either single mutant, undergoing right-looping growth and severe axial twisting instead of waving on tilted hard-agar surfaces. Protein interaction assays revealed that EB1b and SPR1 bind each other and tubulin heterodimers, which is suggestive of a microtubule loading mechanism. EB1b and SPR1 show antagonistic association with microtubules in vitro. Surprisingly, our combined analyses revealed that SPR1 can load onto microtubules and function independently of EB1 proteins, setting SPR1 apart from most studied +TIPs in animals and fungi. Moreover, we found that the severity of defects in microtubule dynamics in spr1 eb1b mutant hypocotyl cells correlated well with the severity of growth defects. These data indicate that SPR1 and EB1b have complex interactions as they load onto microtubule plus ends and direct polar cell expansion and organ growth in response to directional cues.
INTRODUCTION
The directed and coordinated regulation of cell expansion in plants is essential for proper plant morphogenesis and the optimal positioning of roots and shoots in response to environmental cues. Pharmacological and genetic evidence strongly suggests that the microtubule (MT) cytoskeleton, in particular the cortical MT array, is centrally involved (Hamada, 2007; Ishida et al., 2007a).
In elongating cells, such as those found in roots and etiolated hypocotyls, cortical MTs become arranged parallel to each other and transverse to the longitudinal axis of the cell (Sedbrook and Kaloriti, 2008). This organization guides the positioning and trajectories of cellulose synthase complexes as they are delivered to the plasma membrane and deposit cellulose microfibrils around the cell circumference (Baskin, 2001; Paredez et al., 2006, 2008; Crowell et al., 2009, 2010; Gutierrez et al., 2009; Chan et al., 2010, 2011; Chen et al., 2010; Lloyd, 2011; Fujita et al., 2011, 2012). The positions, lengths, and orientations of these microfibrils are thought to confer organizational and mechanical anisotropy (polar cell expansion) to the cell wall. Upon deposition, cellulose microfibrils become cross-linked with hemicelluloses and other molecules that structurally constrain turgor driven cell expansion (Szymanski and Cosgrove, 2009). The extent to which the MT cytoskeleton is involved in coordinating the vesicular deposition and cross-linking of these polysaccharides and other molecules is unclear.
Treatments with anti-MT drugs as well as mutations in specific Microtubule-Associated Proteins (MAPs) result in twisted growth of the root and shoot axes (Thitamadee et al., 2002; Ishida et al., 2007a, 2007b; Kaloriti et al., 2007). Twisted growth habit also occurs naturally as plant organs avoid obstacles (e.g., roots encountering rocks in soil) or wrap around objects for support (e.g., twining vines). The twisted growth of Arabidopsis thaliana roots is easily visualized and studied in seedlings grown on the surface of tilted hard-agar medium. Under this condition, root tips systematically change growth trajectories and axial twisting handedness, resulting in roots taking on a sinusoidal wave pattern, a process called root waving (Figure 1). Root waving is thought to be an integrated growth response to gravity, touch, and circumnutation (Okada and Shimura, 1990; Rutherford and Masson, 1996; Oliva and Dunand, 2007; Migliaccio et al., 2013).
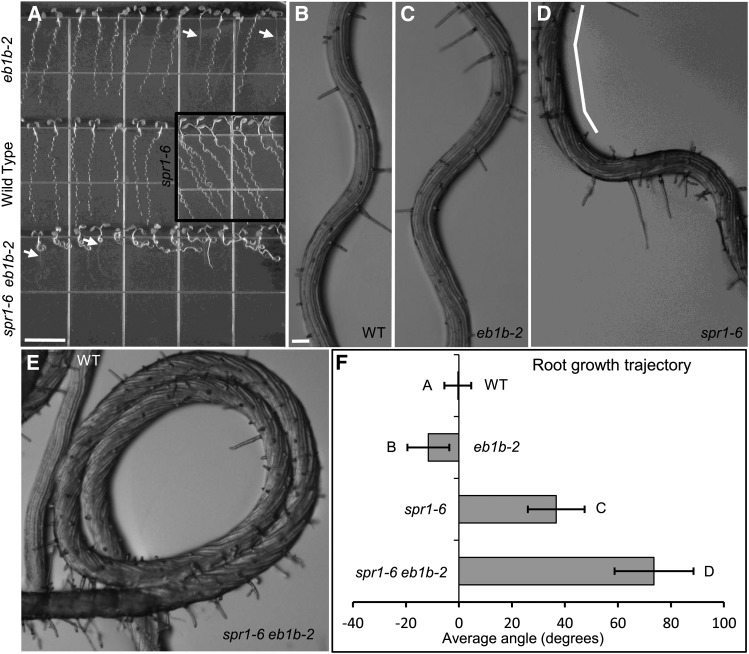
Figure 1.
Root Waving Growth Phenotypes.
Pictured are wild-type ([A] and [B]), eb1b-2 ([A] and [C]), spr1-6 ([A] and [D]), and spr1-6 eb1b-2 ([A] and [E]) seedlings. Both mutants and the wild type are in the Col-0 ecotype.
(A) Seedlings grown for 7 d on 1.5% agar-solidified GM tilted at 40° from the vertical (wave assay) and imaged from above the agar surface. Arrows delineate roots that had penetrated the agar surface, growing vertically until reaching the plate-agar interface, at which point skewed growth reoccurred. Bar = 1 cm.
(B) to (E) Closeup views of the roots of seedlings put through the wave assay. Note the prolonged right-handed axial twisting of the spr1-6 root (D) and of the spr1-6 eb1b-2 double mutant root (E), as revealed by the twisted epidermal cell files (demarcated by the white line in [D]). Bar in (B) = 0.1 mm for (B) to (E).
(F) Average trajectories of roots put through the wave assay. Negative and positive angles are leftward and rightward slants, respectively, as viewed from above the agar. Error bars indicate sd. Different letters represent significant differences based on single-factor ANOVA; 37 < n < 41.
Twisted growth is often associated with rearrangements of cortical MTs to oblique instead of transverse alignments. These helical rearrangements have been proposed to drive cell elongation of the opposite handedness (Ishida et al., 2007b). However, organ twisting also occurs when root cell cortical MTs are oriented transversely, suggesting that factors besides MT array organization are involved (Sedbrook et al., 2004).
SPIRAL1 (SPR1) was one of the first proteins identified to be involved in determining twisted growth. Loss of SPR1 function results in marked right-handed twisting of the plant axis. Moreover, when grown on tilted hard-agar surfaces, spr1 roots wave and skew in a rightward direction instead of waving straight downward like wild-type roots (Figure 1A). SPR1 is specific to plants and algae, belonging to a six-member gene family in Arabidopsis that is predicted to encode small structural proteins with overlapping functions (Nakajima et al., 2004, 2006; Sedbrook et al., 2004). SPR1:EGFP (for enhanced green fluorescent protein) translational fusion proteins localize preferentially in a shallow gradient at the growing ends of MTs, identifying them as plus-end tracking proteins, or +TIPs (Sedbrook et al., 2004).
In addition to the plant-specific SPR +TIPs, plants express End Binding (EB) proteins, which are evolutionarily and structurally conserved +TIPs present in plants, animals, and fungi. EB proteins are thought of as master regulators of MT plus ends (Vaughan, 2005; Slep, 2010). The genome of Arabidopsis encodes three EB proteins named EB1a, EB1b, and EB1c. Fluorescent protein fusions with EB1a and EB1b exquisitely label growing cortical MT plus ends (Chan et al., 2003; Mathur et al., 2003). Disruption of EB1c function affects spindle and phragmoplast alignment and confers anti-MT drug hypersensitivity (Komaki et al., 2010). Subtle cell expansion defects were found associated with mutations in all three Arabidopsis EB1 genes, with loss of EB1b function causing the most notable root directional growth phenotypes (Bisgrove et al., 2008).
Many +TIPs accumulate only when the MTs are polymerizing (Bisgrove et al., 2004; Akhmanova and Steinmetz, 2008; Galjart, 2010) and have been found to regulate MT dynamic instability, recruit other proteins to the MT cytoskeleton, act as sensors of determinants at the cell cortex and other locations, and assist in the delivery of proteins to specific destinations (Lansbergen and Akhmanova, 2006; Jaworski et al., 2008; Galjart, 2010). Given that SPR1 and EB1 proteins reside at MT plus ends in close proximity to each other and that both affect cell expansion processes, we set out to identify possible genetic and physical interactions. Although SPR1 was previously shown to localize to MTs, it was not clear if SPR1 protein bound directly to the MT lattice or instead was recruited by a MAP. Here, we show that SPR1 and EB1b interact genetically to affect directional cell expansion in Arabidopsis. In addition, SPR1 and EB1b proteins interact physically with each other and directly with MT lattice. We propose a model where SPR1 and EB1b either compete for MT binding sites or otherwise regulate each other’s activity in a complex interplay directing polar cell expansion and organ growth chirality, constitutively and in response to environmental stimuli.
RESULTS
EB1b Affects Root Directional Cell Expansion and Interacts Genetically with SPR1
Previous analysis of root waving, which involves the growth of seedling roots on tilted 1.5% agar-solidified growth medium (GM), a condition that confers directional touch stimulation to the roots, demonstrated that spr1-6 mutant roots displayed sinusoidal growth with a strong rightward trajectory relative to the sinusoidal but straight downward trajectory of growth observed in wild-type Columbia-0 (Col-0) ecotype seedling roots (Sedbrook et al., 2004; Figure 1). To determine if disruption of EB1b function also affected root waving, we obtained an Arabidopsis eb1b T-DNA insertional mutant from the Wisconsin Knockout Facility (Col-0 ecotype background, line WiscDSLox331A08, herein designated as eb1b-2; Woody et al., 2007). PCR analysis was performed to confirm the intragenic location of the T-DNA and to track its segregation (Supplemental Figure 1; see Methods).
By performing the root waving assay, it was found that homozygous but not heterozygous eb1b-2 roots slanted modestly to the left (Figure 1A). This and other related phenotypes described below cosegregated with the T-DNA insertion in a 3:1 Mendelian fashion. While the homozygous eb1b-2 root slanting was subtle, the average root growth trajectories were significantly different from the average growth trajectories of wild-type Col-0 roots (Figure 1F; two-tailed Student’s t test, P < 1 × 10−4). This left-slanting root growth phenotype of eb1b-2 seedlings was comparable to that reported for the eb1b-1 mutant allele, whose phenotype was rescued by the introduction of a wild-type copy of the EB1b gene (Bisgrove et al., 2008; Gleeson et al., 2012).
To determine if any EB1b transcript could be detected in eb1b-2 seedlings, RT-PCR analyses were performed. A primer pair designed to detect a portion of the EB1b transcript located downstream of the T-DNA insertion produced no PCR product, whereas a primer pair located upstream of the T-DNA insertion did produce a PCR product (Supplemental Figures 1A and 1B), indicating that a truncated EB1 transcript was generated in homozygous eb1b-2 plants. To confirm the loss of full-length EB1 protein, we generated affinity-purified antibodies against full-length Escherichia coli-expressed EB1b. As shown by protein gel blot analysis, the affinity-purified anti-EB1b antibody displayed cross-reactivity to purified E. coli-expressed EB1b and EB1a (expected molecular masses of 32.9 and 31.1 kD, respectively), which share 79% amino acid identity, but not EB1c (52% identity with EB1b; Supplemental Figure 1D). Likewise, the affinity-purified anti-EB1b antibody detected two polypeptides in wild-type total protein extracts. However, only the 31.1-kD EB1a polypeptide was detected in homozygous eb1b-2 protein extracts, indicating that eb1b-2 mutants lack full-length EB1b (Supplemental Figure 1C). Since it is possible that the polyclonal mix of EB1b antibodies contains no antibodies that could readily detect the truncated portion of the EB1b protein if it was in fact being made and was partially functional, it remains ambiguous whether the eb1b-2 mutant is a null or instead a hypomorph.
To investigate the genetic relationship between SPR1 and EB1b, we generated a spr1-6 eb1b-2 double mutant plant line and analyzed its growth characteristics. Both spr1-6 and eb1b-2 are in the Col-0 ecotype background. On tilted hard-agar GM, roots of spr1-6 eb1b-2 double mutant seedlings grew shorter than either parent or the wild type and were severely hampered in root waving, typically exhibiting looped or coiled growth instead of wavy growth (Figure 1A). Closeup examinations of spr1-6 eb1b-2 coiled root growth revealed severe and prolonged right-handed axial twisting (Figure 1E). For spr1-6 eb1b-2 roots that looped instead of forming coils, overall growth trajectories could be measured, which showed an average rightward trajectory significantly greater than that for spr1-6 mutant roots (Figure 1F).
We also examined the phenotypes of 7-d-old seedlings, whose roots were allowed to grow into 0.8% agar-solidified GM, a condition that confers uniform touch stimulation to the roots. Wild-type, eb1b-2, spr1-6, and spr1-6 eb1b-2 roots grew comparably straight along the gravity vector (average root tip trajectories ± sd: wild type = 0.4° ± 7.2°, eb1b-2 = −0.8° ± 5.0°, spr1-6 = 1.0° ± 6.3°, spr1-6 eb1b-2 = 0.9° ± 10.7°; no significant differences based on single-factor ANOVA; 28 < n < 36). However, the length of 7-d-old spr1-6 eb1b-2 double mutant roots was found to be significantly shorter than that of either parent, and the root epidermal cells displayed a right-handed axial twisting phenotype considerably more severe than that of the spr1-6 mutant parent (Figure 2). eb1b-2 mutant roots conversely grew to be slightly longer than the wild type and had a left-handed axial twisting bias that was significantly different from that of the wild type (Figure 2).
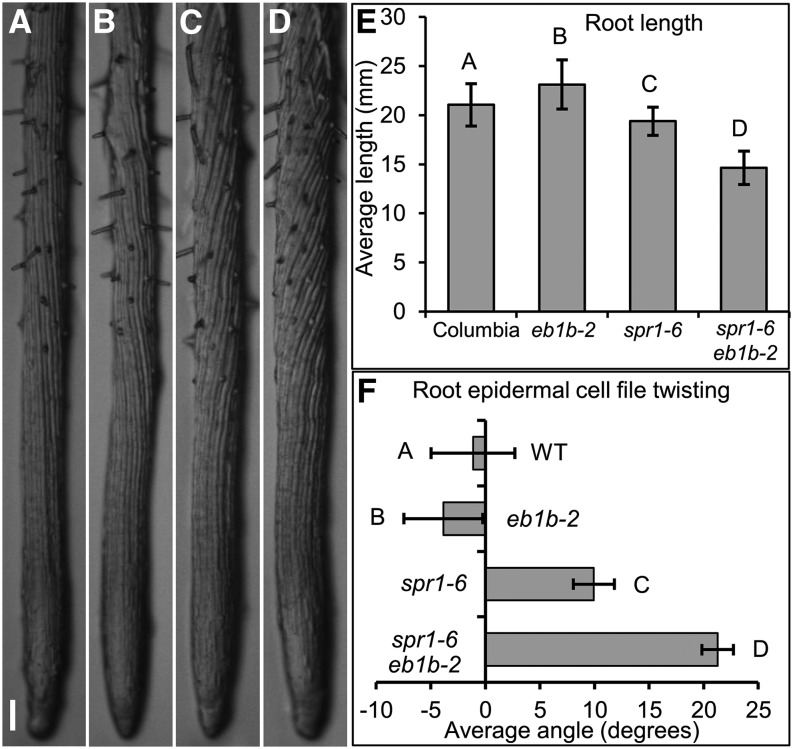
Figure 2.
Phenotypes of Roots Grown Embedded in 0.8% Agar-Solidified GM.
(A) to (D) Pictured are wild-type (A), eb1b-2 (B), spr1-6 (C), and spr1-6 eb1b-2 (D) roots. Note the left-handed (eb1b-2) and right-handed (spr1-6 and spr1-6 eb1b-2) root twisting as revealed by the twisted epidermal cell files. Bar in (A) = 0.1 mm for (A) to (D).
(E) Average root lengths after 7 d of growth.
(F) Average angles of root epidermal cell files relative to the longitudinal axes of the roots. Positive and negative values are representative of the degrees of left-handed and right-handed root twisting, respectively.
Error bars in (E) and (F) represent sd. Different letters represent significant differences based on single-factor ANOVA. For root lengths, 28 < n < 45; for epidermal twisting, 19 < n < 30.
We were interested to see if the reduction in spr1-6 eb1b-2 root lengths was associated with alterations in root cell dimensions, which would indicate a defect in polar cell expansion. To assess this, we stained roots grown in 0.8% agar-solidified GM with propidium iodide, a fluorescent dye that stains cell walls to allow for visualization of cell shapes. Confocal microscopic examination of cell shapes and sizes revealed that spr1-6 eb1b-2 double mutant root cells were significantly shorter and bulged in shape when compared with the wild type or either single mutant parent (Figures 3A to 3D, 3I, and 3J). We also observed shorter, bulged, and twisted cells in spr1-6 eb1b-2 dark-grown (etiolated) hypocotyls (Figures 3E to 3H). Taken together, these data suggest that SPR1 and EB1b act collaboratively to facilitate polar cell elongation and antagonistically to direct organ growth in both uniform and directional touch stimulation environments.
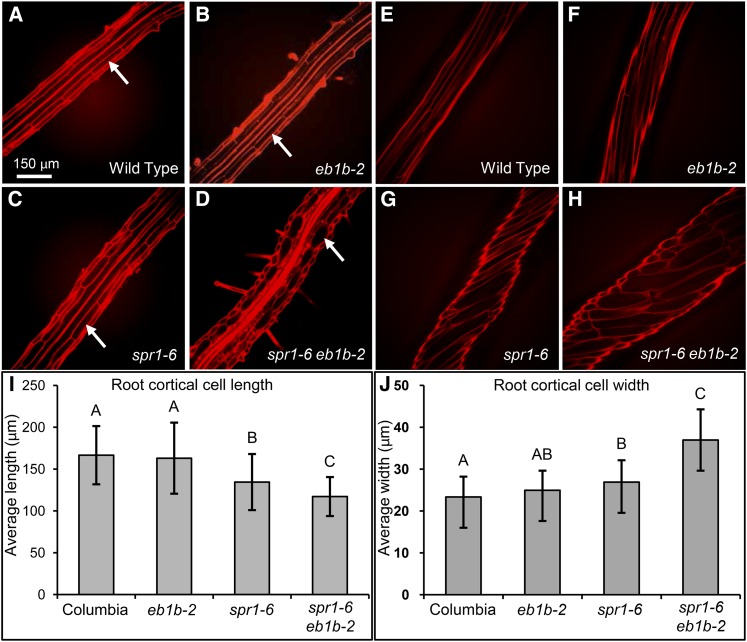
Figure 3.
Comparisons of Root and Etiolated Hypocotyl Cell Sizes and Axial Twisting.
(A) to (H) Confocal image projections of propidium iodide-stained roots ([A] to [D]) and etiolated hypocotyls ([E] to [H]) showing relative cell sizes and organ axial twisting. Arrows in (A) to (D) delineate root cortical cells. Bar in (A) = 150 μm for (A) to (H).
(I) and (J) Bar graphs of average root cortical cell lengths (I) and average root cortical cell widths (J). Error bars represent sd. Different letters represent significant differences based on single-factor ANOVA. For root lengths, 28 < n < 45; for cortical cell lengths, 77 < n < 106; for cortical cell widths, 102 < n < 125.
Altered MT Dynamic Instabilities Correlate with Anisotropic Growth Defects
Transversely oriented and organized cortical MT arrays are essential for proper anisotropic cell expansion in actively elongating root and etiolated hypocotyl cells (Wasteneys and Fujita, 2006; Fu et al., 2009). To determine if the pronounced spr1-6 eb1b-2 cell expansion defects might be a consequence of disorganized MTs, we performed whole-mount immunolocalization using an anti-tubulin antibody and confocal microscopy on roots grown on vertically oriented 0.8% agar-solidified GM. Figures 4A to 4D show that, while the cortical MT arrays in spr1-6 eb1b-2 elongating root cells were sometimes slightly obliquely oriented, the MTs had a parallel organization and density comparable to the wild type and the single mutants. Cortical MT organization in etiolated hypocotyl cells of the various mutants also appeared similar to that in the wild type (Figures 4E to 4H), suggesting that the severe cell expansion phenotypes were primarily due to something other than cortical MT array organization.
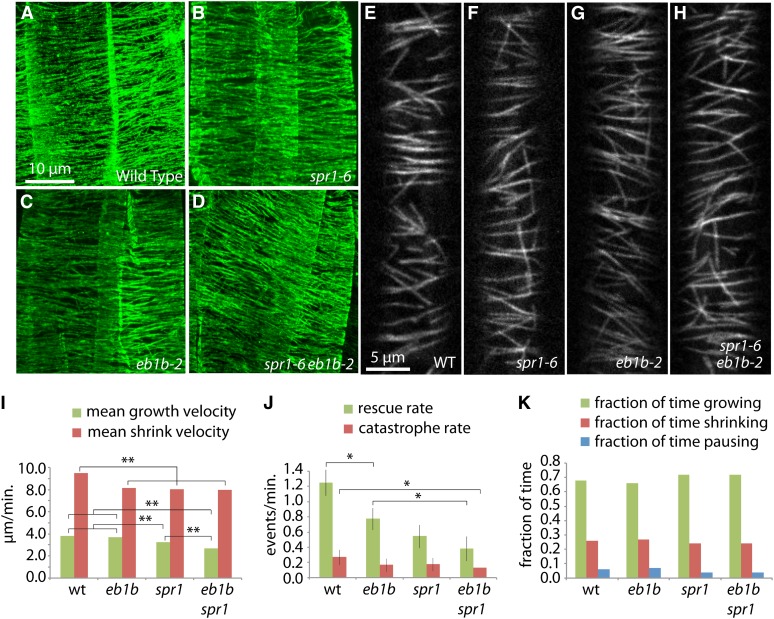
Figure 4.
MT Organization in the Root Elongation Zone and Etiolated Hypocotyl Cells of Mutants versus the Wild Type.
(A) to (H) Confocal microscopic images of MTs in root elongation zone epidermal cells ([A] to [D]) and etiolated hypocotyl epidermal cells ([E] to [H]). (A) to (D) show confocal projections of immunolocalized MTs detected with anti-tubulin antibody and a fluorescein isothiocyanate fluorescent dye-labeled secondary antibody. (E) to (H) show confocal image slices of EYFP-TUA5-labeled MTs. Wild type ([A] and [E]), spr1-6 ([B] and [F]), eb1b-2 ([C] and [G]), and spr1-6 eb1b-2 double mutant ([D] and [H]) seedlings are shown. Note that the MT organization is comparable between the mutants and the wild type. Bar in (A) = 10 μm for (A) to (D); bar in (E) = 5 μm for (E) to (H).
(I) to (K) Measurements of MT dynamics. Parameters of MT plus-end dynamics were acquired from time-series images of epidermal cells in the upper hypocotyl expressing YFP:TUA5 in 4-d-old dark grown seedlings.
(I) Velocities of growth and shrinkage. **P < 0.001, Mann-Whitney U test.
(J) Catastrophe and rescue rates. *P < 0.05, one-tailed C test; whiskers indicate se.
(K) Fractions of time spent in polymerization states. The numbers of measured excursions used to acquire each data set were 431, 470, 445, and 371 for the wild type, eb1b-2, spr1-6, and eb1b-2 spr1-6, respectively. Five cells and plants were sampled per data set.
In other Arabidopsis mutants, alterations in MT dynamic instability parameters have been reported to correlate with defects in polar cell expansion (Ishida et al., 2007a; Yao et al., 2008; Wang et al., 2012). To determine if cortical MT dynamic instability parameters were aberrant in elongating hypocotyl cells of the spr1-6 and eb1b single and double mutants, we measured the polymerization dynamics of MT plus ends in dark-grown hypocotyl cells (upper hypocotyl, 4 d after germination) expressing EYFP:TUA5 in the wild type and spr1-6, eb1b-2, and spr1-6 eb1b-2 mutant backgrounds. Compared with the wild type, the double mutant showed significantly reduced values for the velocities of both growth and shortening as well as catastrophe and rescue rates (Figures 4I to 4K; Supplemental Figure 2 and Supplemental Movie 1). The effects on rescue rate were especially dramatic, with the double mutant showing approximately one-third the rate as the wild type as well as a reduction relative to the eb1b single mutant parent. The amount of time spent in each polymerization mode, growth, shrink, or pause, was similar in all three mutants. The individual mutants were sometimes deficient in these values as compared with the wild type (Figures 4I to 4K; Supplemental Figure 2). Shrinking velocities were similarly defective (slower) in the double spr1 eb1b mutants as well as single eb1b and spr1 mutant lines relative to the wild type, while growth was slower in spr1 but not measured to be different from the wild type in eb1b. Rescue rates were lower in both eb1b and spr1, while catastrophe rates were not measured to be significantly different from those in the wild type. Overall, changes in polymerization dynamics were often more severe in the double mutant than in individual mutants, with cortical MTs in the double mutant being significantly less dynamic than those in the wild type; the severity of the cell expansion phenotypes correlated with the degree by which MT dynamics were reduced.
SPR1 and EB1b Proteins Bind Directly to MTs and Tubulin Heterodimers in Vitro
Previously, we found that the expression of SPR1:EGFP under the control of its native upstream regulatory sequences rescued the spr1-6 mutation and labeled MTs throughout the cell cycle, including the cortical MT array, preprophase band, phragmoplast, and spindle (Sedbrook et al., 2004). However, the question remained whether SPR1 localized to MTs by binding directly to the MT lattice or through interactions with other MT binding proteins. To address this, we expressed and purified from E. coli an SPR1 fusion protein containing a C-terminal tandem glutathione S-transferase (G) and 6xHis (H) epitope tag (SPR1-G-H). Affinity-purified SPR1-G-H protein was incubated with taxol-stabilized MTs assembled from bovine tubulin, BSA as a control, or no tubulin at all. Binding was assessed by cosedimentation analysis. We found that sedimentation of the SPR1-G-H fusion protein was promoted in the presence of MTs (Figure 5A), suggesting a direct interaction between SPR1 and polymerized tubulin. Similarly, we examined the MT binding properties of EB1b, which, like SPR1, has been shown to preferentially localize to the growing ends of MTs (Mathur et al., 2003). Consistent with previous reports that EB proteins bind directly to MTs (Berrueta et al., 1998; Tirnauer et al., 2002; Busch and Brunner, 2004; Komaki et al., 2010), E. coli-expressed and -purified N-terminal 6xHis-tagged EB1b fusion protein (H-EB1b) was found to cosediment with taxol-stabilized MTs when centrifuged at 25,000g, while less H-EB1b protein sedimented in the absence of MTs (Figure 5A).
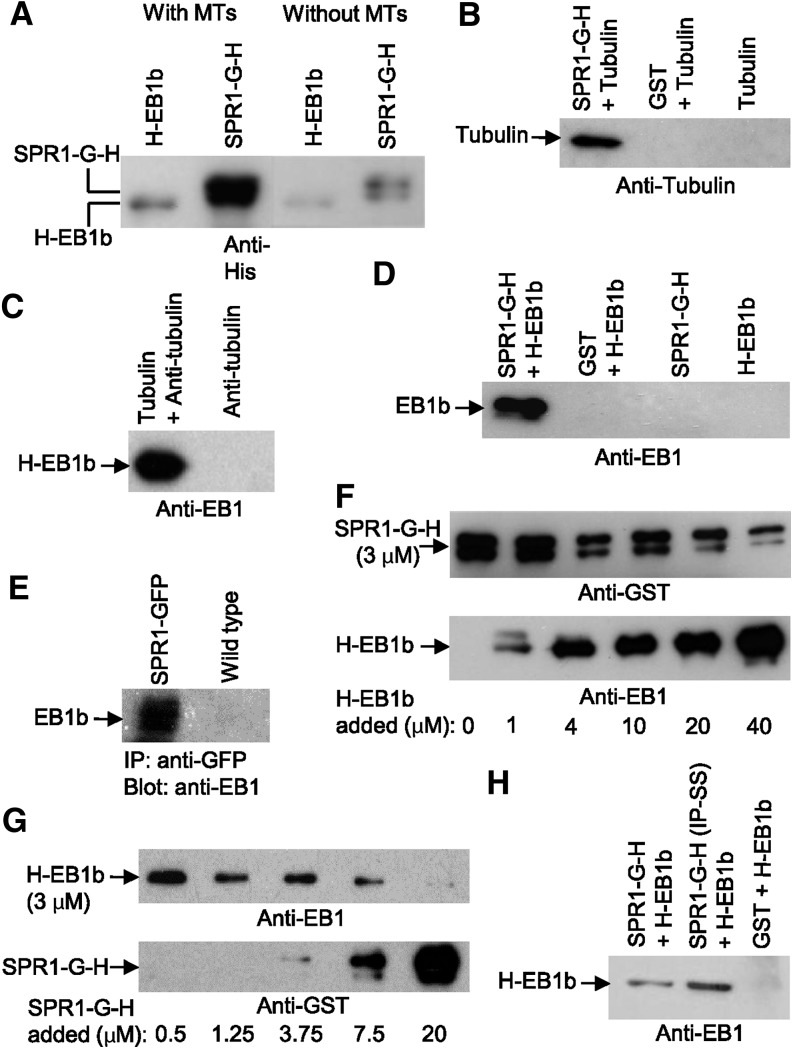
Figure 5.
Biochemical Determinations of SPR1 and EB1b Protein Binding to MTs, Tubulin Heterodimers, and Each Other.
All images are immunoblots probed with the antibody listed below the blot image. G = GST tag; H = 6His tag.
(A) Cosedimentation assay showing the amounts of SPR1-G-H and H-EB1b protein that pelleted at 25,000g in the presence (with MTs) or absence (without MTs) of MTs.
(B) GST pull-out assay showing the amount of unpolymerized tubulin pulled out of solution with glutathione resin and SPR1-G-H, GST, or no other protein.
(C) Immunoprecipitation assay showing the amount of H-EB1b protein pulled out of solution by Protein A-Sepharose beads precoated with anti-tubulin antibody plus or minus unpolymerized tubulin.
(D) GST pull-out assay showing the amount of H-EB1b protein pulled out of solution with glutathione resin and SPR1-G-H, GST, or no other protein.
(E) Immunoprecipitation (IP) assay showing the amount of H-EB1b protein pulled out of either a SPR1-GFP-expressing plant extract (SPR1-GFP) or a wild-type plant extract using an anti-GFP antibody and Protein A-Sepharose beads.
(F) and (G) Cosedimentation assays showing the relative amounts of SPR1-G-H and H-EB1b proteins that pelleted at 25,000g in the presence of MTs.
(F) Each sample contained a 3 μM concentration of SPR1-G-H protein and increasing amounts of EB1b protein (from no H-EB1b in sample 1 to 40 μM H-EB1b in sample 6; amounts are listed below the blot images).
(G) Each sample contained a 3 μM concentration of H-EB1b protein and increasing amounts of SPR1-G-H protein. The images are of the same protein gel blot first probed with an anti-GST antibody to detect SPR1-G-H, stripped, and then reprobed with the anti-EB1 antibody.
(H) GST pull-out assay showing the amount of H-EB1b protein pulled out of solution with glutathione resin and SPR1-G-H (lane 1), SPR1-G-H with Iso70-Pro71 changed to Ser70-Ser71 (SPR1-G-H [IP-SS]; lane 2), or GST protein (lane 3).
To determine if SPR1 protein also binds to tubulin heterodimers, we mixed SPR1-G-H protein with bovine brain tubulin at concentrations below that required for MT polymerization and in the absence of polymerization enhancers (no taxol nor glycerol; Andreu et al., 1992). SPR1-G-H was then extracted from the mixture by affinity chromatography using glutathione-Sepharose resin. Figure 5B shows that tubulin copurified together with SPR1-G-H. Binding of tubulin to immobilized glutathione was not detected in the absence of SPR1-G-H or when glutathione S-transferase (GST) protein was substituted for SPR1-G-H (Figure 5B).
To determine if EB1b protein also binds to tubulin heterodimers, we performed a coimmunoprecipitation assay, mixing H-EB1b protein with Protein A-Sepharose beads precoated with anti-tubulin antibody and unpolymerized tubulin protein. As shown in Figure 5C, H-EB1b was immunoprecipitated in the presence but not the absence of purified tubulin protein, indicating that EB1b binds directly to unpolymerized tubulin heterodimers. This result does not appear to be the consequence of nonspecific protein binding, since BSA protein did not coprecipitate when substituted for EB1b (Supplemental Figure 3).
EB1b Binds to SPR1 Protein in Vitro, in a Yeast-Two Hybrid Assay, and in Plant Extracts
Since SPR1 and EB1b bind directly to MTs in vitro and localize to the polymerizing plus ends of cortical MTs in planta, we tested whether these proteins interact physically in vitro and in vivo. As detected by protein gel blot analysis (Figure 5D), bacterially expressed H-EB1b protein copurified with SPR1-G-H protein via glutathione-Sepharose affinity chromatography. Conversely, H-EB1b did not bind to glutathione-Sepharose resin in the presence of purified GST protein (negative control). We also conducted a yeast two-hybrid analysis, finding that EB1b fused to the Gal4 DNA binding domain interacted with SPR1 fused to the Gal4 activation domain, as determined by yeast growth on corresponding dropout medium (Supplemental Figure 4).
To assess whether SPR1 and EB1b interact in vivo, we examined if EB1 coimmunoprecipitated with SPR1:EGFP. As shown in Figure 5E, EB1 was found to copurify specifically with anti-GFP immunopurified SPR1:EGFP from protein extracts of transgenic lines expressing SPR1:EGFP, but not from wild-type plants, as determined by protein gel blot analysis using affinity-purified anti-EB1b antibody. As shown in Supplemental Figure 1D, the affinity-purified anti-EB1b antibody cross-reacts with EB1b and to a lesser extent with EB1a but not with bacterially expressed EB1c protein. Thus, it is likely that at least a portion of the detected plant-extract purified protein was that of EB1a.
SPR1 and EB1b Affect Each Other’s Binding to MTs
To investigate whether SPR1 and EB1b influence each other’s binding to MTs, we performed MT binding assays, mixing constant amounts of SPR1-G-H and polymerized bovine brain-derived taxol-stabilized MTs with increasing amounts of H-EB1b protein. The data in Figure 5F show that increasing amounts of H-EB1b protein led to decreasing amounts of SPR1-G-H protein cosedimenting with MTs in conjunction with increasing amounts of H-EB1b cosedimenting. The opposite result was observed when mixing constant amounts of H-EB1b and polymerized MTs with increasing amounts of SPR1 protein (more and more SPR1-G-H cosedimented with MTs in conjunction with less and less H-EB1b cosedimenting; Figure 5G). These results suggest that SPR1 and EB1b compete for the same binding sites on MTs or that the binding of SPR1 and EB1b to each other mutually affects their abilities to bind to MTs. In the future, protein binding affinities must be determined, along with a quantification of the extents to which labeled SPR1 and EB1b proteins bind along the MT lattice, in order to get a clearer picture of how SPR1 and EB1b affect each others’ MT binding. This experiment is complicated by the fact that, at least in planta, SPR1 protein localizes to a greater (and varying) extent of the MT lattice compared with EB1b (Supplemental Figure 5). Moreover, it is unclear if MT plus ends (and thus binding sites) are limiting in the pull-out assays, given that it is unknown how many ends form under these polymerization conditions and if the MT lengths and number of ends change with the addition of the SPR1 and/or EB1b proteins.
Fluorescence Recovery after Photobleaching Analysis Suggests That SPR1 Loads onto Polymerizing MT Tips
Our in vitro interaction studies indicated that SPR1 interacts directly with MTs and free tubulin dimers. To probe the dynamics of its association with MT ends, we performed fluorescence recovery after photobleaching (FRAP) analysis of MT growing ends in transgenic lines that express SPR1:EGFP under the control of its native promoter. However, despite showing a clear comet-like distribution at MT growing ends, the SPR1:EGFP signal was too weak and sparse to permit fast FRAP analysis of small regions just behind the MT tips, as was performed previously for EB1 (Dragestein et al., 2008). To overcome this problem, we took advantage of the fact that, relative to 35S-expressed mCherry:EB1b (Supplemental Figure 5; Chan et al., 2003; Mathur et al., 2003; Kirik et al., 2007), SPR1:EGFP is localized in long MT-associated tails in Arabidopsis cells (Sedbrook et al., 2004). These longer tails enabled us to photobleach larger regions and to determine if these longer tails recovered rapidly, consistent with rapid lateral exchange of SPR1 with the MT lattice, or if these tails were relatively slow to recover or did not recover at all. Using a point scanning confocal microscope, we bleached SPR1:EGFP in hypocotyl cells within 3- × 10-μm rectangles that aligned with the net orientation of the cortical array as determined by imaging of SPR1:EGFP. The recovery in the first frames after bleaching was observed only along the portion of the MT assembled after bleaching (Figure 6A). Extended tails of SPR1:EGFP were only observed after further MT growth, confined to the portion that grew after bleaching (Figure 6A). To quantify this observation, we measured SPR1:EGFP signal along the MT axis using a three-pixel-wide lane, both in the penultimate frame before bleaching and 4 s after bleaching (16 plus ends in 10 cells). The mean peak signal at the very tip in the frame just prior to bleaching was 65.9 ± 18 arbitrary density units (ADU); in the second frame following bleaching (4 s later), this value was 42.2 ± 10.3 ADU. During recovery, this peak value is measured on newly assembled lattice, thus containing no dark protein present on the lattice at the time of bleaching. The observed diminution in peak signal, therefore, is likely due to bleaching of free SPR1:EGFP in the vicinity of the tip and dilution of the labeled pool available for TIP interaction.
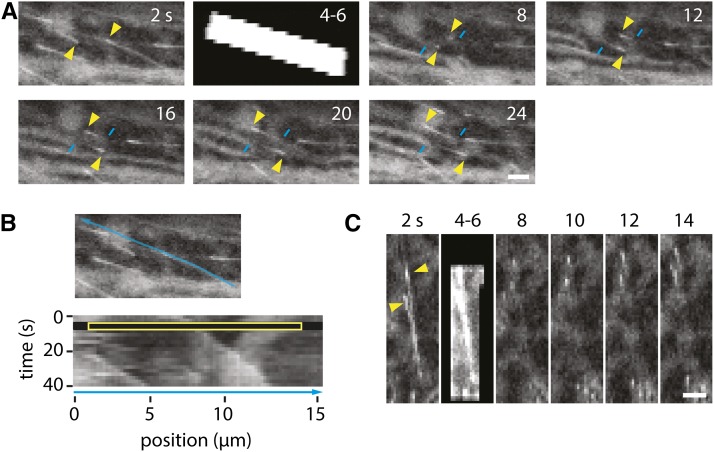
Figure 6.
Dynamics of SPR1:EGFP Localization.
(A) FRAP experiment in a hypocotyl epidermal cell expressing SPR1:EGFP. Photobleaching energy was applied to the area indicated in the second panel. The arrowheads indicate the growing ends of MTs in the photobleached region. Note the long tails of label in the prebleach frame and the much shorter region of recovered tip signal immediately after bleaching. Longer tails of SPR1:EGFP are observed after further MT tube growth. Similar results were observed in 51 of 51 MTs. Bar = 2 μm.
(B) Kymograph analysis of the two bleached MTs shown in (A). The yellow box indicates the bleached region.
(C) Selected images of SPR1:EGFP-labeled MTs shown 2 s before and 4 s after bleaching. Bar = 2 μm.
When we measured the recovery of label only on lattice already assembled at the time of bleaching (see Methods), we measured no detectible recovery over the same interval. The signal immediately behind the tip prior to photobleaching was 45.8 ± 26.6 ADU above the local mean background, while 4 s after bleaching, signal at the same absolute location was only 3.2 ± 11.1 ADU above local background (not distinguishable from background statistically). By comparison, the mean signal at this same position relative to the tip prior to bleaching was 31.9 ± 21.5 ADU. Furthermore, we observed examples in long SPR1:EGFP tails where bleach boundaries were maintained without detectible recovery in the bleached areas over at least 12 s (Supplemental Figure 6). Thus, the observed recovery of SPR1:EGFP on the lattice appears to be more slow than was observed previously for EB proteins and the +TIP CLIP-170, both of which have been observed to exchange rapidly along the lattice, with half-times of recovery of a couple of seconds or less (Busch et al., 2004; Bieling et al., 2007: Dragestein et al., 2008). These results are consistent with a mechanism in which a significant portion of SPR1 is loaded by copolymerization, which then dissociates stochastically over time, or alternatively, by a mechanism in which SPR1 recognizes a feature concentrated at the plus end but where lateral exchange is relatively limited compared with previous observations of EB1 and CLIP-170 in other systems. Transport as a tip concentration mechanism is argued against by the observation of stable and sharp bleach edges in long SPR1 tails.
Hypothetical SPR1 Isoleucine-Proline Motif Is Not Required for EB1b Binding
Honnappa et al. (2005, 2009) showed that a Ser-x-Ile-Pro (SxIP) sequence motif present in a variety of +TIPs was essential for binding of those proteins to a highly conserved C-terminal region of human EB1 protein. Although SPR1 lacks the canonical EB1 SxIP sequence-interacting motif, we found that SPR1 does contain an Iso70-Pro71 dipeptide sequence (Figure 7A). To test if Iso70-Pro71 was essential for SPR1 binding to Arabidopsis EB1b in vitro, we expressed and purified a mutant form of SPR1 Ser70-Ser71 lacking the Iso70-Pro71 dipeptide and compared the binding of wild-type and mutant SPR1 to H-EB1b using the glutathione-Sepharose resin pull-out assay. Figure 5H shows that H-EB1b protein copurified with SPR1-G-H (Ser70-Ser71) protein as it did with wild-type SPR1-G-H protein. Therefore, loss of the Iso70-Pro71 motif did not preclude the binding of SPR to EB1, suggesting that an SxIP motif is not essential for their interaction.
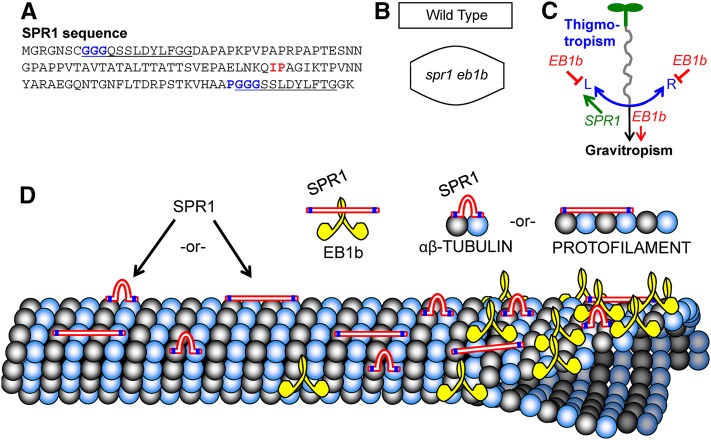
Figure 7.
Model of SPR1 and EB1b Interactions.
(A) SPR1 amino acid sequence, with the predicted MT binding motifs (GGG and PGGG) highlighted in blue within the direct repeat sequences (underlined). The Iso70-Pro71 (IP) motif that was tested for a possible role in SPR1 protein binding to EB1B is highlighted in red.
(B) Depictions of wild-type and spr1-6 eb1b-2 root and etiolated hypocotyl cell shapes to point out that double mutant cells are shorter than wild-type cells and bulge abnormally, possibly due to defects in the localization of SPR1- and EB1B-related cell expansion determinants.
(C) Working model of EB1B and SPR1 involvement in mechanical force-based directional growth responses (gravitropism and thigmotropism). SPR1 likely participates in thigmotropism process L (left root skewing), which is counter to thigmotropism process R (right root skewing). Process L is slightly stronger than process R in Col-0 roots. EB1B negatively regulates processes L and R and positively regulates gravitropism. With the loss of EB1b function, the gravitropism process is less effective at counteracting the thigmotropism processes, resulting in whatever imbalance there may be in processes L and R to become accentuated, resulting in root tip deviation from the downward growth direction.
(D) Illustrations of SPR1, EB1b, tubulin, and cortical MT protein interactions. Locations of the GGG and PGGG motifs in SPR1 are marked in blue. Note that the spacing of these motifs is such that if SPR1 lays unfolded along the MT lattice (as do MAP2/Tau family proteins, which contain comparably spaced PGGG motifs; Al-Bassam et al., 2002), SPR1 would span four tubulin proteins. Alternatively, if the SPR1 GGG and PGGG motifs bind adjacent tubulin proteins, the internal linker portion of SPR1 would loop out. Note that the C termini of two EB1b proteins interact to form a dimer (Komaki et al., 2010). While our data show that SPR1 interacts physically with EB1b, it is unclear exactly where; the interaction is likely not facilitated by an Iso70-Pro71 motif used by other proteins to bind EB1.
DISCUSSION
+TIPs are a diverse class of proteins characterized by their preferential localization to MT plus ends. +TIPs in animals have been shown to facilitate mitotic spindle formation/function as well as to regulate brain and muscle development, cell differentiation, cell migration including protrusion shape, and cell polarity (Barth et al., 2008; Jaworski et al., 2008; Geraldo and Gordon-Weeks, 2009; Minc et al., 2009; Morrison, 2009; Zhang et al., 2009; Brüning-Richardson et al., 2011; Gierke and Wittmann, 2012; Schober et al., 2012). EB1b and SPR1 are two +TIPs found to regulate directional organ growth in the model plant Arabidopsis (Nakajima et al., 2004; Sedbrook et al., 2004; Bisgrove et al., 2008). Here, we report that SPR1 acts in concert with EB1b in regulating polar cell expansion and organ growth both constitutively and in response to touch stimulation. Our data show that SPR1 binds directly to MTs in vitro, indicating that it is a bona fide MAP (Lloyd and Hussey, 2001). Moreover, SPR1 and EB1b proteins interact with each other and may compete for MT binding sites. Together, these genetic and biochemical interactions indicate that SPR1 and EB1b are key components of a dynamic MT-based modulator that regulates growth responses to mechanical stimuli (touch and gravity).
To explain the spr1-6 and eb1b-2 mutant root-skewing data, we propose a model where three processes act competitively in regulating root directional growth on inclined impenetrable surfaces. For simplicity sake, we will call these processes process L (L stands for left), process R (R stands for right), and gravitropism (Figure 7C). In this model, process L directs touch-induced leftward directional growth accompanied by left-handed axial twisting, process R directs touch-induced rightward directional growth accompanied by right-handed axial twisting, and gravitropism maintains the overall root growth trajectory in the downward direction. When roots are growing on inclined agar surfaces, gravitropism results in the root tip bending downward and pushing against the inclined surface, which enhances directional touch stimulation and resultant root waving. The balance between the L and R processes then determines the waving growth chirality.
Our data suggest that SPR1 is an important component of process L. In spr1 loss-of-function mutants, process L is compromised, thereby disrupting the balance between processes R and L, resulting in the roots taking on a rightward growth trajectory and right-handed axial twisting bias. Gravitropism counters the rightward skewing, resulting in spr1 roots maintaining an overall straight growth trajectory or a set point angle. If the gravity vector were to be randomized with the use of a clinostat, the spr1 roots would instead grow into coils, as is observed with other root-skewing mutants grown on clinostats (Sedbrook et al., 2002; Migliaccio et al., 2013).
At first glance, one would think that the main role of EB1b is enhancing process R, given that eb1b loss-of-function mutant roots exhibit a leftward slant and left-handed axial twisting bias. However, if this were the case, then with this model, eb1b-2 spr1-6 double mutant roots would be expected to have a rightward growth trajectory less severe than that of spr1-6 mutant roots. Instead, eb1b-2 spr1-6 double mutant roots skew even more strongly to the right than spr1-6 roots, to the point of forming coils (Figure 1A). Therefore, it seems likely that EB1b suppresses processes L and R and enhances gravitropism (Figure 7C). With EB1b function disrupted, whatever imbalance exists between processes L and R is accentuated. Gleeson et al. (2012) proposed a similar role for EB1b to explain their findings that eb1b-1 mutant roots appeared to be hypersensitive to touch and were slow in responding to gravity.
We observed that, under our growth conditions, wild-type Col-0 roots typically have a very slight overall root growth trajectory and axial twisting bias to the left, which suggests that the strength of process L is slightly greater than that of process R in this ecotype (Figures 1F and 2E). As such, with this model, eb1b-2 loss of function in this ecotype background would be expected to result in an accentuation of this bias, as observed.
However, our finding that overexpression of an EYFP:EB1b fusion protein resulted in seedling roots skewing to the right during the wave assay (Supplemental Figure 7A) would appear to be inconsistent with our model. Indeed, if the role of the EB1b protein is to enhance the gravity response and suppress processes L and R, then one would expect EB1b overexpression to cause roots to follow the gravity vector more closely. To explain this result in a way that is consistent with our model, we speculate that the EYFP:EB1b fusion protein disrupts the function of SPR1 and/or another protein functioning in process L as a consequence of the extra EB1b protein taking up MT binding sites and/or the EYFP protein hindering the access of other proteins, thereby resulting in the observed right skewing. Interestingly, ectopic expression of the MT binding domain of the mouse MICROTUBULE ASSOCIATED PROTEIN4 (MAP4) fused to GFP (MAP4:GFP) in Arabidopsis seedlings also causes root right skewing, possibly due to a disruption of process L (Supplemental Figure 7B; discussed in more detail below).
At least two observations support the idea that EB1b acts both as a repressor and enhancer. Our data show that when roots and etiolated hypocotyls are growing in a uniform touch or no-touch environment (embedded in solid agar or growing in midair), loss of EB1b function results in an accentuation of the organ-twisting bias (e.g., accentuation of left-handed twisting in the Col-0 ecotype background and right-handed twisting in the spr1 mutant background; Figure 2). These data suggest that EB1b acts to repress organ axial twisting independent of touch and gravistimulation (in other words, constitutively). On the other hand, Bisgrove et al. (2008) found that eb1b-1 mutant roots that grew past a barrier on vertically oriented agar exhibited a delayed bending response toward the gravity vector, a result that suggests that EB1b acts to enhance the gravity response.
These genetic data, together with the evidence for biochemical interaction and antagonism between SPR1 and EB1b, indicate that SPR1 and EB1b act as part of an MT-based modulator to regulate processes L, R, and growth curvature in response to mechanical stimuli from the environment. This mechanism might underlie the findings by Massa and Gilroy (2003) that root cap touch stimulation modulates the graviresponse. The idea that +TIPs, and SPR1 in particular, may be targets of environmental signaling is supported further by studies of salt stress. Wang et al. (2011) found that salt stress led to the 26S proteasome-based degradation of SPR1 protein, while Shoji et al. (2006) found that salt stress affected cortical MT array organization and suppressed the right-handed helical growth phenotype of spr1 mutant roots.
In animals and fungi, EB proteins have been shown to bind to other +TIPs and recruit an assortment of proteins to MTs via a C-terminal cargo binding domain, carrying out a variety of functions affecting cell division, differentiation, and polarity (Ligon et al., 2006; Honnappa et al., 2009; Galjart, 2010). It may be that SPR1 and EB1 together act to recruit proteins to MTs necessary for the establishment of MT-membrane connections and/or domains involved in cell expansion-related signaling or trafficking. With respect to competition, SPR1 and EB1b may compete in recruiting cell expansion determinants to MTs that promote left-handed and right-handed cell expansion and organ directional growth.
Our data also show that both SPR1 and EB1b promote array dynamics, enhancing MT polymerization, depolymerization, catastrophe rate, and especially rescue rate. The fact that a range of mutants altered in MT stability exhibit defects in cell growth (Ishida et al., 2007a; Perrin et al., 2007; Wang et al., 2007, 2012; Yao et al., 2008; Li et al., 2011a) suggests that proper MT dynamics and stability may be key to proper cell expansion. Given our data showing that the cortical MT arrays in spr1-6 eb1b-2 expanding root and etiolated hypocotyl cells have relatively normal organization (Figure 4), it seems unlikely that the severe anisotropic expansion defects of these cells is due to the orientation and ordering of cortical MTs and, by extension, oriented cellulose deposition. One possible explanation for the defects observed is that proper polymerization dynamics allow for dynamic restructuring of the cortical array as it organizes and mediates events at the cell membrane and wall during wall synthesis and expansion. That is, a cortical array that is oriented but too static may be compromised in function. An analogy may exist in bacterial cells, where cortical cytoskeleton MreB filaments (MreB is an actin homolog) appear to regulate cell wall synthesis and growth pattern through interactions with biosynthetic proteins; MreB movements undergo circumferential rotation dependent on wall synthesis. Modeling studies suggest that this rotation may act to distribute peptidal glycan insertion to ensure the creation of a robust cell shape (van Teeffelen et al., 2011; Huang et al., 2012).
In addition to maintaining cytoskeleton dynamics necessary for proper cell expansion, SPR1 and EB1b may regulate cell expansion by recruiting and organizing cell expansion determinants at the cell cortex. These cell expansion determinants could be cell signaling components and/or proteins involved in the targeted secretion and/or endocytosis of membrane and/or cell wall materials and proteins.
Data from animal and fungal studies suggest that EB proteins are central adaptors that mediate MT binding and plus-end tracking of a variety of other proteins (Vaughan, 2005; Kumar and Wittmann, 2012). EBs have been shown to autonomously bind to MTs, while many other +TIPs require EBs for proper MT binding and/or tip localization (Dixit et al., 2009; Akhmanova and Steinmetz, 2010; Slep, 2010; Li et al., 2011b). While we found SPR1 to behave like other +TIPs in that it physically interacts with EB1b, our data show that SPR1 can function, at least in part, autonomously of EB1b. We found that SPR1 protein can bind to MTs independently of EB1b (Figure 5A; Supplemental Figure 8). Moreover, SPR1 retains in planta function in the absence of EB1b, as demonstrated by the fact that the eb1b single mutant directional growth phenotype is considerably more subtle and opposite to that of the spr1-6 single and spr1-6 eb1b-2 double mutant phenotypes (Figure 1). If SPR1 were completely dependent upon EB1b to function, then one would expect an epistatic interaction, with spr1 mutant roots behaving similarly to eb1b mutant roots.
Our data also suggest that the SPR1 protein binds to EB1b differently from most other +TIPs. The majority of +TIPs studied bind EBs through a short hydrophobic (Ser/Thr)x(Ile/Leu)Pro sequence motif, designated the SxIP motif or the MT tip localization signal (Buey et al., 2012; Kumar and Wittmann, 2012). Substituting Ser-Ser for Iso-Pro in these proteins abolished their interactions with EBs (Honnappa et al., 2005, 2009). The SPR1 protein does not harbor the canonical SxIP motif, although it does harbor an Iso-Pro dipeptide sequence within a Lys-Gln-Ile/Val-Pro-Ala-Gly motif conserved in SPR1 and two of the five SPR1-like proteins in Arabidopsis (Figure 7; Sedbrook et al., 2004). Substituting Ser-Ser for Iso70-Pro71 in SPR1 did not reduce its binding to EB1b in vitro. It remains to be seen if these substitutions affect SPR1 function in planta.
We previously found that SPR1:EGFP exhibited MT +TIP localization in planta (Sedbrook et al., 2004), but in relatively long and shallow gradients (Supplemental Figure 5). +TIPs have been proposed to localize to MT plus ends by a variety of mechanisms, including transport by motors or one-dimensional diffusion (Bisgrove et al., 2004; Galjart, 2010), direct binding to structural features of the MT lattice concentrated at the plus end, hitchhiking on other proteins that bind to MTs, and coassembly with tubulin dimers. Our FRAP analysis of SPR1:EGFP dynamics argues against a tip concentration mechanism by transport (Supplemental Figure 6), instead revealing evidence that lateral exchange of SPR1 with the MT lattice is limited in comparison with previous analyses of EB proteins and CLIP-170 (Dragestein et al., 2008; Galjart, 2010). The slow rate of recovery and long comet tails are consistent with a low off rate for SPR1, which may be loaded by copolymerization (consistent with our observations that SPR1 can bind tubulin dimers in vitro) or which may recognize binding sites concentrated at the tip but which decay in number over time. Wang et al. (2011) recently found that salt stress accelerates SPR1 degradation by the 26S proteasome. It will be interesting to determine if salt stress and/or other environmental stimuli directly affect SPR1 residence time on MTs.
Like other animal and fungal EBs, plant EBs harbor a calponin homology domain, which is likely responsible for MT binding. On the other hand, the MT binding domains for SPR1 are likely two nearly identical direct repeat sequences at the N-terminal and C-terminal regions of the protein (Figure 7). These sequences are conserved in all SPR1-like proteins and have been shown to be sufficient for MT localization in planta (Nakajima et al., 2004). One of the repeats contains a Gly-Gly-Gly (GGG) motif while the other contains a Pro-Gly-Gly-Gly (PGGG) motif. PGGG motifs have been shown to enhance MT binding in animal MAP2, MAP4, and tau proteins, occurring repetitively in combinations of three or four direct repeat sequences (Olson et al., 1995; Fauquant et al., 2011). The direct repeat portions of MAP2 and tau proteins have been shown by electron microscopy to lie unfolded and lengthwise along the outer ridges of protofilaments in the MT lattice, where they may function in part to stimulate/stabilize MT assembly (Al-Bassam et al., 2002; Dehmelt and Halpain, 2005). No MAP2, MAP4, or tau homologs exist in plants.
The PGGG motifs in the four tau protein repeats are spaced ∼32 amino acids apart. The SPR1 GGG motif and PGGG motif are spaced 98 amino acids apart, which is comparable to the 101-amino acid spacing between the first and fourth tau PGGG motifs (Figure 7). As such, one can speculate that the SPR1 protein, which is also predicted to take on an unfolded configuration (e.g., the PYRE2 online tool predicts 91% of SPR1 protein to be disordered; Kelley and Sternberg, 2009), also lies unfolded along the MT lattice, where it stimulates and/or stabilizes MT assembly, acts as a binding partner for other proteins, and/or inhibits the binding of other MAPs, including EB1b, to the MT lattice (Figure 7). Interestingly, the expression of a mouse MAP4 MT binding domain:GFP fusion protein (Marc et al., 1998) in Arabidopsis results in roots slanting strongly to the right on tilted agar surfaces (Supplemental Figure 7B), a phenotype similar to that observed with spr1 mutants. One possibility is that this fusion protein is interfering with SPR1 MT binding and function. If so, that would suggest that SPR1 function involves the recruitment of cell expansion determinants to MTs as opposed to generally enhancing MT assembly or blocking MAPs binding to MTs, although the latter two functions may require MT binding dynamics unique to SPR1, which the MAP4 fusion protein may lack. Interestingly, one function of Caenorhabditis elegans Tau/PTL-1 is to regulate the relative mobility of MT plus-end-directed kinesins and minus-end-directed dyneins (Telley et al., 2009; Tien et al., 2011).
Further experiments will be necessary to test if more SPR1 protein binds to MTs in an eb1b mutant background and vice versa, as suggested by our in vitro protein binding results. While our preliminary analyses suggest that this may not be the case (Supplemental Figure 8), a complication to this experiment is that Arabidopsis harbors three EB1 proteins and a family of six SPR1-like proteins with overlapping function (Nakajima et al., 2006). As such, other EB1 and SPR1 family members could facilitate or otherwise influence SPR1 and EB1b protein binding and function.
In conclusion, our biochemical and genetic analyses have uncovered an intimate relationship between EB1b and the plant-specific protein SPR1, the latter of which has structural similarities to MAP2/Tau proteins present in animals but absent in plants. Our data support a model where SPR1 and EB1b are key components of an MT-related mechanism that maintains proper polar cell elongation and potentially integrates environmental cues into optimal directional growth responses. The identification of EB1b and SPR1 binding partners, as well as an exploration of the link between SPR1, EB1b and the actin cytoskeleton, should help to flesh out related molecular pathways and shed important light on fundamental processes of morphogenesis, growth, and environmental responsiveness.
METHODS
Plant Material
Arabidopsis thaliana wild-type seeds of ecotype Col-0 were obtained from the ABRC. spr1-6 is a loss-of-function mutant (insertion in At2g03680) isolated from a T-DNA activation-tagged population in the Col-0 background (Sedbrook et al., 2004). The eb1b-2 T-DNA insertional mutant (WiscDsLox331A08; insertion in At5g62500) was obtained from the Arabidopsis Knockout Facility (University of Wisconsin Biotechnology Center; Woody et al., 2007).
Plant Growth and Analysis
Arabidopsis seeds were sterilized by rinsing with 70% ethanol, followed by surface sterilization with 30% bleach and 0.01% SDS for 15 min at room temperature, followed by three rinses with sterile deionized water.
For the root waving assay, seeds were plated on 1.5% agar-solidified GM, which contained 1.5% type E agar (Sigma-Aldrich), 1.5% sucrose, and half-strength Murashige and Skoog salts (Caisson Laboratories), and vernalized at 4°C in the dark for 3 d. Thereafter, plates were aligned vertically on edge for 3 d in a Percival incubator at 22°C under 16-h-light and 8-h-dark conditions. On day 4, plates were tilted to a 40° angle, and seedlings were grown for another 4 d. For measuring root lengths, seeds were germinated and grown embedded in 0.8% agar-solidified GM on vertically oriented plates. After 3 d of growth, the locations of root tips were marked, and seedlings were grown for another 4 d. Images were then taken, and root lengths (distance from the 3-d marks to the root tips) and tip angles were measured using NIH Image 1.62 software.
For all growth measurements, statistical analyses were performed using SAS version 9.3 by one-factor fixed-effect ANOVA using PROC glm. Data sets met the assumptions for ANOVA with the exception of root cortical length data, which were analyzed after performing a log10 transformation.
PCR to Confirm T-DNA Presence, and RT-PCR to Assess EB1b Transcripts
To confirm the chromosomal location and track the presence of the EB1b-2 T-DNA in plants, PCR was performed using alkali-prepped seedling cotyledon extracts (Klimyuk et al., 1993) as templates and the following primer pair to amplify a portion of the T-DNA and flanking genomic sequences: EB1BF3 (5′-GACCACTAGACAATTTGGAGTTTCTTCAA-3′) and WiscDSLoxP745 (5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′) (product size of 600 bp). This PCR product was DNA-sequenced, revealing a T-DNA left border sequence located in intron 6 of the EB1b gene (At5g62500; Supplemental Figure 1A) (junction sequence with uppercase being genomic DNA and lowercase being T-DNA: 5′-CTCTTAACCATTCGGTTTCATTCTTTTATTcaggatatattgtggtgtaaacaaattgac-3′). To identify plant chromosomes not harboring the eb1b-2 T-DNA, the following primer pair was used (the primers flank the T-DNA site location): EB1BF3 (sequence above) and EB1BR3 (5′-CGGAAGATCATCGAGTTCAGGAGTCTG-3′) (product size of 700 bp). To confirm and track the presence of the spr1-6 T-DNA, PCR was performed using the following two primer pairs: SKI15RDIF (5′-GACGGATCGTAATTTGTCGTTTTATCAA-3′) and SPR1R1 (5′-GGACACGAAAACGTACAGTGAGGA-3′) (product size of 250 bp) identify the spr1-6-specific T-DNA; and SPR1F1 (5′-TTGACGTTGCTACTATTCTGATTGTG-3′) and SPR1R1 (product size of 460 bp) identify genomic DNA lacking the spr1-6 T-DNA. Using PCR with the above-noted primers on DNA isolated from F2 and F3 generation segregants, we confirmed that the left-slanting root phenotype of eb1b-2 seedlings as well as the unique spr1-6 eb1b-2 double mutant phenotypes were present only in plants homozygous for the T-DNA insertions in the EB1b and SPR1 genes.
RT-PCR analysis was performed to assess whether EB1b mRNA was produced in eb1b-2 mutant seedlings (Supplemental Figure 1B). Total RNA was isolated from whole seedling tissues using the TRIZOL method (Life Technologies). Reverse transcription was performed using total RNA as a template, M-MLV reverse transcriptase (Promega), and an oligo(dT) primer, as per the manufacturer’s instructions. PCR amplification was performed using primer pairs EB1BF1 (5′-GGATCAATGACCGCCTTCATCTCAATC-3′) and EB1BR1 (5′-CTCCAAATTGTCTAGTGGTCGGCC-3′) (product size of 249 bp) and EB1BF2 (5′-GTGGCTACTTCCAACAAACCAGCT-3′) and EB1BR2 (5′-CCTCAAGCCCTAGAGACTGGTTTA-3′) (product size of 316 bp). The primer pairs were designed to flank introns, so as to have a control for possible PCR amplification of contaminating genomic DNA. As an internal control, the TUBULIN ALPHA-5 (TUA5; At5g19780) reverse transcription product was amplified using primers TUA5F1 (5′-ATGACATTTGCCGCAGATCCCT-3′) and TUA5R1 (5′-ATCAAACTTGTGGTCGATCCGTGAG-3′) (product size of 560 bp).
Propidium Iodide Staining of Cell Walls
Arabidopsis seedlings were grown embedded in 0.8% agar-solidified GM on vertically oriented plates for 5 d. Roots were excised from these seedlings and mounted on cover glasses in 10 μg/mL propidium iodide (Sigma-Aldrich). For etiolated hypocotyl analyses, seedlings were germinated and grown in constant darkness on the surface of 0.8% agar-solidified GM medium on vertically oriented plates for 5 d, followed by mounting on cover glasses in 10 μg/mL propidium iodide. Images were collected using a Leica SP2 confocal microscope (Leica Microsystems) using a 561-nm diode laser for excitation and an emission band of 570 to 700 nm. All images were acquired with a 63× 1.4 numerical aperture oil-immersion objective in a 1024 × 1024 format with no scanning zoom.
Imaging Experiments
Most imaging was performed on a spinning disk confocal microscope based on a Leica DM600B stand with adaptive focus and a Leica 100× 1.4 numerical aperture oil-immersion objective. Excitation energy was supplied by either a 443-nm diode laser (Coherent) or a 515-nm DPSS laser (Cobolt). In all cases, excitation power was shuttered and attenuated by an acousto-optical tunable filter (Crystal Technology) to ∼3.5 mW at the end of the fiber delivering energy to the Yokogawa CSU-X confocal head. Semrock filters were employed: 405/442/514 dichroic, 540/60 bandpass, and 480/40 bandpass. Images were acquired with a Photometrics Evolve EMCCD camera operated at an intensification setting of 300, and instrumentation was driven using Slidebook software (3i).
Measurements of MT dynamics were made from time-series images acquired from seedlings expressing 35S:EYFP:TUA5, 3-d-old seedlings germinated in the dark, and epidermal cells in the growth zone of the upper hypocotyl. Images were acquired at 1-s intervals, and the first 100 s of each series was analyzed. Images of the trajectories of individual MTs were traced and converted into kymographs using tools in ImageJ. Polymerization velocities were determined from the slopes of the positions of MT ends in the kymographs, and transition rates were determined by counting the numbers of peaks (catastrophe) or valleys (rescues) and dividing by the total time of growing plus any pause (catastrophe) or shrinking plus any pause (rescue). Statistical comparison of these rates was performed by the C-test for comparison of two estimated Poisson means (Przyborowski and Wilenski, 1940). For each genotype, we analyzed movies from five cells in five different plants.
FRAP experiments were performed on a Leica SP5 AOBS point scanning confocal microscope. Excitation was provided by a 514 line from an argon ion laser, attenuated to 3% of its peak output. Frames were acquired at 2-s intervals using a reduced frame size, 0.19-μm pixel size, and bidirectional scanning. After collecting two baseline frames, bleaching was performed in one pass at 100% power (514 nm), and postbleaching images were collected at 2-s intervals using 10% power. Long tails of SPR1:EGFP observed before bleaching were reduced to background by the bleaching step (51 of 51 observations), and initial recovery of label was typically detectible at the tip in the first postbleaching frame (45 of 51 observations). Images were analyzed using ImageJ software and the Multiple Kymograph plugin.
For measurements of signal in FRAP experiments, a three-pixel-wide lane was drawn using ImageJ down the axis of each SPR1 comet. Profiles of 16 comets just prior to bleaching were aligned to the leak value, and means at each pixel position were calculated. The local background was defined by the four pixels in front of the comet after signal dropped to a steady baseline value. The signal along the MT lattice was corrected for this background value. The mean background just prior to photobleaching and 4 s after photobleaching were not significantly different (Student’s t test).
Plasmid Construction, Cloning, and Analysis
To generate the pTrcHis2B-SPR1-GST-6His construct (SPR1-G-H), the SPR1 open reading frame (ORF) was PCR-amplified to have NcoI and NheI-EcoRI linkers at the 5′ and 3′ ends, respectively, using the following primer pair: SPR1BAMNCOF1 (5′-CCCGGGGGATCCATGGGTCGTGGAAACAG-3′) and SPR1NHEECOR1 (5′-CCCGGGGAATTCTTGCTAGCTTGCCACCAGTGAAG-3′), then cloned into the NcoI and EcoRI restriction sites of pTrcHis2B (Invitrogen). The GST ORF was PCR-amplified from pGEX-4T-3 (GE Healthcare Life Sciences) to have NheI and XbaI linkers at the 5′ and 3′ ends, respectively, using the following primer pair: GSTHINHENTEROF1 (5′-CCCGGGAAGCTTGCTAGCCGATGACGATGACAAGATGTCCCCTATACTAGGT-3′) and GSTBAMXBAR1 (5′-ATTCGGGGATCCATCTAGAACCAGATCCGATTTTGG-3′), then cloned into the NheI and XbaI restriction sites of the SPR1 ORF-containing pTrcHis2B clone. The completed clone was verified by DNA sequencing.
To generate the SPR1-G-H (Ser70-Ser71) DNA clone, a modified QuikChange Site-Directed Mutagenesis protocol (Agilent Technologies) was employed. PCR amplification was performed using the pTrcHis2B SPR1-G-H clone as a template, Phusion DNA polymerase (New England Biolabs), and the following two overlapping primers containing base changes to generate Ser70-Ser71 codons to replace the Iso70-Pro71 codons in the SPR1 ORF: SPR1IP-SSF1 (5′-CTTAACAAGCAGAGTTCTGCTGGTATCAAAACT-3′) and SPR1IP-SSR1 (5′-AGTTTTGATACCAGCAGAACTCTGCTTGTTAAG-3′; PCR elongation duration of 11 min). The PCR product was digested with DpnI (cuts the methylated bacteria-derived template DNA but not the PCR-derived DNA) and then transformed into Escherichia coli followed by DNA sequencing of the ensuing plasmid prep DNA to confirm that the desired mutations were present and no other alterations were inadvertently introduced into the clone.
The full-length EB1b cDNA was obtained by RT-PCR from Arabidopsis T87 suspension-cultured cell total RNA as described (Kang et al., 2001) using the following primer pair: SB420 (5′-ATGGCGACGAACATTGGGATGATGGAT-3′) and SB231 (5′-GGTTTTCCCAGTCACGACTTTTTTTTTTTT-3′). The amplified product was cloned into pGEM-T Easy (Promega) and verified by sequencing using T7 promoter (5′-GTAATACGACTCACTATAGGGC-3′) and SP6 promoter (5′-ATTTAGGTGACACTATAGAATAC-3′) primers. To make E. coli expression vectors for EB1b, the ORF was PCR-amplified from the previous full-length cDNA clone, cloned into pGEM-T Easy, and verified by sequencing as above. Primers used for EB1b ORF amplification were SB420 (5′-ATGGCGACGAACATTGGGATGATGGAT-3′) and SB421 (5′-TGTCACAACTTTAAGTTTGGGTCTCTGCA-3′).
E. coli expression vectors for a GST-EB1 translational fusion were made by subcloning an EcoRI fragment from pGEMT-easy-EB1 ORFs into EcoRI-digested pGEX4T3-Tev (Rancour et al., 2004) and expression screened by small-scale induction in DH5α to verify the clones. 6xHis-tagged fusions of EB1b were made by subcloning the coding sequences into BamHI/XhoI restriction sites of the pET28a expression vector (Novagen).
Plasmid constructs were transformed into DH5α E. coli cells, propagated, purified, and sequenced to check for errors using standard procedures. Binary vector constructs were transformed into and propagated in Agrobacterium tumefaciens GV3101 using standard procedures, then used to transform Arabidopsis plants using the floral dip method (Clough and Bent, 1998).
Protein Expression and Purification
pTrcHis2B-SPR1-GST-6His (SPR1-G-H), pGEX 4T-3-EB1b (EB1b-G), and pET28a-EB1b (EB1b-H) clones were transformed into the BL21 (DE3) plysS strain of E. coli using standard procedures. Overnight cultures of 20-mL volume with respective transformants were grown in Luria-Bertani broth and inoculated into 500 mL of Luria-Bertani broth in 1000-mL Erlenmeyer flasks. After the cells had reached an OD600 in the range of 0.6 to 0.8, they were induced for protein expression with 1 mM isopropylthio-β-galactosidase for 6 h at 37°C. Cells were pelleted by centrifugation at 4000 rpm at 4°C for 15 min. The cell pellet was resuspended in 15 mL of binding buffer (GST bind buffer: 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.3; His bind buffer: 320 mM NaCl, 20 mM Tris-HCl, and 5 mM imidazole, pH 7.9). To prevent protein degradation by proteases, a protease inhibitor cocktail (Calbiochem protease inhibitor cocktail set I) at 1 mM final concentration was added. Cells were lysed by sonication (∼15 pulses, each lasting 15 s and spaced apart by 45 s using a sonicator [Branson Ultrasonic] set at control 5 and 50% duty cycle). Sonication was performed on the ice. The lysed culture was subjected to centrifugation at 10,000 rpm for 20 min at 4°C, and the resulting supernatant was applied to either 1 mL of the settled nickel-charged His-Bind resin (Novagen) or the GST-Bind resin (Novagen) depending on the protein epitope tag. The batch method of purification was performed as follows. Briefly, the resin was first equilibrated with bind buffer for 30 min (20 mM Tris-HCl, 500 mM NaCl, and 5 mM imidazole, pH 7.9, for His-Bind resin; 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, and 2.7 KCl, pH 7.3, for GST-bind resin) and then the cell lysate for 1 h. Subsequently, the resin was washed with wash buffer (20 mM Tris-HCl, 500 mM NaCl, and 50 mM imidazole, pH 7.9, for His-Bind resin; 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.3, for GST-bind resin) three times for 10 min each. The bound protein was eluted with an elute buffer (20 mM Tris-HCl, 300 mM imidazole, and 500 mM NaCl, pH 7.9, for His-tagged protein; 10 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0, for GST-tagged protein). The eluted protein fractions were diafiltrated and concentrated with 25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT, and 1 mM EDTA for GST pull-down experiments and coimmunoprecipitation experiments and with 80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA, and 20% (v/v) glycerol for MT cosedimentation assays, using Amicon Ultra-15 centrifugal filter devices (Millipore) spun in a Beckman tabletop centrifuge with a swinging-bucket rotor at 4000 rpm.
Protein Gel Blotting and Antibodies
Polyclonal rabbit antibodies were generated against full-length E. coli-expressed GST-EB1b protein using standard protocols. Serum was cleared of anti-GST antibodies, and affinity purification of anti-EB1 antibodies against the full-length GST-EB1b antigens was performed as described (Rancour et al., 2010). Protein gel blotting and analyses were performed using standard procedures. EB1b protein was detected with rabbit polyclonal antibodies (1:5000 dilution) specific for EB1a and EB1b proteins. His-tagged and GST-tagged SPR1 and EB1b were detected using anti-His (1:250 dilution) and anti-GST (1:1500 dilution) antibodies, respectively (Santa Cruz Biotech). Horseradish peroxidase-conjugated goat anti-rabbit antibody (Immunopure antibody; Pierce) was used as the secondary antibody at a dilution of 1:7500, followed by detection using the Pierce ECL protein gel blotting substrate and film autoradiography.
GST Pull-Down Assay
The GST pull downs were performed by binding 100 μg of GST fusion proteins to glutathione-Sepharose beads (Novagen) and incubating them with 25 μg of His-tagged proteins in a bind buffer containing 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5% Nonidet P-40. After an incubation of 1 h at room temperature, beads were washed three times with the wash buffer containing 25 mM Tris-HCl, pH 8.0, 150 to 300 mM NaCl, and 0.5% Nonidet P-40. Proteins were then eluted from the washed beads with glutathione and analyzed by protein gel blotting.
SPR1:GFP Coimmunoprecipitation
Coimmunoprecipitation experiments using plant extracts were performed as described by Serino and Deng (2007). About 3-week-old transgenic plants expressing SPR1:GFP driven off of the endogenous SPR1 promoter (Sedbrook et al., 2004) were harvested and ground to a fine powder in liquid nitrogen and further in cold grinding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM phenylmethylsulfonyl flouride, and 1× protease inhibitor cocktail [Calbiochem]) and then spun at 16,000g for 10 min at 4°C. The extract was precleared with one-tenth volume of Protein A-Sepharose beads (GE Healthcare) by shaking for 1 h at 4°C. The precleared lysate was incubated with 3 μg of anti-GFP antibody (Invitrogen) and one-tenth volume of Protein A-Sepharose beads (added together) overnight at 4°C. Beads were washed three times with 5 volumes of grinding buffer supplemented with 0.1% Nonidet P-40. Immunoprecipitation complexes were eluted by boiling the beads in 20 μL of SDS-PAGE loading buffer for 5 min and then electrophoresed on an SDS-PAGE gel followed by blotting on polyvinylidene difluoride membrane and protein gel blot analysis.
MT Cosedimentation Assay
A 10-μL bovine brain tubulin protein aliquot (5 μg/μL; Cytoskeleton) stored at −80°C was defrosted at room temperature and polymerized at 37°C for 10 min in an MT stabilization buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 20% [v/v] glycerol, and 20 μM taxol). Solutions of recombinant proteins to be added to MTs were centrifuged at 100,000g for 20 min at 4°C to remove any aggregates. Recombinant proteins were then added to 10 μg of taxol-stabilized MTs (50 μL total volume, final tubulin concentration of 3.64 μM). After 30 min of incubation at room temperature, samples were overlaid on a cushion buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 50% glycerol, and 20 μM taxol) and centrifuged at 25,000g for 30 min to pellet MTs. Proteins in the pellet and the supernatant fractions were analyzed by SDS-PAGE and protein gel blot analysis. Intensities of Pierce ECL-detected bands on autoradiographic film from protein gel blots were imaged on a flatbed scanner and quantified using NIH Image 1.62 software. For the SPR1 and EB1b cosedimentation assays, proteins were added consecutively to the taxol-stabilized MTs. For example, for the assay shown in Figure 5F, for each tube, SPR1 was added first, immediately followed by the addition of EB1b.
Tubulin Coimmunoprecipitation
Ten micrograms of bovine brain tubulin protein (Cytoskeleton) and 3 μL of anti-tubulin antibody (Sigma-Aldrich) were allowed to interact for 30 min before incubating the mixture with Protein A-Sepharose beads. The beads were washed three times with TNT buffer (40 mM Tris-HCl, 150 mM NaCl, and 0.1% Nonidet P-40). Twenty-five microliters of precoated and washed beads was incubated with EB1b or SPR1 proteins expressed in and purified from E. coli for 1 h at room temperature. The beads were subsequently washed three times with the wash buffer and boiled at 80°C for 5 min in the SDS loading buffer to elute and denature proteins.
We note that all of the protein interaction experiments described in this article were reproducibly repeated at least three times.
Yeast Two-Hybrid Analyses
The ORFs of SPR1 and EB1b were cloned into Gateway versions of the pACT2 and pAS2-1 vectors, respectively (Ketelaar et al., 2004). The corresponding SPR1 and EB1b fusion constructs were cotransformed into the yeast strain Mav203, as per instructions for the Saccharomyces cerevisiae EasyComp Transformation Kit (Invitrogen). Preparation of selective media, yeast growth techniques, protein interaction analyses, and autoactivation testing were performed following the ProQuest Two-Hybrid System with Gateway Technology manual (Invitrogen).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: SPR1 (At2g03680), EB1a (At3g47690), EB1b (At5g62500), and EB1c (At5g67270).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Depiction of the eb1b-2 T-DNA Location and Determination of Related eb1b-2 Gene Product Formation.
Supplemental Figure 2. Histograms of Measured Growth Velocities of Microtubule Plus Ends.
Supplemental Figure 3. In Vitro Pullout of EB1b Protein by Tubulin Heterodimers.
Supplemental Figure 4. Yeast Two-Hybrid Analysis of EB1b by SPR1 Protein Interaction.
Supplemental Figure 5. Colocalization of pSPR1:SPR1:EGFP and p35S:mCherry:EB1b.
Supplemental Figure 6. Stable Boundary of SPR1:EGFP at the Edge of the Bleached Region.
Supplemental Figure 7. Images of 7-d-Old Arabidopsis Seedlings on 1.5% Agar-Solidified GM Put through the Wave Assay.
Supplemental Figure 8. SPR1:EGFP Localization in an eb1b-2 Hypocotyl Cell versus the Wild Type.
Supplemental Movie 1. Microtubule Dynamics in an spr1-6 eb1b-2 Etiolated Hypocotyl Cell versus the Wild Type.
Supplementary Material
Acknowledgments
We thank Andrei Smertenko for technical advice on performing protein interaction experiments and Cynthia Cass for help with statistical analyses. This work was supported by grants from the National Science Foundation to J.C.S. (Grant 0524355), D.W.E. (Grant 0524334), and S.Y.B. (Grant 0446157) and by an E.L. Mockford Fellowship and an R.D. Weigel Research Grant from the Beta Lambda Chapter of Phi Sigma to C.G.
AUTHOR CONTRIBUTIONS
All authors designed aspects of the research. J.C.S., D.W.E., S.Y.B., and P.J.H. supervised the research. C.G. and J.C.S. built many of the constructs, generated transgenics, and performed the biochemical and genetic analyses including root measurements and imaging. V.K. and D.W.E. performed the fluorescent protein localization and FRAP analyses. J.J.L. and D.W.E. performed the MT dynamics analyses and contributed the hypocotyl MT array images and movie. D.M.R. generated the EB1 cDNA clones, GST-EB1 fusion expression systems, and the anti-EB1 antibodies. D.K. and P.J.H. contributed the yeast two-hybrid data. J.C.S., D.W.E., and C.G. wrote the article. All authors edited the article.
Glossary
- MT
microtubule
- GM
growth medium
- Col-0
Columbia-0
- FRAP
fluorescence recovery after photobleaching
- ADU
arbitrary density units
- ORF
open reading frame
Footnotes
Online version contains Web-only data.
References
- Akhmanova A., Steinmetz M.O. (2008). Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9: 309–322. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M.O. (2010). Microtubule +TIPs at a glance. J. Cell Sci. 123: 3415–3419. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., Ozer R.S., Safer D., Halpain S., Milligan R.A. (2002). MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 157: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu J.M., Bordas J., Diaz J.F., García de Ancos J., Gil R., Medrano F.J., Nogales E., Pantos E., Towns-Andrews E. (1992). Low resolution structure of microtubules in solution. Synchrotron x-ray scattering and electron microscopy of taxol-induced microtubules assembled from purified tubulin in comparison with glycerol and MAP-induced microtubules. J. Mol. Biol. 226: 169–184. [DOI] [PubMed] [Google Scholar]
- Barth A.I., Caro-Gonzalez H.Y., Nelson W.J. (2008). Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin. Cell Dev. Biol. 19: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171. [DOI] [PubMed] [Google Scholar]
- Berrueta L., Kraeft S.K., Tirnauer J.S., Schuyler S.C., Chen L.B., Hill D.E., Pellman D., Bierer B.E. (1998). The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc. Natl. Acad. Sci. USA 95: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450: 1100–1105. [DOI] [PubMed] [Google Scholar]
- Bisgrove S.R., Hable W.E., Kropf D.L. (2004). +TIPs and microtubule regulation. The beginning of the plus end in plants. Plant Physiol. 136: 3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove S.R., Lee Y.-R.J., Liu B., Peters N.T., Kropf D.L. (2008). The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. Plant Cell 20: 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüning-Richardson A., Langford K.J., Ruane P., Lee T., Askham J.M., Morrison E.E. (2011). EB1 is required for spindle symmetry in mammalian mitosis. PLoS ONE 6: e28884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey R.M., et al. (2012). Sequence determinants of a microtubule tip localization signal (MtLS). J. Biol. Chem. 287: 28227–28242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K.E., Brunner D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14: 548–559. [DOI] [PubMed] [Google Scholar]
- Busch K.E., Hayles J., Nurse P., Brunner D. (2004). Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6: 831–843. [DOI] [PubMed] [Google Scholar]
- Chan J., Calder G.M., Doonan J.H., Lloyd C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5: 967–971. [DOI] [PubMed] [Google Scholar]
- Chan J., Crowell E., Eder M., Calder G., Bunnewell S., Findlay K., Vernhettes S., Höfte H., Lloyd C. (2010). The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 123: 3490–3495. [DOI] [PubMed] [Google Scholar]
- Chan J., Eder M., Crowell E.F., Hampson J., Calder G., Lloyd C. (2011). Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls. J. Cell Sci. 124: 1088–1094. [DOI] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Gonneau M., Stierhof Y.D., Höfte H., Vernhettes S. (2010). Regulated trafficking of cellulose synthases. Curr. Opin. Plant Biol. 13: 700–705. [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Halpain S. (2005). The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 6: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Barnett B., Lazarus J.E., Tokito M., Goldman Y.E., Holzbaur E.L. (2009). Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. USA 106: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragestein K.A., van Cappellen W.A., van Haren J., Tsibidis G.D., Akhmanova A., Knoch T.A., Grosveld F., Galjart N. (2008). Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J. Cell Biol. 180: 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquant C., Redeker V., Landrieu I., Wieruszeski J.M., Verdegem D., Laprévote O., Lippens G., Gigant B., Knossow M. (2011). Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J. Biol. Chem. 286: 33358–33368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Xu T., Zhu L., Wen M., Yang Z. (2009). A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 19: 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Himmelspach R., Hocart C.H., Williamson R.E., Mansfield S.D., Wasteneys G.O. (2011). Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J. 66: 915–928. [DOI] [PubMed] [Google Scholar]
- Fujita M., Lechner B., Barton D.A., Overall R.L., Wasteneys G.O. (2012). The missing link: Do cortical microtubules define plasma membrane nanodomains that modulate cellulose biosynthesis? Protoplasma 249 (suppl. 1): S59–S67. [DOI] [PubMed] [Google Scholar]
- Galjart N. (2010). Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20: R528–R537. [DOI] [PubMed] [Google Scholar]
- Geraldo S., Gordon-Weeks P.R. (2009). Cytoskeletal dynamics in growth-cone steering. J. Cell Sci. 122: 3595–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierke S., Wittmann T. (2012). EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr. Biol. 22: 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson L., Squires S., Bisgrove S.R. (2012). The microtubule associated protein END BINDING 1 represses root responses to mechanical cues. Plant Sci. 187: 1–9. [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hamada T. (2007). Microtubule-associated proteins in higher plants. J. Plant Res. 120: 79–98. [DOI] [PubMed] [Google Scholar]
- Honnappa S., et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138: 366–376. [DOI] [PubMed] [Google Scholar]
- Honnappa S., John C.M., Kostrewa D., Winkler F.K., Steinmetz M.O. (2005). Structural insights into the EB1-APC interaction. EMBO J. 24: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.C., Ehrhardt D.W., Shaevitz J.W. (2012). The molecular origins of chiral growth in walled cells. Curr. Opin. Microbiol. 15: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kaneko Y., Iwano M., Hashimoto T. (2007a). Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Thitamadee S., Hashimoto T. (2007b). Twisted growth and organization of cortical microtubules. J. Plant Res. 120: 61–70. [DOI] [PubMed] [Google Scholar]
- Jaworski J., Hoogenraad C.C., Akhmanova A. (2008). Microtubule plus-end tracking proteins in differentiated mammalian cells. Int. J. Biochem. Cell Biol. 40: 619–637. [DOI] [PubMed] [Google Scholar]
- Kaloriti D., Galva C., Parupalli C., Khalifa N., Galvin M., Sedbrook J.C. (2007). Microtubule associated proteins in plants and the processes they manage. J. Integr. Plant Biol. 49: 1164–1173. [Google Scholar]
- Kang B.-H., Busse J.S., Dickey C., Rancour D.M., Bednarek S.Y. (2001). The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 126: 47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Voss C., Dimmock S.A., Thumm M., Hussey P.J. (2004). Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 567: 302–306. [DOI] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J.C., Ehrhardt D.W., Hülskamp M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120: 4416–4425. [DOI] [PubMed] [Google Scholar]
- Klimyuk V.I., Carroll B.J., Thomas C.M., Jones J.D. (1993). Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 3: 493–494. [DOI] [PubMed] [Google Scholar]
- Komaki S., Abe T., Coutuer S., Inzé D., Russinova E., Hashimoto T. (2010). Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 123: 451–459. [DOI] [PubMed] [Google Scholar]
- Kumar P., Wittmann T. (2012). +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22: 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Akhmanova A. (2006). Microtubule plus end: a hub of cellular activities. Traffic 7: 499–507. [DOI] [PubMed] [Google Scholar]
- Li J., Wang X., Qin T., Zhang Y., Liu X., Sun J., Zhou Y., Zhu L., Zhang Z., Yuan M., Mao T. (2011a). MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23: 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Miki T., Watanabe T., Kakeno M., Sugiyama I., Kaibuchi K., Goshima G. (2011b). EB1 promotes microtubule dynamics by recruiting sentin in Drosophila cells. J. Cell Biol. 193: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon L.A., Shelly S.S., Tokito M.K., Holzbaur E.L. (2006). Microtubule binding proteins CLIP-170, EB1, and p150Glued form distinct plus-end complexes. FEBS Lett. 580: 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C. (2011). Dynamic microtubules and the texture of plant cell walls. Int. Rev. Cell Mol. Biol. 287: 287–329. [DOI] [PubMed] [Google Scholar]
- Lloyd C., Hussey P. (2001). Microtubule-associated proteins in plants—Why we need a MAP. Nat. Rev. Mol. Cell Biol. 2: 40–47. [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao T., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa G.D., Gilroy S. (2003). Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J. 33: 435–445. [DOI] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kernebeck B., Srinivas B.P., Hülskamp M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13: 1991–1997. [DOI] [PubMed] [Google Scholar]
- Migliaccio F., Tassone P., Fortunati A. (2013). Circumnutation as an autonomous root movement in plants. Am. J. Bot. 100: 4–13. [DOI] [PubMed] [Google Scholar]
- Minc N., Bratman S.V., Basu R., Chang F. (2009). Establishing new sites of polarization by microtubules. Curr. Biol. 19: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.E. (2009). The APC-EB1 interaction. Adv. Exp. Med. Biol. 656: 41–50. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Kawamura T., Hashimoto T. (2006). Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol. 47: 513–522. [DOI] [PubMed] [Google Scholar]
- Okada K., Shimura Y. (1990). Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250: 274–276. [DOI] [PubMed] [Google Scholar]
- Oliva M., Dunand C. (2007). Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol. 176: 37–43. [DOI] [PubMed] [Google Scholar]
- Olson K.R., McIntosh J.R., Olmsted J.B. (1995). Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J. Cell Biol. 130: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Perrin R.M., Wang Y., Yuen C.Y., Will J., Masson P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49: 961–971. [DOI] [PubMed] [Google Scholar]
- Przyborowski J., Wilenski H. (1940). Homogeneity of results in testing samples from Poisson series. Biometrika 31: 313–323. [Google Scholar]
- Rancour D.M., Backues S.K., Bednarek S.Y. (2010). Protein antigen expression in Escherichia coli for antibody production. Methods Mol. Biol. 657: 3–20. [DOI] [PubMed] [Google Scholar]
- Rancour D.M., Park S., Knight S.D., Bednarek S.Y. (2004). Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. J. Biol. Chem. 279: 54264–54274. [DOI] [PubMed] [Google Scholar]
- Rutherford R., Masson P.H. (1996). Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol. 111: 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober J.M., Kwon G., Jayne D., Cain J.M. (2012). The microtubule-associated protein EB1 maintains cell polarity through activation of protein kinase C. Biochem. Biophys. Res. Commun. 417: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Kaloriti D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13: 303–310. [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Ehrhardt D.W., Fisher S.E., Scheible W.R., Somerville C.R. (2004). The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino G., Deng X.W. (2007). Protein coimmunoprecipitation in Arabidopsis. CSH Protoc. 2007: pdb.prot4683. [DOI] [PubMed] [Google Scholar]
- Shoji T., Suzuki K., Abe T., Kaneko Y., Shi H., Zhu J.K., Rus A., Hasegawa P.M., Hashimoto T. (2006). Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 47: 1158–1168. [DOI] [PubMed] [Google Scholar]
- Slep K.C. (2010). Structural and mechanistic insights into microtubule end-binding proteins. Curr. Opin. Cell Biol. 22: 88–95. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Cosgrove D.J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19: R800–R811. [DOI] [PubMed] [Google Scholar]
- Telley I.A., Bieling P., Surrey T. (2009). Obstacles on the microtubule reduce the processivity of Kinesin-1 in a minimal in vitro system and in cell extract. Biophys. J. 96: 3341–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196. [DOI] [PubMed] [Google Scholar]
- Tien N.-W., Wu G.-H., Hsu C.-C., Chang C.-Y., Wagner O.I. (2011). Tau/PTL-1 associates with kinesin-3 KIF1A/UNC-104 and affects the motor’s motility characteristics in C. elegans neurons. Neurobiol. Dis. 43: 495–506. [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., Canman J.C., Salmon E.D., Mitchison T.J. (2002). EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell 13: 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S., Wang S., Furchtgott L., Huang K.C., Wingreen N.S., Shaevitz J.W., Gitai Z. (2011). The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. USA 108: 15822–15827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K.T. (2005). TIP maker and TIP marker: EB1 as a master controller of microtubule plus ends. J. Cell Biol. 171: 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kurepa J., Hashimoto T., Smalle J.A. (2011). Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 23: 3412–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang J., Yuan M., Ehrhardt D.W., Wang Z., Mao T. (2012). Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24: 4012–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhu L., Liu B., Wang C., Jin L., Zhao Q., Yuan M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G.O., Fujita M. (2006). Establishing and maintaining axial growth: Wall mechanical properties and the cytoskeleton. J. Plant Res. 119: 5–10. [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165. [DOI] [PubMed] [Google Scholar]
- Yao M., Wakamatsu Y., Itoh T.J., Shoji T., Hashimoto T. (2008). Arabidopsis SPIRAL2 promotes uninterrupted microtubule growth by suppressing the pause state of microtubule dynamics. J. Cell Sci. 121: 2372–2381. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zaal K.J., Sheridan J., Mehta A., Gundersen G.G., Ralston E. (2009). Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J. Cell Sci. 122: 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.