ATL1 is a ubiquitin ligase that promotes cell death, but this activity is inhibited by the protein kinase EDR1, which physically associates with ATL1. Loss of EDR1 function enhances cell death in response to biotic and abiotic stresses; thus, stress-induced cell death signaling is regulated by two major and interlinked posttranslational modification mechanisms: phosphorylation and ubiquitination.
Abstract
Loss-of-function mutations in the Arabidopsis thaliana ENHANCED DISEASE RESISTANCE1 (EDR1) gene confer enhanced programmed cell death under a variety of abiotic and biotic stress conditions. All edr1 mutant phenotypes can be suppressed by missense mutations in the KEEP ON GOING gene, which encodes a trans-Golgi network/early endosome (TGN/EE)-localized E3 ubiquitin ligase. Here, we report that EDR1 interacts with a second E3 ubiquitin ligase, ARABIDOPSIS TOXICOS EN LEVADURA1 (ATL1), and negatively regulates its activity. Overexpression of ATL1 in transgenic Arabidopsis induced severe growth inhibition and patches of cell death, while transient overexpression in Nicotiana benthamiana leaves induced cell death and tissue collapse. The E3 ligase activity of ATL1 was required for both of these processes. Importantly, we found that ATL1 interacts with EDR1 on TGN/EE vesicles and that EDR1 suppresses ATL1-mediated cell death in N. benthamiana and Arabidopsis. Lastly, knockdown of ATL1 expression suppressed cell death phenotypes associated with the edr1 mutant and made Arabidopsis hypersusceptible to powdery mildew infection. Taken together, our data indicate that ATL1 is a positive regulator of programmed cell death and EDR1 negatively regulates ATL1 activity at the TGN/EE and thus controls stress responses initiated by ATL1-mediated ubiquitination events.
INTRODUCTION
Arabidopsis thaliana plants containing loss-of-function mutations in the ENHANCED DISEASE RESISTANCE1 (EDR1) gene exhibit enhanced host cell death and enhanced resistance during infection by the biotrophic fungal pathogen Golovinomyces cichoracearum, the causal agent for powdery mildew disease on Arabidopsis (Frye and Innes, 1998). The edr1 mutant also displays enhanced cell death in response to many abiotic stress conditions such as drought, ethylene treatment, and aging (Frye et al., 2001; Tang et al., 2005a, 2005b). Thus, EDR1 has been considered as a negative regulator of programmed cell death. EDR1 encodes a protein kinase similar to CONSTITUTIVE TRIPLE RESPONSE1, a negative regulator of the ethylene response pathway. Transcriptome and genetic analyses indicate that the SA signaling pathway is essential for edr1-mediated phenotypes (Frye et al., 2001; Tang et al., 2005b; Christiansen et al., 2011), as is the case for many other cell death-promoting mutants (Rate et al., 1999; Brodersen et al., 2002; Tang et al., 2005a, 2006).
To investigate the specific signaling pathway(s) in which EDR1 participates, extensive genetic screens have been performed to identify second site suppressors of edr1. From these screens, KEEP ON GOING (KEG) was identified as a key player. Specific missense mutations in KEG have been identified that fully suppress all known edr1 mutant phenotypes (Wawrzynska et al., 2008). KEG is a RING-finger E3 ligase that has been previously shown to regulate abscisic acid signaling through mediating degradation of the transcription factors ABSCISIC ACID INSENSITIVE5 (ABI5), ABA BINDING FACTOR1 (ABF1), and ABF3 (Stone et al., 2006; Liu and Stone, 2010; Chen et al., 2013). We have previously shown that KEG physically associates with the kinase domain of EDR1 on trans-Golgi network/early endosome (TGN/EE) vesicles (Gu and Innes, 2011), while Liu and Stone (2013) have recently shown that KEG physically associates with ABI5 on TGN/EE vesicles. Together, these data suggest that EDR1 may function at the TGN/EE and regulate the E3 ligase activity of KEG.
Here, we report that EDR1 interacts with another RING-finger E3 ligase ARABIDOPSIS TOXICOS EN LEVADURA1 (ATL1), which contains a predicted transmembrane domain and localizes to endosomes and the plasma membrane. However, the interaction between EDR1 and ATL1 is restricted to the TGN/EE. In contrast to the negative role of EDR1 in regulating cell death, we found that ATL1 promotes cell death during transient expression in Nicotiana benthamiana. Furthermore, overexpression of ATL1 in transgenic Arabidopsis caused severe dwarfing and cell death. These phenotypes were dependent on the RING E3 ligase activity of ATL1. Coexpression of EDR1 with ATL1 in N. benthamiana suppressed ATL1-induced cell death, and this suppression required EDR1 kinase activity. Knockdown of ATL1 using RNA interference suppressed edr1 associated cell death phenotypes, suggesting that edr1-mediated phenotypes are caused in part by hyperactivity of ATL1. Furthermore, ATL1 knockdown plants were hypersusceptible to powdery mildew infection, indicating that ATL1 plays a central role in regulating plant defense responses and programmed cell death.
RESULTS
EDR1 Physically Associates With ATL1 in the TGN/EE
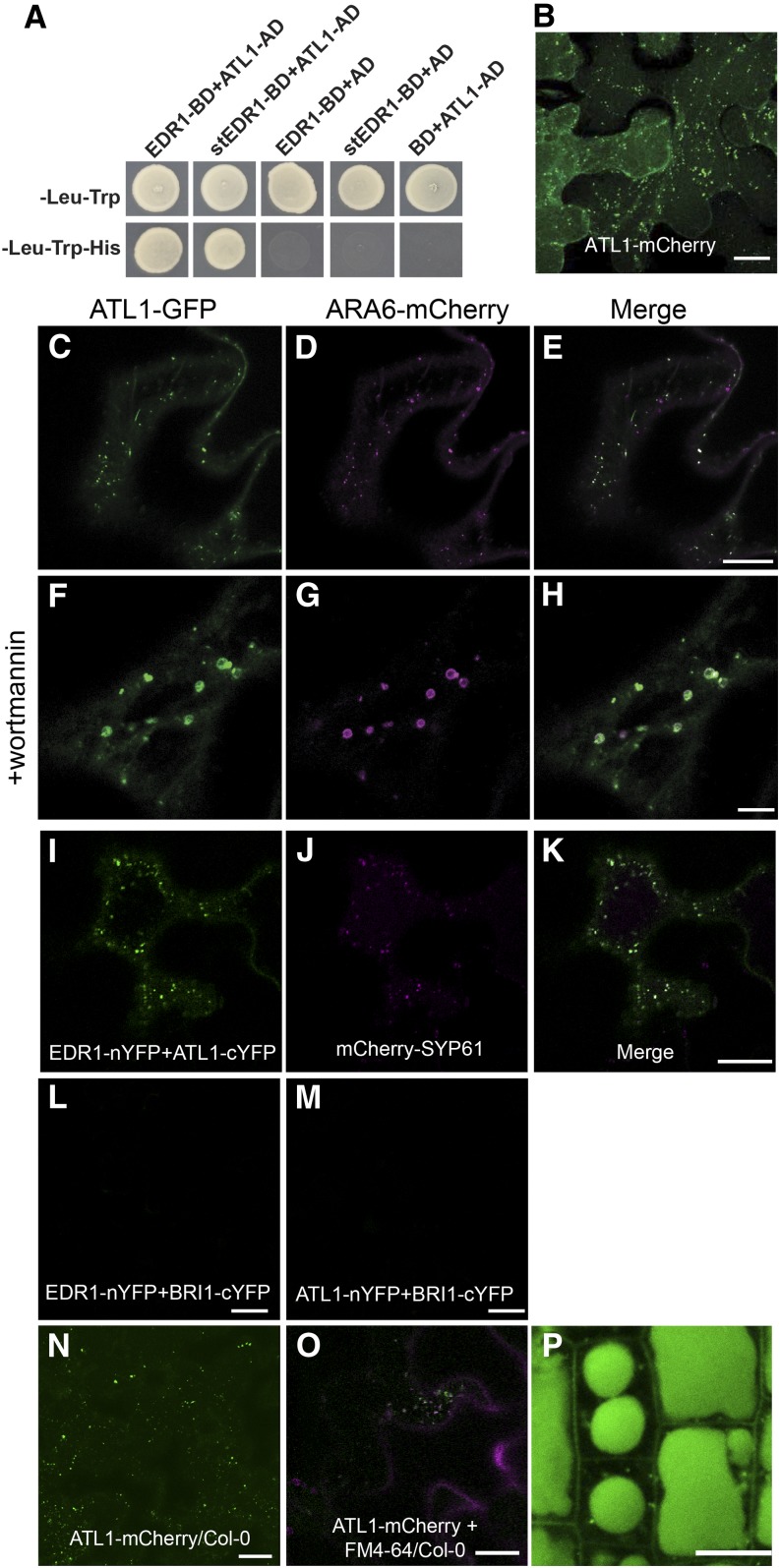
To search for components involved in EDR1-mediated signaling pathways, we conducted a yeast two-hybrid screen using full-length wild-type EDR1 as bait. ATL1 (ARABIDOPSIS TOXICOS EN LEVADURA; “toxic in yeast”; At1g04360) was one putative EDR1-interacting protein identified in this screen. To confirm the EDR1-ATL1 interaction in yeast, we obtained a full-length cDNA clone of ATL1. As shown in Figure 1A, full-length ATL1 interacted with full-length wild-type EDR1 in a yeast two-hybrid assay, whereas ATL1 and EDR1 did not interact with the empty vector controls. Notably, ATL1 also interacted with a substrate trap form of EDR1 (stEDR1; Gu and Innes, 2011), which contains a D810A substitution in the phosphotransfer domain. The equivalent substitution in other kinases has been shown to stabilize interactions with substrates (Gibbs and Zoller, 1991). To confirm that ATL1 forms a complex with EDR1 in plant cells, we performed a coimmunoprecipitation analysis using transient expression of epitope-tagged proteins in Arabidopsis protoplasts. Although EDR1 accumulated very poorly in these assays, we were able to immunoprecipitate EDR1 and observed a clear coimmunoprecipitation with ATL1 (Supplemental Figure 1A).
Figure 1.
ATL1 Interacts with EDR1 and Localizes to Endomembrane Structures.
(A) EDR1 and ATL1 interact in a yeast two-hybrid assay. Yeast strains expressing the indicated constructs were plated on double dropout (-Leu-Trp) media to select for the bait and prey plasmids and triple dropout (-Leu-Trp-His) media to select for interaction between the indicated proteins. Empty vectors containing the yeast GAL4 activation domain (AD) or DNA binding domain (BD) were used as negative controls.
(B) ATL1-mCherry localizes to the plasma membrane and intracellular puncta. An N. benthamiana leaf was transiently transformed with a 35S:ATL1-mCherry construct and imaged by confocal microscopy at 40 h after Agrobacterium infiltration. Image is a Z stack composed of eight optical sections.
(C) to (E) ATL1-eGFP colocalizes with the MVB/LE marker ARA6-mCherry. Epidermal cells of N. benthamiana were cotransformed with ATL1-eGFP and the indicated organelle markers fused with mCherry. Cells were imaged using confocal laser scanning microscopy, and single optical sections are shown.
(F) to (H) Leaves from (C) to (E) were infiltrated with 33 μM wortmannin for 1 h to induce MVB dilation.
(I) to (M) EDR1 and ATL1 interact on TGN/EE vesicles in a BiFC assay. EDR1-nYFP and ATL1-cYFP were transiently coexpressed in N. benthamiana with the TGN/EE marker mCherry-SYP61. The BiFC signal is shown in green (I) and the mCherry-SYP61 signal in magenta (J). As negative controls, EDR1-nYFP and BRI1-cYFP were coexpressed (L), as well as BRI1-nYFP and ATL1-cYFP (M).
(N) ATL1-mCherry localizes to the plasma membrane and intracellular puncta in transgenic Arabidopsis. Wild-type Col-0 plants were transformed with a 35S:ATL1-mCherry construct and leaf epidermal cells of 4-week old plants were imaged using CSLM. Image is a Z stack.
(O) ATL1-mCherry colocalizes with the endocytic tracer dye FM4-64 in stable transgenic Arabidopsis lines. Image was taken 15 min poststaining.
(P) ATL1-mCherry accumulates inside the lytic vacuoles of root epidermal cells.
Bars = 10 μm.
ATL1 belongs to a family of RING finger E3 ubiquitin ligases that consists of 91 members in Arabidopsis (Salinas-Mondragón et al., 1999; Morris et al., 2010; Aguilar-Hernández et al., 2011; Guzmán, 2012). Members of the ATL family all contain an N-terminal transmembrane domain followed by a RING-H2 type E3 ligase domain (Aguilar-Hernández et al., 2011; Guzmán, 2012). Functional analyses of several ATL family members in Arabidopsis indicate that they play important roles in diverse biological processes, including defense responses, growth phase transitions, endosperm development, regulation of cell death during root development, and photoperiod response (Serrano and Guzmán, 2004; Pagnussat et al., 2005; Lin et al., 2008; Sato et al., 2009; Berrocal-Lobo et al., 2010; Morris et al., 2010; Guzmán, 2012). However, there have been no prior publications describing a function for ATL1.
To study the function of ATL1 and potential links between ATL1 and EDR1 function, we first determined the subcellular localization of ATL1. To avoid potential interference with membrane targeting, fluorescent protein was fused to the C terminus of ATL1. Two different fluorescent proteins, enhanced green fluorescent protein (eGFP) and mCherry, were used to exclude fluorophore-specific artifacts. In particular, mCherry was used to avoid quenching of fluorescence in the acidic vacuole. These fusion constructs were placed under the control of a 35S promoter and transiently expressed in N. benthamiana leaves by Agrobacterium tumefaciens-mediated transformation. We found that for both fluorescent protein fusion constructs, ATL1 was predominantly localized to punctate vesicular structures, although there was also a weak plasma membrane signal visible in three-dimensional reconstructions (Figure 1B).
To further investigate the identity of ATL1-labeled intracellular vesicles, we coexpressed the ATL1-eGFP construct with the Golgi cisternae marker GmMan49-mCherry (Nelson et al., 2007), TGN/EE marker VHA-a1-RFP (red fluorescent protein) (Dettmer et al., 2006), and two multivesicular body/late endosome (MVB/LE) markers ARA6-mCherry (Ueda et al., 2004; Haas et al., 2007) and mCherry-SYP21 (Ueda et al., 2004; Uemura et al., 2004). We found that ATL1-eGFP partially colocalized with VHA-a1-RFP (Supplemental Figures 1B to 1D) and predominantly overlapped with ARA6-mCherry (Figures 1C to 1E) and mCherry-SYP21 (Supplemental Figures 1E to 1G). No colocalization was observed between ATL1 and Golgi (Supplemental Figures 1H to 1J). To further validate the primary localization of ATL1 as MVB/LE vesicles, we treated cells with the pharmacological trafficking inhibitor wortmannin, which induces dilation of MVB/LE vesicles (Tse et al., 2004; Jaillais et al., 2006; Lam et al., 2007; Silady et al., 2008; Wang et al., 2009). As shown in Figures 1F to 1H, wortmannin induced small ring-like structures that were colabeled by ARA6-mCherry and ATL1-eGFP, confirming that ATL1 accumulated mostly in MVB/LE vesicles upon transient expression.
EDR1 and KEG have previously been shown to interact on TGN/EE vesicles (Gu and Innes, 2011). Given that ATL1 is distributed to multiple locations within the endomembrane system, we investigated where in the endomembrane system EDR1 was interacting with ATL1. This was accomplished using a bimolecular fluorescence complementation (BiFC) assay in which EDR1-nYFP (EDR1 fused with N-terminal half of super yellow fluorescent protein) was coexpressed with ATL1-cYFP (ATL1 fused with C-terminal half of sYFP) in N. benthamiana. As a negative control, we used BRASSINOSTEROID INSENSITIVE1 (BRI1), which distributes throughout the entire endomembrane system. We observed a punctate fluorescence pattern with EDR1-nYFP and ATL1-cYFP (Figure 1I), but not with EDR1-nYFP and BRI1-cYFP, or BRI1-nYFP and ATL1-cYFP (Figures 1L and 1M), confirming that EDR1 and ATL1 interact. To determine the identity of these fluorescent puncta, we coexpressed the ATL1/EDR1 BiFC constructs with the TGN/EE marker mCherry-SYP61 (Figure 1J) (Uemura et al., 2004) and the MVB/LE marker ARA6-mCherry (Supplemental Figure 1L). As shown in Figure 1K, the BiFC signal colocalized with mCherry-SYP61, but not with ARA6-mCherry (Supplemental Figure 1M), demonstrating that the EDR1-ATL1 interaction occurs primarily on TGN/EE vesicles.
ATL1 Undergoes Constitutive Internalization and Vacuolar Degradation
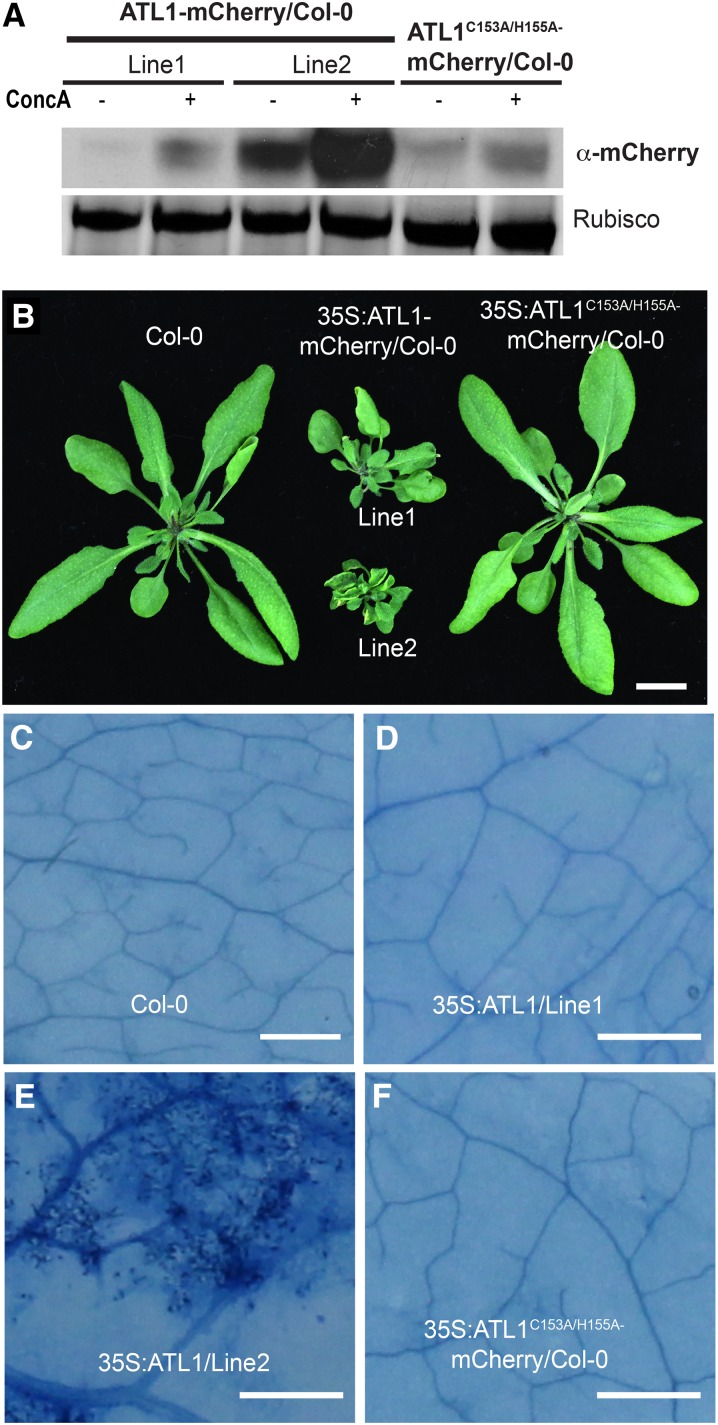
To avoid potential artifacts associated with heterologous transient expression, we generated stable transgenic Arabidopsis lines expressing 35S:ATL1-mCherry. ATL1 localization was analyzed in leaf epidermal cells of T3 transgenic seedlings. Consistent with our results in N. benthamiana, ATL1-mCherry was observed at the plasma membrane and in puncta (Figure 1N). To assess whether these puncta were endosomes, we stained root tips of these transgenic plants with the endocytic marker dye FM4-64 for 15 min and observed partial colocalization of the mCherry and FM4-64 fluorescent signals (Figure 1O). Significantly, we also observed a strong mCherry signal inside the lytic vacuoles of root epidermal cells (Figure 1P), suggesting that ATL1 may be actively degraded in the lytic vacuole. To test this hypothesis, we used concanamycin A, which inhibits vacuolar protein degradation by increasing the internal vacuolar pH, thus inhibiting vacuolar proteases (Tamura et al., 2003; Kleine-Vehn et al., 2008). Treatment of stable transgenic Arabidopsis lines expressing 35S:ATL1-mCherry with concanamycin A resulted in increased accumulation of ATL1-mCherry compared with mock-treated control plants (Figure 2A), demonstrating that ATL1 is degraded in the central vacuole.
Figure 2.
Overexpression of ATL1 Causes Dwarfing and Spontaneous Cell Death in Transgenic Arabidopsis.
(A) ATL1 protein accumulation in Arabidopsis increases in the presence of concanamycin A. Total protein was extracted from the ATL1 transgenic lines shown in (B) that were treated with 0.5 μM concanamycin A or mock treated for 16 h and immunoblotted with anti-mCherry.
(B) Overexpression of wild-type ATL1, but not a RING finger mutant form, induces dwarfing in Arabidopsis. Wild-type Col-0 plants were transformed with 35S:ATL1-mCherry or 35S:ATL1C153A/H155A-mCherry and grown for 4 weeks under short-day conditions. Bar = 1 cm.
(C) to (F) Overexpression of ATL1 induces cell death in Arabidopsis. Leaves from the plants shown in (B) were excised and stained with trypan blue. Dark staining in (E) indicates cell death. Bars = 25 μm.
The localization patterns of ATL1 in both transient and stable expression systems mirror those of a typical plasma membrane protein that undergoes constitutive endocytic trafficking, such as BRI1 (Geldner et al., 2007; Geldner and Robatzek, 2008; Kleine-Vehn et al., 2008; Gu and Innes, 2012). Therefore, we conclude that ATL1 is a membrane-associated E3 ligase that constitutively cycles between the plasma membrane and TGN/EE and after internalization can follow a retrograde transport pathway to the MVB/LE and eventually to the lytic vacuole for turnover.
Overexpression of ATL1 Inhibits Growth in Transgenic Arabidopsis and Promotes Cell Death in N. benthamiana
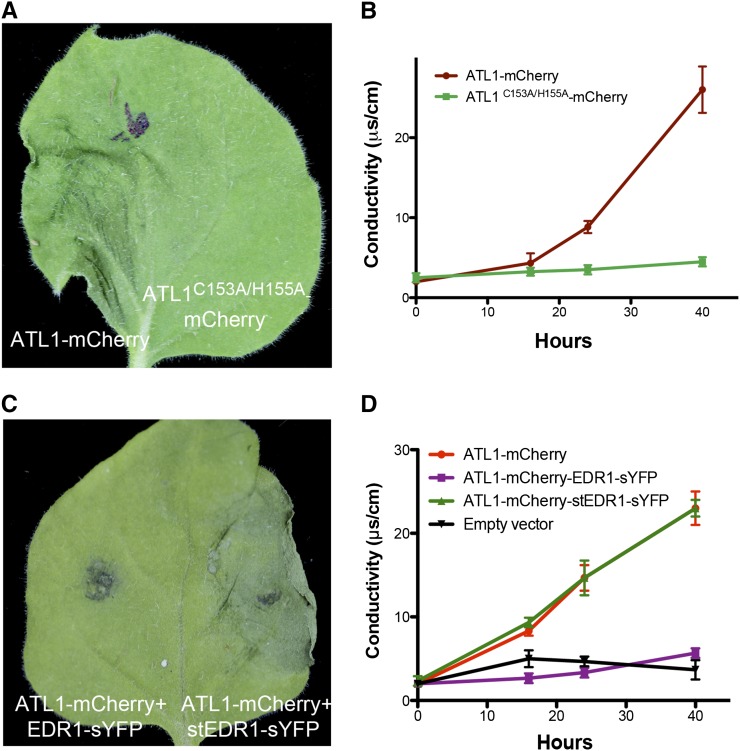
The 35S:ATL1-mCherry plants used for the subcellular localization analyses described above displayed a severe dwarf phenotype (Figure 2B). This growth inhibitory effect was observed in independent transgenic lines and appeared to correlate with ATL1 expression level (Figure 2A). In addition, we observed patches of dead cells in the leaves of the most severely dwarfed lines, indicating that ATL1 overexpression induces cell death (Figures 2C to 2F). These observations indicate that the ATL1 protein levels must be tightly controlled under normal growth conditions, which is further supported by the vacuolar signal observed in 35S:ATL1-mCherry plants (Figure 1P). Overexpression of ATL1 also caused cell death during transient expression in N. benthamiana. We found that 40 to 48 h postinfiltration (hpi) of Agrobacterium carrying a 35S:ATL1-mCherry construct, infiltrated leaves displayed macroscopic tissue collapse and electrolyte leakage indicative of programmed cell death (Figures 3A and 3B).
Figure 3.
ATL1-Induced Cell Death Is Dependent on the RING Domain and Is Suppressed by Coexpression with EDR1.
(A) and (B) Overexpression of ATL1 induces cell death in N. benthamiana, which is dependent on a functional RING domain. The ATL1-mCherry constructs were expressed under control of a 35S promoter, while the EDR1-sYFP constructs were under control of a dexamethasone-inducible promoter. Dexamethasone was applied 16 to 18 h after infiltration and photographs were taken 48 h after infiltration. (B) shows ion leakage data. Error bars represent sd; n = 3. This experiment was repeated four times with similar results.
(C) and (D) ATL1-mCherry-induced cell death in N. benthamiana is suppressed by coexpression with EDR1-sYFP, but not with the substrate trap mutant version, stEDR1-sYFP. (D) shows ion leakage data. Infiltration of Agrobacterium carrying an empty 35S promoter vector was used as the negative control and is shown in (D) only.
EDR1 Specifically Antagonizes ATL1 Signaling
The physical interaction between EDR1 and ATL1 suggested a functional link between these two proteins. EDR1 has previously been shown to function as a negative regulator of stress-induced cell death responses (Frye et al., 2001; Tang and Innes, 2002; Tang et al., 2005b; Christiansen et al., 2011). We thus tested whether overexpression of EDR1 could suppress ATL1-induced cell death. A dexamethasone-inducible EDR1-sYFP construct was cotransformed with a 35S:ATL1-mCherry construct into N. benthamiana leaves using Agrobacterium. EDR1-sYFP expression was induced by spraying leaves with 50 μM dexamethasone 16 to 18 hpi. As shown in Figures 3C and 3D, coexpression of EDR1-sYFP with ATL1-mCherry suppressed both the tissue collapse and electrolyte leakage induced by ATL1-mCherry. We also observed suppression of cell death when ATL1-mCherry was coexpressed with EDR1-Myc instead of EDR1-sYFP, indicating that suppression was not a property of the epitope tag (Supplemental Figure 2). This cell death suppression was dependent on EDR1-Myc expression, as no suppression was observed in the absence of dexamethasone application (Supplemental Figures 2A and 2C). Consistent with this finding, we observed that overexpression of ATL1-mCherry in an edr1 mutant background caused more severe developmental defects than in a wild-type background (Supplemental Figures 3A and 3B). In fact, none of the T1 35S:ATL1-mCherry edr1 lines survived to the bolting stage, while all T1 lines in the wild-type Columbia-0 (Col-0) background produced T2 progeny.
We next tested whether the kinase activity of EDR1 was required for suppression of the ATL1 overexpression phenotype. The substrate trap mutant stEDR1-sYFP, which has a mutation in the phosphotransfer site of EDR1 (Gu and Innes, 2011), was cloned into the same dexamethasone-inducible vector and coinfiltrated with ATL1-mCherry into N. benthamiana leaves. Unlike EDR1-sYFP, stEDR1-sYFP was unable to suppress ATL1-induced cell death (Figures 3C and 3D), although it accumulated to higher levels than wild-type EDR1 (Supplemental Figure 2D). ATL1-mCherry protein levels were not significantly affected by EDR1 kinase activity (Supplemental Figure 2E), indicating that cell death suppression was not caused by EDR1-induced degradation of ATL1. Immunoblot analysis of ATL1-mCherry in total protein extracts of transiently transformed N. benthamiana leaves showed a strong band corresponding in size to free mCherry, indicating that ATL1-mCherry was being actively degraded, but not in an EDR1-dependent manner. This is consistent with active degradation of ATL1 in transgenic lines (Figure 1P). The fact that the EDR1 substrate trap mutation did not disrupt the interaction between EDR1 and ATL1 (Figure 1A) but blocked the suppression of ATL1-mediated cell death suggests that the mechanism of this suppression may involve direct phosphorylation of ATL1 by EDR1.
Lastly, we tested whether EDR1’s ability to suppress cell death was specific to ATL1. We coinfiltrated EDR1-Myc with an autoactive mutant form of the disease resistance protein RPS5 (D266E), which has previously been shown to induce a cell death response upon overexpression in N. benthamiana (Ade et al., 2007; Qi et al., 2012). In addition, we coinfiltrated ATL1-mCherry with Arabidopsis BAX inhibitor-1 (BI-1), an endoplasmic reticulum-localized protein that has been shown to suppress BAX-, pathogen-, and abiotic stress-induced cell death (Watanabe and Lam, 2008). In both cases, no obvious suppression of cell death was observed (Supplemental Figures 2F to 2I), suggesting that EDR1 and ATL1 define a specific signaling pathway to regulate cell death that is distinct from disease resistance proteins and BI-1-mediated pathways. Taken together, these data suggest that EDR1 specifically antagonizes ATL1 functions in vivo and its kinase function is required during this process.
To test whether these effects of ATL1 overexpression were specific to ATL1 or were shared with other ATL family members, we obtained a cDNA clone of ATL16, the closest paralog of ATL1 in Arabidopsis (Aguilar-Hernández et al., 2011). ATL16 is 60% identical to ATL1 over its entire length and 95% identical in the RING domain. ATL16-mCherry completely colocalized with ATL1-eGFP upon transient coexpression (Supplemental Figures 4A to 4C). However, overexpression of ATL16-mCherry in N. benthamiana failed to induce cell death (Supplemental Figures 4D and 4E), suggesting that the toxicity of ATL1 is not an artifact of overexpressing ATL family proteins, but more likely is related to the specific function of ATL1.
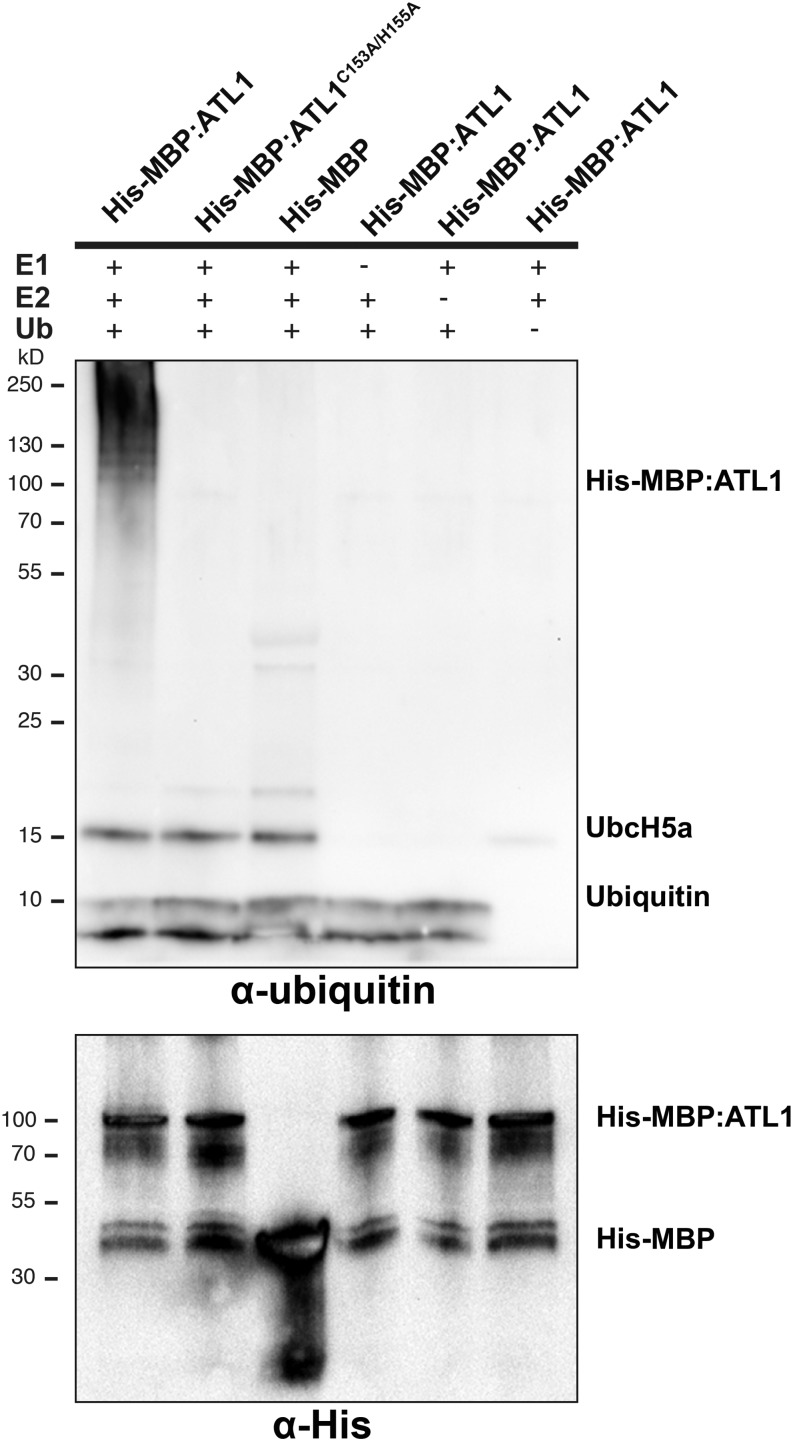
ATL1 Has Ubiquitin Ligase Activity
Other members of the ATL1 family have been shown to possess E3 ubiquitin ligase activity (Salinas-Mondragón et al., 1999; Serrano and Guzmán, 2004; Berrocal-Lobo et al., 2010). To determine whether ATL1 might function as an E3 ligase, ATL1 activity was analyzed using an in vitro ubiquitination assay (Figure 4). A histidine:maltose binding protein fusion (His:MBP:ATL1) was affinity purified and its E3 ligase activity was assayed in vitro. Multiple forms of autoubiquitinated ATL1 were detected in the assay using an antiubiquitin antibody (Figure 4, lane 1). When the conserved zinc binding residues Cys-153 and His-155 were mutated to Ala (ATL1C153A/H155A; Aguilar-Hernández et al., 2011) or when any of the components of the ubiquitination pathway (E1, E2, E3, or ubiquitin) were omitted from the assay, a loss of protein polyubiquitination was observed (Figure 4, lanes 2 to 6). These results confirmed that ATL1 has ubiquitin ligase activity.
Figure 4.
ATL1 Possesses E3 Ubiquitin Ligase Activity.
ATL1 autoubiquitinates in vitro. Recombinant ATL1 protein was used for in vitro ubiquitination assays in the presence of the E1 protein Uba2, the E2 protein UbcH5a, and Mg-ATP. Multiple ubiquitinated protein forms were detected when wild-type ATL1 was used (lane 1). No autoubiquitination was detected when the ATL1 RING mutant, ATL1C153A/H155A (lane 2), or His-MBP tag (lane 3) was used. No autoubiquitination was detected when ATL1 was incubated in a reaction lacking the E1 protein (lane 4), the E2 protein (lane 5), or ubiquitin (lane 6). The presence of recombinant protein was detected using antihistidine antibody.
E3 Ligase Activity Is Essential for ATL1 Function
We next tested whether the toxic effect of overexpressing ATL1 required its E3 ligase activity. To answer this question, we fused an ATL1C153A/H155A cDNA clone with mCherry and used this clone for transient expression assays in N. benthamiana and to generate stable transgenic Arabidopsis. In contrast to the wild-type ATL1-mCherry construct, expression of ATL1C153A/H155A-mCherry did not cause cell death in N. benthamiana (Figures 3A and 3B) and did not cause growth inhibition in transgenic wild-type or edr1 mutant Arabidopsis lines (Figure 2B; Supplemental Figure 3C). The subcellular localization pattern and accumulation of ATL1C153A/H155A in N. benthamiana was similar to wild-type ATL1 (Supplemental Figure 5); thus, the failure to cause cell death was not due to protein instability or mislocalization. Taken together, these data indicate that ATL1 contains a functional E3 ligase domain and its toxicity upon overexpression may result from uncontrolled E3 ligase activity.
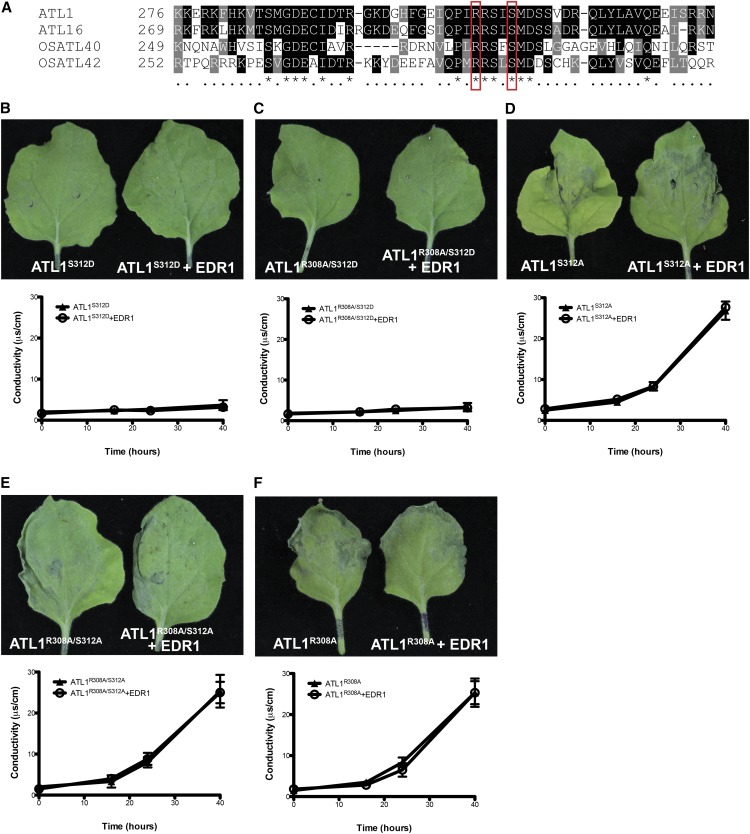
Mutations in an RxxxS Motif of ATL1 Block ATL1-Induced Cell Death
To identify potential conserved regulatory elements in ATL1, we aligned the ATL1 amino acid sequence with its closest paralog in Arabidopsis, ATL16, and with its two closest homologs in rice (Oryza sativa; ATL40 and ATL42; Serrano et al., 2006). We found a highly conserved region between amino acids 285 and 327 of ATL1. A search for functional motifs within this region detected a recognition motif for cGMP-dependent protein kinases defined as RxxxS spanning from R308 to S312 (Bensmihen et al., 2002; Figure 5A). Given that we had established that EDR1 kinase activity was required to suppress ATL1-mediated cell death, we investigated whether S312 might be required for EDR1-mediated suppression of ATL1 activity. We thus constructed ATL1 missense mutations that block phosphorylation on S312 (S312A) and that mimic phosphorylation (S312D). We also generated missense mutations in the conserved arginine (R308A) as well as the double mutants (R308A/S312A and R308A/S312D). All of these constructs were then tested for their ability to induce cell death by transient expression in N. benthamiana leaves, both in the presence and absence of EDR1. The phosphomimic ATL1S312D and ATL1R308A/S312D mutants did not produce any signs of cell death at 48 hpi (Figures 5B and 5C). The inability of these mutants to induce cell death was not the result of protein destabilization or mislocalization, as these proteins displayed the same pattern of fluorescence and accumulated to comparable levels in N. benthamiana cells (Supplemental Figure 6). In contrast to ATL1S312D, the ATL1S312A and ATL1R308A/S312A mutants induced cell death, and this cell death was not blocked by coexpression with EDR1 (Figures 5D and 5E). Collectively, these results indicate that the phosphorylation status of S312 plays a central regulatory role in ATL1 activity and is likely a target for EDR1.
Figure 5.
Mutations in the RxxxS Motif of ATL1 Eliminate EDR1-Mediated Suppression of ATL1-Induced Cell Death.
(A) Amino acid sequence alignment of ATL1 with its closest paralog in Arabidopsis, ATL16, and with its two closest homologs in rice (OsATL40 and OsATL42). Black and gray shading indicates conserved amino acids. Red boxes indicate conserved Arg and Ser residues that were mutagenized to Ala and/or Asp in the different ATL1 mutants.
(B) and (C) ATL1S312D and ATL1R308A/S312D do not induce cell death in N. benthamiana.
(D) to (F) ATL1S312A-, ATL1R308A-, and ATL1R308A/S312A-induced cell death in N. benthamiana cannot be suppressed by coexpression with EDR1-sYFP. Ion leakage measurements are shown for each mutant. Error bars represent sd; n = 3. All ATL1 constructs were expressed using a 35S promoter, and EDR1 was expressed under a dexamethasone-inducible promoter. Photographs were taken 48 hpi of Agrobacterium. The entire experiment was repeated twice with similar results.
The above analyses also revealed that the ATL1 R308A mutant induced cell death, even in the presence of EDR1 (Figure 5F). This prompted us to test whether EDR1 was able to interact with ATL1R308A using yeast two-hybrid and BiFC analysis. The R308A substitution did not eliminate the EDR1-ATL1 interaction nor did the S312A substitution (Supplemental Figure 7). These data indicate that the RxxxS motif is required for regulation by EDR1 but not for interaction with EDR1. Given that the S312D phosphomimic substitution prevented ATL1-induced cell death, these data suggest that the RxxxS motif is a target for phosphorylation, either directly by EDR1 or indirectly by a second kinase that is itself regulated by EDR1, and that the R308A substitution prevents such phosphorylation.
A Phosphomimic Substitution at S312 Inhibits Its E3 Ubiquitin Ligase Activity
Since we determined that ATL1-induced cell death requires ATL1 ubiquitin ligase activity (Figure 3B) and that mimicking phosphorylation of ATL1 on S312 inhibits its ability to induce cell death (Figure 5B), we hypothesized that phosphorylation of S312 blocks ATL1 E3 ligase activity. To test this hypothesis, we performed in vitro ubiquitination assays using recombinant ATL1S312D and ATL1S312A proteins. As predicted, ATL1S312D lost ubiquitin ligase activity (Figure 6, lane 3), while ATL1S312A retained activity (Figure 6, lane 4). However, we were unable to test for direct phosphorylation of ATL1 by EDR1 in vitro due to our inability to purify active full-length EDR1 protein.
Figure 6.
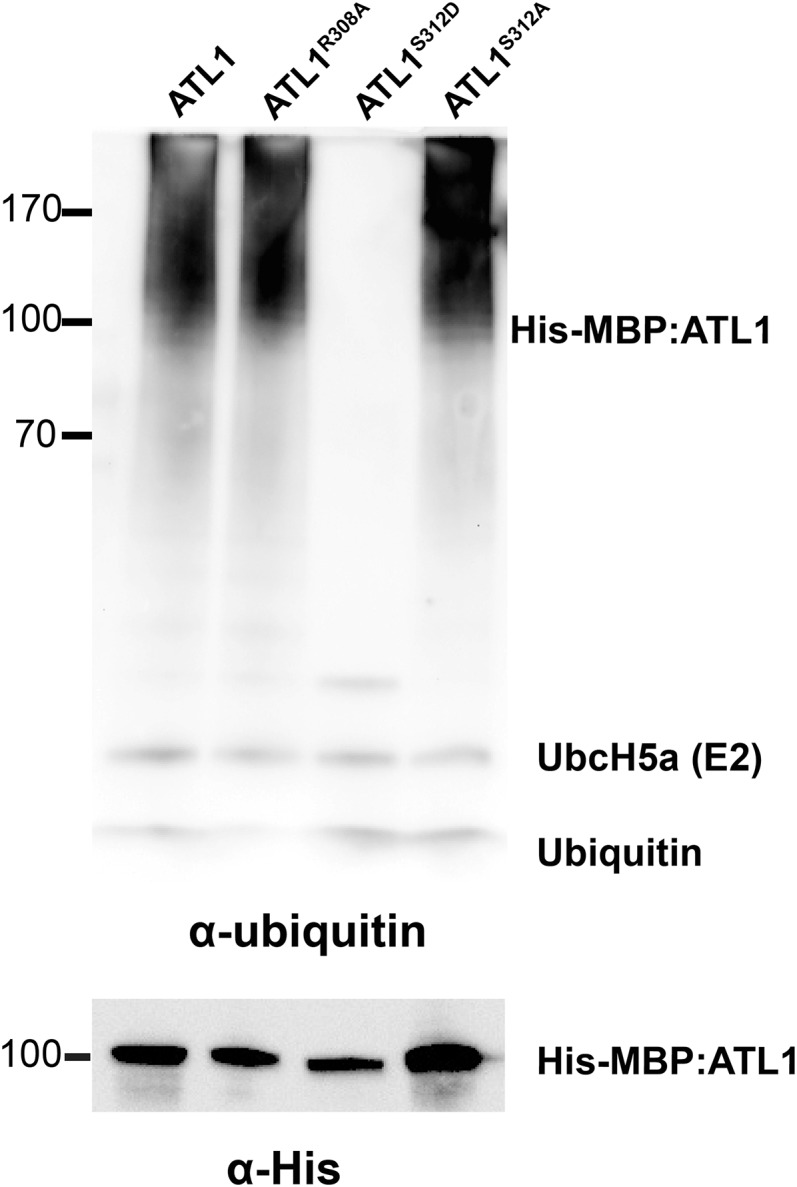
Mimicking Phosphorylation of ATL1 on S312 Inhibits Its in Vitro Ubiquitination Activity.
In vitro ubiquitination assays were performed in the presence of the E1 protein Uba2, the E2 protein UbcH5a, and Mg-ATP, and ATL1 autoubiquitination was detected using an antiubiquitin antibody. Multiple ubiquitinated protein forms were detected when wild-type ATL1 was used (lane 1). No ubiquitination activity was detected using ATL1S312D (lane 3). ATL1R308A (lane 2) and ATL1S312A (lane 4) retained ubiquitination activity.
Knockdown of ATL1 Suppresses edr1-Mediated Enhanced Resistance to Powdery Mildew
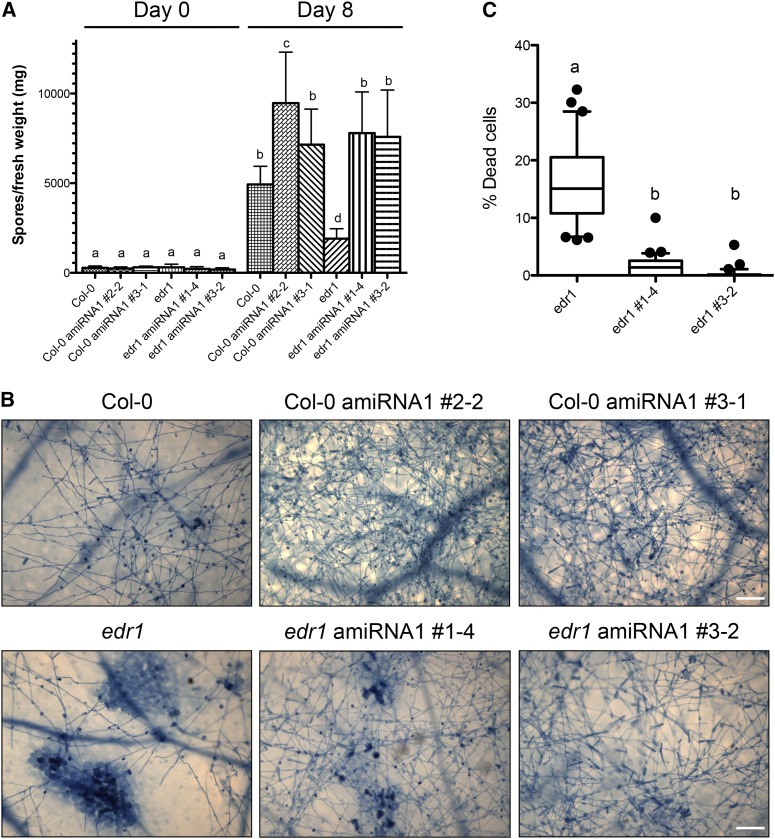
The above analyses indicate that EDR1 functions as a negative regulator of ATL1 and that loss of EDR1 function may lead to enhanced ATL1 activity. We therefore tested whether edr1 mutant phenotypes are caused in part by enhanced ATL1 activity. Unfortunately, putative T-DNA insertion lines obtained from the ABRC and from other sources were found to lack insertions in the ATL1 coding region, suggesting that the insertions were lost during propagation. We thus generated transgenic ATL1 knockdown lines using an artificial microRNA (amiRNA) construct expressed from a constitutive 35S promoter (Schwab et al., 2006). T2 generation plants carrying the amiRNA construct from three independent transformants in both the wild-type Col-0 and edr1 mutant backgrounds were selected for analysis of ATL1 transcript levels by quantitative RT-PCR. All plants tested showed a significant reduction in ATL1 transcript abundance, while the transcript level of the closest ATL1 homolog, ATL16, was unaltered (Supplemental Figure 8). Interestingly, the nontransgenic edr1 mutant was found to have an elevated level of ATL1 transcript (∼2.5-fold), suggesting that EDR1 may also negatively regulate ATL1 at the level of transcription. T3 seeds were collected from T2 plants homozygous for the amiRNA transgene and that showed the lowest ATL1 transcript levels. These plants appeared phenotypically wild-type in terms of plant morphology when grown under short days (9 h of light) but were slightly smaller when grown under long days (16 h of light). Two T3 families from each genotype were then assessed for resistance to infection by powdery mildew (Golovinomyces golovinomyces) under short-day conditions. All four amiRNAi lines showed enhanced susceptibility compared with their nontransgenic parents as measured by the abundance of conidiospores 8 d after inoculation (Figure 7). In addition, the large patches of dead mesophyll cells observed in the edr1 parent at this time point were reduced or absent in the lines (Figure 7B), indicating that knockdown of ATL1 suppresses edr1-mediated cell death and disease resistance. Furthermore, the enhanced susceptibility observed in the Col-0 wild-type parent indicates that ATL1 contributes to basal resistance to this pathogen.
Figure 7.
Knockdown of ATL1 Enhances Susceptibility to Powdery Mildew and Suppresses edr1-Associated Cell Death.
(A) Quantitative analysis of powdery mildew conidia (asexual spores) on ATL1 amiRNAi transgenic Arabidopsis lines. The indicated Arabidopsis lines were inoculated with powdery mildew and conidium production determined 8 d later. Bars indicate the mean of n ± sd (n = 3). Lowercase letters indicate values that are significantly different (P < 0.01; two-way ANOVA test followed by a Bonferroni post-hoc test).
(B) Trypan blue staining of powdery mildew-infected ATL1 amiRNA lines. The indicated lines were assessed for leaf mesophyll cell death and conidium production at 8 d after infection using trypan blue staining. Only the nontransformed edr1 mutant showed significant patches of cell death. Bars = 100 μm.
(C) Quantification of cell death in powdery mildew infected ATL1 amiRNA lines. The trypan blue-stained area in 30 leaves was measured for each genotype using ImageJ and divided by the total leaf area. Results are provided as means with 10th and 90th percentiles (box) and range (whiskers). Statistical outliers are shown as circles. Lowercase letters indicate values that are significantly different (P < 0.0001; two-way ANOVA followed by a Bonferroni post-hoc test).
DISCUSSION
In this work, we identified the Arabidopsis ATL1 protein as a potential substrate of the EDR1 protein kinase, which prior work has shown to be involved in regulation of endomembrane trafficking and cell death (Gu and Innes, 2011, 2012). ATL1 is a membrane-localized E3 ubiquitin ligase that contains a RING-H2 variant of the canonical RING finger domain (Aguilar-Hernández et al., 2011). We found that overexpression of ATL1 induces cell death, that ATL1 physically associates with EDR1 on TGN/EE vesicles, and that ATL1-induced cell death can be suppressed by overexpression of EDR1. Most significantly, knockdown of ATL1 suppressed edr1-mediated resistance to powdery mildew and also enhanced susceptibility of wild-type Arabidopsis to powdery mildew infection (Figure 7), indicating that ATL1 is required for edr1-mediated resistance and that EDR1 negatively regulates ATL1 activity.
EDR1 also physically and genetically interacts with another TGN/EE resident RING E3 ligase, KEG, which was recently demonstrated to regulate multiple intracellular trafficking processes and play a role in fungus-host interactions (Wawrzynska et al., 2008; Gu and Innes, 2011, 2012). KEG also mediates the ubiquitination and degradation of at least three transcription factors involved in abscisic acid-induced responses, ABI5, ABF1, and ABF3, with ABI5 ubiquitination likely occurring on TGN/EE vesicles (Stone et al., 2006; Liu and Stone, 2010). Thus, EDR1 appears to modulate stress-induced pathways through regulating the activities of multiple E3 ubiquitin ligases at the TGN/EE.
EDR1 has also been shown to physically associate with the mitogen-activated protein kinase kinases MKK4 and MKK5 via the N-terminal nonkinase domain of EDR1 (Zhao et al., 2014). This association appears to affect the protein stability of MKK4/5, as EDR1 overexpression causes a reduction in MKK4/5 protein level, while edr1 mutation causes an increase. Furthermore, mutation of either MKK4 or MKK5 partially suppresses edr1 mutant phenotypes, including enhanced resistance to powdery mildew (Zhao et al., 2014). However, the latter result is difficult to interpret because mkk4 and mkk5 mutations by themselves display enhanced susceptibility to powdery mildew, and in double mutations with edr1, this enhanced susceptibility is reduced, suggesting that edr1-mediated resistance functions at least partially independent of MKK4/5 signaling. We have not yet assessed whether ATL1 knockdown affects MKK4 or MKK5 protein levels, but if it does, the mechanism would likely be indirect since ATL1 knockdown causes enhanced susceptibility and, thus, likely reduced levels of MKK4/5.
Although we have not yet demonstrated that EDR1 directly phosphorylates ATL1, our data are consistent with such a model (Figure 8). Mutation of Ser-312 of ATL1 to aspartate (mimicking phosphorylation) blocks ATL1 autoubiquitination activity (Figure 6), while mutation of Ser-312 to alanine does not affect in vitro autoubiquitination but instead blocks the ability of EDR1 to suppress ATL1-induced cell death in vivo (Figure 5). The simplest model to explain these observations is that EDR1 regulates ATL1 E3-ligase activity by phosphorylating ATL1 on Ser-312.
Figure 8.
Model Showing ATL1 Dynamics and Regulation by EDR1.
ATL1 is a plasma membrane-localized E3 ubiquitin ligase that constitutively undergoes endocytosis, cycling between the plasma membrane and TGN/EE. ATL1 can interact in the TGN/EE with EDR1, which inactivates ATL1, possibly by direct phosphorylation. By contrast, active ATL1 targets negative regulators of stress responses (X), ubiquitination of which leads to cell death.
In addition to regulation by phosphorylation, the activity of ATL1 is presumably regulated by protein turnover via targeting to the lytic vacuole (Figures 1P and 2A). Our data indicate, though, that EDR1 is not required for such turnover; thus, if vacuolar targeting is regulated (as opposed to occurring constitutively), such regulation would occur independently of EDR1 function. The ability to regulate ATL1 activity by two independent processes (phosphorylation and protein turnover) would enable the plant to fine tune ATL1 activity in response to multiple environmental inputs. Given that both protein degradation and phosphorylation inhibit ATL1 activity, ATL1 activity is likely very low under stress-free conditions. Under this model, environmental stresses would lead to a reduction in ATL1 protein turnover and/or phosphorylation and thus increase its activity.
Regulation of ubiquitin ligase enzymatic activity by direct phosphorylation of an E3 ligase has not been reported previously in plants. In humans, the RING E3 ubiquitin ligase MDM2, which ubiquitinates the p53 transcription factor, is phosphorylated by the protein kinase ATM (Cheng et al., 2011). This phosphorylation event inhibits E3 ubiquitin ligase activity, likely by inhibiting oligomerization of the RING domain, which, in turn, prevents degradation of p53 via the proteasome. ATM-mediated phosphorylation of MDM2 is thus a key regulator or p53 levels and, hence, central to maintaining genome integrity. However, outside the RING domain, ATL1 and MDM2 display no obvious similarity. It is thus difficult to predict whether phosphorylation of ATL1 inhibits E3 ligase activity by the same mechanism. A second example of phosphorylation-mediated regulation of E3 ubiquitin ligase activity is phosphorylation of the human RING E3 ligase Cbl-b, which is a negative regulator of T cell activation and has been linked to autoimmune diseases such as diabetes and multiple sclerosis (Kobashigawa et al., 2011). In this example, however, phosphorylation promotes rather than inhibits E3 ligase activity, with phosphorylation on Tyr-363 blocking an autoinhibitory interdomain interaction.
Although there are no prior reports of phosphorylation-mediated regulation of E3 ligase activity in plants, there is one well-characterized example of phosphorylation of RING E3 ligases promoting interaction with substrates, thus enhancing ubiquitination of the substrate. In this example, the substrate is Arabidopsis FLS2, which is a pattern recognition receptor that is activated by bacterial flagellin (Gómez-Gómez et al., 2001; Chinchilla et al., 2006). Following activation, FLS2 signaling is attenuated by endocytosis (Robatzek et al., 2006; Beck et al., 2012b). The FLS2 attenuation process is dependent on BRI1 ASSOCIATED KINASE1-dependent phosphorylation of two U-box E3 ubiquitin ligases, PLANT U-BOX 12 and 13 (PUB12 and PUB13). Phosphorylation of PUB12 and PUB13 promotes their association with FLS2 and, hence, ubiquitination of FLS2 (Lu et al., 2011). PUB12 and PUB13 thus function in a negative feedback loop to downregulate FLS2 signaling following activation.
In addition to its role in regulating FLS2 signaling, PUB13 regulates numerous defense responses (Li et al., 2012). Loss-of-function mutations in PUB13 cause an increase in salicylic acid levels, the accumulation of hydrogen peroxide, spontaneous cell death, and enhanced resistance to biotrophic pathogens, but increased susceptibility to necrotrophic pathogens. Mutations in the rice ortholog of PUB13, Spl11, cause similar phenotypes, including upregulation of numerous defense and oxidative stress genes and enhanced resistance to biotrophic pathogens (Zeng et al., 2004). Spl11 and PUB13 are thus negative regulators of defense responses, including cell death, and act antagonistically to ATL1.
Regulation of ATL1 activity by EDR1 appears to occur on TGN/EE vesicles, which represent the endosomal population derived immediately from internalized plasma membrane (Figure 8). Ubiquitin modifications on TGN/EE-localized proteins may generate binding surfaces for recruitment of endosomal specific signal transduction components. In this scenario, overexpression of ATL1 could result in an increase in endosomal signaling events, including stress-induced responses. EDR1 negatively regulates this process through downregulating ATL1’s ubiquitin ligase activity. Consistent with this model, the Arabidopsis Membrane-Protein Interaction Network Database indicates that ATL1 interacts with several membrane-localized leucine-rich repeat protein kinases, including the defense-related WALL ASSOCIATED KINASE3. Previous work on ubiquitination of leucine-rich repeat kinases has indicated that ubiquitination is associated primarily with termination of receptor signaling (Acconcia et al., 2009; Beck et al., 2012a, 2012b). In these examples, ubiquitinated plasma membrane receptors are targeted to the lytic vacuole for degradation. Although this model appears inconsistent with ATL1 overexpression phenotypes, it is plausible that ATL1 may negatively regulate other ubiquitinating enzymes such as PUB13 that directly mediate ubiquitination and attenuation of stress signaling receptors. Thus overexpression of ATL1 could lead to inactivation of another E3 ligase, which is itself directly involved in attenuating stress responses (Figure 8).
It has been recently shown that edr1-mediated defense responses require RPN1a, a subunit of the 26S proteasome (Yao et al., 2012), suggesting that EDR1 may be regulating ubiquitination of proteins that are then targeted to the proteasome. Under this model, loss of EDR1 function leads to hyperactivity of the E3 ubiquitin ligases KEG and ATL1, which then leads to excess proteasomal degradation of their targets. Inhibition of the proteasome can thus suppress edr1 phenotypes.
Ubiquitination also regulates internalization and degradation of plasma membrane transporters to maintain levels of hormones, metals, and nutrients appropriate for growth and development (Barberon et al., 2011; Kasai et al., 2011; Leitner et al., 2012). We do not exclude the possibility that the detrimental effects of overexpressing ATL1 may involve misregulation of surface transporter abundance.
Taken together, our data suggest that stress-induced cell death signaling is regulated by two major and interlinked posttranslational modification mechanisms: phosphorylation and ubiquitination.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana accession Col-0 and edr1 mutant (Frye and Innes, 1998) were used in this study. Seeds were surface sterilized with 50% (v/v) bleach and planted on one-half-strength Murashige and Skoog plates supplemented with 0.8% agar. Plates were placed at 4°C for 72 h for stratification and then transferred to a growth room set to 23°C and 9 h light (150 μE m−2 s−1)/15 h dark cycle. Seven-day-old seedlings were transplanted to MetroMix 360 (Sun Gro Horticulture) and grown for three additional weeks.
Nicotiana benthamiana plants used for transient protein expression were grown under the same conditions, except that seeds were planted directly into MetroMix 360.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The EDR1-Myc, GmManI49-mCherry, ARA6-mCherry, mCherry-SYP21, mCherry-SYP61, and RPS5-D266E-Myc constructs have been described previously (Ade et al., 2007; Gu and Innes, 2011, 2012). Fusion of epitope tags and fluorescent proteins to ATL1, EDR1, BI-1, and BRI1 was accomplished using a multisite Gateway cloning strategy (Invitrogen) described previously (Gu and Innes, 2011). Briefly, the full-length open reading frames of ATL1 (At1g04360), EDR1 (At1g08720), AtBI-1 (At5g47120), and BRI-1 (At4g39400) were amplified by PCR from a cDNA template and recombined into the multisite Gateway entry vector pBSDONR P1-P4. eGFP (Cormack et al., 1996), mCherry (Shaner et al., 2004), 5xMyc, N-terminal sYFP, and C-terminal sYFP (Kremers et al., 2006) were cloned into the entry vector pBSDONR P4r-P2. To fuse ATL1, EDR1, AtBI-1, and BRI1 with the C-terminal epitope tags, the P1-P4 clones were mixed with corresponding P4r-P2 tag and the desired destination vectors and recombined using Gateway LR Clonase II (Invitrogen). For EDR1-Myc and stEDR1-Myc, the above pBSDONR constructs were recombined with the steroid-inducible destination vector pTA7002-GW (Aoyama and Chua, 1997; McNellis et al., 1998); for ATL1-mCherry, ATL1-eGFP, BI-1-Myc, organelle marker constructs, and BiFC constructs, the constructs were recombined with the destination vector pEarleyGate100 (Earley et al., 2006), which drives transgene expression using a cauliflowers mosaic virus 35S promoter. For ATL1 recombinant expression in Escherichia coli, the corresponding pBSDONR carrying ATL1 without transmembrane domain (from amino acids 67 to 381) was recombined with the expression vector pHMGWA (Busso et al., 2005). The RING E3 ubiquitin ligase domain mutants, as well as the different mutants of the RxxxS domain of ATL1, were generated using a QuikChange II site-directed mutagenesis kit (Agilent Technologies).
An ATL1-specific amiRNA construct was created by PCR amplification of an ATL1-specific region following the procedures of Schwab et al. (2006), which included insertion of the ATL1 sequence between flanking regions of the MIR319 microRNA. This amiRNA was then cloned into pEarlyGate100 (Earley et al., 2006) and transformed into Agrobacterium tumefaciens GV3101 (pMP90).
All clones were verified for correct construction using DNA sequencing. The primers used to create the above constructs are listed in Supplemental Table 1.
Plasmids were transformed into Agrobacterium strain GV3101 (pMP90) by electroporation with selection on Luria-Bertani plates containing 50 μg/mL kanamycin sulfate (Sigma-Aldrich) and 20 μg/mL gentamycin (Gibco). For generating ATL1 amiRNAi, 35S:ATL1-mCherry and 35S:ATL1C153A/H155A-mCherry transgenic lines, Col-0 wild-type and edr1 mutant Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). T1 generation plants containing the transgene were selected by spraying 1-week-old seedlings with 300 μM BASTA (Finale) three times in 2-d intervals.
Quantitative RT-PCR Analysis of amiRNA Lines
Assessment of ATL1 silencing in transgenic Arabidopsis was performed by quantitative RT-PCR as follows: RNA was extracted from 4-week-old plants using the Spectrum plant total RNA kit (Sigma-Aldrich). cDNA was produced from 1 μg total RNA using the Verso cDNA synthesis kit (Thermo Scientific). Relative RNA amounts were determined by quantitative RT-PCR using the SYBR Premix Ex Taq II kit (Takara). A comparative Ct method was used to determine relative quantities (Schmittgen and Livak, 2008). UBIQUITIN10 was used for normalization (Czechowski et al., 2005). Quantitative RT-PCR analyses was performed in triplicate on four individual plants for each line.
Yeast Two-Hybrid Assays
The pGADT7 vector containing the GAL4 activation domain and the pGBKT7 vector containing the GAL4 DNA binding domain were obtained from Clontech. The full-length EDR1 open reading frame, stEDR1 (D810A), and RKA domain-encoding region of KEG were cloned into pGBKT7 and subsequently transformed into yeast strain Y187 (Clontech) by electroporation and selected on synthetic dextrose (SD)-Trp medium. The full-length open frames encoding ATL1, ATL1R308A, ATL1S312A, and ATL1S312D were cloned into pGADT7, transformed into yeast strain AH109 (Clontech) by electroporation, and selected on SD-Leu medium. Matings between the Y187 and AH109 strains carrying the appropriate constructs were performed in yeast peptone dextrose medium at 30°C for 16 h. Mating cultures were then diluted and plated on SD-Trp-Leu and SD-Trp-Leu-His.
Transient Protein Expression in N. benthamiana
Agrobacterium cultures carrying corresponding constructs were grown and resuspended in water at OD600 0.8 (Wroblewski et al., 2005). For coexpression of multiple constructs, suspensions were mixed in equal ratios. Bacterial suspension mixtures were infiltrated using a needleless syringe into expanding leaves of 4-week-old N. benthamiana plants. When needed, protein expression was induced by spraying the leaves with 50 μM dexamethasone (Sigma-Aldrich) 16 to ∼18 h after injection. Samples were collected for protein extraction, microscopy imaging, or ion leakage analysis at the time points indicated in figure legends. All transient expression experiments were performed a minimum of three times and gave consistent results.
For total protein extraction, four leaves of infiltrated N. benthamiana were collected and ground in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, and Plant Proteinase Inhibitor Cocktail [Sigma-Aldrich]). Samples were centrifuged at 9300g at 4°C for 5 min, and supernatants were transferred to new tubes. Total proteins were mixed with 4× SDS loading buffer at a ratio of 3:1 and boiled for 5 min before loading. Total proteins were separated by electrophoresis on a 4 to 20% gradient Tris-HEPES-SDS polyacrylamide gel (Thermo Scientific). Proteins from duplicate gels were transferred to a nitrocellulose membrane and probed with anti-mCherry antibody (Abcam ab125096). The anti-mCherry primary antibody was detected using goat anti-mouse IgG (H+L) conjugated to horseradish peroxidase (HRP) (Pierce catalog number 31430). SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used to detect HRP-conjugated antibody complexes. Anti-GFP antibody (Sigma-Aldrich catalog number G1546-200UL) was used to detect proteins fused to sYFP, employing the same secondary antibody, but detecting HRP with an ImmunoStar HRP substrate kit (Bio-Rad).
Transient Transfection of Arabidopsis Protoplasts and Immunoprecipitations
The isolation and transient transfection of leaf mesophyll cell protoplasts from Arabidopsis plants (4 weeks old) was performed at room temperature following published procedures (Yoo et al., 2007). A total of 10 μg plasmid DNA was used for each transfection experiment and plasmids were mixed in an equal ratio for cotransfections. Protoplasts were harvested by centrifugation for 1 min at 20,000g and resuspended in IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, and Plant Proteinase Inhibitor Cocktail [Sigma-Aldrich]). HA-tagged proteins were immunoprecipitated by incubating the samples with Anti-HA Agarose (Pierce Biotechnology) for 3 h at 4°C. Samples were washed five times in IP buffer, resuspended in 1× SDS loading buffer, and boiled for 5 min before loading. Immunocomplexes were separated by electrophoresis on a 4 to 20% gradient Tris-HEPES-SDS polyacrylamide gel (Thermo Scientific). Proteins from duplicate gels and filters were transferred to a nitrocellulose membrane and probed with anti-HA:HRP antibody (Sigma-Aldrich H6533-1VL) or anti-mCherry antibody (Abcam ab125096), with HRP detected using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific). Immunoprecipitation experiments were replicated four times with similar results.
Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy was performed on a Leica SP5 AOBS inverted confocal microscope (Leica Microsystems) equipped with a 63×, 1.2-numerical aperture, water objective. eGFP fusions were excited with a 488-nm argon laser and detected using a 505- to 530-nm band-pass emission filter. sYFP fusions were excited with a 514-nm argon laser and detected using a 522- to 545-nm band-pass emission filter. mCherry fusions were excited using a 561-nm He-Ne laser and detected using a custom 595- to 620-nm band-pass emission filter. FM4-64 (Life Technologies) was excited using a 488-nm argon laser and detected at >640 nm.
Recombinant Protein Expression in E. coli and Purification of MBP-ATL1 Constructs
Expression constructs were transformed into E. coli BL21.AI cells (Invitrogen). Cultures (15 mL) were inoculated and cultured overnight at 37°C and shaken at 200 rpm and then transferred to 300 mL of fresh Luria-Bertani. The cultures were incubated for 1 h at 37°C at 200 rpm, and protein expression was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside and 0.2% l-arabinose. Cultures were incubated for 3 h at 30°C at 200 rpm. Cells were harvested by centrifugation (15 min; 5000g), and cell pellets were resuspended in 10 mL column buffer (20 mM Tris, pH 7.8, 200 mM NaCl, and 1 mM DTT) plus protease inhibitor cocktail (Roche complete) and 0.1 mg/mL lysozyme. Cells were lysed by sonication and the lysates were centrifuged at 5000g for 20 min at 4°C. Supernatants were loaded onto a gravity column with 1 mL Amylose resin (NEB). After washing with 20 mL column buffer, proteins were eluted in five fractions with 0.5 mL column buffer containing 10 mM maltose.
In Vitro Ubiquitination Assay
In vitro ubiquitination assays were performed as described by Sato et al. (2009) with minor modifications. Briefly, reactions (30 μL) containing 40 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 50 ng yeast Uba2 (BostonBiochem), 250 ng human UbcH5a (BostonBiochem), 500 ng purified ATL1, and 9 μg ubiquitin were incubated at 30°C for 1.5 h. Reactions were stopped by adding 4× SDS loading buffer and analyzed by electrophoresis on a 4 to 20% gradient Tris-HEPES-SDS polyacrylamide gel (Thermo Scientific). Proteins from duplicate gels and filters were transferred to a polyvinylidene fluoride or nitrocellulose membrane and probed with anti-Ub:HRP antibody (P4D1; Santa Cruz Biotechnology catalog number sc-8017 HRP) or anti-His:HRP antibody (Abcam catalog number ab1187). The ImmunoStar HRP substrate kit (Bio-Rad) was used for detecting antibody complexes.
Electrolyte Leakage Analyses
Cell death progression in transient assays was assayed by measuring ion leakage. Each sample contained ten leaf discs collected from five different N. benthamiana leaves at 16 h after infiltration. Leaf discs were washed three times each for 10 min and then immersed in 4 mL deionized water in a tissue culture plate and gently rocked. The conductivity of the water was measured using a conductivity meter at the indicated time points (model 604; VWR Scientific). Measurements for each time point were performed on three samples to obtain a mean and sd.
Quantifying Powdery Mildew Sporulation
Golovinomyces cichoracearum strain UCSC1 was maintained on hypersusceptible Arabidopsis pad4-2 mutant plants. Four-week-old plants were inoculated using a settling tower ∼0.8 m tall. Plants to be inoculated were placed at the bottom of the tower, which contained a Nitex mesh screen at the top. Twelve pad4-2 mutant plants with heavy powder were passed over the mesh to transfer the spores to the plants below. Spores were counted as described by Weßling and Panstruga (2012) with some modifications. After infection, the spores were allowed to settle and three leaves per genotype were harvested, weighed, and transferred to 1.5-mL microcentrifuge tubes. Water (500 μL) was added and spores were liberated by vortexing 30 s at max speed. Leaves were removed and spores were concentrated by centrifugation at 4000g for 5 min. For each sample, spores were counted in eight 1-mm2 fields of a Neubauer-improved hemocytometer (Marienfeld). Spore counts were normalized to the initial weight of seedlings and results were averaged. The same procedure was repeated 8 d postinoculation. This method of quantifying infection levels was found to be more sensitive than previously used visual assays, in which infection was estimated by comparing levels of visible white powder (Wawrzynska et al., 2008). This entire experiment was repeated three times with similar results.
Trypan Blue Staining
Staining with trypan blue was performed essentially as described by Serrano et al. (2010). Arabidopsis plants were inoculated with G. cichoracearum, and leaves were collected at 5 d postinoculation and then boiled in alcoholic lactophenol (ethanol:lactophenol 1:1 v/v) containing 0.1 mg mL−1 trypan blue (Sigma-Aldrich) for 1 min. Leaves were then destained using a chloral hydrate solution (2.5 mg mL−1) at room temperature overnight. Samples were observed under a Zeiss Axioplan microscope. To quantify cell death, total leaf area and trypan-stained area were measured using ImageJ, and the percentage (area of cell death/total leaf area) was calculated. Mean cell death was quantified using 30 images derived from two independent experiments with three biological replicates each.
Accession Numbers
Arabidopsis sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: ARA6 (At3g54840), AtBI1 (At5g47120), ATL1 (At1g04360), ATL5 (At3g62690), ATL16 (At5g43420), BRI1 (At4g39400), EDR1 (At1g08720), KEG (At5g13530), RPS5 (At1g12220), SYP21 (At5g16830), SYP61 (At1g28490), and VHA-a1 (At2g28520). Rice sequence data from this article can be found in the Rice Genome Annotation Project database under the following accession numbers: ATL40 (Os02g57460) and ATL42 (Os03g05560).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ATL1 Coimmunoprecipitates with EDR1 and Localizes to Endomembrane Structures.
Supplemental Figure 2. EDR1 Specifically Antagonizes ATL1-Induced Cell Death in N. benthamiana.
Supplemental Figure 3. ATL1 Causes Severe Dwarfing When Overexpressed in the Arabidopsis edr1 Mutant.
Supplemental Figure 4. ATL16 Does Not Induce Cell Death When Overexpressed in N. benthamiana
Supplemental Figure 5. Comparison of ATL1-mCherry and ATL1C153A/H155A-mCherry Expression Levels Using Fluorescence Quantification.
Supplemental Figure 6. Mutations in the ATL1 RxxxS Motif Do Not Alter ATL1 Localization.
Supplemental Figure 7. Mutations in the RxxxS Motif of ATL1 Do Not Alter Its Interaction With EDR1.
Supplemental Figure 8. ATL1 amiRNAi Lines Have Reduced Levels of ATL1 mRNA.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We acknowledge the Indiana University Light Microscopy Imaging Center for access to the Leica SP5 confocal microscope. The ABRC at Ohio State University provided the ATL1 and ATL16 cDNA clones. We acknowledge the National Institute of General Medical Sciences of the National Institutes of Health (Grant R01 GM063761 to R.W.I.) and the Indiana University Bridge Program for funding.
AUTHOR CONTRIBUTIONS
I.S., Y.G., and R.W.I. designed the research. I.S., Y.G., D.Q., and U.D. performed the experiments. I.S., Y.G., and R.W.I. analyzed the data and wrote the article.
Glossary
- TGN/EE
trans-Golgi network/early endosome
- MVB/LE
multivesicular body/late endosome
- BiFC
bimolecular fluorescence complementation
- hpi
hours postinfiltration
- Col-0
Columbia-0
- amiRNA
artificial microRNA
- SD
synthetic dextrose
- HRP
horseradish peroxidase
Footnotes
Online version contains Web-only data.
References
- Acconcia F., Sigismund S., Polo S. (2009). Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315: –1618. [DOI] [PubMed] [Google Scholar]
- Ade J., DeYoung B.J., Golstein C., Innes R.W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Hernández V., Aguilar-Henonin L., Guzmán P. (2011). Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments. PLoS ONE 6: e23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Heard W., Mbengue M., Robatzek S. (2012a). The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15: 367–374. [DOI] [PubMed] [Google Scholar]
- Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S. (2012b). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Stone S., Yang X., Antico J., Callis J., Ramonell K.M., Somerville S. (2010). ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 5: e14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Odum N., Jørgensen L.B., Brown R.E., Mundy J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso D., Delagoutte-Busso B., Moras D. (2005). Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343: 313–321. [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Liu H., Stone S., Callis J. (2013). ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 75: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Cross B., Li B., Chen L., Li Z., Chen J. (2011). Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol. Cell. Biol. 31: 4951–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen K.M., Gu Y., Rodibaugh N., Innes R.W. (2011). Negative regulation of defence signalling pathways by the EDR1 protein kinase. Mol. Plant Pathol. 12: 746–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia R.H., Falkow S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173: 33–38. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Frye C.A., Innes R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol. 147: 1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs C.S., Zoller M.J. (1991). Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem. 266: 8923–8931. [PubMed] [Google Scholar]
- Gómez-Gómez L., Bauer Z., Boller T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2011). The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2012). The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24: 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P. (2012). The prolific ATL family of RING-H2 ubiquitin ligases. Plant Signal. Behav. 7: 1014–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T.J., Sliwinski M.K., Martínez D.E., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109. [DOI] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y., Tomitaka A., Kumeta H., Noda N.N., Yamaguchi M., Inagaki F. (2011). Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. USA 108: 20579–20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G.-J., Goedhart J., van Munster E.B., Gadella T.W.J. Jr. (2006). Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580. [DOI] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., et al. (2012). The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-S., Martin R., Mongrand S., Vandenabeele S., Chen K.-C., Jang I.-C., Chua N.-H. (2008). RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 56: 550–561. [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor ABSCISIC ACID INSENSITIVE 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288: 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., Mudgett M.B., Li K., Aoyama T., Horvath D., Chua N.-H., Staskawicz B.J. (1998). Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 14: 247–257. [DOI] [PubMed] [Google Scholar]
- Morris K., Thornber S., Codrai L., Richardson C., Craig A., Sadanandom A., Thomas B., Jackson S. (2010). DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 22: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.-J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.-F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614. [DOI] [PubMed] [Google Scholar]
- Qi D., DeYoung B.J., Innes R.W. (2012). Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D.N., Cuenca J.V., Bowman G.R., Guttman D.S., Greenberg J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Mondragón R.E., Garcidueñas-Piña C., Guzmán P. (1999). Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol. Biol. 40: 579–590. [DOI] [PubMed] [Google Scholar]
- Sato T., et al. (2009). CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 60: 852–864. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano I., Pelliccione S., Olmedilla A. (2010). Programmed-cell-death hallmarks in incompatible pollen and papillar stigma cells of Olea europaea L. under free pollination. Plant Cell Rep. 29: 561–572. [DOI] [PubMed] [Google Scholar]
- Serrano M., Guzmán P. (2004). Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Parra S., Alcaraz L.D., Guzmán P. (2006). The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J. Mol. Evol. 62: 434–445. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Silady R.A., Ehrhardt D.W., Jackson K., Faulkner C., Oparka K., Somerville C.R. (2008). The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J. 53: 29–41. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.-I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Tang D., Innes R.W. (2002). Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J. 32: 975–983. [DOI] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2006). A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2005a). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Christiansen K.M., Innes R.W. (2005b). Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 138: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283: 3200–3210. [DOI] [PubMed] [Google Scholar]
- Wawrzynska A., Christiansen K.M., Lan Y., Rodibaugh N.L., Innes R.W. (2008). Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148: 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R., Panstruga R. (2012). Rapid quantification of plant-powdery mildew interactions by qPCR and conidiospore counts. Plant Methods 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T., Tomczak A., Michelmore R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273. [DOI] [PubMed] [Google Scholar]
- Yao C., Wu Y., Nie H., Tang D. (2012). RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J. 71: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zeng L.-R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.-L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Nie H., Shen Q., Zhang S., Lukowitz W., Tang D. (2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10: e1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.