Abstract
Coated vesicles provide a major mechanism for the transport of proteins through the endomembrane system of plants. Transport between the endoplasmic reticulum and the Golgi involves vesicles with COPI and COPII coats, whereas clathrin is the predominant coat in endocytosis and post-Golgi trafficking. Sorting of cargo, coat assembly, budding, and fission are all complex and tightly regulated processes that involve many proteins. The mechanisms and responsible factors are largely conserved in eukaryotes, and increasing organismal complexity tends to be associated with a greater numbers of individual family members. Among the key factors is the class of ENTH/ANTH/VHS domain-containing proteins, which link membrane subdomains, clathrin, and other adapter proteins involved in early steps of clathrin coated vesicle formation. More than 30 Arabidopsis thaliana proteins contain this domain, but their generally low sequence conservation has made functional classification difficult. Reports from the last two years have greatly expanded our knowledge of these proteins and suggest that ENTH/ANTH/VHS domain proteins are involved in various instances of clathrin-related endomembrane trafficking in plants. This review aims to summarize these new findings and discuss the broader context of clathrin-dependent plant vesicular transport.
INTRODUCTION
As in all eukaryotes, plant cells contain membrane-bound subcompartments with specialized functions that are mainly defined by the proteins residing inside the compartments or at their membranes. The correct transport and localization of such proteins is therefore of crucial importance to the cell, and given the variety of cargo and target compartments, it is not surprising that intracellular protein transport is a complex affair involving different mechanisms and a plethora of proteins. A main means of protein transport between compartments are vesicles, which bud off from a donor membrane, are transported along the cytoskeleton, and eventually fuse with the target membrane to release their contents. Vesicle formation is a stepwise process and typically involves a protein coat. COAT PROTEIN I (COPI) and COPII coats function in transport between the endoplasmic reticulum and the Golgi, while clathrin coats participate in endocytosis at the plasma membrane (PM) and post-Golgi traffic such as secretion and vacuolar transport. In the last few years, knowledge about protein transport through the plant endomembrane system has been increasing, and several recent articles give a comprehensive general overview of our current understanding (Reyes et al., 2011; Park and Jürgens, 2012; Baisa et al., 2013). In this review, we focus on a group of accessory proteins that play roles in early steps of clathrin-coated vesicle (CCV) formation. These proteins share a similar domain organization, with an ENTH, ANTH, or VHS domain at their N terminus for interaction with endomembranes and a relatively unstructured C-terminal part required for interaction with other components of the vesicle generating machinery, such as clathrin. Until recently, only few of them had been characterized in plants, and it was therefore not clear whether the structurally similar ENTH, ANTH, and VHS domains distinguish these proteins functionally. However, the last two years have seen considerable progress in the functional and biochemical understanding of these proteins, and it is now possible to suggest a classification based on structure and function.
Historically, cell biology knowledge in yeast and animals has been advanced compared with that in plants, and much of the cell biological research in plants has been driven by seeking out (and sometimes discarding) similarities to functional counterparts from other kingdoms. Therefore, we begin this review by describing prototypical ENTH, ANTH, and VHS proteins from yeast and animals. Then, we take a phylogenetic approach to group the plant proteins, using Arabidopsis thaliana as the focus species. Reviewing the current literature, it seems possible to assign for each group a specific functional context within the plant endomembrane system. Nevertheless, there are still many unresolved questions in the field and we conclude with some of them.
ENTH DOMAIN
Although mammalian Epsin1 is the best characterized protein of this class, the first isolated protein with this domain came from plants (Jones et al. 1997) two years before the domain was biochemically described and termed the Epsin N-Terminal Homology (ENTH) domain (Rosenthal et al. 1999). The ENTH domain forms a compact solenoid of eight α-helices, comprising roughly 130 to 150 amino acids. For simplicity, we will use the term “ENTH protein” for proteins that contain an ENTH domain, which includes the family of Epsin homologs. The ENTH domain of mammalian Epsin1 binds phosphoinositides (PtdInsPs) that are present in low amounts in the membranes, with a preference for PdtIns(4,5)P2 (Itoh et al., 2001). The amino acids required for this activity are often conserved between ENTH-containing proteins, yet phosphoinositide binding has not been shown for all ENTH proteins, and not all ENTH proteins have a preference for PtdIns(4,5)P2 (see below). The interaction with PtdIns(4,5)P2 was speculated to result in a wedge-like insertion into the cytosolic leaflet of the phospholipid bilayer, thus mechanically facilitating the bulging of incipient vesicles (Ford et al., 2002). In most ENTH domain proteins, the part C-terminal to the ENTH domain (which typically makes up more than two thirds of the entire protein) is remarkably unstructured. However, that region contains many short sequence motifs, which are required for interaction with clathrin and with the so-called ear domain of the large subunits of heterotetrameric clathrin adaptor complexes (APs). Mammalian Epsin1 binds the α-ear domain of AP-2, a complex involved in formation of CCVs in endocytosis, in agreement with the enrichment of the PM for PtdIns(4,5)P2.
A similar ENTH protein, mammalian EpsinR/CLINT/Enthoprotein (reviewed in Legendre-Guillemin et al., 2004), instead binds the γ-ear domain of AP-1, which is involved in CCV formation at the trans-Golgi network (TGN) and has high affinity for PtdIns(4)P, in line with the enrichment of this phosphoinositide at the TGN (Mills et al., 2003). Therefore, it seems that slight variations in the phosphoinositide binding properties of the ENTH domain are associated with guiding Epsin activity to different subcellular locations.
A key question is what, precisely, constitutes Epsin activity? An affinity for membrane subdomains, APs, and clathrin, along with protein transport defects in Epsin mutants, leave no doubt that Epsins, and presumably other ENTH domain-containing proteins, are important factors in CCV generation. McMahon and coworkers (Ford et al., 2002) suggested that Epsins induce the membrane curvature required for vesicle bulging, or at least reduce the amount of energy required for forming a membrane bulge. This view has been recently challenged by the finding that clathrin alone is sufficient to induce spherical bulbs in membranes, in the absence of any membrane reshaping proteins (Dannhauser and Ungewickell, 2012). If Epsins indeed do not induce membrane curvature under physiological conditions, then their role would be mainly that of an early adaptor, linking membrane subdomains with APs and clathrin. An interesting alternative hypothesis arises from the recent finding that Epsin1 is required for dynamin-dependent membrane scission at the neck of the budding CCV, which is a late step in CCV generation (Boucrot et al., 2012). This is supported by an analysis of the temporal succession of events in CCV formation, showing that mammalian Epsin2 gradually accumulates at the site of CCV generation right up to the point of scission, a pattern identical to that of clathrin itself (Taylor et al., 2011), whereas AP-2 levels decreased before scission. A role in scission of the budding CCV is also supported by the finding that Epsin seems to concentrate toward the neck of a forming vesicle, whereas AP-2 is found in the opposite, oldest part (Saffarian et al., 2009). Finally, mouse cells with near complete loss of all Epsin activity exhibit failure in CCV scission at the PM (Messa et al., 2014).
Whether the requirement for Epsins is restricted to the late events of vesicle scission or if they also are needed to facilitate the earlier steps of inducing membrane curvature remains to be determined. Recent reports suggest that Epsins link vesicle budding sites with the cytoskeleton. The yeast epsin Ent1 directly interacts with actin via a phospho-regulated binding domain (Skruzny et al., 2012), and a similar acting binding activity has been found in mammalian Epsin1 (Messa et al., 2014). Thus, the membrane destabilizing properties of the ENTH domain, combined with mechanical force introduced by the actin cytoskeleton might lead to membrane bulging and, eventually, vesicle fission. Apart from their function in vesicle assembly, Epsins and other ENTH proteins may have additional roles in recruiting cargo: Several lines of evidence, including identification of the actual interaction surface, demonstrate that Epsin can interact with soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptors (SNAREs) that are important for fusion of the vesicle with its target membrane (Chidambaram et al., 2004; Miller et al., 2007; Messa et al., 2014).
ANTH DOMAIN
The brain-enriched AP180 protein and its non-neuronal homolog Clathrin Assembly Lymphoid Myeloid (CALM) both contain an AP180 N-Terminal Homology (ANTH) domain. Although it shares very little sequence identity with the ENTH domain, the ANTH domain also forms a compact solenoid of α-helices, some of which overlap well with the structure of the ENTH domain (Ford et al., 2001). However, the ANTH domain contains 9 to 10 helices and is considerably longer, at ∼250 to 300 amino acids. The ANTH domain also binds PtdIns(4,5)P2, but the binding surface is different from that of ENTH. A short conserved motif K[X]9[K/R][H/Y] between helices 1 and 2 is required for the binding activity (Ford et al., 2001), but unlike the ENTH binding, no insertion into the membrane takes place. This is consistent with the observation that ANTH proteins do not cause liposome tubulation in in vitro assays, indicating that, in contrast to ENTH proteins, they are not directly associated with membrane-shaping properties. However, they link clathrin to PtdIns(4,5)P2-enriched sites at the PM and thus might indirectly cause membrane curvature and vesicle formation, since clathrin assembly alone is sufficient to induce membrane curvature in vitro, as mentioned above (Dannhauser and Ungewickell, 2012). The canonical and well-described role of ANTH proteins is in endocytosis at the PM; however, there are also hints that CALM may play a role in CCV formation at endosomal structures, presumably the TGN (Borner et al., 2006; Bushlin et al., 2008). Similarly to Epsin, CALM was found to be involved in direct interaction with SNAREs (VAMP2, VAMP3, and VAMP8) to recruit them into endocytic vesicles (Miller et al., 2011).
VHS DOMAIN
The Vps-27, Hrs, and STAM (VHS) domain is structurally very similar to the ENTH domain, and the ENTH, ANTH, and VHS proteins are often treated as a superfamily (Interpro IPR008942=ENTH/VHS). Similar to the ENTH domain, the VHS domain contains eight α-helices and is ∼150 amino acids in length (Mao et al., 2000). Proteins with a VHS domain often have other functionally important structured domains and can be grouped accordingly (for an overview, see Lohi et al., 2002). Members of one such group contain the Fab-1, YGL023, Vps27, and EEA1 (FYVE) domain, which is required for PtdIns(3)P binding at the animal early endosome (EE). FYVE-containing proteins act as sorting receptors for ubiquitinated cargo to target it for degradation via the Endosomal Sorting Complexes Required for Transport (ESCRT) pathway, which is based on the sequential action of ESCRT subcomplexes termed ESCRT-0 to ESCRT-III. Two interacting members of the FYVE group, Hrs and Vps27, form the ESCRT-0 complex in yeast. Proteins of another group lack the FYVE domain and instead contain a Golgi-localized, γ-ear-containing, ADP-ribosylation-factor binding, and Tom1 (GAT) domain that interacts with ubiquitinated cargo and thus may perform a function similar to ESCRT-0 components. A third important group consists of the Golgi-localizing, γ-adaptin ear homology domain, ARF binding (GGA) proteins, which are involved in vesicle formation at the TGN (Puertollano et al., 2001; Demmel et al., 2008) and also at the PM (Puertollano and Bonifacino, 2004). In contrast to the ENTH domain, no mechanism for physical insertion of the VHS domain into the membrane has been reported. Therefore, it seems more likely that these proteins function in cargo recruitment and interfacing with subsequent components for either incorporation into CCVs or the ESCRT-dependent degradation pathway.
PHYLOGENETIC ANALYSIS OF ENTH/ANTH/VHS PROTEINS
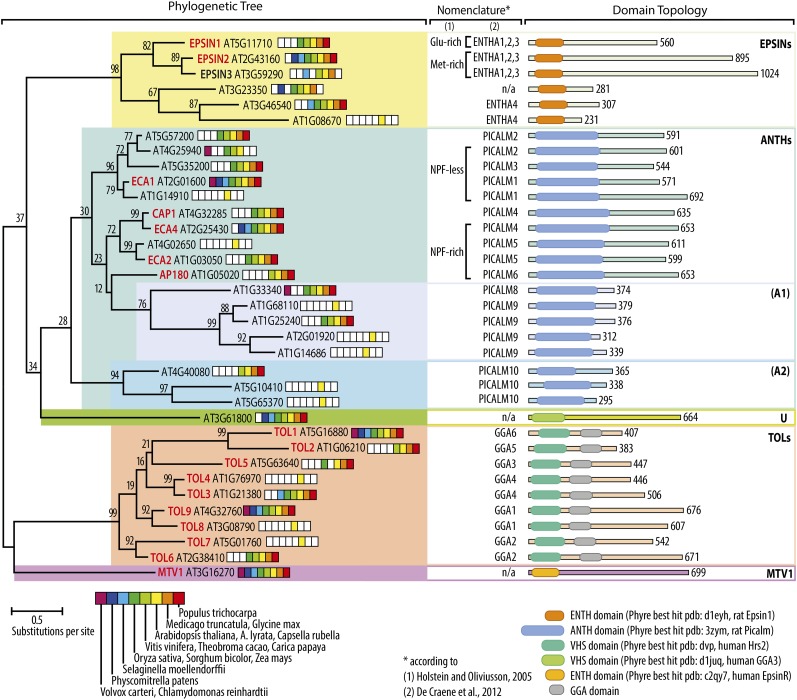
The TAIR10 database lists 43 proteins with the automatically generated Interpro IPR008942=ENTH/VHS annotation, which also includes ANTH domain proteins. We performed an initial analysis for motifs using SMART v7 (Letunic et al., 2012) and identified eight proteins that are likely not involved in vesicular biogenesis (see Methods for their AGIs). The ENTH/VHS domain in these eight proteins is not at the N terminus, and all of them contain an RPR domain (PDB ID: 2diw), which is found in proteins related to RNA processing. Indeed, one of the eight proteins, RRC1, has been recently functionally characterized as a splice factor (Shikata et al., 2012). We performed phylogenetic analysis of the remaining 35 proteins (Figure 1; Supplemental Data Set 1). We also analyzed the presence/absence of homologs in other, selected plant species by reciprocal BLAST (Figure 1; see Methods for details). Based on the tree topology and other factors, such as predicted structure (using the Protein Homology/analogY Recognition Engine V 2.0 [Phyre2]; Kelley and Sternberg, 2009) and protein size, we propose classification into three main groups, which reflect the ENTH, the ANTH, and the VHS domains. The ENTH group splits by tree topology, predicted structure, and protein size into two groups of three proteins each, in agreement with an earlier analysis of eukaryotic ENTH/ANTH/VHS proteins by De Craene et al. (2012). For consistency, their nomenclature (ENTH1,2,3 and ENTH4) is indicated as well. The ANTH group is the largest, comprising the eight ANTH proteins mentioned in an earlier report (Holstein and Oliviusson, 2005) along with another 10 members. We further subdivided this group into three subgroups mainly based on protein size, although it is unclear whether this reflects any functional difference. The third main group contains proteins that have an N terminus with structural similarity to the VHS domain of human Hrs1 and additionally contain a GGA domain. In addition to these three main groups, there are two outliers: AT3G61800 and the recently described MTV1/AT3G16270 (Sauer et al., 2013). These are single genes in Arabidopsis, but they are conserved throughout the plant lineage with homologs found even in ancestral plants. A tree based on alignment of only the first 200 amino acids (where the ENTH/ANTH/VHS domain lies) has an almost identical topology (Supplemental Figure 1 and Supplemental Data Set 2), confirming that the C-terminal part of the proteins is generally poorly structured and as such not useful to determine phylogeny. Our analysis goes beyond two earlier analyses of this group of proteins (Holstein and Oliviusson, 2005; De Craene et al., 2012) in that it includes all annotated Arabidopsis ENTH/ANTH/VHS proteins and further provides information on the presence/absence of homologs in 15 other plant species. It uses data on protein length and information on the predicted structure to define additional subgroups. The overall tree topology agrees well with the aforementioned studies, and we included their nomenclature for consistency (Figure 1).
Figure 1.
Phylogenetic Analysis of Arabidopsis ENTH/ANTH/VHS Domain-Containing Proteins.
Midpoint-rooted maximum likelihood tree of 35 Arabidopsis ENTH/ANTH/VHS domain-containing genes. See Methods for details of analysis. Common names for genes described in the literature in bold, preceding the AGI. Segments of the rainbow-colored bar indicate the presence/absence of a clear homolog in at least one of the species indicated. For consistency, features and domains referred to in previous works are also indicated in columns (1) and (2). Rough domain topology is shown on the right, with approximate length and placement of N-terminal domains. The structurally most similar protein is also given, according to the Phyre2 prediction.
TRANSCRIPTIONAL REGULATION OF ENTH/ANTH/VHS PROTEINS
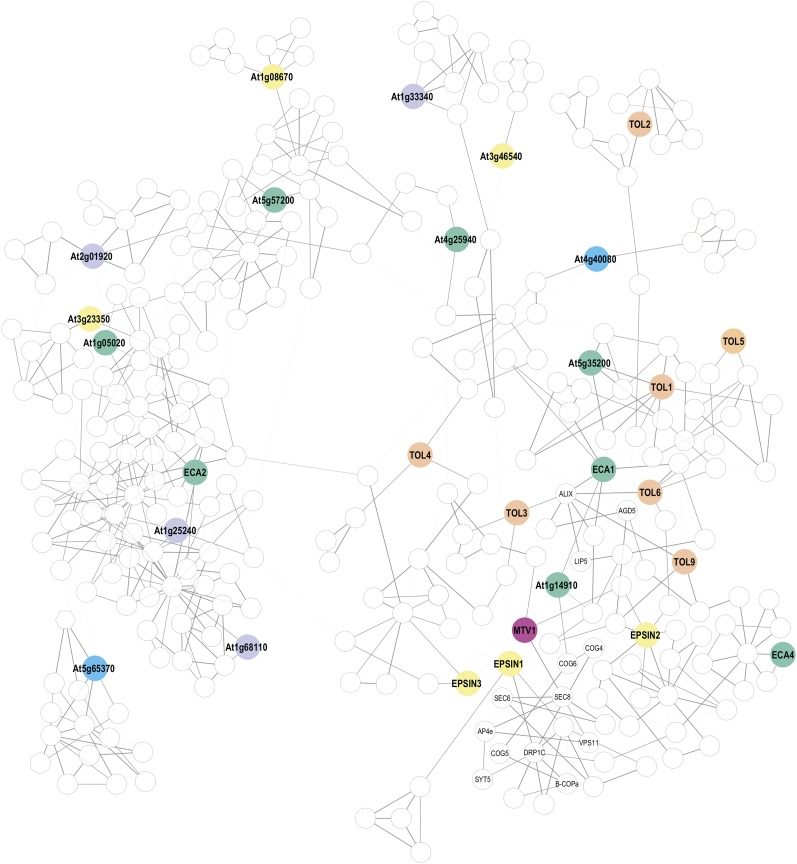
Generally, not much is known about the transcriptional regulation of factors required for protein transport, and the ENTH/ANTH/VHS proteins are no exception. In the few cases where expression patterns have been analyzed, the expression was ubiquitous. Arabidopsis coexpression databases allow for visualization of gene-to-gene relationships in gene networks by connecting the most correlated genes, which may suggest potential protein-protein interactions or underlying biological networks. Therefore, this type of analysis can help to identify genes that might be central for a given physiological process. To implement this approach, we took advantage of the ATTED-II database of gene coexpression in Arabidopsis (Obayashi et al., 2009) and constructed an undirected gene coexpression network (Figure 2; see Methods for details). The network contained 297 nodes (i.e., genes) and 606 edges (Supplemental Data Set 3 and Supplemental Figure 2) and was further analyzed using the Cytoscape network analysis framework to identify and visualize clusters of genes whose expression is correlated and therefore may form a functional group. A scale-free biological network, such as a gene coexpression network, shows the distribution of a node connectivity that follows a power law and as a consequence, some nodes have many locally connected neighbors and are characterized by high degree centrality (DC). In addition, nodes that are part of the shortest route between two arbitrary nodes in the network often control the communication between network modules and are represented by high between-ness centrality (BC) measures. Therefore, we can differentiate between local and global importance of a particular node by analyzing its centrality. Out of the 35 analyzed ENTH genes, EPSINs 1, 2, and 3 and ECA1 were the only ones with high centrality measures. EPSIN2 and ECA1 were among the 10 genes with the highest DC, but they showed low BC values, suggesting that they may function locally inside larger modules (Yu et al., 2007). Conversely, BC has been described as a common attribute of globally essential nodes in biological networks (Joy et al., 2005; Yu et al., 2007). Among the 10 genes with the highest BC, we identified MTV1, EPSIN1, and EPSIN3. None of these nodes showed high DC, suggesting that these genes may act as major intersections between network clusters and mediate crosstalk (Joy et al., 2005).
Figure 2.
Transcriptional Network Analysis of Arabidopsis ENTH/ANTH/VHS Genes.
Graphical representation of the coexpression network analysis. See text for details and Supplemental Figure 2 and Supplemental Data Set 3 for gene identifiers.
EPSIN1 formed a coexpression cluster with genes encoding members of the conserved oligomeric Golgi complex, the exocyst complex (SEC6 and SEC8), and other known membrane trafficking genes (DYNAMIN-LIKE 1C, VPS11, B-COPa, SYT5, and AP-4e) (Figure 2; for gene identifiers, see Supplemental Data Set 3). The proximal coexpression cluster contained two members of the ESCRT complex (ALIX and LIP5), the ADP ribosylation factor GTPase-activating protein AGD5, and several members of the TOL gene family. Both modules were connected via MTV1, the node with the highest value of between-ness centrality among the analyzed ENTH genes. Thus, the unique topological features of the MTV1 node may contribute to the striking ubiquitous phenotype of the mtv1 agd5 double mutant (Sauer et al., 2013).
Using the AmiGO annotation and ontology toolkit (Carbon et al., 2009), we analyzed the gene ontology (GO) term enrichment for the 297 nodes of the coexpression network. Among the enriched Biological Process GO terms, we identified GO:0009827 (plant-type cell wall modification, 16 times above background) and GO:0048868 (pollen tube development, 12 times above background). This is in agreement with the fact that secretion and endocytosis are key processes for cell wall formation and pollen tube outgrowth. Interestingly, the network contained 35 kinase genes that made the GO:0016301 (kinase activity) the most enriched GO term in the Molecular Function category. As many of these kinases are directly coexpressed with particular ENTH genes and there is evidence for regulation of Epsin-like proteins by phosphorylation (see below), they are interesting targets for future studies.
EPSINs AND OTHER ENTHs: FUNCTIONALLY DIVERSE IN POST-GOLGI TRAFFIC
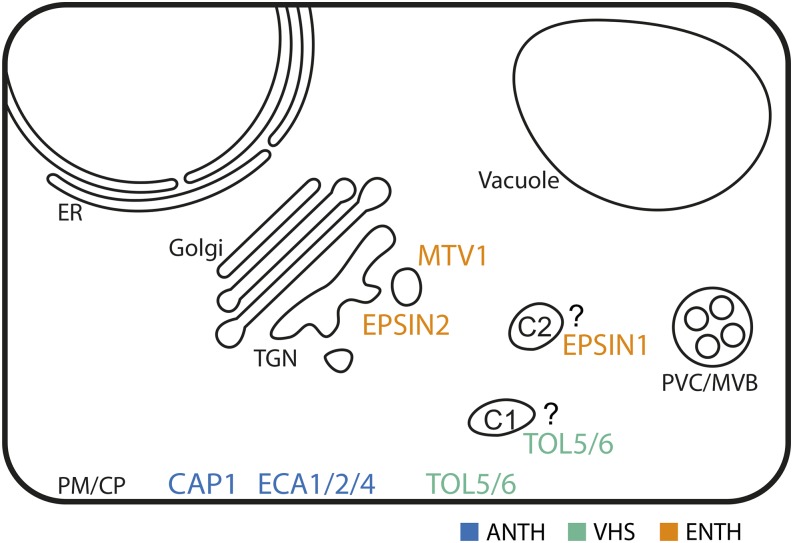
Of the six plant proteins in the ENTH domain group, two have been functionally characterized, and despite their similarity, they appear to be involved in distinct processes. Song et al. (2006) generated antibodies against Arabidopsis EPSIN1/EPSIN1R (AT5G11710) to examine patterns of localization and confirmed that EPSIN1 is a soluble protein that is expressed in a wide range of tissue types. Its subcellular localization was studied in transiently transformed protoplasts, where a punctate and (in unfixed samples) a network-like pattern was observed. Treatment with the actin-depolymerizing drug Latrunculin B led to loss of the network pattern and loss of colocalization with an actin marker; this might hint at association with actin filaments. Whether the punctate pattern indeed reflects a partial Golgi and prevacuolar compartment (PVC) localization is not entirely clear. Song et al. observed partial colocalizations with the Golgi marker ST-GFP (green fluorescent protein), the TGN marker vSNARE VTI11, and the late endosome (LE)/PVC marker SYP21/PEP12p:HA and also show colocalization with clathrin, which by current knowledge is expected at neither the Golgi nor the LE/PVC. It was not indicated whether clathrin heavy or light chain (CHC or CLC) were detected, but both localize mainly to PM and TGN, with differences in dynamics and localization patterns (C. Wang et al., 2013). Since the EPSIN1 constructs were not tested for functionality, and only transient expression assays were used, the definitive localization of endogenous EPSIN1 cannot be inferred from these data. It is possible that the observed punctate localization corresponds to a yet uncharacterized endomembrane compartment between the TGN and the LE/PVC or subspecies of the TGN or LE/PVC that are not labeled by conventional TGN or LE/PVC markers, respectively (Figure 3). However, biochemical evidence corroborates both VTI11 and clathrin interactions (Table 1 lists all known interactors from the plant literature). Epsin1 binds to clathrin through its C-terminal domain, where a canonical clathrin binding motif LIDL is required for the interaction (Song et al., 2006). This motif is conserved in all tested homologs from the species analyzed in Figure 1. The C terminus also binds specifically and directly to the ear domain of γ-Adaptin1 of the AP-1 complex. Three recent publications place AP-1 firmly in the late secretory pathway at the TGN, and loss of function of its central μ1 subunit AP1M leads to strong defects in delivery of vacuolar cargo and cell plate material (Park et al., 2013; Teh et al., 2013; J.-G. Wang et al., 2013). This is in line with the earlier finding that the μ1 subunit of AP-1 interacts with VACUOLAR SORTING RECEPTOR (VSR) (Happel et al., 2004). Correspondingly, a partial loss-of-function epsin1 mutant showed defects in delivery of vacuolar cargo (Sporamin-GFP; Song et al., 2006). Taken together, it appears that Arabidopsis EPSIN1 is an accessory protein for AP1-dependent post-Golgi trafficking events. Whether it is involved in a functional spectrum as broad as that of AP-1 (vacuolar sorting, secretion, and cytokinesis) or rather is specialized for vacuolar trafficking events remains to be seen. Also unclear is whether EPSIN1 is essential because data from a full loss-of-function mutant is lacking.
Figure 3.
Known Subcellular Localizations of ENTH/ANTH/VHS Proteins
Proteins are color-coded according to their N-terminal domain. C1 stands for a hypothetical endomembrane compartment involved in sorting cargo for degradation, and C2 is a not fully characterized compartment where EPSIN1 localizes, apparently distinct from TGN and PVC/MVB (Song et al., 2006). ER, endoplasmic reticulum; PM/CP, plasma membrane/cell plate.
Table 1. Known Interactors of Arabidopsis ENTH/ANTH/VHS Proteins.
| Name | Interactor | Technique | Localization | Reference |
|---|---|---|---|---|
| AP180 | AT5G22780 α adaptin (AP-2) | Pull-down | n.d. | Barth and Holstein (2004) |
| AT1G05020 | ||||
| ECA1 AT2G01600 | TML | Affinity purification + MS, CoIP | n.d. | Gadeyne et al. (2014) |
| AT5G57460 | ||||
| Clathrin heavy chain | Pull-down (specific for C-terminus of ECA1) | PM, early CP, TGN | Song et al. (2012) | |
| ECA2 | n.d. | n.a. | PM, endosomes | Song et al. (2012) |
| AT1G03050 | ||||
| ECA4 | n.d. | n.a. | PM, CP, endosomes | Song et al. (2012) |
| AT2G25430 | TML | Affinity purification + MS, CoIP | n.d. | Gadeyne et al. (2014) |
| AT5G57460 | ||||
| CAP1 AT4G32285 | TML | Affinity purification + MS, CoIP | n.d. | Gadeyne et al. (2014) |
| AT5G57460 | ||||
| EPSIN1 (EPSINR1) | Clathrin heavy chain, γ-ADR (γ-AP-1 homolog), VTI11, VSR1 | Pull-down (specific for C terminus) | Punctate structures | Song et al. (2006) |
| AT5G11710 | VTI11, VSR1 | Pull-down (specific for ENTH domain) | ||
| KRP4 AT2G32710 | TAP-Tag | n.d. | Van Leene et al. (2010) | |
| EPSIN2 (EPSIN2R) | Clathrin heavy chain (weakly with AP-2 α-adaptin) | Pull-down (specific for two domains in the C terminus) | Punctate structures | Lee et al. (2007) |
| AT2G43160 | AP-3 δ-adaptin, VTI12 | Pull-down (specific for the ENTH domain) | ||
| Ubiquitin3 | Affinity-capture MS | Igawa et al. (2009) | ||
| TOL2 | Ubiquitin | Pull-down | n.d. | Korbei et al. (2013) |
| AT1G06210 | ||||
| TOL6 | Ubiquitin | Pull-down | n.d. | Korbei et al. (2013) |
| AT2G38410 | ||||
| MTV1 | Clathrin heavy chain | Pull-down (specific for C terminus of MTV1) | TGN, CCVs | Sauer et al. (2013) |
| AT3G16270 |
n.d., not determined; n.a., not applicable; CP, cell plate; MS, mass spectrometry; CoIP, coimmunoprecipitation.
The other characterized member is Arabidopsis EPSIN2/EPSINR2 (AT2G43160), which was shown to bind PI(3)P in a lipid overlay experiment (Lee et al., 2007). This is surprising because this activity depended on conserved amino acids (R69 and K82) that also are strictly conserved in mammalian Epsins (rEpsin1, hEpsin1, and 2), where they are critical for binding PI(4,5)P2 (Ford et al., 2002). EPSIN2 binds clathrin via a motif (LADVF) that resembles a degenerate clathrin box, and multiple additional binding sites on its Met-rich C-terminal side. Interestingly, the LADVF motif is not strictly conserved in EPSIN2 homologs from other plant species, but it is unknown whether the degenerate motifs of other species are functionally equivalent. Phylogenetic footprinting might be a straightforward approach to enhance our understanding of such degenerate motifs and help to predict protein function. EPSIN2 binds to the δ-subunit of the AP-3 complex and, to a much lesser degree, to the α-subunit of the AP-2 complex (Table 1). Consistent with the AP-3 binding data, EPSIN2 colocalizes with δ adaptin on punctate structures. Little to no colocalization of these punctae was observed for VSR1, Golgi, or the EE markers Ara6 or endocytosed FM4-64 (after 3 h). Although colocalization with clathrin and a subset of VTI12 might suggest a localization at the TGN, the steady state distribution of VTI12 likely labels a TGN/PVC continuum.
More recent studies clearly implicate the AP-3 complex in post-Golgi transport to the vacuole (Niihama et al., 2009; Feraru et al., 2010) and moreover suggest an interaction with clathrin (Zwiewka et al., 2011). Interestingly, mutants in AP-3 function (pat2 in AP-3 β, pat4 in AP-3 δ, or dominant-negative AP-3 μ) only affect part of the vacuolar traffic, as transport of lytic, but not storage vacuolar cargos, is impaired (Feraru et al., 2010; Zwiewka et al., 2011). In addition to soluble cargo that is delivered to the lumen of the vacuole, tonoplast-resident membrane proteins also are differentially dependent on AP-3 function: The sucrose transporter SUC4 is blocked at the Golgi under AP-3 deficiency, while the inositol transporter INT4 is still properly transported (Wolfenstetter et al., 2012). Based on the current data regarding EPSIN2 and AP-3, it seems plausible that they interact at a Golgi-derived endosome, which may simply represent a subpopulation of the TGN, but could be a more separate entity (Figure 3). The PI3P binding of EPSIN2 seems surprising at first, since PI3P in plants so far has been associated with LE/PVC, and neither EPSIN2 or AP-3 colocalize with commonly used markers for the LE/PVC. However, mammalian AP-3 also preferentially binds PI3P (Baust et al., 2008); therefore, it would be worthwhile to see whether a discrete, AP-3-, EPSIN2-, and clathrin-dependent vacuolar trafficking pathway starts at a PtdIns(3)P-enriched endomembrane compartment or subdomain relatively near the Golgi.
EPSIN3 (AT3G59290) is similar to EPSIN2 (>60% sequence identity), but no functional analysis has been performed to date. Our preliminary analysis suggests that it might be absent in monocotyledons (Figure 1). The other three members of the ENTH group remain uncharacterized. It is striking that they are very short proteins, containing not much more than the ENTH domain itself. It is therefore unclear whether their C termini are sufficient to function as adaptors in CCV formation or whether they might adopt other, perhaps regulatory roles by outcompeting “true” EPSINs at phosphoinositide-rich sites in the membrane.
ANTHs: A LARGE GROUP INVOLVED IN ENDOCYTOSIS
As noted above, our phylogenetic analysis shows that the ANTH group has the most members (Figure 1). Based on topology and additional criteria, we define a core ANTH group that contains the eight ANTH genes previously identified in the phylogenetic analysis of Holstein and Oliviusson (2005), who further subdivided those proteins into two subclades based on the presence or absence of a functionally important NPF domain (in plants, this is a DPF motif). This core group contains all ANTH proteins that have been characterized to date. We define two additional groups “A1” and “A2,” mainly based on protein length, but none of them has been characterized in further detail, and three of the five members seem to be restricted to the Brassicaceae. At-AP180 (AT1G05020) was the first plant ANTH protein identified (Barth and Holstein, 2004). It was isolated due to its high similarity to the mammalian AP180 homolog CALM and binds to the ear region of the AP-2 α-subunit with high affinity (Table 1). Binding requires the DPF motif in the C terminus, which is conserved in its homologs. It also binds clathrin through uncharacterized motifs on the C terminus. Clathrin reassembly experiments show that the C terminus of At-AP180 is sufficient to drive formation of regular clathrin cages and that a conserved DLL motif is required for this function. So far, there is no direct data about At-AP180 subcellular localization, phosphoinositide binding, or in vivo function, but from its clathrin binding and association with AP-2, it is no far stretch to assume that At-AP180 plays a role in clathrin-dependent endocytosis at the PM: AP-2 is an endocytic adaptor in animals (reviewed in Robinson 2004) and AP-2 homologs are found throughout the eukaryotes. AP-2 is not always strictly required for clathrin-mediated endocytosis (CME): It is dispensable in yeast (Huang et al., 1999) but essential in mammalian cells. In plants, the functional characterization of AP-2 has been reported only recently by several independent groups. Various mutants in AP-2 subunits show impaired CME and developmental aberrations that presumably are the consequence of these cellular defects. These results, plus the dynamic spatiotemporal localization of AP-2 components at the PM, support the notion that AP-2 is an early adaptor complex for clathrin recruitment/assembly (Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013) in plant endocytosis.
For another three ANTH proteins, involvement in CCV formation at the cell plate has been postulated (Song et al., 2012). GFP-tagged ECA1 (AT2G01600), ECA2 (AT1G03050), and ECA4 (AT2G25430) (and ECA1-specific antibody signal) localize to the PM and the nascent cell plate and to some extent to punctate structures (Figure 3). In the case of ECA1, these punctae were further identified as EEs, mainly via colocalization with early endocytosed FM4-64 and colocalization also after treatment with Brefeldin A (BFA; Song et al., 2012). Analysis of spatiotemporal dynamics of ECA1 labeling at the nascent cell plate revealed a pattern that is suggestive of a role in endocytosis from the forming cell plate (reviewed in Jürgens, 2005). ECA1 arrives somewhat later than the fusogenic factors KNOLLE and DRP1C and also later than membrane material of incoming vesicles labeled with FM-4-64. However, it arrives earlier than the clathrin light chain, with which it colocalizes perfectly later on. Further corroborating a role in CME at the maturing cell plate, pull-down assays showed that ECA1 binds clathrin via a yet unidentified motif(s) on its C terminus (Table 1). No direct association with the alpha AP-2 adaptin AP2A2 subunit was found in this work, which might seem in conflict with later findings (see below), but more evidence is needed to determine whether there is indeed no involvement in AP-2 related CME. Currently, there is no direct data on the physiological function of ECA1, ECA2, or ECA4. It is likely that higher order loss-of-function mutant combinations would need to be tested in order to overcome an expected functional redundancy.
Evidence for a role of ECA4 and another related ANTH protein, CAP1 (AT4G32285), in CME comes from the recent identification of the TPLATE complex (Gadeyne et al., 2014), a tightly interacting multiprotein complex that includes ECA4 and CAP1. The eponymous TPLATE protein was originally identified as a factor in cytokinesis and pollen development with some similarity to other adaptor proteins (Van Damme et al., 2006). It is now clear that the TPLATE complex is essential for CME in plants both at the PM as well as the cell plate. It operates in concert with the AP-2 complex; indeed, AP2A1 and AP2S form part of the TPLATE complex network. However, AP-2 and TPLATE are not unvariably linked, and there are also isolated subpopulations of TPLATE as well as AP-2 at the PM, which indicate that each may serve a unique, nonoverlapping role. It seems that ECA4 and CAP1 interact directly with TML, one of the core components of the TPLATE complex (Gadeyne et al., 2014) (Table 1); however, an interaction study in the absence of any remaining bridging factors, such as clathrin, has not been performed to date. It will be interesting to see whether CAP1 and ECA4 indeed are specific accessory proteins for the TPLATE complex or if they can also associate with AP-2 and with what affinity. Even more telling would be a similar analysis for ECA1, which is phylogenetically far more distant from the former two (Figure 1). A more systematic analysis of ANTH interactions and their physiological roles would contribute to our understanding of CME in plants, as well as the bigger picture of evolution and specialization of CME in eukaryotes. Although the TPLATE complex has no clear homologs in animals and fungi, a recent analysis found the homologous TSET complex in a wide range of eukaryotic taxa, so it appears TPLATE/TSET is an evolutionarily ancestral complex that has been lost secondarily in some clades (Hirst et al., 2014).
As described above, all ANTH proteins characterized to date play a role in endocytosis. Most likely, the phosphoinositide binding properties of the ANTH domain itself are responsible for a strong affinity to the PM and nascent cell plate. Whether endocytosis at the PM and the cell plate are identical in terms of machinery is currently not known, but so far there is no evidence to suggest that this not the case. Whether ANTH proteins have additional non-PM-associated functions or if plant ENTH proteins under certain circumstances play a role in endocytosis is not known. In pull-down experiments using AP-2 α as bait, no plant EPSIN was found using mammalian anti-Epsin antibodies (Barth and Holstein, 2004), which hints to exclusive use of ANTHs in AP-2 related endocytosis, but more detailed studies would be needed for a conclusive answer.
In this respect, the outlier AT3G61800 (which we placed in group “U” on its own; Figure 1) is interesting because in this tree topology it is placed in between the ANTH and the ENTH group, though the branch is not supported with high confidence. No data about its localization, function, or gene expression are available. Structural analysis by Phyre2 reveals that its N terminus closely resembles that of the VHS domain of the human Golgi-localized, γ ear-containing, ARF binding protein 3 (hsGGA3), which is involved in the recycling of cargo to the PM from a sorting endosome (Parachoniak et al., 2011). Whether in Arabidopsis AT3G61800 is involved in a recycling pathway to the PM or, as suggested by other studies on human GGA (Puertollano et al., 2001), in vacuolar transport is currently not known.
TOLs: COUPLING ENDOCYTOSIS AND LYSOSOMAL DEGRADATION
LE/PVCs are membrane-delineated structures that contain small internal membrane-clad vesicles and are therefore called multivesiculate bodies (MVBs). The formation of the intraluminal vesicles topologically is entirely different from endocytosis or vesicle formation at the TGN because it requires negative membrane curvature (i.e., away from the cytosol). Correspondingly, the machinery involved in membrane reshaping is very different from that which drives the positive curvature in COP or CCV formation and will not be covered here (Wollert and Hurley, 2010; for an overview, see Henne et al., 2011). However, MVB formation also initiates with cargo recognition and recruitment of the remodeling machinery; in this aspect, it is not different from other vesicle-generating mechanisms.
The stepwise process of cargo recognition to intraluminal vesicle generation is performed by a series of protein complexes termed ESCRT-0 to ESCRT-III. The process and the machinery are highly conserved between the eukaryotic kingdoms, with one notable exception: ESCRT-0, which recognizes and sorts cargo and recruits ESCRT-I, is absent in plants. Instead, there is evidence that a group of proteins termed TARGET OF RAPAMYCIN LIKEs (TOLs) carries out a function comparable to that of ESCRT-0 (Korbei et al., 2013; for a mini review on the topic, see Sauer and Friml, 2014). This divergence regarding ESCRT-0 function is linked to another difference between plants and animals, with respect to the location of MVB formation: In animals, this process occurs on EEs that are commonly conceptualized as a sorting station for endocytosed cargo. The major phosphoinositide on the bounding membrane is PtdIns(3)P (Gillooly et al., 2000). The animal ESCRT-0 constituent HRS (Vps27 in yeast) binds PtdIns(3)P with high affinity through its FYVE domain, and is thus recruited to the EE. In plants, however, the sorting functions of the EE are performed by the TGN (Dettmer et al., 2006; Viotti et al., 2010), which is poor in PtdIns(3)P. Correspondingly, plant TOLs lack a FYVE domain.
The location of the interaction between TOLs and endocytosed polyubiquitinated membrane proteins destined for degradation is not clear at present. Interestingly, Mayers et al. (2013) identified a FEI complex that seems to recruit ESCRT-0 to a subset of clathrin coated pits on the plasma membrane in Caenorhabditis elegans. Loss of the complex does not affect the overall rate of clathrin-mediated endocytosis but specifically inhibits degradation of lysosomal cargo. Thus, the cargo-ESCRT-0 interaction is expected to begin at the PM, before reaching the endosome, and could play a role in concentrating cargo. It is unknown whether in plants the TOL-cargo interaction also occurs at the PM, but the data so far do not exclude this possibility. Further studies on phosphoinositide binding and interactors and detailed data on localization and dynamics should answer these questions. Interestingly, some of the results of Korbei et al. (2013) suggest that TOLs themselves might be subject to dynamic spatiotemporal regulation of their abundance at or close to the PM, which could be a means to modulate the endocytosis and degradation of PM effector proteins, such as channels, transporters, or receptors. The underlying mechanisms, however, are currently not known.
MTV1: A PLANT OUTSIDER?
Phylogenetic analysis shows MODIFIED TRANSPORT TO VACUOLE1 (MTV1) (AT3G16270) as the only member of a very distant clade (Figure 1) that seems to be conserved throughout the plant kingdom. Its sequence is so divergent that it has not been included in the largest phylogenetic analysis of the ENTH/ANTH/VHS superfamily in eukaryotes (De Craene et al., 2012). Nevertheless, its N terminus bears all hallmarks of an ENTH domain; indeed, before its functional characterization, MTV1 was subjected to structural NMR analysis (López-Méndez et al., 2004), and a comparison to known protein structures using Phyre2 reveals human Enthoprotein as the best scoring hit. A screen for mutants impaired in vacuolar transport (Sanmartín et al., 2007) identified a mtv1 loss-of-function mutant, and MTV1 was characterized functionally and biochemically (Sauer et al., 2013). Loss of MTV1 leads to mis-sorting of several endogenous and chimeric vacuolar marker proteins, which instead are secreted to the extracellular space. MTV1 localizes to the TGN, where it colocalizes with SYP61, VHA-a1, and CLC (Figure 3). It is also strongly enriched on purified CCVs, together with VSR1 and the γ-subunit of AP-1. Biochemical data show that it binds CHC via a single or multiple unknown motifs in the C-terminal region (Table 1). A candidate could be a LIDTG motif, which is highly conserved among all plant homologs, but this is yet unproven.
Another player in vacuolar transport was identified from the same screen (Sanmartín et al., 2007), the ARF-GAP5 NEVERSHED (AGD5/NEV) protein, which was previously identified as a factor required for floral organ abscission (Liljegren et al., 2009). Intriguingly, we found MTV1 and AGD5/NEV highly coexpressed in our network analysis (see above), hinting at a physiological link. Indeed, a mtv1 agd5/nev double mutant shows a dramatically enhanced overall phenotype (dwarfism) and strongly enhanced defects in vacuolar sorting, suggesting that MTV1 and AGD5/NEV act in a common pathway. This notion is supported by virtually identical expression patterns and complete colocalization at the TGN. Treatment of Arabidopsis cells with the trafficking inhibitor Brefeldin A causes the TGN to aggregate in so-called BFA bodies (Langhans et al., 2011). Intriguingly, both MTV1 and AGD5/NEV localize to the periphery of the BFA body, which may indicate the presence of different types of TGN. However, at present, this is speculation, and it may be that this localization simply reflects TGN from which CCVs are budding. Indeed, clathrin itself shows similar behavior upon BFA treatment (Ito et al., 2012). Both MTV1 and AGD5/NEV were shown by immunogold labeling to reside on purified CCVs. Currently, there is no way to determine the origin of these CCVs, but it seems reasonable to assume that they formed at the TGN, and the fact that loss of function of either MTV1 or AGD5/NEV leads to defective vacuolar transport suggests that their destination is the vacuole or the prevacuolar compartment. There is now evidence that, like in animals, the plant LE/PVC matures from the TGN (Scheuring et al., 2011). It was therefore suggested that vacuolar transport from the TGN does not involve CCVs. This is currently an ongoing debate (reviewed in Robinson and Pimpl, 2014) with experimental evidence on both sides. For definitive proof, CCVs would have to be traced from their origin to their destination, an experimentally daunting task, especially given the fact that the clathrin coat disassembles prior to fusion with the target membrane. Unless definite evidence is gathered, it seems prudent to assume that both TGN maturation as well as CCVs are mechanisms of vacuolar transport.
OPEN QUESTIONS
Functional Redundancy: Robustness versus Specificity
In the phylogenetic analysis, it is striking that some of the subgroups contain many members among the vascular plants (namely, the ANTHs and the TOLs), but only few of them have clear homologs in the more distantly related bryophyte and algal lineages. From a functional perspective, this expansion in the vascular plants could signify either specialization or increased robustness. Both ANTHs and TOLs are likely to be involved in processes at the PM, controlling the amounts of proteins by endocytosis and targeting for degradation. Since many PM proteins are regulators that interface with the environment (receptors, channels, transporters, etc.), it is conceivable that along with a more complex architecture and lifestyle, higher demands on a robust interaction with the environment have evolved. This seems to be the case for the TOLs where there is significant functional overlap: Only a quintuple mutant shows a phenotype and the patterns of expression broadly overlap (Korbei et al., 2013). This is consistent with increased robustness, reflecting on the importance of this pathway. For the ANTHs, insufficient functional data are available and further studies, including higher order mutants and expression analyses, are needed to elucidate functional specificity in this group.
Posttranslational Regulation
In animals, Epsins are phosphorylated during mitosis, resulting in lower affinity toward adaptor proteins (Chen et al., 1999; Kariya et al., 2000). In plants, to date, there is no evidence for phosphorylation, although it was speculated that At-AP180 is regulated via phosphorylation by a casein kinase because of the presence of four conserved casein kinase substrate sites in its primary sequence (Barth and Holstein, 2004). EPSIN1 was found as an interactor of the cyclin-dependent kinase inhibitor KIP RELATED PROTEIN4 (KRP4) in a TAP-Tag experiment (Van Leene et al., 2010), but whether this is related to a role of EPSIN1 in the cell cycle or a KRP4-dependent modulation of EPSIN1 activity has not been addressed experimentally to date. In animals, GGA proteins (which are similar to plant TOLs) can be phosphorylated by casein kinases associated with AP-1, and this phosphorylation modulates the affinity for membranes (Doray et al., 2002). Whether plant TOLs undergo a similar mechanism of regulation is unknown. In summary, the posttranslational modulation of ENTH/ANTH/VHS proteins is largely uncharted territory, and we expect important insights from its study in the future.
Phosphoinositide Binding
Vesicle formation is influenced by membrane phosphoinositide composition, yet our understanding of composition and also of phosphoinositide-specific interactions is limited in plants. Independently of the ENTH/ANTH/VHS proteins, it would be desirable to have a better understanding of how membrane phosphoinositide composition influences and guides the activity of regulatory effector proteins. Of all proteins covered in this review, phosphoinositide binding data are available only for EPSIN2, which predominantly binds PtdIns(3)P (Lee et al., 2007). Interestingly, Lee et al. (2007) indicate that they could not demonstrate phosphoinositide binding activity for EPSIN1, which might suggest that the commonly used lipid overlay (“fat blot”) technique is not the most suitable for this type of analyses. For mammalian Epsin, a high-resolution crystal structure in presence of d-myo-inositol-1,4,5-triphosphate has identified several key amino acids forming an interaction site that can accommodate PtdIns(4,5)P2 (Ford et al., 2002). Given that the ENTH domains are relatively well conserved between animals and plants, it might be possible to predict whether a similar binding interface is present in one of the plant ENTH proteins. Indeed, high affinity for PtdIns(4,5)P2 in any of the known plant Epsins would be a surprise, since this should go along with localization in the PM, in contrast to the currently reported localizations.
SUMMARY
Based on our phylogenetic analysis and by extrapolating findings from the literature, we propose a functional classification in which ENTH-containing proteins such as EPSINs and MTV1 play roles in post-Golgi (presumably vacuolar) transport, while ANTH-containing proteins are involved in endocytosis, and TOLs are required for the selection of PM-derived cargo for vacuolar degradation (Figure 3). We believe this classification is the most plausible based on the current knowledge. Because the data are still limited and many open questions remain, this classification should be taken as a work in progress.
METHODS
Phylogenetic Analysis
From the TAIR10 database, all proteins with an Interpro domain IPR008942=ENTH/VHS were selected. This group includes also proteins with a VHS-like domain in the center, rather than at the N terminus (see main text), which were excluded from further analysis. AGIs of excluded proteins are AT2G36480, AT2G36485, AT3G26990, AT4G04885, AT5G10060, AT5G10800, AT5G25060, and AT5G65180. Alignments of the remaining 35 proteins were performed using the ProbCons algorithm (Do et al., 2005), with two passes of consistency transformation, no pretraining, and 100 passes of iterative refinement. Phylogenetic analysis was performed using MEGA6 (Tamura et al., 2013), using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992) and 1000 bootstrap replicates. A midpoint-rooted tree with the highest log likelihood (−7744.0359) is shown in Figure 1. Bootstrap values are indicated above the branches. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with <95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 124 positions in the final data set. For the presence/absence analysis of homologs in other species (Volvox carteri, Chlamydomonas reinhardtii, Physcomitrella patens, Sellaginella moellendorffii, Oryza sativa, Sorghum bicolor, Zea mays, Vitis vinifera, Teobroma cacao, Carica papaya, Arabidopsis thaliana, Arabidopsis lyrata, Capsella rubella, Medicago truncatula, Glycine max, and Populus trichocarpa), we obtained sequences from the suggested protein homologs of Phytozome v10.0.2. The top three hits were chosen for subsequent reciprocal BLAST analysis, plus any other hit with a similarity greater than 50%. Splice isoforms, fragments, and duplicate entries were discarded. Reciprocal BLAST was performed locally using PrfctBlast v.2 with the TAIR10 database (Santiago-Sotelo and Ramirez-Prado, 2012). Only proteins that returned the original Arabidopsis query as first hit were retained for further analysis.
Transcriptional Network Analysis
From the manually curated list of the 35 Arabidopsis ENTH-domain containing genes, seven loci (AT1G14686, AT3G08790, AT3G61800, AT4G02650, AT4G32285, AT5G01760, and AT5G10410) are not represented in the microarray database and therefore could not be included in our analysis. The remaining genes were used as seeds in the ATTED-II (Obayashi et al., 2009) database, using default conditions. We chose to restrict the protein-protein interaction data to reduce a possible bias toward better characterized genes. Node centrality analysis was performed in the Cytoscape framework environment using the Hubba plug-in (Shannon et al., 2003; Lin et al., 2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/TAIR data libraries under the following accession numbers: ECA2, AT1G03050; AP180, AT1G05020; TOL2, AT1G06210; AT1G08670; AT1G14686; AT1G14910; TOL3, AT1G21380; AT1G25240; AT1G33340; AT1G68110; TOL4, AT1G76970; ECA1, AT2G01600; AT2G01920; ECA4, AT2G25430; AT2G36480; AT2G36485; TOL6, AT2G38410; EPSIN2, AT2G43160; TOL8, AT3G08790; MTV1, AT3G16270; AT3G23350; AT3G26990; AT3G46540; EPSIN3, AT3G59290; AT3G61800; AT4G02650; AT4G04885; AT4G25940; CAP1, AT4G32285; TOL9, AT4G32760; AT4G40080; TOL7, AT5G01760; AT5G10060; AT5G10410; AT5G10800; EPSIN1, AT5G11710; TOL1, AT5G16880; AT5G25060; AT5G35200; AT5G57200; TOL5, AT5G63640; AT5G65180; AT5G65370.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of Only the First 200 Amino Acids.
Supplemental Figure 2. Transcriptional Network Analysis as in Figure 2 with All Gene Identifiers Displayed.
Supplemental Data Set 1. Amino Acid Sequence Alignment for Phylogenetic Tree Shown in Figure 1.
Supplemental Data Set 2. Amino Acid Sequence Alignment for Phylogenetic Tree Shown in Supplemental Figure 1.
Supplemental Data Set 3. Genes Identified in the Expression Network Analysis with Gene Description and Results of Network Analysis.
Supplementary Material
Acknowledgments
We thank C. Alonso Blanco for helpful discussions on the phylogenetic aspects of this work. J.Z. is supported by the Ramón y Cajal program of the Spanish Ministerio de Economía y Competitividad (MINECO).
AUTHOR CONTRIBUTIONS
J.Z. performed the network analysis. M.S. performed the phylogenetic analysis. J.Z. and M.S. wrote the article.
Footnotes
Online version contains Web-only data.
References
- Baisa G.A., Mayers J.R., Bednarek S.Y. (2013). Budding and braking news about clathrin-mediated endocytosis. Curr. Opin. Plant Biol. 16: 718–725. [DOI] [PubMed] [Google Scholar]
- Barth M., Holstein S.E.H. (2004). Identification and functional characterization of Arabidopsis AP180, a binding partner of plant alphaC-adaptin. J. Cell Sci. 117: 2051–2062. [DOI] [PubMed] [Google Scholar]
- Baust T., Anitei M., Czupalla C., Parshyna I., Bourel L., Thiele C., Krause E., Hoflack B. (2008). Protein networks supporting AP-3 function in targeting lysosomal membrane proteins. Mol. Biol. Cell 19: 1942–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H.H., Antrobus R., Hirst J., Bhumbra G.S., Kozik P., Jackson L.P., Sahlender D.A., Robinson M.S. (2012). Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197: 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Pick A., Çamdere G., Liska N., Evergren E., McMahon H.T., Kozlov M.M. (2012). Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149: 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushlin I., Petralia R.S., Wu F., Harel A., Mughal M.R., Mattson M.P., Yao P.J. (2008). Clathrin assembly protein AP180 and CALM differentially control axogenesis and dendrite outgrowth in embryonic hippocampal neurons. J. Neurosci. 28: 10257–10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub, Web Presence Working Group (2009). AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Slepnev V.I., Di Fiore P.P., De Camilli P. (1999). The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals. J. Biol. Chem. 274: 3257–3260. [DOI] [PubMed] [Google Scholar]
- Chidambaram S., Müllers N., Wiederhold K., Haucke V., von Mollard G.F. (2004). Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 279: 4175–4179. [DOI] [PubMed] [Google Scholar]
- Dannhauser P.N., Ungewickell E.J. (2012). Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat. Cell Biol. 14: 634–639. [DOI] [PubMed] [Google Scholar]
- De Craene J.-O., Ripp R., Lecompte O., Thompson J.D., Poch O., Friant S. (2012). Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics 13: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmel L., et al. (2008). The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol. Biol. Cell 19: 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S., et al. (2013). The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do C.B., Mahabhashyam M.S.P., Brudno M., Batzoglou S. (2005). ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 15: 330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B., Ghosh P., Griffith J., Geuze H.J., Kornfeld S. (2002). Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297: 1700–1703. [DOI] [PubMed] [Google Scholar]
- Fan L., Hao H., Xue Y., Zhang L., Song K., Ding Z., Botella M.A., Wang H., Lin J. (2013). Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837. [DOI] [PubMed] [Google Scholar]
- Feraru E., Paciorek T., Feraru M.I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., Friml J. (2010). The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G.J., Pearse B.M.F., Higgins M.K., Vallis Y., Owen D.J., Gibson A., Hopkins C.R., Evans P.R., McMahon H.T. (2001). Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291: 1051–1055. [DOI] [PubMed] [Google Scholar]
- Ford M.G., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J., Evans P.R., McMahon H.T. (2002). Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366. [DOI] [PubMed] [Google Scholar]
- Gadeyne A., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704. [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.-M., Parton R.G., Stenmark H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19: 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.-M., Paris N., Robinson D.G., Holstein S.E.H. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693. [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Emr S.D. (2011). The ESCRT pathway. Dev. Cell 21: 77–91. [DOI] [PubMed] [Google Scholar]
- Hirst J., Schlacht A., Norcott J.P., Traynor D., Bloomfield G., Antrobus R., Kay R.R., Dacks J.B., Robinson M.S. (2014). Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife 3: e02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein S.E.H., Oliviusson P. (2005). Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: membrane tethers of the clathrin-dependent vesicle budding machinery. Protoplasma 226: 13–21. [DOI] [PubMed] [Google Scholar]
- Huang K.M., D’Hondt K., Riezman H., Lemmon S.K. (1999). Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 18: 3897–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Fujiwara M., Takahashi H., Sawasaki T., Endo Y., Seki M., Shinozaki K., Fukao Y., Yanagawa Y. (2009). Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J. Exp. Bot. 60: 3067–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., Nakano A. (2012). Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69: 204–216. [DOI] [PubMed] [Google Scholar]
- Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. (2001). Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Jones H.D., Holdsworth M.J., Hooley R. (1997). A full-length cDNA, AflO (Accession No. U80041), isolated from wild oat (Avena fatua) aleurone encodes a protein with similarity to clathrin assembly proteins (PGR 97-018). Plant Physiol. 113: 665. [Google Scholar]
- Joy M.P., Brock A., Ingber D.E., Huang S. (2005). High-betweenness proteins in the yeast protein interaction network. J. Biomed. Biotechnol. 2005: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. (2005). Cytokinesis in higher plants. Annu. Rev. Plant Biol. 56: 281–299. [DOI] [PubMed] [Google Scholar]
- Langhans M., Förster S., Helmchen G., Robinson D.G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62: 2949–2957. [DOI] [PubMed] [Google Scholar]
- Lee G.-J., Kim H., Kang H., Jang M., Lee D.W., Lee S., Hwang I. (2007). EpsinR2 interacts with clathrin, adaptor protein-3, AtVTI12, and phosphatidylinositol-3-phosphate. Implications for EpsinR2 function in protein trafficking in plant cells. Plant Physiol. 143: 1561–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V., Wasiak S., Hussain N.K., Angers A., McPherson P.S. (2004). ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 117: 9–18. [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Chin C.H., Wu H.H., Chen S.H., Ho C.W., Ko M.T. (2008). Hubba: hub objects analyzer—a framework of interactome hubs identification for network biology. Nucleic Acids Res. 36: W438-W443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren S.J., Leslie M.E., Darnielle L., Lewis M.W., Taylor S.M., Luo R., Geldner N., Chory J., Randazzo P.A., Yanofsky M.F., Ecker J.R. (2009). Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 136: 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O., Poussu A., Mao Y., Quiocho F., Lehto V.-P. (2002). VHS domain — a longshoreman of vesicle lines. FEBS Lett. 513: 19–23. [DOI] [PubMed] [Google Scholar]
- López-Méndez B., et al. (2004). NMR assignment of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana. J. Biomol. NMR 29: 205–206. [DOI] [PubMed] [Google Scholar]
- Kariya K., Koyama S., Nakashima S., Oshiro T., Morinaka K., Kikuchi A. (2000). Regulation of complex formation of POB1/epsin/adaptor protein complex 2 by mitotic phosphorylation. J. Biol. Chem. 275: 18399–18406. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Xu Z.-Y., Song K., Kim D.H., Kang H., Reichardt I., Sohn E.J., Friml J., Juergens G., Hwang I. (2013). Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbei B., Moulinier-Anzola J., De-Araujo L., Lucyshyn D., Retzer K., Khan M.A., Luschnig C. (2013). Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 23: 2500–2505. [DOI] [PubMed] [Google Scholar]
- Mao Y., Nickitenko A., Duan X., Lloyd T.E., Wu M.N., Bellen H., Quiocho F.A. (2000). Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100: 447–456. [DOI] [PubMed] [Google Scholar]
- Mayers J.R., Wang L., Pramanik J., Johnson A., Sarkeshik A., Wang Y., Saengsawang W., Yates J.R. III, Audhya A. (2013). Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc. Natl. Acad. Sci. USA 110: 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messa M., Fernández-Busnadiego R., Sun E.W., Chen H., Czapla H., Wrasman K., Wu Y., Ko G., Ross T., Wendland B.. andCamilli P.D. (2014). Epsin deficiency impairs endocytosis by stalling the actin-dependent invagination of endocytic clathrin-coated pits. eLife Sciences 3: e03311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.E., Collins B.M., McCoy A.J., Robinson M.S., Owen D.J. (2007). A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450: 570–574. [DOI] [PubMed] [Google Scholar]
- Miller S.E., Sahlender D.A., Graham S.C., Höning S., Robinson M.S., Peden A.A., Owen D.J. (2011). The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell 147: 1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills I.G., Praefcke G.J.K., Vallis Y., Peter B.J., Olesen L.E., Gallop J.L., Butler P.J.G., Evans P.R., McMahon H.T. (2003). EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M., Takemoto N., Hashiguchi Y., Tasaka M., Morita M.T. (2009). ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 50: 2057–2068. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachoniak C.A., Luo Y., Abella J.V., Keen J.H., Park M. (2011). GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev. Cell 20: 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Jürgens G. (2012). Membrane traffic and fusion at post-Golgi compartments. Front. Plant Sci. 2: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Song K., Reichardt I., Kim H., Mayer U., Stierhof Y.-D., Hwang I., Jürgens G. (2013). Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc. Natl. Acad. Sci. USA 110: 10318–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Randazzo P.A., Presley J.F., Hartnell L.M., Bonifacino J.S. (2001). The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105: 93–102. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J.S. (2004). Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 6: 244–251. [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673. [DOI] [PubMed] [Google Scholar]
- Robinson M.S. (2004). Adaptable adaptors for coated vesicles. Trends Cell Biol. 14: 167–174. [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Pimpl P. (2014). Clathrin and post-Golgi trafficking: a very complicated issue. Trends Plant Sci. 19: 134–139. [DOI] [PubMed] [Google Scholar]
- Rosenthal J.A., Chen H., Slepnev V.I., Pellegrini L., Salcini A.E., Di Fiore P.P., De Camilli P. (1999). The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J. Biol. Chem. 274: 33959–33965. [DOI] [PubMed] [Google Scholar]
- Saffarian S., Cocucci E., Kirchhausen T. (2009). Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 7: e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín M., Ordóñez A., Sohn E.J., Robert S., Sánchez-Serrano J.J., Surpin M.A., Raikhel N.V., Rojo E. (2007). Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Sotelo P., Ramirez-Prado J.H. (2012). prfectBLAST: a platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques 53: 299–300. [DOI] [PubMed] [Google Scholar]
- Sauer M., Delgadillo M.O., Zouhar J., Reynolds G.D., Pennington J.G., Jiang L., Liljegren S.J., Stierhof Y.-D., De Jaeger G., Otegui M.S., Bednarek S.Y., Rojo E. (2013). MTV1 and MTV4 encode plant-specific ENTH and ARF GAP proteins that mediate clathrin-dependent trafficking of vacuolar cargo from the trans-Golgi network. Plant Cell 25: 2217–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Friml J. (2014). Plant biology: gatekeepers of the road to protein perdition. Curr. Biol. 24: R27–R29. [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H., Shibata M., Ushijima T., Nakashima M., Kong S.-G., Matsuoka K., Lin C., Matsushita T. (2012). The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70: 727–738. [DOI] [PubMed] [Google Scholar]
- Skruzny M., Brach T., Ciuffa R., Rybina S., Wachsmuth M., Kaksonen M. (2012). Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. USA 109: E2533–E2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Lee M.H., Lee G.-J., Yoo C.M., Hwang I. (2006). Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18: 2258–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Jang M., Kim S.Y., Lee G., Lee G.-J., Kim D.H., Lee Y., Cho W., Hwang I. (2012). An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol. 159: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.J., Perrais D., Merrifield C.J. (2011). A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9: e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.-K., Shimono Y., Shirakawa M., Fukao Y., Tamura K., Shimada T., Hara-Nishimura I. (2013). The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol. 54: 838–847. [DOI] [PubMed] [Google Scholar]
- Van Damme D., Coutuer S., De Rycke R., Bouget F.-Y., Inzé D., Geelen D. (2006). Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18: 3502–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yan X., Chen Q., Jiang N., Fu W., Ma B., Liu J., Li C., Bednarek S.Y., Pan J. (2013). Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25: 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-G., Li S., Zhao X.-Y., Zhou L.-Z., Huang G.-Q., Feng C., Zhang Y. (2013). HAPLESS13, the Arabidopsis μ1 adaptin, is essential for protein sorting at the trans-Golgi network/early endosome. Plant Physiol. 162: 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenstetter S., Wirsching P., Dotzauer D., Schneider S., Sauer N. (2012). Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell 24: 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J.H. (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464: 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S., Shimono Y., Shirakawa M., Fukao Y., Kawase T., Hatsugai N., Tamura K., Shimada T., Hara-Nishimura I. (2013). Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25: 2958–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Kim P.M., Sprecher E., Trifonov V., Gerstein M. (2007). The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLOS Comput. Biol. 3: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Möller B., Hwang I., Feraru M.I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21: 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.