RDO5 encodes a phosphatase that suppresses transcript levels of several genes encoding members of the PUF family of RNA binding proteins and plays a positive role during seed dormancy.
Abstract
Seed dormancy determines germination timing and contributes to crop production and the adaptation of natural populations to their environment. Our knowledge about its regulation is limited. In a mutagenesis screen of a highly dormant Arabidopsis thaliana line, the reduced dormancy5 (rdo5) mutant was isolated based on its strongly reduced seed dormancy. Cloning of RDO5 showed that it encodes a PP2C phosphatase. Several PP2C phosphatases belonging to clade A are involved in abscisic acid signaling and control seed dormancy. However, RDO5 does not cluster with clade A phosphatases, and abscisic acid levels and sensitivity are unaltered in the rdo5 mutant. RDO5 transcript could only be detected in seeds and was most abundant in dry seeds. RDO5 was found in cells throughout the embryo and is located in the nucleus. A transcriptome analysis revealed that several genes belonging to the conserved PUF family of RNA binding proteins, in particular Arabidopsis PUMILIO9 (APUM9) and APUM11, showed strongly enhanced transcript levels in rdo5 during seed imbibition. Further transgenic analyses indicated that APUM9 reduces seed dormancy. Interestingly, reduction of APUM transcripts by RNA interference complemented the reduced dormancy phenotype of rdo5, indicating that RDO5 functions by suppressing APUM transcript levels.
INTRODUCTION
The moment when a seed germinates is a crucial decision during the life cycle of plants because it determines all subsequent events. Seed dormancy is defined as the failure of a viable seed to germinate under favorable conditions (Bewley, 1997) and plays an adaptive role in nature by optimizing germination at the most suitable time. Seed dormancy is induced during maturation of seeds on the mother plant and released by dry storage (after-ripening) or imbibition at low temperatures. A tight control of dormancy is important in many crop species, including rapeseed (Brassica napus), wheat (Triticum spp), barley (Hordeum vulgare), and rice (Oryza sativa). Low levels of dormancy can cause preharvest spouting, which can cause economic losses to cereal production including reductions in seed quantity and quality. On the other hand, low seed dormancy is required for uniform and fast germination after sowing (Gubler et al., 2005; Holdsworth et al., 2008; Graeber et al., 2012). An improved control of seed dormancy and germination requires understanding of its molecular mechanisms.
Several genes regulating dormancy have been identified during the last decades. Many of these are involved in the biosynthesis and signaling of hormones among which abscisic acid (ABA) plays a major role (Gubler et al., 2005; Holdsworth et al., 2008; Graeber et al., 2012). Mutant analyses of ABA biosynthesis and metabolism genes showed the essential role of ABA in the induction and maintenance of seed dormancy. Seeds of ABA-deficient mutants have lower ABA and dormancy levels than those of the wild type (Koornneef et al., 1982; Lefebvre et al., 2006). By contrast, seeds of ABA catabolic pathway mutants accumulate higher ABA levels than those of the wild type and exhibit enhanced dormancy levels (Millar et al., 2006; Okamoto et al., 2006). Genes involved in the ABA signal transduction pathway also influence seed dormancy (Colucci et al., 2002; Shen et al., 2006; Raghavendra et al., 2010). In particular, members of the Protein Phosphatase 2C (PP2C) family were identified as components of ABA signaling from work with the aba insensitive1-1 (abi1-1) and abi2-1 mutants (Koornneef et al., 1984; Rodriguez et al., 1998). At least four Arabidopsis thaliana PP2Cs, including ABI1, ABI2, ABA HYPERSENSITIVE GERMINATION1 (AHG1), and HIGHLY ABA-INDUCED PP2C GENE2 (HAI2), are key regulators of ABA signaling and function as negative regulators of seed dormancy (Rodriguez et al., 1998; Gosti et al., 1999; Nishimura et al., 2007; Kim et al., 2013). Members of the PYR/PYL/RCAR family of ABA receptors bind and inactivate these PP2C proteins in an ABA-dependent manner, allowing SNF1-related protein kinase subfamily 2 (SnRK2) protein kinases to phosphorylate downstream substrates, thereby acting as positive regulators of seed dormancy (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Soon et al., 2012).
Besides ABA, gibberellins (GAs), ethylene, strigolactone, and brassinosteroid also influence seed dormancy and germination (Steber and McCourt, 2001; Gubler et al., 2005; Finkelstein et al., 2008; Nelson et al., 2011; Graeber et al., 2012). In general, these hormones reduce seed dormancy and promote seed germination. For example, mutants severely defective in GA biosynthesis such as ga requiring1 show deep seed dormancy and fail to germinate in the absence of exogenous GA (Debeaujon and Koornneef, 2000; Ogawa et al., 2003). Ethylene can regulate dormancy by affecting ABA levels and signal transduction (Linkies et al., 2009).
Apart from plant hormones, several chromatin factors have recently been shown to be involved in the regulation of seed dormancy. HISTONE MONOUBIQUITINATION1 and REDUCED DORMANCY2 have been identified in a screen for reduced dormancy mutants (Peeters et al., 2002). Both proteins probably influence seed dormancy by regulating transcription elongation of seed dormancy genes during maturation (Liu et al., 2007, 2011). Two other chromatin factors, SIN3-LIKE1 (SNL1) and SNL2, positively regulate seed dormancy by modifying the ABA-ethylene antagonism through histone acetylation (Z. Wang et al., 2013). A role for histone acetylation in seed dormancy was recently also demonstrated by the reduced dormancy phenotype of the histone deacetylase9 mutant (van Zanten et al., 2014).
Finally, modifications of proteins and transcripts influence dormancy and germination. Oxidative processes affecting both proteins and transcripts during the storage of dry seeds have been shown to influence the release of dormancy (Oracz et al., 2007; Bazin et al., 2011; Gao et al., 2013). Posttranslational modifications during seed imbibition, such as oxidation, phosphorylation/dephosphorylation, ubiquitination, and acetylation, affect protein function and are probably involved in the control of germination (Le et al., 1998; Buchanan and Balmer, 2005; Lu et al., 2008; Arc et al., 2011).
The above-mentioned regulators are not specific for dormancy but also influence other plant traits. By contrast, DELAY OF GERMINATION1 (DOG1) has been identified as a dormancy-specific gene in Arabidopsis (Bentsink et al., 2006). DOG1 encodes a protein with unknown function and is part of a small gene family. DOG1 homologs have been found in both monocots and dicots and for several of them a conserved function in seed dormancy has been demonstrated (Ashikawa et al., 2013; Graeber et al., 2014). The abundance of DOG1 in freshly harvested seeds highly correlates with dormancy levels. DOG1 becomes modified during seed storage, leading to its reduced functionality (Nakabayashi et al., 2012).
Most lab accessions of Arabidopsis have low seed dormancy levels, decreasing the efficiency of mutagenesis screens for reduced dormancy. Here, we describe the isolation of the reduced dormancy5 (rdo5) mutant in a screen of a highly dormant Arabidopsis genotype containing a strong DOG1 allele. RDO5 encodes a PP2C protein that positively regulates seed dormancy without influencing ABA and DOG1 levels. The rdo5 mutant phenotype requires expression of the Arabidopsis PUMILIO9 (APUM9) and APUM11 genes that encode RNA binding proteins. RDO5 represents a novel dormancy pathway that probably regulates seed dormancy at the posttranscriptional level.
RESULTS
RDO5 Is Required for Seed Dormancy
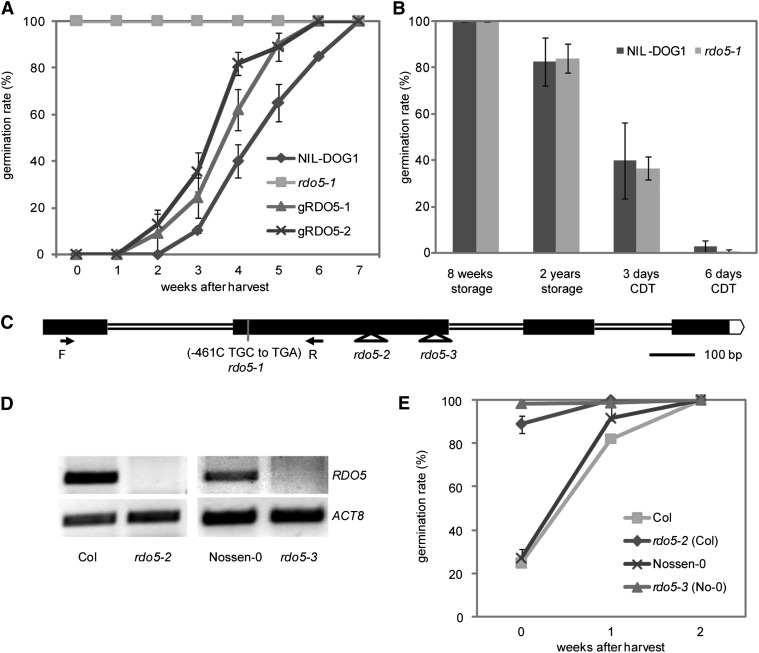
To identify genes with a role in seed dormancy, the highly dormant near isogenic line NIL-DOG1 was mutagenized by γ-irradiation. The genotype of NIL-DOG1 consists of the Arabidopsis accession Landsberg erecta (Ler) containing a 4.5-Mb introgression from the Cape Verde Islands (Cvi) accession in chromosome 5, including a strong DOG1 allele (Alonso-Blanco et al., 2003). DOG1 has been identified as a major regulator of seed dormancy in Arabidopsis (Bentsink et al., 2006). Several mutants with abolished or reduced seed dormancy were isolated in this mutagenesis screen, among them a nondormant mutant that was later identified as dog1-1 (Bentsink et al., 2006). Another isolated mutant with strongly reduced seed dormancy was named rdo5. We analyzed the germination of seeds from homozygous rdo5 mutant plants in detail and confirmed their reduced dormancy phenotype (Figure 1A). Besides the absence of dormancy, the dog1 mutant is also characterized by a reduction in seed longevity (Bentsink et al., 2006). Analysis of the germination of rdo5 seeds after controlled deterioration or long-term storage showed a similar seed longevity as wild-type seeds (Figure 1B). In addition, rdo5 mutant plants did not show any other obvious pleiotropic phenotypes (Supplemental Figure 1), suggesting that the function of RDO5 is specific for seed dormancy.
Figure 1.
The rdo5 Mutant Shows Reduced Dormancy.
(A) Germination after different periods of dry storage of rdo5-1, its wild-type background NIL-DOG1, and two complementation lines containing a pRDO5:RDO5 insertion in the rdo5-1 background. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(B) Seed longevity phenotype of rdo5-1 and NIL-DOG1 after long time storage and controlled deterioration (CDT). Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(C) Schematic diagram of the RDO5 gene structure including the position of the rdo5-1 mutation and the two T-DNA insertions. Exons are shown as black boxes and introns as lines. The positions of the primers used for RT-PCR analysis are indicated below the structure (arrows).
(D) RT-PCR analysis of the RDO5 transcripts in freshly harvested seeds of wild-type and T-DNA insertion mutants. ACTIN8 (ACT8) was used as a loading control.
(E) Germination after different periods of dry storage of the T-DNA insertion lines rdo5-2 and rdo5-3 and their respective wild-type backgrounds Col and Nossen. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
RDO5 Encodes a PP2C Phosphatase
To identify the RDO5 gene, we followed a map-based cloning approach. The rdo5 mutant was crossed with the Columbia (Col) accession to produce a mapping population. Plants containing the Cvi DOG1 allele were selected in the F2 progeny of this cross and separated in a dormant and a nondormant group according to their germination phenotype. Hybridization of the bulk DNA of these two groups to the Arabidopsis SNP Genotyping and Tiling Array showed that RDO5 is localized on chromosome 4 (Supplemental Figure 2A). Further fine mapping narrowed down the location of RDO5 to a 17-kb region corresponding to the BAC clone T22B4 containing five predicted open reading frames. We sequenced this region in wild-type NIL-DOG1 and the rdo5 mutant and detected a single base pair deletion in the rdo5 ORF4, corresponding to the gene At4g11040. This mutation was located in the second exon of RDO5 and resulted in a stop codon (Supplemental Figure 2B). At4g11040 encodes a member of the PP2C gene family in Arabidopsis. This family consists of 10 clades named A to J. Most of the proteins belonging to clade A are involved in ABA signaling and seed dormancy regulation. However, RDO5 has a unique composition and could not be clustered within one of the clades (Schweighofer et al., 2004).
To confirm that At4g11040 is RDO5, we complemented the rdo5 mutant with a 6-kb Ler genomic fragment including a 2.7-kb promoter sequence, the At4g11040 coding sequence, and a 0.9-kb sequence after the stop codon. T3 seeds from two independent homozygous single insertion lines showed similar dormancy levels as seeds from wild-type NIL-DOG1 plants, indicating that the 6-kb fragment is sufficient to complement the reduced dormancy phenotype of the rdo5 mutant (Figure 1A).
We named our original mutant allele rdo5-1 and obtained additional insertion mutant alleles for RDO5 in the Col and Nossen-0 background that were named rdo5-2 and rdo5-3, respectively. Both insertions are located in the second exon of RDO5 as shown in Figure 1C. RT-PCR analysis, using RDO5 gene-specific primers, showed that RDO5 transcript could not be detected in the two insertion mutants (Figure 1D). Similar to the rdo5-1 allele, rdo5-2 and rdo5-3 showed strongly reduced seed dormancy levels compared with their corresponding wild-type Col and Nossen-0 (Figure 1E).
RDO5 Is a Seed-Specific Protein That Is Located in the Nucleus
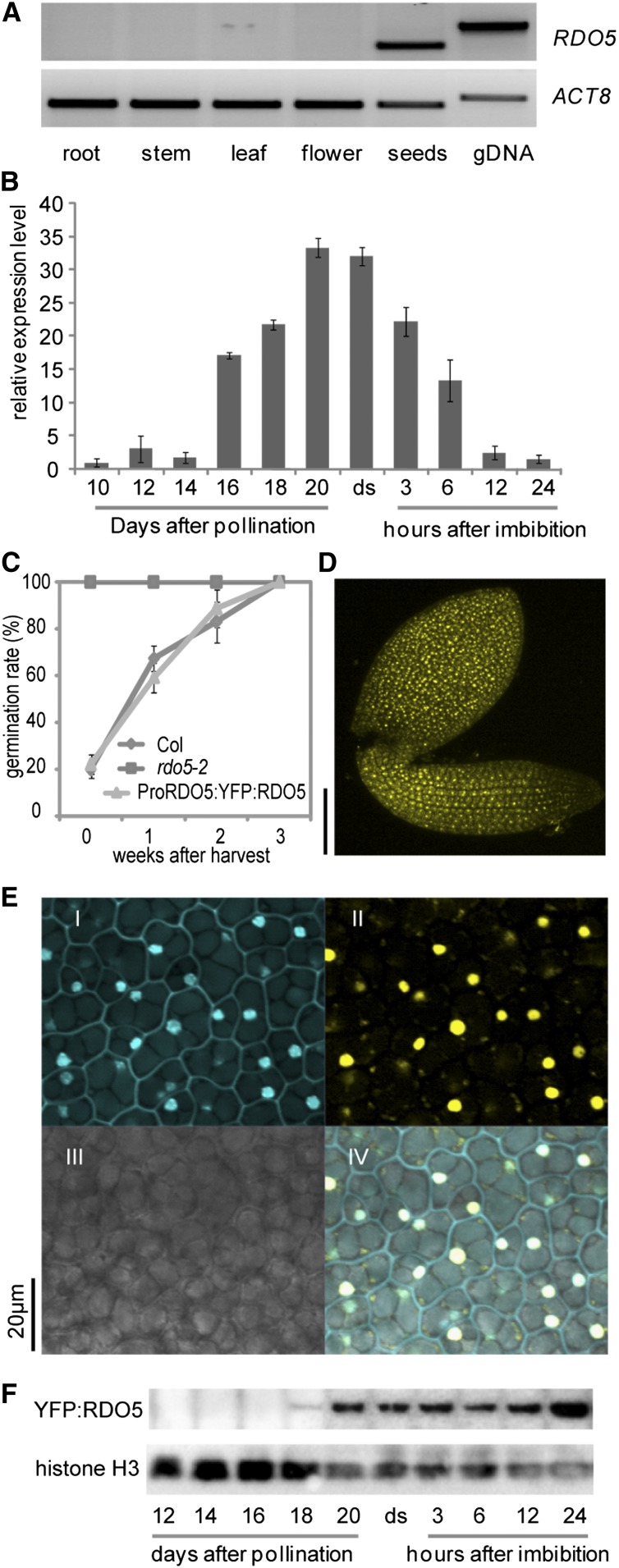
The identification of RDO5 as a dormancy gene suggests that its transcript and protein can be found in seeds. Public data about RDO5 transcript levels are not available because this gene is not represented on the ATH1 microarray. RT-PCR analysis confirmed the presence of RDO5 transcript in dry seeds, but it could not be detected in root, stem, leaf, or flower tissues (Figure 2A). The seed-specific transcript pattern of RDO5 is consistent with the location of its protein, which was only detected in seeds in a genome-scale proteomics analysis (http://fgcz-atproteome.unizh.ch/; Baerenfaller et al., 2008).
Figure 2.
RDO5 Transcripts Are Seed-Specific and Its Encoded Protein Is Localized in the Nucleus.
(A) RT-PCR analysis of RDO5 transcript levels in various tissues. ACT8 was used as a loading control.
(B) qRT-PCR analysis of RDO5 transcript levels during seed maturation and imbibition. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(C) Germination after different periods of dry storage of wild-type Col, rdo5-2, and a transgenic rdo5 line containing the ProRDO5:YFP:RDO5 construct. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(D) Localization of YFP:RDO5 in an embryo of a freshly harvested seed from a transgenic rdo5 plant containing the ProRDO5:YFP:RDO5 construct. Bar = 100 μm.
(E) Subcellular localization of YFP:RDO5 in embryo cells from a transgenic rdo5-2 seed containing the ProRDO5:YFP:RDO5 construct. I to IV show 4′,6-diamidino-2-phenylindole signal, YFP signal, transmission, and merged images, respectively. Bar = 20 μm.
(F) Immunoblot analysis of YFP:RDO5 protein accumulation in the rdo5-2 complementation line during seed maturation and imbibition. Histone H3 was used as loading control. ds, freshly harvested dry seeds.
The temporal pattern of RDO5 transcript levels in seeds was analyzed in detail by quantitative RT-PCR (qRT-PCR). RDO5 transcript levels strongly increased during seed maturation from ∼16 d after pollination and reached a maximum in freshly harvested dry seeds. The transcript levels quickly decreased during imbibition of seeds and became very low after 24 h (Figure 2B).
The abundance and localization of the RDO5 protein was studied using transgenic plants in an rdo5-2 mutant background containing the RDO5 cDNA fragment with an N-terminal yellow fluorescent protein (YFP) tag driven by the RDO5 native promoter. One homozygous single insertion line that complemented the rdo5 reduced dormancy phenotype was selected (Figure 2C). The localization of RDO5 in freshly harvested seeds was studied by detection of the YFP:RDO5 protein with the confocal microscope. YFP fluorescence could be observed in the entire embryo and was confined to nuclei, indicating that RDO5 is a nuclear protein (Figures 2D and 2E). An immunoblot analysis showed that RDO5 protein started to accumulate at 16 d after pollination until the end of seed maturation, which is similar to the dynamics of RDO5 transcript. In contrast to its transcript, protein levels slightly increased during imbibition, indicating that the RDO5 protein is very stable (Figure 2F).
RDO5 Expression Correlates with Seed Dormancy Levels
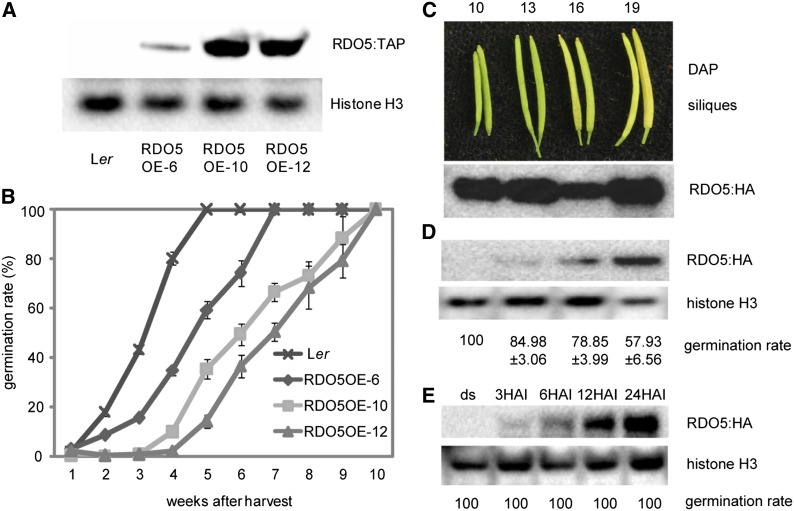
The phenotype of the rdo5 mutant alleles showed that the absence of RDO5 leads to strongly reduced seed dormancy. This raised the question of whether an increase in RDO5 could enhance dormancy. We constructed transgenic Ler plants containing the RDO5 genomic fragment with a C-terminal tandem affinity purification (TAP) tag driven by the 35S promoter. Three independent single insertion lines (RDO5OE-6, RDO5OE-10, and RDO5OE-12) were obtained that accumulated varying amounts of RDO5:TAP in their seeds (Figure 3A). All three lines showed enhanced seed dormancy levels that correlated with the amount of RDO5:TAP protein that they accumulated (Figure 3B).
Figure 3.
Constitutive Expression of RDO5 During Seed Maturation Enhances Seed Dormancy.
(A) Immunoblot analysis of RDO5:TAP protein accumulation in seeds of three independent RDO5:TAP overexpression transgenic lines.
(B) Germination after different periods of dry storage of RDO5:TAP overexpression transgenic lines and wild-type Ler. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(C) Immunoblot analysis of RDO5:HA protein accumulation of transgenic rdo5-2 lines, containing RDO5 driven by the heat shock promoter (ProHS). Protein was isolated from siliques after they had received 3 h of heat induction (37°C) during three subsequent days that started at 10, 13, 16, or 19 d after pollination. The top panel shows representative siliques of the four stages.
(D) Immunoblot analysis of RDO5:HA protein in freshly harvested seeds from siliques that had been heat treated at different stages after pollination as indicated in (C). The germination percentage (averages ± se) of seeds from six independent batches for each genotype is shown at the bottom.
(E) Immunoblot analysis of RDO5:HA protein in after-ripened seeds after 6 h of heat induction that was given at 3, 6, 12, and 24 h after imbibition. The germination percentage (averages ± se) of seeds from six independent batches for each genotype is shown at the bottom.
[See online article for color version of this figure.]
The influence of RDO5 on seed dormancy was analyzed in more detail by temporal induction of RDO5 expression. For this purpose, we created transgenic rdo5-2 plants containing a single insertion of a construct consisting of the RDO5 cDNA fused with a C-terminal HA tag, driven by a heat shock promoter. Plants were grown at 16/14°C to reduce background activity of the heat shock promoter. RDO5 expression was induced at different times during seed maturation by applying a 3-h heat shock at 37°C for three consecutive days. An immunoblot analysis that was performed directly after the treatment showed high accumulation of RDO5 protein at all silique stages (Figure 3C). These high amounts of RDO5 protein were not stable, and freshly harvested seeds only showed high levels of RDO5 protein when this was induced at the later stages of seed maturation (Figure 3D). The germination rate of seeds harvested from these siliques showed an inverse correlation with the abundance of RDO5 protein, indicating that high levels of RDO5 induced at the end of seed maturation prevent germination (Figure 3D). By contrast, induction of RDO5 in after-ripened seeds during imbibition could not prevent germination of these seeds (Figure 3E). These results suggest that the RDO5 protein is functional at the later stages of seed maturation and in dry seeds, which is consistent with the temporal pattern of RDO5 protein accumulation in wild-type seeds (Figure 2C). The presence of RDO5 protein at the early stages of seed maturation or during seed imbibition does not prevent germination (Figures 3C to 3E).
RDO5 Does Not Influence ABA and DOG1 Levels
The plant hormone ABA and the dormancy protein DOG1 have been identified as major factors for the induction of seed dormancy in Arabidopsis (Graeber et al., 2012). We analyzed whether the reduced seed dormancy phenotype of the rdo5-1 mutant is associated with changes in ABA or DOG1.
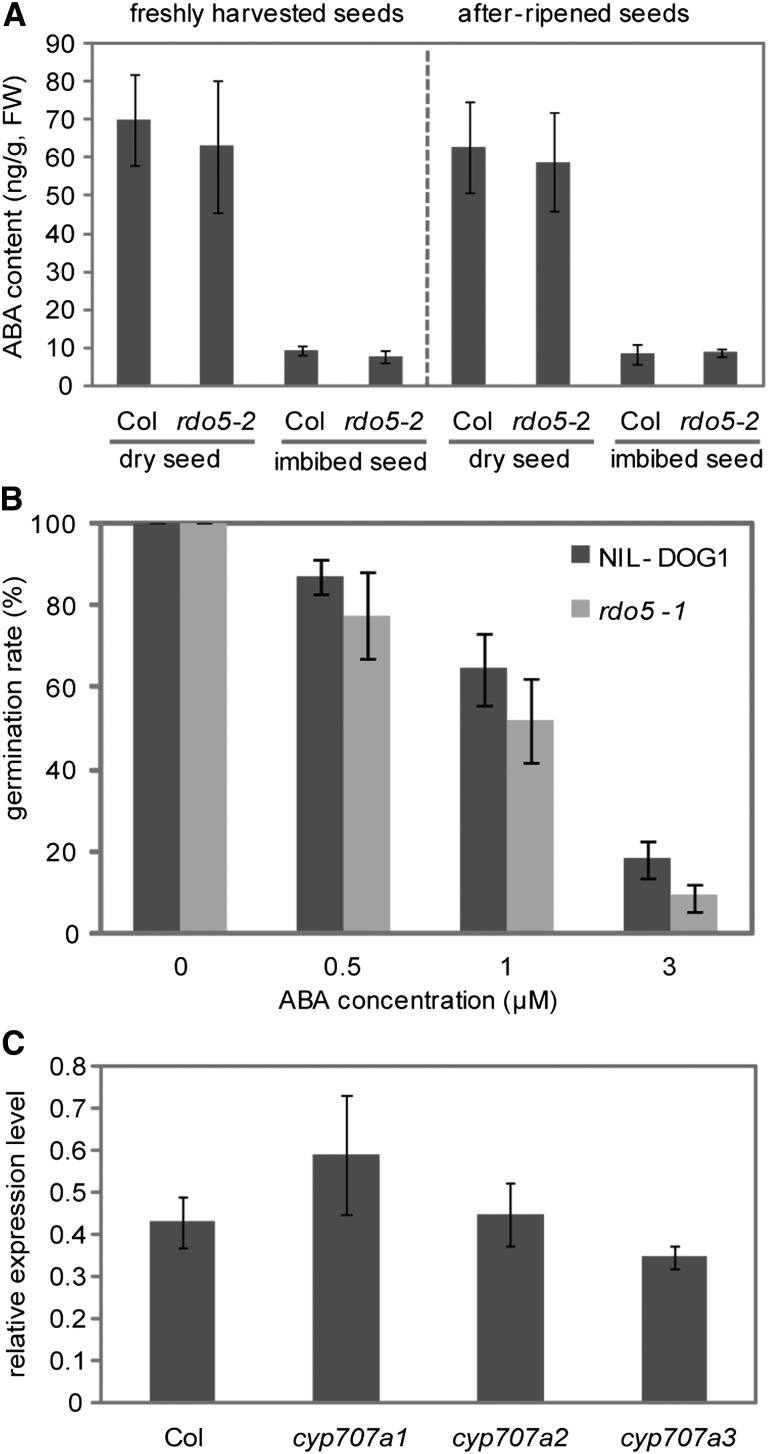
The ABA levels in rdo5-2 are very similar to those of wild-type Col in freshly harvested and after-ripened seeds, as well as in dry and imbibed seeds (Figure 4A). Reduced dormancy can also be associated with decreased sensitivity to ABA. However, Figure 4B shows that the ABA sensitivity of rdo5-1 seeds is not significantly different from wild-type NIL-DOG1 seeds. In addition, we analyzed whether enhanced ABA levels in seeds can influence RDO5 transcript levels by studying the cyp707a1, cyp707a2, and cyp707a3 mutants. CYP707A1, 2, and 3 are ABA 8’-hydroxylases with a key role in ABA catabolism. Mutations in these genes cause enhanced ABA levels and dormancy phenotypes in different degrees (Okamoto et al., 2006). RDO5 transcript levels in freshly harvested seeds of these mutants did not significantly differ from those of the wild type (Figure 4C). Taken together, our results indicated that RDO5 and ABA control seed dormancy independently.
Figure 4.
RDO5 and ABA Do Not Influence Each Other’s Levels.
(A) ABA contents in dry and 36-h imbibed wild-type Col and rdo5-2 seeds that were either freshly harvested or after-ripened. Shown are averages ± se calculated from three biological repeats.
(B) Germination rate of after-ripened wild-type NIL-DOG1 and rdo5-1 seeds sown on wet filter paper containing different concentrations of ABA. Shown are averages ± se of more than eight independent batches of seeds for each genotype.
(C) qRT-PCR analysis of RDO5 transcript levels in freshly harvested seeds of wild-type Col and the cyp707a1-a3 mutants. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
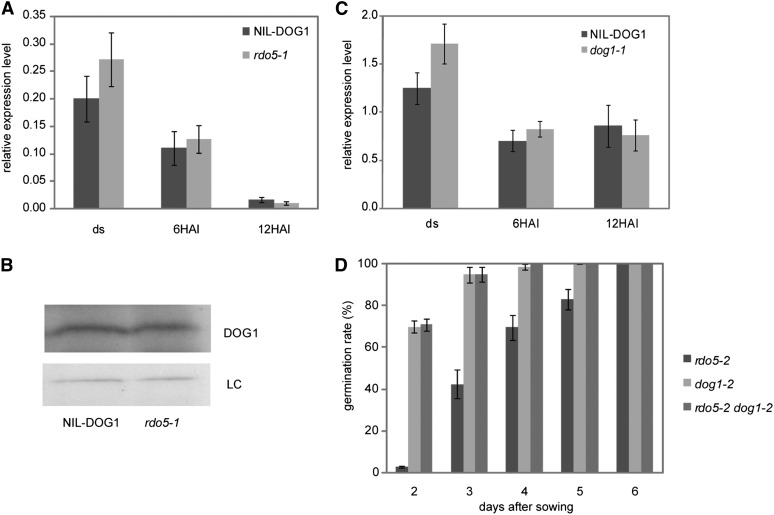
We previously showed that the levels of DOG1 transcript and protein in freshly harvested seeds are highly correlated with dormancy (Nakabayashi et al., 2012). This correlation is lost in the rdo5-1 mutant, which has very low dormancy levels but high DOG1 transcript and protein levels similar to those of wild-type NIL-DOG1 (Figures 5A and 5B). This suggests that RDO5 does not regulate DOG1 levels, but DOG1 requires RDO5 to induce seed dormancy. DOG1 does not influence RDO5 transcript levels because they are similar in the dog1-1 mutant and wild-type NIL-DOG1 seeds (Figure 5C). The relation between DOG1 and RDO5 was further studied by analysis of the rdo5-2 and dog1-2 single and double mutants. As expected, these mutants are all nondormant. Interestingly, a detailed analysis showed that the dog1-2 mutant germinates faster than the rdo5-2 mutant (Figure 5D). Faster germination is usually correlated with reduced dormancy, indicating that dog1 has a stronger influence on seed dormancy than rdo5. The double mutant rdo5-2 dog1-2 showed a similar germination speed as the single dog1-2 mutant, indicating that the high germination speed of dog1-2 cannot be further enhanced by the rdo5-2 mutation.
Figure 5.
RDO5 and DOG1 Do Not Influence Each Other’s Levels.
(A) qRT-PCR analysis of DOG1 transcript levels in freshly harvested and imbibed seeds of NIL-DOG1 and rdo5-1. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(B) Immunoblot analysis of DOG1 protein in NIL-DOG1 and rdo5-1 freshly harvested seeds. DOG1 was detected using DOG1-specific antibody. A nonspecific band of around 60 kD is used as loading control (LC).
(C) qRT-PCR analysis of RDO5 transcript levels in freshly harvested and imbibed seeds of NIL-DOG1 and dog1-1. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(D) Germination percentages of freshly harvested seeds from rdo5-2, dog1-2, and rdo5-2 dog1-2 at 2 to 6 d after sowing. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
RDO5 Influences the Transcriptome during Seed Imbibition
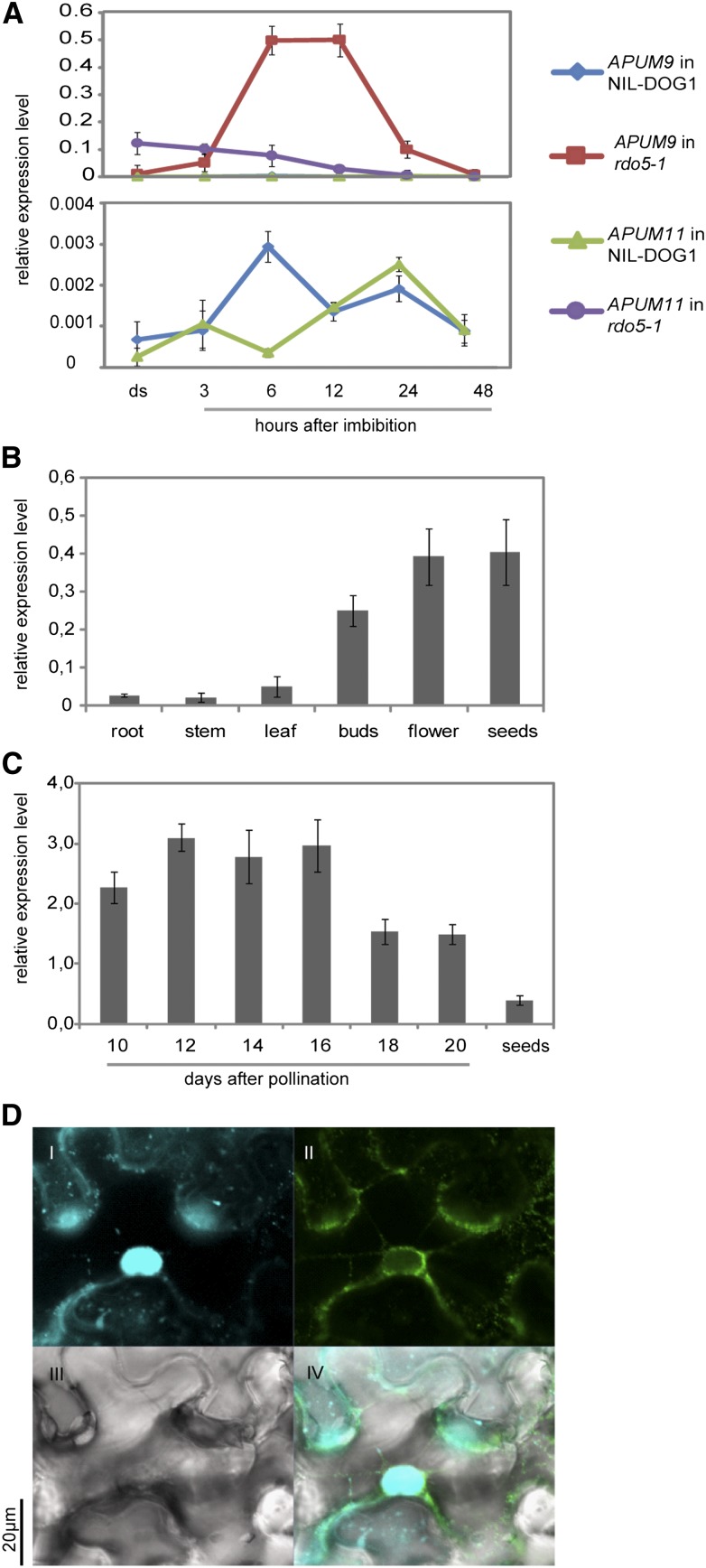
We identified RDO5 as a phosphatase that promotes seed dormancy, most likely independent from ABA and DOG1. To obtain an indication about the genetic and molecular pathways by which RDO5 functions, we performed a transcriptomic comparison between rdo5-1 and its wild-type NIL-DOG1 at different seed stages, using the Arabidopsis Affymetrix AGRONOMICS1 tiling microarrays (Rehrauer et al., 2010). Relatively few genes showed differential transcript levels between rdo5-1 and NIL-DOG1 in siliques at 14 d after pollination and in freshly harvested dry seeds. However, 123 genes are upregulated in rdo5-1 seeds after 6 h of imbibition (Supplemental Data Set 1). None of these genes are known to be involved in dormancy or ABA signaling. Interestingly, three genes belonging to the same family of RNA binding proteins, APUM9, APUM11, and APUM19, were upregulated in rdo5-1 seeds that undergone 6 h of imbibition. APUM9 showed the greatest increase in expression of all upregulated genes and expression was 43 times higher in rdo5-1 than in wild-type NIL-DOG1. APUM9 was also upregulated at 14 d after pollination and APUM11 was upregulated under all three conditions. A qRT-PCR analysis confirmed the microarray data and showed that APUM9 and APUM 11 transcript levels are greatly increased in rdo5-1 compared with the wild type during early imbibition (Figure 6A). A similar, but less dramatic, upregulation of APUM9 and APUM11 was observed in the rdo5-2 mutant (Supplemental Figure 3). The smaller increase in transcript levels in rdo5-2 compared with rdo5-1 could be related to the dormancy level of their background genotypes, which is much higher in rdo5-1 (NIL-DOG1) than in rdo5-2 (Col) (Figures 1A and 1E). The highly enhanced transcript levels in the rdo5 mutant specifically for APUM9 and APUM 11 suggest a role for these genes in the mechanism by which RDO5 controls seed dormancy.
Figure 6.
Expression Pattern and Protein Localization of APUM9.
(A) qRT-PCR analysis of APUM9 and APUM11 transcript levels in dry and imbibed rdo5-1 and wild-type NIL-DOG1 seeds during imbibition. The bottom panel shows the transcript levels for APUM9 and APUM11 in NIL-DOG1 in more detail at a smaller scale. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(B) and (C) qRT-PCR analysis of APUM9 transcript levels in different tissues (B) and during seed maturation (C). The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(D) Protein subcellular localization of APUM9 in N. benthamiana cells transiently expressing Pro35S:GFP:APUM9. I to IV show 4′,6-diamidino-2-phenylindole signal, GFP signal, transmission, and merged images, respectively.
APUM Genes Are Involved in Seed Dormancy
APUM9, APUM 11, and APUM19 are predicted to encode PUF proteins, which are characterized by the presence of a conserved Pumilio homology domain (PUM-HD) that binds to RNA with sequence specificity (Francischini and Quaggio, 2009; Tam et al., 2010). These proteins have been well characterized in animal and fungal systems, but little is known about their function in plants. The Arabidopsis PUF family has 25 members, among which APUM9 and APUM11 cluster together. The APUM9 and APUM11 proteins have 47% identity. APUM9 has a tandem duplicate gene, APUM10, which has 74% identity at the protein level (Tam et al., 2010; Supplemental Figure 4). APUM9, APUM10, and APUM11 are expressed at a low level in all tissues (Supplemental Figure 5). A detailed qRT-PCR analysis showed that the highest transcript levels of APUM9 can be found in reproductive tissues (Figure 6B). APUM9 transcript levels are especially high during the first half of seed maturation, after which they slowly reduce (Figure 6C). This transcription pattern during seed maturation is opposite to that of RDO5. Transient expression of GFP:APUM9 in Nicotiana benthamiana leaves showed localization of the GFP:APUM9 protein in the cytoplasm, especially at the nuclear periphery (Figure 6D). RDO5 was found in the nucleus (Figure 2E), indicating that it is not colocalized with APUM9. Therefore, RDO5 probably regulates APUM9 indirectly.
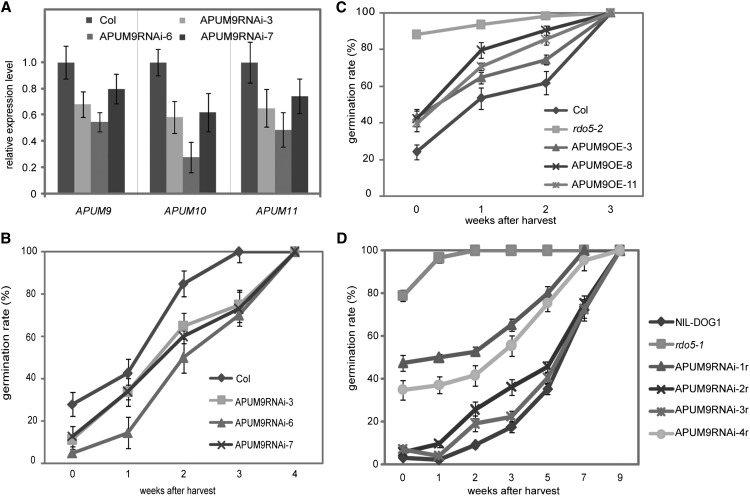
Insertion mutants of APUM9, APUM10, and APUM11 were obtained from the NASC seed stock center. The apum9-1 mutant contains an insertion in the third exon and full-length APUM9 transcript could not be detected by RT-PCR (Supplemental Figures 6A and 6B). The apum10-1 and apum11-1 mutants contain insertions in the promoter regions of APUM10 and APUM11, which do not cause a decrease in their transcript levels compared with wild-type Col (Supplemental Figures 6A and 6C). An analysis of germination during extended seed storage showed that these three insertion lines have normal dormancy levels both in the wild-type and rdo5-2 mutant background (Supplemental Figure 7). This lack of a dormancy phenotype could be due to redundancy (Tam et al., 2010) and (especially for APUM10 and APUM11) by the absent or limited influence of the insertion on the transcript. To reduce the expression of all three APUM genes, an RNA interference (RNAi) construct was made based on a DNA fragment that exhibits high degrees of sequence similarity in all three genes. Three independent single insertion transgenic lines containing this construct were obtained that showed significantly lower APUM9, APUM10, and APUM11 transcript levels compared with the wild-type control (Figure 7A). These three lines all showed enhanced dormancy levels compared with wild-type Col (Figure 7B).
Figure 7.
APUM9 Reduces Seed Dormancy.
(A) qRT-PCR analysis of APUM9, APUM10, and APUM11 transcript levels in 24-h imbibed seeds of three independent APUM9 RNAi lines. The expression values were normalized using ACT8 as control. n = 3 biological replicates; error bars represent se.
(B) Germination after different periods of dry storage of three independent APUM9 RNAi lines in the Col background. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(C) Germination after different periods of dry storage of three independent APUM9 overexpression lines in the Col background. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
(D) Germination after different periods of dry storage of four independent APUM9 RNAi lines in the rdo5-1 background. Germination of the dormant line NIL-DOG1 is shown for comparison. Shown are averages ± se of six to eight independent batches of seeds for each genotype.
In addition, transgenic plants were created in which APUM9 was constitutively expressed using the 35S promoter (Supplemental Figure 8). Three independent single insertion lines all showed reduced seed dormancy compared with the Col background (Figure 7C). These results indicate a negative relation of APUM9 transcript levels with seed dormancy, which is consistent with the high APUM9 and APUM11 transcript levels in the nondormant rdo5-1 mutant.
The Reduced Dormancy Phenotype of rdo5 Requires APUM9 and APUM11
The above described results suggest that the very low dormancy levels of rdo5 seeds could be caused by enhanced transcript levels of APUM9 and APUM11. This hypothesis was tested by downregulation of APUM9, APUM10, and APUM11 in the rdo5-1 background using the same RNAi constructs as described above. We obtained four independent single insertion transgenic lines. These lines complemented the rdo5-1 phenotype. All four lines had strongly enhanced dormancy levels and two of them showed full complementation with dormancy levels similar to wild-type NIL-DOG1 (Figure 7D). We conclude that RDO5 regulates germination potential during seed imbibition by suppressing transcript levels of APUM9 and APUM11.
DISCUSSION
We identified RDO5 as a protein belonging to the PP2C phosphatases that regulates seed dormancy. RDO5 is a seed dormancy specific gene because we did not observe additional phenotypes in the rdo5 mutant. In accordance, RDO5 is only expressed in seeds and exhibits peak transcript levels in freshly harvested dry seeds. Other genes that have been described to influence seed dormancy also influence other plant traits, including the key regulator of dormancy DOG1 whose mutants are reduced in seed longevity (Bentsink et al., 2006). RDO5 could be a suitable target gene to alter seed dormancy levels in crop plants without affecting other plant traits.
RDO5 Does Not Influence the ABA and DOG1 Dormancy Pathways
RDO5 encodes a PP2C protein that has not been previously analyzed. The Arabidopsis genome contains 76 PP2Cs, which are divided into 10 clades, except for six members that could not be clustered (Schweighofer et al., 2004). Several PP2C members belonging to clade A have been characterized as negative regulators of ABA signaling and seed dormancy, such as ABI1, ABI2, AHG1, and HAI2 (Rodriguez et al., 1998; Gosti et al., 1999; Nishimura et al., 2007; Kim et al., 2013). ABA binding to RCAR/PYR/PYL proteins promotes their association with the clade A PP2Cs, leading to the inhibition of phosphatase activity (Ma et al., 2009; Park et al., 2009). The clade A PP2C proteins have similar structures with a catalytic C-terminal domain and a noncatalytic N-terminal extension. The RDO5 protein lacks the N-terminal extension and only contains the catalytic domain. RDO5 and the clade A PP2Cs both control seed dormancy, but the role of RDO5 is different because it acts as a positive regulator of dormancy. Furthermore, several observations indicated that RDO5 is not regulated by ABA. Mutations in RDO5 neither affected ABA levels in dry and imbibed seeds nor ABA sensitivity during germination (Figures 4A and 4B). In addition, RDO5 transcript levels were not affected by enhanced endogenous ABA (Figure 4C). Finally, we have not detected any ABA signaling genes that are differentially expressed between wild-type and rdo5 mutant seeds during maturation and imbibition (Supplemental Table 1). Overall, we assume that the role and regulation of RDO5 is different from that of the clade A PP2Cs.
Although (loss-of-function) rdo5 mutants show strongly reduced seed dormancy, several of our germination experiments revealed a low level of residual dormancy in freshly harvested rdo5 seeds. This suggests that the influence of RDO5 on seed dormancy is weaker than that of DOG1, whose loss-of-function mutants are always completely nondormant. In accordance, dog1 mutant seeds germinate faster than rdo5 mutant seeds (Figure 5D). RDO5 and DOG1 both have a major role in seed dormancy and are only expressed in seeds. DOG1 transcript levels peak half way through the seed maturation phase, whereas those of RDO5 have the highest level in dry seeds. DOG1 and RDO5 are localized in the nucleus and are highly abundant in freshly harvested dry seeds. In addition, both proteins are stable during seed imbibition (Figure 2F; Nakabayashi et al., 2012). These similarities suggest that DOG1 and RDO5 might function in the same pathway. However, we have not found any indication for this. First, RDO5 and DOG1 do not influence each other at the expression level. Transcript levels of DOG1 are similar in wild-type and rdo5 mutant seeds, whereas RDO5 transcript levels are similar in wild-type and dog1 mutant seeds (Figures 5A and 5C). In addition, DOG1 levels and migration speed are not affected in the rdo5 mutant (Figure 5B), suggesting that the phosphatase RDO5 does not affect DOG1 protein. Taken together, our results suggest that RDO5 regulates seed dormancy independently of the ABA and DOG1 pathways.
A Potential Indirect Role for RDO5 in Posttranscriptional Regulation during Seed Imbibition by Suppression of APUM Genes
A transcriptome analysis showed that relatively few genes are differentially expressed in the rdo5 mutant compared with its wild type during seed maturation and imbibition (Supplemental Data Sets 1 to 6). This suggests that RDO5 does not have a major role in transcriptional regulation. Most differentially expressed genes were identified in seeds that had been imbibed for 6 h, among which APUM9 and APUM11 showed dramatically increased transcript levels in the rdo5-1 background. Transgenic plants with enhanced or reduced expression levels of APUM9 and APUM11 showed respectively a decrease and increase in seed dormancy levels (Figure 7). This indicates that these two APUM proteins negatively regulate seed dormancy. Interestingly, transgenic rdo5-1 plants, in which APUM transcripts are repressed by an RNAi construct, showed wild-type dormancy levels (Figure 7D). This strongly suggests that RDO5 regulates seed dormancy by controlling APUM transcripts.
PUF proteins are a class of RNA binding protein containing a PUM-HD. These domains contain multiple tandem repeats that bind to specific recognition sequences in the 3′ untranslated regions (UTRs) of mRNAs to control their stability and translation. PUF proteins are involved in posttranscriptional regulation, including mRNA repression, activation, and localization (Quenault et al., 2011). The PUF proteins are highly conserved in eukaryotes and are implicated in important biological processes in yeast, Drosophila, humans, and other organisms (Kusz et al., 2007; Ariz et al., 2009; Kedde et al., 2010; Chen et al., 2012; Shigunov et al., 2012; Weidmann and Goldstrohm, 2012). The family of PUF proteins is expanded in Arabidopsis compared with vertebrates and yeast. The Arabidopsis genome contains 25 APUMs, of which most are uncharacterized. Several APUMs represent examples of gene duplication events, such as APUM1/APUM2/APUM3, APUM9/APUM10, and APUM18/APUM19. These APUM gene pairs could have redundant functions (Tam et al., 2010), which could explain the lack of phenotype in the apum9-1 single mutant.
A few Arabidopsis APUMs have recently been described and showed different functions in plant growth. APUM23 is a constitutively expressed gene that is required for normal growth patterning in Arabidopsis. Analysis of rRNA processing showed that APUM23 is involved in the processing and/or degradation of 35S pre-rRNA (Abbasi et al., 2010). APUM5 was isolated in a mutant screen for reduced susceptibility to Cucumber mosaic virus infection. APUM5 directly binds viral RNAs and works as a negative regulator for Cucumber mosaic virus replication (Huh et al., 2013). Additionally, APUM5 was shown to be involved in the abiotic stress response and bound to the 3′ UTR of abiotic stress-responsive genes (Huh and Paek, 2014). All the APUMs that have been characterized in Arabidopsis are therefore involved in different processes. This diversion in function of individual APUMs could be caused by their specific binding to UTRs of different mRNAs as well as by their expression patterns.
The Function of RDO5 and APUMs in the Regulation of Seed Dormancy and Germination
Microarray analyses have revealed that more than 10,000 different mRNA species are present in Arabidopsis dry seeds, including genes for all functional categories (Nakabayashi et al., 2005; Dekkers et al., 2013). In addition, mass spectrometry analyses revealed the presence of more than 3700 proteins in mature seeds (Baerenfaller et al., 2008). The functions of these stored mRNAs and proteins in dry seeds are still not clear. It has been demonstrated that germination of Arabidopsis seeds can be inhibited by a cycloheximide treatment, which is an inhibitor of protein synthesis, but not by α-amanitin, which blocks Pol II-mediated transcription. These experiments indicated that transcription is dispensable for germination and that translatable mRNAs and proteins stored in dry seeds are sufficient for germination (Rajjou et al., 2004; Kimura and Nambara, 2010). A recent study of the translatome in dormant and nondormant sunflower (Helianthus annuus) embryos during imbibition demonstrated a selective recruitment of mRNAs to polysomes (Layat et al., 2014). In our study, we identified two seed dormancy regulators, the phosphatase RDO5 and the RNA binding APUM proteins (in particular APUM9 and 11) that could be part of the molecular bases of the selective association of transcripts with polysomes during imbibition. Based on the predicted function of their homologs in other species and their enhanced transcription levels during imbibition, we hypothesize that the APUMs are involved in the translation efficiency of specific stored mRNAs in seeds during imbibition. The subsequently produced proteins could determine whether a seed will germinate or not. We conclude that RDO5 regulates this process because our genetic and transgenic analyses indicated that RDO5 requires the APUM genes for its function. However, RDO5 probably has an indirect role because its protein is located in the nucleus (Figure 2E), while the APUM9 protein was detected in the cytoplasm and around the nucleus (Figure 6D). In addition, RDO5 and APUM9 seem to function at different times. Inducible expression of RDO5 with the heat shock promoter indicated that RDO5 functions at the end of seed maturation and in dry seeds (Figures 3D and 3E), whereas the enhanced transcript levels of APUM9 and APUM11 in the rdo5 mutant were mainly observed during seed imbibition (Figure 6A). However, the RDO5 protein remained stable during seed imbibition (Figure 2F) and could therefore function by repressing APUM transcripts during early imbibition in wild-type plants.
Protein phosphatases like RDO5 dephosphorylate proteins, leading to their activation or inactivation. In the ABA signaling transduction pathway, PP2C phosphatases from clade A can physically interact with subclass III SnRK2s in an ABA-independent manner. PP2Cs inactivate SnRK2 kinase activity by direct dephosphorylation. In the presence of ABA, the ABA receptors inhibit PP2Cs, leading to the activation of SnRK2 kinases, which in turn activate transcription factors and some other targets (Umezawa et al., 2009; P. Wang et al., 2013). Interestingly, in human cancer cells, it was shown that Pumilio-1 (PUM1) is upregulated and phosphorylated for optimal induction of its RNA binding activity toward the tumor suppressor, p27-3′ UTR (Kedde et al., 2010). This suggests that the activity of APUM proteins could be regulated by phosphorylation. Most likely, the phosphatase RDO5 regulates APUM proteins indirectly because we have not detected any physical interaction and the proteins are not located in the same cell compartment. In analogy to the clade A PP2Cs, the relation between RDO5 and the APUM proteins could involve a kinase phosphorylating the APUM proteins and regulating their activity.
METHODS
Plant Materials and Growth Conditions
Plant materials used in this study were all derived from the Arabidopsis thaliana accessions Ler and Col. NIL-DOG1 is derived from the Ler background with an introgression on chromosome 5, containing the DOG1 gene from Cvi (Alonso-Blanco et al., 2003). RDO5 and APUM insertion lines were obtained from the NASC and RIKEN collections with the following seed stock numbers: rdo5-2, SALK_073656; rdo5-3, pst18302; apum9-1, SALK_028441; apum10-1, SALK_022907; and apum11-1, SALK_089493. The gene-specific primer sequences were obtained from the SALK SIGnAL database. RT-PCR or qRT-PCR with RNA isolated from dry or imbibed seeds was performed to confirm the homozygous knockout lines. All used primers are listed in Supplemental Data Set 7.
Seeds were sown on soil and plants grown in a growth chamber with a 16-h-light/8-h-dark cycle (22°C/16°C) or in a greenhouse where the temperature was maintained close to 23°C and 16 h of light was provided daily. Freshly harvested seeds were immediately used for experiments or stored under constant conditions (21°C, 50% humidity, in the dark) for after-ripening treatment.
Plants for the heat shock induction treatment were grown in a chamber with a 16-h-light/8-h-dark cycle (16°C/14°C) during seed maturation and induced by transfer to 37°C for 3 h per day. For heat induction during seed imbibition, imbibed seeds were exposed to a 37°C heat shock for 3 h per day.
Seed Dormancy and Longevity Measurements
Approximately 50 to 100 seeds of individually harvested plants were sown on filter paper, placed into transparent moisturized containers, and incubated in a germination cabinet in long-day conditions (12 h light at 25°C, followed by 12 h darkness at 20°C). After 7 d of incubation, the germination percentages were analyzed.
Seed longevity was determined as germination ability after long-time storage in natural conditions or after a controlled deterioration test. For controlled deterioration, we stored seeds above a saturated KCl solution (80% relative humidity) in a closed container at 37°C for 3 to 6 d, after which germination assays were performed as described above.
For ABA responsiveness tests, after-ripened seeds with fully released dormancy were sown on filter paper imbibed with water containing different ABA concentrations. For each germination test at least six to eight biological replicates were performed.
Fine Mapping and Identification of RDO5
The rdo5-1 mutant was isolated in a γ-irradiation mutagenesis screen of NIL-DOG1 (Bentsink et al., 2006). RDO5 was identified by map-based cloning in a cross with Col. F2 progeny with a homozygous Cvi-DOG1 background were selected using derived cleaved amplified polymorphic sequence markers and phenotyped for their dormancy level. After phenotyping F2 individuals, genomic DNA of highly dormant and nondormant plants was bulked in two groups. These genomic DNA bulks were hybridized with the Arabidopsis SNP Genotyping and Tiling Array (Affymetrix).
For RDO5 fine mapping, plants were genotyped with simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers, designed based on the Monsanto Arabidopsis polymorphism database. The interval was narrowed down to 17 kb on chromosome 4. Finally, the complete 17-kb genomic region of NIL-DOG1 and rdo5-1 was sequenced.
Constructs and Plant Transformation
All the binary constructs were prepared using Gateway technology (Invitrogen). For rdo5 complementation, a 6-kb Ler genomic fragment, including a 2.7-kb promoter sequence, RDO5 coding sequence, and 0.9-kb sequence after the stop codon, was amplified from Ler using gene-specific primers, cloned into the pDONR207 vector, and transferred into pGWB401 (Nakagawa et al., 2007) after sequencing. The RDO5 cDNA was amplified from Ler with gene-specific primers, cloned into pDONR207, and transferred into pGWB429 (Nakagawa et al., 2007) after sequencing for RDO5 overexpression in fusion with the TAP tag. APUM9 cDNA was amplified using gene-specific primers, cloned into pDONR207, and transferred into pFASTR02 (Shimada et al., 2010) after sequencing. The genomic fragment for APUM-RNAi was amplified with APUM9-specific primers, cloned into pDONR207, and transferred into pFASTR03 (Shimada et al., 2010). The binary clone for RDO5-inducible expression was constructed by cloning of the RDO5 cDNA sequence from the donor clone into the pLEELA-HSP vector, which contains the promoter from the soybean (Glycine max) Gmhsp 17.6L heat shock protein gene (Severin and Schöffl, 1990). The ProRDO5:YFP:RDO5 construct was made by recombining the RDO5 cDNA entry clone with pENSG-YFP. The 35S promoter was subsequently replaced with the RDO5 native promoter with an AscI and XhoI digestion. Finally, the APUM9:GFP construct was made by transfer of APUM9 cDNA from an entry clone into pFAST-R05 (Shimada et al., 2010). All used primers are listed in Supplemental Data Set 7.
All the binary constructs were introduced by electroporation into Agrobacterium tumefaciens strain GV3101 or GV3101 carrying the helper plasmid pMP90RK, which was subsequently used for transformation by floral dipping (Clough and Bent, 1998). All the transgenic lines were first selected based on their antibiotics resistance or fluorescence. T3 homozygous lines containing single insertion events were used for expression level detection and phenotyping.
Confocal Microscopy Analysis
For the analysis of RDO5 localization, embryos of freshly harvested seeds from transgenic plants containing YFP:RDO5 driven by the RDO5 promoter were dissected from the testa after a short imbibition time. YFP fluorescence was observed using a Zeiss LSM 700 confocal microscope. The APUM9 subcellular localization was studied after transient expression of Pro35S:GFP:APUM9 in Nicotiana benthamiana leaves.
RNA Extraction and Expression Studies
Total RNA was extracted from developing Arabidopsis siliques or imbibed seeds using RNAqueous columns and an RNA isolation aid (Ambion) as described previously (Kushiro et al., 2004). cDNA was synthesized from 1 μg of total RNA using the QuantiTect reverse transcription kit (Qiagen). qRT-PCR was performed with QuantiTect SYBR Green PCR (Qiagen) on a Mastercycler Realplex2 system (Eppendorf) with gene-specific primer sets. The PCR program was as follows: 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 20 s at 60°C, and 20 s at 72°C. The relative expression value for each gene was quantified using the delta Ct valve method and normalized to the ACTIN8 control. At least three biological replicates were analyzed. All primers are listed in Supplemental Data Set 7.
Transcriptome Analysis
RNA was extracted from siliques at 14 d after pollination, freshly harvested seeds, and seeds that had been imbibed for 6 h. The transcriptome analysis was performed using Affymetrix microarrays (Arabidopsis AGORONOMICS1 genome array) containing more than 30,000 probe sets. Labeling and hybridization were performed according to the manufacturer's instructions at the Max Planck Genome Centre Cologne. Three hybridizations of independent biological replicates were performed. Normalization, processing, and statistical analysis of the raw data were performed using RankProd (Hong et al., 2006) in R programming. Genes were only considered differentially expressed between the mutant and wild type when having a significant expression difference of at least 1.5-fold with false discover rate < 0.05.
ABA Measurements
Extraction, purification, and measurement of ABA were performed as described previously with some modifications (Zhou et al., 2003). About 10 mg seed material was used for ABA measurements. Metabolites were extracted from ground seeds using an acetone:water:acetic acid (80:19:1) solution with the addition of ABA-d6 as internal standards for quantification. Samples were subsequently purified with 1 mL Oasis HLB column by gravity flow. After drying, samples were resuspended in water:acetic acid:formic acid (94.9:5:0.1) and analyzed by liquid chromatography-tandem mass spectrometry.
Protein Extraction and Immunoblotting
Twenty milligrams of developing siliques or 10 mg of dry seeds was ground in liquid nitrogen and then extracted with a buffer containing 6 M urea, 2 M thiourea, 0.2% (v/v) Triton X-100, 0.2% (w/v) sarcosyl, and 2 mM DTT in 100 mM Tris-Cl, pH 7.5. After two cycles of 30 min shaking the supernatant was collected by centrifugation at 4°C, protein concentration was determined by Bradford dye reagent (Bio-Rad) using BSA as a standard. Thirty micrograms of protein samples were separated by NuPAGE 4-12% Bis-Tris protein gels (Life Technologies). Traditional wet transfer was performed using the XCell II blot module and immunological reaction was performed. Protein accumulation was analyzed by immunoblotting using GFP- (Roche), HA- (Covance), or TAP-tag (Thermo) antibodies. Histone H3 antibodies (Abcam) were used for the loading control.
Accession Numbers
The microarray data have been submitted at the Gene Expression Omnibus database (accession number GSE62578). Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RDO5 (At4G11040), DOG1 (AT5G45830), APUM1 (At2g29200), APUM2 (At2g29190), APUM3 (At2g29140), APUM9 (At1G35730), APUM10 (At1g35750), APUM11 (AT4G08840), APUM18 (At5g60110), APUM19 (At5g60180), CYP707A1 (At4g19230), CYP707A2 (At2G29090), CYP707A3 (At5G45340), and ACTIN8 (At1G49240).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotype of 4-Week-Old NIL-DOG1 and rdo5-1 Plants.
Supplemental Figure 2. Map-Based Cloning of RDO5.
Supplemental Figure 3. Analysis of APUM9 and APUM11 Transcript Levels.
Supplemental Figure 4. Amino Acid Sequence Alignment of APUM9, APUM10, and APUM11.
Supplemental Figure 5. Expression Profiles of APUM9, APUM10, and APUM11.
Supplemental Figure 6. Insertion Site Predictions and Transcript Levels of APUM9, APUM10, and APUM11 T-DNA Insertion Lines.
Supplemental Figure 7. Germination Phenotypes of the apum Insertion Mutants in Col and rdo5-2 Background.
Supplemental Figure 8. APUM9 Transcript Levels in APUM9 Overexpression Lines.
Supplemental Data Set 1. Upregulated Genes in rdo5-1 Siliques at 14 Days after Pollination Compared with NIL-DOG1.
Supplemental Data Set 2. Downregulated Genes in rdo5-1 Siliques at 14 Days after Pollination Compared with NIL-DOG1.
Supplemental Data Set 3. Upregulated Genes in rdo5-1 Freshly Harvested Seeds Compared with NIL-DOG1.
Supplemental Data Set 4. Downregulated Genes in rdo5-1 Freshly Harvested Seeds Compared with NIL-DOG1.
Supplemental Data Set 5. Upregulated Genes in rdo5-1 Seeds That Had Been Imbibed for 6 h Compared with NIL-DOG1.
Supplemental Data Set 6. Downregulated Genes in rdo5-1 Seeds That Had Been Imbibed for 6 h Compared with NIL-DOG1.
Supplemental Data Set 7. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Christina Philipp and Kerstin Guhl for technical assistance and Elmon Schmelzer and Quan Wang for help with the confocal microscope. We also thank Tsuyoshi Nakagawa for the pGWB429 vector. This work was supported by the Max Planck Society (Y.X. and W.J.J.S.) and by the Dutch Technology Foundation of the Netherlands Organization for Scientific Research and the Technology Program of the Ministry of Economic Affairs (L.B.).
AUTHOR CONTRIBUTIONS
Y.X. and W.J.J.S. designed the research. Y.X., K.N., and L.B. performed research. Y.X., K.N., F.H., and J.D. analyzed data. Y.X. and W.J.J.S. wrote the article.
Glossary
- ABA
abscisic acid
- GA
gibberellin
- Ler
Landsberg erecta
- Cvi
Cape Verde Islands
- Col
Columbia
- qRT-PCR
quantitative RT-PCR
- RNAi
RNA interference
- UTR
untranslated region
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Bentsink L., Hanhart C.J., Blankestijn-de Vries H., Koornneef M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc E., Galland M., Cueff G., Godin B., Lounifi I., Job D., Rajjou L. (2011). Reboot the system thanks to protein post-translational modifications and proteome diversity: How quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11: 1606–1618. [DOI] [PubMed] [Google Scholar]
- Ariz M., Mainpal R., Subramaniam K. (2009). C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa I., Abe F., Nakamura S. (2013). DOG1-like genes in cereals: investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 208: 1–9. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941. [DOI] [PubMed] [Google Scholar]
- Bazin J., Langlade N., Vincourt P., Arribat S., Balzergue S., El-Maarouf-Bouteau H., Bailly C. (2011). Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23: 2196–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B.B., Balmer Y. (2005). Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56: 187–220. [DOI] [PubMed] [Google Scholar]
- Chen D., Zheng W., Lin A., Uyhazi K., Zhao H., Lin H. (2012). Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr. Biol. 22: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colucci G., Apone F., Alyeshmerni N., Chalmers D., Chrispeels M.J. (2002). GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99: 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Koornneef M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B.J.W., et al. (2013). Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 163: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415. [DOI] [PubMed] [Google Scholar]
- Francischini C.W., Quaggio R.B. (2009). Molecular characterization of Arabidopsis thaliana PUF proteins—binding specificity and target candidates. FEBS J. 276: 5456–5470. [DOI] [PubMed] [Google Scholar]
- Gao F., Rampitsch C., Chitnis V.R., Humphreys G.D., Jordan M.C., Ayele B.T. (2013). Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnol. J. 11: 921–932. [DOI] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A.A.R., Vartanian N., Giraudat J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W.J.J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35: 1769–1786. [DOI] [PubMed] [Google Scholar]
- Graeber K., et al. (2014). Delay of germination 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 111: E3571–E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Millar A.A., Jacobsen J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187. [DOI] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54. [DOI] [PubMed] [Google Scholar]
- Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L., Chory J. (2006). RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827. [DOI] [PubMed] [Google Scholar]
- Huh S.U., Paek K.H. (2014). APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 14: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.U., Kim M.J., Paek K.H. (2013). Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 110: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R. (2010). A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Kim W., Lee Y., Park J., Lee N., Choi G. (2013). HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 54: 555–572. [DOI] [PubMed] [Google Scholar]
- Kimura M., Nambara E. (2010). Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol. Biol. 73: 119–129. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Reuling G., Karssen C.M. (1984). The isolation and characterization of abscisic-acid insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61: 377–383. [Google Scholar]
- Koornneef M., Jorna M.L., Brinkhorst-van der Swan D.L., Karssen C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 61: 385–393. [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23: 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusz K., Ginter-Matuszewska B., Ziolkowska K., Spik A., Bierla J., Jedrzejczak P., Latos-Bielenska A., Pawelczyk L., Jaruzelska J. (2007). Polymorphisms of the human PUMILIO2 gene and male sterility. Mol. Reprod. Dev. 74: 795–799. [DOI] [PubMed] [Google Scholar]
- Layat E., Leymarie J., El-Maarouf-Bouteau H., Caius J., Langlade N., Bailly C. (2014). Translatome profiling in dormant and nondormant sunflower (Helianthus annuus) seeds highlights post-transcriptional regulation of germination. New Phytol. 204: 864–872. [DOI] [PubMed] [Google Scholar]
- Le H., Browning K.S., Gallie D.R. (1998). The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J. Biol. Chem. 273: 20084–20089. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45: 309–319. [DOI] [PubMed] [Google Scholar]
- Linkies A., Müller K., Morris K., Turecková V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., Leubner-Metzger G. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Geyer R., van Zanten M., Carles A., Li Y., Hörold A., van Nocker S., Soppe W.J.J. (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE 6: e22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.C., Meng L.B., Yang C.P., Liu G.F., Liu G.J., Ma W., Wang B.C. (2008). A shotgun phosphoproteomics analysis of embryos in germinated maize seeds. Planta 228: 1029–1041. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Jacobsen J.V., Ross J.J., Helliwell C.A., Poole A.T., Scofield G., Reid J.B., Gubler F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 45: 942–954. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41: 697–709. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Bartsch M., Xiang Y., Miatton E., Pellengahr S., Yano R., Seo M., Soppe W.J.J. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24: 2826–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., Hirayama T. (2007). ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50: 935–949. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., T, Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz K., Bailly C., Gniazdowska A., Côme D., Corbineau F., Bogatek R. (2007). Induction of oxidative stress by sunflower phytotoxins in germinating mustard seeds. J. Chem. Ecol. 33: 251–264. [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters A.J.M., Blankestijn-De Vries H., Hanhart C.J., Léon-Kloosterziel K.M., Zeevaart J.A.D., Koornneef M. (2002). Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant. 115: 604–612. [DOI] [PubMed] [Google Scholar]
- Quenault T., Lithgow T., Traven A. (2011). PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21: 104–112. [DOI] [PubMed] [Google Scholar]
- Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15: 395–401. [DOI] [PubMed] [Google Scholar]
- Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004). The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134: 1598–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer H., et al. (2010). AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol. 152: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P.L., Benning G., Grill E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421: 185–190. [DOI] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9: 236–243. [DOI] [PubMed] [Google Scholar]
- Severin K., Schöffl F. (1990). Heat-inducible hygromycin resistance in transgenic tobacco. Plant Mol. Biol. 15: 827–833. [DOI] [PubMed] [Google Scholar]
- Shen Y.Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826. [DOI] [PubMed] [Google Scholar]
- Shigunov P., Sotelo-Silveira J., Kuligovski C., de Aguiar A.M., Rebelatto C.K., Moutinho J.A., Brofman P.S., Krieger M.A., Goldenberg S., Munroe D., Correa A., Dallagiovanna B. (2012). PUMILIO-2 is involved in the positive regulation of cellular proliferation in human adipose-derived stem cells. Stem Cells Dev. 21: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Soon F.F., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C.M., McCourt P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125: 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P., Barrette-Ng I.H., Simon D.M., Tam M.W., Ang A.L., Muench D.G. (2010). The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Zöll C., Wang Z., Philipp C., Carles A., Li Y., Kornet N.G., Liu Y., Soppe W.J. (2014). HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 80: 475–488. [DOI] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.K. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cao H., Sun Y., Li X., Chen F., Carles A., Li Y., Ding M., Zhang C., Deng X., Soppe W.J.J., Liu Y.X. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann C.A., Goldstrohm A.C. (2012). Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol. Cell. Biol. 32: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Squires T.M., Ambrose S.J., Abrams S.R., Ross A.R., Cutler A.J. (2003). Rapid extraction of abscisic acid and its metabolites for liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 1010: 75–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.