Abstract
Recent clinical trials of chemotherapeutics for advanced bladder cancer (BC) have shown limited benefits. Therefore, new prognostic markers and more effective treatment strategies are required. One approach to achieve these goals is through the analysis of RNA networks. Our recent studies of microRNA (miRNA) expression signatures revealed that the microRNA-23b/27b (miR-23b/27b) cluster is frequently downregulated in various types of human cancers. However, the functional role of the miR-23b/27b cluster in BC cells is still unknown. Thus, the aim of the present study was to investigate the functional significance of the miR-23b/27b cluster and its regulated molecular targets, with an emphasis on its contributions to BC oncogenesis and metastasis. The expression levels of the miR-23b/27b cluster were significantly reduced in BC clinical specimens. Restoration of mature miR-23b or miR-27b miRNAs significantly inhibited cancer cell migration and invasion, suggesting that these clustered miRNAs function as tumor suppressors. Gene expression data and in silico analysis demonstrated that the genes coding for the epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (c-Met) were potential targets of the miR-23b/27b cluster. Luciferase reporter assays and western blotting demonstrated that EGFR and c-Met receptor trypsine kinases were directly regulated by these clustered miRNAs. We conclude that the decreased expression of the tumor-suppressive miR-23b/27b cluster enhanced cancer cell proliferation, migration and invasion in BC through direct regulation of EGFR and c-Met signaling pathways. Our data on RNA networks regulated by tumor-suppressive miR-23b/27b provide new insights into the potential mechanisms of BC oncogenesis and metastasis.
Keywords: bladder cancer, microRNA, miR-23b, miR-27b, tumor suppressor, EGFR, c-Met
Introduction
In developed contries, bladder cancer (BC) is the fifth most commonly diagnosed tumor and the second most common cause of death in patients with genitourinary tract malignancies (1). BCs can be classified into two categories: non-muscle-invasive tumors and muscle-invasive tumors. The 5-year survival frequency for patients with non-muscle-invasive BC is close to 90%, whereas patients with muscle-invasive tumors have 5-year survival frequencies of ~60% (2). Patients with non-muscle-invasive BC tend to have a high rate of recurrence. Moreover, some patients are found to have muscle-invasive BC at recurrence (3). Recent clinical trials of chemotherapeutics for advanced BC have shown limited benefits. Therefore, new prognostic markers and more effective treatment strategies are required. One approach to achieve these goals is through the analysis of RNA networks.
Recent studies have demonstrated the importance of non-coding RNAs (ncRNAs). Participation of these RNAs in human diseases, including cancer, is now apparent (4). For example, microRNAs (miRNAs) are small ncRNA molecules (19–22 bases in length) that regulate protein-coding gene expression by repressing translation or cleaving RNA transcripts in a sequence-specific manner (5). Numerous recent studies have reported that miRNAs are aberrantly expressed in many human cancers. In fact, miRNAs play significant roles in the initiation, development and metastasis of human cancers (6).
Important new information has been gained through the analysis of the cancer-related miRNA networks. We have used our miRNA expression signatures to investigate several tumor-suppressive miRNAs and their regulated cancer pathways (7–11). Notably, some miRNAs are located in close proximity in the human genome; these are termed clustered miRNAs. We previously reported that miR-1/133a, miR-29s, miR-143/145 and miR-195/497 formed clusters and that these clusters function as tumor suppressors, targeting several oncogenic genes in human cancers, including BC (7,12–21).
Previously, our miRNA expression signatures revealed that miR-23b/27b clustered miRNAs were significantly reduced in several cancer tissues (8,9). Our deep sequencing miRNA signature of BC also annotated downregulation of miR-23b in cancer tissues (7). In contrast, Jin et al showed that the expression of miR-23b and miR-27b was highly upregulated in human breast cancer, and knockdown of these miRNAs substantially repressed breast cancer growth (22). Thus, the expression status of the miR-23b/27b cluster is not consistent among different types of cancers. Importantly, the functional roles of the miR-23b/27b cluster have not been fully investigated in BC.
The aim of the present study was to investigate the functional significance of miR-23b/27b clustered miRNAs and to identify the molecular targets regulated by these miRNAs in BC cells. We found that restoration of miR-23b or miR-27b mature miRNAs significantly inhibited cancer cell migration and invasion. Gene expression data and in silico analysis demonstrated that epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (c-Met) were potential targets of the miR-23b/27b cluster. Elucidation of the cancer pathways and targets regulated by tumour-suppressive miR-23b/27b cluster will provide new insights into the potential mechanisms of oncogenesis and metastasis of BC.
Materials and methods
Clinical specimens
A total of 58 BC and 25 normal bladder specimens were collected from patients who underwent cystectomy or transurethral resection of bladder tumors (TUR-BT) at the Kagoshima University Hospital. The 25 normal bladder specimens were derived from patients without BC. Samples were processed and stored in RNAlater® (Qiagen, Valencia, CA, USA) at −20°C until RNA extraction. The samples were staged in accordance with the tumor-node-metastasis classification system of the American Joint Committee on Cancer/Union Internationale Contre le Cancer (UICC), and they were histologically graded. Written informed consent was obtained from all patients and the present study was approved by the Bioethics Committee of Kagoshima University. The patients’ backgrounds and clinicopathological characteristics are summarized in Table I.
Table I.
Patient characteristics.
| Characteristics | Data |
|---|---|
| Bladder cancer | |
| Total number | 58 |
| Median age (range) (years) | 71 (47–91) |
| Gender | |
| Male | 45 (78%) |
| Female | 13 (22%) |
| Pathological tumor stage | |
| pTis | 2 (3%) |
| pTa | 7 (12%) |
| pT1 | 10 (17%) |
| pT2 | 15 (26%) |
| pT3 | 7 (12%) |
| pT4 | 5 (9%) |
| Unknown | 12 (21%) |
| Grade | |
| G1 | 2 (3%) |
| G2 | 29 (50%) |
| G3 | 21 (36%) |
| Unknown | 6 (1%) |
| Operation | |
| Cystectomy | 23 (40%) |
| TUR-BT | 35 (60%) |
| Normal bladder epithelium | 25 |
Cell culture and RNA extraction
We used the human BC cell lines BOY and T24. BOY was established in our laboratory from a male Asian patient, 66-years old, who was diagnosed with stage III BC with lung metastasis. T24 was obtained from the ATCC. The cell lines were incubated in minimum essential medium (MEM) supplemented with 10% fetal bovine serum and maintained in a humidified incubator (5% CO2) at 37°C. Total RNA was extracted as previously described (23).
Quantitative real-time RT-PCR
Stem-loop RT-PCR for miR-23b (P/N 000400; Applied Biosystems, Foster City, CA, USA) and miR-27b (P/N 000409; Applied Biosystems) were used to quantitate miRNAs according to previously published conditions (11). To normalize data for quantifying the miRNAs, we used RNU48 (P/N 001006; Applied Biosystems). The δ-δ threshold count method was used to calculate the fold-change.
Mature miRNA transfection
As previously described (11), the BC cell lines were transfected with Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA) and Opti-MEM (Invitrogen) with 10 nM mature miRNA molecules. As the negative control, Pre-miR miRNA precursor (P/N AM17111; Applied Biosystems) was used in gain-of-function experiments.
Cell proliferation, migration and invasion assays
Cell proliferation was determined using an XTT assay performed according to the manufacturer’s instructions. Cell migration activity was evaluated with a wound healing assay and cell invasion assays were done using modified Boyden chambers as previously described (23). All experiments were performed in triplicate.
Putative miR-23b and miR-27b target gene pathway analysis and expression
To obtain putative miR-23b- and miR-27b-regulated genes, we used the TargetScan database (Release 6.2, http://www.targetscan.org/). To identify signaling pathways regulated by the miR-23b/27b cluster, in silico and gene expression data were analyzed in the Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway categories using the GeneCodis program. We performed gene expression analyses for all candidate genes involved in each of the pathways identified by GeneCodis3 software analysis using microarray expression data. The data were approved by the Gene Expression Omnibus (GEO) and were assigned GEO accession numbers (GSE11783 and GSE31684). For microarray expression data, we examined 90 BCs and 6 normal bladder epithelium collected from patients, none of whom had been exposed to chemotherapy before surgery. The data were normalized and analyzed with GeneSpring (Agilent Technologies, Santa Clara, CA, USA). Statistical analyses were conducted using the Mann-Whitney U test.
Western blotting
Three days after transfection, protein lysates (20 μg) were separated in NuPAGE on 4–12% Bis-Tris gels (Invitrogen) and transferred to polyvinylidene fluoride membranes as previously described (23). Antibodies against EGFR and c-Met were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against GAPDH were purchased from Chemicon (Temecula, CA, USA). Specific complexes were visualized with an echochemiluminescence (ECL) detection system (GE Healthcare, Little Chalfont, UK).
Plasmid construction and dual-luciferase reporter assays
miRNA target sequences were inserted between the XhoI-PmeI restriction sites in the 3′-untranslated region (UTR) of the hRluc gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). miRNA target sequences targeted by miR-23b and miR-27b are summarized in Table II.
Table II.
Insert 3′-UTR sequence of EGFR and c-Met.
| Gene | Target sites | 3′-UTR position | Sequence (5′-3′) | |
|---|---|---|---|---|
| EGFR | miR-27b | 200–207 | Wild | gccaggaagtacttccacctcgggcacattttgggaagttgcattcctttgtcttcaaactgtgaagcatttacagaaacgcatccagcaag aatattgtccctttgagcagaaatttatctttcaaagaggtatatttgaaaaaaaaaaaaagtatatgtgaggatttttattgattgg |
| Mutation | gccaggaagtacttccacctcgggcacattttgggaagttgcattcctttgtcttcaatgacacttgcatttacagaaacgcatccagcaag aatattgtccctttgagcagaaatttatctttcaaagaggtatatttgaaaaaaaaaaaaagtatatgtgaggatttttattgattgg |
|||
| 430–436 | Wild | ggatcttggagtttttcattgtcgctattgatttttacttcaatgggctcttccaacaaggaagaagcttgctggtagcacttgctaccctg agttcatccaggcccaactgtgagcaaggagcacaagccacaagtcttccagaggatgcttgattccagtggttctgcttcaaggctt |
||
| Mutation | ggatcttggagtttttcattgtcgctattgatttttacttcaatgggctcttccaacaaggaagaagcttgctggtagcacttgctaccctg agttcatccaggcccatgacactgcaaggagcacaagccacaagtcttccagaggatgcttgattccagtggttctgcttcaaggctt |
|||
| c-Met | miR-23b | 1019–1026 | Wild | cattaagaaaatttgtatgaaataatttagtcatcatgaaatatttagttgtcatataaaaacccactgtttgagaatgatgctactctgat ctaatgaatgtgaacatgtagatgttttgtgtgtatttttttaaatgaaaactcaaaataagacaagtaatttgttgataaatatttt |
| Mutation | cattaagaaaatttgtatgaaataatttagtcatcatgaaatatttagttgtcatataaaaacccactgtttgagaatgatgctactctgat ctaatgttacacttcatgtagatgttttgtgtgtatttttttaaatgaaaactcaaaataagacaagtaatttgttgataaatatttt |
|||
| 2065–2072 | Wild | gaactcggggaaacatcccatcaacaggactacacacttgtatatacattcttgagaacactgcaatgtgaaaatcacgtttgctatttata aacttgtccttagattaatgtgtctggacagattgtgggagtaagtgattcttctaagaattagatacttgtcactgcctatacctgc |
||
| Mutation | gaactcggggaaacatcccatcaacaggactacacacttgtatatacattcttgagaacactgcttacacttaatcacgtttgctatttata aacttgtccttagattaatgtgtctggacagattgtgggagtaagtgattcttctaagaattagatacttgtcactgcctatacctgc |
|||
| miR-27b | 1564–1571 | Wild | taactggttttgtcgacgtaaacatttaaagtgttatattttttataaaaatgtttatttttaatgatatgagaaaaattttgttaggccac aaaaacactgcactgtgaacattttagaaaaggtatgtcagactgggattaatgacagcatgattttcaatgactgtaaattgcgata |
|
| Mutation | taactggttttgtcgacgtaaacatttaaagtgttatattttttataaaaatgtttatttttaatgatatgagaaaaattttgttaggccac aaaaacactgctgacacttcattttagaaaaggtatgtcagactgggattaatgacagcatgattttcaatgactgtaaattgcgata |
T24 cells were transfected with 50 ng of the vector and 10 nM miRNA using Lipofectamine 2000 (Invitrogen). The activities of firefly and Renilla luciferases in cell lysates were determined with a dual-luciferase assay system (E1910; Promega). Normalized data were calculated as the ratio of Renilla/firefly luciferase activities.
Statistical analysis
Relationships between 2 or 3 variables and numerical values were analyzed using the Mann-Whitney U test or Bonferroni adjusted Mann-Whitney U test. Spearman’s rank test was used to evaluate the correlation between the expressions of miR-23b and miR-27b. Expert StatView, version 4 was used in these analyses.
Results
Expression levels of miR-23b/27b cluster in BC clinical specimens
We evaluated the expression levels of miR-23b and miR-27b in BC tissues (n=58) and normal bladder specimens (n=25). The expression levels of miR-23b and miR-27b were significantly lower in tumor tissues than in corresponding non-cancerous tissues (both P<0.0001; Fig. 1A). Spearman’s rank test showed a positive correlation between the expression of miR-23b and that of miR-27b (r=0.966 and P<0.0001; Fig. 1B). These results suggested that miR-23b and miR-27b were significantly downregulated in BC and could represent putative tumor suppressors in BC.
Figure 1.
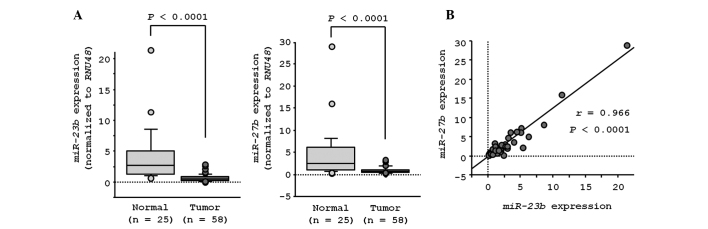
Expression levels of miR-23b/27b in clinical bladder specimens. (A) miR-23b/27b expression levels were significantly lower in 58 BC clinical specimens than in 25 normal bladder specimens. (B) The expression of miR-23b and miR-27b was positively correlated.
Effects of restoring miR-23b and miR-27b expression on cell proliferation, migration and invasion activities in cancer cell lines
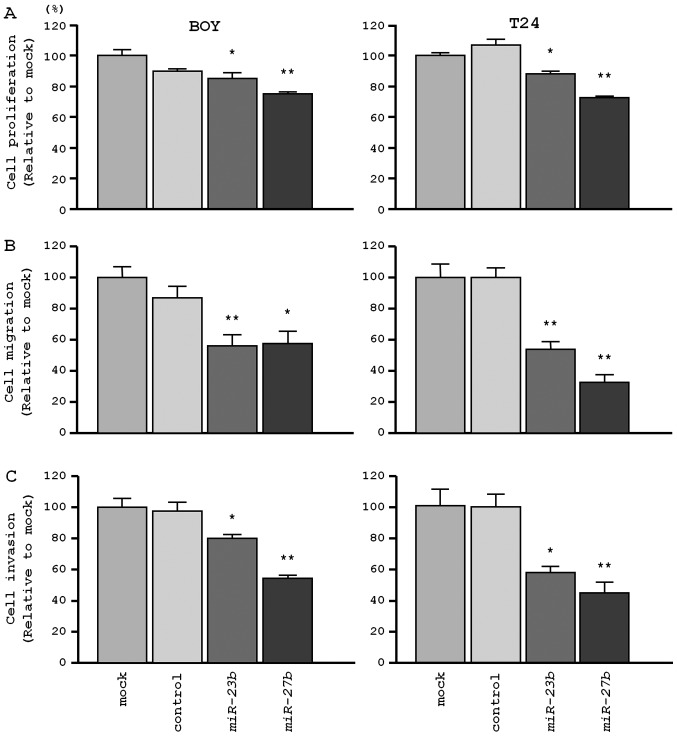
To examine the functional roles of miR-23b and miR-27b, we performed gain-of-function studies using miRNA transfection into BOY and T24 cells. XTT assays revealed significant inhibition of cell proliferation in BOY and T24 cells transfected with miR-23b and miR-27b in comparison with mock-transfected cells and control transfectants (BOY: P=0.0011 and P<0.0001, respectively; T24: P=0.0035 and P<0.0001, respectively) (Fig. 2A).
Figure 2.
Effects of miR-23b/27b transfection into BC cell lines BOY and T24. (A) Cell proliferation determined by XTT assay. *P<0.005. **P<0.0001. (B) Cell migration activity determined with the wound healing assay. *P<0.0005. **P<0.0001. (C) Cell invasion activity determined with the Matrigel invasion assay. *P<0.01. **P<0.0001.
Moreover, wound healing assays demonstrated significant inhibition of cell migration was observed in BOY and T24 cells transfected with miR-23b and miR-27b (BOY: P<0.0001 and P=0.0001, respectively; T24: both P<0.0001) (Fig. 2B).
Similarly, Matrigel invasion assays revealed that transfection with these miRNAs reduced cell invasion. Indeed, the number of invading cells was significantly decreased in BOY and T24 cells transfected with miR-23b and miR-27b (BOY: P<0.0024 and P<0.0001, respectively; T24: P<0.0075 and P<0.0001, respectively) (Fig. 2C).
Identification of targets pathways and genes regulated by the miR-23b/27b cluster in BC cells
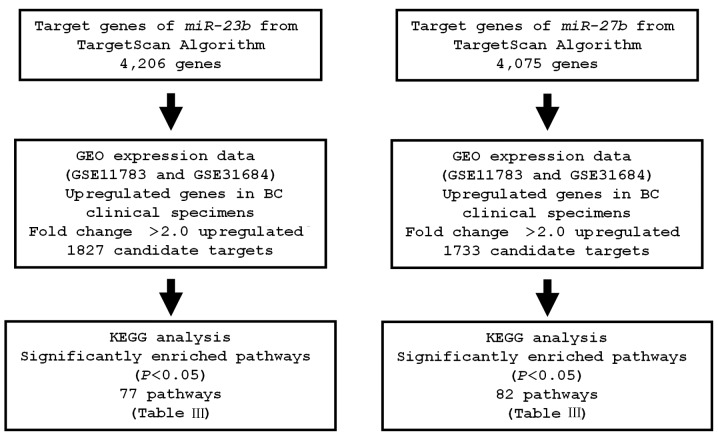
To gain further insight into the molecular mechanisms and pathways regulated by the tumor-suppressive miR-23b/27b cluster in BC, we performed a combination of gene expression and in silico analyses. The strategy for selecting miR-23b/27b cluster-regulated pathways is shown in Fig. 3.
Figure 3.
The strategy for selecting target pathways regulated by the miR-23b/27b cluster.
The TargetScan program showed that 4206 and 4075 genes had putative target sites for miR-23b and miR-27b-, respectively, in their 3′-UTR regions. To confirm the expression levels of these genes in clinical BC tissues, GEO database (GEO accession number: GSE11783 and GSE31684) analysis was performed. The data showed that 1827 and 1733 genes were 2.0-fold or more upregulated in BC tissues compared to normal tissues for miR-23b and miR-27b target genes, respectively. KEGG analysis revealed that the top ‘pathway’ (greatest number of genes) was ‘Pathways in cancer’ (Table III). Several common putative target genes were included in this pathway (Table IV). We focused on EGFR, RET and c-Met genes that coded for tyrosine kinase receptors.
Table III.
Top 10 enriched pathways in miR-23b and miR-27b.
| Annotations | No. of genes | P-value |
|---|---|---|
| miR-23b | ||
| Pathways in cancer | 50 | 5.09E-15 |
| Neuroactive ligand-receptor interaction | 39 | 5.52E-11 |
| MAPK signaling pathway | 24 | 4.60E-04 |
| Cytokine-cytokine receptor interaction | 23 | 8.97E-04 |
| Endocytosis | 22 | 6.98E-05 |
| Focal adhesion | 21 | 2.03E-04 |
| Regulation of actin cytoskeleton | 21 | 3.72E-04 |
| Calcium signaling pathway | 20 | 1.35E-04 |
| Glutamatergic synapse | 19 | 6.90E-06 |
| Chemokine signaling pathway | 19 | 5.79E-04 |
| miR-27b | ||
| Pathways in cancer | 42 | 2.14E-10 |
| MAPK signaling pathway | 30 | 1.30E-06 |
| Neuroactive ligand-receptor interaction | 29 | 4.67E-06 |
| Calcium signaling pathway | 24 | 1.23E-06 |
| Axon guidance | 23 | 2.23E-08 |
| Regulation of actin cytoskeleton | 22 | 1.42E-04 |
| Endocytosis | 20 | 3.85E-04 |
| Glutamatergic synapse | 19 | 3.35E-06 |
| Chemokine signaling pathway | 18 | 1.34E-03 |
| Cytokine-cytokine receptor interaction | 18 | 2.10E-02 |
Table IV.
miR-23b/27b common target genes highly expressed in bladder cancer.
| Clinical BCs | ||||
|---|---|---|---|---|
|
|
||||
| Change | P-value | Entrez gene ID | Symbol | Description |
| 19.55 | 4.67E-05 | 7849 | PAX8 | Paired box 8 |
| 6.79 | 1.14E-04 | 1021 | CDK6 | Cyclin-dependent kinase 6 |
| 5.98 | 1.64E-04 | 1956 | EGFR | Epidermal growth factor receptor |
| 5.00 | 2.65E-04 | 2034 | EPAS1 | Endothelial PAS domain protein 1 |
| 4.74 | 1.21E-04 | 208 | AKT2 | v-akt murine thymoma viral oncogene homolog 2 |
| 4.60 | 5.91E-04 | 2246 | FGF1 | Fibroblast growth factor 1 (acidic) |
| 4.20 | 8.23E-04 | 861 | RUNX1 | Runt-related transcription factor 1 |
| 3.96 | 3.16E-03 | 2250 | FGF5 | Fibroblast growth factor 5 |
| 3.91 | 3.75E-04 | 7170 | TPM3 | Tropomyosin 3 |
| 3.75 | 4.46E-04 | 4089 | SMAD4 | SMAD family member 4 |
| 3.75 | 5.59E-04 | 3918 | LAMC2 | Laminin, γ 2 |
| 3.42 | 7.80E-05 | 862 | RUNX1T1 | Runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| 3.31 | 6.45E-05 | 5979 | RET | Ret proto-oncogene |
| 3.09 | 1.75E-02 | 4286 | MITF | Microphthalmia-associated transcription factor |
| 2.87 | 8.69E-04 | 4233 | c-Met | Met proto-oncogene |
| 2.83 | 1.15E-02 | 2259 | FGF14 | Fibroblast growth factor 14 |
| 2.83 | 1.14E-04 | 3845 | KRAS | Kirsten rat sarcoma viral oncogene homolog |
| 2.68 | 2.32E-02 | 7188 | TRAF5 | TNF receptor-associated factor 5 |
| 2.63 | 2.73E-03 | 2932 | GSK3B | Glycogen synthase kinase 3 β |
| 2.60 | 1.82E-03 | 5594 | MAPK1 | Mitogen-activated protein kinase 1 |
| 2.40 | 1.48E-03 | 4193 | MDM2 | MDM2 oncogene, E3 ubiquitin protein ligase |
| 2.37 | 1.40E-03 | 5579 | PRKCB | Protein kinase C, β |
EGFR and c-Met were regulated by miR-23b and miR-27b
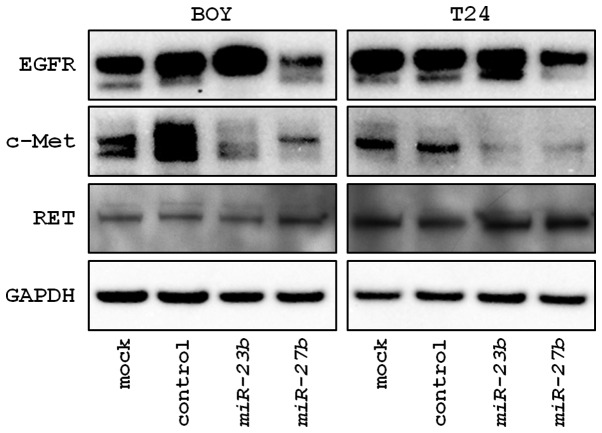
We performed western blot analysis of BOY and T24 cells to investigate whether EGFR, RET and c-Met expression were downregulated by restoration of miR-23b and miR-27b. Expression of EGFR protein was significantly repressed in miR-27b transfectants in comparison with mock or miR-control transfectants (Fig. 4). The protein expression level of c-Met was significantly repressed in miR-23b and miR-27b transfectants ( Fig. 4). RET protein expression was not repressed in either miR-23b or miR-27b transfectants (Fig. 4).
Figure 4.
EGFR and c-Met protein expression levels were suppressed by miR-23b/27b transfection in BOY and T24 cells. Expression of EGFR, c-Met and RET protein as revealed by western blot analysis. GAPDH was used as a loading control.
EGFR and c-Met were directly targeted by miR-23b and miR-27b
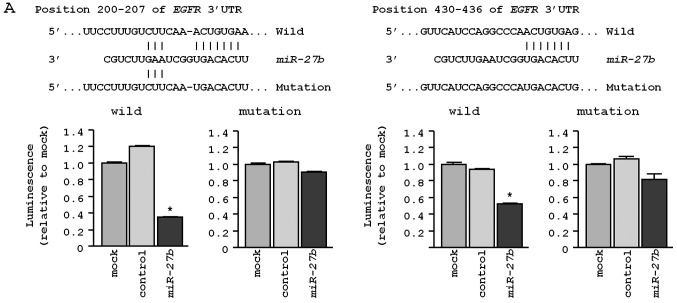
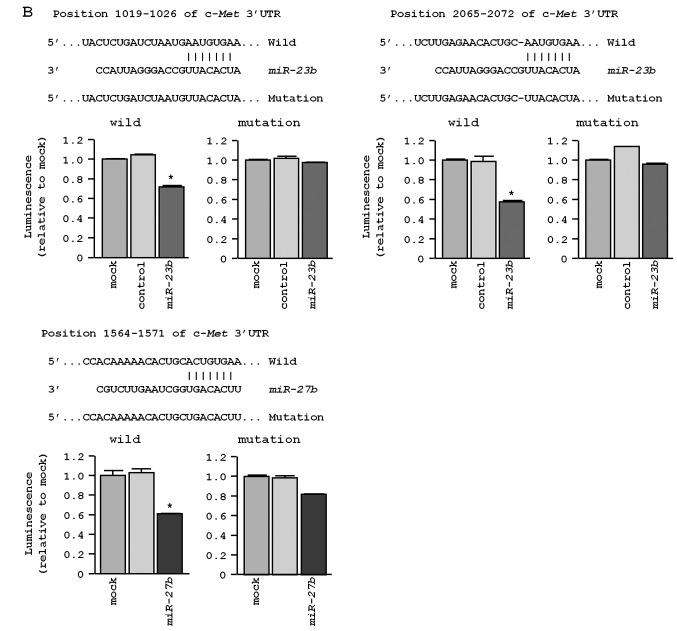
We performed a luciferase reporter assay in T24 to determine whether EGFR and c-Met had target sites for miR-23b and miR-27b. The TargetScan database predicted that two putative miR-27b binding sites existed in the 3′-UTR of EGFR (positions 200–207 and 430–436; Fig. 5A). The database showed that one putative miR-23b binding site existed in the 3′-UTR of EGFR. However, EGFR protein expression was not repressed in miR-23b transfectants (Fig. 4). Therefore, we performed a luciferase reporter assay to determine whether EGFR had target sites for miR-27b. The database also predicted that two putative miR-23b binding sites and one putative miR-27b binding site existed in the c-Met 3′-UTR (positions 1019–1026, 2065–2072 and 1564–1571, respectively; Fig. 5B). We used wild-type and mutant vectors encoding either the partial sequence of the 3′-UTRs of EGFR and c-Met, including the predicted miR-23b and miR-27b target sites.
Figure 5.
(A) Luciferase reporter assays using vectors encoding putative target sites in the 3′-UTR. T24 cells were transiently transfected with Pre-miR miRNA precursor or negative control, followed by transient transfection with wild-type 3′-UTR reporter plasmids or mutated 3′-UTR plasmids. Renilla luciferase activity was measured 24 h after transfection. The results are normalized to firefly luciferase values. *P<0.0001. (B) Luciferase reporter assays using vectors encoding putative target sites in the c-Met 3′-UTR.
We found that the luminescence intensity was significantly reduced by transfection of miR-27b with the wild-type vector carrying the 3′-UTR of EGFR (position 200–207: P<0.0001; position 430–436: P<0.0001; Fig. 5B), whereas transfection with a mutant vector showed no decrease in luminescence.
With regards to c-Met, the luminescence intensity was significantly reduced by transfection of miR-23b with vectors carrying a portion of the 3′-UTR of c-Met (position 1019–1026: P<0.0001; position 2065–2072: P<0.0001; Fig. 5B) and the luminescence intensity was also reduced by transfection of miR-27b with the vector carrying the 3′-UTR of c-Met (position 1564–1571: P<0.0001; Fig. 5B), whereas transfection with a mutant vector failed to decrease luminescence.
Discussion
Aberrant expression of the miR-23b/27b cluster has been reported in several types of human cancers; however, the expression status varies according to the cancer type. Decreased expression of the miR-23b/27b cluster has been observed in castration-resistant prostate cancer (24) and drug-resistant Ehrlich ascites tumor (25). In contrast, upregulation of the miR-23b/27b cluster was reported in breast cancer (22) and chemoresistant ovarian cancer cells (26). In the present study, our data demonstrated that miR-23b and miR-27b expression was significantly downregulated in BC clinical specimens. Functional analysis demonstrated that restoration of miR-23b and miR-27b in BC cells inhibited cancer cell proliferation, migration and invasion. Those results suggest that the miR-23b/27b cluster functions as a tumor suppressor and may contribute to metastasis in BC. Of particular interest, our recent study showed that miR-24-1, which is located close to the miR-23b/27b cluster, was downregulated in BC tissues and functioned as a tumor suppressor targeting FOXM1 (27). Thus, our data suggest that the miR-23b/27b cluster, including miR-24-1, functions as a tumor suppressor and significantly contributes to BC oncogenesis and metastasis.
Next, we investigated the pathways/targets that were regulated by the tumor-suppressive miR-23b/27b cluster in BC cells. To identify tumor-suppressive, miRNA-regulated molecular pathways, we used a combination of expression data and in silico database analysis. Using this strategy, we have identified molecular targets and pathways in several types of cancer that are regulated by tumor-suppressive miRNAs, including BC (7–11). In the present study, ‘pathways in cancer’, the ‘MAPK signaling pathway’ and ‘cytokine-cytokine receptor interaction’ were significantly selected as candidate pathways regulated by the miR-23b/27b cluster in BC cells. Among these pathways, we focused attention on ‘pathways in cancer’ and searched for putative targets of miR-23b/27b regulation. We focused on tyrosine kinase receptors such as EGFR, RET and c-Met genes because molecularly targeted therapies aimed at inhibiting their activities have been developed recently for several types of cancer (28). Our results showed that EGFR was directly regulated by miR-27b and that c-Met was directly regulated by both miR-23b and miR-27b. Unfortunately, RET was not controlled by either miRNA.
EGFR is the cell-surface receptor for members of the epidermal growth factor family of extracellular protein ligands (29). Receptor activation initiates several signal transduction cascades, including the MAPK, Akt and JNK pathways, leading to DNA synthesis and cell proliferation (30). Previous studies showed that overexpression of EGFR occurred in BC and the expression level correlated with tumor grade, stage and survival (31–33).
Another receptor, c-Met (hepatocyte growth factor receptor) activates multiple signal transduction pathways such as those involving RAS, PI3K-Akt, STAT and β-catenin (34–37). The expression of c-Met and phosphorylated c-Met are positively correlated with tumor grade, stage, tumor size and survival of several types of cancers. It is likely that c-Met could be a promising therapeutic target in disease (38,39). The amplification frequency of c-Met is approximately 20% in patients who have acquired resistance to EGFR tyrosine kinase inhibitors (40). Therefore, inhibition of EGFR and c-Met and their associated signaling pathways could be a potent strategy for cancer therapy. Studies of dual tyrosine kinase inhibitors are underway (41). Several laboratories have shown that miRNAs directly inhibit EGFR or/and c-Met expression, such as miR-7, miR-146a, miR-574-3p, miR-34a, miR-130a and miR-1/206 (42–47). In addition, a recent study demonstrated that miR-27a regulated both EGFR and c-Met in non-small cell lung cancer (48). In the present study, the miR-23b/27b cluster regulated EGFR and c-Met in BC cells. Therefore, inhibition of two separate tyrosine kinases via the tumor-suppressive miR-23b/27b cluster represents an attractive possibility in the development of new treatment options in cancer.
In conclusion, downregulation of the miR-23b/27b cluster is a frequent event in BC. Moreover, tumor-suppressive miR-23b and miR-27b directly regulated tyrosine kinase receptor genes EGFR and c-Met. Identification of molecular targets regulated by tumor-suppressive miRNAs will provide insights into the potential mechanisms of BC oncogenesis and metastasis, facilitating the development of novel therapeutic strategies for the treatment of BC.
Acknowledgements
The present study was supported by the KAKENHI (B), 26293354; and (C), 25462490.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Luke C, Tracey E, Stapleton A, Roder D. Exploring contrary trends in bladder cancer incidence, mortality and survival: implications for research and cancer control. Intern Med J. 2010;40:357–362. doi: 10.1111/j.1445-5994.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- 3.Zuiverloon TC, Nieuweboer AJ, Vekony H, Kirkels WJ, Bangma CH, Zwarthoff EC. Markers predicting response to bacillus Calmette-Guerin immunotherapy in high-risk bladder cancer patients: a systematic review. Eur Urol. 2012;61:128–145. doi: 10.1016/j.eururo.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Itesako T, Seki N, Yoshino H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014;9:e84311. doi: 10.1371/journal.pone.0084311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuse M, Kojima S, Enokida H, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 9.Hidaka H, Seki N, Yoshino H, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;3:44–57. doi: 10.18632/oncotarget.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nohata N, Hanazawa T, Kikkawa N, et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011;105:833–841. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 12.Yoshino H, Chiyomaru T, Enokida H, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa R, Goto Y, Kojima S, et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45:401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
- 14.Kojima S, Enokida H, Yoshino H, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita T, Nohata N, Hanazawa T, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshino H, Enokida H, Itesako T, et al. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567–1574. doi: 10.1111/cas.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamasaki T, Yoshino H, Enokida H, et al. Novel molecular targets regulated by tumor suppressors microRNA-1 and microRNA-133a in bladder cancer. Int J Oncol. 2012;40:1821–1830. doi: 10.3892/ijo.2012.1391. [DOI] [PubMed] [Google Scholar]
- 18.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima S, Chiyomaru T, Kawakami K, et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami K, Enokida H, Chiyomaru T, et al. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer. 2012;48:827–836. doi: 10.1016/j.ejca.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Nohata N, Hanazawa T, Kikkawa N, et al. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol. 2011;39:1099–1107. doi: 10.3892/ijo.2011.1096. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Wessely O, Marcusson EG, Ivan C, Calin GA, Alahari SK. Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-alpha in breast cancer. Cancer Res. 2013;73:2884–2896. doi: 10.1158/0008-5472.CAN-12-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiyomaru T, Enokida H, Tatarano S, et al. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishteiwy RA, Ward TM, Dykxhoorn DM, Burnstein KL. The microRNA-23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS One. 2012;7:e52106. doi: 10.1371/journal.pone.0052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husted S, Sokilde R, Rask L, et al. MicroRNA expression profiles associated with development of drug resistance in Ehrlich ascites tumor cells. Mol Pharm. 2011;8:2055–2062. doi: 10.1021/mp200255d. [DOI] [PubMed] [Google Scholar]
- 26.Park YT, Jeong JY, Lee MJ, et al. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J Ovarian Res. 2013;6:18. doi: 10.1186/1757-2215-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoguchi S, Seki N, Chiyomaru T, et al. Tumour-suppressive microRNA-24-1 inhibits cancer cell proliferation through targeting FOXM1 in bladder cancer. FEBS Lett. 2014;588:3170–3179. doi: 10.1016/j.febslet.2014.06.058. [DOI] [PubMed] [Google Scholar]
- 28.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 30.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100014. 2005.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow NH, Liu HS, Lee EI, et al. Significance of urinary epidermal growth factor and its receptor expression in human bladder cancer. Anticancer Res. 1997;17:1293–1296. [PubMed] [Google Scholar]
- 32.Mellon K, Wright C, Kelly P, Horne CH, Neal DE. Long-term outcome related to epidermal growth factor receptor status in bladder cancer. J Urol. 1995;153:919–925. doi: 10.1016/S0022-5347(01)67604-3. [DOI] [PubMed] [Google Scholar]
- 33.Bue P, Wester K, Sjostrom A, et al. Expression of epidermal growth factor receptor in urinary bladder cancer metastases. Int J Cancer. 1998;76:189–193. doi: 10.1002/(SICI)1097-0215(19980413)76:2<189::AID-IJC4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE. ERK and MMPs sequentially regulate distinct stages of epithelial tubule development. Dev Cell. 2004;7:21–32. doi: 10.1016/j.devcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Graziani A, Gramaglia D, Cantley LC, Comoglio PM. The tyrosine-phosphorylated hepatocyte growth factor/scatter factor receptor associates with phosphatidylinositol 3-kinase. J Biol Chem. 1991;266:22087–22090. [PubMed] [Google Scholar]
- 36.Boccaccio C, Ando M, Tamagnone L, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 37.Monga SP, Mars WM, Pediaditakis P, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- 38.Cheng HL, Trink B, Tzai TS, et al. Overexpression of c-met as a prognostic indicator for transitional cell carcinoma of the urinary bladder: a comparison with p53 nuclear accumulation. J Clin Oncol. 2002;20:1544–1550. doi: 10.1200/JCO.20.6.1544. [DOI] [PubMed] [Google Scholar]
- 39.Miyata Y, Sagara Y, Kanda S, Hayashi T, Kanetake H. Phosphorylated hepatocyte growth factor receptor/c-Met is associated with tumor growth and prognosis in patients with bladder cancer: correlation with matrix metalloproteinase-2 and -7 and E-cadherin. Hum Pathol. 2009;40:496–504. doi: 10.1016/j.humpath.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 41.Goldman JW, Laux I, Chai F, et al. Phase 1 dose-escalation trial evaluating the combination of the selective MET (mesenchymalepithelial transition factor) inhibitor tivantinib (ARQ 197) plus erlotinib. Cancer. 2012;118:5903–5911. doi: 10.1002/cncr.27575. [DOI] [PubMed] [Google Scholar]
- 42.Kefas B, Godlewski J, Comeau L, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Vandenboom TG, II, Wang Z, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiyomaru T, Yamamura S, Fukuhara S, et al. Genistein up-regulates tumor suppressor microRNA-574-3p in prostate cancer. PLoS One. 2013;8:e58929. doi: 10.1371/journal.pone.0058929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acunzo M, Visone R, Romano G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan D, da Dong XE, Chen X, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taulli R, Bersani F, Foglizzo V, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acunzo M, Romano G, Palmieri D, et al. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci US A. 2013;110:8573–8578. doi: 10.1073/pnas.1302107110. [DOI] [PMC free article] [PubMed] [Google Scholar]