Abstract
RNA-arbitrarily primed PCR techniques have been applied for the first time to identify differential gene expression in black band disease (BBD), a virulent coral infection that affects reef ecosystems worldwide. The gene activity for the BBD mat on infected surfaces of the brain coral Diploria strigosa was compared with that for portions of the BBD mat that were removed from the coral and suspended nearby in the seawater column. The results obtained indicate that three genes (DD 95-2, DD 95-4, and DD 99-9) were up-regulated in the BBD bacterial mat on the coral surface compared to the transcript base levels observed in the BBD mat suspended in seawater. Clone DD 95-4 has homology with known amino acid ABC transporter systems in bacteria, while clone DD 99-9 exhibits homology with chlorophyll A apoprotein A1 in cyanobacteria. This protein is essential in the final conformation of photosystem I P700. DD 95-2, the only gene that was fully repressed in the BBD mat samples suspended in seawater, exhibited homology with the AraC-type DNA binding domain-containing proteins. These transcriptional activators coordinate the expression of genes essential for virulence in many species of gram-negative bacteria.
Coral reefs are important reservoirs of biodiversity in oceanic environments and thus are used as a sensitive indicator of environmental change (23, 39). The number and incidence of diseases that affect corals around the world have dramatically increased over the last four decades (21). As evidence of this worldwide degradation of coral reef ecosystems mounts, interest in identifying the etiological agents of infectious disease in scleractinian corals continues to intensify (21, 39)
Black band disease (BBD) is among the most important of the globally distributed coral diseases known today, yet the environmental conditions leading to BBD infection are still not well understood (34). BBD results from the rapid migration of a complex mat of microorganisms across the surface of scleractinian coral colonies; this mat lyses healthy coral tissue and leaves dead coral skeleton behind (2, 34). BBD preferentially affects massive-framework-building coral species that serve as the ecological cornerstone of reefs. Therefore, BBD is an important agent in altering coral reef ecosystems.
Although BBD is a polymicrobial disease (34, 35), the population of bacteria growing in the BBD biofilm is composed primarily of large filamentous cyanobacteria previously optically identified as the single species Phormidium corallyticum and recently identified as multiple species by molecular techniques (14, 18). Several previous studies have suggested that these cyanobacteria are the BBD pathogens (40). However, the culturing of these cyanobacteria has proved difficult (23, 33), thus preventing tests to fulfill Koch's postulates to determine BBD pathogenicity (34).
The aim of the present study was to better understand BBD pathogenesis by analyzing bacterial gene expression in the BBD mat consortia by RNA-arbitrarily primed PCR (RAP-PCR) (5, 9, 16, 43). This analysis was done by characterizing abundant mRNAs expressed by bacteria inhabiting the BBD mat on infected surfaces of the brain coral Diploria strigosa and comparing the resulting expression profile with that for samples of the BBD mat that were removed from the infected coral and suspended nearby in the open seawater column. These results demonstrate the great potential of the use of mRNA methodologies, typically reserved for pure cultures, to unravel differential gene expression in complex mixed bacterial communities and to identify differentially expressed genes under different environmental conditions. At least three genes were up-regulated in the BBD bacterial mat on the coral surface compared to the transcript base levels observed in the BBD mat suspended in seawater.
MATERIALS AND METHODS
Sample collection.
Sampling was conducted by using standard scuba techniques on the leeward coral reefs of the island of Curaçao, Netherlands Antilles. Colonies of D. strigosa exhibiting the distinct BBD ring were sampled at water depths of approximately 3 m in the back reef facies of the water plant reef (17, 18). Portions of active BBD mats were physically peeled off the infected coral surfaces with forceps and placed in 50-ml Falcon tubes filled with seawater. An additional sample of BBD mat was then placed into either a 50-ml perforated polypropylene centrifuge tube or a 18-cm-long polyvinyl chloride pipe (inside diameter, 5.2 cm) with a solid cap at one end and a 20-μm-mesh screen at the other end to allow the free flow of water. The tube was suspended in seawater 30 cm to 1 m from the coral head from which samples were taken and oriented with the transparent screen facing slayward to allow light into the tube. This sample tube was left suspended for 3 days. Immediately upon reaching the surface, the in situ and seawater-suspended BBD mat samples were decanted to remove the seawater and the mat samples were immersed in RNAlater (Ambion, Austin, Tex.). For each pairing of samples, the exposure was carried out simultaneously. All samples were then immediately frozen at −20°C. Subsamples of the bacterial mats growing on corals and suspended in seawater were preserved in 80% ethanol at −20°C for use in the analysis of bacterial communities.
Analysis of bacterial communities during infection and after suspension in seawater.
Differences found in the mRNA profiles of two complex microbial communities could arise either from changes in gene expression levels or from differences in community compositions. To characterize and compare the composition of bacteria inhabiting BBD infectious in situ mat communities with that of suspended BBD mat communities, we used terminal-restriction fragment length polymorphism (T-RFLP) analysis. The DNA extraction and T-RFLP protocols, according to those presented by Frias-Lopez et al. (18, 19), are briefly described here. The universal bacterial oligonucleotide primers used in the PCR amplifications were obtained from Integrated DNA Technologies (Coralville, Iowa). These primers were Univ9F (5′-GAGTTTGATYMTGGCTC) and the reverse primer Univ1509R (5′-GYTACCTTGTTACGACTT). Reaction mixtures included 20 μl of TaqMaster buffer (an agent which improves the thermostability and processivity of the polymerase), 10 μl of Taq Reaction buffer containing 15 mM MgCl2 (Eppendorf, Westbury, N.Y.), 8 μl of deoxynucleoside triphosphate (dNTP) mix (each dNTP at 2.5 mM; Gibco/BRL, Rockville, Md.), 1 μl each of forward and reverse primers (200 ng each), 5 to 30 μl of the sample preparation, and water to bring the total volume to 99.5 μl. Some reactions were layered beneath 50 μl of mineral oil (Sigma, St. Louis, Mo.). An initial denaturation-hot start of 5 min at 95°C was followed by the addition of 0.5 μl (approximately 2 U) of MasterTaq polymerase (Eppendorf) or Taq DNA polymerase (Gibco/BRL). The hot start was followed by 30 or 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. A final soak at 72°C for 5 min concluded the reaction.
PCR products were cleaned up with the Wizard PCR Prep kit (Promega, Madison, Wis.) and eluted with 30 μl of water. Ten microliters of clean DNA was aliquoted into each of three tubes, and restriction digests were performed with RsaI, HhaI, or MspI. The final volume of each digest mixture was 20 μl. At least 300 ng of total DNA per digestion was used. Tubes were incubated overnight at 37°C and then covered with aluminum foil and kept at −20°C.
Prior to loading on an acrylamide gel, 1 μl of sample was added to a loading cocktail (1.25 μl of deionized formamide, 0.25 μl of blue loading dye, 0.3 μl of size standard TAMRA 2500). This procedure was repeated for each sample in separate tubes. These preparations were mixed and then denatured at 95°C for 3 min. Samples where then loaded onto a 5% Long Ranger (proprietary acrylamide formula from BioWhittaker) and 7 M urea gel and separated by electrophoresis by using an Applied Biosystems, Inc. (ABI), 377-XL sequencer and a run time of 5 h. Results were analyzed with the ABI GeneScan software.
RNA isolation.
BBD mat samples initially preserved in RNA were later transferred to a 2-ml screw-cap tube containing 1 ml of Trizol reagent (Invitrogen, Carlsbad, Calif.) and 300 μl of 0.1-mm-diameter zirconia-silica beads. The samples were kept on ice during the entire procedure to prevent RNA degradation. Samples were mechanically disrupted in a Mini Beadbeater-8 (Biospec Products, Bartlesville, Okla.) at maximum speed for 30 s, followed by three cycles of freezing and thawing at −80 and 65°C, respectively.
After disruption, samples were vortexed and incubated on ice for 5 min. Two hundred microliters of chloroform was added to the homogenate, and the tubes were shaken vigorously for 15 s and then incubated on ice for 3 min. Samples next underwent centrifugation at 14,000 × g for 15 min at 4°C. The aqueous phase was transferred to a new tube, and the same volume of isopropanol was added. Samples were incubated for 1 h at room temperature, then centrifuged at 14,000 × g for 15 min at 4°C, and then washed once with 75% ethanol. Total RNA was resuspended in diethyl pyrocarbonate water and RNAsecure reagent (Ambion). The isolate total RNA was treated with the DNAfree kit according to the manufacturer's instructions (Ambion) and checked for the presence of remaining DNA. RNA samples were run in a formaldehyde gel to check the integrity of the RNA. Samples with DNA or degraded RNA were excluded from further analysis.
Arbitrarily primed PCR.
cDNA synthesis was carried out immediately after RNA isolation. First-strand synthesis was performed by using ImPromII (Promega), random hexamers (Promega), and random decamers RETROscript (Ambion) as primers. The protocol was performed according to the manufacturer's instructions. Between 500 ng and 2 μg of sample was used, depending on the experiment. Primers (random hexamers or random decamers) were added at 0.5 μg up to a final volume of 5 μl. Samples were incubated in a Mastercycler gradient thermocycler (Eppendorf) at 70°C for 5 min followed by a quick chill at 4°C for 5 min. This 5-μl reaction mixture was used in a 20-μl (total volume) cDNA synthesis reaction mixture comprising 4 μl of Improm-II 5X reaction buffer (Promega), 2.4 μl MgCl2 at 25 mM (final concentration, 3 mM), 1 μl of dNTP mix (each dNTP at 10 mM; final concentration, 0.5 mM), 0.5 μl of Rnasin RNase inhibitor (Promega), 0.5 μl of DNAsecure reagent (Ambion), and 1 μl of Improm-II reverse transcriptase (Promega). The synthesis reactions were performed with a Mastercycler gradient thermocycler (Eppendorf) by using annealing at 25°C for 5 min and extension of the first strand for 60 min at 37°C. Parallel negative controls consisted of exactly the same mix except that water was substituted for the RNA sample. One microliter of the first-strand cDNA was then used to perform the PCR amplification. Oligonucleotides were obtained from Integrated DNA Technologies (Table 1). Reaction mixtures included 5 μl of Taq reaction buffer, 1.5 μl of 25 mM magnesium acetate (Eppendorf), 4 μl of dNTP mix (each dNTP at 2.5 mM; Gibco/BRL), 2 μl of the random primer (200 ng), 1 μl of the cDNA sample, and water to bring the total volume to 50 μl. The hot start was followed by between 45 and 50 cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min. A final soak at 72°C for 5 min concluded the reaction. In all cases, reactions were performed in duplicate.
TABLE 1.
Random primers used for RAP-PCR analysis of BBD samples
| Sequence | Primer name | Reference |
|---|---|---|
| CTGCTTGATGAAA | R1 | J. T. Flemming, personal communication |
| CTGCTTGATGAAC | R2 | J. T. Flemming, personal communication |
| CTGCTTGATGAAG | R3 | J. T. Flemming, personal communication |
| TGAGCGGACA | Lnr81 | 38 |
| TCGCCGCAAA | Lnr97 | 38 |
| CAGCCCAGAG | Lnr95 | 38 |
| TCGTGCGGGT | Lnr99 | 38 |
| AAGCTTTGGTCAG | H-AP3 | 7 |
| AAGCTTAGTAGGC | H-AP5 | 7 |
| GCCGGAGCTCNNNN | RP-1 | This study, based on reference 20 |
| GCCGGAGCTCNNN | RP-2 | This study, based on reference 20 |
| GCCGGAGCTCNN | RP-3 | This study, based on reference 20 |
Identification of differentially expressed genes.
Differences in gene expression for in situ and suspended samples of BBD mats were detected by comparing side-by-side samples of RAP-PCR products. These products were electrophoresed either in 2.0% high-resolution agarose Agarose-HR (Ambion) or in a 6% polyacrylamide gel under native conditions. Only bands present in two separate RAP-PCRs were analyzed. Bands obtained from agarose gels were passed through a 250- to 300-μm-diameter glass bead (Sigma) column and purified by using the PCR Prep kit (QIAGEN, Alameda, Calif.). DNA from bands excised from the polyacrylamide gel was eluted as follows. Two hundred microliters of crush-and-soak solution (0.5 M ammonium acetate, 0.1% sodium dodecyl sulfate, 0.1 mM EDTA) was added, and the acrylamide was crushed and incubated overnight at 37°C. The following day, tubes were spun down for 10 min at 14,000 rpm. The supernatant was removed, another 200 μl of crush-and-soak solution was added, the spin was repeated, and the recovered supernatant was pooled with the initial supernatant fraction. DNA was then precipitated and recovered. In order to clone the eluted bands into the pGEM-T vector, the DNA was incubated for 10 min at 72°C in the presence of Taq polymerase and dATP (2.5 mM). The gel-purified PCR product was cloned into the pGEM-T vector (Promega) and transformed into calcium chloride-competent DH5αMCR Escherichia coli cells by using the manufacturer's instructions and standard techniques (41). Clones were checked for the presence of the insert by PCR with the universal primers M13 (position −21) and T7 (position −26). An (RFLP) analysis of the products was performed to determine identical patterns. PCR products were digested with MspI and HinP1 enzymes and analyzed in a 1.6% Metaphor agarose gel. Clones selected for sequence analysis were patched onto Luria broth agar petri dishes supplemented with 100 μg of ampicillin/ml (Roche Molecular Biochemicals, Indianapolis, Ind.) and incubated overnight at 37°C.
Inoculations, cultures, DNA preparations, and sequence reactions were performed in the High Throughput Laboratory of the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. Plates were used to inoculate 2-ml 96-well culture blocks containing Circle Grow media (Bio100) supplemented with ampicillin. Plasmid template DNA was purified from the cultures by using an automated system and the QIAwell 96 Turbo prep BioRobot kit (QIAGEN, Valencia, Calif.). The first round of sequencing was completed by using the T7 (position −26) primer. Plasmid templates were prepped on a QIAGEN Bio Robot 9600, and Big Dye Terminator Chemistry (version 2.0) from ABI was used for sequencing reactions. Sequencing was performed with an ABI 3700 capillary sequencer, followed by processing at the Bioinformatics Unit of the W. M. Keck Center.
Confirmation of differential gene expression by semiquantitative RT-PCR.
In order to verify the differences in gene expression of the genes identified by RAP-PCR, we performed reverse transcription-PCR (RT-PCR) with the original samples of RNA used for RAP-PCR. First-strand cDNA was obtained as described above by using random primers (Promega) and 1 μg of total RNA. One microliter of cDNA was used as a template for the PCR, with specific primers being used for each target gene. Different numbers of cycles were used to avoid saturation of the PCR products. Only products obtained in a number of cycles below the saturation point were used for relative quantification analysis. Analysis of the relative levels of expression of each of the bands obtained for the different genes was performed by using the program ImageJ 1.30v (Wayne Rasband, National Institutes of Health, Bethesda, Md.), obtained in the public domain (http://rsb.info.nih.gov/ij/Java 1.3.1). Analyses were performed in three different independent experiments and in triplicate RAP-PCRs.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences generated in this study are AY494600 through AY494604.
RESULTS
Bacterial community analysis.
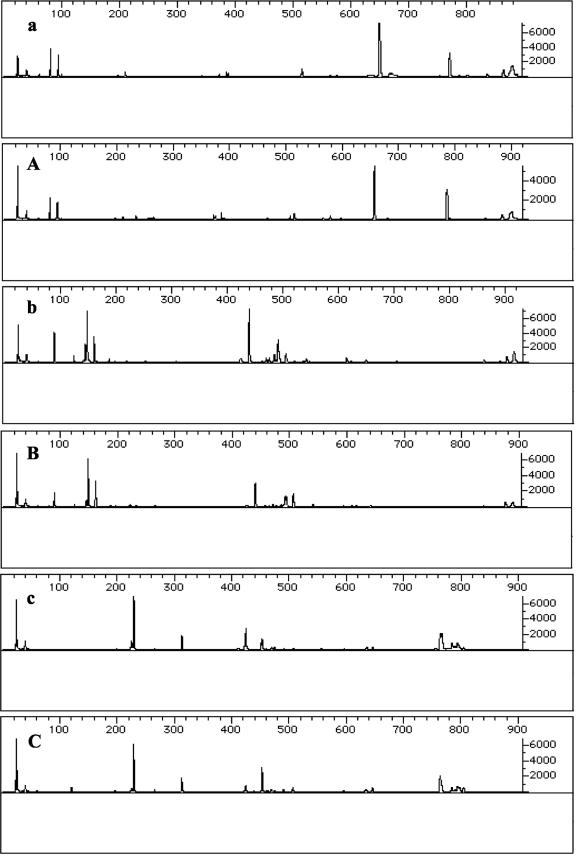
Prior to the evaluation of differential gene expression, the compositions of the microbial consortia comprising the in situ coral surface and water column-suspended samples of the BBD bacterial mats were determined. This was an important starting point, since BBD is a polymicrobial disease, and therefore RNA was extracted from a mixed population of bacteria. In both the in situ and the suspended samples of BBD mats, we obtained the same T-RFLP profile. This finding indicates that the most abundant bacteria in both types of BBD mat samples were the same (Fig. 1). Therefore, the differences between RAP-PCR fingerprinting profiles may be attributable to a change in gene expression rather than to changes in the composition of the bacterial communities comprising each BBD mat sample in our controlled experiment.
FIG. 1.
T-RFLP profiles of the BBD mat bacterial communities. (a and A) HhaI-digested PCR products from BBD mat samples taken from coral and BBD mat samples suspended in seawater, respectively. (b and B) MspI-digested PCR products from BBD mat samples taken from coral and BBD mat samples suspended in seawater, respectively. (c and C) RsaI-digested PCR products from BBD mat samples taken from coral and BBD mat samples suspended in seawater, respectively.
RAP-PCR fingerprinting of RNA.
Different authors have recently used RAP-PCR to analyze differences in gene expression in prokaryotes. However, all of those experiments used cultures grown under laboratory conditions (5, 9, 16, 43). Because our samples were collected directly from the ocean environment, we had to use RNAlater to preserve them before analysis. The RNA obtained from the environmental samples was of good quality, as shown in Fig. 2. The first cDNA strand was obtained by using either random hexamers or random decamers as primers. In all cases, the shorter random primers provided cDNA for a larger number of different genes. In combination with the cDNA products, a total of 12 different random primers were used (Table 1). Preliminary results showed that the use of primers Lnr95 and Lnr99 provided a larger number of DNA bands as well as a more consistent pattern, and therefore these primers were used in subsequent experiments.
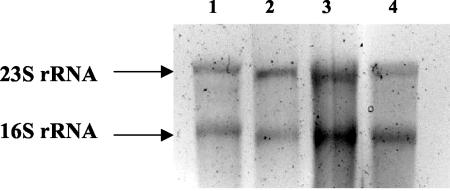
FIG. 2.
Formaldehyde agarose gel showing RNA extractions of BBD microbial mats growing in situ on coral surfaces (lanes 1 and 2) and suspended in seawater (lanes 3 and 4).
Nonradioactive methods have previously been used successfully to analyze differences in gene expression (1, 6, 10, 38). Both a high-resolution agarose and a 6% polyacrylamide gel were used with good results to analyze RAP-PCR fingerprinting results for our samples (Fig. 3). Nevertheless, polyacrylamide gels were more sensitive, showing a larger number of different bands. All sequences analyzed except one came from bands obtained from BBD mats growing on the coral during infection. The number of genes up-regulated during infection was always higher than the number of genes down-regulated, and a total of seven bands were finally chosen for subsequent analysis. These bands were cloned into the pGEM-T vector and sequenced. BLASTX analysis for the fragments of DNA sequenced gave significant hits for five out of six sequences. One of the bands was excluded from further analysis because it was too short and did not give any significant hits in the database. The results obtained are shown in Table 2. Two of the bands (clones DD 95-4 and DD 99-8) had homology with proteins involved in amino acid transporters. DD 95-4 had 51% identity (68% positives) with a hypothetical amino acid ABC transporter from Shewanella oneidensis, while DD 99-8 had 46% homology (70% positives) with a glutamine-binding protein of a member of Archaea, Methanosarcina mazei. Nonetheless, the rest of the hits for DD 99-8 had homology with amino acid ABC transporters in bacteria with homologies higher than 35%. Another clone, DD 95-1, had 47% homology (64% positives) with a hypothetical protein from Vibrio parahaemolyticus. Clone DD 99-9 had 93% homology (97% positives) with a protein that is part of photosystem I in cyanobacteria. Since the filamentous cyanobacteria are the most abundant microorganisms present in the infectious mat, the sequence from clone DD 99-9 is probably derived from these cyanobacteria. Finally, the best hit for clone DD 95-2 was an AraC-type DNA-binding domain protein with a homology of 25% (68% positives). Finally, the best hit for clone DD 95-2 was an AraC-type DNA-binding domain protein with a homology of 25% (68% positives).
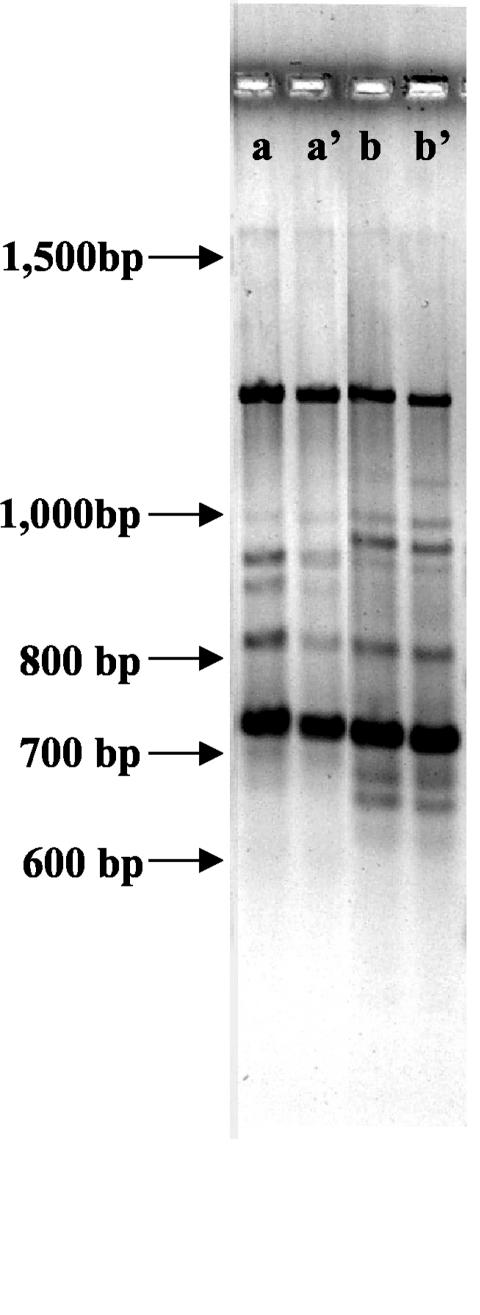
FIG. 3.
Example of RAP-PCR results obtained by using a high-resolution agarose gel and the Lnr95 primer. Samples a and a' are duplicates from the same sample of RNA extracted from an in situ BBD mat growing on the coral surface. Samples b and b' are duplicates from the same sample of RNA extracted from a BBD mat suspended in seawater. Arrows indicate the positions of molecular weight standards not visible in the photo.
TABLE 2.
Best BLASTX hits for the sequences obtained from RAP-PCR profiles
| Clone | Accession no. | Fragment size (bp) | BLASTX results
|
||
|---|---|---|---|---|---|
| Accession no. | Protein encoded by gene | Organism | |||
| DD 95-1 | AY494600 | 670 | gi|28898568| | Conserved hypothetical protein | V. parahaemolyticus |
| DD 95-4 | AY494601 | 568 | gi|24372841| | Hypothetical amino acid ABC transporter | S. oneidensis |
| DD 99-8 | AY494602 | 482 | gi|21228041| | Glutamine-binding protein | M. mazei |
| DD 95-2 | AY494603 | 410 | gi|23137865| | AraC-type DNA binding domain-containing proteins | C. hutchinsonii |
| DD 99-9 | AY494604 | 417 | gi|97659| | Photosystem I P700 PsaA | Synechocystis sp. (strain PCC 6803) |
Semiquantitative RT-PCR analysis.
The final step was to verify that the characterized genes were actually differentially expressed during infection of the coral. To confirm this possibility, semiquantitative RT-PCR was performed with the original samples of RNA, in situ and suspended, from three different experiments. Semiquantitative RT-PCR analysis has previously been used to confirm differences in gene expression (15, 24, 26). Specific primers were designed to analyze the levels of expression of the genes presented in Table 2. During the amplification of the genes, different numbers of cycles were used to avoid PCR saturation. Samples from three independent experiments were analyzed. Figure 4 shows an example of results obtained from these experiments, and Table 3 shows results for the relative differential expression of the genes analyzed. These results come from at least three different RT-PCRs and from three different samples.
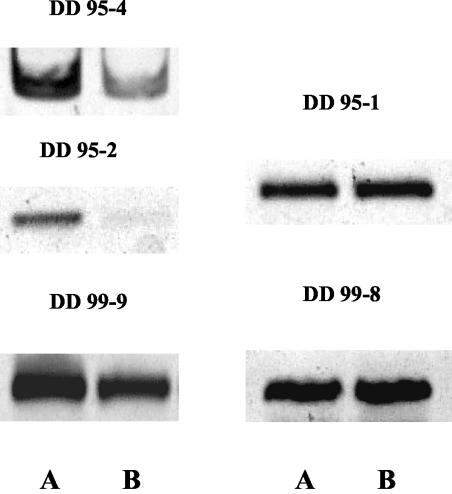
FIG. 4.
RT-PCR analysis to assess relative differences in gene expression. Clone designations are given above the pictures. (A) BBD mat growing in situ on coral; (B) BBD mat in seawater.
TABLE 3.
Results for semiquantitative RT-PCR products analyzed by densitometry
| Clone | RT-PCR result fora:
|
Differential expression range of up- or down-regulation (%) | |
|---|---|---|---|
| BBD mat growing on coral | BBD mat in seawater | ||
| DD 95-1 | ++ | ++ | 0 |
| DD 95-4 | ++ | + | 31-40 |
| DD 99-8 | ++ | ++ | 0 |
| DD 95-2 | ++ | − | 100 |
| DD 99-9 | ++ | + | 9-34 |
−, no expression detected; +, low levels of transcript expression; ++, high levels of transcript expression.
Two out of the five genes analyzed did not show differences in gene expression with RT-PCR even when a low number of PCR cycles was used. This result is not surprising, as one of the problems that RAP-PCR has, in addition to differential display, is the high proportion of false-positive results among clones obtained from differentially expressed bands (25). A major concern is the fact that one band may actually be composed of cDNAs derived from multiple genes (8, 25). Some of these genes may be differentially expressed, but others may be constitutively expressed. This was the case for clones DD 95-1 and DD 99-8. However, three of the clones analyzed did have differences in their levels of expression depending on whether they came from BBD mats infecting coral in situ or from BBD mats suspended in seawater. In all cases, these genes were up-regulated when the bacterial mat was infecting the coral.
Clone DD 95-4 had 51% homology with amino acid ABC transporter systems in bacteria and was up-regulated between 31 and 40% (Table 3) during infection. In one sample, we found almost 100% up-regulation. Clone DD 99-9 was up-regulated between 9 and 34% (Table 3) during infection. As with DD 95-4, in one case there was almost complete transcriptional inhibition of gene DD 99-9 when the bacterial mat was living in seawater. This sequence presented high homology (93% identity) with photosystem I P700 chlorophyll A apoprotein A1 (PsaA) in cyanobacteria. Finally, the last gene that showed up-regulation during infection, clone DD 95-2, had 25% homology with AraC-type DNA binding domain-containing proteins. All samples analyzed were completely inhibited for RNA synthesis of the DD 95-2 gene when the bacterial mat was removed from the infection site (Table 3). PCR analysis of the DNA obtained from these samples showed that the gene was present in the bacterial community. Therefore, the complete repression of DD 95-2 expression was not due to the absence of bacteria harboring this gene in the samples suspended in seawater.
DISCUSSION
Due to the difficulties in studying differences in gene expression in prokaryotes, most studies using RAP-PCR have been done only in recent years and have been restricted to laboratory conditions (5, 9, 16, 43). With the present study, we demonstrated that RAP-PCR may be used successfully to analyze differential gene expression in environmental samples. The differences in the gene expression of infectious BBD bacterial mats, which cause the death of a large number of coral worldwide (31), have been analyzed. The final goal of analyzing differences in gene expression is to determine the mechanism or mechanisms of pathogenesis and to look for clues about the environmental factors that lead to disease.
Significant differences between transcription levels of a set of different genes in the in situ and in the suspended BBD mat samples were observed. When removed from the coral tissue for 3 days, the BBD mat genes that were up-regulated during infection should be turned off or at least show a decreased level of activity. The short distances maintained between the infected bacterial mat and the samples suspended in seawater guaranteed identical environmental conditions (salinity, turbidity, light intensity, etc.) during the experiments. One of the major problems of analyzing differences in gene expression in bacteria comes from the fact that mRNA in prokaryotes does not have a poly(A) tail and that random primers therefore have to be used in order to synthesize cDNA. Those primers are likely to anneal to rRNA, which constitutes the largest fraction of RNA in the samples, thus preventing efficient conversion of the small portion of mRNA into cDNA. It has been reported that when prokaryotic RNA was used, up to 40% of differentially expressed bands identified using differential display analysis were of ribosomal origin (30, 31). However, the results obtained by using RAP-PCR analysis show that this method successfully overcomes that problem. None of the cloned differentially expressed genes was a 16S rRNA gene, probably because bands that appear in all samples and at a high concentration are most likely to be the ones that come from rRNA.
A possible mechanism of pathogenesis for BBD in which an anoxic environment and the production of sulfide are toxic for the coral tissue and result in coral death has been proposed (37). However, there is no definitive proof that this is the real mechanism for tissue destruction (36), and nothing is known about what conditions lead to disease. The photosynthetic filamentous cyanobacterium P. corallyticum is the most conspicuous bacterium in BBD, and it is not known what its the role during infection is. It is not yet understood why a photosynthetic organism is infecting a coral; perhaps this species requires something from the coral to survive or to obtain a growth advantage through the infectious process.
One of the sequences was up-regulated during infection; clone DD 95-4 had homology with amino acid ABC transporter systems in bacteria. Amino acids are important in both carbon and nitrogen metabolism of bacteria; for this reason, the ABC transport systems of bacteria have been widely studied (22). Nevertheless, there is no clear connection between the up-regulation of this ABC transporter and pathogenesis. In the case of BBD, the observed up-regulation of this gene could be due to the presence of amino acids available for consumption as a consequence of coral tissue destruction.
Of equal importance are the results obtained for the other two cDNAs analyzed. Clone DD 99-9 was up-regulated during infection and presented 100% homology with photosystem I P700 PsaA in cyanobacteria. Photosystem I is a membrane complex of 12 different proteins that produces the reduced NADPH needed to reduce CO2 in the reactions of the Calvin cycle (48). PsaA along with PsaB coordinates the two chlorophylls forming the dimer in P700 (42, 48). Studies of differential expression in Synechocystis spp. under different environmental conditions have previously been reported (4, 28, 46). The level of expression of psaA is controlled by several environmental factors, such as day-night light cycles (13, 29). Singh and Sherman, using a customized amplification library, have shown that the expression of psaA increases when iron is available and decreases with iron depletion (44). Iron is an essential trace element for most bacteria and a limiting factor for primary production in seawater (12, 27, 47). During BBD infection, with the destruction of coral tissue and the death of the symbiotic dinoflagellates (Symbiodinium spp.) living inside the coral, the availability of iron increases at levels that allow the staggering growth of P. corallyticum. The low levels of available iron in seawater could explain, at least in part, why P. corallyticum is so difficult to detect in samples other than that of the BBD infectious mat (18, 36). Finally, the last gene that showed up-regulation during infection, clone DD 95-2, had 25% identity with AraC-type DNA binding domain-containing proteins. All samples analyzed showed complete inhibition of RNA synthesis for the DD 95-2 gene when the bacterial mat was removed from the infection site on the coral tissue surface. In animal pathogens, AraC-like transcriptional activators coordinate the expression of type III secretion genes (17), which are essential for the virulence of a large number of gram-negative pathogens. Moreover, some plant pathogens also utilize this kind of signal regulator (17). Most type III secretion systems have been found in Proteobacteria. However, DD 95-2 had homology to a gene present in Cytophaga hutchinsonii. Previous works have shown that members of the Cytophaga-Flexibacter-Bacteroides group are present in large numbers in the BBD bacterial mat (14, 19). Therefore, it is reasonable to hypothesize that these bacteria play an important role in the disease. Moreover, this group of bacteria includes microorganisms that are well-known pathogens in a variety of organisms that inhabit a wide variety of environments (3, 11, 32, 45).
Conclusions.
RAP-PCR has been successfully used to detect genes that are differentially expressed under different environmental conditions in complex bacterial communities. This work has identified genes that are involved in the pathogenesis of BBD, one of the most important infectious coral diseases. Three genes that are up-regulated when a BBD bacterial mat is infecting coral have been identified. One of the cDNAs, DD 95-4, had homology with amino acid ABC transporter systems in bacteria but no clear link with the mechanism of pathogenesis of the disease.
Conversely, the other two cDNAs analyzed may have a more direct link to the pathogenesis of BBD. DD 99-9 was up-regulated during infection and presented homology with photosystem I P700 PsaA in cyanobacteria. It has been shown that the expression of psaA increases when iron is available and decreases with iron depletion (44). The up-regulation of this gene could be linked to the increased levels of iron available due to coral tissue destruction. Finally, DD 95-2 was completely repressed when the bacterial mat was not in contact with the coral tissue. Furthermore, DD 95-2 has homology with AraC-type DNA binding domain-containing proteins, which are important regulators for expression of genes involved in virulence in gram-negative bacteria.
Acknowledgments
We thank the Office of Naval Research (ONR-N00014-00-1-0609) for support of this research.
We also thank J. Klaus for his thorough review of the manuscript.
The conclusions of this study are those of the authors and do not necessarily reflect those of the funding agencies.
REFERENCES
- 1.Ahmed, N., A. A. Siddiqui, and A. Ahmed. 2000. DDRT-PCR: use of agarose gels for detection of amplified products. Mol. Vis. 6:144-147. [PubMed] [Google Scholar]
- 2.Antonius, A. 1981. The “band” diseases in coral reefs, p. 7-14. In Fourth International Coral Reef Symposium, vol. 2. E. D. Gomez et al. (ed.), Proceedings of the University of the Philippines Marine Center, Manila, Philippines. [Google Scholar]
- 3.Bader, J. A., C. A. Shoemaker, and P. H. Klesius. 2003. Rapid detection of columnaris disease in channel catfish (Ictalurus punctatus) with a new species-specific 16-S rRNA gene-based PCR primer for Flavobacterium columnare. J. Microbiol. Methods 52:209-220. [DOI] [PubMed] [Google Scholar]
- 4.Bhaya, D., D. Vaulot, P. Amin, A. W. Takahashi, and A. R. Grossman. 2000. Isolation of regulated genes of the cyanobacterium Synechocystis sp. strain PCC 6803 by differential display. J. Bacteriol. 182:5692-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidle, K. A., and D. H. Bartlett. 2001. RNA arbitrarily primed PCR survey of genes regulated by ToxR in the deep-sea bacterium Photobacterium profundum strain SS9. J. Bacteriol. 183:1688-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockelmann, R., B. Bonnekoh, and H. Gollnick. 1999. Optimized visualization and PCR reamplification of differentially displayed cDNA bands detected by silver staining in polyacrylamide gels as established in the model of dithranol-treated keratinocytes. Skin Pharmacol. Appl. Skin Physiol. 12:54-63. [DOI] [PubMed] [Google Scholar]
- 7.Bosch, I., H. Melichar, and A. B. Pardee. 2000. Identification of differentially expressed genes from limited amounts of RNA. Nucleic Acids Res. 28:E27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callard, D., B. Lescure, and L. Mazzolini. 1994. A method for the elimination of false positives generated by the mRNA differential display technique. BioTechniques 16:1096-1907. [PubMed] [Google Scholar]
- 9.Chakrabortty, A., S. Das, S. Majumdar, K. Mukhopadhyay, S. Roychoudhury, and K. Chaudhuri. 2000. Use of RNA arbitrarily primed-PCR fingerprinting to identify Vibrio cholerae genes differentially expressed in the host following infection. Infect. Immun. 68:3878-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. J., and K. Peck. 1996. Non-radioisotopic differential display method to directly visualize and amplify differential bands on nylon membrane. Nucleic Acids Res. 24:793-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cipriano, R. C., L. A. Ford, and J. D. Teska. 1995. Association of Cytophaga psychrophila with mortality among eyed eggs of Atlantic salmon (Salmo salar). J. Wildl. Dis. 31:166-171. [DOI] [PubMed] [Google Scholar]
- 12.Coale, K. H., K. S. Johnson, S. E. Fitzwater, R. M. Gordon, S. Tanner, F. P. Chavez, L. Ferioli, C. Sakamoto, P. Rogers, F. Millero, P. Steinberg, P. Nightingale, D. Cooper, W. P. Cochlan, and R. Kudela. 1996. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature 383:495-501. [DOI] [PubMed] [Google Scholar]
- 13.Colon-Lopez, M. S., and L. A. Sherman. 1998. Transcriptional and translational regulation of photosystem I and II genes in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 180:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooney, R. P., O. Pantos, M. D. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 15.Deora, R. 2002. Differential regulation of the Bordetella bipA gene: distinct roles for different BvgA binding sites. J. Bacteriol. 184:6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du, L. D., and P. E. Kolenbrander. 2000. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect. Immun. 68:4834-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 18.Frias-Lopez, J., G. T. Bonheyo, Q. Jin, and B. W. Fouke. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl. Environ. Microbiol. 69:2409-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froussard, P. 1992. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 20:2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvell, C. D., K. Kim, J. M. Burkholder, R. R. Colwell, P. R. Epstein, D. J. Grimes, E. E. Hofmann, E. K. Lipp, A. D. Osterhaus, R. M. Overstreet, J. W. Porter, G. W. Smith, and G. R. Vasta. 1999. Emerging marine diseases—climate links and anthropogenic factors. Science 285:1505-1510. [DOI] [PubMed] [Google Scholar]
- 22.Hosie, A. H., and P. S. Poole. 2001. Bacterial ABC transporters of amino acids. Res. Microbiol. 152:259-270. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, T. P., A. H. Baird, D. R. Bellwood, M. Card, S. R. Connolly, C. Folke, R. Grosberg, O. Hoegh-Guldberg, J. B. Jackson, J. Kleypas, J. M. Lough, P. Marshall, M. Nystrom, S. R. Palumbi, J. M. Pandolfi, B. Rosen, and J. Roughgarden. 2003. Climate change, human impacts, and the resilience of coral reefs. Science 301:929-933. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. J., I. Estrada, Z. Shen, S. Ress, P. Willcox, M. J. Colston, and G. Kaplan. 1998. Differential gene expression in response to adjunctive recombinant ruman interleukin-2 immunotherapy in multidrug-resistant tuberculosis patients. Infect. Immun. 66:2426-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, F., E. S. Barnathan, and K. Kariko. 1994. Rapid method for screening and cloning cDNAs generated in differential mRNA display: application of Northern blot for affinity capturing of cDNAs. Nucleic Acids Res. 22:1764-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macaluso, K. R., A. Mulenga, J. A. Simser, and A. F. Azad. 2003. Differential expression of genes in uninfected and Rickettsia-infected Dermacentor variabilis ticks as assessed by differential-display PCR. Infect. Immun. 71:6165-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, J. H., K. H. Coale, K. S. Johnson, S. E. Fitzwater, R. M. Gordon, S. J. Tanner, C. N. Hunter, V. A. Elrod, J. L. Nowicki, T. L. Coley, R. T. Barber, S. Lindley, A. J. Watson, K. Van Scoy, C. S. Law, M. I. Liddicoat, R. Ling, T. Stanton, J. Stocjel, C. Collins, A. Anderson, R. Bidigare, M. Ondrusek, M. Latasa, F. J. Millero, K. Lee, W. Yao, J. Z. Zhang, D. Friederich, C. Sakamoto, F. Chavez, K. Buck, Z. Kolber, R. Greene, P. Falkowski, S. W. Chisholm, F. Hoge, R. Swift, J. Yungel, S. Turner, P. Nightingale, A. Hatton, P. Liss, and N. W. Tindale. 1994. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature 371:123-129. [Google Scholar]
- 28.Mate, Z., L. Sass, M. Szekeres, I. Vass, and F. Nagy. 1998. UV-B-induced differential transcription of psbA genes encoding the D1 protein of photosystem II in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 273:17439-17444. [DOI] [PubMed] [Google Scholar]
- 29.Muramatsu, M., and Y. Hihara. 2003. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216:446-453. [DOI] [PubMed] [Google Scholar]
- 30.Nagel, A., J. T. Fleming, and G. S. Sayler. 1999. Reduction of false positives in prokaryotic mRNA differential display. BioTechniques 26:641-643. [DOI] [PubMed] [Google Scholar]
- 31.Nagel, A. C., J. T. Fleming, G. S. Sayler, and K. L. Beattie. 2001. Screening for ribosomal-based false positives following prokaryotic mRNA differential display. BioTechniques 30:988-990. [DOI] [PubMed] [Google Scholar]
- 32.Nomura, S. 1997. Recent knowledge on fish pathogenic bacteria Aeromonas salmonicida, Listonella anguillara and Cytophaga columnaris, and their virulence factors. Nippon Saikingaku Zasshi 52:393-416. [DOI] [PubMed] [Google Scholar]
- 33.Pandolfi, J. M., R. H. Bradbury, E. Sala, T. P. Hughes, K. A. Bjorndal, R. G. Cooke, D. McArdle, L. McClenachan, M. J. Newman, G. Paredes, R. R. Warner, and J. B. Jackson. 2003. Global trajectories of the long-term decline of coral reef ecosystems. Science 301:955-958. [DOI] [PubMed] [Google Scholar]
- 34.Richardson, L. L. 1998. Coral disease: what is really known? Trends Ecol. Evol. 13:438-443. [DOI] [PubMed] [Google Scholar]
- 35.Richardson, L. L. 1996. Horizontal and vertical migration patterns of Phormidium corallyticum and Beggiatoa spp. associated with black-band disease of corals. Microb. Ecol. 32:323-335. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, L. L. 1997. Occurrence of the black band disease cyanobacterium on healthy corals of the Florida Keys. Bull. Mar. Sci. 61:485-490. [Google Scholar]
- 37.Richardson, L. L., K. G. Kuta, S. Schnell, and R. G. Carlton. 1997. Ecology of the black band disease microbial consortium, p. 597-600. In H. A. Lessios and I. G. Macintyre (ed.), Eighth International Coral Reef Symposium, vol. 1. Smithsonian Tropical Research Institute, Balboa, Panama. [Google Scholar]
- 38.Rompf, R., and G. Kahl. 1997. mRNA differential display in agarose gels. BioTechniques 23:28, 30, 32. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg, E., and Y. Ben-Haim. 2002. Microbial diseases of corals and global warming. Environ. Microbiol. 4:318-326. [DOI] [PubMed] [Google Scholar]
- 40.Rützler, K., and D. L. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of the cyanophyte pathogen. Mar. Ecol. 4:301-319. [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schubert, W. D., O. Klukas, N. Krauss, W. Saenger, P. Fromme, and H. T. Witt. 1997. Photosystem I of Synechococcus elongatus at 4 A resolution: comprehensive structure analysis. J. Mol. Biol. 272:741-769. [DOI] [PubMed] [Google Scholar]
- 43.Shepard, B. D., and M. S. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, A. K., and L. A. Sherman. 2000. Identification of iron-responsive, differential gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803 with a customized amplification library. J. Bacteriol. 182:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toncheva-Panova, T., and J. Ivanova. 2000. Influence of physiological factors on the lysis effect of Cytophaga on the red microalga Rhodella reticulata. J. Appl. Microbiol. 88:358-363. [DOI] [PubMed] [Google Scholar]
- 46.Vinnemeier, J., and M. Hagemann. 1999. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch. Microbiol. 172:377-386. [DOI] [PubMed] [Google Scholar]
- 47.Webb, E., J. Moffett, and J. Waterbury. 2001. Iron stress in open-ocean cyanobacteria (Synechococcus, Trichodesmium, and Crocosphaera spp.): identification of the IdiA protein. Appl. Environ. Microbiol. 67:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webber, A. N., and W. Lubitz. 2001. P700: the primary electron donor of photosystem I. Biochim. Biophys. Acta 1507:61-79. [DOI] [PubMed] [Google Scholar]