Abstract
Individual mutations in the tumor suppressor TP53 alter p53 protein function. Some mutations create a non-functional protein, whereas others confer oncogenic activity, which we term ‘oncomorphic’. Since mutations in TP53 occur in nearly all ovarian tumors, the objective of this study was to determine the relationship of oncomorphic TP53 mutations with patient outcomes in advanced serous ovarian cancer patients. Clinical and molecular data from 264 high-grade serous ovarian cancer patients uniformly treated with standard platinum- and taxane-based adjuvant chemotherapy were downloaded from The Cancer Genome Atlas (TCGA) portal. Additionally, patient samples were obtained from the University of Iowa and individual mutations were analyzed in ovarian cancer cell lines. Mutations in the TP53 were annotated and categorized as oncomorphic, loss of function (LOF), or unclassified. Associations between mutation types, chemoresistance, recurrence, and progression-free survival (PFS) were calculated. Oncomorphic TP53 mutations were present in 21.3% of ovarian cancers in the TCGA dataset. Patients with oncomorphic TP53 mutations demonstrated significantly worse PFS, a 60% higher risk of recurrence (HR=1.60, 95% confidence intervals 1.09, 2.33, p=0.015), and higher rates of platinum resistance (χ2 test p=0.0024) when compared with single nucleotide mutations not categorized as oncomorphic. Furthermore, tumors containing oncomorphic TP53 mutations displayed unique protein expression profiles, and some mutations conferred increased clonogenic capacity in ovarian cancer cell models. Our study reveals that oncomorphic TP53 mutations are associated with worse patient outcome. These data suggest that future studies should take into consideration the functional consequences of TP53 mutations when determining treatment options.
Keywords: oncomorphic p53 mutation, TP53, gain-of-function, ovarian cancer, chemoresistance
Introduction
Epithelial ovarian cancer is the most deadly of the gynecologic malignancies and the fifth leading cause of cancer-related death among women (1). Although there has been an improvement in the 5-year survival of patients diagnosed with advanced disease, the long-term survival rate remains poor at 30% (1). Low survival can be attributed to the insidious nature of ovarian cancer progression, resulting in late diagnosis. Unfortunately, 75% of cases involve metastases to the abdominal cavity (FIGO stages III–IV) at the time of diagnosis (2). An additional complication contributing to low survival is the high rate of chemoresistance (1). The ability to predict the patients at highest risk for rapid disease progression would allow clinicians to optimize therapy up front using more aggressive regimens.
The Cancer Genome Atlas (TCGA) has provided key insight into molecular alterations that are common in ovarian tumors (3). Of note, mutations in a single gene, TP53, were identified in 96% of all serous ovarian tumors (3). TP53 encodes the tumor suppressor protein p53, which acts as the major control center in the cellular response to various stress such as DNA-damaging chemotherapy. Once activated in response to chemotherapy, p53 enhances cell cycle arrest and DNA damage repair, or induces apoptosis and senescence if cellular repair is not possible.
Although almost all serous ovarian cancer patients harbor mutations in TP53, the mutations are extremely heterogeneous and occur at almost every codon in the DNA-binding domain of the gene (4). However, the specific TP53 mutation can drastically alter the function of the mutated protein in a myriad of different ways. For example, studies using biochemical assays, cell models, as well as mouse and rat models have demonstrated that some TP53 mutations abolish the wild-type (WT) function of p53 as well as confer new oncogenic activities (5). We have termed these types of mutations oncomorphic TP53 mutations (6). Studies in cultured cancer cell lines and animal models of cancer demonstrate that oncomorphic TP53 mutations can contribute to chemoresistance and cancer progression. However, the phenomenon has not yet been convincingly demonstrated in patients, partly due to the lack of a study population size with sufficient power to observe significant associations (7). This type of analysis is now achievable through the TCGA with the availability of clinical and genetic data from hundreds of ovarian cancer patients. Using these data, as well as findings from patients at the University of Iowa, we sought to test our hypothesis that oncomorphic TP53 mutations in advanced serous ovarian tumors are associated with worse outcomes.
Using stringent criteria to define oncomorphic TP53 mutations, we evaluated the relationship of oncomorphic p53 expression with progression-free survival (PFS), risk of recurrence, and response to standard platinum and taxane chemotherapy. Our data provide the first evidence that ovarian cancer patients with oncomorphic TP53 mutations have worse clinical outcomes compared to patients with unclassified TP53 mutations, including a shorter PFS and a 60% greater risk of recurrence. These findings have important potential implications for all cancers characterized by mutations in TP53.
Materials and methods
Ovarian cancer cell cultures
Eleven ovarian cancer cell lines were utilized in these studies. ES-2, and SKOV3 cells were cultured as monolayers in McCoy’s 5A medium. Caov3 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM). Ovcar3 and UCI-107 cells were cultured in RPMI-1640 medium. Caov4 and SW626 cells were maintained in Leibovitz’s L-15 medium. TOV112D and OV-90 cells were cultured in a 1:1 mixture of MCDB 105 medium containing 1.5 g/l of sodium bicarbonate and medium 199 containing 2.2 g/l sodium bicarbonate. UWB1.289 cells were grown in a 1:1 mixture of RPMI-1640 and Mammary Epithelial Growth Medium (MEGM) (Clonetics/Lonza). All media conditions were supplemented with 10% fetal bovine serum (FBS) and 1 U/ml penicillin and 10 μg/ml streptomycin and cells were maintained in a humidified incubator with 5% CO2 at 37°C. All cell lines are available from American Type Cell Culture, except UCI-107 cells that were generously gifted from Dr Michael J. Goodheart.
The cell line SKOV3 has a loss of function (LOF) TP53 mutation that results in a lack of p53 protein expression. This cell line was used as a model to study the effects of the most common oncomorphic TP53 mutations by stably expressing the following mutants in TP53: R175H, R248Q, R248Q.P72R, R248W, R273C, R273L, R273S, and Y220C as previously described (8).
Western blot analysis
Analysis of protein expression/phosphorylation was performed as previously described (9) for the following proteins: p53 (sc-126; Santa Cruz Biotechnology, Inc.), p21 (no. 2947), ERCC1 (no. 12345), c-Myc (no. 9402), β-catenin (no. 9582), mammalian target of rapamycin (mTOR) (no. 2983) (all from Cell Signaling Technology, Inc.), p-Rb S807 (no. ab47762; Abcam), and β-actin (no. A1978; Sigma).
Clonogenic survival
Cells were trypsinized and plated in triplicate into 60 mm tissue culture dishes at 800 cells/well. Twenty-four hours later, cells were treated with 1 μM cisplatin or 5 nM taxol for 48 h. Fresh media was added and cells were allowed to grow for 21 days. Viable clones were visualized by staining with crystal violet, and colonies >50 cells were counted. Plating efficiency was calculated by dividing the average number of colonies per plate by the number of cells plated. Surviving fractions were calculated by normalization to the plating efficiency.
Subjects
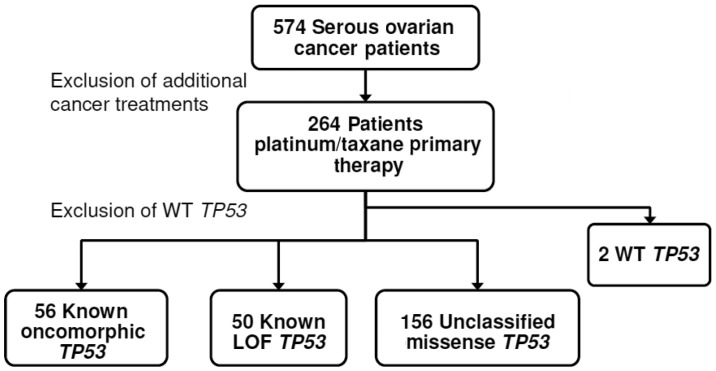
Clinical, genetic, and protein expression data from 264 advanced serous ovarian cancer patients without a previous cancer history were downloaded from the TCGA data portal (accessed 05/06/2013). Analyses were limited to data from those patients who received platinum (carboplatin, cisplatin, or oxaliplatin)- and taxane (Taxotere or Paclitaxel)-based chemotherapy (Fig. 1). Clinical characteristics of the study cohort are listed in Table I. An independent validation patient cohort (n=32) was obtained from the University of Iowa Gynecologic Oncology Tumor Bank. The University of Iowa Institutional Review Board approved these studies. The same inclusion criteria were used for both patient cohorts: patients were of advanced stage (III or IV), specific TP53 sequencing information was available, and clinical outcome was known.
Figure 1.
Inclusion criteria to study the effect of TP53 mutation type on serous ovarian cancer patient outcomes. Out of 574 serous ovarian cancer patients included in The Cancer Genome Atlas (TCGA) dataset, patients included in our study had uniform platinum- and taxane-based primary chemotherapy. Of those, we determined the TP53 mutation based on exon sequencing and discriminated each mutation into four groups: known oncomorphic, known loss of function (LOF), unclassified, or wild-type (WT). Oncomorphic mutations were designated based on in vivo or in vitro evidence of an oncogenic phenotype. LOF mutations contain a nonsense or a frameshift mutation. Remaining TP53 mutations are designated as unclassified mutations, and have unknown functions.
Table I.
Clinical and pathological characteristics of TCGA serous ovarian tumors from patients treated with standard platinum- and taxane-based chemotherapy.
| Characteristic | n | % |
|---|---|---|
| Age at diagnosis | ||
| <60 years | 148 | 56.06 |
| ≥60 years | 116 | 43.94 |
| Vital Status | ||
| Dead | 126 | 47.73 |
| Alive | 138 | 52.27 |
| Tumor grade | ||
| G2 | 21 | 7.95 |
| G3/G4 | 236 | 89.39 |
| Unknown | 7 | 2.65 |
| FIGO stage | ||
| IIIA/B | 21 | 7.95 |
| IIIC | 197 | 74.62 |
| IV | 46 | 17.42 |
| Lymph invasion | ||
| No | 38 | 14.39 |
| Yes | 63 | 23.86 |
| Unknown | 163 | 61.74 |
| Residual disease | ||
| ≤1 cm | 126 | 47.73 |
| >1 cm | 60 | 22.73 |
| Complete removal | 51 | 19.32 |
| Unknown | 27 | 10.23 |
| Clinical response to chemotherapy | ||
| Complete response | 155 | 58.71 |
| Partial response | 24 | 9.09 |
| Stable disease | 19 | 7.20 |
| Progressive disease | 12 | 4.55 |
| No data | 54 | 20.45 |
| Platinum status | ||
| Resistant | 49 | 20.25 |
| Sensitive | 112 | 46.28 |
| Too early | 34 | 14.05 |
| Unknown | 47 | 19.42 |
| p53 mutation type | ||
| LOF | 51 | 19.32 |
| Oncomorphic | 56 | 21.21 |
| Unclassified | 154 | 58.33 |
| WT | 2 | 0.76 |
| Unknown (no sequence information available) | 1 | 0.38 |
TCGA, The Cancer Genome Atlas; LOF, loss of function; WT, wild-type.
Criteria for designating TP53 mutations
TP53 mutations were binned into three categories: oncomorphic, LOF, and unclassified. Oncomorphic mutations were designated based on previously published studies showing that a particular mutation causes an oncogenic phenotype. For example, Hanel et al used a knock-in mouse to determine the function of two common mutations (10). Compared with the p53 null mouse (p53−/−), a mouse carrying a p53 R248Q allele (p53R248Q/−) displayed accelerated tumor onset and shortened survival, but a mouse model carrying a p53 G245S allele (p53G245S/−) showed no differences in survival when compared with the p53−/− mouse (10). These are some of the first data indicating that TP53 mutations vary in function with respect to tumorigenicity. Eight TP53 mutations were considered oncomorphic, and were selected based on previous in vivo and in vitro studies [P151S (11,12), Y163C (13), R175H (14–16), L194R (17), Y220C (18), R248Q (10), R248W (19,20), R273C (21,22), R273H (15,19,23), R273L (24), R282W (13)]. LOF mutations were defined as i) point mutations that create a stop codon (nonsense mutation); or ii) frame shift mutations that cause significant disruptions in the translation of the protein. WT mutations were defined as mutations that do not alter the amino acid sequence. The remaining mutations were single nucleotide substitutions, the function of which is not fully known at this time, but do not meet oncomorphic criteria. These were categorized as ‘unclassified’ mutations. Splice mutations located at the intron-exon borders were categorized into the ‘unclassified’ category due to conflicting studies on their function (25–28).
Defining clinical endpoints
Clinical details available from the TCGA portal were used to document the following clinical endpoints: PFS and platinum status. PFS was defined as the interval between the date of initial surgical removal of the tumor to the date of progression in patients who were not cancer free, or date of recurrence. Chemotherapy details were available that documented the date of last primary platinum treatment. Platinum-free interval was defined as the interval between last primary platinum treatment to the date of progression or recurrence. Platinum status was defined as resistant if the platinum-free interval was <6 months when the patient recurred. Platinum status was defined as sensitive if the interval to recurrence was >6 months, or the follow-up period for those lost to contact was >6 months from the date of the last platinum treatment. Patients who did not progress or have a recurrence were censored in both analyses at the date of the last-known contact.
RPPA protein data
Corrected and normalized reverse phase protein array (RPPA) data were downloaded from the TCGA portal to analyze protein expression differences between patients with oncomorphic, LOF, or unclassified mutations. Detailed information on normalization has been previously reported (3); briefly, the raw data were converted from a log 2 value into an arbitrary linear value and corrected based on the normalization of means among all patient samples.
Statistical analysis
To determine if different mutations confer worse patient outcome, plots of the Kaplan-Meier estimated cumulative probabilities of PFS were constructed. Cox proportional hazard regression was utilized to test for differences in PFS between mutation types using a study endpoint of 60 months, as previously reported (4). To assess for group differences between the mutations on relevant clinical variables, a χ2 test or Fisher’s exact test was utilized where appropriate. A Kruskal-Wallis or Wilcoxon rank sum test was performed to detect differential protein expression between all three mutation groups, or between two groups, respectively. All tests were two sided and tested at the 5% significance level. The data analysis was generated using SAS software, version 9.3 (SAS Institute, Inc.).
Results
Selection of patient population
As shown in Fig. 1, the primary exclusion criterion was patient exposure to treatment beyond adjuvant primary chemotherapy with platinum and taxane. The median PFS for the study population was 13.8 months, and median overall survival was 30.2 months, which is consistent with reported outcomes in the full TCGA ovarian cancer data set (3).
Frequency and spectrum of TP53 mutations
Exon sequencing data were downloaded from the TCGA portal and mutations in TP53 were annotated. Two patients had synonymous missense mutations that retained the integrity of WT p53 protein sequence and were designated as WT. Data for these two patients were excluded due to insufficient sample size (Fig. 1).
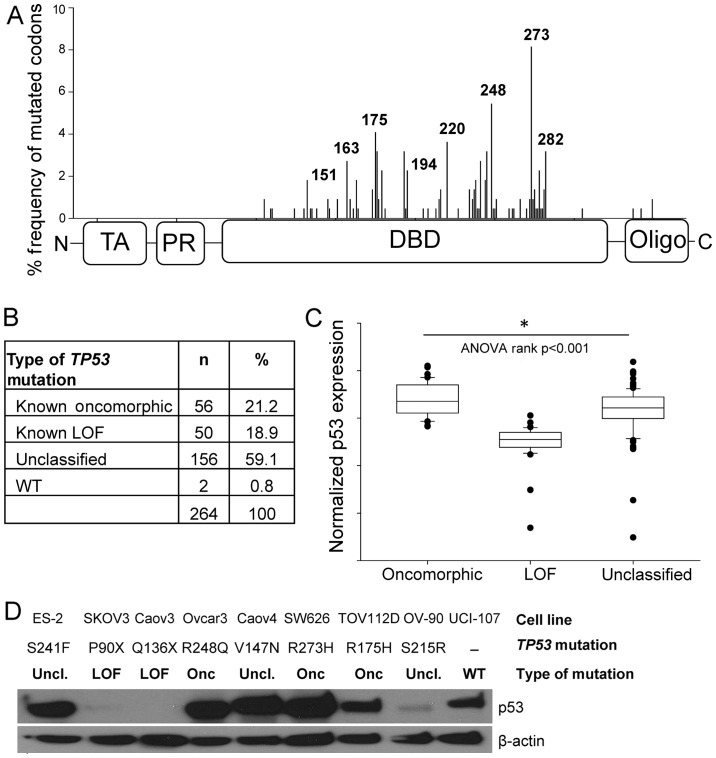
Mutations in TP53 occurred predominately in the DNA-binding domain (Fig. 2A), consistent with a previous report (4). The most common mutations occurred at codons R273 (6.1%), R248 (4.6%), and R175 (3.4%). Oncomorphic mutations comprised 21.2% of the patient population, LOF mutations comprised 18.9%, and the remaining 59.1% were unclassified mutations (Fig. 2B). Splice mutations located at the intron-exon borders were categorized as ‘unclassified’ due to conflicting studies on their function (25–28). Splice mutations occurred in 10% of our study population, a frequency much larger than previously reported (27). We speculate that the advanced technology used to sequence TP53 exons is more sensitive than used previously. The frequency of oncomorphic and LOF mutations in this cohort is similar to that calculated from the International Agency for Research on Cancer p53 database (4,6), thus validating our study population.
Figure 2.
Landscape of TP53 mutations in the study population. (A) TP53 gene structure and frequency of TP53 mutations at individual codons in patients included in this study. Denoted codons are oncomorphic alterations. (B) Number and frequency of TP53 mutations in study cohort categorized by functional consequence. (C) Normalized protein expression of p53 in serous ovarian cancer tumors in the three functional categories of TP53 mutations. *Kruskal-Wallis tests were performed to assess significance. (D) Baseline p53 expression in a panel of nine ovarian cancer cell lines. TA, transactivation domain; PR, proline rich domain; DBD, DNA-binding domain; Oligo, oligomerization domain; LOF, loss of function; Uncl., unclassified.
To confirm our classification of oncomorphic and LOF mutations, we analyzed normalized protein expression of p53 as reported in the RPPA data set. LOF mutations result in loss of p53 protein expression, whereas oncomorphic p53 has been reported to be hyper-stabilized (5). As expected, we detected a significant difference in protein levels of p53 for the oncomorphic, LOF and unclassified mutations (Fig. 2C, p<0.001). Specifically, tumors containing oncomorphic TP53 mutations had the highest p53 protein levels, whereas tumors with LOF TP53 mutations displayed the lowest expression of p53. Tumors with unclassified mutations had a broad range of p53 protein expression.
We utilized a panel of nine ovarian cancer cell lines with various TP53 mutations to characterize expression levels of mutated p53 proteins (Fig. 2D). Three cell lines with oncomorphic TP53 mutations displayed abundant mutated p53 protein expression. Two cell lines with LOF TP53 mutations did not express p53 protein; and cell lines with unclassified TP53 mutations demonstrated a range of p53 protein expression. One cell line, UCI-107, expresses WT TP53.
Oncomorphic mutations in TP53 confer worse patient outcome
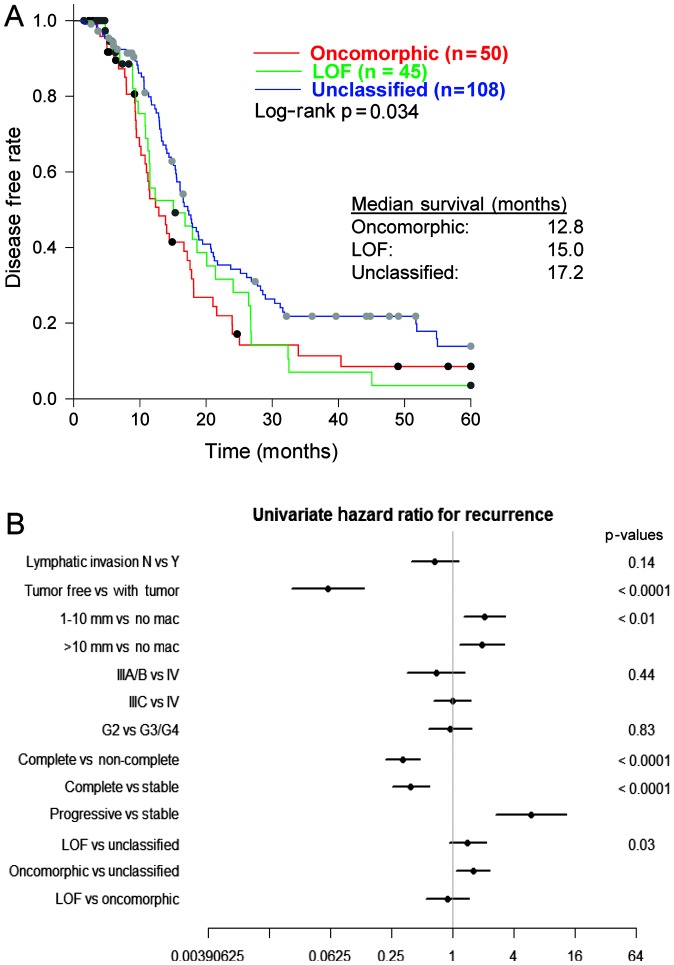
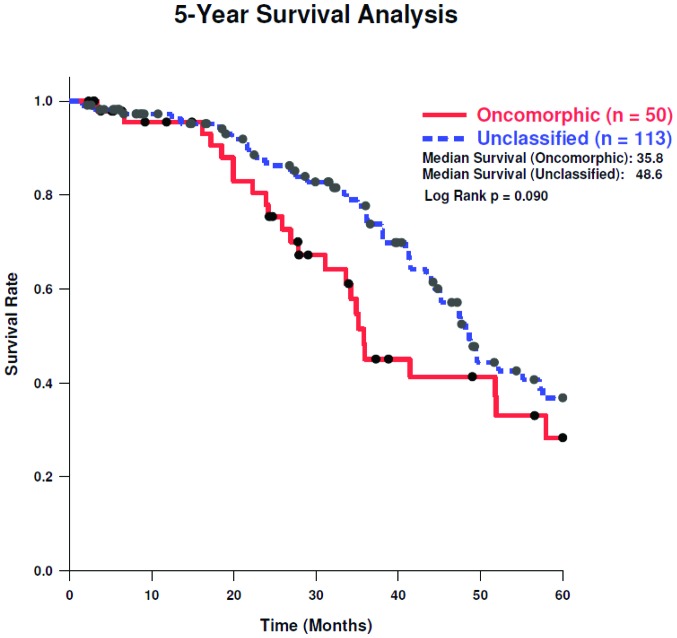
We assessed the association of oncomorphic TP53 mutations with patient outcome, by first calculating PFS among patients with oncomorphic, LOF, or unclassified mutations and found a significant difference between categories (p=0.03). Follow-up pairwise comparisons demonstrated that patients with oncomorphic TP53 mutations showed significantly worse PFS when compared with patients harboring unclassified mutations (p=0.015) (Fig. 3A). The median PFS was 12.8, 15.0, and 17.2 months for patients with oncomorphic, LOF, and unclassified mutations, respectively. Analysis of 5-year survival revealed a trend towards better survival in patients with unclassified mutations as compared to oncomorphic mutations (Fig. 4, log-rank test p=0.11).
Figure 3.
Oncomorphic TP53 mutations are associated with worse progression-free survival (PFS) and increased risk of recurrence. (A) Plot of PFS. Log-rank test was used to assess significance among the three TP53 mutational categories. Median PFS is noted in inset. (B) Hazard ratio plot showing clinical factors associated with recurrence. No mac, no macroscopic disease; G, grade; LOF, loss of function.
Figure 4.
Five-year overall survival rate in patients with oncomorphic and unclassified TP53 mutations. Plots of the Kaplan-Meier estimated cumulative probabilities of overall survival were constructed. Cox proportional hazards regression was utilized to test for differences in progression-free survival (PFS) between mutation types using a study endpoint of 60 months.
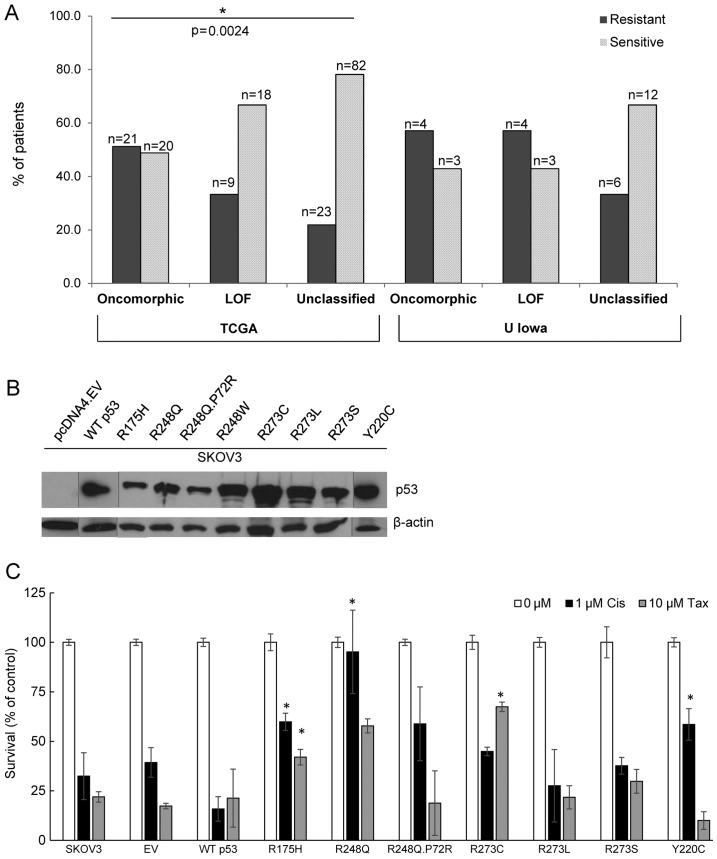
To provide further insight into which clinical factors may be contributing to the differing PFS outcomes between mutational classifications, a univariate comparison of clinical factors was conducted (Table II). Patients with oncomorphic TP53 mutations displayed higher rates of platinum resistance when compared with LOF and unclassified mutants (χ2 test p=0.0024). More than half (51.2%) of patients with oncomorphic mutations displayed platinum resistance, whereas patients with unclassified mutations had the highest rates of platinum sensitivity (Table II). In addition, patients with oncomorphic TP53 mutations had almost 60% higher odds of recurrence (HR=1.60, 95% confidence intervals 1.09, 2.33, p=0.015) when compared to patients with other unclassified mutations (Fig. 3B). We also observed the anticipated associations of recurrence with residual disease and response to therapy (Fig. 3B).
Table II.
Univariate analysis of association of clinical factors with TP53 mutation categories (oncomorphic, LOF, and unclassified) demonstrates that platinum status is significantly different among the three mutation groups.
| Variable | Category | n | p-value χ2 test | ||
|---|---|---|---|---|---|
|
| |||||
| Oncomorphic | LOF | Unclassified | |||
| Lymphatic invasion | No | 13 | 6 | 19 | 0.0767 |
| Yes | 9 | 13 | 39 | ||
| Tumor grade | G2 | 4 | 5 | 12 | 0.8373 |
| G3/G4 | 50 | 43 | 138 | ||
| Cancer status | Tumor free | 15 | 12 | 45 | 0.8439 |
| With tumor | 37 | 33 | 100 | ||
| Residual tumor | ≤1 cm | 28 | 26 | 70 | 0.5075 |
| >1 cm | 11 | 13 | 33 | ||
| No mac | 15 | 6 | 30 | ||
| Tumor stage | IIIA/B | 6 | 4 | 11 | 0.8529 |
| IIIC | 40 | 36 | 117 | ||
| IV | 10 | 10 | 25 | ||
| Vital status | Dead | 28 | 22 | 73 | 0.8234 |
| Alive | 28 | 28 | 80 | ||
| Platinum status | Resistant | 21 | 9 | 23 | 0.0024 |
| Sensitive | 20 | 18 | 82 | ||
| Therapy outcome | Complete response | 33 | 29 | 76 | 0.0970 |
| Progressive disease | 4 | 2 | 4 | ||
| Stable disease | 4 | 14 | 20 | ||
LOF, loss of function; mac, macroscopic disease.
To validate the clinical and genetic data obtained from the TCGA, we determined rates of chemoresistance in patients who were diagnosed with ovarian cancer and had banked tumors at the University of Iowa. Sequencing information on TP53 was available for all tumors. We observed a similar trend towards resistance in tumors with oncomorphic TP53 (Fig. 5A). In addition, patients with unclassified TP53 mutations demonstrated the highest sensitivity to chemotherapy. A p53 null cell line (SKOV3) was utilized to express the most common TP53 oncomorphic mutations (Fig. 5B). Clonogenic survival in response to cisplatin treatment was enhanced by cells expressing R175H, R248Q, and Y220C oncomorphic p53 mutant proteins. In response to taxol chemotherapy, clonogenic survival was enhanced in cells expressing the R175H and R273C p53 mutated proteins (Fig. 5C).
Figure 5.
Tumors with oncomorphic TP53 are more resistant to chemotherapy than patients with loss of function (LOF) or unclassified mutations. Rates of platinum resistance among The Cancer Genome Atlas (TCGA) cohort and a validation cohort from the University of Iowa. Number of patients (n) are noted in the plot. (B) The most common TP53 mutations were expressed in a cell line (SKOV3, p53 null) to examine the ability of oncomorphic p53 mutant proteins to cause chemoresistance to cisplatin or taxol. Western blot images are combined from two separate gels (demarked by the gray lines separating lanes from different gels). (C) Clonogenic survival of cells stably expressing various TP53 mutant proteins after 48 h treatment with 1 μM cisplatin (Cis) or 10 nM taxol (Tax). *P<0.05 vs. empty vector (EV) with the same treatment.
Protein expression differences between oncomorphic mutations and unclassified mutations
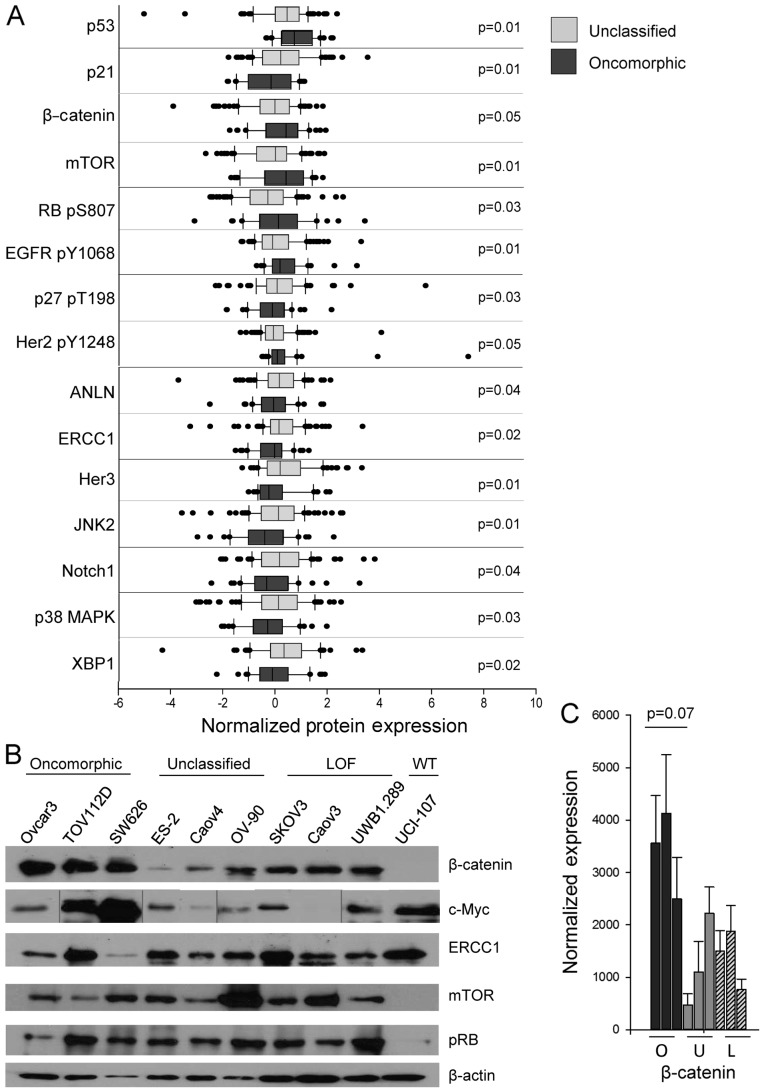
We next interrogated possible mechanisms of chemoresistance in tumors containing oncomorphic mutations by comparing protein expression profiles between oncomorphic and unclassified mutations. Data, which are part of TCGA dataset, were obtained by RPPA, a high-throughput technique for simultaneous measurement of protein expression in a large number of biological samples using antibody-based methods. We observed differential expression of 15 different proteins in tumors with either oncomorphic or unclassified TP53 mutations (Fig. 6A, Table III). In particular, the pro-apoptotic protein BAK and the cell cycle regulator p21 (CIP1/WAF1) were expressed at a significantly lower level in tumors with oncomorphic TP53 mutations. β-catenin, phosphorylated epidermal growth factor receptor (EGFR) (Y1068), and mTOR were significantly elevated in patients with oncomorphic TP53 mutations compared with patients with unclassified mutations. Further evaluation of the RPPA data from the three mutational categories (oncomorphic, LOF and unclassified) revealed differences in tumor protein expression (Table IV). To further define the most significantly altered pathways in cells with oncomorphic TP53, we assessed the expression of the targets identified from the TCGA dataset in representative cell lines (Fig. 6B). The most highly correlated alterations were in the β-catenin pathway, known to be associated with ovarian cancer carcinogenesis and proliferation (29) (Fig. 6C).
Figure 6.
Tumors with oncomorphic TP53 mutations have elevated expression and activity of proteins involved in tumor growth as compared to tumors with unclassified mutations. (A) Reverse phase protein arrays (RPPAs) were used to determine protein expression in The Cancer Genome Atlas (TCGA) analysis. Normalized protein expression was downloaded and compared using a Wilcoxon rank sum test to identify proteins differentially expressed between the two groups. The full dataset is available in Table III. Comparison of RPPA data among oncomorphic, loss of function (LOF), and unclassified TP53 are available in Table IV. (B) Analysis of the expression of proteins identified through the TCGA analysis in a panel of ovarian cancer cell lines. Western blot image of c-Myc contains gray lines demarking re-arrangement of the image, however it is an image of the same blot. (C) Densitometry analysis of β-catenin trended towards higher expression in the panel of cell lines, tested by Kruskal-Wallis test p=0.07.
Table III.
Significant differential protein expression between tumors with oncomorphic versus unclassified TP53 mutations as determined by Wilcoxon rank sum test. Median protein expression is presented.
| Analysis variable | Median expression | p-value Wilcoxon test (two sided) | |
|---|---|---|---|
|
| |||
| Oncomorphic | Unclassified | ||
| ANLN | −0.08 | 0.14 | 0.042568565 |
| β-catenin | 0.40 | −0.03 | 0.047657921 |
| EGFR pY1068 | 0.16 | −0.12 | 0.007949216 |
| ERCC1 | −0.04 | 0.12 | 0.015114876 |
| Her2 pY1248 | 0.06 | −0.08 | 0.049131573 |
| Her3 | −0.26 | 0.17 | 0.010622594 |
| JNK2 | −0.42 | 0.10 | 0.00516184 |
| mTOR | 0.40 | −0.01 | 0.008801626 |
| Notch1 | −0.35 | 0.14 | 0.040835561 |
| p21 | −0.18 | 0.19 | 0.007848037 |
| p27 pT198 | −0.13 | 0.05 | 0.034859604 |
| p38 MAPK | −0.31 | 0.10 | 0.025973549 |
| p53 | 0.71 | 0.43 | 0.008155086 |
| RB pS807 S811 | 0.11 | −0.30 | 0.028065342 |
| XBP1 | −0.13 | 0.32 | 0.020961678 |
EGFR, epidermal growth factor receptor.
Table IV.
Significant differential protein expression among tumors with oncomorphic, LOF and unclassified TP53 mutations as determined by Kruskal-Wallis test. Median protein expression is presented.
| Analysis variable | Median expression | p-value Kruskal-Wallis test | ||
|---|---|---|---|---|
|
| ||||
| Oncomorphic | LOF | Unclassified | ||
| ANLN | −0.08 | −0.09 | 0.14 | 0.044578193 |
| BAX | −0.11 | 0.12 | −0.13 | 0.02924444 |
| Beclin | 0.01 | −0.24 | 0.12 | 0.047399782 |
| CD31 | 0.15 | −0.23 | 0.12 | 0.019310324 |
| CMET pY1235 | −0.16 | −0.26 | 0.09 | 0.040573763 |
| EGFR pY1068 | 0.16 | −0.21 | −0.12 | 0.025684237 |
| Her3 | −0.26 | −0.23 | 0.17 | 0.003727584 |
| JNK2 | −0.42 | 0.26 | 0.10 | 0.002990478 |
| mTOR | 0.40 | 0.07 | −0.01 | 0.030649059 |
| p21 | −0.18 | −0.20 | 0.19 | 0.027093184 |
| p53 | 0.71 | −0.90 | 0.43 | 4.47E-16 |
| PCNA | 0.01 | 0.73 | −0.13 | 0.009614359 |
| RBM3 | −0.09 | 0.60 | 0.08 | 0.018983458 |
| RB pS807 S811 | 0.12 | 0.11 | −0.30 | 0.022229704 |
| XBP1 | −0.13 | −0.18 | 0.32 | 0.000881485 |
LOF, loss of function.
Discussion
Recent advances in cancer biology involve understanding the effects of mutations in TP53 on the function of the mutant protein (5). Many clinical studies have attempted to correlate the presence of a TP53 mutation with patient survival or the development of chemoresistance (7). The results of these studies are conflicting primarily because of indiscriminate grouping of TP53 mutations with different functions (oncomorphic, LOF and unclassified). Given that 21% of all ovarian cancer patients harbor oncomorphic TP53 mutations, studies which take into account the functional implications of these mutations are vital. The availability of a large cohort of ovarian cancer tumors and corresponding clinical data through TCGA has made it possible to address the clinical consequence of oncomorphic mutations in TP53 for the first time and to confirm the mechanistic implications of oncomorphic p53 expression in representative cell models. Thus, the objective of our study was to determine if oncomorphic TP53 mutations are associated with worse patient outcomes. We demonstrate that oncomorphic TP53 mutations predict for worse PFS and higher rates of chemoresistance and recurrence. Preclinical models confirm the oncomorphic function of the identified TP53 mutations and suggest mechanisms by which oncomorphic TP53 drive ovarian cancer cell growth.
Although sequence similarities exist among many p53 mutant proteins, to date only stringent biological, in vivo assays can determine oncomorphic properties (6). Accordingly, a previous study using less stringent criteria to define ‘gain of function’ TP53 mutations did not find a significant relationship between the gain of function mutations and chemoresistance (30). Herein we used more stringent criteria to define oncomorphic mutations and propose that our findings more clearly delineate the impact of these oncogenic proteins. Our criteria required that mutations increase clonogenic potential in vitro or increase tumorigenesis in vivo as compared to TP53-null mice to be considered oncomorphic (10–24,31,32). Using these criteria, we found that the presence of a TP53 oncomorphic mutation in a patient tumor specimen predicts for platinum resistance.
To understand the oncogenic properties of oncomorphic p53 proteins, we analyzed differential protein expression between the TP53 mutation groups. The cell cycle regulator p21, which is induced by p53 and results in cell cycle arrest, was expressed at a low level in tumors containing oncomorphic TP53 mutations. Levels of phosphorylated p27 were also lower in these samples. Conversely, tumors with unclassified TP53 mutations displayed higher p21 expression, suggesting that some of the unclassified mutations may have residual WT p53 functions. Previous studies have demonstrated that positive p21 staining in ovarian tumor specimens correlates with an overall survival advantage (33,34). Our data also indicated that tumors with oncomorphic TP53 have increased expression of activated pro-growth pathways, such as phosphorylation of EGFR, Her2, and retinoblastoma protein (Rb). EGFR phosphorylation at Y1068 is a hallmark for activated EGFR signaling and is the site of Grb2 and Ras binding that perpetuate Ras activation and mitogen-activated protein kinase signaling (35). The proteins mTOR and β-catenin, which are commonly overexpressed in cancer, were also significantly increased in oncomorphic TP53 tumors, indicating enhanced pro-survival signaling, however. Recently, high β-catenin was associated with poor ovarian cancer patient outcome (29). This protein was the most highly altered in our panel of ovarian cancer cell lines as well as in patient tumors. These data correlate well with in vitro studies showing that EGFR is a direct transcriptional target of oncomorphic p53 proteins (36). In addition, others have shown that oncomorphic p53 regulates expression of key cell cycle regulators (37). Understanding the molecular consequences of oncomorphic TP53 mutations has the potential to identify key signaling targets that could be blocked in order to overcome chemoresistance in tumors with these oncogenic mutations.
Patients whose tumors expressed unclassified TP53 mutations made up the majority of the ovarian cancer study population. These patients represent an interesting clinical population since our data demonstrate that patients harboring unclassified mutations are significantly more sensitive to chemotherapy and have lower rates of recurrence. Tumors with unclassified TP53 mutations express the mutated p53 protein at a fairly high level, and it is possible that these proteins have some residual WT p53 function as evidenced by higher expression of p21. The overall survival of patients with unclassified mutations trended towards improved 5-year survival as compared to oncomorphic mutations. Note, however, that overall survival data are not mature for some patients in TCGA dataset; thus, overall survival should be re-examined when these data are complete.
Although two patients with WT TP53 were excluded from our study, and these patients are rare in advanced ovarian cancers, a recent study of 11 ovarian tumors with WT p53 reported a worse overall survival and PFS as compared to a mutated TP53 (38). The study by Wong et al represents a step towards understanding how p53 function affects outcomes, but it remains unclear why the tumors with functional p53 fail to respond to standard DNA-damaging chemotherapy (38). One possibility is that other mutations present in the tumors drive drug resistance; another possibility is that WT p53 enforces cell cycle checkpoints, making the cells less vulnerable to chemotherapeutic agents which act specifically in mitosis (9).
An important aspect of p53 biology is the integrity of the second TP53 allele. Mutant p53 proteins can exert dominant negative activity by inhibiting DNA binding and hence, the tumor-suppressive function of the remaining WT TP53 allele (39). The status of both alleles is necessary to have a complete understanding of the effect of a particular mutation; however this is a limitation of the TCGA data. The use of exon sequencing did not distinguish between loss of heterozygosity (LOH) or tumor heterogeneity (3). Future studies will need to take this into account.
In conclusion, almost all advanced serous ovarian tumors contain TP53 mutations. Understanding the p53 mutational category, which significantly impacts function, is critical to predicting patient outcomes. Specifically, we demonstrate that patients with oncomorphic TP53 mutations are significantly more resistant to chemotherapy, have shorter PFS and a higher risk of recurrence. A recent study in Li-Fraumeni syndrome patients analyzed the individual impact of common TP53 missense mutations and identified a particular mutation (R282W) that results in earlier onset of tumor formation (40). Such patients, and patients identified in our study with oncomorphic TP53 mutations deserve careful follow-up post-therapy and may require novel treatment regimens to improve outcomes. In addition, when studying the impact of new therapies in ovarian cancer, we propose that stratification should be considered based upon p53 mutational category.
Acknowledgements
We are grateful for the continued services provided by the Genomics Division of the Iowa Institute for Human Genetics. This study was supported by NIH R01CA99908 (K.K.L.) and the Department of Obstetrics and Gynecology Research Development Fund (K.K.L.). The agencies had no involvement in study design, collection, analysis and interpretation of data, writing of the report, or the decision to submit the report for publication. D.D. and K.W.T. are owners of Immortagen, L.L.C.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 5.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brachova P, Thiel KW, Leslie KK. The consequence of oncomorphic TP53 mutations in ovarian cancer. Int J Mol Sci. 2013;14:19257–19275. doi: 10.3390/ijms140919257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall J, Paul J, Brown R. Critical evaluation of p53 as a prognostic marker in ovarian cancer. Expert Rev Mol Med. 2004;6:1–20. doi: 10.1017/S1462399404007781. [DOI] [PubMed] [Google Scholar]
- 8.Meng X, Dizon DS, Yang S, et al. Strategies for molecularly enhanced chemotherapy to achieve synthetic lethality in endometrial tumors with mutant p53. Obstet Gynecol Int. 2013;2013:828165. doi: 10.1155/2013/828165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng X, Laidler LL, Kosmacek EA, et al. Induction of mitotic cell death by overriding G2/M checkpoint in endometrial cancer cells with non-functional p53. Gynecol Oncol. 2013;128:461–469. doi: 10.1016/j.ygyno.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanel W, Marchenko N, Xu S, Xiaofeng Yu S, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie TX, Zhou G, Zhao M, et al. Serine substitution of proline at codon 151 of TP53 confers gain of function activity leading to anoikis resistance and tumor progression of head and neck cancer cells. Laryngoscope. 2013;123:1416–1423. doi: 10.1002/lary.23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scian MJ, Stagliano KE, Deb D, et al. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- 14.Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Olive KP, Tuveson DA, Ruhe ZC, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Wang YX, Kotlikoff MI. Inactivation of calcium-activated chloride channels in smooth muscle by calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci USA. 1997;94:14918–14923. doi: 10.1073/pnas.94.26.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko JL, Chiao MC, Chang SL, et al. A novel p53 mutant retained functional activity in lung carcinomas. DNA Repair (Amst) 2002;1:755–762. doi: 10.1016/S1568-7864(02)00094-0. [DOI] [PubMed] [Google Scholar]
- 18.Sproston AR, Boyle JM, Heighway J, Birch JM, Scott D. Fibroblasts from Li-Fraumeni patients are resistant to low dose-rate irradiation. Int J Radiat Biol. 1996;70:145–150. doi: 10.1080/095530096145139. [DOI] [PubMed] [Google Scholar]
- 19.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 20.Krepulat F, Löhler J, Heinlein C, Hermannstädter A, Tolstonog GV, Deppert W. Epigenetic mechanisms affect mutant p53 transgene expression in WAP-mutp53 transgenic mice. Oncogene. 2005;24:4645–4659. doi: 10.1038/sj.onc.1208557. [DOI] [PubMed] [Google Scholar]
- 21.Bergamaschi D, Gasco M, Hiller L, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer cell. 2003;3:387–402. doi: 10.1016/S1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 22.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer cell. 2003;3:403–410. doi: 10.1016/S1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 23.Duan W, Ding H, Subler MA, et al. Lung-specific expression of human mutant p53-273H is associated with a high frequency of lung adenocarcinoma in transgenic mice. Oncogene. 2002;21:7831–7838. doi: 10.1038/sj.onc.1205909. [DOI] [PubMed] [Google Scholar]
- 24.Morselli E, Tasdemir E, Maiuri MC, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 25.Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofstetter G, Berger A, Fiegl H, et al. Alternative splicing of p53 and p73: the novel p53 splice variant p53delta is an independent prognostic marker in ovarian cancer. Oncogene. 2010;29:1997–2004. doi: 10.1038/onc.2009.482. [DOI] [PubMed] [Google Scholar]
- 27.Holmila R, Fouquet C, Cadranel J, Zalcman G, Soussi T. Splice mutations in the p53 gene: case report and review of the literature. Hum Mutat. 2003;21:101–102. doi: 10.1002/humu.9104. [DOI] [PubMed] [Google Scholar]
- 28.Sameshima Y, Akiyama T, Mori N, et al. Point mutation of the p53 gene resulting in splicing inhibition in small cell lung carcinoma. Biochem Biophys Res Commun. 1990;173:697–703. doi: 10.1016/S0006-291X(05)80091-9. [DOI] [PubMed] [Google Scholar]
- 29.Bodnar L, Stanczak A, Cierniak S, et al. Wnt/β-catenin pathway as a potential prognostic and predictive marker in patients with advanced ovarian cancer. J Ovarian Res. 2014;7:16. doi: 10.1186/1757-2215-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang HJ, Chun SM, Kim KR, Sohn I, Sung CO. Clinical relevance of gain-of-function mutations of p53 in high-grade serous ovarian carcinoma. PloS One. 2013;8:e72609. doi: 10.1371/journal.pone.0072609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monti P, Campomenosi P, Ciribilli Y, et al. Characterization of the p53 mutants ability to inhibit p73 beta transactivation using a yeast-based functional assay. Oncogene. 2003;22:5252–5260. doi: 10.1038/sj.onc.1206511. [DOI] [PubMed] [Google Scholar]
- 32.Liu G, McDonnell TJ, Montes de Oca Luna R, et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci USA. 2000;97:4174–4179. doi: 10.1073/pnas.97.8.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose SL, Goodheart MJ, DeYoung BR, Smith BJ, Buller RE. p21 expression predicts outcome in p53-null ovarian carcinoma. Clin Cancer Res. 2003;9:1028–1032. [PubMed] [Google Scholar]
- 34.Schmider A, Gee C, Friedmann W, et al. p21 (WAF1/CIP1) protein expression is associated with prolonged survival but not with p53 expression in epithelial ovarian carcinoma. Gynecol Oncol. 2000;77:237–242. doi: 10.1006/gyno.2000.5748. [DOI] [PubMed] [Google Scholar]
- 35.Rojas M, Yao S, Lin YZ. Controlling epidermal growth factor (EGF)-stimulated Ras activation in intact cells by a cell-permeable peptide mimicking phosphorylated EGF receptor. J Biol Chem. 1996;271:27456–27461. doi: 10.1074/jbc.271.44.27456. [DOI] [PubMed] [Google Scholar]
- 36.Ludes-Meyers JH, Subler MA, Shivakumar CV, et al. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Agostino S, Strano S, Emiliozzi V, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Wong KK, Izaguirre DI, Kwan SY, et al. Poor survival with wild-type TP53 ovarian cancer? Gynecol Oncol. 2013;130:565–569. doi: 10.1016/j.ygyno.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004;23:2330–2338. doi: 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Qian J, Hu Y, et al. Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep. 2014;4:4223. doi: 10.1038/srep04223. [DOI] [PMC free article] [PubMed] [Google Scholar]