SUMMARY
Negative elongation factor (NELF) is known to enforce promoter-proximal pausing of RNA polymerase II (Pol II), a pervasive phenomenon observed across multicellular genomes. However, the physiological impact of NELF on tissue homeostasis remains unclear. Here, we show that whole-body conditional deletion of the B subunit of NELF (NELF-B) in adult mice results in cardiomyopathy and impaired response to cardiac stress. Tissue-specific knockout of NELF-B confirms its cell-autonomous function in cardiomyocytes. NELF directly supports transcription of those genes encoding rate-limiting enzymes in fatty acid oxidation (FAO) and the tricarboxylic acid (TCA) cycle. NELF also shares extensively transcriptional target genes with peroxisome proliferator-activated receptor α (PPARα), a master regulator of energy metabolism in the myocardium. Mechanistically, NELF helps stabilize the transcription initiation complex at the metabolism-related genes. Our findings strongly indicate that NELF is part of the PPARα-mediated transcription regulatory network that maintains metabolic homeostasis in cardiomyocytes.
INTRODUCTION
RNA polymerase II (Pol II) is preferentially accumulated at transcription start sites (TSSs) of a large number of genes in multicellular organisms (Adelman and Lis, 2012; Levine, 2011). Whereas the enrichment of Pol II density at TSS alone is not necessarily indicative of a distinct mode of regulation, whole-genome analysis of nascent transcripts clearly demonstrates that Pol II indeed pauses at a position downstream of TSS after the synthesis of short stretches of RNA (Core et al., 2008). Furthermore, negative elongation factor (NELF) in metazoan is an important regulator of Pol II pausing (Kwak and Lis, 2013; Yamaguchi et al., 1999). Depletion of any of the four NELF subunits results in disintegration of the entire complex and global reduction of Pol II pausing. NELF-mediated Pol II pausing is antagonized by the positive transcription elongation factor, P-TEFb, a cyclin-dependent kinase (Zhou et al., 2012). Whereas NELF was first identified biochemically as an inhibitor of transcription elongation, subsequent studies indicate that NELF-mediated Pol II pausing can lead to both decreased and increased transcription (Adelman and Lis, 2012). The underlying mechanism by which NELF facilitates transcription is not fully understood. However, it has been shown that NELF-mediated Pol II pausing can prevent the encroachment of nucleosomes at the promoter-proximal region, suggesting that NELF may support multiple rounds of transcription in vivo by maintaining a nucleosome-free region at the promoter (Gilchrist et al., 2010; Sun and Li, 2010). In contrast to the extensive in vitro studies, there is a significant gap of knowledge concerning the physiological roles of NELF in mammals.
Cardiomyopathy is characterized by a rigid, thick, and enlarged heart muscle (Cahill et al., 2013). As cardiomyopathy deteriorates, normal cardiac functions (e.g., blood pumping and maintenance of electrical rhythm) are significantly compromised due to myocyte loss and increased fibrosis. This can ultimately result in heart failure, a prevalent and debilitating disease with high morbidity and mortality. At the histological level, hearts with cardiomyopathy manifest with infiltrating inflammatory cells and interstitial collagen accumulation. One of the major causes of cardiomyopathy is inefficient energy production in cardiomyocytes, which results in failure to meet the high demands of energy consumption and compromised intracellular Ca2+ homeostasis for contraction (Frey et al., 2012). In the normal myocardium, cardiomyocytes alternate between carbohydrates and fatty acids as sources of energy, with the latter contributing up to 70% of the energy requirement for an adult heart (Stanley et al., 2005). Energy metabolism is regulated by both acute mechanisms (e.g., allosteric controls and posttranslational modifications) and long-term transcriptional regulation that renders more sustained changes in metabolic rates. Reduced transcription of rate-limiting enzymes involved in cardiac fatty acid metabolism is often associated with heart failure, forcing the cardiac switch to carbohydrates as the main source of energy (Hue and Taegtmeyer, 2009).
Several members of the nuclear receptor superfamily and their coactivators, in particular peroxisome proliferator-activated receptors (PPARs), PPAR gamma coactivator 1 (PGC-1), and estrogen-related receptors (ERRs), are known to play critical roles in controlling energy-metabolism-related transcription in cardiomyocytes (Giguère, 2008; Madrazo and Kelly, 2008; Rowe et al., 2010). Impairment of the transcriptional programs dictated by these critical regulators in humans is often associated with heart failure and myocardial ischemia (Sihag et al., 2009). Consistent with the clinical observations, heart-specific disruption of the mouse counterparts of these transcription factors results in cardiac dysfunction with reduced capacity in fatty acid oxidation (FAO) and mitochondrial ATP production (Huss et al., 2007; Smeets et al., 2008).
To elucidate the physiological role of NELF in homeostasis of adult tissues, we generated both whole-body and cardiomyocyte-specific knockout (KO) mouse models that deleted the B subunit of mouse NELF (NELF-B). By combining transcriptomics and metabolomics approaches, we further investigated the underlying mechanism by which NELF contributes to cardiac functions.
RESULTS AND DISCUSSION
Inducible NELF-B Knockout Animals Develop Cardiomyopathy
Our previous work indicates that mouse NELF-B is essential for early embryogenesis (Amleh et al., 2009). To determine the role of NELF-B in adult tissue, we circumvented embryonic lethality of conventional NELF-B KO by crossing the NELF-Bf/f mice with a whole-body tamoxifen (TAM)-inducible Cre-ER strain (Figure S1A). The resulting NELF-Bf/f, Cre-ER mice were born with the expected Mendelian ratio. NELF-Bf/f, Cre-ER and their control littermates were injected with TAM at 8 weeks of age and confirmed for TAM-induced deletion of the floxed NELF-B allele (Figure S1B). Compared to TAM-treated control mice, NELF-Bf/f, Cre-ER mice with inducible KO of NELF-B had a shortened life span (11 months), resulting from increased incidences of sudden death with no outward signs of gross abnormalities. For this reason, we selected 10 months (40 weeks) as our primary endpoint for the following histological study.
Histological analysis of KO myocardium revealed obvious signs of cardiomyopathy accompanied by infiltrating inflammatory cells including neutrophils (Figures 1A, S1C, and S1D). This was in sharp contrast to the myocardium from age-matched animals carrying one or two alleles of wild-type NELF-B, either with or without Cre-ER (NELF-B+/+, Cre-ER; NELF-Bf/f; and NELF-B+/f, Cre-ER; collectively referred to as the control group hereafter). Immunohistochemistry confirmed efficient depletion of NELF-B in cardiomyocytes of the KO mice (Figure S1E). Inflammatory infiltration was observed in 100% of the KO mice at 32 weeks after TAM-induced NELF-B deletion (40 weeks of age). Thus, the KO mice developed the pathological phenotype with a high penetrance and relatively early onset.
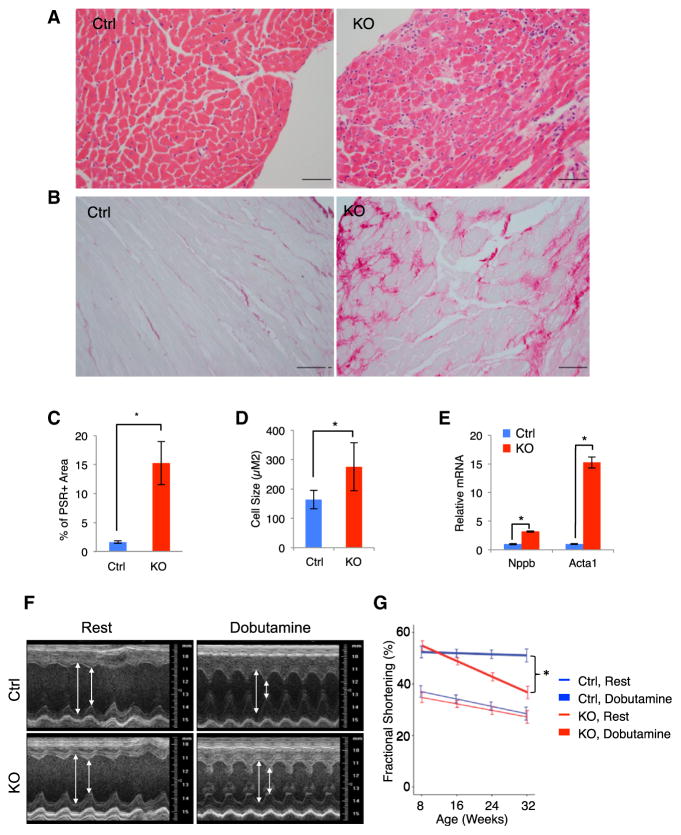
Figure 1. NELF-B KO Leads to Cardiomyopathy.
(A) Hematoxylin and eosin staining of left ventricles from control (Ctrl) and NELF-B knockout (KO) mice. The scale bar represents 50 μm.
(B) Picrosirius red staining for collagen deposition. The scale bar represents 50 μm.
(C) Quantification of picrosirius red staining (n = 5). Here and elsewhere in the figures, *p < 0.05. Error bars represent SEM.
(D) Quantification of cell size of over 100 individual cardiomyocytes (n = 3).
(E) Relative mRNA levels of two hypertrophy markers (n = 6).
(F) Representative left ventricular M-mode echocardiography. The double-headed arrows indicate the left ventricular dimension at diastole (longer arrow) and systole (shorter arrow)
(G) Fractional shortening from the left ventricle (LV) at rest (thin lines) and following dobutamine injection (thick lines). Ctrl: n = 11; KO: n = 8.
Fibrosis is often associated with cardiomyopathy, and we therefore stained heart sections with picrosirius red for collagen deposition. Pronounced interstitial fibrosis was observed in the left ventricles of KO mice (Figures 1B and 1C). Because cardiac hypertrophy is an initiating event in the development of progressive heart failure, we measured myocyte cross-sectional areas. The average individual cardiomyocyte cross-sectional area in the KO mice was significantly larger than their control counterparts (Figure 1D). In addition, expression of two established markers of cardiac hypertrophy, Nppb and Acta1, were markedly elevated in the KO mice (Figure 1E). Taken together, these results are consistent with the cardiomyopathy evaluated at autopsy of the NELF-B KO mice.
To further assess cardiac function of the live KO animals, we performed echocardiography of the left ventricle (LV) from weeks 8 to 32, at 8-week intervals. We evaluated function at rest and following pharmacologically induced cardiac stress. At the 8-week time point prior to TAM-induced NELF-B deletion, the KO mice exhibited comparable LV function as the control littermates (Figures 1F and 1G; Table S1). Following the TAM-induced NELF-B deletion, the control and KO groups of mice at rest displayed no differences for up to the 32-week time point evaluated, indicating that NELF-B deletion did not affect LV function under basal conditions. Dobutamine, a β1-adrenergic agonist and cardiac-stress inducer, is used to evaluate cardiac reserve in experimental animals and in patients who cannot exercise on a treadmill. Stress echocardiography was acquired at 30 min after intraperitoneal injection of dobutamine. In both control and KO mice, dobutamine treatment significantly increased fractional shortening (FS) and decreased LV end systolic and end diastolic dimensions at all age groups measured (Figures 1F and 1G; Table S1). This indicates that both control and KO mice responded to dobutamine stress. However, KO mice at week 32 demonstrated significantly lower FS compared to the control mice treated with dobutamine, suggesting impairment of cardiac reserve in KO mice. The functional cardiac deterioration in KO mice strongly indicates an important role of NELF-B in sustaining normal cardiac contractility under stressed conditions.
Despite the effective depletion of NELF-B in cardiomyocytes (Figure S1E), it remained possible that the cardiac phenotype could be due to NELF-B deletion in other tissue/cell types. We therefore generated a cardiomyocyte-specific inducible knockout mouse model for NELF-B by crossing the floxed NELF-B mice with α-myosin heavy chain promoter-driven, inducible MerCreMer (α-MHC-MCM) transgenic mice. TAM-induced depletion of NELF-B in cardiomyocytes of the resulting MHC-KO animals was confirmed by genotyping and mRNA analysis (Figures S2A and S2B). Similar to the findings of the whole-body KO, the cardiomyocyte-specific KO mice developed prominent signs of cardiomyopathy, excessive cardiac fibrosis, and elevated expression of the hypertrophy marker Acta1 by age of 24 weeks (Figures S2C–S2E). The mRNA increase of Nppb, another hypertrophy marker, in the cardiomyocyte-specific KO was not as significant as that in the whole-body KO (compare Figures 1E and S2E). However, the overall similarity of cardiac defects between the whole-body and cardiomyocyte-specific KO mice strongly indicates a cell-autonomous role of NELF-B in supporting normal myocardial function.
NELF-B KO Reduces Energy-Metabolism-Related Transcription in Cardiomyocytes
Next, we performed gene expression profiling of LV from three pairs of the control and whole-body KO mice. Out of approximately 19,000 unique genes in the mouse genome evaluated, a total of 211 and 253 genes were up- and downregulated, respectively, 1.3-fold or more in KO samples as compared with the control samples (Table S2; GSE54372). In line with our earlier findings (Figure 1D), the hypertrophy marker Acta1 was at the top of the upregulated genes in the KO mice. Normal cardiomyocytes are adapted to high-capacity energy production through efficient FAO and aerobic metabolism in mitochondria. Consistent with the cardiac dysfunction in KO mice, gene ontology indicates that the downregulated genes in LV of KO mice are enriched with those that are involved in energy metabolism including mitochondrial function, FAO, and the tricarboxylic acid (TCA) cycle (data not shown). The changes in mRNA levels of these genes were validated by gene-specific RT-PCR (Figures 2A and 2B). In addition, nine of the ten metabolism-related genes examined were also downregulated in LV of the cardiomyocyte-specific KO mice (Figure S3A), confirming a cell-autonomous function of NELF-B in gene regulation.
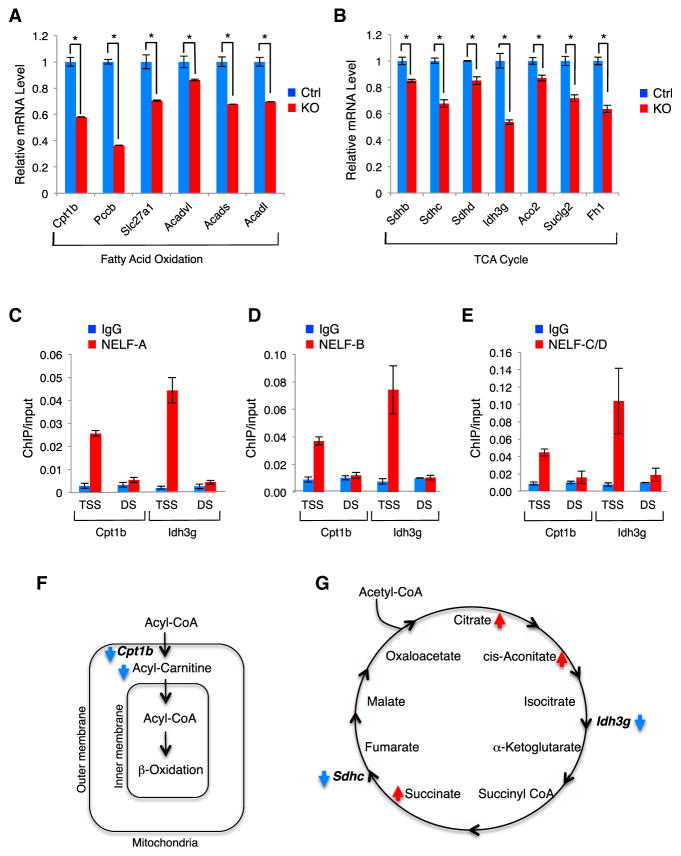
Figure 2. NELF-B Supports Energy-Metabolism-Related Transcription in Cardiomyocytes.
(A and B) Verification of microarray results (n = 3) for genes involved in fatty acid oxidation (FAO) and TCA cycle.
(C–E) ChIP of NELF-A (C), NELF-B (D), and NELF-C/NELF-D (E) at the TSS and 1 kb downstream (DS) region of the Cpt1b and Idh3g genes in wild-type myocardium (n = 3). IgG, immunoglobulin G.
(F and G) Diagrams of the FAO (F) and TCA cycle (G) with indicated changes in metabolic intermediates and enzymes in KO (red for increase and blue decrease).
Among the metabolism-related genes affected in KO myocardium, carnitine palmitoyltransferase 1b (Cpt1b) encodes the rate-limiting enzyme in FAO that is responsible for transporting long-chain fatty acyl-coenzyme A via the carnitine shuttle into the mitochondrial matrix, where they are subsequently oxidized. Isocitrate dehydrogenase 3g (Idh3g), another downregulated gene in KO, encodes one of the rate-limiting enzymes in the TCA cycle. Reduced protein expression of both enzymes in the KO hearts was verified by immunoblotting (Figure S3B). To determine whether the NELF complex acts directly on these genes, we performed chromatin immunoprecipitation (ChIP) for different NELF subunits using myocardium of wild-type mice. NELF-A, -B, and -C/D were all enriched preferentially at the TSSs of the Cpt1b and Idh3g genes as compared to a region 1 kb downstream of TSS (Figures 2C–2E), suggesting a direct role of NELF in activation of these energy-metabolism-related genes.
To explore the functional outcome of the reduced metabolic gene expression in KO mice, we performed metabolomics by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry (HPLC-ESI-MS/MS). Consistent with reduced expression of Cpt1b, the levels of various acyl-carnitine molecules were significantly diminished in the KO heart tissue (Figure 2F; Table S3). In addition, reduced expression of the enzymes in the TCA cycle coincided with accumulation of the corresponding upstream intermediates in the KO samples (Figure 2G; Table S3). This includes citrate and cis-aconitate upstream of Idh3g and succinate upstream of Sdhc. Interestingly, KO hearts also exhibited a significant increase in the levels of ceramides, which have been implicated in cardiomyopathy and cardiac failure (Table S3; Park and Goldberg, 2012). Thus, the combined transcriptomics and metabolomics data strongly suggest that NELF supports the high demand of energy production in cardiomyocytes through sustained metabolic gene expression.
NELF Is Part of the PPARα-Dependent Transcription Regulatory Network in Cardiomyocytes
Gene ontology analysis of the gene expression profile indicates a significant enrichment of genes involved in the PPAR-regulated pathways (data not shown). Among the several PPAR family members, PPARα plays an especially important role in orchestrating the cardiac transcription program required for high-capacity energy production in cardiomyocytes (Giguère, 2008; Madrazo and Kelly, 2008). Comparing the gene expression profile of the NELF-B KO with previously published microarray data from PPARα KO myocardium (Georgiadi et al., 2012), we found that these two KO mouse models shared a substantial overlap in downregulated target genes (p < 1 × 10−5; Figure 3A), 85% of which are metabolism-related (Figure 3B). In contrast, the overlap among the upregulated target genes was statistically much less significant (p = 0.0128; Figure 3A). A similar, albeit weaker, correlation was also observed between NELF-B and ERRα KO (Figure S3C; Dufour et al., 2007).
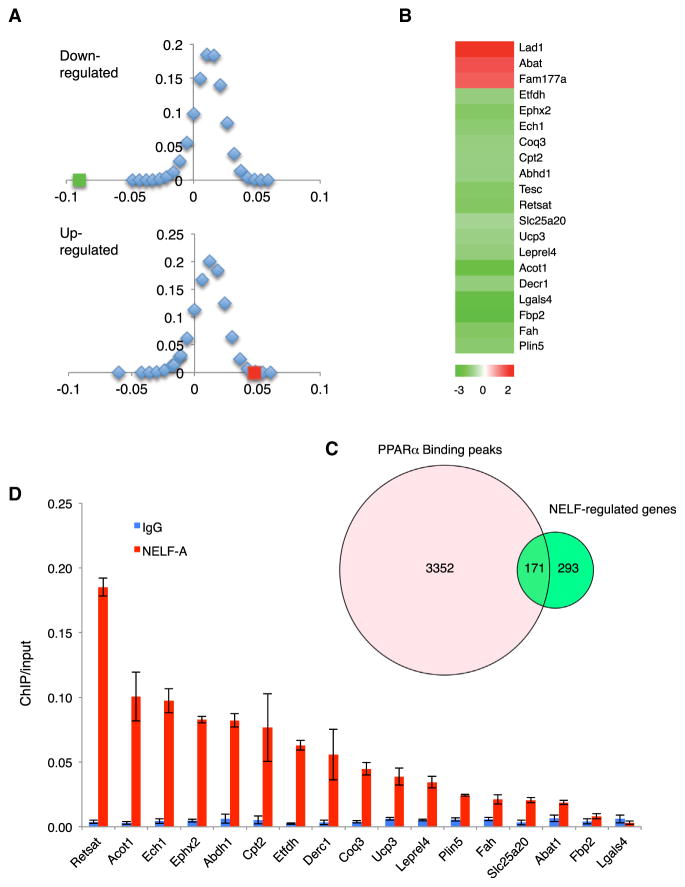
Figure 3. NELF-B Is Part of the PPARα Transcriptional Regulatory Network.
(A) Overlap in down- and upregulated genes in NELF-B and PPARα KO myocardium. The green and red squares denote the enrichment score (ES)0 values of down- and upregulated genes, respectively. The diamond-dotted curve is the frequency distribution of ES values for sets of randomly selected genes.
(B) Heatmap for the overlapped genes in the NELF-B and PPARα KO mouse myocardium.
(C) Venn diagram illustrating the presence of PPARα binding sites at the NELF target genes in myocardium.
(D) NELF-A ChIP at the NELF-B and PPARα common target genes listed in (B) (n = 3).
The NELF-B KO myocardium did not have reduced expression of PPARα, its DNA-binding partner retinoid X receptor α (RXRα), or its coactivator PGC-1α (Table S2; Figure S3D). Furthermore, expression of Cdk9 and cyclin T1, two components of P-TEFb, was comparable between the control and KO myocardium (Figure S3E). We therefore favored the possibility that NELF directly supports transcription of the PPARα target genes. Using a recently published PPARα ChIP sequencing data set (Boergesen et al., 2012), we found that 36.9% of NELF-regulated genes in myocardium have PPARα binding peak(s) within 10 kb of TSS, in comparison with 12.1% of the genes in the entire mouse genome (Figure 3C; p = 2.2 × 10−16). Moreover, ChIP confirmed chromatin association of NELF with the TSS region at 16 out of the 17 downregulated genes shared by NELF-B and PPARα KO (Figure 3D). These findings further support the notion that NELF is part of the PPARα-mediated transcription regulatory circuitry in cardiomyocytes.
NELF Facilitates Occupancy of the Transcription Initiation Complex at the Metabolism-Related Promoters
To obtain more insight into NELF-mediated transcription, we examined chromatin occupancy of NELF and Pol II. As expected, the TSS-associated NELF-A ChIP signals were significantly diminished in NELF-B KO myocardium (Figure S4A), corroborating the action of the different NELF subunits as an integrative complex in intact animal tissue. Consistent with the reduced mRNA levels of these metabolic genes in KO myocardium, the ChIP signals of total Pol II at the TSS were reduced substantially for four of the five metabolism-related genes in the NELF-B KO myocardium (Figure 4A). A similar observation was made for the phosphorylated-serine 5 Pol II (Figure S4B). We also examined Pol II intensity at gene-body regions downstream of TSS and calculated the ratio of TSS over gene-body ChIP signals. Surprisingly, none of the five NELF-regulated metabolic genes showed altered ratio in KO myocardium (Figure 4B), suggesting that NELF-B KO resulted in proportional reduction in Pol II density at TSS and gene body of these metabolic genes. In contrast, Lpl and Myl3, two NELF-associated genes whose expression was not affected by NELF-B KO, exhibited a preferential decrease of Pol II density at TSS in KO (Figures S4C and S4D). Thus, the effect of NELF on mRNA expression does not necessarily correlate with its impact on TSS-specific Pol II density.
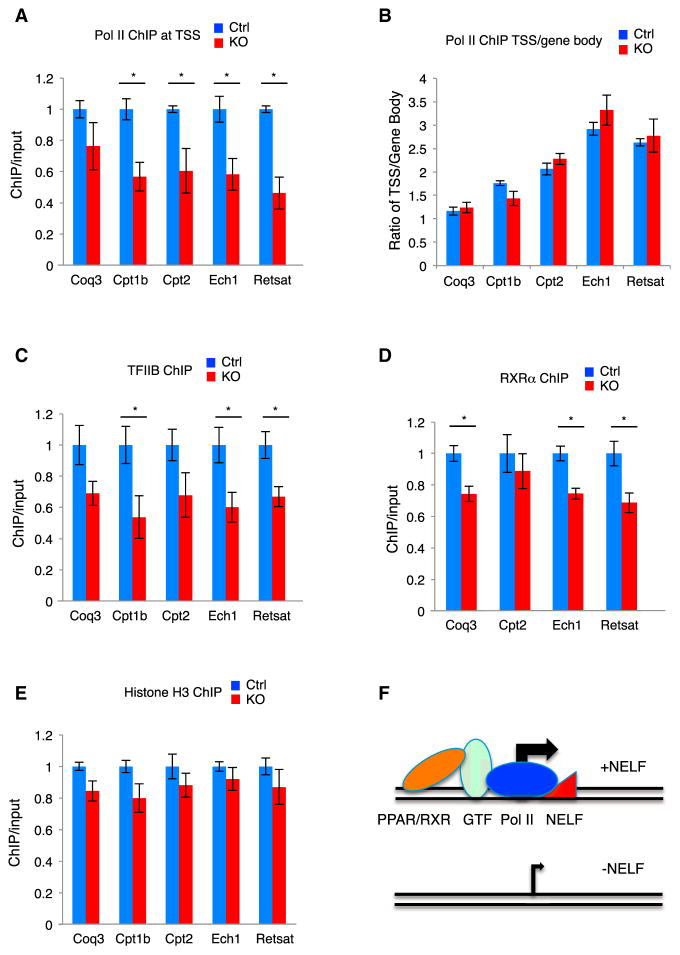
Figure 4. NELF Facilitates Occupancy of the Transcription Initiation Complex.
(A) ChIP of total Pol II at TSS.
(B) Ratio of total Pol II ChIP at TSS over gene body.
(C) TFIIB ChIP at TSS.
(D) RXRα ChIP at PPARα binding sites (PBS).
(E) Histone H3 ChIP at TSS (n = 3).
(F) A proposed role of NELF in controlling chromatin occupancy of the transcription initiation complex.
Next, we assessed promoter occupancy of transcription initiation factors. The KO myocardium exhibited substantially reduced chromatin binding of the general transcription factors TFIIB (Figure 4C) and TFIID (TBP; Figure S4E) at the NELF-regulated metabolic genes, but not a number of metabolism-unrelated genes whose transcription was not affected by NELF-B KO (Figure S4F). Chromatin binding of RXRα, the PPAR DNA-binding partner (Boergesen et al., 2012), was also reduced at three of the four common target promoters for NELF and PPARα (Figure 4D). On the other hand, the density of histone H3, which was used for assessing nucleosome density (Gilchrist et al., 2010; Sun and Li, 2010; Figure S4G), was not significantly affected at any of the NELF-regulated promoters in the KO myocardium (Figure 4E), suggesting that the reduced chromatin occupancy of the initiation factors is unlikely due to nucleosome encroachment. Taken together, these results indicate that NELF plays a role in facilitating association of transcription initiation factors with the promoters of the metabolism-associated genes.
The NELF-B knockout animals developed in the current study provide a genetic model for dissecting the physiological impact of this critical Pol II-pausing factor in mammals. Importantly, our in vivo work uncovers a role of NELF-mediated Pol II pausing in energy-metabolism-related gene activation in cardiomyocytes. Furthermore, our findings strongly suggest that NELF acts as part of the PPARα-dependent regulatory network. The organ-selective dysfunction and distinctive transcriptomics of the NELF-B KO mice clearly point to a context-dependent role of NELF in transcriptional regulation.
NELF-B depletion led to diminished promoter occupancy of transcription initiation factors, indicating that NELF function in transcription may not be restricted to Pol II pausing during early elongation. How a Pol II-pausing factor influences transcription initiation remains to be elucidated. Notably, a recent study using computational simulation suggests a complex and reciprocal influence between recruitment and movement of Pol II on the ultimate transcriptional efficiency (Ehrensberger et al., 2013). We propose that NELF acts to forestall precocious release of the paused transcription apparatus, which in turn ensures efficient and sustained occupancy of transcription initiation factors for the subsequent rounds of transcription at the energy-metabolism-related genes in myocardium (Figure 4F). Thus, transcriptional control at the initiation and early elongation steps can be bidirectional for the metabolism-related genes in cardiomyocytes.
Given the essential role of NELF-B in early embryogenesis, it is somewhat surprising that NELF-B deletion in adult mice did not lead to acute mortality. The chronic deterioration of the cardiac function of the KO mice strongly suggests that NELF is particularly important to support transcriptional program that meets the heightened needs of energy production in the myocardium. Deregulation of transcriptional elongation has been previously implicated in pathologic cardiac hypertrophy (Anand et al., 2013; Espinoza-Derout et al., 2009; Sano et al., 2002). The link between energy metabolism and NELF raises the distinct possibility that pharmacological agents aimed at enhancing Pol II pausing may overcome stress-triggered metabolic deficiency and cardiac abnormality in humans.
EXPERIMENTAL PROCEDURES
Animals
NELF-B flox/flox mice (Amleh et al., 2009) were backcrossed to C57BL/6J inbred mice for over ten generations. The CAG-Cre-ER transgenic mice and α-MHC-MerCreMer transgenic mice were purchased from the Jackson Laboratory.
Echocardiography
Transthoracic echocardiography was conducted using a Vevo 770 system (VisualSonics) with a 30 MHz image transducer to assess function of the LV. The same cohort of mice at weeks 8, 16, 24, and 32 was used for serial echocardiography with or without dobutamine administration.
Metabolomic Profiling
HPLC-ESI-MS/MS analyses of the LV samples were conducted on a Thermo Fisher Q Exactive mass spectrometer with online separation using either a Thermo Fisher/Dionex RSLC nano HPLC (for lipids) or an Ultimate 3000 HPLC (for polar metabolites).
Statistical Analysis
For analysis of the echocardiography results, a linear regression model was fitted to the fractional shortening data with age, genotype, sex, and dobutamine (i.e., whether the measurement was taken before or after administration) as the explanatory variables. Random animal effect was included in the model along with a random animal-specific component to age-related change. Bidirectional stepwise model selection was performed, yielding the final model for fractional shortening presented in Figure 1G. Multiple group comparisons were performed using the one-way ANOVA, followed by the Student-Newman-Keuls posttest (when the Bartlett variation test passed), or using the nonparametric Kruskal-Wallis test followed by Dunn posttest (when the Bartlett variation test did not pass). Statistical analysis of all the other experimental data in the current study was performed by a Student’s t test. p values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Wesley Lowell for technical assistance and Dr. Daniel Kelly for reagents. The work was supported by grants to H.P. from DOD (BC100388); to D.W. from NIH/NLM (R01LM010212); to M.L.L. from NIH/NHLBI HHSN 268201000036C (N01-HV-00244), HL051971, R01 HL075360, and from VA 5I01BX000505; to G.J.A. from NIH/NCRR and NCATS (8UL1TR000149); to R.L. from the San Antonio Nathan Shock Center; and to S.T.W. from NIH/NCRR (1S10OD016417-01).
Footnotes
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus accession number for the microarray data reported in this paper is GSE54372.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.02.028.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland AE, Hasty P, Li R. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS ONE. 2009;4:e5034. doi: 10.1371/journal.pone.0005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesbøll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, Ashrafian H, Watkins H. Genetic cardiomyopathies causing heart failure. Circ Res. 2013;113:660–675. doi: 10.1161/CIRCRESAHA.113.300282. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguère V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ehrensberger AH, Kelly GP, Svejstrup JQ. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Espinoza-Derout J, Wagner M, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MA. Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circ Res. 2009;104:1347–1354. doi: 10.1161/CIRCRESAHA.108.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N, Luedde M, Katus HA. Mechanisms of disease: hypertrophic cardiomyopathy. Nat Rev Cardiol. 2012;9:91–100. doi: 10.1038/nrcardio.2011.159. [DOI] [PubMed] [Google Scholar]
- Georgiadi A, Boekschoten MV, Müller M, Kersten S. Detailed transcriptomics analysis of the effect of dietary fatty acids on gene expression in the heart. Physiol Genomics. 2012;44:352–361. doi: 10.1152/physiolgenomics.00115.2011. [DOI] [PubMed] [Google Scholar]
- Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297:E578–E591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguère V, Murphy E, Kelly DP. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Park TS, Goldberg IJ. Sphingolipids, lipotoxic cardiomyopathy, and cardiac failure. Heart Fail Clin. 2012;8:633–641. doi: 10.1016/j.hfc.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets PJ, Teunissen BE, Willemsen PH, van Nieuwenhoven FA, Brouns AE, Janssen BJ, Cleutjens JP, Staels B, van der Vusse GJ, van Bilsen M. Cardiac hypertrophy is enhanced in PPAR alpha−/−mice in response to chronic pressure overload. Cardiovasc Res. 2008;78:79–89. doi: 10.1093/cvr/cvn001. [DOI] [PubMed] [Google Scholar]
- Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.