Abstract
Transcription elongation is interrupted by sequences that inhibit nucleotide addition and cause RNA polymerase (RNAP) to pause. Here, by use of native-elongating-transcript sequencing (NET-seq) and a variant of NET-seq that enables analysis of mutant RNAP derivatives in merodiploid cells (mNET-seq), we analyze transcriptional pausing genome-wide in vivo in Escherichia coli. We identify a consensus pause-inducing sequence element, G−10Y−1G+1 (where −1 corresponds to the position of the RNA 3′ end). We demonstrate that sequence-specific interactions between RNA polymerase core enzyme and a core recognition element (CRE) that stabilize transcription initiation complexes also occur in transcription elongation complexes and facilitate pause read-through by stabilizing RNAP in a post-translocated register. Our findings identify key sequence determinants of transcriptional pausing and establish that RNAP-CRE interactions modulate pausing.
Regulation of gene expression during transcription elongation often involves sequences in DNA that cause the transcription elongation complex (TEC) to pause. Pausing can impact gene expression by facilitating engagement of regulatory factors, influencing formation of RNA secondary structures, and enabling synchronization of transcription and translation.
Several lines of evidence suggest that pausing involves specific sequence signals that inhibit nucleotide addition (1-11). To define key sequence determinants for pausing we utilized native-elongating transcript sequencing (NET-seq), which permits occupancies of TECs to be mapped genome-wide with base-pair resolution (12-13; Fig. S1). The occupancy of the TEC at a given position is correlated with the tendency of the TEC to pause at the position. Accordingly, NET-seq analysis enables identification of pause sites. To perform NET-seq in Escherichia coli, cells carrying a chromosomal rpoC-3xFLAG gene, encoding RNAP β′ subunit with C-terminal 3xFLAG tag were grown to mid-exponential phase; cells were flash-frozen and lysed; 3xFLAG-tagged TECs were immunoprecipitated with anti-FLAG; RNAs were extracted from TECs, and RNA 3′ ends were converted to cDNAs and analyzed using high-throughput sequencing. We defined pause sites as positions where TEC occupancy exceeds TEC occupancy at each position 25 bp upstream and downstream. We identified 15,553 pause sites, which corresponds to ∼19,800 total pause sites, given the estimated ∼78% saturation of the analysis (Tables S1-S7). Alignment of pause-site sequences revealed a clear consensus pause element (PE): G−10Y−1G+1, where position −1 corresponds to the position of the RNA 3′ end (Figs. 1A,S2). Of the identified pause sites ∼35% exhibited a 3-of-3 match to the consensus PE, and ∼42% exhibited a 2-of-3 match to the consensus PE (Tables S4,S5). Comparing the total number of pause sites with a 3-of-3 match (∼6,900) to the total number of sequences in the transcribed portion of the genome with a 3-of-3 match (∼43,500-58,000; 14) indicates that, under these conditions, functional pausing occurs at ∼12-16% of sequences with a 3-of-3 match.
Fig. 1. Identification of consensus PE.

A. Sequence logo for consensus PE from NET-seq. Red, bases with ≥0.2 bit sequence-information content.
B. In vitro transcription assays with consensus PEs and mutant PEs. Top, templates. Bottom, results. +29, RNA before addition of UTP; +46, RNA in TEC at PE; RT, read-through RNA; red, consensus PE bases.
To validate the NET-seq results, we selected two consensus PEs for analysis in vitro. One, located in yrbL, has −1C, and the other, located in gltP, has −1T (Figs. 1B,S3). Transcription assays show that the yrbL PE and gltP PE induce pausing in vitro (Figs. 1B,S4), and show that consensus base pairs at positions +1, −1, and −10 are required for efficient pause capture in vitro (Fig. 1B).
We next determined the contribution of individual bases of the consensus PE to pausing at the yrbL PE (Fig. 2A). The results indicate that introduction of each of the three non-consensus base pairs at position +1 (C, A, and T) reduces pausing, introduction of each of the two non-consensus base pairs at position −1 (A and G) reduces pausing, and introduction of two of three non-consensus base pairs at position −10 (A and T) reduces pausing (Fig. 2A). In each case, the effect on pausing is manifest at the level of pause capture efficiency.
Fig. 2. Contributions of individual base pairs of consensus PE.

A. In vitro transcription assays with yrbL PE derivatives. Red, consensus PE bases.
B. Schematic representation of TEC in pre-translocated state at consensus PE (top) and TEC in post-translocated state at consensus PE (bottom). White boxes, DNA; gray boxes, RNA; gray shading, RNAP; red, consensus PE bases; i and i+1, RNAP active-center i and i+1 sites.
The consensus PE comprises sequence determinants that previously have been implicated as important for pausing: G at +1 (10,15), T or C at −1 (3,5,6,10), and G at −10 (4-10). The consensus PE is especially similar to a consensus pause-inducing sequence identified by Herbert et al. in single-molecule studies (10).
The sequence determinants in the consensus PE (G−10Y−1G+1) can be understood in terms of the structure and mechanism of the TEC (Fig. 2B). In each nucleotide-addition cycle in transcription elongation, RNAP translocates between a “pre-translocated state,” in which the RNAP active-center “i” and “i+1” sites interact with RNA positions −2 and −1, and a “post-translocated state,” in which the RNAP i site interacts with RNA position −1 and the RNAP i+1 site is unoccupied and available for binding of an NTP (1,2,16). Translocation requires breaking the DNA base pair at position +1 and breaking the RNA-DNA base pair at position −10 (17-18). Because the DNA bp at position +1 must be broken for forward translocation, G/C at +1 will disfavor forward translocation relative to the less stable A/T. Similarly, because the RNA-DNA bp at position −10 must be broken for forward translocation, G/C at −10 will disfavor forward translocation relative to the less stable A/T. Furthermore, because 5′-rG-rN-3′/3′-dC-dN-5′ is more stable than 5′-rC-rN-3′/3′-dG-dN-5′ (19), rG:dC at position −10 also will disfavor translocation over rC:dG at −10. Translocation requires movement of the template-strand DNA nucleotide and base-paired RNA nucleotide at position −1 from the RNAP active-center i+1 site to the i site and movement of the template-strand DNA nucleotide at position +1 to the i+1 site. Because available evidence indicates that the RNAP active-center i and i+1 sites preferentially interact with 5′-rR-rY-3′/3′-dY-dR-5′ (3), a nontemplate-strand Y at position −1 and nontemplate-strand G at position +1 will disfavor forward translocation relative to all other sequences (the former by stabilizing the pre-translocated state; the latter by destabilizing the post-translocated state). Thus, each position of the consensus sequence is predicted to favor the pre-translocated state over the post-translocated state (−10 through effects on duplex stability, −1 through effects on active-center binding, and +1 through both). Accordingly, each position of the consensus PE would be predicted to increase the opportunity for the TEC to enter an “elemental pause” state (a state that, according to one view, is accessed from the pre-translocated state and serves as an obligatory intermediate for pausing; 7,8,10-11,20) or a “backtracked” state (a state that, according to another view, is accessed from the pre-translocated state and serves as the primary state for pausing; 2,16).
A TEC in a post-translocated state at a PE will contain an unpaired G at the downstream end of the nontemplate strand of the unwound “transcription bubble” (Fig. 3A). In transcription initiation complexes, RNAP core enzyme makes sequence-specific interactions with an unpaired G at the downstream end of the transcription bubble (“core recognition element” CRE; 21; Fig. 3A). In transcription initiation, interaction between RNAP and the unpaired G of the CRE (GCRE) facilitates promoter unwinding to form a stable, catalytically competent, RNAP-promoter open complex (RPo; 21). It has been proposed that RNAP-GCRE interaction may be functionally important in transcription elongation as well as in transcription initiation (21), but this has not previously been documented.
Fig. 3. Sequence-specific RNAP-GCRE interactions modulate pausing.

A. Structural organization of TEC in post-translocated state at consensus PE (left) and RPo in transcription initiation at promoter containing consensus CRE (right; 21). The presence in each case of an unpaired G at downstream end of nontemplate strand of transcription bubble suggests the possibility of equivalent sequence-specific interactions between RNAP core and the G. Red at left, bases of the consensus PE; red at right, GCRE.
B. Results of fluorescence-detected equilibrium assays (top) and kinetic (bottom) assays of interactions of RNAP derivatives with nucleic-acid scaffolds containing G, A, T, or an abasic site (X) at position corresponding to GCRE. ntDNA, nontemplate-strand DNA; tDNA template-strand DNA.
C. Sequence logos for consensus PE with RNAP-βWT (top; 0.4 bit sequence-information content at GCRE) and RNAP-βD446A (bottom; 1 bit of sequence-information content at GCRE), as defined by mNET-seq. Red, bases with ≥0.2 bit sequence information content.
D. Pause-capture efficiencies of RNAP βWT (left panels) and RNAP βD446A (right panels) at yrbL PE and gltP PE in vitro. +29, RNA before addition of UTP; +46, RNA in TEC at PE; asterisks, RNA in TECs at additional sites where RNAP βD446A exhibits higher pause-capture efficiency than RNAP βWT; RT, read-through RNA; red, consensus PE bases.
The observation that the consensus PE contains the sequence feature required for establishment of RNAP-GCRE interaction raises the possibility that RNAP-GCRE interaction may mediate or modulate pausing. To test this possibility, we constructed and analyzed a mutant RNAP defective in sequence-specific recognition of GCRE. The crystal structure of RPo shows that RNAP βD446 makes H-bonds with Watson-Crick atoms of GCRE and suggests that D446 reads the identity of GCRE (21). Substitution of βD446 by alanine results in the loss of the ability to distinguish G, A, T, or an a basic site at the position corresponding to GCRE (Figs. 3B,S5), confirming that βD446 reads the identify of GCRE and providing a reagent to assess functional significance of sequence-specific RNAP-GCRE interactions.
To establish whether RNAP-CRE interactions affect pausing, we used a variant of NET-seq, “merodiploid NET-seq” (mNET-seq), that enables analysis of mutant RNAP derivatives, including mutant RNAP derivatives that do not support viability in haploid (Fig. S1). mNET-seq employs merodiploid cells containing a plasmid-encoded, epitope-tagged RNAP and a chromosomally-encoded, untagged RNAP, and involves selective analysis of transcripts associated with epitope-tagged RNAP in the presence of a mixed population of epitope-tagged RNAP and untagged RNAP. We introduced into cells a plasmid encoding 3xFLAG-tagged wild-type RNAP β subunit (βWT) or 3xFLAG-tagged RNAP β subunit containing D446A (βD446A), we isolated RNAs associated with RNAP-βWT or RNAP-βD446A by immunoprecipitation, and we identified pause sites. For RNAP-βWT, alignment of pause sites revealed a consensus sequence matching the consensus PE, validating mNET-seq as an effective system for analysis of pausing (Figs. 1A,3C,S2,S6; Tables S4-S9). For RNAP-βD446A we identified a ∼60-90% higher number of pause sites than with RNAP-βWT (Tables S4-S10). Alignment of the pause sites revealed a ∼30% higher proportion of the pause sites carried a G at position +1 (Figs. 3C,S6; Table S6). We conclude that RNAP-βD446A is more susceptible than RNAP-βWT to pausing at sites with G at position +1.
We next compared pausing properties of RNAP-βD446A and RNAP-βWT in vitro, using templates carrying the yrbL PE and gltP PE (Figs. 3D,S7). RNAP-βD446A enhances pausing at yrbL PE and gltP PE. RNAP-βD446A also enhances pausing at other positions where the next nucleotide to be added to the transcript is G (positions with asterisks in Fig. 3D). The results indicate that a substitution that disrupts sequence-specific RNAP-CRE interaction increases pausing, both in vivo and in vitro, at positions where the post-translocated state contains GCRE. We conclude that sequence-specific RNAP-GCRE interaction occurs during elongation and counteracts pausing.
To explore why disruption of sequence-specific RNAP-GCRE interaction enhances pausing we assessed whether RNAP-GCRE interactions affect the translocational register of the TEC by assessing sensitivity of TECs to pyrophosphorolysis (Fig. 4). Sensitivity to pyrophosphorolysis provides a measure of TEC translocation because a TEC in a pre-translocated state is sensitive to pyrophosphorolysis but a TEC in a post-translocated state is resistant (Fig. 4A; 3). We performed assays with RNAP-βWT or RNAP-βD446A on templates containing G or T at position +1 (Fig. 4B). We found that TECs with RNAP-βWT were ∼5fold more sensitive to pyrophosphorolysis when the template contained +1G than when the template contained +1T, indicating that a greater proportion of TECs on templates containing +1G were in a pretranslocated state than of TECs on templates containing +1T (Fig. 4C). The results directly demonstrate the effect of G at position +1 on translocation bias. TECs with RNAP-βD446A were ∼4-fold more sensitive to pyrophosphorolysis than TECs with RNAP-βWT when the template contained +1G (Fig. 4C), whereas, in contrast, TECs with RNAP-βD446A and RNAP-βWT exhibited identical sensitivities to pyrophosphorolysis when the template contained +1T (Fig. 4C). Therefore, on templates containing +1G, a greater proportion of TECs with RNAP-βD446A are in a pre-translocated state as compared to TECs with RNAP-βWT. We conclude that sequence-specific RNAP-GCRE interactions stabilize the TEC post-translocated state, providing a mechanistic explanation for the finding that RNAP-GCRE interactions counteract pausing.
Fig. 4. Sequence-specific RNAP-GCRE interactions modulate translocation bias.
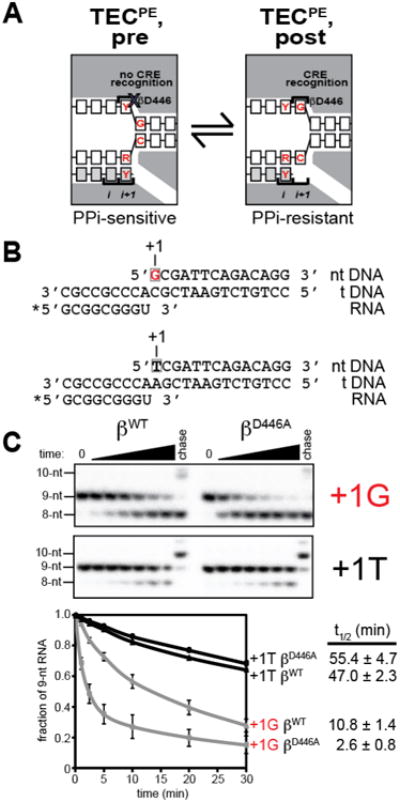
A. Structural organization of TEC in pre-translocated state at consensus PE (left; unfavorable RNAP-CRE interaction) and TEC in post-translocated state at consensus PE (right; favorable RNAP-CRE interaction). PPi, pyrophosphate.
B. Nucleic-acid scaffolds used for translocation-bias assays. Asterisk, radiolabel on RNA 5′ end; box, position corresponding to GCRE; red, consensus PE base.
C. Translocational bias for RNAP βWT and RNAP βD446A on nucleic-acid scaffolds containing G or T at position corresponding to GCRE. Gel images show pyrophosphorolysis reaction progress from 0-30 min. 9-nt, scaffold; 8-nt, product of pyrophosphorolysis; 10-nt, product of “chase” reaction with GTP (+1G template) or UTP (+1T template). Graph shows fraction of unaltered scaffold (mean±SEM; 3 measurements) as function of time.
Our findings define the key sequence determinants of transcriptional pausing as G−10Y−1G+1 (Figs. 1-2). The consensus PE promotes pausing by disfavoring translocation of the TEC to the post-translocated state (Fig. 2B), thereby increasing the opportunity for the TEC to enter an “elemental pause” state and/or a “backtracked” state. We further show that sequence-specific RNAP-GCRE interactions counteract pausing by stabilizing the TEC in a post-translocated state (Figs. 3-4). Because residues of RNAP core that mediate sequence-specific RNAP-CRE interaction are conserved in RNAP from all living organisms, we suggest that RNAP-CRE interaction counteracts pausing in all multisubunit RNAPs. The consensus PE will, on average, be encountered by RNAP every ∼32 bp during transcription elongation for organisms, such as E. coli, with ∼50% G/C content and will be encountered even more frequently for organisms with higher G/C content. RNAP-GCRE interactions may help overcome a barrier to forward translocation that occurs each time RNAP encounters a G10Y1G+1 sequence during transcription elongation. A major function of RNAP-GCRE interactions may be to suppress noise during transcription elongation by smoothing the sequence dependent energy landscape for transcription elongation.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM041376 (RHE), GM088343 (BEN), and GM096454 (BEN). Reads are deposited in the Sequence Read Archive (accession SRP039384).
Footnotes
References and Notes
- 1.Larson M, Landick R, Block S. Single-molecule studies of RNA polymerase. Mol Cell. 2011;41:249–262. doi: 10.1016/j.molcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C. Molecular mechanisms of transcription through single-molecule experiments. Chem Rev. 2014;114:3203–3223. doi: 10.1021/cr400730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hein P, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochem. 2011;50:7002–7014. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyzer S, Ha K, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 5.Chan C, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 6.Aivazashvili V, Bibilashvili R, Vartikian R, Kutateladze T. Effect of the primary structure of RNA on the pulse character of RNA elongation in vitro by Escherichia coli RNA polymerase. Mol Biol. 1981;15:915–929. [PubMed] [Google Scholar]
- 7.Weixlbaumer A, Leon K, Landick R, Darst S. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landick R. Transcriptional pausing without backtracking. Proc Natl Acad Sci USA. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kireeva M, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbert K, et al. Sequence resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuman K, Abbondanzieri E, Landick R, Gelles J, Block S. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 12.Churchman L, Weissman J. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Materials and Methods are available as supplementary materials on Science Online
- 14.Haas B, Chin M, Nusbaum C, Birren B, Livny J. How deep is deep enough for RNA-seq profiling of bacterial genomes. BMC Genomics. 2012;13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- 16.Dangkulwanich M, et al. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. eLife. 2013;2:e00971. doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai L, Shundrovsky A, Wang M. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 18.Tadigotla V, et al. Thermodynamic and kinetic modeling of transcriptional pausing. Proc Natl Acad Sci USA. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto N, et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochem. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Ha K, La Porta A, Landick R, Block S. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol Cell. 2011;44:635–646. doi: 10.1016/j.molcel.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


