Abstract
In aflatoxin biosynthesis, the pathway for the conversion of 1-hydroxyversicolorone to versiconal hemiacetal acetate (VHA) to versiconal (VHOH) is part of a metabolic grid. In the grid, the steps from VHA to VHOH and from versiconol acetate (VOAc) to versiconol (VOH) may be catalyzed by the same esterase. Several esterase activities are associated with the conversion of VHA to VHOH, but only one esterase gene (estA) is present in the complete aflatoxin gene cluster of Aspergillus parasiticus. We deleted the estA gene from A. parasiticus SRRC 2043, an O-methylsterigmatocystin (OMST)-accumulating strain. The estA-deleted mutants were pigmented and accumulated mainly VHA and versicolorin A (VA). A small amount of VOAc and other downstream aflatoxin intermediates, including VHOH, versicolorin B, and OMST, also were accumulated. In contrast, a VA-accumulating mutant, NIAH-9, accumulated VA exclusively and neither VHA nor VOAc were produced. Addition of the esterase inhibitor dichlorvos (dimethyl 2,2-dichlorovinylphosphate) to the transformation recipient strain RHN1, an estA-deleted mutant, or NIAH-9 resulted in the accumulation of only VHA and VOAc. In in vitro enzyme assays, the levels of the esterase activities catalyzing the conversion of VHA to VHOH in the cell extracts of two estA-deleted mutants were decreased to approximately 10% of that seen with RHN1. Similar decreases in the esterase activities catalyzing the conversion of VOAc to VOH were also obtained. Thus, the estA-encoded esterase catalyzes the conversion of both VHA to VHOH and VOAc to VOH during aflatoxin biosynthesis.
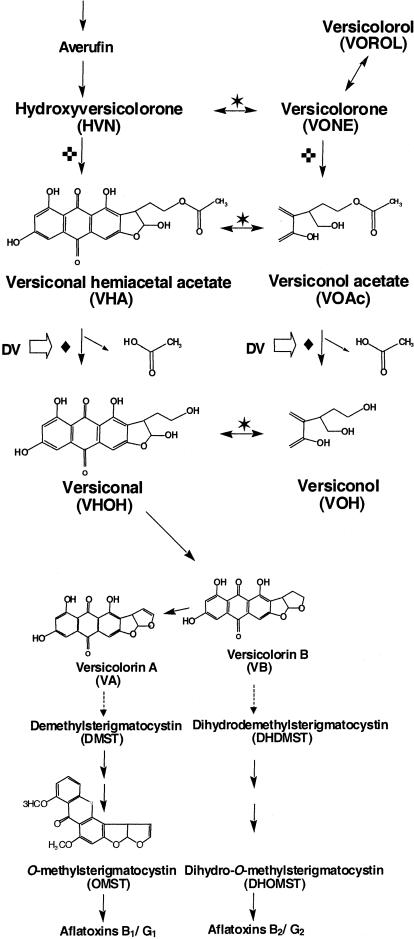
Aflatoxins are potent carcinogens produced by fungi in the Aspergillus flavus group, such as Aspergillus parasiticus and some isolates of Aspergillus flavus. Aflatoxin contamination of agricultural commodities both pre- and postharvest is a serious food safety issue (13) and a significant economic concern (3). The biosynthesis of aflatoxin B1 is a multistep process, and most of the aflatoxin pathway genes belong to a 70-kb gene cluster (6, 10, 21, 23, 25, 34-36, 38; reviewed in references 20, 27, and 33). The first stable intermediate, norsolorinic acid (NOR), is formed by NOR synthase (NorS), a complex containing a polyketide synthase and a specialized pair of fatty acid synthases (26). The two carbons of the terminal acetate in the C6 side chain of NOR are not exchangeable with acetates in the metabolic pool (24). Of the six carbons in the NOR side chain, only four remain in aflatoxin B1. The terminal acetate in the oxidized product of averufin, i.e., 1-hydroxyversicolorone (HVN), is esterified by a monooxygenase-catalyzed Baeyer-Villiger rearrangement reaction to generate versiconal hemiacetal acetate (VHA) (29). The esterified acetate is supposedly released from the side chain of VHA by an esterase as acetate to give versiconal (VHOH) (Fig. 1).
FIG. 1.
Metabolic grids and intermediates involved in the conversion of averufin to VA and the late stage of aflatoxin biosynthesis. Reactions catalyzed by the same enzyme are indicated by the same symbol: ✶, VHA reductase; ♦, VHA esterase; ✜, monooxygenase. The open arrows indicate reactions inhibited by dichlorvos.
VHA-associated esterase activities have been confirmed in A. parasiticus cell extracts (2, 12), and the reactions were inhibited by dichlorvos (dimethyl 2,2-dichlorovinylphosphate) (22, 32). Yabe et al. (28) described a metabolic grid containing VHA, versiconol acetate (VOAc), VHOH, and versiconol (VOH) and suggested that the reactions from VHA to VHOH and from VOAc to VOH are catalyzed by the same enzyme. Kusumoto and Hsieh (17) showed that at least three esterases are present in the cell extract of A. parasiticus ATCC 15517 and that a partially purified VHA esterase has a molecular mass of 32 kDa. More recently, Yabe et al. (29) identified another metabolic grid containing HVN, versicolorone (VONE), VOAc, and VHA that also is involved in aflatoxin biosynthesis (Fig. 1).
Aspergillus nidulans produces sterigmatocystin (ST), which is a penultimate intermediate of aflatoxin biosynthesis. The pathway of ST biosynthesis up to the formation of ST is identical to that of aflatoxin biosynthesis, and the genes involved also form a gene cluster (4, 14-16). The A. nidulans stcI gene of the ST gene cluster encodes an esterase (4). Recently, we found the estA gene, an stcI gene homolog, in the complete aflatoxin gene cluster of A. parasiticus. The estA gene is constitutively expressed under either aflatoxin-conducive or aflatoxin-nonconducive conditions (37). A second copy of estA (estA2) is present in the partially duplicated aflatoxin gene cluster of A. parasiticus SU-1 but appears to be nonfunctional (8). The stcI and estA genes are believed to be involved in the conversion of VHA to VHOH.
Our objective was to determine the role of the estA gene in aflatoxin biosynthesis. We expected the loss of estA in the O-methylsterigmatocystin (OMST)-accumulating strain A. parasiticus SRRC 2043 to affect the conversions of VHA to VHOH and VOAc to VOH. This work provides new insights into how structurally related intermediates in the metabolic grid are formed during aflatoxin biosynthesis.
MATERIALS AND METHODS
Fungal strains, media, and metabolites.
A. parasiticus SRRC 2043, a strain that accumulates OMST, was maintained on V8 agar plates (5% V8 vegetable juice [Campbell Soup Company, Camden, N.J.] and 2% agar, pH 5.2). A. parasiticus RHN1, which is a NiaD− (nitrate reductase) mutant derived from SRRC 2043, was used as the recipient strain for transformation. Czapek solution agar (Difco, Detroit, Mich.) supplemented with 0.6 M KCl was used as the protoplast regeneration medium. For examination of the aflatoxin intermediates and pigment(s), nitrate-utilizing transformants were grown on potato dextrose agar (PDA; Difco) plates. A. parasiticus estA-D1 to estA-D4 were estA-deleted mutants isolated from this work. Adye and Mateles medium (1) was used to grow submerged estA-deleted cultures for the analysis of accumulated aflatoxin intermediates and for the preparation of genomic DNA for Southern blot hybridization and PCR analyses. A. parasiticus NIAH-9, a versicolorin A (VA)-accumulating mutant derived from A. parasiticus NRRL 2999, was used for preparation of VHA and VOAc (28) and also for investigation of the effect of dichlorvos. Conidiospores were collected from the isolate, which had been grown on a PDA slant at 28°C for 1 week. Aflatoxin intermediate standards, OMST, VHOH, versicolorol (VOROL), VONE, VOAc, VHA, versicolorin B (VB), and VA, were prepared in collaboration with H. Nakajima, Tottori University, Tottori, Japan (28, 29, 31).
Construction of the estA replacement vector and fungal transformation.
The estA gene replacement vector, pestDV, was constructed in a three-step procedure. First, a 1.8-kb XbaI-SphI fragment containing the complete estA gene of A. parasiticus SU-1 was cloned into pUC18. Second, the 1.2-kb BglII-EcoRV fragment, within the cloned 1.8-kb XbaI-SphI fragment and containing the estA coding region, was removed and the ends were filled with Pfu polymerase (Stratagene, La Jolla, Calif.) at 72°C for 30 min in a Perkin-Elmer 2400 thermal cycler. Third, the 6.7-kb XbaI fragment containing the A. parasiticus niaD gene (7) was blunt-ended and then ligated into the linear vector obtained in the second step. The final step gave the replacement vector pestDV. Before fungal transformation, which was carried out as previously described (11), pestDV was linearized with XbaI and SphI to release the portion derived from pUC18. This gene replacement strategy forces the linearized deletion vector to integrate at either a targeted gene locus or the selectable marker niaD locus via double-crossover recombination as shown previously (10).
Determination of estA deletion by Southern blot hybridization and PCR.
Genomic DNA of A. parasiticus RHN1 and putative estA-deleted mutants was prepared using a GenElute plant genomic DNA miniprep kit (Sigma, St. Louis, Mo.). Approximately 1.0 μg of DNA was digested with EcoRI or KpnI, separated in a 0.8% agarose gel, and transferred onto a GeneScreen Plus membrane (NEW Research Products, Boston, Mass.) by capillary action. Blots were hybridized with digoxigenin (DIG)-labeled DNA probes in DIG Easy Hib hybridization buffer (Roche, Mannheim, Germany) at 42°C overnight. The concentration of the probes added was 10 ng per ml of Easy Hib buffer. The estA probe was a 0.71-kb coding region, and the niaD probe was a 0.54-kb fragment obtained by PCR using a PCR DIG probe synthesis kit (Roche). The primer pairs for generating the DIG-labeled probes were as follows: estF1, 5′-GCTGTAGAGACACTGGCATACT-3′, and estR1, 5′-CTGTCAACTCCGCATTCTCCT-3′, for the estA probe; niaDInt3, 5′-GAAGATGCTGCGACACACCC-3′, and niaDInt6, 5′-TACGTACAATATTTATGACC-3′, for the niaD probe. The PCR conditions consisted of 30 cycles of a denaturation step at 94°C for 30 s, an annealing step at 50°C for 30 s, and an extension step at 72°C for 2 min. After hybridization, blots were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 5 min at room temperature and then washed twice in 0.1× SSC-0.1% sodium dodecyl sulfate for 15 min at 68°C under constant agitation. Detection of hybridization signals was carried out using a DIG nucleic acid detection kit (Roche). The detection method is based on an enzyme-catalyzed color reaction with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium salts that produces an insoluble blue precipitate on the blots.
Gene-specific oligoprimer pairs also were used to confirm the deletion of estA in the recipient genome. The primer pairs were as follows: pair 1, estF (5′-CCTATCCTGAAGCTGCCGTTAC-3′) and niaDXV1 (5′-CTGTTTCGGACTCTCTTCTG-3′); pair 2, estF3 (5′-GATGTCCAGTTGGATGGACTGT-3′) and estR (5′-CTCTCGCATAAGGATATAAGCATG-3′); pair 3, estF2 (5′-GTTGACAGCAGAGGACATACC-3′) and estR2 (5′-GATAGCGAGTCTCAGGAGCCTT-3′); pair 4, niaDXV2 (5′-TCTCTTCCACTGTGCTATCCA-3′) and norA63 (5′-GGGGTAAGTCCTTGGCTCG-3′).
TLC.
For semiquantitative thin-layer chromatography (TLC) analysis of aflatoxin intermediates (9), fungal cultures were grown on PDA plates for 5 days at 30°C. Two agar plugs, 1 cm from the inoculation site, were cored with Transfertubes (Spectrum, Houston, Tex.) and placed in a 1.5-ml microcentrifuge tube and extracted with a mixture of acetone and chloroform (1:1, vol/vol). The extracts were separated on a silica gel TLC plate (J. T. Baker, Inc., Phillipsburg, N.J.) with a solvent system composed of toluene-ethyl acetate-glacial acetic acid (TEA; 50:30:4, vol/vol/vol). For the scale-up preparation of extracts, 4-day-old cultures grown in 100 ml of Adye and Mateles medium (1) at 30°C were extracted with acetone until they were colorless. Chloroform was added to the combined acetone extracts (1:1 ratio), and the golden-yellow pigments were extracted from the acetone phase twice. The chloroform layers were combined and concentrated to 2 ml at room temperature. Extracts were separated on analytical TLC plates, and a desired compound was scraped from the silica gel matrix. The scraped compound was rechromatographed one or two times until only a single band was obtained. The single band was scraped and collected in a microcentrifuge tube, and the compound was eluted by immersing the silica gel matrix in acetone for 30 min. An eluted compound was subjected to another round of TLC separation with a developing solvent system composed of chloroform-ethyl acetate-90% (wt/wt) formic acid (CEF; 6:3:1, vol/vol/vol) to check the purity. To identify the compounds in the extracts, standards of known aflatoxin intermediates, NOR, averantin, averufin, VHA, VOAc, VA, and OMST, were spotted along with each eluted compound on a TLC plate. A compound was tentatively identified on the basis of whether it had the same Rf value and color as the standard on TLC with the TEA and CEF developing solvent systems. The putative aflatoxin intermediates, VHA and VOAc, were analyzed by high-performance liquid chromatography (HPLC) to confirm their identities.
Tip culture.
To monitor the effect of dichlorvos on accumulation of aflatoxin intermediates in mycelia, the tip culture method was used (28, 29, 31). This method employs a 1-ml Pipetman tip (Gilson, Middleton, Wis.) as a culture vessel for liquid stationary culture. With this procedure, aflatoxin intermediates in the mycelia and aflatoxins excreted into the medium are easily measured quantitatively or semiquantitatively. A 5-μl spore suspension of the A. parasiticus estA-deleted mutant, the transformation recipient RHN1, or NIAH-9 was inoculated into 200 μl of YES medium (2% [wt/vol] yeast extract and 20% [wt/vol] sucrose) supplemented with or without 80 ppm dichlorvos. After 4 days of incubation at 28°C, aflatoxin intermediates in the mycelia were extracted with 1 ml of acetone, dried, and then resolubilized in 100 μl of methanol. Two microliters of the resultant solution was analyzed by HPLC as described below.
Enzyme assay.
Cell extracts were prepared from the mycelia of A. parasiticus estA-deleted mutants and the recipient RHN1, which had been cultured in YES medium at 28°C for 4 days (27). To remove contaminating pigments, the cell extracts were further purified with a Sephadex G-26 column (PD-10; Pharmacia LKB Biotechnology, Uppsala, Sweden), which had been equilibrated and eluted with a solution containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.4 mM EDTA, 1 mM mercaptoethanol, and 10% glycerol. The protein concentration in the cell extract was determined by using a protein assay solution (Bio-Rad, Hercules, Calif.). For the esterase enzyme assay of the conversion of VHA to VHOH, the cell extract (0.2 mg of protein ml−1) was incubated with 10 μM VHA in a reaction mixture composed of 90 mM potassium phosphate (pH 7.50) and 10% (vol/vol) glycerol at 30°C. For the assay of the conversion of VOAc to VOH, VOAc was used as the substrate instead of VHA. The total volume of each reaction mixture in a 0.5-ml microcentrifuge tube was 50 μl. After incubation at 30°C for either 5 or 15 min, the reaction was terminated by the addition of 70 μl of ethyl acetate and mixing with a Vortex mixer. The tubes were centrifuged at 10,000 × g for 2 min, and an aliquot of the upper ethyl acetate layer was transferred to a new microcentrifuge tube. The ethyl acetate layer was dried at room temperature in the dark by keeping the tube lid open. Residues were dissolved in methanol before HPLC analyses. For the determination of VHOH cyclase activity involved in the reaction of VHOH to VB, the cell extract was incubated with 0.4 μM VHOH by using the same reaction mixture and production of VB was then measured by HPLC. VHOH was prepared by incubation of porcine esterase with VHA (30).
HPLC analysis.
HPLC was performed at 35°C at a flow rate of 1 ml min−1 by using a Shimadzu HPLC apparatus (LC-6A) equipped with an octyldecyl silane HPLC column measuring 0.46 by 15 cm with a guard column measuring 0.46 by 1 cm (Inertsil ODS-2; Shimadzu Co. Ltd., Kyoto, Japan). Absorbance at 290 nm was monitored. A solvent system consisting of acetonitrile-tetrahydrofuran-water (25:20:55, vol/vol/vol) was used in HPLC to identify the spots isolated from TLC plates. The retention times of the aflatoxin intermediate standards used were as follows: 3.6 min for VHOH, 6.2 min for VOAc, 9.0 min for VHA, and 11.4 min for versicolorin C (VC). The putative VHOH was further treated with 1 N HCl at room temperature for 5 min, extracted with ethyl acetate, dried, and resolubilized in methanol before HPLC analysis. For the analysis of aflatoxin intermediates accumulated in the tip cultures and reaction products in the enzyme assays, the solvent system used consisted of acetonitrile-tetrahydrofuran-water (18:17:65, vol/vol/vol). A standard curve of VHA was used to estimate the amount of each intermediate except for OMST. The amount of OMST was estimated based on an OMST standard curve. The typical retention times of aflatoxin intermediate standards were as follows: 5.2 min for OMST, 6.5 min for VHOH, 7.4 min for VOROL, 8.0 min for VONE, 9.6 min for VOAc, 14.9 min for VHA, 23.5 min for VB, and 31.4 min for VA.
RESULTS
Isolation of estA-deleted mutants.
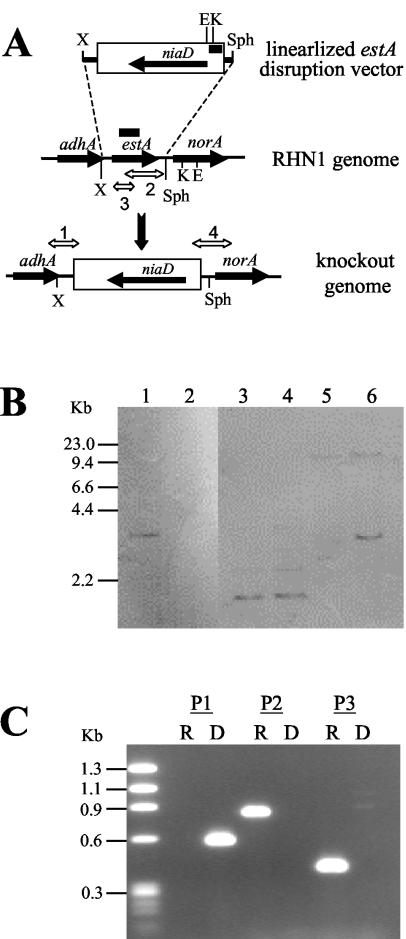
Transformation of A. parasiticus RHN1 with pestDV, which had been digested with XbaI and SphI (Fig. 2A), produced 137 transformants on Czapek regeneration plates from two independent experiments. Seventy-five arbitrarily selected transformants were further screened for accumulation of aflatoxin intermediates on PDA plates. Four of the transformants accumulated yellow pigment(s) on PDA plates (data not shown), which suggested that the functional estA gene in these transformants had been deleted. Southern hybridization was used to analyze the genomes of the recipient RHN1 and two putative estA-deleted mutants, estA-D2 and estA-D3. RHN1 contained only a single copy of the estA gene since the estA probe hybridized to a 3.0-kb EcoRI genomic DNA fragment (Fig. 2B, lane 1). Restriction site analysis confirmed that the A. parasiticus estA gene resides in a 3.0-kb EcoRI genomic DNA fragment (37). The 0.71-kb estA probe did not hybridize with the genomic DNA of estA-D2 and estA-D3, which suggested that their estA genes had been deleted (Fig. 2B, lane 2). We used a probe specific to the 5′ flanking region of the niaD marker gene and compared the restriction patterns of RHN1 and estA-D2 (Fig. 2B, lanes 3 to 6) to determine whether targeted estA deletion had occurred. Only a single hybridization signal was observed from the KpnI-digested patterns. This was likely due to the recombination of the 0.9-kb KpnI-SphI fragment in the linearized estA disruption vector with a 0.8-kb SphI-KpnI fragment in the norA gene, leading to a result indistinguishable from the resident 1.6-kb KpnI genomic DNA in the 5′ flanking region of the niaD gene (7). The EcoRI pattern of estA-D2 showed that besides a >10-kb EcoRI fragment, another 3.0-kb EcoRI fragment was present. This >10-kb DNA fragment was the genomic fragment containing the 5′ flanking region of the resident niaD gene because it was also present in RHN1. The 3.0-kb EcoRI fragment was generated after the linearized estA disruption vector had recombined with the genome of RHN1. This fragment, as expected, resulted from the combination of the 1.3-kb EcoRI-SphI fragment located in the linearized estA disruption vector and the portion from the SphI site to the first EcoRI site in the norA gene, which is about 1.7 kb in size (8).
FIG. 2.
(A) Schematic diagram depicting replacement of estA by niaD through a double-crossover recombination. Restriction sites: E, EcoRI; K, KpnI; X, XbaI. The bold lines shown above the estA gene and within the niaD marker gene represent the DIG-labeled 0.71-kb estA and 0.54-kb niaD probes, respectively, used in Southern hybridization analysis. (B) Southern blot hybridization of RHN1 and estA-D2 with the estA probe (lanes 1 and 2) and the niaD probe (lanes 3 to 6). The genomic DNAs and restriction enzymes used are as follows: lane 1, RHN1 and EcoRI; lane 2, estA-D2 and EcoRI; lane 3, RHN1 and KpnI; lane 4, estA-D2 and KpnI; lane 5, RHN1 and EcoRI; lane 6, estA-D2 and EcoRI. (C) Representative PCR profiles generated from genomic DNA of RHN1 and estA-deleted mutants. The regions amplified by primer sets P1 (estF-niaDXV1), P2 (estR-estF3), and P3 (estF2-estR2) are shown in panel A. The PCR product amplified by primer set 4, niaDXV2-norA63, is not shown. The molecular size markers are HaeIII-digested φχ174 DNA fragments. R, RHN1; D, estA-deleted mutant.
Deletion of the estA gene in the four yellow-pigmented transformants, D1 to D4, also was confirmed with primers specific to the estA-coding region, the niaD downstream and upstream regions, and regions beyond the expected recombination regions (Fig. 2A). The genomic DNA of estA-deleted transformants yielded an expected 0.6-kb PCR fragment when the estF-niaDXV1 primer pair, which amplifies a region beyond the XbaI site and the downstream region in the niaD-containing fragment, was used. No PCR products were generated from genomic DNA of the recipient RHN1. Only the estA-specific estR-estF3 and estF2-estR2 primer pairs yielded a 0.8- and a 0.4-kb PCR fragment, respectively, from RHN1 genomic DNA (Fig. 2C). The niaDXV2-norA63 primer pair also yielded a 1.0-kb PCR fragment, expected to be present in another flanking region after targeted deletion of estA, from the estA-deleted transformants but not from RHN1 (data not shown). These results indicate that the niaD selectable marker was inserted into the estA locus and replaced the resident estA gene.
Use of TLC to identify aflatoxin intermediates produced by estA-deleted mutants.
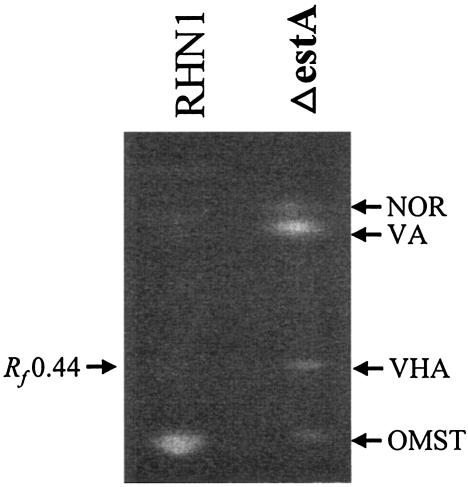
Semiquantitative TLC analysis showed that the estA-deleted mutants produced less than 20% of the OMST produced by RHN1 on PDA plates and that they also accumulated NOR, VA, and a major compound with an Rf value of 0.44 when resolved with the TEA solvent system (Fig. 3). This compound had the same Rf value when resolved by the CEF solvent system and the same retention time of 9.0 min as the VHA standard when analyzed by HPLC. The metabolite profiles from Adye and Mateles medium and PDA were similar except that NOR was reduced in the former. When larger amounts of the extracts from Adye and Mateles medium were resolved by TLC, three distinct metabolites, in addition to VHA, with Rf values of 0.14, 0.27, and 0.32 were found. The compounds were cleaned up by repeated TLC with TEA and further resolved by TLC with CEF. The compound with an Rf value of 0.14 was a mixture, and the compound with an Rf value of 0.27 had a retention time of 3.6 min, which was identical to that of the VHOH standard when analyzed by HPLC. Furthermore, when 1 N HCl was added to this compound or to the VHOH standard, a significant portion of each was converted to VC, as confirmed by HPLC (data not shown). The compound with an Rf value of 0.32 had the same Rf values as VOAc when resolved by TEA and CEF, and it also had the same retention time of 6.2 min as the VOAc standard.
FIG. 3.
Representative semiquantitative TLC of aflatoxin intermediates produced by RHN1 and estA-deleted mutants on PDA plates. Metabolites were extracted from 5-day-old cultures grown at 30°C in the dark and resolved by the TEA solvent system. The positions of NOR, VA, VHA (Rf = 0.44), and OMST are indicated. RHN1, the recipient strain; ΔestA, estA-deleted mutants derived from RHN1.
Effects of dichlorvos on production of aflatoxin intermediates in the tip cultures.
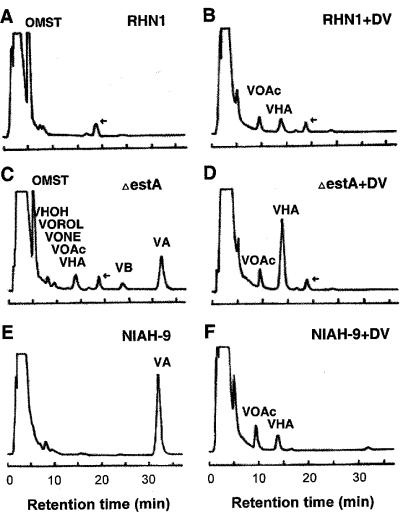
RHN1 produced only OMST as its end product (Fig. 4A), but the estA deleted-mutant accumulated significant amounts of VHA and VOAc, suggesting that the estA-encoded esterase catalyzes the conversions from VHA to VHOH and from VOAc to VOH. The estA-deleted mutant also produced small amounts of VOROL and VONE. Although the estA-deleted mutant did not have the estA gene (Fig. 2B), it also accumulated VHOH, VA, VB, and OMST (Fig. 4C). The amount of OMST in the estA-deleted mutant decreased to 42% of that in RHN1. These results suggested that another esterase(s) could complement the function of EstA to form downstream intermediates such as VHOH, VA, VB, and OMST (Fig. 4C).
FIG. 4.
Accumulation of aflatoxin intermediates by A. parasiticus isolates with or without the addition of dichlorvos. RHN1, the transformation recipient strain; ΔestA, estA-deleted mutant derived from RHN1; NIAH-9, VA-accumulating strain derived from NRRL 2999. RHN1 (A and B), estA-deleted mutant (C and D), and NIAH-9 (E and F) strains were cultured in the absence or presence of dichlorvos. A peak for an unknown substance at around 19 min is indicated by an arrow. Retention times of the authentic aflatoxin intermediate standards are described in Materials and Methods. No significant difference in the mycelial weights was found among the cultures.
The estA-deleted mutant accumulated a substantial amount of VA and some VB. We compared the profile of the accumulated aflatoxin intermediates in the estA-deleted mutant with that in NIAH-9, in which the process is blocked at the step just after VA production. NIAH-9 accumulated only VA, which indicates that the blocked step in the estA-deleted mutant is different from that in NIAH-9. Although the amount of VA was higher than the amount of VB in the mycelia, the ratio of VA to VB was reversed in the culture medium (data not shown).
The addition of dichlorvos blocked OMST synthesis by RHN1 and resulted in accumulation of VHA and VOAc (Fig. 4B). Dichlorvos also inhibited production of VA in NIAH-9 and resulted in the accumulation of VHA and VOAc (Fig. 4E and F). When dichlorvos was added to the estA-deleted mutant, VB and VA production were inhibited but VHA accumulation was enhanced. These results are consistent with the hypothesis that the estA-encoded esterase as well as other esterases is effectively inhibited by dichlorvos. The unknown substance, which eluted at around 19 min, was produced by RHN1 and the estA-deleted mutant irrespective of the presence of dichlorvos. This substance was not produced by NIAH-9 (Fig. 4E and F) or by an aflatoxigenic A. parasiticus SYS-4 strain (data not shown). This substance probably is not related to aflatoxin biosynthesis but instead is specific to SRRC 2043-derived strains.
Esterase activities of RHN1 and estA-deleted mutants.
To confirm that the estA-encoded esterase is directly involved in the steps from VHA to VHOH and from VOAc to VOH, we measured the esterase activities of the cell extracts prepared from RHN1 and two independent estA-deleted mutants, estA-D2 and estA-D3. When VHA was added as the substrate to the reaction mixture containing the cell extract of RHN1, 9.2 ± 1.8 pmol of VB and 11.7 ± 1.6 pmol of VHOH mg of protein−1 were produced after 15 min of incubation. Since the VHOH cyclase activity converting VHOH to VB (18) was also present in the cell extracts and was not affected by the estA deletion (data not shown), a portion of the resulting VHOH was converted to VB. Therefore, the total amount of VHOH and VB was measured to estimate esterase activity. The reactions were linear until 15 min under the experimental conditions, indicating that the esterase-specific activity of the RHN1 extract was 1.4 ± 0.2 pmol of VHOH (plus VB) mg−1 min−1. In contrast, the esterase-specific activities for estA-D2 and estA-D3 were 0.10 ± 0.03 and 0.12 ± 0.02 pmol of VHOH (plus VB) mg−1 min−1, respectively. Therefore, about 90% of the esterase activity of RHN1 was lost after the estA gene was deleted. Similar results were obtained when VOAc was used as the substrate instead of VHOH. The esterase activity in the RHN1 cell extract converting VOAc to VOH was 0.51 ± 0.07 pmol of VOAc mg−1 min−1. In contrast, the activities for estA-D2 and estA-D3 were 0.09 ± 0.05 and 0.06 ± 0.00 pmol of VOAc mg−1 min−1, respectively. Thus, about 10% of the conversion in both reactions was carried out by an esterase(s) other than the estA-encoded esterase.
DISCUSSION
The estA gene is the only predicted esterase-encoding gene found in the 70-kb aflatoxin pathway gene cluster (37). Southern and PCR analyses also indicated that no other estA homolog(s) is present in the genome of SRRC 2043 (Fig. 2B and C). Deletion of the estA gene did not completely block formation of OMST and the aflatoxin intermediates after the esterase step (Fig. 4C), hence another esterase(s) could complement the function of EstA. In the in vitro enzyme assays, EstA accounts for about 90% and the unidentified esterase(s) accounts for about 10% of the total esterase activity. Although EstA efficiently converts VHA to VHOH and VOAc to VOH in vitro, this result does not mirror the conversions in vivo observed under YES medium growth conditions (Fig. 4 A to D). This discrepancy may be due to reduced specific activities of the unidentified esterases in the 4-day-old cultures or because the in vitro reaction conditions do not allow full activation of the esterases that are active in the cell. Nonetheless, results from the PDA solid cultures, the YES liquid cultures, and the enzyme assays clearly show that EstA converts VHA to VHOH and VOAc to VOH.
An esterase with activity similar to that predicted for EstA is common in various Aspergillus strains. For example, an esterase activity catalyzing the conversion of VHA to VHOH was found even when A. parasiticus NIAH-26 was cultured in aflatoxin-noninducible YEP medium (28). Aspergillus sojae strain 477, a nonaflatoxigenic fungus, commonly believed to be a domesticated strain of A. parasiticus, had 29% of the esterase activity associated with the conversion of VOAc to VOH when compared to the activity in A. parasiticus NIAH-26 (19) but lacked all other enzyme activities for aflatoxin biosynthesis (19). Also, the porcine esterase used in this study can convert VHA to VHOH, which suggests that many esterases may use VHA and probably VOAc as a substrate. Thus, an aflatoxin pathway-nonspecific esterase(s) may be involved in aflatoxin biosynthesis. EstA (estimated size, 34 kDa) may correspond to the VHA esterase partially purified by Kusumoto and Hsieh (17) which consists of two 30-kDa isomeric subunits. The other esterase activities identified by Kusumoto and Hsieh (17) also may convert VHA to VHOH or VOAc to VOH.
Secondary metabolic enzymes often are only relatively specific and may catalyze analogous reactions of structurally related metabolites, resulting in the generation of a metabolic grid (4, 5). EstA can convert VHA to VHOH and VOAc to VOH (Fig. 1). The pathway from HVN to VHA to VHOH is the major route for aflatoxin biosynthesis because it is the shortest path from HVN to VHOH. However, the side pathway from VONE to VOH also functions. Some HVN may be converted to VONE by the VHA reductase, and the resultant VONE can be converted either to VOROL by a reductase or to VOAc by a monooxygenase (29). This hypothesis is consistent with the deletion of estA in A. parasiticus, which results in a higher level of VHA than VOAc and the concomitant accumulation of VONE and VOROL (Fig. 4C).
Dichlorvos inhibits all esterase activities irrespective of the enzyme source. All A. parasiticus strains accumulated VHA and VOAc (Fig. 4). The amounts of VHA and VOAc accumulated by RHN1 and the estA-deleted mutant were similar, but more VOAc than VHA was accumulated by NIAH-9. The same VHA reductase can also catalyze the conversions of HVN to VONE, VHA to VOAc (29), and VHOH to VOH (K. Yabe, unpublished data). The VHA reductase activity of A. parasiticus NIAH-9 thus may be stronger than that of A. parasiticus SRRC2043. These results suggest that the contributions of the side pathway, VONE→VOAc→VOH, to the overall aflatoxin biosynthesis may depend on the activity of the VHA reductase.
The estA-deleted mutant also accumulated significant amounts of other intermediates, such as VHOH, VB, VA, and OMST, downstream of the proposed block in the aflatoxin biosynthetic pathway. Accumulation of VHOH might be due to the racemization of VHA or VHOH. If (2′S)-HVN is produced from the natural averufin enantiomer, (1′S, 5′S)-AVR, by the microsome enzyme (29), it could be rapidly converted to (2′S)-HVN, to (2′S)-VHA, and to (2′S)-VHOH via the main pathway of the metabolic grid because the resultant (2′S)-VHOH can be converted to VB by VHOH cyclase. The cyclase is specific for the 2′S) isomer of VHOH (30). However, if most of the esterase activity is lost due to the estA gene deletion, the flow of the main pathway from HVN to VHOH would be significantly slowed. In such cases, some of the accumulated (2′S)-HVN, (2′S)-VHA, and (2′S)-VHOH could racemize to (2′R) intermediates. The (2′R) intermediates could proceed through the side pathway VONE→VOAc→VOH. (2′R)-VHOH, however, would transiently accumulate in the mycelia since the VHOH cyclase could not use it as its substrate. The (2′R) intermediates would revert to (2′S) intermediates by nonenzymatic racemization. Thus, intermediates past the block resulting from the deletion of estA could still be produced and accumulated (Fig. 1 and 4C).
Accumulation of VA, which is independent of medium type, in the estA-deleted mutant cannot be easily explained. The fact that RHN1 accumulates neither VA nor VB suggests that EstA might be involved in the step from VA to demethylsterigmatocystin or from VB to dihydrodemethylsterigmatocystin (Fig. 1) although only Baeyer-Villiger oxidation, nuclear rearrangement, reduction, and decarboxylation have previously been proposed to be involved (20, 27). However, no enzyme activities associated with these steps have been found in cell extracts (K. Yabe, unpublished) and the enzymatic mechanisms responsible for these conversions remain to be elucidated. Temporal delay of normal metabolic flux due to the loss of EstA could be a reason for the observed accumulation.
It is not uncommon for more than one enzyme to catalyze an identical step during aflatoxin biosynthesis (6, 10, 25). The present study suggests that another esterase(s) may play as important a role as EstA in aflatoxin biosynthesis. Thus, the hypothesis that a gene cluster exclusively encodes all of the enzyme activities for the aflatoxin biosynthetic pathway may not be true. Functional studies of the clustered pathway genes would shed more light on the role of nonclustered genes in fungal secondary metabolism.
Acknowledgments
We thank H. Nakajima, Tottori University, for supplying aflatoxin precursors and H. Arai and H. Hatabayashi at the National Food Research Institute, Tsukuba, Japan, and L. Scharfenstein at the Southern Regional Research Center for technical assistance.
This work was supported in part by a grant-in-aid (Bio-Design-Program) from the Ministry of Agriculture, Forestry and Fisheries (BDP-03-VI-1-9).
REFERENCES
- 1.Adye, J., and R. I. Mateles. 1964. Incorporation of labeled compounds into aflatoxins. Biochim. Biophys. Acta 86:418-420. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. A., and C. H. Chung. 1990. Conversion of versiconal acetate to versiconal and versicolorin C in extracts from Aspergillus parasiticus. Mycopathologia 110:31-35. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar, D., T. E. Cleveland, R. L. Brown, J. W. Cary, J. Yu, and P.-K. Chang. 1997. Preharvest aflatoxin contamination: elimination through biotechnology, p. 100-129. In G. S. Dhaliwal, R. Arora, N. S. Randhawa, and A. R. Dhawan (ed.), Ecological agriculture and sustainable development: bio/technology in agriculture. Chaman Enterprises, New Delhi, India.
- 4.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bu'Lock, J. D. 1965. The biogenesis of natural products. McGraw-Hill, London, England.
- 6.Cary, J. W., M. Wright, D. Bhatnagar, R. Lee, and F. S. Chu. 1996. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl. Environ. Microbiol. 62:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P.-K., K. C. Ehrlich, J. E. Linz, D. Bhatnagar, T. E. Cleveland, and J. W. Bennett. 1996. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet. 30:68-75. [DOI] [PubMed] [Google Scholar]
- 8.Chang, P.-K., and J. Yu. 2002. Characterization of a partial duplication of the aflatoxin gene cluster in Aspergillus parasiticus ATCC 56775. Appl. Microbiol. Biotechnol. 58:632-636. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P.-K., J. Yu, D. Bhatnagar, and T. E. Cleveland. 1999. Repressor-AFLR interaction modulates aflatoxin biosynthesis in Aspergillus parasiticus. Mycopathologia 147:105-112. [DOI] [PubMed] [Google Scholar]
- 10.Chang, P.-K., J. Yu, K. C. Ehrlich, S. M. Boue, B. G. Montalbano, D. Bhatnagar, and T. E. Cleveland. 2000. adhA in Aspergillus parasiticus is involved in conversion of 5′-hydroxyaverantin to averufin. Appl. Environ. Microbiol. 66:4715-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horng, J. S., P.-K. Chang, J. J. Pestka, and J. E. Linz. 1990. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. 224:294-296. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh, D. P. H., C. C. Wan, and J. A. Billington. 1989. A versiconal hemiacetal acetate converting enzyme in aflatoxin biosynthesis. Mycopathologia 107:121-126. [DOI] [PubMed] [Google Scholar]
- 13.Jelinek, C. F., A. E. Pohland, and G. E. Wood. 1989. Worldwide occurrence of mycotoxins in foods and feeds—an update. J. Assoc. Off. Anal. Chem. 72:223-230. [PubMed] [Google Scholar]
- 14.Kelkar, H. S., T. W. Skloss, J. F. Haw, N. P. Keller, and T. H. Adams. 1997. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J. Biol. Chem. 272:1589-1594. [DOI] [PubMed] [Google Scholar]
- 15.Keller, N. P., S. Segner, D. Bhatnagar, and T. H. Adams. 1995. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans. Appl. Environ. Microbiol. 61:3628-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, N. P., C. M. Watanabe, H. S. Kelkar, T. H. Adams, and C. A. Townsend. 2002. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl. Environ. Microbiol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusumoto, K., and D. P. H. Hsieh. 1996. Purification and characterization of the esterases involved in aflatoxin biosynthesis in Aspergillus parasiticus. Can. J. Microbiol. 42:804-810. [DOI] [PubMed] [Google Scholar]
- 18.Lin, B. K., and J. A. Anderson. 1992. Purification and properties of versiconal cyclase from Aspergillus parasiticus. Arch. Biochem. Biophys. 293:67-70. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima, K., K. Yashiro, Y. Hanya, K. Abe, K. Yabe, and T. Hamasaki. 2001. Absence of aflatoxin biosynthesis in koji mold (Aspergillus sojae). Appl. Microbiol. Biotechnol. 55:771-776. [DOI] [PubMed] [Google Scholar]
- 20.Minto, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2556. [DOI] [PubMed] [Google Scholar]
- 21.Motomura, M., N. Chihaya, T. Shinozawa, T. Hamasaki, and K. Yabe. 1999. Cloning and characterization of the O-methyltransferase I gene (dmtA) from Aspergillus parasiticus associated with the conversions of demethylsterigmatocystin to sterigmatocystin and dihydrodemethylsterigmatocystin to dihydrosterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 65:4987-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroeder, H. W., R. S. Cole, R. D. Grigsby, and H. J. Hein. 1974. Inhibition of aflatoxin production and tentative identification of an aflatoxin intermediate “versicolor acetate” from treatment with dichlorvos. Appl. Microbiol. 27:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skory, C. D., P.-K. Chang, J. Cary, and J. E. Linz. 1992. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 58:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Townsend, C. A., S. B. Christensen, and K. Trautwein. 1984. Hexanoate as a starter unit in polyketide biosynthesis. J. Am. Chem. Soc. 106:3868-3869. [Google Scholar]
- 25.Trail, F., P.-K. Chang, J. W. Cary, and J. E. Linz. 1994. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microbiol. 60:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe, C. M., and C. A. Townsend. 2002. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem. Biol. 9:981-988. [DOI] [PubMed] [Google Scholar]
- 27.Yabe, K. 2002. Pathway and genes of aflatoxin biosynthesis, p. 227-251. In F. Fierro and J. Francisco (ed.), Microbial secondary metabolites: biosynthesis, genetics and regulation. Research Signpost, Kerala, India.
- 28.Yabe, K., Y. Ando, and T. Hamasaki. 1991. A metabolic grid among versiconal hemiacetal acetate, versiconol acetate, versiconol and versiconal during aflatoxin biosynthesis. J. Gen. Microbiol. 137:2469-2475. [DOI] [PubMed] [Google Scholar]
- 29.Yabe, K., N. Chihaya, S. Hamamatsu, E. Sakuno, T. Hamasaki, H. Nakajima, and J. W. Bennett. 2003. Enzymatic conversion of averufin to hydroxyversicolorone and elucidation of a novel metabolic grid involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 69:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yabe, K., and T. Hamasaki. 1993. Stereochemistry during aflatoxin biosynthesis: cyclase reaction in the conversion of versiconal to versicolorin B and racemization of versiconal hemiacetal acetate. Appl. Environ. Microbiol. 59:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao, R. C., and D. P. H. Hsieh. 1974. Step of dichlorvos inhibition in the pathway of aflatoxin biosynthesis. Appl. Microbiol. 27:2852-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J., D. Bhatnagar, and K. C. Ehrlich. 2002. Aflatoxin biosynthesis. Rev. Iberoam. Micol. 19:191-200. [PubMed] [Google Scholar]
- 34.Yu, J., J. W. Cary, D. Bhatnagar, T. E. Cleveland, N. P. Keller, and F. S. Chu. 1993. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, B. Montalbano, J. M. Dyer, D. Bhatnagar, and T. E. Cleveland. 1998. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 64:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, J., P.-K. Chang, D. Bhatnagar, and T. E. Cleveland. 2000. Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:583-590. [DOI] [PubMed] [Google Scholar]
- 37.Yu, J., P.-K. Chang, D. Bhatnagar, and T. E. Cleveland. 2002. Cloning and functional expression of an esterase gene in Aspergillus parasiticus. Mycopathologia 156:227-234. [DOI] [PubMed] [Google Scholar]
- 38.Yu, J., C. P. Woloshuk, D. Bhatnagar, and T. E. Cleveland. 2000. Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene 248:157-167. [DOI] [PubMed] [Google Scholar]