Abstract
The Roseobacter clade of marine bacteria is often found associated with dinoflagellates, one of the major producers of dimethylsulfoniopropionate (DMSP). In this study, we tested the hypothesis that Roseobacter species have developed a physiological relationship with DMSP-producing dinoflagellates mediated by the metabolism of DMSP. DMSP was measured in Pfiesteria and Pfiesteria-like (Cryptoperidiniopsis) dinoflagellates, and the identities and metabolic potentials of the associated Roseobacter species to degrade DMSP were determined. Both Pfiesteria piscicida and Pfiesteria shumwayae produce DMSP with an average intracellular concentration of 3.8 μM. Cultures of P. piscicida or Cryptoperidiniopsis sp. that included both the dinoflagellates and their associated bacteria rapidly catabolized 200 μM DMSP (within 30 h), and the rate of catabolism was much higher for P. piscicida cultures than for P. shumwayae cultures. The community of bacteria from P. piscicida and Cryptoperidiniopsis cultures degraded DMSP with the production of dimethylsulfide (DMS) and acrylate, followed by 3-methylmercaptopropionate (MMPA) and methanethiol (MeSH). Four DMSP-degrading bacteria were isolated from the P. piscicida cultures and found to be taxonomically related to Roseobacter species. All four isolates produced MMPA from DMSP. Two of the strains also produced MeSH and DMS, indicating that they are capable of utilizing both the lyase and demethylation pathways. The diverse metabolism of DMSP by the dinoflagellate-associated Roseobacter spp. offers evidence consistent with a hypothesis that these bacteria benefit from association with DMSP-producing dinoflagellates.
Pfiesteria piscicida, Pfiesteria shumwayae, and Pfiesteria-like organisms, such as Cryptoperidiniopsis sp., are estuarine, heterotrophic dinoflagellates with a global distribution (reviewed in reference 31). Reports have implicated Pfiesteria sp. as the causative agent of massive fish deaths along the Atlantic Coast of the United States, especially in the estuaries of Pamlico Sound, N.C., and the Chesapeake Bay of Maryland and Virginia (6). Certain Pfiesteria species are thought to kill fish by excreting potent ichthyotoxins, similar to those of other well-characterized dinoflagellate species (13). However, no toxins from this organism have been identified (32), and recent reports show that P. shumwayae can kill fish by consuming epithelial cells, a process that does not require toxin production (5, 43). While the toxicity of Pfiesteria species is an important question, other physiological aspects of these dinoflagellates deserve further attention.
Dimethylsulfoniopropionate (DMSP) is the major source of organic sulfur in the world's oceans and plays a significant role in the global sulfur cycle (reviewed in reference 45). During blooms of marine unicellular algae, cellular DMSP is released due to algal senescence, predation, or stress and is degraded by both algal and bacterial enzymes. In marine environments, dinoflagellates and prymnesiophytes are the major producers of DMSP, with intracellular concentrations as high as 0.5 M (45). Although the exact function of DMSP is unclear, it has possible roles in osmoprotection (9, 40) and cryoprotection (23), antiherbivory (44), and protection from oxidative stress (37).
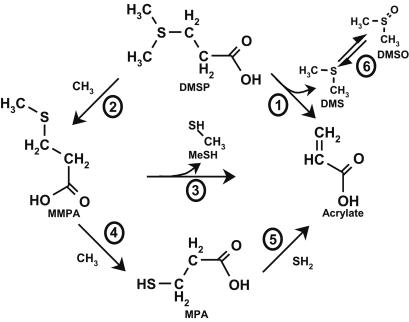
Bacteria and certain species of phytoplankton produce an enzyme, dimethylpropiothetin dethiomethylase (DMSP lyase) (EC 4.4.1.3) (Fig. 1, reaction 1), which degrades DMSP to produce dimethylsulfide (DMS) and acrylate (46). As shown in Fig. 1, DMS produced from this reaction is oxidized by some bacterial species to form dimethyl sulfoxide (Fig. 1, reaction 6) (39). Acrylate is readily consumed by bacteria and is converted to β-hydroxypropionate by α- and γ-proteobacterial species (2, 3). Bacteria may also demethylate DMSP at the DMS moiety (Fig. 1, reaction 2), producing 3-methylmercaptopropionate (MMPA) (20), which may be further demethylated to 3-mercaptopropionate (MPA; Fig. 1, reaction 4) (41) or demethiolated to produce acrylate (Fig. 1, reaction 3) and methanethiol (MeSH); (38). The products of these reactions provide a rich source of carbon and sulfur for bacterial production (26, 42). It is estimated that ∼15% of the DMSP produced by marine phytoplankton is degraded by the lyase pathway and 85% or more is degraded by bacterial demethylation of DMSP, suggesting an essential bacterial role in controlling DMS emissions from the world's oceans (33, 47).
FIG. 1.
Pathways involved in the catabolism of DMSP. Degradation of DMSP can occur by the lyase pathway, which involves the hydrolysis of the C-3 carbon of DMSP, producing acrylate and DMS (reaction 1), or by a series of demethylation steps (reactions 2 to 5). The first step in the demethylase pathway is demethylation of the DMS moiety of DMSP, producing MMPA (reaction 2). MMPA may be further demethylated to MPA (reaction 4), followed by the elimination of hydrogen sulfide (reaction 5), yielding acrylate, or in a demethiolation reaction (reaction 3), producing acrylate and MeSH. In some cases, the DMS produced by the lyase pathway may be oxidized to dimethyl sulfoxide (reaction 6).
In marine surface waters, α-proteobacteria phylogenetically related to Roseobacter spp. are predominantly responsible for the degradation of DMSP, its catabolites, and other sulfonium compounds (15). Although Roseobacter spp. are cosmopolitan in nature, their production and activity are significantly correlated with DMSP-producing algae, including dinoflagellates and prymnesiophytes (16, 47). Furthermore, some Roseobacter spp. exhibit close physical or physiological relationships with toxic, DMSP-producing dinoflagellates, including Prorocentrum spp. (30), Alexandrium spp. (8, 12), and Pfiesteria spp. (1).
In the environment, dinoflagellates coexist and interact with a diverse community of bacteria and other microorganisms. These interactions can be studied in monocultures of dinoflagellates obtained from environmental samples. Within these cultures, bacteria native to the algal niche assimilate dinoflagellate-derived nutrients and are intrinsically propagated with the dinoflagellates in continuous subcultures. In a recent study, we characterized the bacterial community inhabiting several Pfiesteria dinoflagellate cultures isolated from the Chesapeake Bay, Md. (1). All of the dinoflagellate cultures examined contained one or more Roseobacter spp. representing the second most abundant clone obtained from 16S ribosomal DNA (rDNA) clone libraries. In addition, several bacteria were found attached to these dinoflagellates by using fluorescent in situ hybridization and confocal scanning laser microscopy. After a stringent washing procedure to remove unattached bacteria, the predominant bacterial species present was a bacterium closely related to Sulfitobacter pontiacus, a Roseobacter clade organism. Also, a Roseobacter sp. was found to be necessary for the growth of Pfiesteria in culture (1).
In this study, we measured the production of DMSP by P. piscicida and P. shumwayae and then assessed DMSP catabolism by the Pfiesteria cultures and the bacterial communities associated with them. New dinoflagellate-associated roseobacters capable of DMSP degradation by both the lyase and demethylation pathways were isolated and identified.
MATERIALS AND METHODS
Dinoflagellate strains and culturing.
Dinoflagellate cultures of P. piscicida CCMP1830, CCMP1921, and CCMP1834; Cryptoperidiniopsis sp. strain CCMP1829; and P. shumwayae CCMP2089 (Provasoli-Guillard National Center for Culture of Marine Phytoplankton) were grown as previously described (1). The dinoflagellates were fed the prey alga Rhodomonas sp. strain CCMP768 continuously as needed. Rhodomonas sp. was grown in 35 practical salinity units (psu) F/2 medium lacking silica at 20°C under a 14-h light, 10-h dark cycle (18). For all assays, the dinoflagellates were grown to a maximum cell density of ∼105 per ml, whereupon feeding was stopped for 36 h, which allowed complete removal of the Rhodomonas algae (monitored by inverted microscopy).
Bacterial strains and media.
Bacteria were isolated from P. piscicida CCMP1830 culture by first spreading a 10-fold dilution series of the dinoflagellate culture on 0.5× Zobell marine agar 2216 (18.7 g of Difco marine broth 2216, 15 g of Difco Bacto Agar, and 1,000 ml of distilled H2O), hereafter referred to as marine agar. After 5 to 7 days of incubation at 30°C, colonies with unique morphologies were picked at random and streaked to purity on marine agar, resulting in the strains TM1034 to TM1042. The bacterial cultures were used after incubation in marine broth (same as marine agar but lacking the agar) at 30°C in a shaking water bath for 1 to 3 days. Escherichia coli INVαF′ was grown in Luria-Bertani (LB) broth (4) or on LB agar containing 1.5% Bacto Agar (Becton Dickinson, Franklin Lakes, N.J.).
Chemicals.
DMSP was synthesized from acrylate and DMS according to the method of Chambers et al. (7). The purity of the resulting DMSP was confirmed by chemical analyses, including flash point, melting point, and total C, H, O, N, S, and Cl (Galbraith Laboratories, Inc., Knoxville, Tenn.). MMPA was synthesized by alkaline hydrolysis of its methyl ester, methyl-3-(methylthio)propionate (Aldrich, Milwaukee, Wis.) (21). Other organic sulfur compounds were purchased from Aldrich. All chemicals used were of the highest purity commercially available.
DMSP content and metabolism.
To measure the DMSP contents of the dinoflagellates P. piscicida CCMP1830 and P. shumwayae CCMP2089 and the prey alga Rhodomonas sp. strain CCMP768, cells were grown to a maximum density of ∼105 per ml in 500-ml batch cultures, except for Rhodomonas sp., which was grown in 1-liter batch cultures to a maximum density of 106 cells per ml. The abundance and cellular volume of the cells were measured in the 7- to 20-μm-diameter particle range from three or more 1-ml samples of culture using a Coulter Multisizer II particle counter (Becton-Dickinson). Because the particle counter does not distinguish between Pfiesteria dinoflagellates and Rhodomonas prey algae, all dinoflagellate cultures were starved to reduce the Rhodomonas population to below detectable limits (as described above).
DMSP was measured in 2-ml whole-culture aliquots and in concentrated cell lysates. To obtain concentrated cell lysates, 200-ml culture aliquots were centrifuged at 4,000 × g for 15 min and the cell pellets were resuspended in 2 ml of sterile distilled water on ice. For Rhodomonas sp. strain CCMP768, multiple cell pellets were combined and resuspended in a final 2-ml aliquot of sterile distilled water. A cell homogenate of each sample was then obtained using a Sonic Dismembrator sonicator (Fisher, Hampton, N.H.). DMSP was measured in 2-ml samples as described in “Analytical techniques” below.
To measure the degradation of DMSP by dinoflagellate and prey algal cultures, cells were grown as described above and the cell density was normalized across all cultures to 104 per ml by diluting the cultures with sterile medium. DMSP was added to the culture from a sterile neutralized stock at a final concentration of 200 μM, and its degradation was measured at intervals throughout the duration of the experiment, as described in “Analytical techniques” below.
The catabolism of DMSP by the bacterial component of each culture was measured in suspensions containing a mixture of dinoflagellate-associated bacteria that were isolated as follows. A 10-fold dilution series of each dinoflagellate culture at peak dinoflagellate density (∼105 cells per ml) was spread on marine agar and incubated at 30°C for 5 days. The resulting colonies from plates containing 50 to 200 colonies were resuspended from the agar surface using sterile 10-psu artificial seawater (Instant Ocean, Mentor, Ohio) and washed twice by centrifugation at 14,000 × g, whereupon the optical density was normalized to 0.6 at 600 nm for each suspension. An aliquot of DMSP was then added from a sterile neutralized stock to a final concentration of 1 mM, and DMS, MeSH, MMPA, and acrylate were measured as described in “Analytical techniques” below.
DMSP catabolism was also measured in four bacterial strains (TM1035, TM1038, TM1040, and TM1042) isolated from the dinoflagellate culture. Each strain was grown in a 50-ml marine broth culture amended with 1 mM DMSP to induce the production of enzymes necessary for DMSP catabolism. The cultures were grown to an optical density at 600 nm of 0.6 and washed twice with sterile 10-psu artificial seawater. DMSP was added to a final concentration of either 0.1 or 1 mM, and 1-ml aliquots of the bacterial cultures were dispersed into 26-ml serum bottles. The bottles were immediately capped with a butyl rubber septum and incubated at 30°C with shaking. At intervals throughout the experiment, samples were sacrificed for measurement of DMSP, DMS, MeSH, acrylate, and MMPA as described below.
Analytical techniques.
DMSP was measured as DMS following alkaline hydrolysis. An aliquot of the sample was added to a 26-ml serum bottle with the addition of an equal volume of either 5 M NaOH or distilled water, and the bottle was capped with a butyl rubber septum. Solutions of pure DMSP at 1 to 500 μM dissolved in distilled water were prepared in exactly the same manner in parallel with each experiment. After overnight incubation, DMS resulting from alkaline hydrolysis of DMSP was measured in 500 μl of headspace gas using a Hewlett-Packard 5890 gas chromatograph equipped with flame ionization detection (GC-FID). DMS produced without alkaline hydrolysis was subtracted from the total, and the result was compared to DMS produced from the hydrolysis of pure DMSP standards to obtain the final molar concentration of DMSP in the unknown sample. The retention time of DMS was determined by injecting 50 μl of headspace gas from a capped serum bottle containing 5 μl of pure DMS that had completely volatilized.
DMS and MeSH production in the cultures was measured by direct sampling of 500 μl of headspace gas without prior alkaline hydrolysis of the sample. The concentration of DMS was determined using standard curves generated from known concentrations of DMS produced by complete alkaline hydrolysis of known amounts of DMSP. The concentrations of gaseous MeSH in the cultures were determined from standard curves using a dilution series of pure MeSH gas.
The presence and concentrations of acrylate and MMPA in the cultures were measured by high-performance liquid chromatography (Agilent [Palo Alto, Calif.] 1100 equipped with diode array detection) and a Zorbax XDB C18 column (2.1 by 150 mm; 5-μm pore size) (Agilent) according to the method of Ansede et al. (2).
Statistics.
The Mann-Whitney test for two independent samples was used to compare the DMSP contents of P. piscicida CCMP1830 and P. shumwayae CCMP2089. The apparent first-order rate constants for DMSP degradation and catabolite production by bacterial strains TM1035, TM1038, TM1040, and TM1042 was calculated using a linear least-squares regression analysis of the data, where the slope of the line equals the first-order rate constant (r > 0.90).
DNA methods.
Chromosomal DNA was extracted from bacterial cells by routine methods (36) and used as a template in a PCR to amplify the near-full-length (∼1,300-bp) 16S rDNA gene. The PCR conditions were as previously described (1). The resulting PCR products were analyzed by electrophoresis using a 1.0% agarose gel in 1× TAE (4) to confirm the presence of a single 1,300-bp product, which was then excised from the gel using a sterile razor blade, purified using the QIAGEN gel extraction kit, and cloned into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.) under the ligation conditions recommended by the manufacturer. Plasmid DNA was transformed into E. coli INVαF′ competent cells (Invitrogen). Transformants were selected and screened for DNA insertion using LB agar containing kanamycin (80 μg per ml) plus X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg per ml). White colonies, i.e., those harboring recombinant plasmids, were picked at random and grown overnight with antibiotic selection. Plasmid DNA was extracted by alkaline lysis and purified by standard methods using a cesium chloride gradient (4). The presence of a near-full-length 16S rDNA insert was confirmed by agarose gel electrophoresis analysis of EcoRI-digested plasmid DNA. The nucleotide sequence of each 16S rDNA was determined as previously described (1).
Nucleotide sequence analysis and phylogenetic-tree construction.
The construction of phylogenetic trees was done as described by Alavi et al. (1). Briefly, evolutionary trees were generated using the neighbor-joining (35), Fitch-Margoliash (11), and maximum-parsimony (29) algorithms in the PHYLIP package (10). Evolutionary-distance matrices for the neighbor-joining and Fitch-Margoliash methods were generated as described by Jukes and Cantor (22). The confidence in tree topology was evaluated after 1,000 bootstrap resamplings of the neighbor-joining data, and only values of ≥500 were shown on the tree.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rDNA sequences used to generate phylogenetic trees are as follows: Roseovarius tolerans, Y11551; Roseovarius sp. strain DFL-24, AJ534215; marine bacterium ATAM407-61, AF359525; Sagittula stellata, U58356; α-proteobacterium GMD29C12, AY162070; Roseovarius nubinhibens, AF098495; uncultured Rhodobacter LA1-B32N, AF513928; Roseobacter sp. strain LA7, AF513438; marine bacterium HP29w, AY239008; α-proteobacterium MBIC1887, AB026492; Silicibacter lacuscaerulensis, U77644; Silicibacter pomeroyi, AF098491; marine bacterium P20, AY082668; Reugeria atlantica, AF124521; Reugeria algocolus, X78313; Roseobacter gallaciensis, Y13244; Sulfitobacter mediterraneus, Y17387; Roseobacter denitrificans, X69159; Roseobacter litoralis, X78312; Sulfitobacter sp. strain GAI-37, AF007260; Sulfitobacter sp. strain GAI-21, AF007257; Sulfitobacter pontiacus, Y13155; and Sulfitobacter sp. strain EE36, AF007254. The nucleotide sequences incorporating 1,300 bp of the 16S rDNA gene from strains TM1035, TM1038, TM1040, and TM1042 have been deposited in the GenBank database under accession numbers AY332660, AY332661, AY332662, and AY332663, respectively.
RESULTS
DMSP content of P. piscicida and P. shumwayae.
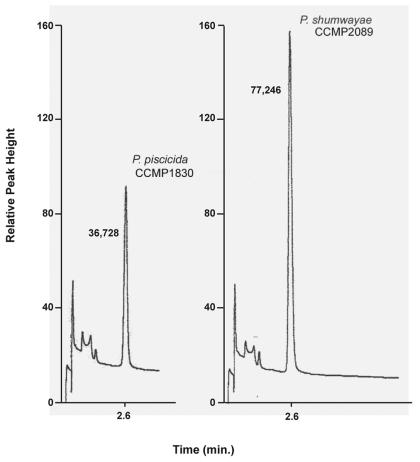
Figure 2 shows representative chromatograms of DMSP in cell lysates from each Pfiesteria species. The internal cell volumes for P. piscicida and P. shumwayae were determined to be 0.69 and 0.55 nl, respectively; thus, the average intracellular DMSP concentration of P. piscicida is ∼3.44 μM, while that of P. shumwayae is estimated to be 4.25 μM (Table 1). Statistical analyses show that the mean DMSP concentrations and the mean intracellular volumes for the species are not significantly different (P > 0.05).
FIG. 2.
Representative GC-FID chromatograms of DMS. DMSP in Pfiesteria dinoflagellates was detected as DMS, the major peak in each chromatogram (retention time, 2.6 min), after alkaline hydrolysis of DMSP. The numbers to the left of the DMS peak represent the peak area. The concentration of DMSP, as DMS, was determined by measurement of the peak area and comparison to a set of known standard concentrations of DMSP. The peaks shown were obtained from cultures containing 37,642 P. piscicida cells per ml and 50,985 P. shumwayae cells per ml. The data, normalized for cell density and cell volume, are presented in Table 1. The minor peaks that display shorter retention times than DMS are not associated with DMSP, DMS, or other DMSP catabolites.
TABLE 1.
Intracellular DMSP contents of Pfiesteria species
| Species | Strain | Intracellular DMSP (pg per cell) | Cell vol (nl) | DMSP (μM) |
|---|---|---|---|---|
| P. piscicida | 1830 | 0.41 | 0.69 ± 0.12 | 3.44 ± 1.00 |
| P. shumwayae | 2089 | 0.40 | 0.55 ± 0.02 | 4.25 ± 1.47 |
Intracellular DMSP was not detected in the Rhodomonas prey algal cultures, even when 1,000-fold more cells were used for the analysis. Thus, Rhodomonas sp. strain CCMP768 is not a significant source of DMSP in the Pfiesteria culture. While DMSP was readily detectable in Pfiesteria cell lysates, it was not found in either supernatants or whole-cell samples.
Degradation of DMSP by dinoflagellate cultures.
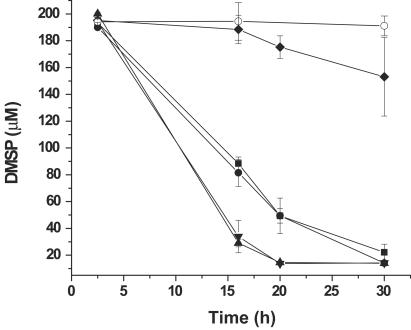
Cultures of P. piscicida, P. shumwayae, and a taxonomically similar dinoflagellate, Cryptoperidiniopsis sp., lacking Rhodomonas, as well as a culture of the prey algae, were analyzed for the ability to degrade DMSP. While Rhodomonas cultures failed to degrade DMSP (data not shown), P. piscicida and Cryptoperidiniopsis cultures degraded exogenously added DMSP within 20 to 30 h of incubation (Fig. 3). In contrast, the P. shumwayae culture was much slower to degrade DMSP, requiring >72 h to achieve complete degradation of the DMSP (data not shown). P. piscicida CCMP1830, Cryptoperidiniopsis sp. strain CCMP1829, and P. shumwayae CCMP2089 were chosen for further analysis.
FIG. 3.
Degradation of DMSP by Pfiesteria and Pfiesteria-like dinoflagellate cultures. DMSP was added to dinoflagellate cultures that contained both the dinoflagellates and their associated bacteria, and the loss of DMSP was measured over time using GC-FID. The results are presented for Cryptoperidiniopsis sp. strain CCMP1829 (•), P. piscicida CCMP1830 (▪), P. piscicida CCMP1921 (▴), P. piscicida CCMP1834 (▾), and P. shumwayae CCMP2089 (♦), and the negative control was medium alone (○). The error bars represent the standard errors in three separate experiments with each culture.
DMSP catabolism by the dinoflagellate-associated bacterial consortium.
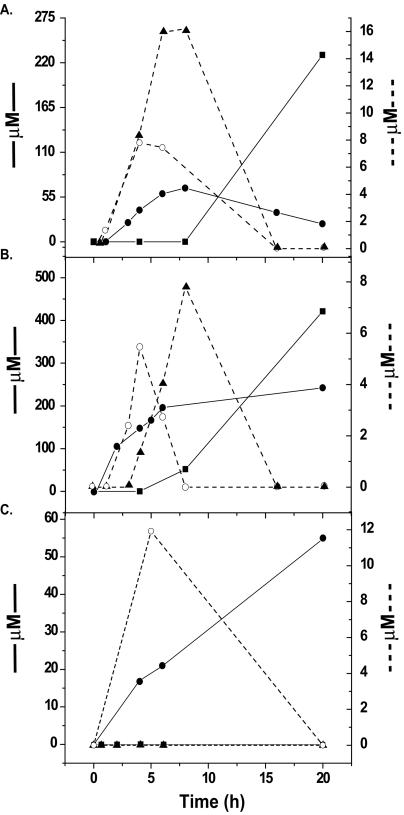
The contribution of the bacterial community in the dinoflagellate cultures to degrading DMSP was assessed. Three separate mixed communities of culturable heterotrophic bacteria from cultures of P. piscicida CCMP1830, Cryptoperidiniopsis sp. strain CCMP1829, and P. shumwayae CCMP2089 were isolated, and their abilities to degrade DMSP were measured. The bacterial suspensions from the P. piscicida and Cryptoperidiniopsis cultures catabolized DMSP, initially producing DMS and acrylate, followed by the production of MMPA and MeSH (Fig. 4A and B). The concentrations of MeSH and DMS were consistently much higher (∼100-fold) than the concentration of either MMPA or acrylate. In general, the concentrations of the DMSP catabolites eventually decreased over 20 h, except for MeSH gas, which continued to increase throughout the period. In contrast to these results, the bacterial suspension obtained from the P. shumwayae culture produced only DMS and acrylate and failed to produce either MMPA or MeSH. These data demonstrate that one or more species of DMSP-degrading bacteria are associated with the P. piscicida and Cryptoperidiniopsis dinoflagellates and that both the demethylase and lyase pathways are active. Since the bacterial communities from both the P. piscicida CCMP1830 and Cryptoperidiniopsis sp. strain CCMP1829 cultures showed similar profiles of DMSP catabolism, only bacteria from the P. piscicida CCMP1830 culture were selected for further analysis.
FIG. 4.
Degradation of DMSP by mixed communities of culturable heterotrophic bacteria obtained from either Pfiesteria or Pfiesteria-like dinoflagellate cultures. Production of the DMSP catabolites, acrylate, DMS, MeSH, and MMPA was measured after 1 mM DMSP was added to the mixed bacterial suspensions. Shown are the mixed bacterial communities obtained from P. piscicida CCMP1830 (A), Cryptoperidiniopsis sp. strain CCMP1829 (B), and P. shumwayae CCMP2089 (C). The production of DMSP catabolites, MeSH (▪) and DMS (•), was measured using GC-FID (left axis), while the production of MMPA (▴) and acrylate (○) was measured using high-performance liquid chromatography (right axis). These experiments were repeated three or more times with similar results.
DMSP catabolism by isolated dinoflagellate-associated bacteria.
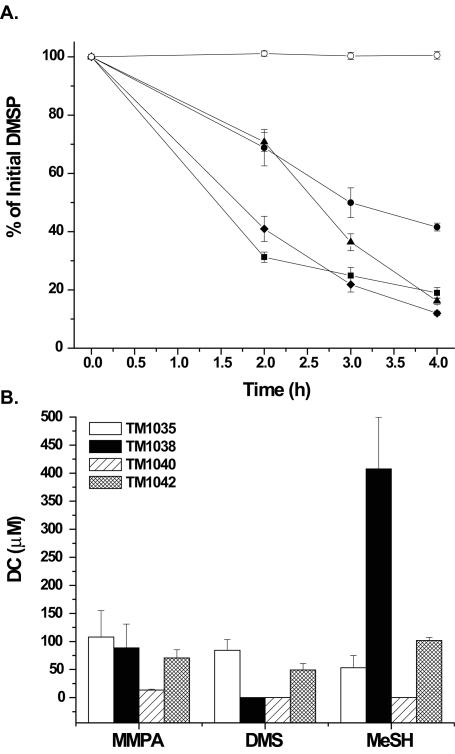
Using inocula from the P. piscicida CCMP1830 culture, nine bacterial strains were isolated, representing a range of colony phenotypes. An analysis of the rate of DMSP degradation showed that four of the nine strains degraded DMSP within a 4-h period (Fig. 5A) while the other five showed little or no ability to degrade DMSP (data not shown). Strain TM1040 appeared to degrade DMSP fastest, followed by strains TM1035 and TM1042, with strain TM1038 being the slowest DMSP degrader. Three distinct colony morphologies were apparent in the four DMSP-degrading strains. Strains TM1035 and TM1042 were similar and produced small (1- to 1.5-mm-diameter), translucent, smooth colonies with a light-pink pigment. Strain TM1038 also produced light-pink colonies with a translucent appearance, but they were much smaller (0.2- to 0.7-mm diameter). TM1040, on the other hand, gave rise to colonies that were larger (4- to 6-mm diameter), translucent, and smooth with a brownish-yellow pigment that diffused throughout the agar medium.
FIG. 5.
The degradation of DMSP by four pure-culture isolates of bacteria obtained from cultures of P. piscicida CCMP1830. (A) The data are presented as the percentages of 100 μM DMSP remaining after incubation with either bacterial strain TM1035 (▪), TM1038 (•), TM1040 (⧫), or TM1042 (▴), or the control (○), 100 μM DMSP without inoculum. (B) Comparison of the production of the DMSP catabolites (DC), MMPA, DMS, and MeSH, by the four bacterial strains after 3 h of incubation in the presence of 1 mM DMSP. The error bars represent the average standard error from three separate experiments.
The production of DMSP catabolites by each of the four bacterial isolates was assessed 3 h after addition of exogenous DMSP (Fig. 5B). All four strains produced the primary demethylation product of DMSP, MMPA, while three strains (TM1035, TM1038, and TM1042) also produced the secondary demethiolation product, MeSH. Among the three MeSH-producing strains, TM1038 produced significantly more MeSH (Fig. 5B), while TM1035 and TM1042, but not TM1038, produced DMS from DMSP in addition to MMPA. These data indicate that TM1035 and TM1042 possess both the DMSP lyase and demethylation pathways.
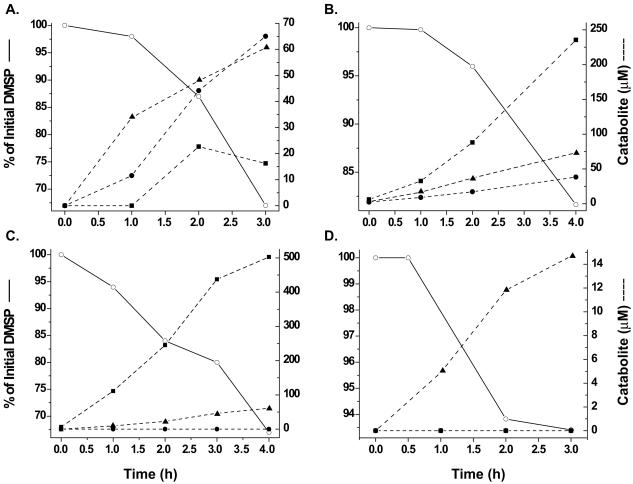
The kinetics of DMSP degradation and catabolite production were examined in greater detail (Fig. 6). Strains TM1035 and TM1042 have similar colony morphologies (medium size; pink), suggesting that they may be taxonomically closely related. The two strains also produce some of the same DMSP catabolites; however, they differ in both the rate of DMSP degradation and the rate of production of DMS (Fig. 6A and B). Strain TM1035 removed ∼33% (330 μM) of the added DMSP during a 3-h period and produced ∼65 μM DMS, 60 μM MMPA, and 16 μM MeSH as a result (Fig. 6A). Production of DMS and MMPA occurred within 30 min, with MeSH production following 1 h later. On the other hand, strain TM1042 removed 19% (190 μM) of the added DMSP during the same 3-h period, producing 236 μM MeSH, 38 μM DMS, and 73 μM MMPA (Fig. 6B). Production of these compounds occurred simultaneously and within 30 min. The first-order rate constants for DMSP degradation and catabolite production were calculated from linear regions of Fig. 6 and are presented in Table 2. TM1035 and TM1042 produced MMPA at similar rates (19.5 and 17.5 μM per h, respectively). MeSH production by strain TM1035 was highly variable across replicate experiments. Therefore, it was not possible to compare the rates of MeSH production in these two strains, although TM1042 always produced higher levels of MeSH (Fig. 5B). TM1042 also had a lower rate of DMSP degradation (32.6 μM per h) than TM1035 (95 μM per h) and a higher rate of DMS production (59.0 μM per h) than TM1035 (22.7 μM per h). These data suggest that, while they produce similar colony phenotypes, TM1035 and TM1042 are physiologically unique, at least in their metabolism of DMSP.
FIG. 6.
Kinetics of DMSP metabolism in four DMSP-degrading bacterial isolates from cultures of P. piscicida 1830. The symbols represent the amounts of DMSP (○), MMPA (▴), MeSH (▪), and DMS (•) measured in the cultures of TM1035 (A), TM1042 (B), TM1038 (C), and TM1042 (D). The graphs represent an average data set whose mean was reproducible over several repeated experiments.
TABLE 2.
Rates of DMSP metabolism of four bacterial strains isolated from P. piscicida CCMP1830 culture
| Strain | Rate of production (μM/h)a
|
Rate of DMSP degradation (μM/h)a | ||
|---|---|---|---|---|
| MMPA | MeSH | DMS | ||
| TM1035 | 19.5 | NAb | 22.7 | 95 |
| TM1038 | 15.9 | 132 | NPc | 22.3 |
| TM1040 | 5.1 | NP | NP | 91 |
| TM1042 | 17.9 | 9.1 | 59 | 32.6 |
Rates of DMSP degradation and production of DMSP catabolites calculated based upon the slope of the linear portion of each curve from Fig. 6.
NA, r < 0.90.
NP, catabolite not detected or produced by this strain.
Strain TM1038 produced both the demethylation and demethiolation products MMPA and MeSH, respectively, but no products of the lyase pathway. The strain removed ∼33% (330 μM) DMSP during a 4-h period, producing 502 μM MeSH and 61 μM MMPA (Fig. 6C). Of the four strains, TM1038 was slowest in degrading DMSP (22.3 μM per h) but had the highest rate of MeSH production (132 μM per h), which was at least an order of magnitude above that of TM1042 (Table 2). The rate of production of MMPA by TM1038 (15.9 μM per h) was quite similar to those of TM1035 (19.5 μM per h) and TM1042 (17.5 μM per h).
The fourth strain, TM1040, catabolized DMSP to produce only MMPA. The cells removed ∼7% (70 μM) DMSP in a 3-h period while producing only 15 μM MMPA (Fig. 6D). Thus, strain TM1040 has a high rate of DMSP degradation (91 μM per h), but a very low rate of MMPA production (5.1 μM/h), compared to the other three DMSP-degrading strains (Table 2).
Taxonomic identification of DMSP-degrading bacteria.
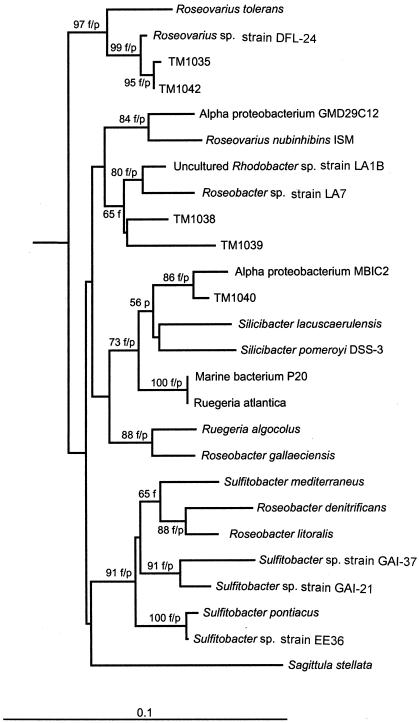
A taxonomic analysis of the DMSP-metabolizing strains placed the four isolates in the α-Proteobacteria, closely related to the Roseobacter clade (Fig. 7). Strains TM1042 and TM1035 are closely related to each other (99% identity; 1,306 of 1,310 bp) and cluster with another dinoflagellate-associated bacterium, strain ATAM407_61, isolated from the dinoflagellate Alexandrium lustanicum (19). Both TM1035 and TM1042 are also more distantly related to R. tolerans from Ekho Lake (96% [1,246 of 1,294 bp] and 96% [1,248 of 1,294 bp] identity, respectively), which, like strains TM1035 and TM1042, is capable of producing both DMS and MeSH from DMSP (14).
FIG. 7.
Taxonomic analysis of the four DMSP-degrading bacteria isolated from P. piscicida. The phylogenetic tree was inferred from comparative sequence analysis, using 1,047 bp of 16S rDNA, generated by the neighbor-joining method and the Jukes-Cantor distance algorithm. The resulting tree shows the relationships among the Roseobacter clade bacteria associated with P. piscicida CCMP1830; strains TM1035, TM1038, TM1040, and TM1042; and nucleotide sequences of other known Roseobacter clade bacteria. Bootstrap values (n = 1,000 replicate resamplings) are indicated for the neighbor-joining method where values are >500. An ′f' or ′p' indicates that the Fitch or parsimony method (respectively) is in agreement with the neighbor-joining tree. Bar = 0.1 units of evolutionary distance.
The sequences of TM1038 and TM1040 16S rDNAs show the greatest similarity to 16S rDNA sequences obtained from bacteria within the Roseobacter clade, yet they did not cluster well with any cultured organisms within this clade and are unrelated to any of the well-characterized Roseobacter species listed in GenBank. As shown in Fig. 7, the 16S rDNA from TM1038 was 95% (1,230 of 1,282 bp) homologous to rDNA obtained from an uncharacterized bacterium isolated from marine snow, HP29w (17). Similarly, strain TM1040 grouped with an uncharacterized α-proteobacterium, MBIC1887 (99% sequence identity; 1,275 of 1,284 bp), and to a lesser extent showed some relatedness to two well-characterized Silicibacter species, S. pomeroyi and S. lacuscaerulensis (Fig. 7). Both of these Silicibacter species are capable of DMSP degradation (14).
DISCUSSION
In this study, Pfiesteria dinoflagellates and members of the bacterial community cooccurring with them were used as a model system to study DMSP production and degradation as it may influence prokaryote-eukaryote interactions. Pfiesteria dinoflagellates are mainly heterotrophic organisms that acquire nutrients through the consumption of algal prey and at certain times may utilize prey chloroplasts for energy. Not only have Pfiesteria species been implicated in causing human illness and fish mortality, these dinoflagellates are dominant species in nutrient-rich estuaries and are thus important to the greater understanding of nutrient cycling and microbial interactions in coastal marine habitats (31).
The results show that both P. piscicida and P. shumwayae contain significant levels of DMSP, which to our knowledge is the first report of DMSP in Pfiesteria and only the second observation of DMSP in a heterotrophic dinoflagellate (28). The DMSP contents of the two Pfiesteria species (3.44 to 4.25 μM) (Table 1) are similar to those measured in other dinoflagellates. For example, the intracellular DMSP concentrations in photosynthetic species, such as Prorocentrum, Gymnodinium, and Amphidinium species, are reported to be 1 to 10 μM (24). Interestingly, the concentration of DMSP in another heterotrophic dinoflagellate, Crypthecodinium cohnii, has been reported to be 10 pg per cell (24), a value that is much higher than those observed in either Pfiesteria species (∼0.4 pg per cell). This difference undoubtedly reflects physiological and taxonomic differences between Pfiesteria and Crypthecodinium and underscores the difficulty in making general statements about DMSP physiology among taxonomically diverse dinoflagellate species.
Both P. piscicida and P. shumwayae contain significant levels of DMSP, yet the P. piscicida cultures that include dinoflagellates plus associated bacteria degrade DMSP at significantly higher rates (Fig. 3). Previous data have demonstrated differences in the bacterial flora associated with Pfiesteria cultures (1). Thus, it is conceivable that differences in the compositions of the bacterial communities may affect the rates of DMSP decomposition in P. piscicida and P. shumwayae cultures. The data in Fig. 4 showing the difference between DMSP catabolite kinetics in the mixed culturable heterotrophic bacteria support this idea. Other factors may also be important, including the possibility that slow-growing bacteria in the P. shumwayae culture may have a low rate of DMSP degradation or that the concentration of DMSP used may have a deleterious effect specific to the P. shumwayae bacterial community.
Four DMSP-degrading bacterial isolates were obtained from P. piscicida cultures and were found to be phylogenetically related to members of the Roseobacter clade (Fig. 7). As shown in the taxonomic tree, two of the isolates (TM1035 and TM1042) are most closely related to Roseovarius species, while TM1038 and TM1040 are unique Roseobacter species not related to known roseobacters. Despite their taxonomic differences, all four bacteria shared the common trait of demethylating DMSP to MMPA, while strains TM1035 and TM1042 further metabolize DMSP to produce DMS, indicating that demethylation is a major pathway by which Pfiesteria-associated roseobacters degrade DMSP. Equally interesting are the DMSP demethylation pathways used by these strains. As an example, TM1038 demethylates DMSP to produce MMPA and MeSH, a pathway that appears to be commonly used by marine bacteria (25). In contrast, TM1040 strictly demethylates DMSP to produce MMPA without MeSH, a pathway reported to be used by one other aerobic marine bacterium, strain BIS-6, isolated from Biscayne Bay, Fla. (41). This bacterium demethylates DMSP to MMPA (Fig. 1, reaction 2). followed by a further demethylation to MPA (Fig. 1, reaction 4). Although not measured in this study, TM1040 is likely also to produce MPA instead of MeSH, as has been observed for BIS-6.
The two Roseovarius-related strains, TM1035 and TM1042, are capable of both demethylation and lyase cleavage of DMSP. The presence of dual demethylation-lyase pathways in the same organism is a recently discovered phenomenon. Gonzalez et al. (15) reported that 5 out of 15 DMSP-catabolizing bacteria isolated from Georgia coastal seawater and the Caribbean Sea catabolized DMSP to produce both DMS and MeSH, as well as converted MMPA to MeSH. One of these five isolates was taxonomically identified as R. nubinhibens ISM (15). The capacity to use both DMSP pathways may provide these bacteria with a survival advantage, especially in environments where DMSP concentrations are high, such as the phycosphere surrounding DMSP-producing dinoflagellates. Bacteria that utilize the lyase cleavage pathway are capable of growing on DMSP as a sole carbon source (45). In contrast, while the demethylation pathway does not always lead to increased growth (20), much of the sulfur obtained from this pathway is utilized for protein synthesis and seems to be preferred over other sources of sulfur abundant in seawater (27). Thus, coupling of both DMSP-degradative pathways in the same organism may satisfy both the carbon and sulfur requirements of these dinoflagellate-associated marine bacteria.
In analyses of DMSP catabolism, it was occasionally observed that the sum of the DMSP catabolites produced did not always equal the amount of DMSP lost from the culture. There are several possible explanations for this. First, bacterial enzymes may have degraded the DMSP catabolites shortly after they were produced. This is a strong possibility in light of the high reaction rates observed. A good example supporting this is the failure to detect acrylate production in either TM1035 or TM1042, despite the presence of detectable levels of DMS, which constitutes the other half of lyase cleavage of DMSP. The absence of acrylate is most likely due to rapid conversion, a finding that was also noted by Ansede et al. (2), who, using nuclear magnetic resonance analysis, were unable to detect acrylate production by a Roseobacter species even though DMS was produced. The present results also agree with environmental studies that show rapid degradation of acrylate by bacterial communities associated with algal cells or debris (34).
Another possibility to explain the imbalance in DMSP catabolites is that these chemicals may have been degraded or lost due to abiotic factors, such as oxidation of a compound or the adherence of a catabolite to inanimate surfaces. For example, MeSH is readily oxidized to form dimethyl disulfide and may be lost from water samples due to binding with humic acids (25). In our experiments, a minor loss of MeSH due to sticking to inanimate surfaces was observed (data not shown) and may have resulted in a slight overestimation of total MeSH gas production, which in turn may have contributed to the imbalance between MeSH produced and DMSP degraded.
Pfiesteria and other heterotrophic dinoflagellates are in intimate association with a community of bacteria, many of which are members of the Roseobacter clade, which interact in a myriad of ways with their eukaryotic partner. Roseobacter clade bacteria have been observed attached to or physically associated with dinoflagellate cells, while other Roseobacter species are required for dinoflagellate growth (1). It is tantalizing to think that DMSP is involved in these associations, particularly in view of the diversity of DMSP pathways and the high rates of reactions seen in the Pfiesteria cultures and the Roseobacter isolates obtained from them. These results also bring up new questions about roseobacters, DMSP, and dinoflagellate interactions. Are Roseobacter clade bacteria attracted to DMSP or one of its catabolites, which may bring them into close proximity to dinoflagellate cells? Do the catabolites of DMSP have physiological functions in dinoflagellate metabolism, behavior, or growth? In the long term, answers to these questions will provide significant clues about the molecular and cellular natures of the interactions between bacteria, dinoflagellates, and other single-cell eukaryotes.
Acknowledgments
We thank Russell T. Hill and Kevin Sowers for the use of their analytical equipment and the members of the Belas laboratory for their suggestions and comments regarding the manuscript.
This work was supported by grants from NOAA ECOHAB (NA86OP0492) and NIH NIEHS (PO1-ES9563).
REFERENCES
- 1.Alavi, M., T. Miller, K. Erlandson, R. Schneider, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 2.Ansede, J., P. J. Pellechia, and D. C. Yoch. 1999. Metabolism of acrylate to β-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl. Environ. Microbiol. 65:5075-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansede, J. H., P. J. Pellechia, and D. C. Yoch. 2001. Nuclear magnetic resonance analysis of [1-13C]dimethylsulfoniopropionate (DMSP) and [1-13C]acrylate metabolism by a DMSP lyase-producing marine isolate of the α-subclass of Proteobacteria. Appl. Environ. Microbiol. 67:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M. 2001. Current protocols in molecular biology. J. Wiley, New York, N.Y.
- 5.Berry, J. P., K. S. Reece, K. S. Rein, D. G. Baden, L. W. Haas, W. L. Ribeiro, J. D. Shields, R. V. Snyder, W. K. Volgelbein, and R. E. Gawley. 2002. Are Pfiesteria species toxicogenic? Evidence against production of icthyotoxins by Pfiesteria shumwayae. Proc. Natl. Acad. Sci. USA 99:10970-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, J. M., E. J. Noga, C. H. Hobbs, H. B. Glasgow, Jr., and S. A. Smith. 1992. New ′phantom' dinoflagellate is the causative agent of major estuarine fish kills. Nature 358:407-410. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, S. T., C. M. Kunin, D. Miller, and A. Hamada. 1987. Dimethylthetin can substitute for glycine betaine as an osmoprotectant molecule for Escherichia coli. J. Bacteriol. 169:4845-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dantzer, W. R., and R. E. Levin. 1997. Bacterial influence on the production of paralytic shellfish toxins by dinoflagellate algae. J. Appl. Microbiol. 83:464-469. [DOI] [PubMed] [Google Scholar]
- 9.Dickson, D. M. J., and G. O. Kirst. 1986. The role of β-dimethylsulfoniopropionate, glycine betaine and homarine in the osmoacclimation of Platymonas subcordiformes. Planta 167:536-543. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. Phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 11.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 12.Gallacher, S., K. J. Flynn, J. M. Franco, E. E. Brueggemann, and H. B. Hines. 1997. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp. (Dinophyta) in culture. Appl. Environ. Microbiol. 63:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasgow, H. B., Jr., J. M. Burkholder, D. E. Schmechel, P. A. Tester, and P. A. Rublee. 1995. Insidious effects of a toxic estuarine dinoflagellate on fish survival and human health. J. Toxicol. Environ. Health 46:501-522. [DOI] [PubMed] [Google Scholar]
- 14.González, J. M., J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., DMSP demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gram, L., H. P. Grossart, A. Schlingloff, and T. Kiorboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates. Plenum Press, New York, N.Y.
- 19.Hold, G. L., E. A. Smith, M. S. Rappe, E. W. Maas, E. R. B. Moore, C. Stroempl, J. R. Stephen, J. I. Prosser, T. H. Birkbeck, and S. Gallacher. 2001. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 37:161-173. [Google Scholar]
- 20.Jansen, M., and T. A. Hansen. 2001. Non-growth-associated demethylation of dimethylsulfoniopropionate by (homo)acetogenic bacteria. Appl. Environ. Microbiol. 67:300-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen, M., and T. A. Hansen. 1998. Tetrahydrofolate serves as a methyl acceptor in the demethylation of dimethylsulfoniopropionate in cell extracts of sulfate-reducing bacteria. Arch. Microbiol. 169:84-87. [DOI] [PubMed] [Google Scholar]
- 22.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 23.Karsten, U., K. Kuck, C. Vogt, and G. O. Kirst. 1996. Dimethylsulfoniopropionate production in phototrophic organisms and its physiological function as a cryoprotectant, p. 143-153. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, N.Y.
- 24.Keller, M. D., and W. Korjeff-Bellows. 1996. Physiological aspects of the production of dimethylsulfoniopropionate (DMSP) by marine phytoplankton, p. 131-153. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, N.Y.
- 25.Kiene, R. P. 1996. Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54:69-83. [Google Scholar]
- 26.Kiene, R. P., and L. J. Linn. 2000. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45:849-861. [Google Scholar]
- 27.Kiene, R. P., L. J. Linn, J. Gonzalez, M. A. Moran, and J. A. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitaguchi, H., A. Uchida, and Y. Ishida. 1999. Purification and characterization of l-methionine decarboxylase from Crypthecodinium cohnii. Fisheries Sci. 65:613-617. [Google Scholar]
- 29.Kluge, A. G., and F. S. Farris. 1969. Quantitative phyletics and the evolution of annurans. Syst. Zool. 18:1-32. [Google Scholar]
- 30.Lafay, B., R. Ruimy, C. R. de Traubenberg, V. Breittmayer, M. J. Gauthier, and R. Christen. 1995. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 31.Miller, T. R., and R. Belas. 2003. Pfiesteria piscicida, P. shumwayae, and other Pfiesteria-like dinoflagellates. Res. Microbiol. 154:85-90. [DOI] [PubMed] [Google Scholar]
- 32.Moeller, P. D., S. L. Morton, B. A. Mitchell, S. K. Sivertsen, E. R. Fairey, T. M. Mikulski, H. Glasgow, N. J. Deamer-Melia, J. M. Burkholder, and J. S. Ramsdell. 2001. Current progress in isolation and characterization of toxins isolated from Pfiesteria piscicida. Environ. Health Perspect. 109:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niki, T., M. Kunugi, and A. Otsuki. 2000. DMSP-lyase activity in five marine phytoplankton species: its potential importance in DMS production. Mar. Biol. 136:759-764. [Google Scholar]
- 34.Osinga, R., K. de Vries, W. Lewis, W. van Raaphorst, L. Dijkhuizen, and F. van Duyl. 1997. Aerobic degradation of phytoplankton debris dominated by Phaeocystis sp. in different physiological stages of growth. Aquat. Microb. Ecol. 12:11-19. [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sunda, W., D. J. Kieber, R. P. Kiene, and S. Huntsman. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317-320. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, B. F., and D. Gilchrist. 1991. New routes for aerobic biodegradation of dimethylsulfoniopropionate. Appl. Environ. Microbiol. 57:3581-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, B. F., and P. T. Visscher. 1996. Metabolic pathways involved in DMSP degradation, p. 265-276. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, N.Y.
- 40.Vairavamurthy, A., M. O. Andreae, and R. L. Iverson. 1985. Biosynthesis of dimethylsulfide and dimethylpropiothetin by Hymenemonas carterae in relation to sulfur source. Limnol. Oceanogr. 30:59-70. [Google Scholar]
- 41.Visscher, P. T., and B. F. Taylor. 1994. Demethylation of dimethylsulfoniopropionate to 3-mercaptopropionate by an aerobic marine bacterium. Appl. Environ. Microbiol. 60:4617-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visscher, P. T., B. F. Taylor, and R. P. Kiene. 1995. Microbial consumption of dimethyl sulfide and methanethiol in marine coastal sediments. FEMS Microbiol. Ecol. 18:145-154. [Google Scholar]
- 43.Vogelbein, W. K., V. J. Lovko, J. D. Shields, K. S. Reece, P. L. Mason, L. W. Haas, and C. C. Walker. 2002. Pfiesteria shumwayae kills fish by micropredation not exotoxin secretion. Nature 418:967-970. [DOI] [PubMed] [Google Scholar]
- 44.Wolfe, G., M. Steinke, and G. Kirst. 1997. Grazing-activated chemical defence in a unicellular marine alga. Nature 387:894-897. [Google Scholar]
- 45.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoch, D. C., J. H. Ansede, and K. S. Rabinowitz. 1997. Evidence for intracellular and extracellular dimethylsulfoniopropionate (DMSP) lyases and DMSP uptake sites in two species of marine bacteria. Appl. Environ. Microbiol. 63:3182-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microb. 3:304-311. [DOI] [PubMed] [Google Scholar]