Abstract
Cancer is a leading cause of death worldwide. Cancer cells proliferate uncontrollably and, many cases, spread to other parts of the body. A protein historically involved in cancer is protein kinase CK2. CK2 is a serine-threonine kinase that has been involved in cell growth, cell proliferation and cell apoptosis. CK2 functions as an oncogene when overexpressed in mouse tissues, and can synergize with known oncogenes, such as ras, to induce cell transformation in cells in culture. CK2, typically the CK2α protein, is found elevated in a number of human tumors. However, we have little information on CK2α' and CK2β proteins, and scarce information on CK2 gene transcript expression. Here, we explore the expression of CK2 transcripts in primary tumor tissues using the database Oncomine in the six cancers with the highest mortality in the U.S.A. In addition, we studied the correlation between CK2 expression and overall survival using the Kaplan-Meier Plotter database in breast, ovarian, and lung cancers. We found widespread upregulation in the expression of CK2 genes in primary tumor tissues. However, we found underexpression of CK2α' transcripts in some tumors, increased CK2β transcripts in some invasive tumors, and deregulation of CK2 transcripts in some tumor precursors. There was also correlation between CK2 expression levels and patient survival. These data provides additional evidence for CK2 as a biomarker for cancer studies and as a target for cancer therapy.
Introduction
Cancer prevalence
Cancer remains a leading cause of morbidity and mortality worldwide, and it is predicted to be of more importance in the next few decades. According to the World Health Organization, if global cancer rates remain unchanged, by 2030, new cancer cases will reach 21.4 million—a rise from 12.7 million new cancer cases in 2008 [1]. According to the United States Cancer Statistics, the top ten cancers by incidence rate (Age-Adjusted Invasive Cancer Incidence Rates for the 10 Primary Sites with the Highest Rates) from 2005–2011 are cancers of the prostate, breast, lung and bronchus, colorectal, corpus and uterus, urinary bladder, melanomas of the skin, non-Hodgkin lymphomas, cancers of the kidney and renal pelvis, thyroid [2]. Among them, the cancers with the highest death rates in the USA from 2005–2010 include lung and bronchus, prostate, breast, colon and rectum, ovarian and pancreatic cancers [2]. These cancers were the focus of our analysis.
General properties and features of CK2
Protein kinase CK2 is a highly conserved serine/threonine kinase linked to diseases such as cancer [3], cardiac hypertrophy [4], multiple sclerosis [5], and inflammation [6]. Furthermore, it has been shown to be critical in embryonic development [7], and circadian rhythms [8].
CK2 regulates essential cellular processes that are characteristic to cancer development. A number of excellent publications review the role of CK2 in cancer. This include, the regulation of cell growth [9], cell proliferation [10], [11], cell survival [12], [13], cell morphology [14], cell transformation [3], signaling pathway activation [15], and angiogenesis [16], [17].
CK2 is encoded in mammals by two separate genes (CK2α and CK2α'); CK2α has broader tissue expression and higher-level expression compared to the CK2α' [18]. CK2 proteins can function as monomeric kinases or within a tetrameric complex of two CK2 kinase units and two units of the regulatory protein CK2β. In this tetrameric form, CK2β confers different substrate specificity to CK2α [19].
CK2α is an oncogene in mouse transgenic models, as CK2α overexpression in mammary gland and lymphoid compartment leads to tumor formation [20], [21]. In accordance, CK2 is overexpressed in proliferating cells [22]. In addition, there is a link between CK2 expression and environmental toxins. In animal models, CK2 is overexpressed in response to aromatic hydrocarbons [23], and is upregulated in chemically-induced liver carcinogenesis [24].
A limited number of studies testing CK2 protein and activity in human tumors found that CK2 activity and protein expression is upregulated in human cancers [25], [26] (reviewed in [22]). Although we have some knowledge on the expression of CK2α protein, we have little information on the expression of CK2α' or CK2β. Compared to protein expression levels, only a few studies test the expression levels of CK2 gene transcripts [25]. In fact, it is thought that CK2 protein and not CK2 gene transcript are elevated in cancer [22], [27]. In addition, only recently it has been shown that there is deregulated expression of the CK2α pseudogene [28], a non-protein coding CK2α sequence [29]. CK2 can serve as a prognostic marker [12], and can strengthen the multi-drug-resistant phenotype in adult cancer tissues [30], indicating the importance of studying the expression levels of CK2 genes.
Therefore, we extracted data on transcript expression for CK2α, CK2α pseudogene (CK2αP), CK2α' and CK2β from the database Oncomine for lung and bronchus, prostate, breast, colon and rectum, ovarian and pancreatic cancers, focusing on clinical specimens of cancer vs. normal patient datasets, and separated by subtype when possible (Fig. 1). We also tested for the effect of deregulation of CK2 expression on patient overall survival using Kaplan-Meier Plotter. Here we present a summary of the results we obtained. The tables associated with the data include statistical significance, fold change, patient number and the dataset from which the information was obtained.
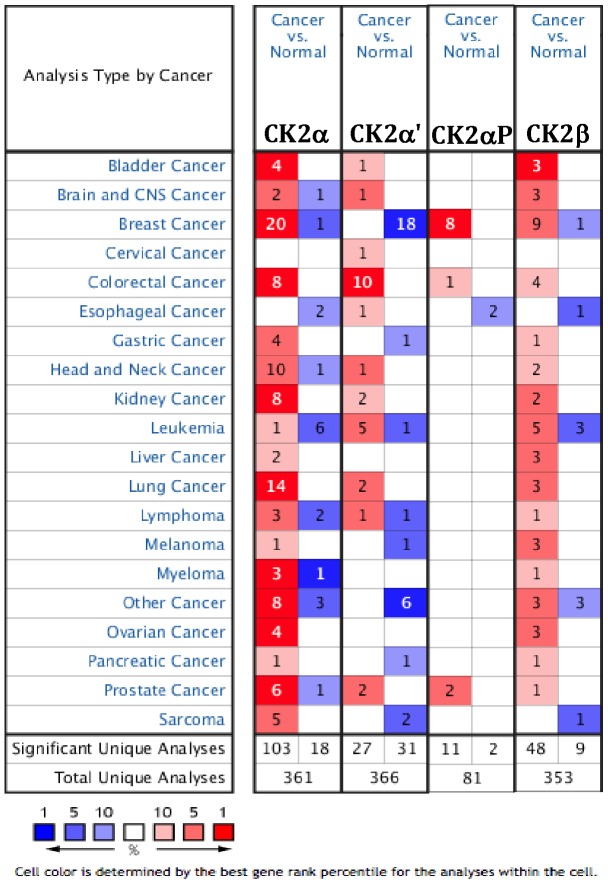
Figure 1. CK2 mRNA Expression in different tumor types.
This graphic compares the number of datasets that had significant mRNA overexpression (left column, red) and underexpression (right column, blue) of the specified gene in cancer versus normal tissue. The datasets were obtained with the following parameters: p-value threshold of 0.01.
Methods
Oncomine analysis
The expression level of CK2 genes in the selected cancers was analyzed using Oncomine [31]. For this, we compared clinical specimens of cancer vs. normal patient datasets. In order to reduce our false discovery rate, we selected p<0.01 as a threshold. We analyzed the results for their p-values, fold change, and cancer subtype. In many instances we found several significant correlations in different tumor types, but we showed only one to three of them.
Kaplan-Meier plotter analysis
The prognostic value of the CK2 genes in ovarian, breast and lung cancer was analyzed using Kaplan-Meier Plotter (http://kmplot.com/analysis/), a database that integrates gene expression data and clinical data [32]. To date, Kaplan-Meier Plotter contains information on 22,277 genes and their effect on survival in 2,977 breast, 1,464 ovarian and 1,715 lung cancer patients. We focused our analysis on overall survival patient information. The patient samples have been split into two groups. The two patient groups (higher and lower expression levels) were compared using a Kaplan-Meier survival plot. The hazard ratio with 95% confidence intervals and log rank p value was calculated. We analyzed the best specific probes (JetSet probes) that recognized CK2α, CK2αP, CK2α', and CK2β. In order to reduce our false discovery rate, we selected p<0.01 as a threshold.
Results and Discussion: CK2 Transcript Expression by Cancer Type
Lung cancer
Lung cancer has the highest mortality of all cancers in both men and women. The NCI predicts that in 2014, approximately 27% of all cancer deaths would be due to lung cancer. The NCI (http://www.cancer.gov/) [33] predicts that there would be over 224,000 new cases of lung cancer (about 14% of cancer diagnoses) in 2014, albeit a decrease in lung cancer incidence over the past few years (particularly in men). Lung cancers are divided into three groups. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer (85% of lung cancers) and includes subtypes: squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (LCLC). Small Cell Lung Cancer (SCLC) (10%–15% of lung cancers) quickly divides and metastasizes. Lung Carcinoid Tumors (5% of lung cancers), a type of neuroendocrine tumor, grow slowly and rarely spread. Advances in surgical and combined therapeutical approaches have increased the one-year survival by 7% (to 44% by 2005–08); however, the five-year survival rate for the combination of all stages is still low (16%); with small cell lung cancer having the lowest five-year survival rate (6%) compared to non-small cell lung cancer (18%).
CK2 in lung and bronchus cancer
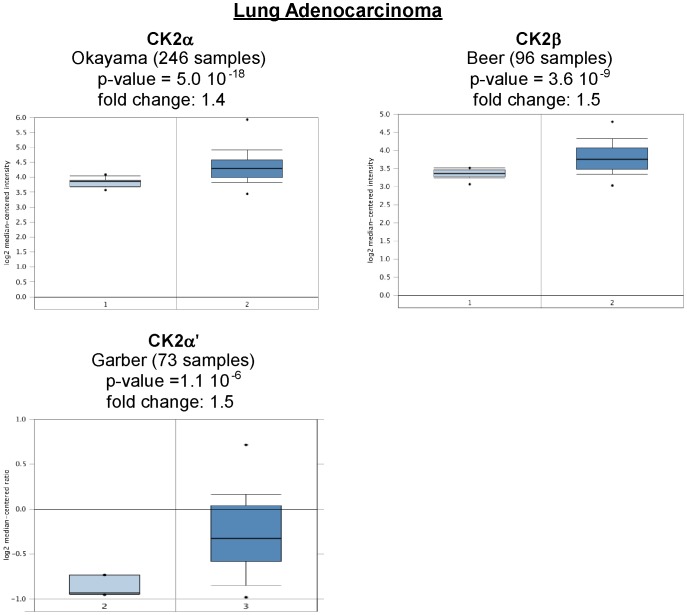
Oncomine analysis of neoplastic vs. normal tissue showed that CK2α, CK2α', CK2β and CK2αP were significantly overexpressed in different types of lung cancer in different datasets (Table 1, Fig. 2). Overexpression of CK2α and CK2α' was found in squamous cell carcinoma and adenocarcinoma. In contrast, LCLC and SCLC had CK2α overexpression but not CK2α'. CK2β was upregulated in adenocarcinoma and LCLC but was downregulated in lung carcinoid. These data are in line with published articles on CK2 expression. For example, CK2α (CSNK2A1) transcript, protein and/or activity are elevated in lung cancer neoplastic tissues such as those found in squamous cell carcinomas of the lung [34], [35], [36], and in NSCLC cell lines [28], [34]. In addition, the CK2α pseudogene (CSNK2A1P) is amplified in non-small cell lung cancer cell lines and lung cancer tissues [28].
Table 1. Changes in CK2 gene expression in lung cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Squamous Cell Carcinoma | ||||||
| CK2α | 1.5 10−7 | 18.9 | 1% | Bhattacharjee | 203 | [58] |
| 2.3 10−13 | 2.2 | 2% | Hou | 156 | [59] | |
| CK2α' | 5.2 10−6 | 1.8 | 2% | Garber | 73 | [60] |
| 0.01 | 2.4 | 10% | Bhattarcharjee | 203 | [58] | |
| 9.9 10−4 | 1.5 | 4% | Yamagata | 31 | [61] | |
| CK2αP | 0.01 | 2.1 | 15% | Garber | 73 | [60] |
| Adenocarcinoma | ||||||
| CK2α | 0.001 | 3.3 | 3% | Bhattarcharjee | 203 | [58] |
| 5.0 10−18 | 1.4 | 1% | Okayama | 246 | [62] | |
| 3.1 10−13 | 1.6 | 2% | Hou | 156 | [59] | |
| CK2α' | 1.1 10−6 | 1.5 | 1% | Garber | 73 | [60] |
| CK2β | 3.6 10−9 | 1.5 | 2% | Beer | 96 | [63] |
| 8.5 10−6 | 1.6 | 13% | Talbot | 93 | [64] | |
| LCLC | ||||||
| CK2α | 2.4 10−9 | 2.5 | 1% | Hou | 156 | [59] |
| CK2β | 8.2 10−5 | 1.5 | 11% | Hou | 96 | [59] |
| SCLC | ||||||
| CK2α | 4.3 10−4 | 6 | 3% | Bhattarcharjee | 203 | [58] |
| Lung Carcinoid | ||||||
| CK2α | 1.2 10−5 | 6.6 | 3% | Bhattarcharjee | 203 | [58] |
| CK2β | 0.002 | −2.6 | 39% | Bhattarcharjee | 203 | [58] |
Different subtypes of lung cancer are analyzed and p-values, fold changes, and datasets are included.
Figure 2. CK2 gene analysis in lung cancer (Oncomine database).
Box plots derived from gene expression data in Oncomine comparing expression of a specific CK2 gene in normal (left plot) and lung cancer tissue (right plot). Only lung adenocarcinoma is shown.
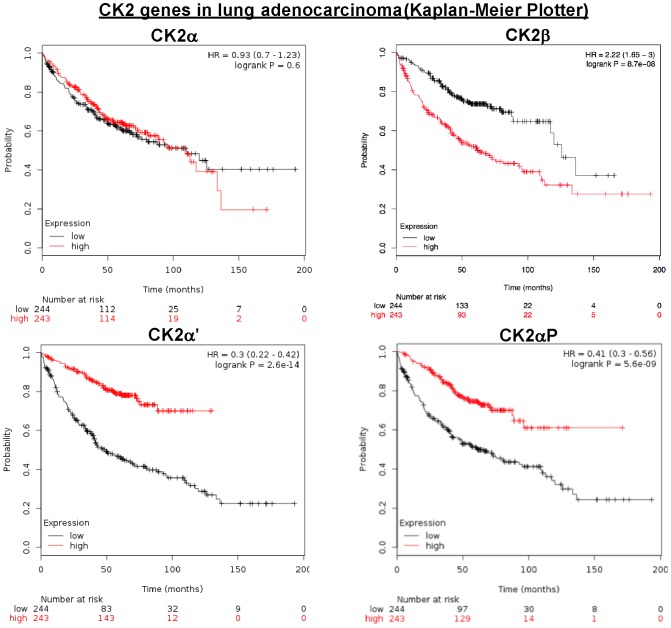
Kaplan-Meier plotter analysis in overall lung cancer showed correlation between overexpression of CK2β and overall lower survival rates, and the opposite for CK2α' and CK2αP, and no significance for CK2α expression (Fig. 3). When we restricted the analysis by tumor type, no significant differences in overall survival were observed for the expression of CK2 genes in squamous cell carcinoma samples. However, in adenocarcinoma there were significant differences for CK2αP, CK2α', and CK2β mirroring the data on overall lung cancer. This indicates that the usage of CK2 gene expression as a predictor of outcome may be cancer subtype dependent (Table 1). Even though our Kaplan-Meier analysis of squamous cell carcinoma showed no significance for CK2α (p = 0.012), a published study identified CK2α (CSNK2A1) overexpression as one of two independent predictors of outcome (prognostic markers) in 21 patients with squamous cell carcinoma of the lung [34]. In particular, this was one of 11 genes specific of the poor prognosis cluster group (cluster II) of non-small cell lung cancers that had corresponding chromosomal aberrations. The authors validated these results independently in an additional set of 45 patients.
Figure 3. CK2 genes in lung adenocarcinoma (Kaplan-Meier Plotter).
Kaplan-Meier plots showing overall survival in lung adenocarcinoma. In red: patients with expression above the median and in black, patients with expressions below the median. CK2α, p = 0.6008; CK2αP, p = 5.6 10−9, CK2α', p = 2.6 10−14; CK2β, p = 8.7 10−8.
Breast cancer
Excluding cancers of the skin, breast cancer is the most frequently diagnosed cancer in women. According to the NCI, an estimated 40,000 breast cancer deaths in both women and men are expected in 2014 (6.8% of all cancers) [33]. Breast cancer ranks second as a cause of cancer death in women (after lung cancer). An estimated 232,670 new cases of breast cancer are expected to be diagnosed in women in the US during 2014 (14% of all cancers). Breast cancer can happen in the ducts, the lobules and in the stroma. The most common breast cancers are ductal carcinoma (70% of cases) and lobular carcinoma (10% of cases). These can be subdivided into non-invasive (in situ) and invasive. Mucinous breast carcinoma is a subtype of ductal carcinoma (2–5% incidence) Breast cancer treatment can involve surgery, radiation therapy, chemotherapy, hormone therapy and/or targeted therapy. The overall 5-year survival rate is 89.2% in localized or low-grade breast cancer. Early detection and novel treatments have increased the survival rate for localized breast cancer to 98.6% and for regional breast cancer to 84.4%.
CK2 in Breast Cancer
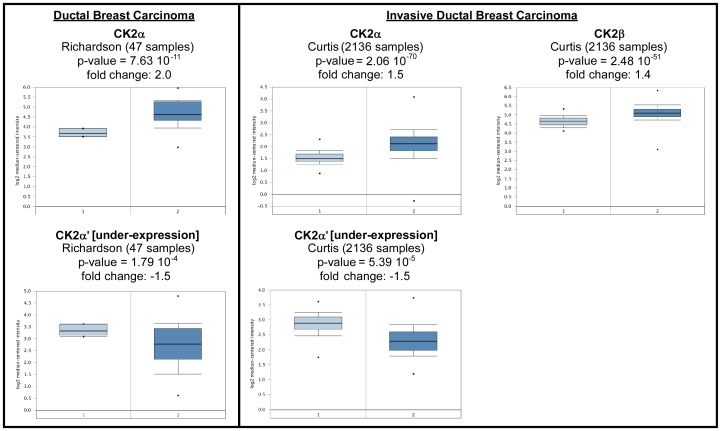
Overall, Oncomine analysis showed significant levels of both CK2α and CK2β overexpression in most breast cancer types, but strong CK2α' underexpression in all breast cancer types (Table 2, Fig. 4). A difference was found between ductal and lobular breast cancers, where ductal, but not lobular, expressed high levels of CK2α. A further difference was found in between non-invasive and invasive breast cancers. Both showed CK2α' underexpression, but in invasive breast cancer, CK2β was also very significantly overexpressed. Since CK2β protein is stabilized post-translationally by CK2α [9], increased CK2β transcript expression in CK2α overexpressing cancers, may contribute to further increase CK2β protein expression.
Table 2. Changes in CK2 gene expression in breast cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Ductal Breast Carcinoma | ||||||
| CK2α | 7.43 10−11 | 2 | 1% | Richardson 2 | 47 | [65] |
| CK2α' | 2.05 10−4 | −1.6 | 3% | Sorlie | 85 | [66] |
| 9.15 10−4 | −1.6 | 6% | Sorlie Breast 2 | 167 | [67] | |
| 1.79 10−4 | −1.5 | 10% | Richardson 2 | 47 | [65] | |
| Invasive Ductal Breast Carcinoma | ||||||
| CK2α | 8.09 10−28 | 1.6 | 4% | TCGA Breast | 593 | [68] |
| 2.06 10−70 | 1.5 | 4% | Curtis | 2136 | [69] | |
| CK2α' | 4.04 10−20 | −1.5 | 10% | TCGA Breast | 593 | [68] |
| 5.39 10−50 | −1.5 | 5% | Curtis | 2136 | [69] | |
| 1.08 10−4 | −1.6 | 4% | Zhao | 64 | [70] | |
| CK2β | 2.48 10−51 | 1.4 | 8% | Curtis | 2136 | [69] |
| Lobular Breast Carcinoma | ||||||
| CK2α' | 6.37 10−4 | −1.5 | 4% | Zhao | 64 | [70] |
| Invasive Lobular Breast Carcinoma | ||||||
| CK2α' | 3.75 10−11 | −1.6 | 5% | TCGA Breast | 593 | [68] |
| 4.51 10−43 | −1.6 | 3% | Curtis | 2136 | [69] | |
| CK2β | 8.00 10−39 | 1.4 | 2% | Curtis | 2136 | [69] |
| Mucinous Breast Carcinoma | ||||||
| CK2α | 4.33 10−5 | 1.5 | 2% | TCGA Breast | 593 | [68] |
| 1.39 10−16 | 1.4 | 3% | Curtis | 2136 | [69] | |
| Invasive Mucinous Breast Carcinoma | ||||||
| CK2β | 2.48 10−51 | 1.4 | 8% | Curtis | 2136 | [69] |
| Breast Carcinoma | ||||||
| CK2α' | 3.09 10−5 | −1.7 | 8% | Curtis | 2136 | [69] |
| Invasive Breast Carcinoma | ||||||
| CK2α | 3.58 10−15 | 1.5 | 6% | TCGA Breast | 593 | [68] |
| Invasive Breast Carcinoma Stroma | ||||||
| CK2α | 1.2 10−28 | −2.2 | 2% | Finak Breast | 59 | [71] |
| CK2α' | 7.45 10−12 | −6.8 | 15% | Finak Breast | 59 | [71] |
| CK2β | 3.02 10−20 | −5.7 | 7% | Finak Breast | 59 | [71] |
| Tubular Breast Carcinoma | ||||||
| CK2α' | 3.22 10−36 | −1.8 | 2% | Curtis | 2136 | [69] |
| Invasive Medullary Carcinoma | ||||||
| CK2αP | 1.2 10−9 | 1.5 | 5% | Curtis | 2136 | [69] |
| CK2β | 1.4 10−10 | 1.6 | 4% | Curtis | 2136 | [69] |
Different subtypes of breast cancer are analyzed and p-values, fold changes, and datasets are included.
Figure 4. CK2 gene analysis in breast cancer (Oncomine database).
Box plots derived from gene expression data in Oncomine comparing expression of a specific CK2 gene in normal (left plot) and lung cancer tissue (right plot). Invasive and non-invasive ductal breast carcinomas are featured in the box plots.
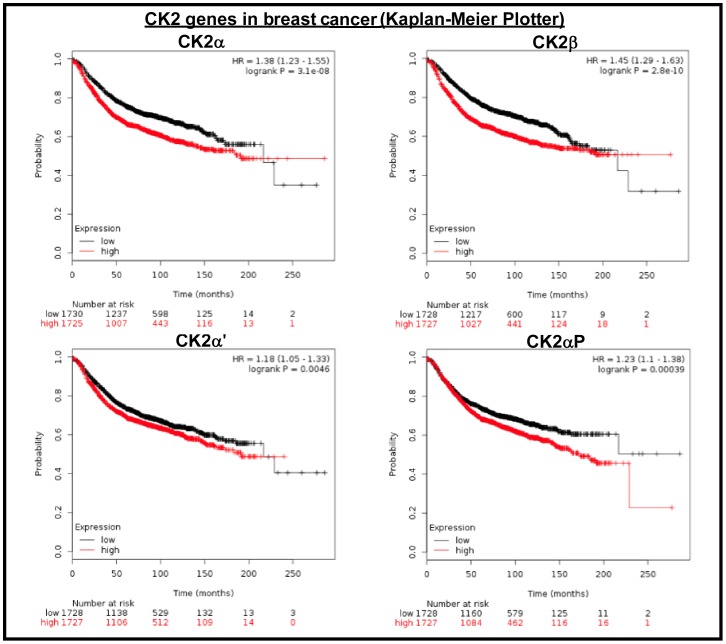
Kaplan-Meier analysis revealed that in overall breast cancer high levels of expression of CK2α and CK2β correlate with lower survival rates (Fig. 5). CK2α' expression had no significant influence in survival in all breast cancers (Fig. 5). This is in line with a published study, CK2α (CSNK2A1) was found as part of a 186-gene “invasiveness” gene signature in breast cancer. This “invasiveness” gene signature was associated with overall survival and metastasis-free survival in patients with breast cancer, and when combined with the NIH prognostic criteria, the “invasiveness” gene signature was used to stratify patients with high-risk early breast cancer. This “invasiveness” gene signature was associated with overall survival and metastasis-free survival in patients with medulloblastoma, lung cancer and prostate cancer [37]. In addition, it has been found that unbalanced expression of CK2 genes is able to promote epithelial to mesenchymal transition in cell lines [38]. These studies emphasize the importance of analysis of the expression of CK2 genes during cancer progression.
Figure 5. CK2 genes in breast cancer (Kaplan-Meier Plotter).
Kaplan-Meier plots showing overall survival in lung adenocarcinoma. In red: patients with expression above the median and in black, patients with expressions below the median. CK2α, p = 3.1 10−8; CK2αP, p = 3.9 10−4; CK2α', p = 4.6 10−3; CK2β, p = 2.8 10−10.
Colon cancer
Colon cancer is the third leading cause of cancer related deaths when men and women are considered separately, and the second leading cause when both considering both sexes. The NCI predicts that in 2014 there would be an estimated 50,310 deaths in men and women, which would be 8.6% of all cancer deaths [33]. They have estimated 136,830 new cases of colorectal cancer in 2014. When combined, colorectal cancers make up 8.2% of all new cancer cases. The most common type of colorectal cancers is adenocarcinoma (95%). Its precursor, adenomatous polyps (adenomas) are polyps that are considered to be a pre-cancerous condition. Treatments of this cancer type include surgery, chemotherapy, biological therapy, and radiation therapy. Early detection of colorectal cancers can increase survival rate when comparing localized colorectal cancers (90.3%) and regional colorectal cancers (70.4%). The relative 1-year relative survival rate is 84% and the relative 5-year survival rate for colorectal cancer is 64.9%.
CK2 in Colon cancer
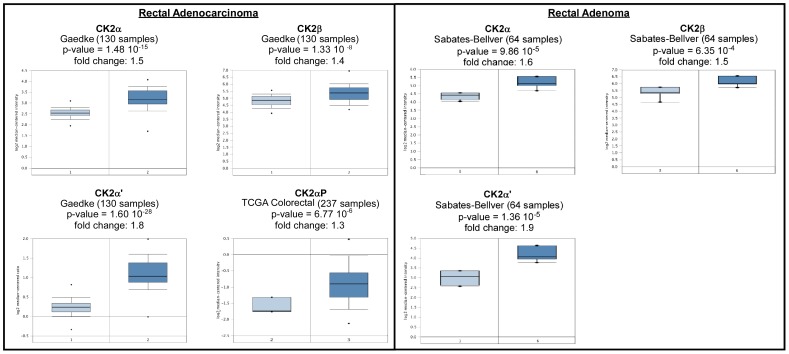
Oncomine analysis revealed many cases of overexpression for all three CK2 genes and the CK2α pseudogene in adenocarcinomas (Table 3, Fig. 6). We found a statistically significant increase in CK2α in colon adenocarcinoma. In agreement with this finding, a published study shows that expression levels of CK2α transcripts are elevated in 10 pairs of colorectal tumor tissues compared to non-tumor tissues [39]. Oncomine analysis found that there is also overexpression of CK2 genes in adenomas (Table 4). This is relevant, because CK2 has been shown to regulate Wnt/β-catenin signaling [7]. Wnt/β-catenin signaling is major player in colorectal cancer progression as it is proposed to be the first hit to be deregulated in colon cancer progression [15].
Table 3. Changes in CK2 gene expression in colon cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Colon Carcinoma | ||||||
| CK2α | 8.38 10−7 | 2.0 | 8% | Skrzypczak | 40 | [72] |
| CK2α' | 6.63 10−10 | 2.3 | 2% | Skrzypczak Colorectal 2 | 40 | [72] |
| 6.17 10−15 | 1.5 | 6% | TCGA Colorectal | 237 | [68] | |
| 2.28 10−19 | 2.6 | 1% | Hong Colorectal | 82 | [73] | |
| CK2αP | 4.12 10−9 | 1.5 | 15% | TCGA Colorectal | 237 | [68] |
| CK2β | 2.30 10−10 | 1.4 | 6% | Ki Colon | 123 | [74] |
| 2.71 10−7 | 1.9 | 7% | Skrzypczak Colorectal 2 | 40 | [72] | |
| Rectal Adenocarcinoma | ||||||
| CK2α | 1.48 10−15 | 1.5 | 9% | Gaedke | 130 | [75] |
| CK2α' | 1.60 10−28 | 1.8 | 2% | Gaedke | 130 | [75] |
| CK2αP | 6.77 10−6 | 1.3 | 22% | TCGA Colorectal | 237 | [68] |
| CK2β | 1.33 10−8 | 1.4 | 20% | Gaedke | 130 | [75] |
| Cecum Adenocarcinoma | ||||||
| CK2α | 4.29 10−7 | 1.7 | 12% | TCGA Colorectal | 237 | [68] |
| CK2α' | 8.74 10−8 | 1.5 | 10% | TCGA Colorectal | 237 | [68] |
| CK2αP | 2.48 10−6 | 1.7 | 14% | TCGA Colorectal | 237 | [68] |
| CK2β | 2.95 10−5 | 1.4 | 19% | TCGA Colorectal | 237 | [68] |
Different subtypes of colon cancer are analyzed and p-values, fold changes, and datasets are included.
Figure 6. CK2 gene analysis in colorectal cancer (Oncomine database).
Box plots derived from gene expression data in Oncomine comparing expression of specific CK2 genes in in normal plot (left plot) and in colorectal cancer tissue (right plot). Oncomine box plots were retrieved from rectal adenomas and adenocarcinomas.
Table 4. Changes in CK2 gene expression in non-neoplastic colon adenomas.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Colon Adenoma | ||||||
| CK2α | 2.32 10−5 | 2.4 | 7% | Skrzypczak | 40 | [72] |
| CK2α' | 2.43 10−10 | 1.6 | 8% | Sabates-Bellver | 64 | [76] |
| CK2β | 1.31 10−7 | 1.4 | 14% | Sabates-Bellver | 64 | [76] |
| Rectal Adenoma | ||||||
| CK2α | 9.86 10−5 | 1.6 | 11% | Sabates-Bellver | 64 | [76] |
| CK2α' | 1.36 10−5 | 1.9 | 6% | Sabates-Bellver | 64 | [76] |
| CK2β | 6.35 10−4 | 1.5 | 18% | Sabates-Bellver | 64 | [76] |
Different subtypes of colon adenomas are analyzed and p-values, fold changes, and datasets are included.
Ovarian cancer
Ovarian cancer ranks fifth in cancer death among women (3% of all cancers among women), accounting for more deaths than any other cancer of the female reproductive system. According to the NCI in 2014, there would be an estimated 14,270 deaths due to ovarian cancer, accounting for 2.4% of all cancer deaths and 7.5% of cancer deaths among women [33]. There would be approximately 21,980 new cases of ovarian cancer in the USA in 2014, accounting for 1.3% of all new cancer cases. The most common type of ovarian cancer is epithelial ovarian carcinoma, which accounts for 85% to 90% of all ovarian cancers. Epithelial ovarian cancer is divided into other subtypes such as serous, which accounts for 80–85%, endometrioid, which accounts for ∼10%, and clear cell, which accounts for ∼5% of all epithelial ovarian cancers. The less-common types of ovarian cancers include germ cell tumors (less than 2% of ovarian cancers), and stromal tumors (1% of ovarian cancers). The treatment methods of ovarian cancer include surgery, chemotherapy and radiation. A woman's risk of developing ovarian cancer in her lifetime is 1 in 72, and the age at which she is diagnosed affects survival rate. 56% of women younger than 65 years survive 5 years following diagnosis, while 27% of women older than age 65 survive 5 years following diagnosis. Overall, the 1-year relative survival rate of all ovarian cancer patients is 75% and the 5-year relative survival rate is 44%. If the tumor is localized, there is a 91.9% 5-year relative survival rate, but if the tumor is regional, the survival rate drops to 72.0%.
CK2 in Ovarian cancer
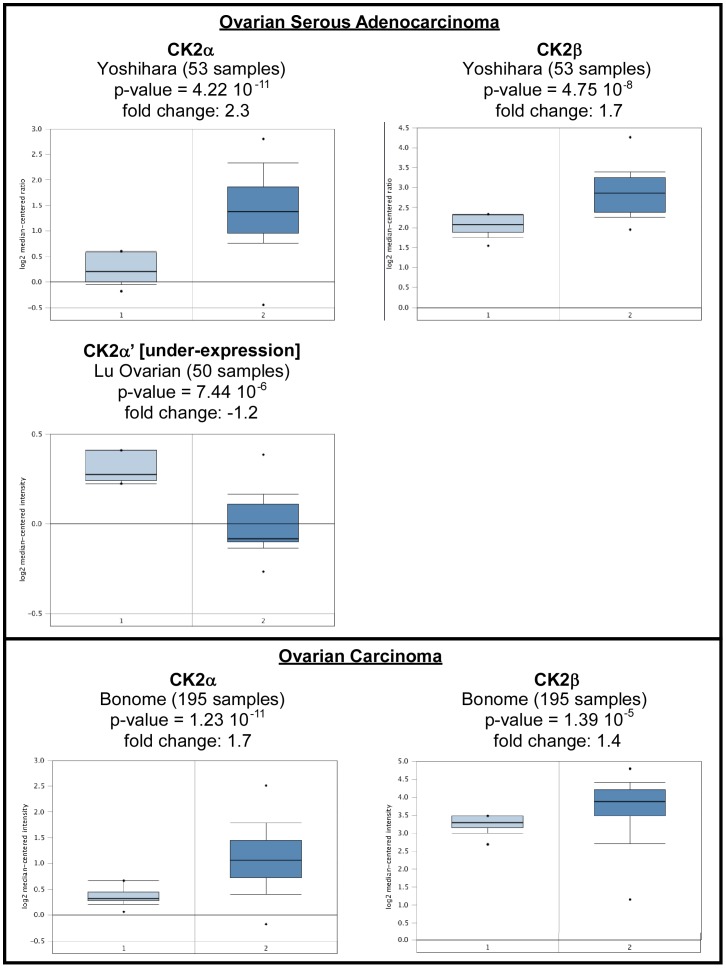
Oncomine analysis of neoplastic vs. normal tissue showed that CK2α and CK2β were significantly upregulated in several ovarian cancer types. CK2α' was found significantly downregulated in two ovarian tumor types. There were no significant results for CK2αP (Table 5, Fig. 7).
Table 5. Changes in CK2 gene expression in ovarian cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | DataSet | #Samples | Reference |
| Ovarian Serous Adenocarcinoma | ||||||
| CK2α | 4.22 10−11 | 2.3 | 1% | Yoshihara | 53 | [77] |
| CK2α' | 7.44 10−6 | −1.2 | 1% | Lu Ovarian | 50 | [78] |
| 0.005 | −1.4 | 6% | Adib | 16 | [79] | |
| CK2β | 4.75 10−8 | 1.7 | 3% | Yoshihara | 53 | [77] |
| Ovarian Carcinoma | ||||||
| CK2α | 1.23 10−11 | 1.7 | 3% | Bonome | 195 | [80] |
| CK2β | 1.39 10−5 | 1.4 | 19% | Bonome | 195 | [80] |
| Ovarian Serous Cystadenocarcinoma | ||||||
| CK2α | 6.51 10−6 | 1.7 | 7% | TCGA Ovarian | 594 | [68] |
| Ovarian Endometroid Adenocarcinoma | ||||||
| CK2β | 0.01 | 1.6 | 11% | Lu Ovarian | 50 | [78] |
| Ovarian Mucinous Adenocarcinoma | ||||||
| CK2α' | 1.82 10−4 | −1.2 | 1% | Lu Ovarian | 50 | [78] |
Different subtypes of ovarian cancer are analyzed and p-values, fold changes, and datasets are included.
Figure 7. CK2 gene analysis in ovarian cancer (Oncomine database).
Box plots derived from gene expression data in Oncomine comparing expression of a specific CK2 gene in normal (left plot) and lung cancer tissue (right plot). Oncomine box plots were retrieved from serous ovarian adenocarcinoma and ovarian carcinoma.
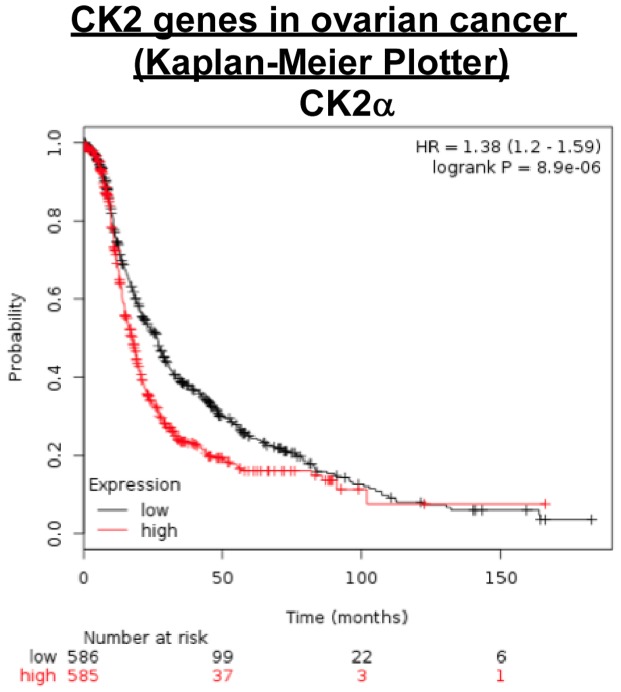
Kaplan-Meier plotter analysis revealed that high-levels of expression of CK2α correlated with lower patients survival rates. In contrast, although CK2α' was significantly downregulated in some adenocarcinomas and CK2β that was upregulated in a few subtypes, Kaplan-Meier plotter analysis did not show any significance of CK2α' and CK2β expression levels in overall survival (Fig. 8).
Figure 8. CK2 genes in ovarian cancer (Kaplan-Meier Plotter).
Kaplan-Meier plots showing overall survival in ovarian cancer. In red: patients with expression above the median and in black, patients with expressions below the median. The CK2 genes and corresponding p-values are listed. CK2α, p = 8.9 10−6; CK2αP, p = 0.034; CK2α', p = 0.45; CK2β, p = 0.12.
Prostate cancer
Prostate Cancer is the most common cancer among men, after skin cancer, according to the NCI. The NCI predicts that in 2014, there would be 233,000 new cases of prostate cancer, accounting for 14% of all new cancer cases [33]. The NCI predicts 29,480 deaths in 2014, which would account for 5% of all cancer deaths. The most common type of prostate cancer is adenocarcinoma, which accounts for about 90% of all prostate cancers. There are subtypes of adenocarcinoma including atrophic, foamy, colloid, and signet ring carcinoma. The remaining 10% of all prostate cancers include transitional cell cancer, squamous cell cancer, carcinoid, small cell cancer, and sarcomas and sarcomatoid cancers. Treatments for prostate cancer include surgery, and different therapies such as radiation, hormone, chemotherapy, chemoradiotherapy, and immunotherapy. There is an extremely high survival rate overall. When caught early and the tumor is either localized or regional, there is a 100.0% chance or a 5-year relative survival. Overall, the relative 5-year relative survival rate for localized or low-grade cancer is 99.2%.
CK2 in Prostate cancer
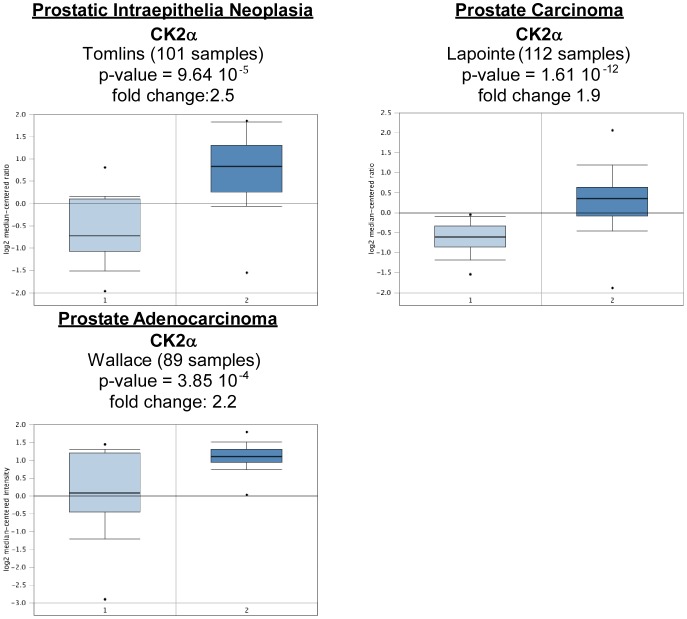
In prostate cancer, Oncomine analysis revealed significant overexpression of CK2α in three subtypes, while CK2α' and CK2β were found only upregulated in one cancer subtype each. There were no significant results for CK2αP (Table 6, Fig. 9). This data are in line with publications showing significant increase in CK2α expression in prostate tumor lesions (p = 0.0224) [40], CK2 activity correlating with Gleason grade [41], and nuclear localization of CK2α associating with poor prognosis [42].
Table 6. Changes in CK2 gene expression in prostate cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Prostatic Intraepithelia Neoplasia | ||||||
| CK2α | 9.64 10−5 | 2.5 | 3% | Tomlins | 101 | [81] |
| CK2α' | 4.4 10−4 | 2 | 5% | Tomlins | 101 | [81] |
| Prostate Adenocarcinoma | ||||||
| CK2α | 3.85 10−4 | 2.2 | 4% | Wallace | 89 | [82] |
| Prostate Carcinoma | ||||||
| CK2α | 1.61 10−12 | 1.9 | 1% | Lapointe | 112 | [83] |
| 3.42 10−5 | 1.9 | 4% | Tomlins | 101 | [81] | |
| 2.57 10−4 | 1.9 | 5% | Singh | 102 | [84] | |
| CK2β | 2.54 10−4 | 1.3 | 8% | Welsh | 34 | [85] |
Different subtypes of prostate cancer are analyzed and p-values, fold changes, and datasets are included.
Figure 9. CK2α gene analysis in prostate cancer (Oncomine database).
Box plots derived from gene expression data in Oncomine comparing expression of CK2α gene in normal (left plot) and various types of prostate cancer tissue (right plot). Oncomine box plots were retrieved from various types of prostate cancer.
Pancreatic cancer
Pancreatic Cancer is the fourth-leading cause of death from cancer in the United States, after lung cancer, colon cancer, and breast cancer, but it is the 12th most common type of cancer in the United States. The NCI estimates that there would be an estimated 39,590 deaths due to pancreatic cancer in 2014, which accounts for 6.8% of all cancer deaths [33]. They estimated that there would be 46,420 new cases, making up 2.8% of all new cancer cases. There are two types of pancreatic cancer cells, pancreatic exocrine tumors and pancreatic neuroendocrine tumors. About 95% of pancreatic cancers are classified as exocrine tumors, which has many different subtypes such as adenocarcinoma, and acinar cell carcinoma. Adenocarcinoma accounts for about 90% of all pancreatic cancers, and the other subtypes are considered to be rare. Pancreatic neuroendocrine tumors account for less than 5% of pancreatic tumors, and have much better survival than pancreatic adenocarcinoma. Treatments for people with adenocarcinomas include surgery, chemotherapy, targeted therapy, and radiation therapy. Despite these treatments, the relative survival rate is fairly low. Even when the tumor is localized or regional, patients have a relatively low 5-year relative survival rate, which is 24.1% and 9.0% respectively. Accounting for all types of pancreatic cancer, there is a 6.0% that a patient can survive 5 years.
CK2 in Pancreatic cancer
Oncomine analysis revealed that only CK2β was significantly upregulated in pancreatic adenocarcinoma. CK2β was downregulated in pancreatic adenocarcinoma while CK2α' was downregulated in pancreatic ductal adenocarcinoma (Table 7). CK2α was overexpressed in both types of pancreatic cancer but not significantly (p-values around 0.02 to 0.03). CK2αP was not significantly changed.
Table 7. Changes in CK2 gene expression in pancreatic cancer.
| Gene | P-Value | Fold Change | Rank (Top %) | Dataset | #Samples | Reference |
| Pancreatic Adenocarcinoma | ||||||
| CK2α | 0.022 | 1.74 | 17% | Iacozb-Donahue | 35 | [86] |
| CK2β | 0.001 | 2.2 | 9% | Logsdon | 27 | [87] |
| Pancreatic Ductal Adenocarcinoma | ||||||
| CK2α | 0.030 | 1.5 | 8% | Grutzmann | 25 | [88] |
| CK2α' | 0.002 | −1.5 | 5% | Buchholz | 38 | [89] |
Different subtypes of pancreatic cancer are analyzed and p-values, fold changes, and datasets are included.
Conclusions
It was reported that CK2α protein but not transcripts are deregulated in cancer [22], [27]. In our analysis of Oncomine we found that not only CK2α, but also CK2α', CK2αP, and CK2β transcripts were deregulated in various cancer types (summarized in Table 8). This discrepancy may be due to the fact that these original publications reported data from a limited number of tumor samples (4–8 tumor samples) from two tumor types [22], [27]. However, more recent publications using larger patient samples show that CK2α transcripts are upregulated in lung and colon cancers [34], [39]. Together with our study, these data suggest that deregulated CK2 gene transcript expression may be a mechanism underlying the increase in CK2 activity and protein levels detected in human tumors. However, posttranscriptional and posttranslational mechanisms may also be involved. In general, there is little data on the regulation of CK2 gene expression, and presumably both genetic and epigenetic mechanism may be involved. A number of studies show that regulation of CK2 transcription happens at the promoter level for CK2α and CK2β [43], [44], [45]. However, there is no data on whether CK2 genes themselves are controlled by epigenetic mechanisms. Still, since CK2 controls epigenetic regulators [46], [47], [48], there is a possibility that CK2 could control itself though epigenetic regulation. Nonetheless, there is no data on whether altered CK2 expression in cancer is due to genetic or epigenetic mechanisms. Identifying the genetic and epigenetic mechanisms that control CK2 expression will help understand the role of CK2 in biological processes and in disease.
Table 8. Overall trends of CK2 mRNA expression levels in the 6 types of cancers.
| Lung | Breast | Colorectal | Ovarian | Prostate | Pancreatic | |
| CK2α | Up | Up | Up | Up | Up | Up |
| CK2α' | Up | Down | Up | Down | Up | Down |
| CK2αP | Up | Up | Up | N/A | N/A | N/A |
| CK2β | Up/down | Up | Up | Up | Up | Up |
Our data indicates that deregulated CK2 expression is an important factor during tumorigenesis. In our Oncomine analysis, the relative fold change in the expression of CK2 genes differs between tumors. In addition, we found a strong correlation between particular CK2 gene expression and certain subtypes of cancer. For example, CK2α' was found underexpressed in breast, ovarian and pancreatic cancers. In addition, while both ductal and lobular breast carcinoma underexpressed CK2α', ductal but not lobular breast carcinoma had upregulation of CK2α. Likewise, the CK2β transcripts were upregulated in both ductal and lobular breast carcinoma when they became invasive. This suggests that CK2β may be required to progress into invasive cancer. Another example is the lack of overexpression of CK2αP in non-neoplastic colon adenomas compared to its overexpression in all three of the colorectal adenocarcinomas.
In contrast with this study on transcript levels, we know relatively little about the expression of CK2 proteins in human cancer and its role in tumorigenesis. A number of publications study CK2 activity and protein levels in a few samples from selected human tumors. As discussed by Tawfic [22], increases on CK2 in cancers based on activity levels alone have to be considered carefully while studies of both protein levels and activity are better indicators of CK2 increases in cancers. In line with our findings, different fold change on the expression of CK2α and CK2β genes is found in kidney tumors [49]. Importantly, in head and neck tumors the level of CK2 correlated with tumor grade and stage [50]. However, a thorough analysis of the specific localization of CK2 transcripts and/or protein is needed to understand the role of CK2 in cancer. In this regard, inmunological studies show CK2 located in the leading edge of the tumor in heterotransplanted human colon tumors in mice [51]. Intriguingly, expression of nuclear CK2α protein was higher in colorectal adenocarcinomas than in adenomas compared to normal tissues, which suggests that CK2α's subcellular localization maybe involved in progression from adenomas to malignant tumors [52].
Our analysis of Kaplan-Meier Plotter also found correlations between CK2 gene deregulation and survival rates (Table 9). Overall, having higher levels of CK2 gene expression led to lower survival amongst patients with lung, breast, and ovarian cancer. Similarly, in head and neck tumors the level of CK2 also correlated clinical outcome [50]. However, in lung adenocarcinoma, displaying a higher level of CK2α' and CK2αP correlates with higher survival rates.
Table 9. Overall Survivability as an effect of High Gene Expression Levels.
| High levels of Expression: | Lung Adenocarcinoma | Breast Cancer | Ovarian Cancer |
| CK2α | Not significant | Lower survival | Lower survival |
| CK2α' | Higher survival | Lower survival | Not Significant |
| CK2αP | Higher survival | Lower survival | Lower survival |
| CK2β | Lower survival | Lower survival | Not significant |
CK2 is an emerging key target in cancer therapy by impacting not only cell growth and proliferation but also anti-apoptotic activity in malignant cells. Indeed, CK2 inhibitors are being tested for their effectiveness in cancer treatment [11], [27], [53], [54]. In addition, CK2 inhibitors are also studied in conjunction with other antitumor agents such as Melphalan and Imatinib [53], [55]. The database mining findings here greatly support the use of CK2-targeted therapies to treat CK2 upregulation in cancer. For example, CK2α,α' and β genes were found significantly deregulated in colorectal adenomas and adenocarcinomas (this study and [39], [52]), therefore CK2 inhibitors could be used to prevent both the start and progression of colon cancer. Our data indicates that, for some types of cancers, a CK2β-dependent CK2 targeted therapy may be more effective. This is the case of breast cancers, where CK2β overexpression correlated with invasive types of breast carcinoma. This property may be exploited to design more effective CK2 inhibitors. In this regard, compounds such as the inhibitor MNA that inhibits more efficiently CK2 holoenzyme (IC50 = 0.3 µM) than monomeric CK2α (IC50 = 2.8 µm) could be an effective CK2β-dependent CK2 targeted therapy [56], [57]. Our data also indicates that for some cancers such as lung adenocarcinoma, the protective effect in patient survival of CK2α' may be counteracted by CK2 inhibitory drugs. In this case, CK2 inhibitors that target specifically CK2α may not affect the protective role of CK2α'.
The importance of our analysis on CK2 gene transcript expression on cancer development, and on cancer patient diagnosis, treatment and survival needs to be further evaluated. First, the correlation between transcript and protein upregulation for different tumor types needs to be researched, as it is protein activity that will be the target of therapy. Second, determining the specific localization of CK2 proteins and transcripts changes in tumor tissues. Third, additional research on the in vitro and in vivo tumorigenic potential the understudied CK2 genes (CK2α', CK2β and CK2αP). Fourth, study the effect on tumorigenesis of different fold changes in the expression of CK2 genes, in particular incorporating the knowledge on CK2 protein localization in tumors for in vivo studies. Fifth, further research of ovarian, prostate and pancreatic cancer as they had a comparatively small number of samples per dataset, and also analyzing other types of cancer. Sixth, additional research on the impact on patient survival of changes in the CK2 genes in additional cancer types. Seventh, adjusting CK2 therapy development to CK2 cancer-specific expression. All together, these studies could lead to cancer diagnostic and predictive tools, and help develop more effective and specific cancer treatments.
Acknowledgments
We would like to thank Drs. David Seldin, Kevan Hartshorn and Marjory Charlot. We will also like to thank the Dominguez lab for discussions and, in particular, Tatyanna Henderson and Mikaela Oliveiro for comments to the manuscript. We would also like to thank the reviewers for thoughtful comments that have improved the manuscript. Data from WHO, NCI, Oncomine and Kaplan Meier Plotter is reproduced with their permission.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
The following resources that were used in this manuscript: buying of computers, maintenance of computers and software, and PI salary. These resource were acquired with funding from the National Institutes of General Medical Sciences; 1R01GM098367 (http://www.nigms.nih.gov/Pages/default.aspx). Therefore, indirectly, this R01 grant supported this work. There are no other sources of funding. The work was performed by two undergraduate students that had no funding, and volunteer their time and effort for this project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. World-Health-Organization (2010) Global status report on noncommunicable diseases. 2010:162. [Google Scholar]
- 2.U.S.-Cancer-Statistics-Working-Group (2014) United States Cancer Statistics - 1999–2011 Incidence and mortality - Web-based Report. 2014.
- 3. Seldin DC, Landesman-Bollag E, Farago M, Currier N, Lou D, et al. (2005) CK2 as a positive regulator of Wnt signalling and tumourigenesis. Mol Cell Biochem 274:63–67. [DOI] [PubMed] [Google Scholar]
- 4. Hauck L, Harms C, An J, Rohne J, Gertz K, et al. (2008) Protein kinase CK2 links extracellular growth factor signaling with the control of p27(Kip1) stability in the heart. Nat Med 14:315–324. [DOI] [PubMed] [Google Scholar]
- 5. Axtell RC, Xu L, Barnum SR, Raman C (2006) CD5-CK2 binding/activation-deficient mice are resistant to experimental autoimmune encephalomyelitis: protection is associated with diminished populations of IL-17-expressing T cells in the central nervous system. Journal of immunology 177:8542–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh NN, Ramji DP (2008) Protein kinase CK2, an important regulator of the inflammatory response? J Mol Med 86:887–897. [DOI] [PubMed] [Google Scholar]
- 7. Dominguez I, Sonenshein GE, Seldin DC (2009) Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kappaB signaling: linking development and cancer. Cellular and molecular life sciences: CMLS 66:1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allada R, Meissner RA (2005) Casein kinase 2, circadian clocks, and the flight from mutagenic light. Molecular and cellular biochemistry 274:141–149. [DOI] [PubMed] [Google Scholar]
- 9. Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. The Biochemical journal 369:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed K, Davis AT, Wang H, Faust RA, Yu S, et al. (2000) Significance of protein kinase CK2 nuclear signaling in neoplasia. Journal of cellular biochemistry Supplement Suppl 35 130–135. [DOI] [PubMed]
- 11. Pinna LA, Meggio F (1997) Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Progress in cell cycle research 3:77–97. [DOI] [PubMed] [Google Scholar]
- 12. Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K (2008) Protein kinase CK2–a key suppressor of apoptosis. Advances in enzyme regulation 48:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahmed K, Gerber DA, Cochet C (2002) Joining the cell survival squad: an emerging role for protein kinase CK2. Trends in cell biology 12:226–230. [DOI] [PubMed] [Google Scholar]
- 14. Canton DA, Litchfield DW (2006) The shape of things to come: an emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cellular signalling 18:267–275. [DOI] [PubMed] [Google Scholar]
- 15. Duncan JS, Litchfield DW (2008) Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochimica et biophysica acta 1784:33–47. [DOI] [PubMed] [Google Scholar]
- 16. Kramerov AA, Saghizadeh M, Caballero S, Shaw LC, Li Calzi S, et al. (2008) Inhibition of protein kinase CK2 suppresses angiogenesis and hematopoietic stem cell recruitment to retinal neovascularization sites. Molecular and cellular biochemistry 316:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruzzene M, Pinna LA (2010) Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochimica et biophysica acta 1804:499–504. [DOI] [PubMed] [Google Scholar]
- 18. Xu X, Landesman-Bollag E, Channavajhala PL, Seldin DC (1999) Murine protein kinase CK2: gene and oncogene. Molecular and cellular biochemistry 191:65–74. [PubMed] [Google Scholar]
- 19. Bibby AC, Litchfield DW (2005) The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. International journal of biological sciences 1:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seldin DC (1995) New models of lymphoma in transgenic mice. Current opinion in immunology 7:665–673. [DOI] [PubMed] [Google Scholar]
- 21. Landesman-Bollag E, Romieu-Mourez R, Song DH, Sonenshein GE, Cardiff RD, et al. (2001) Protein kinase CK2 in mammary gland tumorigenesis. Oncogene 20:3247–3257. [DOI] [PubMed] [Google Scholar]
- 22. Tawfic S, Yu S, Wang H, Faust R, Davis A, et al. (2001) Protein kinase CK2 signal in neoplasia. Histology and histopathology 16:573–582. [DOI] [PubMed] [Google Scholar]
- 23. Currier N, Solomon SE, Demicco EG, Chang DL, Farago M, et al. (2005) Oncogenic signaling pathways activated in DMBA-induced mouse mammary tumors. Toxicologic pathology 33:726–737. [DOI] [PubMed] [Google Scholar]
- 24. Ahmed K (1974) Increased phosphorylation of nuclear phosphoproteins in precancerous liver. Research communications in chemical pathology and pharmacology 9:771–778. [PubMed] [Google Scholar]
- 25.Seldin DC, Landesman-Bollag E (2013) The Oncogenic Potential of CK2; Pinna LA, editor. Ames, Iowa: Wiley-Blackwell. 293–304 p. [Google Scholar]
- 26.Trembley JH, Wu J, Unger GM, Kren BT, Ahmed K (2013) CK2 Suppression of Apoptosis and Its Implication in Cancer Biology and Therapy; PInna LA, editor. Ames, Iowa: Wiley-Blackwell. 219–343 p. [Google Scholar]
- 27. Trembley JH, Chen Z, Unger G, Slaton J, Kren BT, et al. (2010) Emergence of protein kinase CK2 as a key target in cancer therapy. BioFactors 36:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hung MS, Lin YC, Mao JH, Kim IJ, Xu Z, et al. (2010) Functional polymorphism of the CK2alpha intronless gene plays oncogenic roles in lung cancer. PloS one 5:e11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wirkner U, Voss H, Lichter P, Weitz S, Ansorge W, et al. (1992) Human casein kinase II subunit alpha: sequence of a processed (pseudo)gene and its localization on chromosome 11. Biochimica et biophysica acta 1131:220–222. [DOI] [PubMed] [Google Scholar]
- 30. Battistutta R (2009) Protein kinase CK2 in health and disease: Structural bases of protein kinase CK2 inhibition. Cellular and molecular life sciences: CMLS 66:1868–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gyorffy B, Lanczky A, Szallasi Z (2012) Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-related cancer 19:197–208. [DOI] [PubMed] [Google Scholar]
- 33.Howlader N NA, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). 2013) SEER Cancer Statistics Review, 1975–2011 - Web-based Report. posted to the SEER web site, April 2014.
- 34. O-Charoenrat P, Rusch V, Talbot SG, Sarkaria I, Viale A, et al. (2004) Casein kinase II alpha subunit and C1-inhibitor are independent predictors of outcome in patients with squamous cell carcinoma of the lung. Clinical cancer research: an official journal of the American Association for Cancer Research 10:5792–5803. [DOI] [PubMed] [Google Scholar]
- 35. Daya-Makin M, Sanghera JS, Mogentale TL, Lipp M, Parchomchuk J, et al. (1994) Activation of a tumor-associated protein kinase (p40TAK) and casein kinase 2 in human squamous cell carcinomas and adenocarcinomas of the lung. Cancer research 54:2262–2268. [PubMed] [Google Scholar]
- 36. Yaylim I, Isbir T (2002) Enhanced casein kinase II (CK II) activity in human lung tumours. Anticancer research 22:215–218. [PubMed] [Google Scholar]
- 37. Liu R, Wang X, Chen GY, Dalerba P, Gurney A, et al. (2007) The prognostic role of a gene signature from tumorigenic breast-cancer cells. The New England journal of medicine 356:217–226. [DOI] [PubMed] [Google Scholar]
- 38. Deshiere A, Duchemin-Pelletier E, Spreux E, Ciais D, Combes F, et al. (2013) Unbalanced expression of CK2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by Snail1 induction. Oncogene 32:1373–1383. [DOI] [PubMed] [Google Scholar]
- 39. Lin KY, Tai C, Hsu JC, Li CF, Fang CL, et al. (2011) Overexpression of nuclear protein kinase CK2 alpha catalytic subunit (CK2alpha) as a poor prognosticator in human colorectal cancer. PloS one 6:e17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo W, Yu WD, Ma Y, Chernov M, Trump DL, et al. (2013) Inhibition of protein kinase CK2 reduces Cyp24a1 expression and enhances 1,25-dihydroxyvitamin D(3) antitumor activity in human prostate cancer cells. Cancer research 73:2289–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yenice S, Davis AT, Goueli SA, Akdas A, Limas C, et al. (1994) Nuclear casein kinase 2 (CK-2) activity in human normal, benign hyperplastic, and cancerous prostate. The Prostate 24:11–16. [DOI] [PubMed] [Google Scholar]
- 42. Laramas M, Pasquier D, Filhol O, Ringeisen F, Descotes JL, et al. (2007) Nuclear localization of protein kinase CK2 catalytic subunit (CK2alpha) is associated with poor prognostic factors in human prostate cancer. Eur J Cancer 43:928–934. [DOI] [PubMed] [Google Scholar]
- 43. Lupp S, Gumhold C, Ampofo E, Montenarh M, Rother K (2013) CK2 kinase activity but not its binding to CK2 promoter regions is implicated in the regulation of CK2alpha and CK2beta gene expressions. Molecular and cellular biochemistry 384:71–82. [DOI] [PubMed] [Google Scholar]
- 44. Olenkina OM, Egorova KS, Aravin AA, Naumova NM, Gvozdev VA, et al. (2012) Mapping of cis-regulatory sites in the promoter of testis-specific stellate genes of Drosophila melanogaster. Biochemistry Biokhimiia 77:1285–1293. [DOI] [PubMed] [Google Scholar]
- 45. Olenkina OM, Egorova KS, Kibanov MV, Gervaziev YV, Gvozdev VA, et al. (2012) Promoter contribution to the testis-specific expression of Stellate gene family in Drosophila melanogaster. Gene 499:143–153. [DOI] [PubMed] [Google Scholar]
- 46. Eom GH, Cho YK, Ko JH, Shin S, Choe N, et al. (2011) Casein kinase-2alpha1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation 123:2392–2403. [DOI] [PubMed] [Google Scholar]
- 47. Deplus R, Blanchon L, Rajavelu A, Boukaba A, Defrance M, et al. (2014) Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell reports 8:743–753. [DOI] [PubMed] [Google Scholar]
- 48. Costa R, Arrigoni G, Cozza G, Lolli G, Battistutta R, et al. (2014) The lysine-specific demethylase 1 is a novel substrate of protein kinase CK2. Biochimica et biophysica acta 1844:722–729. [DOI] [PubMed] [Google Scholar]
- 49. Stalter G, Siemer S, Becht E, Ziegler M, Remberger K, et al. (1994) Asymmetric expression of protein kinase CK2 subunits in human kidney tumors. Biochemical and biophysical research communications 202:141–147. [DOI] [PubMed] [Google Scholar]
- 50. Gapany M, Faust RA, Tawfic S, Davis A, Adams GL, et al. (1995) Association of elevated protein kinase CK2 activity with aggressive behavior of squamous cell carcinoma of the head and neck. Molecular medicine 1:659–666. [PMC free article] [PubMed] [Google Scholar]
- 51. Seitz G, Munstermann U, Schneider HR, Issinger OG (1989) Characterization of casein kinase II in human colonic carcinomas after heterotransplantation into nude mice. Biochemical and biophysical research communications 163:635–641. [DOI] [PubMed] [Google Scholar]
- 52. Zou J, Luo H, Zeng Q, Dong Z, Wu D, et al. (2011) Protein kinase CK2alpha is overexpressed in colorectal cancer and modulates cell proliferation and invasion via regulating EMT-related genes. Journal of translational medicine 9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarno S, Pinna LA (2008) Protein kinase CK2 as a druggable target. Molecular bioSystems 4:889–894. [DOI] [PubMed] [Google Scholar]
- 54. Cozza G, Pinna LA, Moro S (2013) Kinase CK2 inhibition: an update. Current medicinal chemistry 20:671–693. [DOI] [PubMed] [Google Scholar]
- 55.Drygin D (2013) CK2 as a Logical Target in Cancer Therapy: Potential for Combining CK2 Inhibitors with Various Classes of Cancer Therapeutic Agents; Pinna LA, editor. Ames, Iowa: Wiley-Blackwell. 383–439 p. [Google Scholar]
- 56. Di Maira G, Brustolon F, Bertacchini J, Tosoni K, Marmiroli S, et al. (2007) Pharmacological inhibition of protein kinase CK2 reverts the multidrug resistance phenotype of a CEM cell line characterized by high CK2 level. Oncogene 26:6915–6926. [DOI] [PubMed] [Google Scholar]
- 57. Salvi M, Sarno S, Marin O, Meggio F, Itarte E, et al. (2006) Discrimination between the activity of protein kinase CK2 holoenzyme and its catalytic subunits. FEBS letters 580:3948–3952. [DOI] [PubMed] [Google Scholar]
- 58. Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, et al. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America 98:13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, et al. (2010) Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PloS one 5:e10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, et al. (2001) Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences of the United States of America 98:13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, et al. (2003) A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clinical Cancer Research 9:4695–4704. [PubMed] [Google Scholar]
- 62. Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, et al. (2012) Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-Negative Lung Adenocarcinomas. Cancer Res 72:100–111. [DOI] [PubMed] [Google Scholar]
- 63. Beer DG, Kardia SLR, Huang CC, Giordano TJ, Levin AM, et al. (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8:816–824. [DOI] [PubMed] [Google Scholar]
- 64. Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, et al. (2005) Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res 65:3063–3071. [DOI] [PubMed] [Google Scholar]
- 65. Richardson AL, Wang ZGC, De Nicolo A, Lu X, Brown M, et al. (2006) X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132. [DOI] [PubMed] [Google Scholar]
- 66. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America 98:10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America 100:8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang JJ, Baran J, Cros A, Guberman JM, Haider S, et al. (2011) International Cancer Genome Consortium Data Portal-a one-stop shop for cancer genomics data. Database-the Journal of Biological Databases and Curation. [DOI] [PMC free article] [PubMed]
- 69. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, et al. (2004) Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Molecular Biology of the Cell 15:2523–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, et al. (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14:518–527. [DOI] [PubMed] [Google Scholar]
- 72.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, et al. (2010) Modeling Oncogenic Signaling in Colon Tumors by Multidirectional Analyses of Microarray Data Directed for Maximization of Analytical Reliability. PloS one 5. [DOI] [PMC free article] [PubMed]
- 73. Hong Y, Downey T, Eu KW, Koh PK, Cheah PY (2010) A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clinical & Experimental Metastasis 27:83–90. [DOI] [PubMed] [Google Scholar]
- 74. Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, et al. (2007) Whole genome analysis for liver metastasis gene signatures in colorectal cancer. International Journal of Cancer 121:2005–2012. [DOI] [PubMed] [Google Scholar]
- 75. Gaedcke J, Grade M, Jung K, Camps J, Jo P, et al. (2010) Mutated KRAS Results in Overexpression of DUSP4, a MAP-Kinase Phosphatase, and SMYD3, a Histone Methyltransferase, in Rectal Carcinomas. Genes Chromosomes & Cancer 49:1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sabates-Bellver J, Cattaneo E, Heinimann K, Jiricny J, Marra G (2007) Getting familiar with familial colon cancer. Intestinal Inflammation and Colorectal Cancer 158:27–60. [Google Scholar]
- 77. Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, et al. (2009) Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Science 100:1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, et al. (2004) Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clinical Cancer Research 10:3291–3300. [DOI] [PubMed] [Google Scholar]
- 79. Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, et al. (2004) Predicting biomarkers for ovarian cancer using gene-expression microarrays. British Journal of Cancer 90:686–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, et al. (2008) A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res 68:5478–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tomlins SA, Mehra R, Rhodes DR, Cao XH, Wang L, et al. (2007) Integrative molecular concept modeling of prostate cancer progression. Nature Genetics 39:41–51. [DOI] [PubMed] [Google Scholar]
- 82. Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, et al. (2008) Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res 68:927–936. [DOI] [PubMed] [Google Scholar]
- 83. Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, et al. (2004) Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 101:811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Singh D, Febbo PG, Ross K, Jackson DG, Manola J, et al. (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1:203–209. [DOI] [PubMed] [Google Scholar]
- 85. Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, et al. (2001) Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res 61:5974–5978. [PubMed] [Google Scholar]
- 86. Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, Van Heek NT, et al. (2003) Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. American Journal of Pathology 162:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, et al. (2003) Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 63:2649–2657. [PubMed] [Google Scholar]
- 88. Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R (2004) Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia 6:744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, et al. (2005) Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 24:6626–6636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.