Abstract
Culture-independent molecular analyses of open-sea microorganisms have revealed the existence and apparent abundance of novel eukaryotic lineages, opening new avenues for phylogenetic, evolutionary, and ecological research. Novel marine stramenopiles, identified by 18S ribosomal DNA sequences within the basal part of the stramenopile radiation but unrelated to any previously known group, constituted one of the most important novel lineages in these open-sea samples. Here we carry out a comparative analysis of novel stramenopiles, including new sequences from coastal genetic libraries presented here and sequences from recent reports from the open ocean and marine anoxic sites. Novel stramenopiles were found in all major habitats, generally accounting for a significant proportion of clones in genetic libraries. Phylogenetic analyses indicated the existence of 12 independent clusters. Some of these were restricted to anoxic or deep-sea environments, but the majority were typical components of coastal and open-sea waters. We specifically identified four clusters that were well represented in most marine surface waters (together they accounted for 74% of the novel stramenopile clones) and are the obvious targets for future research. Many sequences were retrieved from geographically distant regions, indicating that some organisms were cosmopolitan. Our study expands our knowledge on the phylogenetic diversity and distribution of novel marine stramenopiles and confirms that they are fundamental members of the marine eukaryotic picoplankton.
Genetic libraries of small subunit (SSU) ribosomal DNA (rDNA) genes constructed from environmental DNA have proven very valuable to study the taxonomic composition of marine prokaryotic plankton (8, 16). Prokaryotic assemblages in the sea appeared to be dominated by novel bacterial and archaeal lineages (17), and this was an important breakthrough for understanding the structure and function of natural assemblages (37). Picoeukaryotes (eukaryotic microbes less than 2 to 3 μm in diameter) are also recognized as fundamental components of marine ecosystems. Phototrophic picoeukaryotes contribute significantly to phytoplankton biomass and primary production (26), and heterotrophic picoeukaryotes, generally small flagellates (5), are the main grazers of bacteria and play key roles within the microbial food web (41). Despite the ecological importance of marine picoeukaryotes, the use of molecular tools to investigate their taxonomic composition, in particular by 18S rDNA libraries, is very recent (9, 28, 34). The most remarkable finding of these studies was the recovery of novel lineages within the stramenopile and alveolate phylogenetic divisions that accounted for a significant fraction of clones in the libraries. These novel lineages represent organisms that must contribute to marine processes but have not been investigated before and thus deserve further detailed phylogenetic and ecological studies.
The stramenopiles (Heterokonta) form one of the eight major phylogenetic groups of eukaryotes (3) and include organisms as diverse as unicellular and multicellular algae, fungus-like cells, and parasitic and free-living flagellates (25, 38). Molecular analyses and the presence of tripartite hairs in the flagella place these heterogeneous groups together. Novel marine stramenopiles from open-sea picoplankton form at least eight independent phylogenetic lineages at the basal part of the stramenopile radiation (31). rDNA probes against two of these lineages showed that the corresponding organisms were small (2 to 5 μm) unpigmented flagellates that could account for a significant fraction of the heterotrophic flagellates (31), an assemblage generally dominated by small and unidentified cells (2, 5). However, only a few samples, and only from the open sea, have been analyzed (31). Whether this group of organisms is equally relevant in other marine environments such as coastal habitats, generally more eutrophic and with terrestrial influence, or anoxic environments, where many organisms cannot live (13), remained unknown.
In order to add picoeukaryotic sequences from coastal habitats to the analysis, we constructed 18S rDNA libraries from four coastal sites and at different seasons over the year. The 19 genetic libraries generated yielded 104 novel stramenopile sequences. Sequences from marine anoxic environments were retrieved from recent studies (7, 11, 42) and analyzed together with our sequences. This substantial increase in phylogenetic information allowed us to address several questions. First, what is the quantitative importance of novel stramenopiles in genetic libraries from the three major marine habitats? Second, what is the overall phylogenetic diversity of novel stramenopiles and how many different clusters can be identified? Finally, which are the main clusters in surface marine waters and how are they biogeographically distributed? Our comparative analysis of novel stramenopile sequences from diverse habitats and geographic origins will allow focusing probe design on the most important clusters, which in turn will allow further progress in the investigation of these novel groups.
MATERIALS AND METHODS
Genetic libraries of coastal picoeukaryotes.
Libraries of 18S rDNAs were constructed from surface coastal picoplankton and were named after the system and the date of sampling. Four libraries were constructed from Blanes Bay samples (BL libraries) in the Mediterranean Sea (41°40′N, 2°48′E). Seven libraries were constructed from a sample obtained off the Roscoff Coast (RA libraries) in the English Channel (48°45′N, 4°00′W), and an additional library (RD010517) was constructed from samples obtained from an estuarine station nearby (48°38′N, 3°51′W). Six libraries were constructed from samples collected near Helgoland (HE libraries) in the North Sea (54°11′N, 7°54′E). Finally, one library was constructed from a sample collected during a cruise in the vicinity of the Orkney Islands (OR000415) in the North Atlantic.
Picoplanktonic biomass (between 0.2 and 3 μm) was collected on filters, and community DNA was extracted. Complete 18S rDNAs were PCR amplified with eukaryote-specific primers, and the PCR products were cloned. Details of the filtering setup, DNA extraction protocol, primers used, and PCR and cloning conditions are described elsewhere (9, 39). Approximately 100 clones in each genetic library were processed by restriction fragment length polymorphism (RFLP), using the restriction enzyme HaeIII. Clones sharing the same RFLP pattern were considered similar phylogenetic entities. At least one clone of each pattern was partially sequenced with the eukaryotic primers 536f and 528f by the QIAGEN Genomics Sequencing Services. Selected clones were completely sequenced by the same service. Sequences were compared by BLAST search (1) and the CHECK_CHIMERA command (29). Results with all picoeukaryotic sequences will be presented separately for the Blanes Bay (R. Massana et al., unpublished data), Roscoff (39), and Helgoland and Orkney (K. Valentin et al., unpublished data) genetic libraries. Here we focus exclusively on novel stramenopile sequences.
Phylogenetic analyses.
Complete 18S rDNA sequences obtained here (15 novel stramenopiles, 3 labyrinthulids, 2 oomycetes, and 1 pirsonid) were aligned by using ClustalW 1.82 (44) with a selection of stramenopile sequences from databases and two dinoflagellate sequences as an outgroup. The stramenopile selection included novel stramenopiles reported so far (7, 31), sequences of basal heterotrophic groups (the most divergent after a database search and separate phylogenetic analyses) including two environmental oomycetes (7, 42), a few sequences from phototrophic groups, and the sequence of Pirsonia diadema (23). Very variable regions of the alignment were automatically removed with Gblocks (6), using parameters optimized for rDNA alignments (minimum length of a block, 5; allowing gaps in half positions), leaving 1,524 informative positions. Maximum-likelihood analysis was carried out with PAUP 4.0b10 (43), with the general time-reversible model assuming a discrete gamma distribution with six rate categories and a proportion of invariable sites. Parameters were estimated from an initial neighbor-joining tree. Bayesian analysis was carried out with MrBayes v3.0B (21), using the same model described above but with four rate categories in the gamma distribution. Bayesian posterior probabilities were computed by running four chains for 1,000,000 generations by using the program default priors on model parameters. Trees were sampled every 100 generations. One thousand or 1,500 trees were discarded as “burn-in” in different runs upon examination of the log likelihood curve of the sampled trees, so that only trees in the stationary phase of the chain were considered. To test for convergence in the Bayesian analysis, four independent runs were performed with the above parameters, obtaining basically the same topologies. Neighbor-joining bootstrap values from 1,000 replicates were calculated with PAUP following the same model used for the maximum-likelihood analysis. Maximum-parsimony bootstrap values from 1,000 replicates were computed with PAUP by using a heuristic search method with a tree bisection-reconnection branch-swapping option with random taxon addition.
Nucleotide sequence accession numbers.
Novel stramenopile sequences have been deposited in GenBank under accession no. AY381156 to AY381219.
RESULTS
Novel stramenopiles in different marine environments.
This study provides a comparative analysis of novel stramenopile sequences retrieved from oceanic (31), coastal (present), and anoxic (7, 11, 42) genetic libraries. Overall we screened 37 libraries (representing more than 2,000 clones) and found 187 novel stramenopile sequences. It is quite remarkable that novel stramenopiles were found in all of these distant and ecologically diverse systems (except in salt marshes), often accounting for a significant fraction of the libraries (Table 1). The relative abundance of novel stramenopile clones was higher in oceanic (around 20%) than in coastal (around 10%) libraries and was variable but generally lower in anoxic libraries. In planktonic open-sea and coastal libraries, most clones affiliated with known phototrophic or heterotrophic groups and unknown groups were represented almost exclusively by novel stramenopiles and novel alveolates (Table 1). Thus, novel stramenopiles appear to be very relevant within the unknown eukaryotic picoplankton, further stressing the interest in focusing on this group. In anoxic habitats, it seems clear that other unknown groups can also be very important (Table 1).
TABLE 1.
Number of libraries analyzed, total clones screened, and percentage of clones affiliating with unknown or known groupsa
| Marine system | No. of libraries | Total no. of clones | % of clones within:
|
Source or reference | ||||
|---|---|---|---|---|---|---|---|---|
| Unknown groups
|
Known groups
|
|||||||
| Novel stramenopiles | Novel alveolates | Other | Phototrophic | Heterotrophic (BHS) | ||||
| Open Sea | ||||||||
| North Atlantic | 2 | 37 | 18.9 | 18.9 | 0 | 48.6 | 13.5 (0) | 9 |
| Mediterranean | 1 | 63 | 19.0 | 19.0 | 0 | 47.6 | 14.3 (0) | 9 |
| Pacific | 1 | 37 | 18.9 | 16.2 | 0 | 51.4 | 13.5 (0) | 34 |
| Antarctica | 2 | 125 | 27.2 | 0 | 3.2 | 59.2 | 10.4 (0) | 9 |
| Coast | ||||||||
| Orkney | 1 | 81 | 17.3 | 6.2 | 6.2 | 14.8 | 55.6 (0) | This study |
| Helgoland | 6 | 633 | 4.7 | 46.8 | 0.6 | 26.7 | 21.1 (1.6) | This study |
| Roscoff | 8 | 367 | 11.7 | 16.3 | 3.8 | 36.2 | 31.9 (2.5) | This study |
| Blanes Bay | 4 | 339 | 10.0 | 41.0 | 0.9 | 35.1 | 10.3 (2.7) | This study |
| Anoxic | ||||||||
| Shallow sediments | 2 | 74 | 14.9 | 0 | 33.8 | 10.8 | 40.5 (2.7) | 7 |
| Hydrothermal vents | 7 | 269 | 6.3 | 18.2 | 11.5 | 19.7 | 44.2 (16.4) | 11 |
| Salt marshes | 3 | 76 | 0 | 0 | 9.2 | 43.4 | 47.4 (1.3) | 42 |
Known groups were separated into those containing mostly phototrophic or heterotrophic organisms. BHS, basal heterotrophic stramenopiles. Deep Antarctic libraries (28) are not included in the table because they were not quantitatively analyzed. The analysis of shallow sediments and salt marshes is semiquantitative (does not take into account the redundancy of identical clones). Metazoan clones have been excluded in all cases.
In general, most clones from open-sea and coastal libraries belonged to phylogenetic groups whose members are mainly phototrophic (Table 1), such as prasinophytes, haptophytes, dinoflagellates, cryptophytes, and diatoms (data not shown). In these libraries, heterotrophic groups are mostly represented by cercomonads, choanoflagellates, acanthareans, fungi, ciliates, and basal heterotrophic stramenopiles. We paid especial attention to basal heterotrophic stramenopiles, such as oomycetes, bicosoecids, and labyrinthulids (percentages shown in Table 1), since their phylogenetic proximity to novel stramenopiles could easily lead to misclassifications. These groups were absent from the open sea and detected at low abundances (0 to 3% of clones) in coastal systems. In anoxic systems, they ranged from almost undetected to rather significant (up to16% in hydrothermal vents).
Phylogenetic diversity of novel stramenopiles.
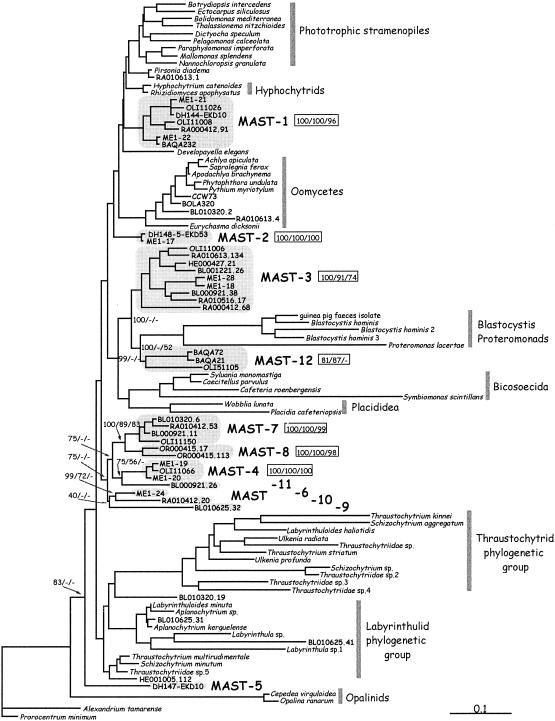
We added 18 novel stramenopile sequences (15 from the coast and 3 from anoxic sediments) to the analysis performed earlier with 16 oceanic sequences (31). These complete 18S rDNA sequences represented the maximal variability observed comparing all partial sequences. Phylogenetic analyses were performed by several methods and summarized in a maximum-likelihood tree with Bayesian posterior probabilities and bootstrap values (neighbor joining and maximum parsimony) in the relevant nodes (Fig. 1). Based on the tree structure and bootstrap values, novel stramenopile sequences were grouped into 12 separate clusters, named MAST (for marine stramenopiles) clusters. Our analysis confirms the eight clusters defined earlier (31) and defines four additional ones. Clusters MAST-1 and MAST-2 appeared as two independent branches (Fig. 1). Our data did not support MAST-1 as a sister group to oomycetes as previously thought (31, 35). MAST-3 and MAST-12 formed two independent lineages related to Blastocystis, bicosoecids, and placididea. Most of the remaining clusters (MAST-4, -6, -7, -8, -9, -10, and -11) formed a large phylogenetic clade without any sequence from known organisms. MAST-8, whose position was previously undetermined (31), appeared confidently related to MAST-7. Finally, MAST-5 was one of the earliest lineages in the stramenopile radiation, appearing just after the Opalinids. This placement was confirmed by separate maximum-likelihood analysis with a selection of eukaryotic sequences (data not shown).
FIG. 1.
Maximum-likelihood phylogenetic tree constructed with complete 18S rDNA sequences of stramenopiles and of two dinoflagellates as an outgroup. Novel marine stramenopile clusters (MAST-1 to MAST-12) are boxed and appear among known heterotrophic basal groups. The scale bar indicates 0.1 substitution per position. Bayesian posterior probabilities (mean of four separate analyses) and neighbor-joining and maximum-parsimony bootstrap values are shown (in that order) to the right of MAST clusters (boxed) and to the left of some other relevant nodes.
Biogeographic distribution of novel stramenopiles.
Separate alignments and maximum-likelihood analyses were performed with partial sequences (550 bp) from MAST-1 (Fig. 2A), MAST-2 (Fig. 2B), MAST-3 (Fig. 2C), MAST-12 (Fig. 2D), and the remaining clusters, except MAST-5 (Fig. 2E). Sequence similarities were calculated with this data set in order to infer the genetic variability within clusters. These similarity values are very close to those obtained by using the complete 18S rDNA (data not shown). The number of clones affiliating with the different MAST clusters was also reported (Table 2). This allowed investigation of the origin of clones inside each cluster and gave us a first picture of the novel stramenopile composition in the different marine environments.
FIG. 2.
Phylogenetic trees constructed with partial 18S rDNA sequences of novel marine stramenopiles from clusters MAST-1 (A); MAST-2 (B); MAST-3 (C); MAST-12 (D); and MAST-4, -6, -7, -8, -9, -10, and -11 (E). We used a closely related sequence as an outgroup (later removed from the tree for clarity). The scale bar indicates 0.1 substitution per position in all trees. Clones are coded according to their origin: North Atlantic (NA), Mediterranean (ME1), Pacific (OLI), Antarctica (ANT and DH), Orkney (OR), Helgoland (HE), Roscoff (RA and RD), Blanes Bay (BL), shallow sediments (BAQA and BAQD), and hydrothermal vents (C1, C2, and C3).
TABLE 2.
Number of clones belonging to different MAST clusters in genetic libraries from the different marine systems
| Marine environment | No. of clones in MAST cluster:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 7 | 8 | 9 | 12 | Othera | |
| Open Sea | |||||||||
| North Atlantic | 0 | 0 | 4 | 1 | 2 | 0 | 0 | 0 | 0 |
| Mediterranean | 2 | 1 | 3 | 4 | 0 | 0 | 0 | 0 | 2 |
| Pacific | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| Antarctic | 4 | 0 | 7 | 0 | 8 | 14 | 0 | 0 | 1 |
| Deep Antarctic | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Coast | |||||||||
| Orkney | 0 | 0 | 1 | 0 | 9 | 4 | 0 | 0 | 0 |
| Helgoland | 0 | 4 | 14 | 4 | 3 | 0 | 0 | 5 | 0 |
| Roscoff | 10 | 1 | 7 | 6 | 11 | 0 | 0 | 3 | 5 |
| Blanes Bay | 0 | 1 | 15 | 7 | 6 | 0 | 2 | 2 | 1 |
| Anoxic | |||||||||
| Shallow sediments | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 |
| Hydrothermal vents | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 1 | 2 |
“Other” includes clusters MAST-5, -6, -10, and -11.
Cluster MAST-1 contained sequences retrieved from most open-sea systems and from a single coastal site (Fig. 2A and Table 2). It appeared to be a planktonic cluster. (BAQA232 retrieved from a shallow anoxic sediment could well have a planktonic origin.) MAST-1 sequences were quite diverse and formed three distinct subclusters (typical similarities within subclusters above 96%). MAST-2 sequences appeared in many systems but always at low clonal abundance (Table 2). These sequences showed a very high similarity (>98.3%) and thus might represent the same (or very similar) organism, found in the Mediterranean, Antarctica, and three coastal sites (Fig. 2B). MAST-1 and MAST-2 sequences dominated in genetic libraries from samples enriched in heterotrophic flagellates from the Norwegian Sea (Massana et al., unpublished). In these enrichments, obtained simply by incubating 3-μm-pore-filtered seawater in the dark, most cells (>80 to 90%) are heterotrophic flagellates. This suggests that MAST-1 and MAST-2 organisms are heterotrophic flagellates
MAST-3 sequences formed the largest cluster (Fig. 2C) and appeared in all open-sea and coastal systems studied (Table 2). This cluster contained a large phylogenetic diversity and distinct subclusters, often composed of sequences retrieved from the same system (one BL, one ANT, and two HE subclusters), suggesting biogeographic adaptation of these organisms. The rDNA probe previously designed for this cluster (NS3) (31) now only targets 25 (out of 38) sequences. Nevertheless, this probe did not match sequences outside the cluster and therefore provided minimal estimates of MAST-3 organisms in fluorescent in situ hybridization (FISH) counts. The application of the NS3 probe to an enrichment (similar to the one described before) from Blanes Bay suggested that MAST-3 organisms are heterotrophic flagellates (31).
MAST-12 was one of the new clusters described here. With the exception of OLI51105 and C2_E027 at the base of the group (Fig. 2D), all clones originated from anoxic shallow sediments and coastal waters (Table 2). This might suggest that MAST-12 organisms were adapted to thrive in anoxic or microoxic environments. Cluster MAST-5 was described from clone DH147-EKD10 retrieved from a depth of 2,000 m in Antarctica (28). Two sequences from deep hydrothermal vents (C1_E032 and C2_E039) are moderately related to this clone (92.4% similarity). No sequences from surface waters affiliated with this cluster, suggesting that MAST-5 organisms could well represent truly deep-sea protists.
The remaining clusters appeared as a large monophyletic group (Fig. 1), and the tree with partial sequences includes all of them (Fig. 2E). This group contained some of the most important marine planktonic clusters (MAST-4, -7, and -8) and clusters formed only by two or three sequences (MAST-6, -10, and -11). The last cluster in this superclade, MAST-9, was described here mostly from hydrothermal vent sequences. Despite containing two Blanes Bay sequences, it is likely that MAST-9 organisms are adapted to anoxic or microoxic habitats.
MAST-4 was formed by a significant number of sequences retrieved from most marine planktonic sites (Table 2). These sequences were remarkably similar (>96%). There were many examples of almost identical sequences (99.5 to 100%) retrieved from distant marine sites, suggesting a cosmopolitan distribution of the corresponding organisms. Probe NS4, previously designed for this cluster (31), targets all new members. In a previous study, we demonstrated that MAST-4 organisms were bacterivorous heterotrophic flagellates that accounted for a significant fraction of natural assemblages (31). MAST-7 sequences were found in significant numbers in all marine planktonic sites (except the Mediterranean), and most of them were rather similar. In some cases, the same sequence was also retrieved from distant sites. Finally, cluster MAST-8 was formed by a relatively large number of clones but retrieved only from two systems (Table 2). MAST-8 sequences were quite diverse and formed three subclusters. Contrary to previous clusters discussed, no MAST-7 and MAST-8 sequences have been retrieved from heterotrophic flagellate enrichments, so the trophic role of these organisms remains unknown. However, given their phylogenetic position, it is reasonable to expect that they are heterotrophic flagellates as well.
DISCUSSION
Libraries of 18S rDNA have revealed the existence and apparent abundance of novel marine picoeukaryotic lineages, mainly within the stramenopiles and alveolates (9, 28, 34). Such a hidden world was striking, given the ecological importance of picoeukaryotes in marine food webs (26, 41). Here we perform a comparative analysis of novel stramenopile sequences from distinct marine habitats. Our phylogenetic reconstruction (Fig. 1) is consistent with previous analyses. First, it shows phototrophic stramenopiles being monophyletic. This was interpreted as stramenopiles being primarily heterotrophic (25) and phototrophic groups resulting from a secondary endosymbiosis event (4). However, recent data showing the monophyly of chromist plastids seem to suggest a chloroplast-containing stramenopile ancestor (46). A potential reconciling view would be that of a heterotrophic stramenopile ancestor and phototrophic groups deriving from a tertiary endosymbiosis with a haptophyte. At any rate, the description of new sequences and organisms will help us to understand the evolution of stramenopiles. Second, relationships among basal heterotrophic groups in Fig. 1 are consistent with previous analyses (18, 36). These groups branch off as separate lineages before the phototrophic radiation: pirsonids, hyphochytrids, Developayella, oomycetes, Blastocystis-bicosoecids-placididea, thraustochytrids-labyrinthulids (separated as “TPG” and “LPG” like in reference 20), and opalinids. Third, novel stramenopiles are placed among the heterotrophic basal groups (31). The 12 MAST clusters add four independent stramenopile lineages and give stability to the Blastocystis-bicosoecid-placididea clade. The placement of MAST clusters among such diverse groups, which include phagotrophs and osmotrophs, free-living and parasitic organisms, does not allow us to infer the type of organisms they represent, except that they are likely heterotrophic. Moreover, given the large phylogenetic distance between MAST clusters, each one could represent organisms with completely different physiological and ecological roles.
The three major marine habitats investigated showed clear differences in the relative abundance of novel stramenopiles (Table 1). Open-sea samples showed the highest representation (around 20% of clones in libraries), with a numerical dominance of clusters MAST-1, -3, -4, and -7. The same MAST clusters dominate in coastal samples, although the overall representation was lower (up to 10% of clones). In coastal samples there was also a moderate representation of labyrinthulids, oomycetes, and bicosoecids, which appeared more common in the coast than in open sea as has already been reported (22). Anoxic environments offered different pictures. In shallow sediments, most MAST clones affiliated with MAST-12, with oomycetes and labyrinthulids as minor components. In hydrothermal vents, many sequences affiliated with oomycetes and a significant proportion of clones (up to 6%) affiliated with MAST-9. Finally, in salt marshes only one clone affiliated with oomycetes and none affiliated with MAST clusters.
Although sampling strategies and seasonal coverage differed, we can draw some general conclusions about the distribution of novel stramenopiles in the marine environment. Clusters MAST-2, -5, -6, -10, and -11 were found sporadically and probably are minor components of marine picoeukaryotic assemblages. Especially interesting among these is MAST-2, which appeared to represent a rare but widespread organism, and MAST-5, which seemed restricted to deep-sea environments. Other clusters, such as MAST-9 and MAST-12, were found predominantly in anoxic environments and probably represent anaerobic or anoxic-tolerant organisms. Similar sequences retrieved from the coastal plankton could represent related organisms with a different niche adaptation, be present within anoxic patches (such as aggregates), or occur following episodic sediment resuspension. Cluster MAST-8 was relatively frequent in two of the open-sea systems analyzed but was absent from the other sites, offering a good example of organisms that are important only locally. Finally, the remaining four clusters, MAST-1, -3, -4, and -7, seem to be truly planktonic, widely distributed, marine aerobic organisms. They were found in almost all open-sea and coastal samples (together accounting for 74% of the novel stramenopile clones) and were never retrieved from anoxic environments. (BAQA232 in MAST-1 is the exception.)
Phylogenetic analyses suggest that the novel stramenopiles are unpigmented heterotrophic cells. It is assumed that marine heterotrophic picoeukaryotes are mostly phagotrophic free-living flagellates (2, 12). Indeed, this is the case at least for MAST-1, -2, -3, and -4 organisms, which developed in heterotrophic flagellate enrichments (31; unpublished data). This was not surprising, since this cell type was also found in phylogenetically related groups, such as bicosoecids, placididea, or Developayella. Heterotrophic flagellates are the primary grazers of marine microbes and play a pivotal role in the microbial food web as a link between picoplankton and larger phagotrophs and as recyclers of inorganic nutrients that sustain primary production (41). Previously, marine pelagic assemblages were considered dominated by chrysomonads, bicosoecids, and choanoflagellates (2, 12, 45), a notion generally derived from studies of enriched (or isolated) organisms and from live counts. The first approach biased flagellate diversity by selecting the easily cultivable organisms (27), whereas live counts often fail to assign in situ organisms even to large taxons (2). The molecular approach shows low clonal abundance of known heterotrophic flagellates and instead suggests that MAST organisms are likely important members of heterotrophic flagellate assemblages.
There is considerable debate about the extent of diversity and biogeographical distribution of microorganisms such as protists. Some authors argue that given the great abundance, small size, and apparent absence of dispersal barriers, most protists must be ubiquitous and the number of protists species relatively low (14, 24). Other authors show locally restricted distributions of protists, claiming that new species will always be found in systems not yet studied (15). Our phylogenetic analysis of novel stramenopile clones from distant marine regions, although including a limited set of samples, provides relevant data to this debate. In most open-sea and coastal systems, MAST clones generally affiliated with the same clusters (MAST-1, -3, -4, and -7), indicating similar organisms in distant assemblages. In addition, some sequences from distant sites were identical (OR000415.39, RA000412.154, BL010320.6, ANT12-6, and NA11-9 within MAST-7), suggesting a cosmopolitan organism. On the other hand, there were also examples of biogeographically restricted organisms, such as those of cluster MAST-8 or system-specific subclusters within MAST-3. In general, our data support the scenario of ubiquitous protist populations, as is generally assumed for marine bacterial (17, 19) and archaeal (30) assemblages, whereas the coexistence of many closely related sequences in the same environment suggests a potentially large diversity. Nevertheless, it is important to note that the 18S rDNA has insufficient resolving power to guarantee the delimitation of species (40). For example within eukaryotic microorganisms, cultures sharing more than 99.5% of the 18S rDNA sequence have been classified as belonging both to a different species (32) and the same (33) species. Therefore, there could be an important cryptic diversity that escapes our molecular analysis.
Novel stramenopile sequences have been retrieved from the picoplankton of all marine systems investigated. They are genetically diverse, with 12 separate lineages detected, but only some of them predominate in open-sea and coastal surface waters. Many MAST organisms are heterotrophic flagellates, being previously unrecognized and likely important components of these assemblages in surface marine waters. This may have implications for the understanding of the functioning of microbial food webs, since the physiology of MAST organisms could differ substantially from that of cultured flagellates used as model organisms (10, 12). The ecological role of each MAST group requires further investigation, specifically on growth and grazing rates, growth efficiencies, food preferences, and nutrient remineralization. Molecular approaches such as FISH targeting specific clusters will soon allow quantitative descriptions of the distribution of these new groups in the sea. Efforts at obtaining MAST organisms in culture should be emphasized, as well as the application of methods to study their activity in situ.
Acknowledgments
This work was supported by EU projects PICODIV (EVK3-CT1999-00021) and BASICS (EVK3-CT-2002-00078).
We appreciate the laboratory assistance of Ramon Terrado and Sònia Garcia Piqué; useful comments from Connie Lovejoy, Jarone Pinhassi, Josep M. Gasol, and Rafel Simó; and phylogenetic advice from José González.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, H., D. Dietrich, B. Auer, E. J. Cleven, T. Gräfenhan, M. Weitere, and A. P. Mylnikov. 2000. Functional diversity of heterotrophic flagellates in aquatic ecosystems, p. 240-268. In B. S. C. Leadbeater and J. C. Green (ed.), The flagellates: unity, diversity and evolution. Taylor & Francis Press, London, United Kingdom.
- 3.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, D., and L. Medlin. 1995. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J. Phycol. 31:489-498. [Google Scholar]
- 5.Caron, D. A., E. R. Peele, E. L. Lim, and M. R. Dennett. 1999. Picoplankton and nanoplankton and their trophic coupling in the surface waters of the Sargasso Sea south of Bermuda. Limnol. Oceanogr. 44:259-272. [Google Scholar]
- 6.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Díez, B., C. Pedrós-Alió, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eccleston-Parry, J. D., and B. S. C. Leadbeater. 1994. A comparison of the growth kinetics of six marine heterotrophic flagellates fed with one bacterial species. Mar. Ecol. Prog. Ser. 105:167-177. [Google Scholar]
- 11.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenchel, T. 1986. The ecology of heterotrophic microflagellates. Adv. Microb. Ecol. 9:57-97. [Google Scholar]
- 13.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 14.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryotic species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 15.Foissner, W. 1999. Protist diversity: estimates of the near-imponderable. Protist 150:363-368. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 18.Guillou, L., M.-J. Chrétiennot-Dinet, S. Boulben, S. Y. Moon-van der Staay, and D. Vaulot. 1999. Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist 150:383-398. [DOI] [PubMed] [Google Scholar]
- 19.Hagström, Å., T. Pommier, F. Rohwer, K. Simu, W. Stolte, D. Svensson, and U. L. Zweifel. 2002. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, D., T. Yokochi, T. Nakahara, S. Raghukumar, A. Nakagiri, K. Schaumann, and T. Higashihara. 1999. Molecular phylogeny of Labyrinthulids and Thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J. Eukaryot. Microbiol. 46:637-647. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, H., T. Fukuba, and T. Naganuma. 1999. Biomass of thraustochytrid protoctists in coastal waters. Mar. Ecol. Prog. Ser. 189:27-33. [Google Scholar]
- 23.Kühn, S., L. K. Medlin, and G. Eller. Phylogenetic position of the parasitoid nanoflagellate Pirsonia inferred from nuclear-encoded small subunit ribosomal DNA and a description of Pseudopirsonia n. gen. and Pseudopirsonia mucosa (Drebes) comb. nov. Protist, in press. [DOI] [PubMed]
- 24.Lee, W. J., and D. J. Patterson. 1998. Diversity and geographic distribution of free-living heterotrophic flagellates—analysis by PRIMER. Protist 149:229-244. [DOI] [PubMed] [Google Scholar]
- 25.Leipe, D. D., S. M. Tong, C. L Goggin, S. B. Slemenda, N. J. Pieniazek, and M. L. Sogin. 1996. 16S-like rDNA sequences from Developayella elegans, Labyrinthuloides haliotidis, and Proteromonas lacertae confirm that the stramenopiles are a primarily heterotrophic group. Eur. J. Protistol. 32:449-458. [Google Scholar]
- 26.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 27.Lim, E. L., M. R. Dennet, and D. A. Caron. 1999. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol. Oceanogr. 44:37-51. [Google Scholar]
- 28.López-García, P., F. Rodríguez-Valera, C. Pedrós-Alió, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massana, R., E. F. DeLong, and C. Pedrós-Alió. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massana, R., L. Guillou, B. Díez, and C. Pedrós-Alió. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medlin, L. K., M. Lange, and M. E. M. Baumann. 1994. Genetic differentiation among three colony-forming species of Phaeocystis: further evidence for the phylogeny of the Prymnesiophyta. Phycologia 33:199-212. [Google Scholar]
- 33.Montresor, M., C. Lovejoy, L. Orsini, and G. Procaccini. 2003. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 26:186-194. [Google Scholar]
- 34.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 35.Moreira, D., and P. López-García. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, M., T. Nakayama, and I. Inouye. 2002. A new class of the stramenopiles, Placidia classis nova: description of Placidia cafeteriopsis gen. et sp. nov. Protist 153:143-156. [DOI] [PubMed] [Google Scholar]
- 37.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 38.Patterson, D. J. 1989. Stramenopiles: chromophytes from a protistan perspective, p. 357-379. In J. C. Green, B. S. C. Leadbeater, and W. L. Diver (ed.), Chromophyte algae: problems and perspectives. Clarendon Press, Oxford, United Kingdom.
- 39.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784-798. [Google Scholar]
- 40.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Ecol. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 41.Sherr, E. B., and B. F. Sherr. 2000. Marine microbes. An overview, p. 13-46. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 42.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong, S. M. 1997. Heterotrophic flagellates and other protists from Southampton water U.K. Ophelia 42:71-131. [Google Scholar]
- 46.Yoon, H. S., J. D. Hackett, G. Pinto, and D. Bhattacharya. 2002. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. USA 99:15507-15512. [DOI] [PMC free article] [PubMed] [Google Scholar]